Abstract
Overexpression of DNA polymerase β (polβ), an error-prone DNA repair enzyme, has been shown to result in mutagenesis, aneuploidy and tumorigenesis. To further investigate the molecular basis leading to cancer-associated genetic changes, we examined whether the DNA polβ could affect homologous recombination (HR). Using mammalian cells carrying an intrachromosomal recombination marker we showed that the DNA polβ overexpression increased the HR mostly by enhancing gene conversion. Concomitantly, we observed the generation of DNA strand breaks as well as a DNA polβ-dependent formation of Rad51 foci. The stimulation of HR was abolished by the coexpression of a dominant negative form of Rad51, suggesting that the Rad51 was involved in the increased HR events. The expression of different DNA polβ mutants lacking polymerase activity did not result in HR stimulation, indicating that the DNA synthesis activity of DNA polβ was related to this phenotype. These results provide new insights into the molecular mechanisms of the genetic instability observed in DNA polβ overexpressing tumour cells.
INTRODUCTION
DNA polymerase β (polβ) is a key enzyme for the protection of the genome against DNA damage via its role in the base excision repair (BER) (1,2). In mammalian cells, most of the BER synthesis is carried out by polβ (1). The polβ not only functions as a DNA polymerase but also catalyses the excision of the deoxyribose phosphate (dRP) (3). The lack of any associated proofreading activity confers to polβ, a highly mutagenic DNA synthesis when copying undamaged DNA (4). When overexpressed in mammalian cells, polβ induces genetic instability and tumorigenesis (5,6). Based on in vitro experiments, the overexpression of polβ has been reported to alter cellular functions that do not normally require it, such as DNA replication (7) and DNA repair via the DNA synthesis step of the nucleotide excision repair pathway (8). Our previous works showed that, in vitro, polβ participated in an error-prone manner in DNA replication essentially during the synthesis of Okazaki fragments on the lagging strand (7). Therefore, we hypothesized that in such a context polβ might generate DNA single-strand stretches or DNA breaks during DNA replication, for several reasons. First, its distributive mode of DNA synthesis may slow down the elongation of the lagging strand into the forks, promoting the uncoupling of the replication of both strands and the persistence of long single-strand DNA. Second, we and others demonstrated the ability of polβ to incorporate efficiently, during DNA synthesis, nucleotide analogues such as oxidized guanosine (8-oxo-G) (7,9), dideoxynucleotides (10), ribonucleotides (11) or 6-thioguanine (12), thereby enhancing the probability to generate repair signals and DNA breaks. All these processes may therefore generate recombination signals since recombination pathways are involved in the repair of DNA strand breaks (13).
Alterations of recombination repair have been associated with human disease [for review see (14)]. Consequences of such defects are chromosomal rearrangements resulting from the loss of fidelity in repairing double-strand breaks (DSBs). These lesions are repaired mainly by two pathways: the non-homologous end joining (NHEJ) and the homologous recombination (HR) (15,16). The absence of either pathway leads to genomic instability and is often characterized by increased radiosensitivity (17,18). Numerous genes and proteins are related to the HR pathway and their alteration is linked to cancer [for review see (14)]. Some of these syndromes are correlated with a decrease in HR activity, as observed following the inactivation of BRCA1 (19) which plays a role in the repair of DSBs (20). In contrast, other syndromes such as Bloom's and Werner's syndromes whose mutated genes, BLM and WRN, are homologous to RecQ helicases are correlated with an elevated HR activity (21). In any case, these genetic disorders predispose to cancer through the chromosome instability.
We previously reported that the polβ overexpressing cells are hypersensitive to ionizing radiations and display elevated levels of chromosomal abnormalities (22). These features are also exhibited by cells defective in one of the recombination pathways (23,24,17). In addition, the overexpression of polβ has been observed in some tumours (prostate, breast and colon) and cancer cell lines (bladder, ovary and glioma) (8,12,25–27). Altogether, these features suggest that the overexpression of polβ could be responsible for the increased genetic instability observed in cancer cells through alteration of HR.
Here, we investigated the HR activity in polβ overexpressing cell lines carrying a unique intrachromosomal tandem repeat recombination substrate (28). We showed that the overexpression of polβ increases the generation of DNA breaks and rad51-dependent HR and that these features are dependent on polβ DNA synthesis activity.
MATERIALS AND METHODS
Chemicals
BrdU, Giemsa, Hoechst, nocodazole and doxycycline were obtained from Sigma (St Quentin Fallavier, France). Geneticin, puromycin and blasticidin were provided by Cayla (Toulouse, France).
Constructs
The polβ expressing vector was created by PCR amplification of the cDNA sequence of the human gene. As 5′ primer, an oligonucleotide was synthesized containing a NheI restriction site and the Kozak sequence GCCACC upstream of the ATG start codon. As 3′ primer, an oligonucleotide containing a stop codon and a second restriction enzyme site for BamHI was synthesized. PCR was performed using the Pfu DNA polymerase (GIBCO). The restriction enzyme sites at each end allowed ligation of the PCR product into the pIRES vector (Clontech). For cloning polβ in the pBLAST vector (InvivoGen, Toulouse, France), the restriction sites at each end of the PCR product were BamHI in 5′ primer and NheI in 3′ primer. Generation of single point (D256A) mutation was obtained by PCR with primers containing the expected mutation using QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions and was checked by sequencing.
Cell lines
The CHO-DRA10 cell line was cultured in α-minimal essential medium (GIBCO BRL) with 10% fetal calf serum and penicillin/streptomycin. The DRA10 cell line (βIND) expressing myc-tagged polβ under the control of the Tet-Off system was cultured in the same medium supplemented with 3 μg/ml of doxycycline.
The DRA10-pIβ4 and DRA10-pIβ15 clones were obtained after transfection with the pIRESpuro2 vector (Clontech) containing the cDNA of the human polβ. The DRA10-pBβ11 and DRA10-pBβ20 clones were obtained after transfection with the pBLAST vector (InvivoGen) containing the cDNA of the human polβ. The DRA10-dRP5 and DRA10-dRP10 clones were obtained after transfection with the pIRES vector (Clontech) containing the cDNA of the first 8 kDa in the N-terminal of polβ (kindly provided by Dr R. Sobol, Research Triangle park, NC) (2). The DRA10-DN14-11 and DRA10-DN14-15 cell lines were obtained after transfection with the pIRES vector (Clontech) containing the cDNA coding for the first 14 kDa in the N-terminal of polβ. The BMUT8 and BMUT9 cell lines were obtained after transfection with the pBLAST vector containing the cDNA of the human polβ bearing the punctual mutation generating a protein (D256A) without polymerase activity (2). Individual clones were obtained after transfection with Lipofectamine (GIBCO BRL) and selection with puromycin (10 μg/ml) or blasticidin (3 μg/ml) for pIRESpuro2 and pBLAST vectors, respectively.
The DRA10 cell line expressing the dominant negative (DN) form of rad51 (F1.4.1) was previously described in (29). The F1.4.1-β2 and F1.4.1-β6 clones were obtained after transfection with the pBLAST vector (InvivoGen) containing the cDNA of the human polβ. Individual clones were obtained after transfection with Lipofectamine (GIBCO BRL) and selection with blasticidin (3 μg/ml).
Generation of polβ inducible expression cells (Tet-Off)
The Tet-Off gene expression system (Clontech) was used to obtain regulated overexpression of polβ. Human polβ cDNA was inserted into the pTRE2pur-myc plasmid (Clontech), which carries the Tet operator sequence, the cytomegalovirus minimal promoter and a protein tag sequence (myc), to obtain the pTRE2pur-myc-polβ plasmid. The DRA10 cells were first transfected according to the manufacturer's protocol (Jet Pei, Qbiogene) with the pTet-Off vector (in which we first replaced neomycin resistance gene by the blasticidin resistance gene) carrying the regulatory protein TetR/HSV-VP16. In the Tet-Off system, the gene expression is turned on when doxycycline is removed from the culture medium. A stable transfectant clone displaying the highest level of induction and the lowest basal level of expression was screened out by transient transfections of 38 independent clones (selected by blasticidin 3 μg/ml) with an inducible pTRE2-luc vector carrying the luciferase gene. The selected clone was then transfected with the pTRE2pur-myc-polβ plasmid, and independent clones were selected by puromycin (10 μg/ml).
Measurement of spontaneous recombination
Determination of recombination was achieved by seeding 5 × 105 cells by plate (5 plates per experiment) growing in the presence of 1 mg/ml of G418. To determine gene conversion, 1 × 106 cells by plate were seeded (5 plates per experiment) in the presence of 1 mg/ml of G418 and 0.5 mg/ml of hygromycin. In the case of the Tet-Off cells (βIND), the polβ expression was induced by removing doxycycline from the culture medium 3 days before plating the cells to determine HR. Doxycycline was maintained in the culture medium during all the selection period for the cells uninduced for polβ expression. Determination of recombination was achieved by seeding 5 × 105 cells by plate (6 plates per experiment) in the presence of 1 mg/ml of G418. To determine gene conversion, 1 × 106 cells by plate were seeded (6 plates per experiment) in the presence of 1 mg/ml of G418 and 0.5 mg/ml of hygromycin. After 9 days of incubation, the plates were rinsed and the cells were fixed and stained with crystal violet. Colonies of more than 50 cells were counted. In parallel, cloning efficiency was evaluated.
Western blot analysis
The cells were washed with phosphate-buffered saline (PBS), scraped and resuspended in lysis buffer [25 mM Tris–HCl, pH 7.5, 1 mM EDTA, 600 mM NaCl, 0.1% Triton X-100 and a cocktail of protease inhibitors (Sigma, France)]. The cell suspension was sonicated and the protein concentration was determined using the Bio-Rad protein assay. An aliquot of 75 μg of cell extracts were separated on a 10% SDS–PAGE. For the determination of the dRP domain expression, the cell extracts were separated on a 12.5% SDS–PAGE. After migration, proteins were transferred to polyvinylidene difluoride membrane and probed with specific antibodies: anti-polβ (18S, Trevigen, Interchim, France), anti-actin (AC10, Sigma, France), anti-rad51 (Ab1, Oncogene Research, VWR, France), anti-FLAG (Sigma), anti-myc (Cell Signaling, Ozyme, France), anti-γ-H2AX (Trevigen). To examine the γ-H2AX expression, cell extracts were prepared by washing cells with PBS and resuspension in Laemmli buffer, and proteins were separated on a 12.5% SDS–PAGE. Equivalent of 3 × 105 cells was loaded in each lane.
Immunofluorescence microscopy
The cells were grown on glass coverslips in 6 well plates at a density of 5000 cells per well. The cells were fixed with 3.6% formaldehyde in PBS for 15 min then permeabilized with Triton X-100 and incubated with anti-rad51 antibody (Ab-1, Oncogene Research) or with anti-γ-H2AX antibody (Trevigen) for 1 h at room temperature, then with an anti-rabbit fluorescein isothiocyanate-conjugated antibody (Molecular Probes) for 1 h at room temperature. The cells were washed 4 times in PBS and mounted using Vectashield mounting medium with DAPI (Vector Laboratories). Fluorescence images were captured using a Zeiss Axioskop fluorescence microscope. For quantitative analysis, the cells presenting foci were counted by eye using a ×100 objective in more than 100 cells.
Single cell gel electrophoresis
DNA breaks were detected by alkaline single cell gel electrophoresis (SCGE). The cells were harvested by trypsinization, embedded in low melting agarose and then spread on microscope slide on ice. The cells were lysed in 250 mM NaCl, 10 mM Tris–HCl, 100 mM EDTA, 1% Triton X-100, 10% dimethyl sulfoxide for 1 h at 4°C. After lysis, the slides were preincubated for 20 min in the electrophoresis buffer (300 mM NaOH, 1 mM EDTA) and then subjected to alkaline gel electrophoresis (300 mA, 25 min). Neutralization of the pH was performed by three washes in Tris–HCl, pH 7.5 (400 mM). The slides were stained with ethidium bromide (0.2 μg/ml) and visualized by fluorescence microscopy. At least 50 cells were scored for each experiment.
RESULTS
HR is increased in polβ overexpressing cells
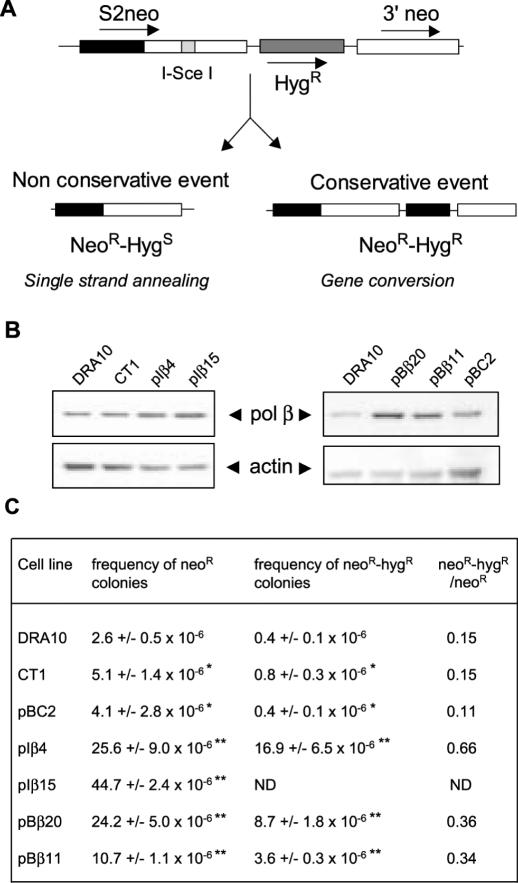
To examine the impact of polβ overexpression on HR, we cloned the human polβ cDNA into either pIRES or pBLAST expression vectors and transfected with either vector the CHO-DRA10 cell line that contains a unique copy of an intrachromosomal recombination substrate composed of two inactive copies of the neomycin-resistance gene (28) (Figure 1A). HR events can restore a functional neomycin gene rendering cells resistant to a selective medium containing G418 (28). Using pIRES vector, we generated independent clones showing a 3- (pIβ4) and 4-fold (pIβ15) polβ overexpression as compared with either the parental cells or empty vector transfected cells (Figure 1B). The clones obtained by transfecting cells with the pBLAST vector showed a 2-(pBβ11) to 3-fold (pBβ20) polβ overexpression (Figure 1B). Analysis of recombination showed that the overexpression of polβ increased the frequency of G418-resistant cells in the four independent clones examined (pIβ4 and pIβ15, pBβ20 and pBβ11) by a factor of 5–10 compared with control cells (Figure 1C). Thus, the overexpression of polβ resulted in a marked increase in spontaneous HR. For qualitative analysis of the type of HR events, we evaluated the double resistance to G418 and hygromycin that only selects gene conversion events (28). The neoR-hygR/neoR ratio allows estimating the balance between gene conversion and single-strand annealing (SSA) (Figure 1A). All the clones overexpressing polβ showed a significant increase in the frequency of neoR-hygR resistant cells, from 3- to 4-fold compared with their respective controls (Figure 1C), together with an increase of the neoR-hygR/neoR ratio. These results indicate that the polβ overexpression stimulates HR and mainly involves the gene conversion mechanism compared with control cells (Figure 1C).
Figure 1.
Intracellular substrate used to measure the recombination events and effect of polβ expression on the frequency of spontaneous recombination events. (A) The DRA10 cell line contains an intrachromosomal substrate represented by a tandem repeat of two inactive sequences of the neomycin resistance gene (S2neo and 3′ neo). S2neo contains a promoter but is inactivated by the insertion of the I-SceI sequence, and 3′ neo is not expressed because of the absence of any promoting sequence. The cells are sensitive to G418 but the recombination between the two inactive neo sequences can re-create a functional neo resistance gene that confers resistance to G418 on the recombinant cell (NeoR) (28). Gene conversion without associated crossing over keeps the hygromycin resistance sequence and the recombinant cell resistant to both G418 and hygromycin (NeoR-HygR). Thus, single G418 resistance (NeoR) monitors all the HR events (gene conversion with or without crossing over + SSA) and double G418 plus hygromycin resistance (NeoR-HygR) gives an estimation of the conservative events specific for gene conversion without crossing over. Not shown are the unequal SCE events that hygromycin resistance gene segregates from the neo resistance gene or gene conversion events without reintegration of the hygromycin resistance gene, giving rise to cells only NeoR (24). (B) Relative amount of polβ was examined by immunoblotting of total cell extracts on SDS–PAGE. Actin was used as internal control of loading. Controls are the parental CHO-DRA10 cell line and CT1 and pBC2 that are CHO-DRA10 cells transfected with the empty vector pIRES and pBLAST, respectively. pIβ4 and pIβ15 are two independent clones overexpressing polβ under the dependence of the pIRES vector. pBβ20 and pBβ11 are two independent overexpressing polβ clones under the dependence of the pBLAST vector. (C) For recombination measurements, the data are expressed as the frequency of neo resistant or neo and hyg resistant colonies observed for 106 viable cells plated. Results are the mean values ± SD of four independent experiments. Asterisk indicates no significant difference between values for DRA10 cells and CT1 or pBC2 cells expressing empty vector, P > 0.05. Double asterisks indicate the difference between values for CT1 or pBC2 cells and their respective transfected polβ overexpressing cells at P < 0.01, as determined by Student's t-test.
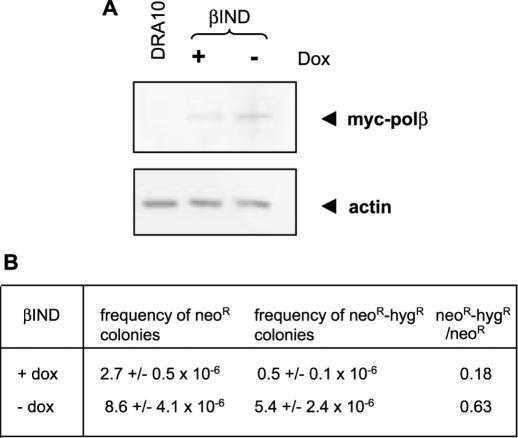
In order to check that the increase of HR was not the consequence of indirect effects accumulated from the time of transfection with polβ, we generated a DRA10 cell line expressing doxycycline-inducible myc-polβ (Tet-Off system). The inducible cells (βIND) expressed the ectopic myc-tagged polβ when doxycycline was removed from the culture medium. Removing doxycycline during 3 days induced a marked increase in the expression of myc-tagged-polβ (Figure 2A). In the presence of doxycycline, the neoR and neoR-hygR frequencies in the βIND cells were similar to those obtained in the DRA10 cell line (compare Figures 1C and 2B). Examination of HR after 3 days of culture in the absence of doxycycline showed both neoR and neoR-hygR increased frequencies (Figure 2B). As observed with the stably transfected clones (Figure 1), induced overexpression of polβ also increased the neoR-hygR/neoR ratio, indicating a specific stimulation of the gene conversion pathway (Figure 2B).
Figure 2.
Effects of the inducible expression of polβ on HR. (A) Examination by western blotting of the induction of polβ in the βIND cells after removing doxycycline from the culture medium during 3 days. Immunodetection was performed with an antibody directed to the myc epitope of tagged polβ expressed with the Tet-Off system. Actin was used as control for equal loading. (B) For recombination measurement, the data are expressed as the frequency of neo resistant or neo and hyg resistant colonies observed for 106 viable cells plated. The data are the results of two independent experiments and are expressed as the mean values ± SD.
Only native polβ overexpression stimulates HR
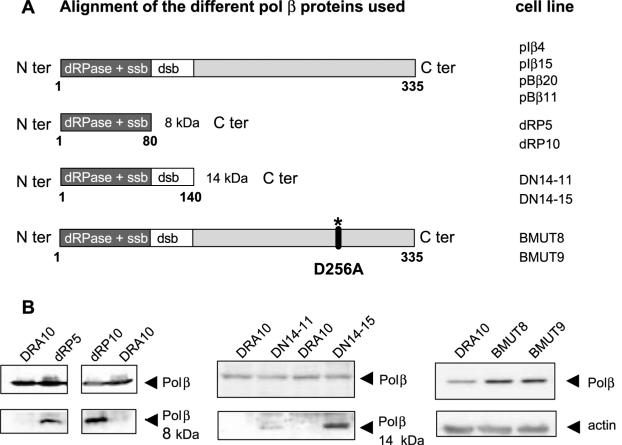
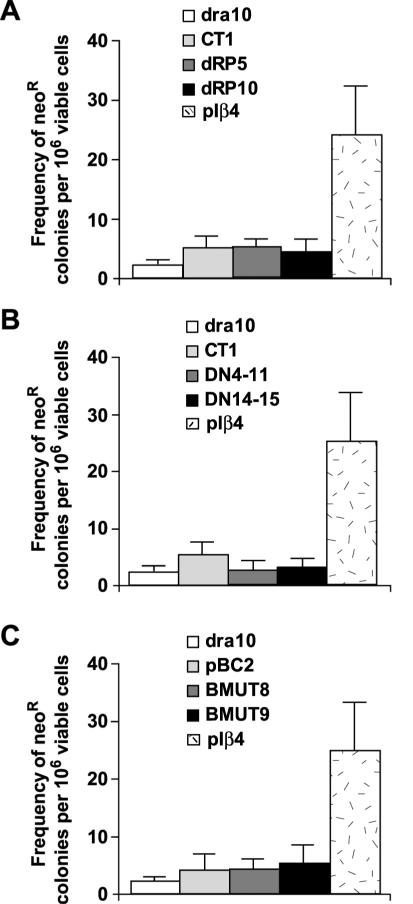
The structure of polβ displays distinct domains. A deoxyribose phosphodiesterase activity carried by the N-terminal 8 kDa domain has been shown to be necessary and sufficient to complement the BER defect observed in polβ null cells (2). In addition, this 8 kDa domain displays a single-strand DNA-binding activity (30) (Figure 3A). Husain et al. (30) also described a fragment of polβ corresponding to the first 14 kDa of the N-terminus that acts as a DN inhibiting the DNA repair activity of polβ in vitro. This 14 kDa domain binds to single- and double-stranded DNA, but lacks DNA polymerase activity (Figure 3A). In order to determine which domain and activity of polβ were responsible for the stimulation of HR, we expressed either the 8 kDa or the 14 kDa domain in the DRA10 cell line (Figure 3B). Neither the 8 kDa nor the 14 kDa domain expression did increase the frequency of HR (Figure 4A and B). To confirm the importance of the catalytic site, we generated cell clones expressing a polymerase active-site point mutant (BMUT8 and BMUT9). The protein generated by the mutation D256A was devoid of polymerase activity (2,31) due to the critical role of this amino acid in the nucleotidyltransferase mechanism (Figure 3A). The two independent cell clones showed no significant increase in the generation of neoR colonies (Figure 4C), demonstrating that the elevated HR observed in the cell lines overexpressing the native form of polβ requires DNA synthesis. These data argue in favour of a direct role for the DNA polymerase activity of polβ in the HR stimulation.
Figure 3.
Generation of cell lines expressing different forms of polβ. (A) Structure of the polβ protein used. The different proteins are expressed in the CHO-DRA10 cell line and the corresponding derivative cell lines are listed on the right. From the top to the bottom, the WT form (pIβ4, pIβ15, pBβ20 and pBβ11), the 8 kDa form (dRP5 and dRP10), the 14 kDa form (DN14-11 and DN14-15) and the full length form with the D256A mutation (BMUT8 and BMUT9) (B) Expression of the different forms of polβ were analysed by western blotting.
Figure 4.
Effect of the expression of the different forms of polβ on recombination frequency. (A) Frequency of neoR events in control cell line (DRA10), cells expressing pIRES empty vector (CT1) and in cell lines expressing the 8 kDa domain (dRP) (dRP5 and dRP10) compared with the polβ overexpressing cell line (pIβ4). Results are the mean values ± SD of three independent experiments. No significant difference between values for DRA10 cells and CT1 and dRP5 and dRP10 cells was observed, P > 0.05 as determined by Student's t-test. (B) Frequency of neoR events in control cell line (DRA10), cells expressing pIRES empty vector (CT1) and in cell lines expressing the 14 kDa domain of polβ (DN14-11) and (DN14-15) compared with the polβ overexpressing cell line (pIβ4). Results are the mean values ± SD of three independent experiments. No significant difference between values for DRA10 cells and DN14-11 and DN14-15 cells was observed, P > 0.05 as determined by Student's t-test. (C) Frequency of neoR events in control cell line (DRA10), cell lines expressing the pBLAST empty vector (pBC2) and in cell lines expressing the mutated form of polβ (BMUT8) and (BMUT9) compared with the polβ overexpressing cell line (pIβ4). Results are the mean values ± SD of three independent experiments. No significant difference between values for DRA10 cells and pBC2, BMUT8 and BMUT9 cells was observed, P > 0.05 as determined by Student's t-test.
Polβ overexpressing cells carry high levels of DNA strand breaks
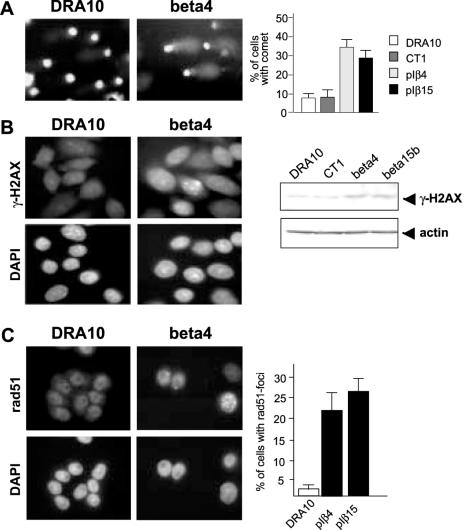
In order to elucidate the cause of HR induction, we investigated whether the cells carried different levels of DNA strand breaks as a potential source of HR substrates. In contrast to control cells that displayed only few DNA strand breaks, a larger part of the polβ overexpressing cells carried DNA strand breaks (Figure 5A). To confirm these data, we investigated the presence of γ-H2AX, the phosphorylated form of the H2AX histone, specifically detected in response to the DNA DSBs (32). Following western blot analysis, polβ overexpressing cells (pIβ4 and pIβ15) displayed a 3-fold higher level of γ-H2AX protein than control cells (DRA10 and CT1) (Figure 5B). Immunofluorescence experiments confirmed this result as 14% of the polβ overexpressing cells displayed γ-H2AX foci compared with only 3% of the control cells, which represented an increase of 4.6-fold. Taken together, these results strongly support the presence of higher levels of DNA strand breaks in polβ overexpressing cells.
Figure 5.
Evaluation of DNA strand breaks and Rad51 foci formation. (A) Detection of DNA strand breaks was performed by SCGE (COMET assay). Slides from two independent experiments were examined, and in each experiment at least 50 cells were observed. Values are the mean values ± SD of two independent experiments. (B) Immunofluorescence detection and western blot analysis of γ-H2AX expression to detect DNA DSBs. (C) Immunofluorescence detection and quantification of the cells displaying Rad51 foci in the different cell populations. Values are the mean values ± SD of three independent experiments. For each experiment, 100 cells were examined.
Polβ overexpressing cells display Rad51 foci
The presence of higher levels of DSB and the stimulation of HR in polβ overexpressing cells led us to examine the expression and function of Rad51, a major component of HR (17,18,33). We examined the presence of Rad51 foci that occurred after the relocalization of the Rad51 protein onto DSB (34). First, we checked the level of Rad51 expression in the different cell lines used. No significant differences in the expression were observed (data not shown). However, we found a 5- to 10-fold increase in the number of cells displaying Rad51 foci in the polβ overexpressing cell population compared with the control cell population (Figure 5C). Therefore, Rad51 foci formation correlates with the HR enhancement observed in the polβ overexpressing cells.
Polβ-dependent HR increase requires intact Rad51 pathway
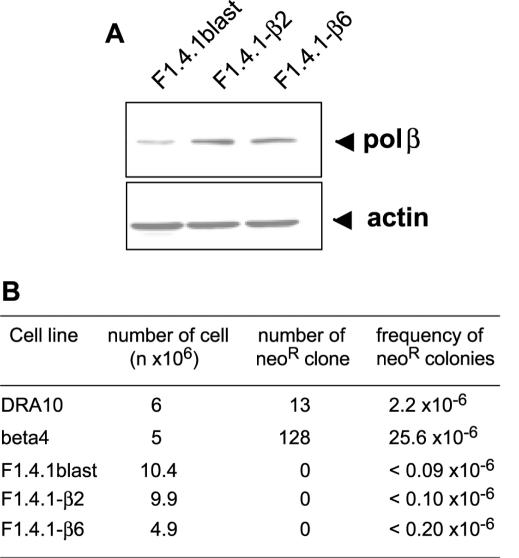
To demonstrate the involvement of the Rad51 pathway in the HR stimulation observed in the polβ overexpressing cells, DRA10 cell lines expressing a DN form of rad51 (F1.4.1) (29) were transfected with polβ vectors (pBLAST-polβ). These cell lines expressed a chimera SMRAD51 corresponding to the fusion of the 55 N-terminal amino acids from the yeast ScRAD51 to MmRAD51. SMRAD51 inhibits spontaneous as well as DSB-induced recombination in CHO-DRA10 cell lines without affecting the cell viability (29). The level of polβ expression after transfection in these cell lines was similar to that obtained in the CHO-DRA10 cell line (Figures 1B and 6A). Co-expression of Rad51 DN form together with the polβ overexpression suppressed the increase in HR observed with overexpression of polβ alone (clone pIβ4) (Figure 6B). No recombinant colonies were detected in cultures expressing the DN form of Rad51 (F1.4.1blast) as well as in cultures of cells expressing both Rad51 DN and ectopic polβ (F1.4.1-β2 and F1.4.1-β6). In order to check that the absence of neoR clones was not due to a defective recombination substrate, we transfected the F1.4.1-β6 cell line with a plasmid containing the rare cutting endonuclease I-SceI gene. The recombination substrate carries a site for I-SceI (Figure 1A) that generates a DSB when cells are transfected. Transient expression of I-SceI has been shown to generate a marked increase (100- to 1000- fold) of the neoR events in the parental DRA10 cell line (28,29). Using I-SceI transfection, we observed the generation of neoR colonies in the F1.4.1-β6 cell line (data not shown) confirming that the recombination substrate was functional, although no clone was detected at the basal level. These results indicate that the increase in HR observed in polβ overexpressing cells is dependent on the Rad51 pathway.
Figure 6.
Effect of the expression of the DN form of Rad51 on recombination in the polβ overexpressing cells. (A) Expression level of polβ evaluated by western blotting in the control cell line expressing DN form of Rad51 (F1.4.1blast) and cell lines expressing both DN Rad51 form and polβ (F1.4.1-β2 and F1.4.1-β6). (B) Evaluation of recombination frequency in the different cell lines. The data are the sum of the recombinants (neoR clones) from all independent cultures (four experiments) related to the total number of cells plated from all the independent cultures.
DISCUSSION
This study was aimed at elucidating the mechanisms involved in the genetic instability resulting from the polβ overexpression (5,6,35). One hypothesis to explain how polβ acts as a genetic instability enhancer was that excess of polβ might disturb the well-defined and accurate specific functions of error-free DNA polymerases (36). In addition, we previously observed that the polβ could play a role in some DNA transactions where it is normally not required, such as genomic replication (7) or nucleotide excision repair (8). The polβ overexpressing cells showed rearranged karyotypes both spontaneously or after genotoxic treatment. The resulting chromosomal alterations included aneuploidy, dicentric and triradial chromosomes (6,22), all being observed in human syndromes displaying altered HR pathway. Here, we examined whether HR could be altered by the overexpression of polβ and found that overexpression of polβ increases both HR and the level of DNA strand breaks. Although HR is considered as an error-free DNA repair system, it can also produce genetic instability when it is upregulated (37). The generation of these DNA strand breaks in vivo could result from the involvement of polβ in DNA replication as demonstrated by in vitro DNA replication assay (7). Servant et al. (7) showed that polβ lowered replication fidelity by its involvement during the DNA synthesis of the lagging strand, a process that may reduce synchronization of DNA synthesis between both strands, slowing down the fork elongation and generating DNA strand breaks. To reinforce this hypothesis, we found here that the expression of the different DNA-binding domains of polβ devoid of its catalytic domain, as well as point mutant deficient for DNA synthesis failed to increase strand breaks and HR. Furthermore, a previous work showed that the inhibition of the elongation of replication produces DNA strand breaks that are processed by HR, leading to increase in the HR activity (38). In addition, the use of an inducible expression system for polβ confirmed that the increase in HR activity was totally dependent on the polβ overexpression and was not the consequence of indirect effects throughout many generations from the time of transfection to the selection of individual clones. The increase in HR activity was specific for polβ since the overexpression of polλ, another error-prone DNA polymerase and the closest homologue of polβ, did not alter HR (data not shown).
Mammalian cells can repair DSB using two distinct pathways: NHEJ and HR. HR can occur through two main mechanisms: gene conversion, i.e. Rad51-dependent and can take place with or without a crossing over, and SSA, i.e. Rad51-independent (29). Analysis of the neoR and neoR-HygR frequencies showed that the polβ overexpression stimulated both (Figures 1C and 2B). However, the ratio neoR-HygR/neoR which is indicative of the balance between SSA and gene conversion was increased in all the cell lines overexpressing polβ, indicating a preferential stimulation of the gene conversion pathway. Gene conversion is the preferred HR-mediated repair pathway for DSBs and maintains chromosomal stability (39). The higher stimulation of gene conversion compared with the SSA (Figures 2 and 3) most likely results from the DSB produced by polβ during DNA replication, giving to the cell an available intact copy to rescue from the DSB. However, we cannot rule out that part of the HR events termed SSA were not SSA events and could result either from gene conversion with crossing over without reintegration of the excised hygromycin resistance gene or from unequal sister chromatid exchange (SCE). Therefore, our results could mean that SSA events were overestimated and even might not be stimulated at all by the overexpression of polβ.
Because DNA strand breaks are potential signals for recombination involving HR (29), we examined whether Rad51 was involved in this increase in HR. Rad51 is a mammalian homologue of the bacterial protein recA essential for the repair of DSB by HR (29,33). Although the level of DNA strand breaks was increased in polβ overexpressing cells, the expression of Rad51 was not affected (data not shown). However, the frequency of Rad51 foci formation was increased in polβ overexpressing cells suggesting that the Rad51-dependent recombination pathway is then stimulated (Figure 6), confirming the observed increase in the frequency of conservative neoR-hygR events. To demonstrate the involvement of the Rad51 pathway in the increase of HR, we used polβ overexpressing cells transfected with a DN form of Rad51 (29). The absence of a functional Rad51 pathway suppressed the increase in HR produced by the overexpression of polβ (Figure 6), demonstrating that the increase in HR observed in polβ overexpressing cells depends on Rad51. As previously shown, deletion of the RAD51 gene has little effect on SSA events (29). The independence of the SSA events on RAD51 argues in favour of either gene conversion without reintegration of the hygromycin resistance gene or unequal SCE rather than the SSA for the events that lose the hygromycin resistance gene. The fact that the inhibition of the RAD51 pathway suppressed all the HR events seems to indicate that the increase in HR produced by the polβ overexpression was mostly due to gene conversion events and that there was little effect on SSA.
HR appears to be a key process in the maintenance of genetic stability [for review see (14,15)], in particular to deal with DSB that arise spontaneously or following genotoxic stress. Defect in such DNA repair pathway promotes genomic instability, one of the hallmarks of cancer cells. Conversely, an increase in HR has been described in the Bloom's syndrome in which cells display an increased frequency of SCE (40). Here, we observed an increase in HR that could have consequences on genetic stability. We conclude that because of the continuous stimulation of HR, errors could arise during the repair of DNA strand breaks and generate rearranged karyotypes because none of the DNA repair pathways is totally accurate. This hypothesis can be related to our previous observations that the overexpression of polβ produces rearranged karyotypes spontaneously (6) or after genotoxic stress (22). In the latter case, dicentric, triradial, quadriradial and ring chromosomes were observed (22) and all of these chromosomal abnormalities were also found in recombinational defective syndromes (14). As observed in the case of cells exhibiting increased HR, resulting from the expression of ATM (41), BCR/ABL (42) and mutated p53 (43,44) a genetic instability including translocations and deletions occurred. Because the human genome contains numerous repetitive sequences ubiquitously dispatched throughout the genome, increased HR activity could generate recombinational events between these repeated sequences leading to genetic instability.
In conclusion, this study shows that the HR can be modulated by a disregulated gene product that is not normally involved in such a DNA repair pathway. Therefore, we also bring new findings on the potential role that the polβ overexpression, observed in some tumours, may play.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Dominique Lautier for her critical reading of the manuscript. Dr Maria Jasin is greatly acknowledged for providing DRA10 cell line. This work was supported by ‘Ligue Nationale contre le Cancer, Equipe labellisée’ and ACI ‘Cancéropôles en émergence’.
REFERENCES
- 1.Sobol R.W., Horton,J.K., Kuhn,R., Gu,H., Singhal,R.K., Prasad,R., Rajewsky,K. and Wilson,S.H. (1996) Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature, 379, 183–186. [DOI] [PubMed] [Google Scholar]
- 2.Sobol R.W., Prasad,R., Evenski,A., Baker,A., Yang,X., Horton,J. and Wilson,S.H. (2000) The lyase activity of the DNA repair protein beta-polymerase protects from DNA-damage-induced cytotoxicity. Science, 405, 807–810. [DOI] [PubMed] [Google Scholar]
- 3.Matsumoto Y. and Kim,K. (1995) Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science, 269, 699–702. [DOI] [PubMed] [Google Scholar]
- 4.Osheroff W., Jung,H., Beard,W., Wilson,S. and Kunkel,T. (1999) The fidelity of DNA polymerase beta during distributive and processive DNA synthesis. J. Biol. Chem., 274, 3642–3650. [DOI] [PubMed] [Google Scholar]
- 5.Canitrot Y., Cazaux,C., Frechet,M., Bouayadi,K., Lesca,C., Salles,B. and Hoffmann,J.S. (1998) Overexpression of DNA polymerase beta in cell results in a mutator phenotype and a decreased sensitivity to anticancer drugs. Proc. Natl Acad. Sci. USA, 95, 12586–12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergoglio V., Pillaire,M.J., Lacroix-Triki,M., Raynaud-Messina,B., Canitrot,Y., Bieth,A., Gares,M., Wright,M., Delsol,G., Loeb,L.A. et al. (2002) Deregulated DNA polymerase beta induces chromosome instability and tumorigenesis. Cancer Res., 62, 3511–3514. [PubMed] [Google Scholar]
- 7.Servant L., Bieth,A., Hayakawa,H., Cazaux,C. and Hoffmann,J.S. (2002) Involvement of DNA polymerase beta in DNA replication and mutagenic consequences. J. Mol. Biol., 315, 1039–1047. [DOI] [PubMed] [Google Scholar]
- 8.Canitrot Y., Hoffmann,J., Calsou,P., Hayakawa,H., Salles,B. and Cazaux,C. (2000) Nucleotide excision repair DNA synthesis by excess DNA polymerase beta: a potential source of genetic instability in cancer cells. FASEB J., 14, 1765–1774. [DOI] [PubMed] [Google Scholar]
- 9.Kamath-Loeb A., Hizi,A., Kasai,H. and Loeb,L. (1997) Incorporation of the guanosine triphosphate analogs 8-oxo-dGTP and 8-NH2-dGTP by reverse transcriptases and mammalian DNA polymerases. J. Biol. Chem., 272, 5892–5898. [DOI] [PubMed] [Google Scholar]
- 10.Bouayadi K., Hoffmann,J.S., Fons,P., Tiraby,M., Reynes,J. and Cazaux,C. (1997) Overexpression of DNA polymerase beta sensitizes mammalian cells to 2′,3′-dideoxycytidine and 3′-azido-3′-deoxythymidine. Cancer Res., 57, 110–116. [PubMed] [Google Scholar]
- 11.Bergoglio V., Ferrari,E., Hubscher,U., Cazaux,C. and Hoffmann,J.S. (2003) DNA polymerase beta can incorporate ribonucleotides during DNA synthesis of undamaged and CPD-damaged DNA. J. Mol. Biol., 331, 1017–1023. [DOI] [PubMed] [Google Scholar]
- 12.Bergoglio V., Canitrot,Y., Hogarth,L., Minto,L., Howell,S., Cazaux,C. and Hoffmann,J. (2001) Enhanced expression and activity of DNA polymerase beta in human ovarian tumor cells: impact on sensitivity towards antitumor agents. Oncogene, 20, 6181–6187. [DOI] [PubMed] [Google Scholar]
- 13.Hoeijmakers J. (2001) Genome maintenance mechanism for preventing cancer. Nature, 411, 366–374. [DOI] [PubMed] [Google Scholar]
- 14.Thompson L. and Schild,D. (2002) Recombinational DNA repair and human disease. Mutat. Res., 509, 49–78. [DOI] [PubMed] [Google Scholar]
- 15.Thompson L. and Schild,D. (2001) Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat. Res., 477, 131–153. [DOI] [PubMed] [Google Scholar]
- 16.Barnes D. (2001) Non-homologous end joining as a mechanism of DNA repair. Curr. Biol., 11, R455–R457. [DOI] [PubMed] [Google Scholar]
- 17.Sonoda E., Takata,M., Yamashita,Y., Morrison,C. and Takeda,S. (2001) Homologous DNA recombination in vertebrate cells. Proc. Natl Acad. Sci. USA, 98, 8388–8394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson S. (2002) Sensing and repairing DNA double-strand breaks. Carcinogenesis, 23, 687–696. [DOI] [PubMed] [Google Scholar]
- 19.Xu X., Weaver,Z., Linke,S., Li,C., Gotay,J., Wang,X., Harris,C., Ried,T. and Deng,C. (1999) Centrosome amplification and G2M cell cycle checkpoint induce genetic instability in BRCA1 exon11 isoform-deficient cells. Mol. Cell, 3, 389–395. [DOI] [PubMed] [Google Scholar]
- 20.Moynahan M., Cui,T. and Jasin,M. (2001) Homology directed repair, mitomycin C resistance and chromosome stability is restored with correction of the Brca1 mutation. Cancer Res., 61, 4842–4850. [PubMed] [Google Scholar]
- 21.Imamura O., Fujita,K., Itoh,C., Takeda,S., Furuichi,Y. and Matsumoto,T. (2002) Werner and Bloom helicases are involved in DNA repair in a complementary fashion. Oncogene, 21, 954–963. [DOI] [PubMed] [Google Scholar]
- 22.Frechet M., Canitrot,Y., Bieth,A., Dogliotti,E., Cazaux,C. and Hoffmann,J.S. (2002) deregulated DNA polymerase beta strengthens ionizing radiation-induced nucleotidic and chromosomal instabilities. Oncogene, 21, 2320–2327. [DOI] [PubMed] [Google Scholar]
- 23.Liu N., Lamerdin,J., Tebbs,R., Schild,D., Tucker,J., Shen,M., Brookman,K., Siciliano,M., Walter,C., Fan,W. et al. (1998) XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol. Cell, 1, 783–793. [DOI] [PubMed] [Google Scholar]
- 24.Delacote F., Han,M., Stamato,T., Jasin,M. and Lopez,B. (2002) An XRCC4 defect or wortmannin stimulates homologous recombination specifically induced by double-strand breaks in mammalian cells. Nucleic Acids Res., 30, 3454–3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomi A., Shinoda,S., Sakai,R., Hirai,H., Ozawa,K. and Masuzawa,T. (1996) Elevated expression of DNA polymerase beta gene in glioma cell lines with acquired resistance to 1-(4-amino-2-methyl-5-pyrimidinyl)methyl-3-(2-chloroethyl)-3-nitrosourea. Biochem. Biophys. Res. Commun., 227, 558–563. [DOI] [PubMed] [Google Scholar]
- 26.Singh S., Scalomogna,D., Xia,H., O'Toole,S., Roy,D., Emerson,E., Gupta,V. and Zaren,H. (1996) Biochemical characterization of a mitomycin C resistant human bladder cancer cell line. Int. J. Cancer, 65, 852–857. [DOI] [PubMed] [Google Scholar]
- 27.Srivastava D., Husain,I., Arteaga,C. and Wilson,S. (1999) DNA polymerase beta expression differences in selected human tumors and cell lines. Carcinogenesis, 20, 1049–1054. [DOI] [PubMed] [Google Scholar]
- 28.Liang F., Han,M., Romanienko,P. and Jasin,M. (1998) Homology-directed repair is a major double-strand-break repair pathway in mammalian cells. Proc. Natl Acad. Sci. USA, 95, 5172–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lambert S. and Lopez,B. (2000) Characterization of mammalian RAD51 double strand break repair using non-lethal dominant negative forms. EMBO J., 19, 3090–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Husain I., Morton,B., Beard,W., Singhal,R., Prasad,R., Wilson,S. and Besterman,J. (1995) Specific inhibition of DNA polymerase beta by its 14 kDa domain: role of single- and double-stranded DNA binding and 5′-phosphate recognition. Nucleic Acids Res., 23, 1597–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menge K., Nodes,B., Hudson,G., Rahmati,S., Moomaw,E., Almassy,R. and Hostomska,Z. (1995) Structure-function analysis of the mammalian DNA polymerase beta active site: role of aspartic acid 256, arginine 254, and arginine 258 in nucleotidyl transfer. Biochemistry, 34, 15934–15942. [DOI] [PubMed] [Google Scholar]
- 32.Mahadevaiah S., Turner,J., Baudat,E., Rogakou,E., de Boer,P., Blanco-Rodriguez,P., Jasin,M., Keeney,S., Bonner,W. and Burgoyne,P. (2001) Recombinational DNA double-strand breaks in mice precede synapsis. Nature Genet., 27, 271–276. [DOI] [PubMed] [Google Scholar]
- 33.Sonoda E., Sasaki,M., Morrison,C., Yamaguchi-Iwai,Y., Takata,M. and Takeda,S. (1999) Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell. Biol., 19, 5166–5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haaf T., Golub,E., Reddy,G., Radding,C. and Ward,D. (1995) Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc. Natl Acad. Sci. USA, 92, 2298–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamada N. and Farber,R. (2002) Induction of a low level of microsatellite instability by overexpression of DNA polymerase beta. Cancer Res., 62, 6061–6064. [PubMed] [Google Scholar]
- 36.Canitrot Y., Frechet,M., Servant,L., Cazaux,C. and Hoffmann,J.S. (1999) Overexpression of DNA polymerase beta: a genomic instability enhancer process. FASEB J., 13, 1107–1111. [DOI] [PubMed] [Google Scholar]
- 37.Purandare S.M. and Patel,P.I. (1997) Recombination hot spots and human disease. Genome Res., 7, 773–786. [DOI] [PubMed] [Google Scholar]
- 38.Saintigny Y., Delacote,F., Varès,G., Petitot,F., Lambert,S., Averbeck,D. and Lopez,B. (2001) Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J., 20, 3861–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson R. and Jasin,M. (2000) Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J., 19, 3398–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chaganti R., Schonberg,S. and German,J. (1974) A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc. Natl Acad. Sci. USA, 71, 4508–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meyn M. (1993) High spontaneous intrachromosomal recombination rates in ataxia-telangiectasia. Science, 260, 1327–1330. [DOI] [PubMed] [Google Scholar]
- 42.Slupianek A., Schmutte,C., Tombline,C., Nieborowska-Skorska,M., Hoser,G., Nowicki,M., Pierce,A., Fishel,R. and Skorski,T. (2001) BCR/ABL regulates mammalian RecA homologs, resulting in drug resistance. Mol. Cell, 8, 795–806. [DOI] [PubMed] [Google Scholar]
- 43.Gebow D., Miselis,N. and Liber,H. (2000) Homologous and non homologous recombination resulting in deletion: effects of p53 status, microhomology, and repetitive DNA length and orientation. Mol. Cell. Biol., 20, 4028–4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bertrand P., Rouillard,D., Boulet,A., Levalois,C., Soussi,T. and Lopez,B. (1997) Increase of spontaneous intrachromosomal homologous recombination in mammalian cells expressing a mutant p53 protein. Oncogene, 14, 1117–1122. [DOI] [PubMed] [Google Scholar]