Abstract
One of the most frequent lesions formed in cellular DNA are abasic (apurinic/apyrimidinic, AP) sites that are both cytotoxic and mutagenic, and must be removed efficiently to maintain genetic stability. It is generally believed that the repair of AP sites is initiated by the AP endonucleases; however, an alternative pathway seems to prevail in Schizosaccharomyces pombe. A mutant lacking the DNA glycosylase/AP lyase Nth1 is very sensitive to the alkylating agent methyl methanesulfonate (MMS), suggesting a role for Nth1 in base excision repair (BER) of alkylation damage. Here, we have further evaluated the role of Nth1 and the second putative S.pombe AP endonuclease Apn2, in abasic site repair. The deletion of the apn2 open reading frame dramatically increased the sensitivity of the yeast cells to MMS, also demonstrating that the Apn2 has an important function in the BER pathway. The deletion of nth1 in the apn2 mutant strain partially relieves the MMS sensitivity of the apn2 single mutant, indicating that the Apn2 and Nth1 act in the same pathway for the repair of abasic sites. Analysis of the AP site cleavage in whole cell extracts of wild-type and mutant strains showed that the AP lyase activity of Nth1 represents the major AP site incision activity in vitro. Assays with DNA substrates containing base lesions removed by monofunctional DNA glycosylases Udg and MutY showed that Nth1 will also cleave the abasic sites formed by these enzymes and thus act downstream of these enzymes in the BER pathway. We suggest that the main function of Apn2 in BER is to remove the resulting 3′-blocking termini following AP lyase cleavage by Nth1.
INTRODUCTION
Apurinic/apyrimidinic (AP) sites are one of the most frequently formed lesions in DNA and arise spontaneously by hydrolysis of the N-glycosylic bond (1) and are induced by the action of DNA glycosylases removing damaged, modified or even normal bases from DNA (2–4). In addition, the AP sites are also induced directly by ionizing radiation, oxygen radical species and to some extent by alkylation as well (5). The AP sites are both cytotoxic and premutagenic, and efficient repair mechanisms are required to prevent the detrimental effects of such damage. DNA replication and transcription are blocked by abasic sites (6,7) whereas bypass by translesion polymerases result in base substitutions and frameshift mutations (8–10).
The major pathway for the repair of AP sites in DNA involves the action of an AP endonuclease followed by a 5′-phoshodiesterase or dRPase leaving a gap in DNA. The gap is filled by a DNA polymerase and the strand sealed by a DNA ligase. Another branch of the base excision repair (BER) pathway repairs abasic sites through the action of an AP endonuclease and a 2–8 nt displacement reaction of the strand containing the 5′-deoxyribose residue (long patch repair). Replication factor C, proliferating cell nuclear antigen and the structure-specific nuclease Fen1 are required for the strand displacement reaction (11–13). In addition to BER, epistatic analysis of mutant survival in Schizosaccharomyces pombe suggests that the nucleotide excision repair (NER) also plays an important role in the repair of abasic sites (14,15).
Several AP endonucleases from different sources have been identified and they can be divided into two major classes as typified by the Escherichia coli enzymes, exonuclease III [Xth (16)] and endonuclease IV [Nfo (17)]. In addition to hydrolysis at the 5′-side of abasic sites (AP endonuclease activity), these enzymes possess 3′-phosphodiesterase and 3′-phosphatase activities that function on the 3′-termini in DNA. Some members of the Xth family also possess a strong 3′–5′ exonuclease activity. Another subfamily of exonuclease III homologues have also been identified, termed as Apn2 in Saccharomyces cerevisiae (18,19) and Ape2 in mammalian cells (20). These proteins exhibit a very weak AP endonuclease activity despite the extensive sequence homology to E.coli Xth and human Hap1/Ape1.
Yet another type of enzymes makes strand breaks at abasic sites in DNA. These are termed as AP lyases that cleave 3′ to AP sites using a β-elimination mechanism, leaving a 5′-phosphate group and a 3′-α,β-unsaturated aldehyde. The AP lyase activity is associated with bifunctional DNA glycosylases that are major repair enzymes existing in all organisms from bacteria to man (21–26). The 3′-deoxyribose phosphate (3′-dRP) termini resulting from an AP lyase cleavage and other related 3′-blocking groups, such as 3′-phosphate and 3′-phosphoglycolate, inhibits DNA polymerases and can be converted into highly cytotoxic double strand breaks during replication (27).
The biological significance of the AP lyase activities is unclear and the classical model for the repair of AP sites via the BER pathway does not involve AP lyases, but proceeds via AP endonucleases (28–30). However, previous work with the fission yeast Sc.pombe has shown that yeast cells lacking the DNA glycosylase Nth1 involved in the repair of oxidative DNA damage are sensitive to methyl methanesulfonate (MMS). This is not because Nth1 removes alkylated bases per se but rather because the AP lyase activity of Nth1 is critical for repair of the MMS-induced AP sites (15). The Sc.pombe genome encodes homologues of E.coli Nfo and Xth termed as Apn1 and Apn2, respectively. However, the apn1 mutant exhibits wild-type resistance to MMS (31), which is in contrast to the strong MMS-sensitive phenotype of the S.cerevisiae apn1 mutant (32), indicating an alternative pathway for the repair of AP sites in Sc.pombe. The Apn2 function in Sc.pombe has so far not been extensively characterized (33), however, a very recent study has indicated that Apn2 is more potent as an AP endonuclease than Apn1 in Sc.pombe (34). Here, we have investigated the BER of abasic sites in Sc.pombe by monitoring AP site incision in cell-free extracts and by analysing mutants' carrying of nth1 and apn2 deletions. Our results show that Nth1 possesses the major incision activity at AP sites in cell-free extracts from Sc.pombe and that Nth1 can act downstream of the monofunctional DNA glycosylases Udg and MutY in BER as well. Furthermore, mutant analysis indicates that Apn2 can act downstream of Nth1 in alkylation repair as the severe MMS sensitivity of the apn2 mutant is partly relieved by the deletion of nth1. Thus, it appears that Nth1 and Apn2 operate sequentially in the repair of abasic sites in Sc.pombe and a possible in vivo function of Apn2 is proposed.
MATERIALS AND METHODS
Yeast strains and media
Schizosaccharomyces pombe wild-type strains were FO656h+ (ura4-D18 leu1-32 his3-D1 arg3-D4) and FO101h− (ura4-D18 leu1-32 his3-D1) and mutant strain FO763h+ (nth1::ura4+ ura4-D18 leu1-32 his3-D1 arg3-D4) (15). Complete yeast medium consists of 0.5% yeast extract and 3% glucose supplemented with 200 mg/l of arginine, leucine, histidine and uracil (YES) and for selection, Edinburgh minimal medium (EMM) with necessary supplements was used (35). For canavanine mutagenesis, glutamate (3.75 g/l) replaced ammonium as the nitrogen source and l-canavanine sulfate (75 mg/l; Sigma) was added to the EMM. The genomic sequence of apn2 (1609 bp) was PCR-amplified using Vent DNA polymerase (New England Biolabs) with start primer 5′-cgggatcctccgaattctaagttggaatgt (lacking atg) and stop primer 5′-aactgcagtcagtcctttgaatttgctct (linker sequences are in bold face letters) and inserted into the BamHI and PstI sites of pT7SCII (Stratagene). To generate an apn2 disruption construct, the BsaBI–SacI fragment of apn2 in pT7 was replaced with the kanMX4 (SmaI–AlwNI) cassette of pCore (36). The apn2::kanMX mutant (RHP103) was made by targeted gene disruption of the apn2 wild-type allele. The apn2kanMX fragment was excised and S.pombe FO101 cells were transformed according to the protocol of the Sc EasyComp Transformation kit (Invitrogen) and plated on YES before replica plating on YES with G418 (100 mg/l; Gibco). The apn2, nth1 double mutant (RHP109) was generated by random spore analysis after crossing FO763 and RHP103. The genotypes of all mutants were verified using PCR or Southern blotting.
Enzymes
Restriction enzymes and uracil DNA glycosylase were from New England Biolabs and E.coli MutY was from Trevigen. Escherichia coli Nth was purified as described previously (24). Escherichia coli Fpg was kindly provided by Lars Eide and S.cerevisiae Apn1 was purified as described previously (37).
Yeast cell-free extract
Schizosaccharomyces pombe cells (400 ml) were grown at 30°C until A600 was 0.6–1.0, harvested by centrifugation, washed once with water and suspended in 5 ml of 50 mM Tris–HCl, pH 8.0, 200 mM NaCl, 0.1 mM EDTA, 10 mM 2-mercaptoethanol and protease inhibitors (1 μg/ml leupeptin, 1 μg/ml pepstatin A, 1 μg/ml antipain, 10 μg/ml benzamidine and 100 mM phenylmethylsulfonyl fluoride). The cells were disrupted using a French Press (Sim Aminco) and the cell debris were collected by centrifugation at 12 000 r.p.m. for 15 min at 4°C.
Assay for enzyme cleavage of DNA substrates containing base damage
Double-stranded DNA substrates containing single base damages as indicated were generated by labelling the 5′ end of the oligonucleotide using T4 polynucleotide kinase (MBI Fermentas) and [γ-32P]ATP (3000 Ci/mmol; Amersham). The labelled oligonucleotides were annealed to complementary strands. The enzyme activities were assayed in a reaction buffer containing 50 mM MOPS, pH 7.5, 1 mM DTT, 1 mM EDTA and 5% glycerol at 37°C for 30 min. When indicated, MgCl2 (2–5 mM) was included in the reactions. The reaction mixtures contained 1–10 fmol substrate and enzymes/protein extracts as indicated in a total volume of 10 μl. To hydrolyse uncleaved abasic sites, 100 mM NaOH was added, and the incubation continued for 10 min at 70°C, followed by neutralization with 100 mM HCl. The reactions were stopped by adding 10 μl formamide DNA loading buffer, the samples were heated for 3 min at 80°C and the reaction products were separated on 7 M urea–20% polyacrylamide gels (LongRanger) in 1× taurin buffer. The radiolabelled fragments were visualized using a PhosphorImager (Molecular Dynamics, 445 SI).
The sequences of the DNA substrates used in this study are as follows: 5′-GGCGGCATGACCCXGAGGCCCATC-3′, where X = uracil and A, C, G or T was in the complementary strand. AP DNA was prepared by the incubation of uracil-containing oligonucleotide with Udg (NEB) for 30 min at 37°C before use. The MutY substrate was 5′-GATGGGCCTCAGGGTCATGCCGCC-3′ and the complementary strand contained 8oxoG opposite ‘A’ (in boldface).
NaCNBH3 mediated trapping of enzyme–substrate intermediates
Endlabelled AP-DNA (10 fmol) was incubated with whole cell extract in the presence of 50 mM sodium cyanoborohydride (NaCNBH3), 50 mM MOPS, pH 7.5, 1 mM DTT, 1 mM EDTA and 5% glycerol in a total volume of 12.5 μl. After incubation for 30 min at 37°C, the reaction was stopped by adding 5 μl of SDS-loading buffer. Samples were boiled, analysed on 10% Tricine–SDS gels and visualized by phosphorimaging.
Yeast survival and mutagenesis
Schizosaccharomyces pombe cells were grown until A600 was 0.6–0.8, washed in water, diluted and spread on agar plates containing up to 0.01% MMS in triplicates to measure cell survival. For canavanine mutagenesis, 6–8 cultures (A600 0.6–1.0) from each strain were plated on canavanine (75 mg/l)/EMM agar plates containing glutamic acid (3.75 g/l) as nitrogen source and incubated at 30°C for 9–10 days before the colony number was counted. Dilutions from the same cultures were plated in duplicate on YES plates to measure the number of viable cells.
RESULTS
apn2 and nth1 mutant analysis
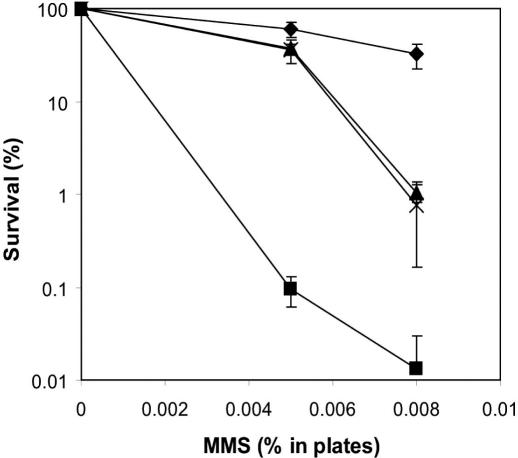
The genome sequence of S.pombe contains two open reading frames encoding putative AP endonucleases, termed Apn1 and Apn2. The apn1 mutant is normally resistant to the alkylating agent and AP site inducer MMS, indicating a limited role if any, for Apn1 in the repair of abasic sites in S.pombe (15,31). To evaluate the role of Apn2, an apn2 mutant was generated. Haploid mutant cells are viable and grow normally, suggesting that Apn2 is not an essential function. However, the apn2 mutant is highly sensitive to MMS exposure (Figure 1). This is in contrast to the apn2 mutant of the budding yeast S.cerevisiae, which exhibits wild-type resistance to MMS (19). Previously, we have shown that the nth1 mutant of S.pombe is also hypersensitive to MMS and proposed that the AP lyase activity of Nth1 is critical for AP site repair in the fission yeast (15). To study if Nth1 and Apn2 acted in different or the same pathway of alkylation or abasic site repair an apn2, nth1 double mutant was generated. The deletion of nth1 in the apn2 background partially relieved the MMS sensitivity of the apn2 single mutant indicating that Apn2 and Nth1 act in the same pathway for the repair of AP sites in S.pombe (Figure 1).
Figure 1.
MMS sensitivity of apn2 and nth1 single and double mutants. S.pombe wild-type (closed diamonds), apn2 (closed squares), nth1 (closed triangles), and apn2, nth1 (crosses) mutant cells were plated on solid media containing increasing doses of MMS and colony survival calculated relative to plates without MMS.
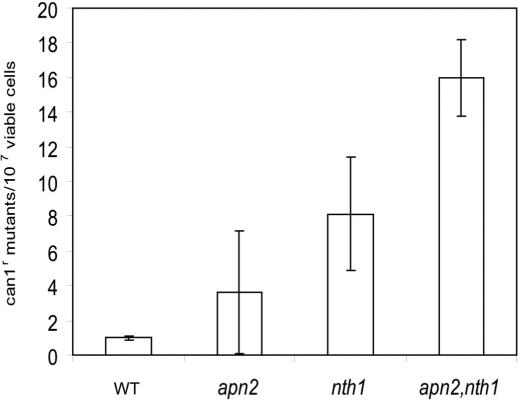
To further characterize the apn2 and nth1 single and double mutants, the spontaneous mutation frequencies were determined (Figure 2). Compared to wild-type, the mutation frequency of the apn2 and nth1 single mutants increased 3–4-fold and 8-fold, respectively, suggesting that both repair enzymes are essential for repairing spontaneous DNA damage. In the apn2, nth1 double mutant, the level of spontaneous mutations is further increased (16-fold) when compared with the wild-type. These findings suggest that the double mutant accumulates abasic sites, which could be bypassed by translesion polymerases, whereas the single mutants predominantly accumulate lethal intermediates of partially processed abasic sites.
Figure 2.
Spontaneous canavanine forward mutations in cells lacking apn2 and nth1. Logarithmically growing S.pombe cells were plated on EMM plates supplemented with l-canavanine sulfate for the determination of spontaneous mutation frequencies to Can1R in wild-type (WT), apn2, nth1 and apn2, nth1 mutant cells.
Nth1 is the major incising enzyme at abasic sites in cell-free extract from S.pombe
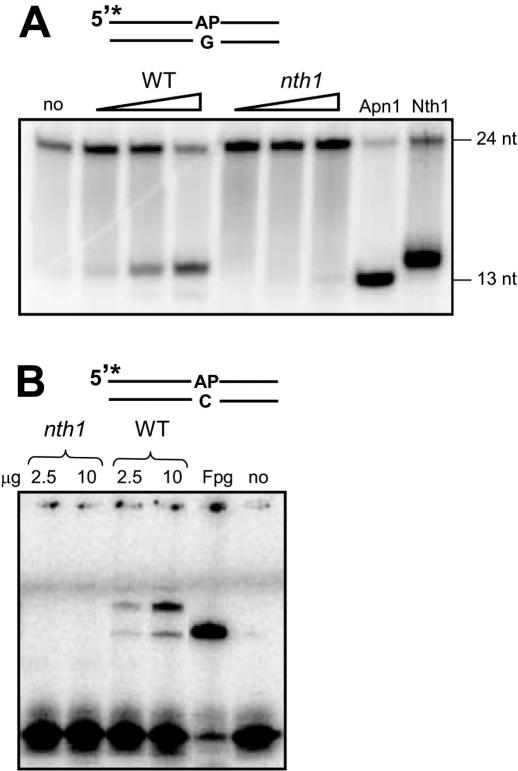
The alkylation sensitive and mutator phenotypes of the apn2 and nth1 deletion strains, suggest that both enzymes are involved in the processing of AP sites, and a biochemical approach was used to study the separate roles of Apn2 and Nth1 in the repair of AP sites. Whole cell extract from S.pombe was incubated with a 5′ endlabelled oligonucleotide substrate containing a single abasic site opposite G and the cleavage products were analysed using denaturing PAGE (Figure 3A). Mg2+ was included in the reaction mixture as many AP endonucleases required Mg2+ for optimal activity. Incision of AP DNA in the wild-type extract yielded a major product corresponding to an AP lyase cleavage (14 nt) and no band corresponding to an AP endonuclease cleavage (13 nt). Experiments with oligonucleotides containing an abasic site opposite A, C or T produced similar results, demonstrating that the AP lyase activity of Nth1 is independent of the base in the complementary strand (data not shown). AP lyase incision was absent in cell-free extract from nth1 mutant cells, showing that the cleavage product observed in wild-type extracts was catalysed by the Nth1 protein. In the nth1 mutant extracts, a faint band of the size of 5′-nicking can be seen, indicating that an AP endonuclease activity is present in the extracts from S.pombe. This band is not always evident in the wild-type extracts suggesting perhaps that the AP endonuclease activity might be upregulated in the nth1 mutant cells. A third faint band, migrating faster than the AP lyase product, was found in both wild-type and nth1 mutant extract. This product might correspond to δ-elimination by Fpg/Nei-like activities. However, no such activity has been described in S.pombe or identified by sequence homology searches of the genome sequence.
Figure 3.
DNA incision and borohydride protein complex trapping at AP sites by cell extracts from wild–type (WT) and nth1 mutant cells. (A) Cleavage of AP DNA in S.pombe cell-free extracts. Duplex 24mer oligodeoxyribonucleotide containing a single abasic site (opposite G) was incubated with 0.1, 0.4 or 1 μg of whole cell extracts from wild-type (FO656) and nth1 mutant (FO763) cells in reaction buffer supplemented with 5 mM MgCl2. Cleaved DNA was separated from intact DNA using PAGE and visualized by phosphorimaging. Purified Apn1 (15 ng) from S.cerevisiae and Nth1 (15 ng) from S.pombe were used as markers of AP endonuclease and AP lyase cleavage, respectively. (B) Probing for covalent enzyme–AP DNA intermediates by NaCNBH3 reduction. An aliquot of 2.5 or 10 μg S.pombe whole cell extract, 20 ng E.coli Fpg or no enzyme was incubated with 10 fmol of a 24mer DNA duplex containing a single AP site opposite C in the presence of 50 mM NaCNBH3. Protein–DNA complexes were separated from DNA by SDS–PAGE.
Bifunctional DNA glycosylases such as Nth1 coordinate the glycosylase/AP lyase reaction through a Schiff base intermediate between the C1 carbon of the sugar moiety and the amino group of a lysine residue. This enzyme–DNA complex can be irreversibly linked in the presence of borohydride and visualized using SDS–PAGE. As shown in Figure 3B, two trapped products were observed in extracts from wild-type cells. Both bands were absent in extracts of the nth1 mutant, suggesting that both products represent complex formation with Nth1 and suggest the presence of two different molecular forms, e.g. both a nuclear and a mitochondrial form. These data further support that there are no other significant AP lyase activities in S.pombe. This is also consistent with searches in the S.pombe genome database, where no other sequences with homology to known AP lyases can be identified.
Nth1 acts downstream of the MutY and Udg DNA glycosylases
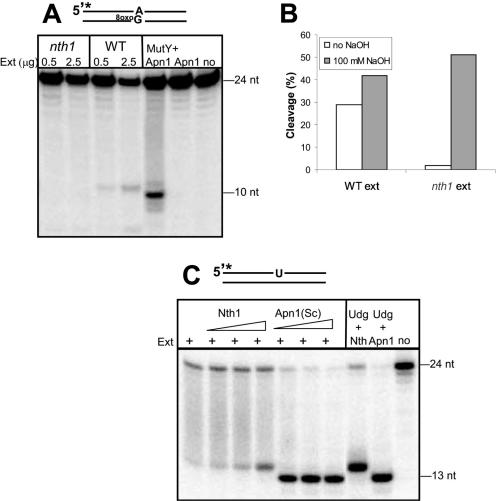
The finding that Nth1 is the major AP site incising activity in cell-free extract from S.pombe was somewhat unexpected, but consistent with the MMS-sensitive phenotype of the nth1 mutant and the apparent lack of a functional Apn1 homologue. To examine whether Nth1 has a more generalized incision role in BER, strand cleavage of DNA oligonucleotides containing base lesions were analysed in cell-free extract. A duplex DNA containing adenine opposite 8oxoG was in the extract cleaved by the Nth1 AP lyase presumably after base removal by MutY, which is known to be present in S.pombe (Figure 4A). Also, in assays with uracil-containing DNA and wild-type extract AP lyase cleavage was evident (Figure 4C, lane 1 from left). AP endonuclease cleavage was not detected indicating that the AP lyase activity of Nth1 processes strand cleavage of abasic site following base removal in S.pombe. In nth1 mutant extract, no strand cleavage of neither A:8oxoG (Figure 4A) nor uracil (data not shown) DNA substrates could be detected, underscoring the fact that Nth1 normally acts downstream of the monofunctional DNA glycosylases. Subsequent treatment of wild-type and nth1 mutant extracts containing A:8oxoG DNA with NaOH demonstrated that the MutY activity is not altered in the nth1 mutant (Figure 4B). To investigate whether strand incision of abasic sites is a rate-limiting step of BER, increasing amounts of either purified Nth1 or S.cerevisiae Apn1 was added to the wild-type extract and nicking of the uracil containing oligonucleotide was monitored. Addition of both AP lyase (Nth1) and the AP endonuclease increased the extent of the strand cleavage of the uracil substrate. Thus, it appears that AP site incision by Nth1 may be a rate-limiting step for the initiation of BER in S.pombe.
Figure 4.
DNA glycosylase activities in cell extracts from S.pombe. (A) Cleavage of A/8oxoG-containing duplex DNA. An aliquot of 0.5 and 2.5 μg of protein extract from wild-type or nth1 cells, 20 ng MutY from E.coli and 10 ng purified Apn1 from S.cerevisiae, or 10 ng Apn1 only, were incubated with 10 fmol of a 24mer32P-labelled oligodeoxyribonucleotide harbouring an A opposite 8oxoG in the presence of 5 mM Mg2+. Strand cleavage was analysed by 20% denaturing PAGE and phosphorimaging. (B) MutY activity in wild-type and nth1 extracts. Protein extracts (2.5 µg) from wild-type or nth1 cells were incubated with A:8oxoG DNA (as in A) and subsequently treated with 100 mM NaOH. Relative cleavage was quantified with the ImageQuaNT software. (C) Uracil removing and nicking activity in wild-type protein extracts of S.pombe. Whole cell extract (0.5 μg) was incubated with 10 fmol of a 24 bp oligodeoxyribonucleotide containing a single uracil residue (opposite A) at position 14 with or without S.pombe Nth1 (2, 5 or 10 ng) or S.cerevisiae Apn1 (100, 250 or 500 pg) for 30 min at 37°C. Similar experiments with Udg (NEB) were also included as indicated. The reaction products were separated on a polyacrylamide gel and bands were detected by phosphorimaging.
DISCUSSION
Abasic sites are one of the most frequent endogenous lesions in cellular DNA and their repair is critical for genomic stability and cellular survival. Here, we have addressed the roles of the DNA glycosylase/AP lyase Nth1 and the putative AP endonuclease Apn2 of S.pombe in the repair of abasic sites. Both mutant analysis and biochemical studies of cell-free extracts indicate that the AP lyase activity of Nth1 is the major activity for cleavage of the abasic sites in S.pombe. Moreover, DNA containing base lesions removed by MutY (A/8oxoG) and Udg (uracil) is also cleaved by the AP lyase activity of Nth1 in the following reaction step. However, the MMS-sensitive phenotype of the apn2 mutant also suggests an important function of Apn2 in the repair of abasic sites. Deletion of the nth1 gene reduced the MMS sensitivity of the apn2 mutant to the level of the nth1 single mutant, indicating that Apn2 further processes the toxic AP lyase product of Nth1. Thus it appears that the Nth1 AP lyase operate downstream of the monofunctional DNA glycosylases and upstream of Apn2 in the BER pathway of S.pombe.
In contrast to many other organisms S.pombe apparently possesses only one AP lyase activity, Nth1. S.cerevisiae has three bifunctional DNA glycosylases Ntg1, Ntg2 and Ogg1, which are all involved in base excision of oxidative damage and 3′ cleavage at abasic sites. Ntg1 and Ntg2 are sequence homologues of Nth1 with enzymatic characteristics similar to the S.pombe enzyme, whereas Ogg1 of S.cerevisiae has no sequence homologue in fission yeast. Similar to our findings with S.pombe, in which nth1 deletion reduces the MMS sensitivity of the apn2 mutant, the deletion of all three AP lyase activities in S.cerevisiae (Ntg1, Ntg2 and Ogg1) partially relieves the MMS hypersensitivity of the AP endonuclease deficient mutant (apn1, apn2) (7). These results suggest that the AP lyase activities of endonuclease III homologues of budding yeast and fission yeast are all involved in BER of alkylation damage. However, in contrast to the MMS sensitivity of the S.pombe nth1 single mutant, the ntg1, ntg2 double mutant (38,39) and ntg1, ntg2, ogg1 triple mutant of S.cerevisiae, (unpublished data) are normally resistant to alkylation. These results indicate that the alternative AP endonuclease-dependent pathway for the repair of abasic sites is absent or less important in S.pombe (15).
Incision of abasic sites appears to be a rate-limiting step of BER initiated by monofunctional glycosylases in S.pombe. This may indicate an important control mechanism to avoid the accumulation of detrimental AP lyase induced strand breaks with 3′-dRP moieties in the genome. The AP lyase activity may be cytotoxic to the cell if subsequent steps of BER do not process the 3′-blocking termini further because this intermediate may be more difficult to process than intact abasic sites. Therefore, we hypothesize that the AP lyase step slows down the BER pathway in S.pombe to ensure the recruitment of downstream BER proteins or direct the process through alternative mechanisms for the repair of intact AP sites. Epistatic analysis suggests that BER and NER are overlapping pathways for the repair of abasic sites in S.pombe (15). In addition, abasic sites are tolerated by translesion synthesis whereas 3′-dRP termini block both replicative polymerases and translesion polymerases. In addition, 3′-dRP termini will cause DNA double strand breaks if present in the template strand during replication. The ultraviolet (UV) endonuclease, Uvde1, was also shown to posses AP endonuclease activity in vitro (40); however, mutant analysis indicate that the physiological significance for the repair of AP sites in vivo is unclear (15).
In the absence of a functional Apn1 protein in S.pombe, Apn2 appears to be the only 3′-phosphodiesterase activity involved in the repair of 3′-dRP termini. The structure-specific 3′-flap endonucleases Mus81/Mms4 heterodimer and Rad1/Rad10 complex of S.cerevisiae overlap the 3′-phosphodiesterase activities of Apn1 and Apn2 in the repair of 3′-dRP residues. However, mutant analysis suggests that the Rad16/Swi10 of S.pombe (S.cervisiae Rad1/Rad10) is not involved in the repair of 3′-dRP residues but overlap with Nth1 in the repair of unprocessed AP sites (15). Further work is required to elucidate whether Mus81/Eme1 (S.cerevisiae Mus81/Mms4) overlap with Apn2 in the repair of 3′-dRP termini.
In vitro analysis of repair synthesis with whole cell extracts or purified enzymes show that multiple pathways of BER exist in prokaryotic and eukaryotic cells (41). The intrinsic properties of the DNA glycosylases are important for selection of the BER branch, in which bifunctional glycosylases most often initiate single nucleotide replacement (short patch), whereas monofunctional glycosylases can initiate both short and long patch repair. The AP lyase activity of bifunctional glycosylases creates a 3′-dRP residue subsequently removed by the phosphodiesterase activity of AP endonucleases. The resulting single nucleotide gap (short patch) is filled in by DNA polymerases. Our results show that the abasic sites are cleaved by the AP lyase activity of Nth1, indicating that single nucleotide replacement may be the major pathway of BER in S.pombe. Moreover, only very weak AP endonuclease activity could be detected in whole cell extracts, implying that long patch repair, initiated by base removal and subsequent AP endonuclease cleavage at the 5′ side of the abasic site might be a less significant pathway of BER in S.pombe.
Finally, Ribar et al. (34) characterized the mutants of S.pombe lacking apn1 and apn2. Similarly as shown here, they also conclude from the genetic analysis that Apn2 rather than Apn1 is the most important protein for abasic site repair in S.pombe. They also describe a very weak AP endonuclease activity associated with the purified protein but have not investigated any connection to Nth1. However, in essence, their results are largely consistent with the results described here.
Acknowledgments
ACKNOWLEDGEMENTS
This research was supported by the Norwegian Research Council and The Norwegian Cancer Society.
REFERENCES
- 1.Lindahl T. and Nyberg,B. (1972) Rate of depurination of native deoxyribonucleic acid. Biochemistry, 11, 3610–3618. [DOI] [PubMed] [Google Scholar]
- 2.Berdal K.G., Johansen,R.F. and Seeberg,E. (1998) Release of normal bases from intact DNA by a native DNA repair enzyme. EMBO J., 17, 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Memisoglu A. and Samson,L. (2000) Base excision repair in yeast and mammals. Mutat. Res., 451, 39–51. [DOI] [PubMed] [Google Scholar]
- 4.Seeberg E., Luna,L., Morland,I., Eide,L., Johnsen,B., Larsen,E., Alseth,I., Dantzer,F., Baynton,K., Aamodt,R., Kristiansen,K.I., Rognes,T., Klungland,A. and Bjoras,M. (2000) Base removers and strand scissors: different strategies employed in base excision and strand incision at modified base residues in DNA. Cold Spring Harb. Symp. Quant. Biol., 65, 135–142. [DOI] [PubMed] [Google Scholar]
- 5.Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature, 362, 709–715. [DOI] [PubMed] [Google Scholar]
- 6.Doetsch P.W. and Cunningham,R.P. (1990) The enzymology ofapurinic/apyrimidinic endonucleases. Mutat. Res., 236, 173–201. [DOI] [PubMed] [Google Scholar]
- 7.Guillet M. and Boiteux,S. (2000) Endogenous DNA abasic sites cause cell death in the absence of Apn1, Apn2 and Rad1/Rad10 in Saccharomyces cerevisiae. EMBO J., 21, 2833–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loeb L.A. (1985) Apurinic sites as mutagenic intermediates. Cell, 40, 483–484. [DOI] [PubMed] [Google Scholar]
- 9.Haracska L., Unk,I., Johnson,R.E., Johansson,E., Burgers,P.M., Prakash,S. and Prakash,L. (2001) Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites. Genes Dev., 15, 945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu S.L., Lee,S.K., Johnson,R.E., Prakash,L. and Prakash,S. (2003) The stalling of transcription at abasic sites is highly mutagenic. Mol. Cell. Biol., 23, 382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsumoto Y. and Kim,K. (1995) Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science, 269, 699–702. [DOI] [PubMed] [Google Scholar]
- 12.Wu X., Li,J., Li,X., Hsieh,C.L., Burgers,P.M. and Lieber,M.R. (1996) Processing of branched DNA intermediates by a complex of humanFEN-1 and PCNA. Nucleic Acids Res., 24, 2036–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klungland A. and Lindahl,T. (1997) Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J., 16, 3341–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Memisoglu A. and Samson,L. (2000) Contribution of base excision repair, nucleotide excision repair, and DNA recombination to alkylation resistance of the fission yeast Schizosaccharomyces pombe. J. Bacteriol., 182, 2104–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osman F., Bjørås,M., Alseth,I., Morland,I., McCready,S., Seeberg,E. and Tsaneva,I. (2003) A new Schizosaccharomyces pombe base excision repair mutant, nth1, reveals overlapping pathways for repair of DNA base damage. Mol. Microbiol., 48, 465–480. [DOI] [PubMed] [Google Scholar]
- 16.Rogers S.G. and Weiss,B. (1980) Cloning of the exonuclease III gene of Escherichia coli. Gene, 11, 187–195. [DOI] [PubMed] [Google Scholar]
- 17.Cunningham R.P., Saporito,S.M., Spitzer,S.G. and Weiss,B. (1986) Endonuclease IV (nfo) mutant of Escherichia coli. J. Bacteriol., 168, 1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson R.E., Torres-Ramos,C.A., Izumi,T., Mitra,S., Prakash,S. and Prakash,L. (1998) Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev., 12, 3137–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bennett R.A. (1999) The Saccharomyces cerevisiae ETH1 gene, an inducible homolog of exonuclease III that provides resistance toDNA-damaging agents and limits spontaneous mutagenesis. Mol. Cell. Biol., 19, 1800–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hadi M.Z. and Wilson,D.M.,III (2000) Second human protein with homology to the Escherichia coli abasic endonuclease exonuclease III. Environ. Mol. Mutagen., 36, 312–324. [PubMed] [Google Scholar]
- 21.Breimer L.H. and Lindahl,T. (1984) DNA glycosylase activities for thymine residues damaged by ring saturation, fragmentation, or ring contraction are functions of endonuclease III in Escherichia coli. J. Biol. Chem., 259, 5543–5548. [PubMed] [Google Scholar]
- 22.Boiteux S., O'Connor,T.R. and Laval,J. (1987) Formamidopyrimidine-DNA glycosylase of Escherichia coli: cloning and sequencing of the fpg structural gene and overproduction of the protein. EMBO J., 6, 3177–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eide L., Bjoras,M., Pirovano,M., Alseth,I., Berdal,K.G. and Seeberg,E. (1996) Base excision of oxidative purine and pyrimidine DNA damage in Saccharomyces cerevisiae by a DNA glycosylase with sequence similarity to endonuclease III from Escherichia coli. Proc. Natl Acad. Sci. USA, 93, 10735–10740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alseth I., Eide,L., Pirovano,M., Rognes,T., Seeberg,E. and Bjoras,M. (1999) The Saccharomyces cerevisiae homologues of endonuclease III from Escherichia coli, Ntg1 and Ntg2, are both required for efficient repair of spontaneous and induced oxidative DNA damage in yeast. Mol. Cell. Biol., 19, 3779–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bjoras M., Luna,L., Johnsen,B., Hoff,E., Haug,T., Rognes,T. and Seeberg,E. (1997) Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic sites. EMBO J., 16, 6314–6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morland I., Rolseth,V., Luna,L., Rognes,T., Bjoras,M. and Seeberg,E. (2002) Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res., 30, 4926–4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuzminov A. (2001) Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc. Natl Acad. Sci. USA, 98, 8241–8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dianov G.L., Sleeth,K.M., Dianova,I.I. and Allinson,S.L. (2003) Repair of abasic sites in DNA. Mutat. Res., 531, 157–163. [DOI] [PubMed] [Google Scholar]
- 29.Slupphaug G., Kavli,B. and Krokan,H.E. (2003) The interacting pathways for prevention and repair of oxidative DNA damage. Mutat. Res., 531, 231–251. [DOI] [PubMed] [Google Scholar]
- 30.Fleck O. and Nielsen,O. (2004) DNA repair. J. Cell. Sci., 117, 515–517. [DOI] [PubMed] [Google Scholar]
- 31.Ramotar D., Vadnais,J., Masson,J.Y. and Tremblay,S. (1998) Schizosaccharomyces pombe apn1 encodes a homologue of the Escherichia coli endonuclease IV family of DNA repair proteins. Biochim. Biophys. Acta, 1396, 15–20. [DOI] [PubMed] [Google Scholar]
- 32.Popoff S.C., Spira,A.I., Johnson,A.W. and Demple,B. (1990) Yeast structural gene (APN1) for the major apurinic endonuclease: homology to Escherichia coli endonuclease IV. Proc. Natl Acad. Sci. USA, 87, 4193–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fraser J.L., Neill,E. and Davey,S. (2003) Fission yeast Uve1 and Apn2 function in distinct oxidative damage repair pathways in vivo. DNA Repair, 2, 1253–1267. [DOI] [PubMed] [Google Scholar]
- 34.Ribar B., Izumi,T. and Mitra,S. (2004) The major role of humanAP-endonuclease homolog Apn2 in repair of abasic sites in Schizosaccharomyces pombe. Nucleic Acids Res., 32, 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guthrie C. and Fink,G. (1991) Methods in Enzymology. Academic Press, San Diego, CA, pp. 795–823. [Google Scholar]
- 36.Storici F., Lewis,L.K. and Resnick,M.A. (2001) In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol., 19, 773–776. [DOI] [PubMed] [Google Scholar]
- 37.Levin J.D., Shapiro,R. and Demple,B. (1991) Metalloenzymes in DNA repair. Escherichia coli endonuclease IV and Saccharomyces cerevisiae Apn1. J. Biol. Chem., 266, 22893–22898. [PubMed] [Google Scholar]
- 38.You H.J., Swanson,R.L., Harrington,C., Corbett,A.H.,Jinks-Robertson,S., Senturker,S., Wallace,S.S., Boiteux,S., Dizdaroglu,M. and Doetsch,P.W. (1999) Saccharomyces cerevisiae Ntg1p and Ntg2p: broad specificity N-glycosylases for the repair of oxidative DNA damage in the nucleus and mitochondria. Biochemistry, 38, 11298–11306. [DOI] [PubMed] [Google Scholar]
- 39.Hanna M., Chow,B.L., Morey,N.J., Jinks-Robertson,S., Doetsch,P.W. and Xiao,W. (2004) Involvement of two endonuclease III homologs in the base excision repair pathway for the processing of DNA alkylation damage in Saccharomyces cerevisiae. DNA Repair, 3, 51–59. [DOI] [PubMed] [Google Scholar]
- 40.Kanno S., Iwai,S., Takao,M. and Yasui,A. (1999) Repair of apurinic/apyrimidinic sites by UV damage endonuclease; a repair protein for UV and oxidative damage. Nucleic Acids Res., 27, 3096–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dogliotti E., Fortini,P., Pascucci,B. and Parlanti,E. (2001) The mechanism of switching among multiple BER pathways. Prog. Nucleic Acid Res. Mol. Biol., 68, 3–27. [DOI] [PubMed] [Google Scholar]