Abstract
Chronic arsenic exposure via drinking water has become a worldwide public health concern. In humans, inorganic arsenic (iAs) is metabolized to monomethylarsonic acid (MMA) and dimethylarsinic acid (DMA) mainly mediated by arsenic (+3 oxidation state) methyltransferase (As3MT). We reported recently that N-6 adenine-specific DNA methyltransferase 1 (N6AMT1) was involved in arsenic metabolism, and examined its interactive effect with As3MT on arsenic metabolism in vitro. To further evaluate the interactive effect of N6AMT1 and As3MT on arsenic biomethylation in humans, we conducted a human population-based study including 289 subjects living in rural villages in Inner Mongolia, China, and assessed their urinary arsenic metabolites profiles in relation to genetic polymorphisms and haplotypes of N6AMT1 and As3MT. Five N6AMT1 single nucleotide polymorphisms (SNPs; rs1003671, rs7282257, rs2065266, rs2738966, rs2248501) and the N6AMT1 haplotype 2_GGCCAT were significantly associated with the percentage of iAs (% iAs) in urine (e.g., for rs7282257, mean was 9.62% for TT, 6.73% for AA). Rs1003671 was also in a significant relationship with urinary MMA and DMA (the mean of %MMA was 24.95% for GA, 31.69% for GG; the mean of % DMA was 69.21% for GA, 59.82% for GG). The combined effect of N6AMT1 haplotype 2_GGCCAT and As3MT haplotype 2_GCAC showed consistence with the additive significance of each haplotype on % iAs: the mean was 5.47% and 9.36% for carriers with both and null haplotypes, respectively. Overall, we showed that N6AMT1 genetic polymorphisms were associated with arsenic biomethylation in the Chinese population, and its interaction with As3MT was observed in specific haplotype combinations.
Keywords: arsenic metabolism, N6AMT1, As3MT, polymorphism, haplotype.
Arsenic compounds are ubiquitous environmental contaminants that humans commonly encounter daily at relatively low levels. However, in some regions of the world, high levels of arsenic naturally exist in drinking water, and consumption of inorganic arsenic (iAs) contaminated drinking water has constituted a worldwide public health concern (Abernathy et al., 1999; Tapio and Grosche, 2006). Hetao Plain in Inner Mongolia, China, is known to be one of the severely arsenic-affected areas in the world (Fujino et al., 2004, 2006; Guo et al., 2001, 2003; Mao et al., 2010). Previous investigations have reported that approximately 80% of tube wells in this area had arsenic concentrations higher than 50 µg/l (Guo et al., 2001, 2007). In 2007, the permitted maximum level of inorganic arsenic in drinking water in China was reduced from 50 to 10 µg/l, which is also the recommended maximum allowance in drinking water by World Health Organization (WHO).
In humans, iAs is metabolized to trivalent and pentavalent mono-methylated (MMA) and di-methylated arsenicals (DMA), which are excreted in the urine (Hopenhayn-Rich et al., 1996; Marapakala et al., 2012). Biomethylation of arsenic is accomplished dominantly by the arsenic (+3 oxidation state) methyltransferase (As3MT) enzyme (Thomas et al., 2007). Studies have reported that genetic polymorphisms of As3MT affect arsenic metabolism, which result in varied urinary arsenical profiles in individuals and among different populations (Agusa et al., 2009; Engström et al., 2011; Valenzuela et al., 2009; Whitbread et al., 2003). Variability in arsenic biomethylation and the resulting changes of arsenic metabolite profiles contribute to the differential susceptibility of individuals to the toxic and carcinogenic activity of arsenic (Drobna et al., 2004; Gomez-Rubio et al., 2010). In addition, biological and demographic characteristics such as gender, age, body mass index (BMI), personal lifestyle (smoking history or alcohol consumption status) could affect arsenic metabolism (Lindberg et al., 2010; Sun et al., 2007; Tseng, 2009; Zhang et al., 2014). Thus, it is important to consider the influence of these potential factors on arsenic metabolism when evaluating the association between genetic variations and arsenic metabolic profiles.
We previously demonstrated that N-6 adenine-specific DNA methyltransferase 1 (N6AMT1), a putative methyltransferase, could selectively participate in the conversion of the toxic MMAIII to less toxic DMAV (Ren et al., 2011), while its effect is relatively minor and limited compared to As3MT (Zhang et al., 2015). An evaluation of the association between polymorphisms in N6AMT1 and the efficiency of arsenic biomethylation suggested that genetic variants of N6AMT1 had a significant effect in the variability of urinary MMA levels among individuals of the Andean women (Harari et al., 2013).
In the present study, we assessed the association of genetic polymorphisms and haplotypes of N6AMT1 and As3MT and urinary arsenic metabolites profiles in a Chinese population, living in Hetao plain of Inner Mongolia, China. Specifically, we were interested in examining the interactive effect of N6AMT1 and As3MT on arsenic biomethylation in humans.
MATERIALS AND METHODS
Study area and population
In 2010, our study population was recruited from 3 arsenic-exposed villages of Wuyuan County, located in the Hetao plain of Inner Mongolia, China. Wuyuan County is one of the heavily arsenic-exposed areas of China, where approximately 10% of villages were found to be arsenic-affected (water arsenic levels > 50 µg/l) (Ma et al., 1996). Most of the residents in this region engaged in agricultural work, and shared similar life style, dietary structures, public health service facilities and socio-economic status. No factories, mines or other industries discharge arsenic into the local air, water or soil. In the past 2 decades, a large number of villagers in this region have started to use a central municipal water supply instead of a water system characterized by numerous widespread individual wells. The 3 villages were selected for this study partly because their villagers continue to rely on private open-tube wells for drinking water need, which contain arsenic with a level higher than 10 µg/l. A total of 450 participants (age ≥ 18 years old) were recruited from a total of 653 residents living in the 3 villages. Pregnant women were excluded for safety reasons; and pregnancy has been suggested to affect arsenic methylation (Gardner et al., 2012). Overall, 203 were excluded due to not meeting inclusion criteria or refusing to participate. The protocol was reviewed and approved by the ethics committee of Wenzhou Medical University, China. The purpose and procedures of the study were carefully explained to all participants. Written informed consent from all of the participants was obtained before initiation of the study. Each recruited participant was interviewed using a standardized questionnaire specifically designed for this study to collect demographic characteristics, such as date of birth, gender, occupation, education, current health status, medical history, alcohol and tobacco consumption history, length of residence in the area, and drinking water sources. Height and weight were measured to the nearest 0.1 cm and 0.1 kg, respectively, and body mass index (BMI) was calculated as weight (kg)/height (meters squared). All surveys, physical examinations and samples collection were carried out by specifically trained staff from Wenzhou Medical University and Wuyuan Centre for Disease Prevention and Control (Wuyuan CDC), China. Water samples from tube wells of each household were collected during the home interview. Inorganic arsenic concentrations in the collected water samples were determined using the silver diethyl-dithiocarbamate spectrometry method (detection limit being 1 µg/l).
Among the 450 participants, 161 individuals were excluded from final analysis due to several reasons: 119 with missing urine or blood samples, 42 with incomplete personal information or lacking of drinking water arsenic level.
Blood and urine collection
Fasting venous blood samples were drawn from each subject with tubes containing ethylenediaminetetraacetic acid. Buffy coat was isolated from blood samples within 30 min. The second or third urine voids (20 ml) of the same day with blood collection were collected and kept immediately in ice. All these biological specimens were transferred to Wuyuan CDC within 2 hours after collection and were stored in freezers at -80°C until analysis.
DNA extraction and genotyping
Genomic DNA was extracted from buffy coat with the QIAGEN FlexiGene DNA kit (QIAGEN, Hilden, Germany) following the manufacturer’s protocol. The genotyping of 8 N6AMT1 SNPs (in 3′–5′ order: rs1006903, rs1003671, rs4816333, rs7282280, rs7282257, rs2065266, rs2738966, rs2248501) and 6 As3MT SNPs (in 5′–3′ order: rs7085104, rs3740400, rs3740393, rs3740390, rs11191439, rs1046778) (Supplementary Table S1) were performed by using iPLEX Gold chemistry on Agena Bioscience MassARRAY® System (Agena Bioscience, San Diego, California) at the Genomic Shared Resources at Roswell Park Cancer Institute. Multiplex assays were designed using Assay Design Suite v2.0 (Agena Bioscience) software and genotyping was performed as per manufacturer protocol. Following a locus-specific PCR amplification, the unincorporated dNTPs were dephosphorylated with shrimp alkaline phosphatase (Agena Bioscience) and the assay probes were extended into SNP sites by single-nucleotide extension with acyclo-NTP termination (iPLEX Pro Reagent Kit, 40 cycles of the extension reaction, Agena Bioscience). Primer sequences of PCR reaction and extension sequencing were provided in Supplementary Information (Supplementary Table S2). The contents of the PCR/extension reactions were desalted with strong cation-exchange resin and transferred onto a SpectroChip® Array (Agena Bioscience) for MALDI-TOF analysis. The genotypes for each SNP were obtained after visual inspection and corrections of the spectra using MassARRAY Typer Analyzer v4 (Agena Bioscience). The SNPs of As3MT were selected for this study according to previous reports that these SNPs are significantly associated with As3MT methylation efficiency in human samples (Hernández et al., 2008; Gomez-Rubio et al., 2010; Schläwicke Engström et al., 2007, 2009; Valenzuela et al., 2009; Wood et al., 2006). Given little evidence of investigation of N6AMT1 SNPs in terms of functional association with its enzyme activity in the Chinese population, we chose SNPs spanning the whole N6AMT1 gene and the extended region from 5′ and 3′ terminals for study.
Tag SNPs selection and haplotype inferring
Tag SNPs for N6AMT1 and As3MT were selected from the genotyped SNPs, and linkage disequilibrium (LD) was estimated using Haploview (version 4.2; Barrett et al., 2005; Barrett, 2009) based on the physical positions on chromosome of selected SNPs and genotyping result of the study population’s DNA. LD values (R2) of 0.8 or higher have been recommended as an acceptable threshold for tag SNPs selection (de Bakker et al., 2005). According to the strength of LD (R2 ≥ 0.8), 6 N6AMT1 SNPs (in 3′–5′ order: rs1006903, rs1003671, rs4816333, rs7282280, rs7282257, rs2065266) and 4 As3MT SNPs (in 5′–3′ order: rs7085104, rs3740400, rs3740390, rs1046778) were picked up for haplotype inferring. Haplotypes were inferred from chosen tag SNPs by PHASE software using a Bayesian method (version 2.1; Stephens and Donnelly, 2003).
Urinary arsenic species profile analysis by high-performance liquid chromatography/inductively coupled plasma-mass spectrometry
The urine samples were thawed to room temperature right before analysis. They were centrifuged at 1200× g for 15 min and diluted 1:10 with mobile phase to minimize matrix effects. Separation of the various species of arsenic was performed using a high-performance liquid chromatography system (Thermo HPLC Spectra System, Thermo Fisher Scientific Inc., U.S.) for separation coupled to an inductively coupled plasma mass spectrometer (Thermo X-Series 2 ICP-MS) for detection. Separation was achieved using an ion-pair chromatography column (Gemini-NX-C18 column, 3 μm × 4.6 mm id × 150 mm) equipped with a guard column (Gemin-NX C18, 4 × 3.0 mm id) from Phenomenex Inc., U.S. The HPLC column outlet was connected to the nebulizer of the ICP-MS by polyethylene tubing. Instrument conditions were modified based on prior reported work (Kalman et al., 2014). The optimized conditions for ICP-MS and HPLC settings, as well as detailed reagents preparation, were given in the supplementary information. HPLC was run using Thermo Xcalibur software, and the ICP-MS data were collected and processed using the Thermo PlasmaLab software package.
Retention time of peaks were identified by mixed arsenic speciation standards. Possible interference with same mass to charge response such as 40Ar35Cl+ was monitored through chromatographic separation to ensure specificity of each interested arsenic species at their retention time. Arsenic species were quantified by external calibration curves (5, 25, 50, 100, 200 µg/l). Detection limit (LOD) of AsIII, DMAV, MMAV, and AsV was 5, 1, 1, and 1 µg/l, respectively. LOD was evaluated by the concentration of compound resulting signal to noise above 3. Stability was determined by the percent relative standard deviation (RSD) of peak area from 5 replicates of 50 µg/l standard solutions. For arsenic species, the peak area reproducibility ranged from 0.3% to 1.0% RSD. The accuracy of the method was demonstrated through the analysis of 50 µg/l spiked urine samples. Percent recovery of 120%, 87%, 107%, and 100% were calculated for AsIII, DMAV, MMAV, and AsV, respectively.
Statistical analysis
The total arsenic (tAs) concentration in urine was calculated by summing up iAs (iAsIII+iAsV), MMA (MMAIII+MMAV) and DMA (DMAIII+DMAV). The percentages of urinary arsenic metabolites (% iAs, % MMA and % DMA) were defined as: iAs/tAs*100%, MMA/tAs*100% and DMA/tAs*100%, respectively. In contrast to urinary arsenic concentrations profiles, 2 indexes, the primary methylation index [PMI = (MMA + DMA)/tAs] and the secondary methylation index [SMI = DMA/(MMA + DMA)], were applied to evaluate As methylation ability at different stages (Sun et al., 2007).
The Hardy-Weinberg equilibrium (HWE) test for each polymorphism was assessed by a Chi-squared analysis. The differences of urinary arsenic metabolites profiles between subgroups of potential confounding risk factors (gender, age, BMI, smoking and alcohol consumption history) were analyzed by Student’s t-test. Genotypes of each polymorphism were arranged as categorical variables (homozygote and heterozygote). Likewise, haplotypes were also modeled as categorical variables (0, 1 and 2 copies of the haplotype). For consideration of increasing statistical analysis power, groups with 1 or 2 (non-null) haplotype copies were pooled together, since there were relatively few subjects with 2 haplotype copies in all groups. Pairwise analysis was conducted to evaluate the joint effect of N6AMT1 and As3MT haplotypes on arsenic metabolism outcomes. Analysis of Covariance (ANCOVA) was applied to compare the differences of urinary arsenic metabolites and methylation indexes (% iAs, % MMA, % DMA, PMI and SMI) among the polymorphism genotype groups, haplotype groups, and haplotypes combination groups, adjusted for potential confounding risk factors, including gender, age, BMI, alcohol consumption and smoking history. Covariates appearing in the model were evaluated at the following values: Gender = 1.4014, Age = 50.529, BMI = 24.616, Smoker = 0.3668, Drinker = 0.3080. Data shown in the text were based on the adjusted model, while crude data were arranged as supplementary materials.
All statistical analysis was carried out using SAS V9.4 (SAS Institute, Cary, North Carolina, U.S.). Statistical tests were 2-sided and considered significant for P < .05.
RESULTS
General Characteristics and Genetic Background
Descriptive characteristics of the study population are presented in Table 1. Among the 289 study subjects included in the analysis, 173 were females (59.9%) and 116 were males (40.1%), with an average age of 50.5 ± 11.4 years old and an average BMI of 24.6 ± 3.3 kg/m2. Among the subjects, 106 (36.7%) and 89 (30.8%) were current or former tobacco users and alcohol users, respectively. The average arsenic concentration in the study population’s drinking water was 92.4 ± 52.4 µg/l. As the outcome of arsenic exposure, average urinary arsenic metabolites (total iAs, MMA, DMA) were (7.67 ± 6.98)%, (26.08 ± 12.52)%, (66.25 ± 14.06)%, respectively, with PMIs of 0.92 ± 0.07 and SMIs of 0.72 ± 0.14 for the methylation capacity.
TABLE 1.
Demographic characteristics of the study population.
| Characteristics | Mean (SD)a, n (%) | Range |
|---|---|---|
| N | 289 | |
| Gender | ||
| Female [n (%)] | 173 (59.9%) | |
| Male [n (%)] | 116 (40.1%) | |
| Age (years) | 50.5 (11.4) | 17–79 |
| Height (cm) | 162.0 (8.2) | 143.3–182.4 |
| Weight (kg) | 64.7 (10.6) | 35.6–110.0 |
| Body mass index (kg/m2) | 24.6 (3.3) | 14.3–37.6 |
| Tobacco users [n (%)] | 106 (36.7%) | |
| Alcohol users [n (%)] | 89 (30.8%) | |
| Water arsenic concentration (µg/l) | 92.4 (52.4) | 10–165 |
| Urinary arsenic metabolites in percentage | ||
| Total iAs (%) | 7.67 (6.98) | 0–34.80 |
| MMA (%) | 26.08 (12.52) | 0–55.57 |
| DMA (%) | 66.25 (14.06) | 27.73–100.00 |
| Urinary arsenic metabolites methylation index | ||
| PMI | 0.92 (0.07) | 0.65–1.00 |
| SMI | 0.72 (0.14) | 0.34–1.00 |
SD: standard deviation.
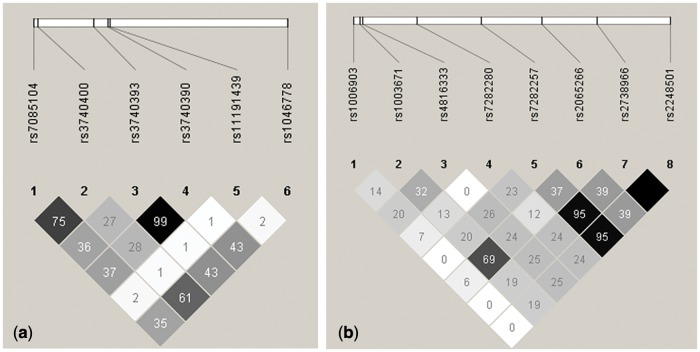
Genotypes of all SNPs were in Hardy-Weinberg equilibrium (P > .05), except rs11191439, which was excluded from the following analysis. Figure 1 showed the linkage disequilibrium (LD) value (R2) for the N6AMT1 SNPs and As3MT SNPs. Among 8 N6AMT1 SNPs, rs7282257 was in LD with rs2738966 (R2= 0.95) and rs2248501 (R2= 0.95), and rs2738966 was in strongest LD with rs2248501 (R2= 1). Rs3740393 showed the strongest LD with rs3740390 (R2= 0.99) among the 6 As3MT SNPs of interest. Based on the LD analysis results, 6 N6AMT1 SNPs (in 3′–5′ order: rs1006903, rs1003671, rs4816333, rs7282280, rs7282257, rs2065266) and 4 As3MT SNPs (in 5′–3′ order: rs7085104, rs3740400, rs3740390, rs1046778) were picked up as tag SNPs for haplotype inferring.
FIG. 1.
LD values (R2) for the SNPs of As3MT (a) and N6AMT1 (b) based on the study population. LD values were estimated using Haploview (version 4.2; Barrett et al., 2005; Barrett, 2009) based on the physical positions on chromosome of selected SNPs and genotyping result of the study population’s DNA. SNPs are shown 5′–3′ order for As3MT and 3′–5′ order for N6AMT1.
Differences of Arsenic Metabolites Profiles Between Subgroups
The differences of urinary arsenic metabolites profiles between subgroups of potential confounding risk factors (gender, age, BMI, smoking and alcohol consumption history) are presented in Table 2. % iAs and % MMA were significantly higher in males than in females, while % DMA was remarkably higher in females, which corresponded to significantly higher PMI and SMI. In the tobacco user subgroup, non-smokers had significantly lower % iAs, relatively lower % MMA, noticeably higher % DMA, PMI and SMI than current or former smokers. No significant differences were observed in any metabolites and methylation indexes between age, BMI or alcohol users subgroups. However, individuals of younger age, higher BMI or non-drinkers demonstrated better arsenic metabolism capacity (relatively lower % MMA, higher % DMA and SMI).
TABLE 2.
Differences of fractions of arsenic metabolites and methylation capacity indexes between subgroups.
| Variables | Subgroups | N | % iAs | % MMA | % DMA | PMI | SMI |
|---|---|---|---|---|---|---|---|
| Gender | Female | 173 | 6.77 (0.51)a | 23.95 (0.96) | 69.28 (1.07) | 0.93 (0.01) | 0.74 (0.01) |
| Male | 116 | 9.02 (0.66) | 29.26 (1.09) | 61.72 (1.19) | 0.91 (0.01) | 0.68 (0.01) | |
| P-value | .007 | <.001 | <.001 | .007 | <.001 | ||
| Age (years) | ≤49.2b | 146 | 7.72 (0.62) | 25.06 (1.03) | 67.22 (1.21) | 0.92 (0.01) | 0.73 (0.01) |
| >49.2 | 143 | 7.63 (0.54) | 27.13 (1.05) | 65.25 (1.12) | 0.92 (0.01) | 0.71 (0.01) | |
| P-value | .909 | .161 | .233 | .909 | .206 | ||
| BMI (kg/m2) | ≤25 | 167 | 7.62 (0.53) | 27.31 (0.92) | 65.07 (0.99) | 0.92 (0.01) | 0.70 (0.01) |
| >25 | 122 | 7.75 (0.64) | 24.40 (1.19) | 67.85 (1.41) | 0.92 (0.01) | 0.73 (0.01) | |
| P-value | .871 | .051 | .098 | .871 | .079 | ||
| Tobacco users | Current or former | 106 | 9.03 (0.69) | 27.79 (1.11) | 63.18 (1.24) | 0.91 (0.01) | 0.69 (0.01) |
| Never | 183 | 6.89 (0.50) | 25.09 (0.96) | 68.02 (1.07) | 0.93 (0.01) | 0.73 (0.01) | |
| P-value | .012 | .077 | .005 | .012 | .032 | ||
| Alcohol users | Current or former | 89 | 8.49 (0.83) | 27.49 (1.29) | 64.01 (1.48) | 0.92 (0.01) | 0.70 (0.01) |
| Never | 200 | 7.31 (0.46) | 25.45 (0.90) | 67.24 (0.99) | 0.93 (0.01) | 0.72 (0.01) | |
| P-value | .156 | .229 | .076 | .156 | .151 |
Mean (Standard Error).
Cut point is median value of the variable.
Arsenic Metabolites Profiles in Relation to Genotypes
Urinary arsenic metabolites concentration and methylation indexes in relation to polymorphism genotypes are shown in Table 3, in the form of categorical variables (homozygote and heterozygote). The genotype of the ancestral allele homozygote in each SNP is denoted first.
TABLE 3.
Influence of N6AMT1 and As3MT genotypes on fractions of arsenic metabolites and methylation capacity indexes.
| Gene | SNPs | Genotypea | N | % iAs | % MMA | % DMA | PMI | SMI |
|---|---|---|---|---|---|---|---|---|
| N6AMT1 | rs1006903 | GG | 170 | 7.69 (0.53)b | 26.32 (0.94) | 66.00 (1.04) | 0.92 (0.01) | 0.71 (0.01) |
| GC | 105 | 7.52 (0.68) | 25.06 (1.20) | 67.42 (1.33) | 0.93 (0.01) | 0.73 (0.01) | ||
| CC | 14 | 8.66 (1.87) | 30.88 (3.28) | 60.47 (3.64) | 0.91 (0.02) | 0.66 (0.04) | ||
| P-value | .848 | .234 | .189 | .848 | .158 | |||
| rs1003671 | AA | 130 | 8.96 (0.60)*c | 26.32 (1.06) | 64.72 (1.17)* | 0.91 (0.01)* | 0.71 (0.01) | |
| GA | 130 | 6.20 (0.60)* | 24.59 (1.07)* | 69.21 (1.17)*† | 0.94 (0.01)* | 0.74 (0.01)* | ||
| GG | 29 | 8.49 (1.27) | 31.69 (2.26)* | 59.82 (2.48)† | 0.92 (0.01) | 0.65 (0.03)* | ||
| P-value | .005 | .018 | .001 | .005 | .007 | |||
| rs4816333 | CC | 107 | 7.32 (0.67) | 26.76 (1.19) | 65.92 (1.31) | 0.93 (0.01) | 0.71 (0.01) | |
| CG | 132 | 7.15 (0.60) | 25.53 (1.07) | 67.33 (1.19) | 0.93 (0.01) | 0.72 (0.01) | ||
| GG | 50 | 9.81 (0.98) | 26.08 (1.74) | 64.11 (1.93) | 0.90 (0.01) | 0.71 (0.02) | ||
| P-value | .055 | .744 | .349 | .055 | .677 | |||
| rs7282280 | CC | 174 | 7.94 (0.53) | 26.21 (0.93) | 65.86 (1.03) | 0.92 (0.01) | 0.72 (0.01) | |
| CT | 101 | 7.17 (0.70) | 25.59 (1.23) | 67.25 (1.37) | 0.93 (0.01) | 0.72 (0.01) | ||
| TT | 14 | 8.03 (1.88) | 28.11 (3.32) | 63.87 (3.68) | 0.92 (0.02) | 0.69 (0.04) | ||
| P-value | .668 | .762 | .584 | .668 | .733 | |||
| rs7282257 | TT | 68 | 9.62 (0.84)* | 24.69 (1.49) | 65.69 (1.66) | 0.90 (0.01)* | 0.73 (0.02) | |
| TA | 143 | 7.26 (0.57) | 26.66 (1.03) | 66.07 (1.14) | 0.93 (0.01) | 0.71 (0.01) | ||
| AA | 78 | 6.73 (0.78)* | 26.23 (1.39) | 67.04 (1.55) | 0.93 (0.01)* | 0.72 (0.02) | ||
| P-value | .026 | .548 | .820 | .026 | .716 | |||
| rs2065266 | CC | 144 | 8.85 (0.57)* | 25.75 (1.02) | 65.40 (1.13) | 0.91 (0.01)* | 0.72 (0.01) | |
| CT | 120 | 6.16 (0.62)* | 26.09 (1.12) | 67.76 (1.24) | 0.94 (0.01)* | 0.72 (0.01) | ||
| TT | 25 | 8.20 (1.38) | 27.94 (2.47) | 63.86 (2.74) | 0.92 (0.01) | 0.70 (0.03) | ||
| P-value | .006 | .716 | .250 | .006 | .692 | |||
| rs2738966 | GG | 67 | 9.49 (0.84)* | 24.43 (1.50) | 66.08 (1.67) | 0.91 (0.01)* | 0.73 (0.02) | |
| GA | 147 | 7.44 (0.57) | 27.16 (1.01) | 65.40 (1.12) | 0.93 (0.01) | 0.71 (0.01) | ||
| AA | 75 | 6.52 (0.80)* | 25.43 (1.42) | 68.05 (1.58) | 0.94 (0.01)* | 0.73 (0.02) | ||
| P-value | .033 | .280 | .391 | .033 | .316 | |||
| rs2248501 | GG | 75 | 6.52 (0.80)* | 25.43 (1.42) | 68.05 (1.58) | 0.94 (0.01)* | 0.73 (0.02) | |
| GT | 147 | 7.44 (0.57) | 27.16 (1.01) | 65.40 (1.12) | 0.93 (0.01) | 0.71 (0.01) | ||
| TT | 67 | 9.49 (0.84)* | 24.43 (1.50) | 66.08 (1.67) | 0.91 (0.01)* | 0.73 (0.02) | ||
| P-value | .033 | .280 | .391 | .033 | .316 | |||
| As3MT | rs7085104 | AA | 80 | 8.09 (0.78) | 24.86 (1.38) | 67.04 (1.54) | 0.92 (0.01) | 0.73 (0.02) |
| AG | 142 | 6.96 (0.58) | 27.68 (1.03) | 65.36 (1.15) | 0.93 (0.01) | 0.70 (0.01) | ||
| GG | 67 | 8.69 (0.87) | 24.14 (1.53) | 67.18 (1.71) | 0.91 (0.01) | 0.73 (0.02) | ||
| P-value | .208 | .091 | .559 | .208 | .184 | |||
| rs3740400 | CC | 84 | 7.75 (0.78) | 24.96 (1.36) | 67.30 (1.51) | 0.92 (0.01) | 0.73 (0.02) | |
| CA | 146 | 7.44 (0.58) | 27.83 (1.01) | 64.73 (1.12) | 0.93 (0.01) | 0.70 (0.01) | ||
| AA | 56 | 8.28 (0.94) | 23.50 (1.64) | 68.23 (1.82) | 0.92 (0.01) | 0.74 (0.02) | ||
| P-value | .746 | .046 | .175 | .746 | .064 | |||
| rs3740393 | CC | 15 | 7.97 (1.79) | 24.82 (3.19) | 67.21 (3.52) | 0.92 (0.02) | 0.73 (0.04) | |
| CG | 117 | 6.68 (0.64) | 25.05 (1.14) | 68.28 (1.26) | 0.93 (0.01) | 0.73 (0.01) | ||
| GG | 157 | 8.39 (0.55) | 26.97 (0.98) | 64.64 (1.09) | 0.92 (0.01) | 0.71 (0.01) | ||
| P-value | .132 | .414 | .092 | .132 | .286 | |||
| rs3740390 | GG | 158 | 8.41 (0.55) | 26.93 (0.98) | 64.67 (1.08) | 0.92 (0.01) | 0.71 (0.01) | |
| GA | 116 | 6.64 (0.64) | 25.09 (1.14) | 68.28 (1.26) | 0.93 (0.01) | 0.73 (0.01) | ||
| AA | 15 | 7.97 (1.79) | 24.83 (3.19) | 67.21 (3.52) | 0.92 (0.02) | 0.73 (0.04) | ||
| P-value | .116 | .441 | .095 | .116 | .305 | |||
| rs1046778 | TT | 83 | 8.70 (0.76) | 26.57 (1.36) | 64.74 (1.50) | 0.91 (0.01) | 0.71 (0.02) | |
| TC | 158 | 7.44 (0.55) | 25.92 (0.98) | 66.64 (1.08) | 0.93 (0.01) | 0.72 (0.01) | ||
| CC | 48 | 6.67 (1.00) | 25.78 (1.78) | 67.55 (1.98) | 0.93 (0.01) | 0.72 (0.02) | ||
| P-value | .230 | .913 | .458 | .230 | .806 |
The genotype of ancestral allele homozygote in each SNP was denoted first.
Mean (Standard Error). ANCOVA were used to adjust for potential confounding variablesat the following values: Gender = 1.4014, Age = 50.529, BMI = 24.616, Smoker = 0.3668, Drinker = 0.3080.
Comparison was done among any 2 groups, and those marked with same symbols (*,†) were significantly different (P < .05).
N6AMT1 SNP rs1003671 was significantly associated with % iAs, % MMA, % DMA, PMI and SMI. Individuals (45.0% of study population) with genotype GA demonstrated lowest % iAs and % MMA, but highest % DMA and methylation capacity at primary and secondary stage. This difference noticed in subjects carrying heterozygote suggested not allele dose dependent. Four SNPs (rs7282257, rs2065266, rs2738966, rs2248501) showed significant association with % iAs and PMI. With the exception of rs2065266, % iAs and PMI altered monotonically according to the dose of alleles for the other 3 SNPs. However, the other metabolites and SMIs did not show statistical significance. No remarkable difference was noted in the remaining 3 SNPs (rs1006903, rs4816333, rs7282280) in relation to any arsenic metabolites or methylation indexes. Though statistical significance of % iAs and % DMA was not reached in all polymorphism groups, it appeared that the N6AMT1 genotypes associated with the lowest % iAs were contributing to the highest % DMA and PMI. In each SNP, the change pattern of % iAs and PMIs was in opposite directions. The trend of significance did not show distinct alteration before or after adjusting for potential confounding variables (age, gender, BMI, alcohol and smoking status) (see supplementary information). Despite the gain or loss of significant difference for some indicators after adjustment, a strong tendency towards statistical significance (P < .05) was still maintained.
None of the 5 investigated As3MT SNPs showed a significant association with arsenic metabolites or methylation capacity indexes before or after adjustment for potential confounding variables. As for rs1046778, % iAs and % MMA changed monotonically according to the pattern of allele dose-dependence, while the other 3 indicators were in the opposite trend, even though they were not significantly different. Its CC genotype suggested the lowest % iAs and % MMA, but the highest % DMA, PMI and SMI.
Arsenic Metabolites Profiles in Relation to Haplotypes
Haplotypes were inferred from tag SNPs of N6AMT1and As3MT by PHASE, respectively. Haplotypes with an estimated frequency of higher than 5.0% were considered common enough and selected for analysis. Based on these criteria, the picked-up haplotypes were: N6AMT1 haplotype 1_GAGCTC, haplotype 2_GGCCAT, haplotype 3_CACCTC, haplotype 4_CACTAC, haplotype 5_GAGTAC (31.8%, 27.2%, 11.3%, 10.0% and 7.5%, respectively); As3MT haplotype 1_AAGT, haplotype 2_GCAC, haplotype 3_GCGT, haplotype 4_GCGC, haplotype 5_ACGC (44.5%, 25.2%, 11.5%, 11.0% and 7.1%, respectively) (Table 4).
TABLE 4.
Influence of N6AMT1 and As3MT haplotypes on fractions of arsenic metabolites and methylation capacity indexes.
| Gene | Haplotype | Frequency (%) | Copies | n | % iAs | % MMA | % DMA | PMI | SMI |
|---|---|---|---|---|---|---|---|---|---|
| N6AMT1 | Haplotype 1_GAGCTC | 31.8 | 0 | 134 | 7.33 (0.60)a | 26.55 (1.06) | 66.12 (1.18) | 0.93 (0.01) | 0.71 (0.01) |
| 1/2 | 125/30 | 7.97 (0.56) | 25.67 (0.98) | 66.36 (1.09) | 0.92 (0.01) | 0.72 (0.01) | |||
| P-value | .435 | .544 | .882 | .435 | .574 | ||||
| Haplotype 2_GGCCAT | 27.2 | 0 | 147 | 8.83 (0.56) | 26.49 (1.03) | 65.49 (1.12) | 0.91 (0.01) | 0.72 (0.01) | |
| 1/2 | 122/20 | 6.48 (0.57) | 25.69 (1.01) | 67.03 (1.14) | 0.94 (0.01) | 0.72 (0.01) | |||
| P-value | .004 | .579 | .336 | .004 | .925 | ||||
| Haplotype3_CACCTC | 11.3 | 0 | 228 | 7.41 (0.46) | 26.06 (0.82) | 66.54 (0.90) | 0.93 (0.01) | 0.72 (0.01) | |
| 1/2 | 57/4 | 8.67 (0.90) | 26.17 (1.59) | 65.16 (1.77) | 0.91 (0.01) | 0.71 (0.02) | |||
| P-value | .215 | .948 | .490 | .215 | .842 | ||||
| Haplotype 4_CACTAC | 10.0 | 0 | 233 | 7.99 (0.45) | 26.07 (0.80) | 65.94 (0.89) | 0.92 (0.01) | 0.72 (0.01) | |
| 1/2 | 54/2 | 6.35 (0.92) | 26.12 (1.64) | 67.54 (1.82) | 0.94 (0.01) | 0.72 (0.02) | |||
| P-value | .111 | .980 | .430 | .111 | .913 | ||||
| Haplotype 5_GAGTAC | 7.5 | 0 | 251 | 7.62 (0.44) | 25.95 (0.77) | 66.43 (0.86) | 0.92 (0.01) | 0.72 (0.01) | |
| 1/2 | 36/2 | 8.03 (1.13) | 26.92 (2.00) | 65.06 (2.22) | 0.92 (0.01) | 0.71 (0.02) | |||
| P-value | .739 | .654 | .566 | .739 | .570 | ||||
| As3MT | Haplotype 1_AAGT | 44.5 | 0 | 85 | 7.72 (0.77) | 24.92 (1.36) | 67.37 (1.51) | 0.92 (0.01) | 0.73 (0.02) |
| 1/2 | 150/54 | 7.66 (0.49) | 26.57 (0.87) | 65.78 (0.96) | 0.92 (0.01) | 0.71 (0.01) | |||
| P-value | .949 | .315 | .383 | .949 | .348 | ||||
| Haplotype 2_GCAC | 25.2 | 0 | 158 | 8.41 (0.55) | 26.93 (0.98) | 64.66 (1.08) | 0.92 (0.01) | 0.71 (0.01) | |
| 1/2 | 116/15 | 6.79 (0.61) | 25.06 (1.08) | 68.15 (1.19) | 0.93 (0.01) | 0.73 (0.01) | |||
| P-value | .051 | .201 | .031 | .051 | .123 | ||||
| Haplotype 3_GCGT | 11.5 | 0 | 227 | 7.25 (0.46) | 25.92 (0.82) | 66.83 (0.91) | 0.93 (0.01) | 0.72 (0.01) | |
| 1/2 | 58/4 | 9.21 (0.89) | 26.67 (1.59) | 64.12 (1.76) | 0.91 (0.01) | 0.71 (0.02) | |||
| P-value | .054 | .680 | .177 | .054 | .456 | ||||
| Haplotype 4_GCGC | 11.0 | 0 | 229 | 7.45 (0.46) | 26.07 (0.81) | 66.49 (0.90) | 0.93 (0.01) | 0.72 (0.01) | |
| 1/2 | 56/4 | 8.54 (0.89) | 26.12 (1.58) | 65.33 (1.76) | 0.92 (0.01) | 0.71 (0.02) | |||
| P-value | .275 | .975 | .560 | .275 | .865 | ||||
| Haplotype 5_ACGC | 7.1 | 0 | 249 | 7.92 (0.44) | 25.79 (0.78) | 66.30 (0.86) | 0.92 (0.01) | 0.72 (0.01) | |
| 1/2 | 39/1 | 6.16 (1.10) | 27.92 (1.94) | 65.92 (2.16) | 0.94 (0.01) | 0.70 (0.02) | |||
| P-value | .137 | .307 | .870 | .137 | .439 |
Mean (Standard Error). ANCOVA were used to adjust for potential confounding variables at the following values: Gender = 1.4014, Age = 50.529, BMI = 24.616, Smoker = 0.3668, Drinker = 0.3080.
Subjects with 1 or 2 copies (non-null) of N6AMT1 haplotype 2_GGCCAT had significantly lower % iAs and higher PMI, compared to those individuals with zero copies. % MMA and % DMA were respectively lower and higher among these haplotype carriers, although the difference was not statistically significant. The difference trend did not change after adjusting for potential confounding variables. Haplotype 4_CACTAC carriers demonstrated similar arsenic metabolism pattern with haplotype 2, but no statistical significance was observed. No remarkable differences were associated with other haplotypes.
In the aspect of As3MT haplotype, % DMA was significantly higher among participants with 1 or 2 copies of haplotype 2_GCAC versus individuals carrying zero copies. % iAs, and PMI of these subjects also showed noticeably lower and higher, respectively, close to being statistically significant (P = .051). Although % iAs within haplotype 3_GCGT groups decreased slightly after adjustment for potential confounding variables, it was still close to significance level (P = .054), showing lower % iAs and higher PMI among subjects with zero copies of haplotype versus non null groups. Other As3MT haplotype groups did not show significant differences in the metabolism pattern.
Joint Effect of N6AMT1 and As3MT Haplotypes on Arsenic Metabolites Profiles
Considering the common frequency and involvement of haplotypes showing significant difference in arsenic species, the haplotypes of the top 3 frequencies in each gene were picked up for pairwise interaction analysis in order to evaluate the combined effect on the arsenic metabolism pattern. Among the N6AMT1 and As3MT haplotypes combinations (Table 5), we found the feature of significance was retained for the haplotypes that had a noticeable influence on the arsenic metabolites profiles, especially for the combinations containing N6AMT1 haplotype 2_GGCCAT. Individuals with 1 or 2 copies of N6AMT1 haplotype 2 and 1 or 2 copies of As3MT haplotype 1 or As3MT haplotype 2 had the lowest % iAs [6.44 ± 0.66 (P = .034), 5.47 ± 0.84 (P = .007), respectively] and highest PMI [0.94 ± 0.01 (P = .034), 0.95 ± 0.01 (P = .007), respectively] in their combination groups, suggesting the additive influence from both haplotypes. Subjects carrying 1 or 2 copies of both N6AMT1 haplotype 2 and As3MT haplotype 2 even showed the lowest % MMA (24.79 ± 1.51), highest % DMA (69.74 ± 1.66) and SMI (0.74 ± 0.02), though the differences were not statistically significant. In contrast, individuals with zero copies of N6AMT1 haplotype 2 and As3MT haplotype 2 demonstrated the highest % iAs (9.36 ± 0.76) and lowest PMI (0.91 ± 0.01). However, some combinations may attenuate the individual feature of the haplotype itself. For example, individuals with 1 or 2 copies of both N6AMT1 haplotype 2 and As3MT haplotype 3 showed neutralized % iAs (8.65 ± 1.16) compared to those having only one of these haplotypes (5.76 ± 0.66, 10.03 ± 1.33). No statistically significant effects were seen for other haplotype combination groups. The significance trend did not change before or after adjusting for potential confounding variables.
TABLE 5.
Influence of N6AMT1 and As3MT haplotypes interaction on fractions of arsenic metabolites and methylation capacity indexes.
| Haplotypes | Copies | n | % iAs | % MMA | % DMA | PMI | SMI |
|---|---|---|---|---|---|---|---|
| N6AMT1 Haplotype 1 + As3MT Haplotype 1 | 1/2 + 1/2 | 105 | 7.86 (0.69)a | 25.69 (1.21) | 66.45 (1.35) | 0.92 (0.01) | 0.72 (0.01) |
| 1/2 + 0 | 50 | 8.21 (1.00) | 25.65 (1.77) | 66.14 (1.96) | 0.92 (0.01) | 0.72 (0.02) | |
| 0 + 1/2 | 99 | 7.45 (0.70) | 27.46 (1.23) | 65.09 (1.37) | 0.93 (0.01) | 0.70 (0.01) | |
| 0 + 0 | 35 | 7.00 (1.19) | 23.95 (2.09) | 69.05 (2.32) | 0.93 (0.01) | 0.74 (0.02) | |
| P-value | .849 | .484 | .533 | .849 | .470 | ||
| N6AMT1 Haplotype 1 + As3MT Haplotype 2 | 1/2 + 1/2 | 68 | 6.87 (0.84) | 24.29 (1.49) | 68.84 (1.65) | 0.93 (0.01) | 0.74 (0.02) |
| 1/2 + 0 | 87 | 8.83 (0.74) | 26.76 (1.32) | 64.41 (1.46) | 0.91 (0.01) | 0.71 (0.01) | |
| 0 + 1/2 | 63 | 6.70 (0.88) | 25.90 (1.56) | 67.41 (1.72) | 0.93 (0.01) | 0.72 (0.02) | |
| 0 + 0 | 71 | 7.88 (0.82) | 27.13 (1.46) | 64.99 (1.61) | 0.92 (0.01) | 0.70 (0.02) | |
| P-value | .206 | .528 | .168 | .206 | .412 | ||
| N6AMT1 Haplotype 1 + As3MT Haplotype 3 | 1/2 + 1/2 | 36 | 9.19 (1.18) | 25.84 (2.10) | 64.98 (2.32) | 0.91 (0.01) | 0.72 (0.02) |
| 1/2 + 0 | 119 | 7.60 (0.64) | 25.62 (1.14) | 66.78 (1.26) | 0.92 (0.01) | 0.72 (0.01) | |
| 0 + 1/2 | 26 | 9.22 (1.36) | 27.81 (2.42) | 62.97 (2.68) | 0.91 (0.01) | 0.69 (0.03) | |
| 0 + 0 | 108 | 6.88 (0.67) | 26.25 (1.18) | 66.87 (1.31) | 0.93 (0.01) | 0.72 (0.01) | |
| P-value | .232 | .872 | .543 | .232 | .744 | ||
| N6AMT1 Haplotype 2 + As3MT Haplotype 1 | 1/2 + 1/2 | 110 | 6.44 (0.66)*b | 27.44 (1.17) | 66.12 (1.30) | 0.94 (0.01)* | 0.71 (0.01) |
| 1/2 + 0 | 32 | 6.63 (1.22) | 23.24 (2.18) | 70.13 (2.42) | 0.93 (0.01) | 0.75 (0.02) | |
| 0 + 1/2 | 94 | 9.08 (0.71)* | 25.52 (1.27) | 65.40 (1.41) | 0.91 (0.01)* | 0.72 (0.01) | |
| 0 + 0 | 53 | 8.37 (0.96) | 25.96 (1.72) | 65.67 (1.91) | 0.92 (0.01) | 0.72 (0.02) | |
| P-value | .034 | .361 | .387 | .034 | .510 | ||
| N6AMT1 Haplotype 2 + As3MT Haplotype 2 | 1/2 + 1/2 | 66 | 5.47 (0.84)* | 24.79 (1.51) | 69.74 (1.66) | 0.95 (0.01)* | 0.74 (0.02) |
| 1/2 + 0 | 76 | 7.36 (0.78) | 27.97 (1.41) | 64.67 (1.55) | 0.93 (0.01) | 0.70 (0.02) | |
| 0 + 1/2 | 65 | 8.14 (0.85) | 25.34 (1.53) | 66.52 (1.69) | 0.92 (0.01) | 0.72 (0.02) | |
| 0 + 0 | 82 | 9.36 (0.76)* | 25.96 (1.36) | 64.68 (1.50) | 0.91 (0.01)* | 0.72 (0.02) | |
| P-value | .007 | .431 | .091 | .007 | .295 | ||
| N6AMT1 Haplotype 2 + As3MT Haplotype 3 | 1/2 + 1/2 | 35 | 8.65 (1.16) | 28.35 (2.09) | 63.00 (2.31) | 0.91 (0.01) | 0.69 (0.02) |
| 1/2 + 0 | 107 | 5.76 (0.66)*† | 25.88 (1.20) | 68.37 (1.32) | 0.94 (0.01)*† | 0.73 (0.01) | |
| 0 + 1/2 | 27 | 10.03 (1.33)* | 24.43 (2.41) | 65.54 (2.66) | 0.90 (0.01)* | 0.73 (0.03) | |
| 0 + 0 | 120 | 8.57 (0.62)† | 25.97 (1.12) | 65.46 (1.24) | 0.91 (0.01)† | 0.71 (0.01) | |
| P-value | .003 | .634 | .178 | .003 | .457 | ||
| N6AMT1 Haplotype 3 + As3MT Haplotype 1 | 1/2 + 1/2 | 45 | 8.74 (1.04) | 25.71 (1.84) | 65.55 (2.05) | 0.91 (0.01) | 0.72 (0.02) |
| 1/2 + 0 | 16 | 8.46 (1.76) | 27.53 (3.11) | 64.01 (3.46) | 0.92 (0.02) | 0.70 (0.03) | |
| 0 + 1/2 | 159 | 7.35 (0.56) | 26.82 (0.99) | 65.84 (1.09) | 0.93 (0.01) | 0.71 (0.01) | |
| 0 + 0 | 69 | 7.55 (0.86) | 24.29 (1.52) | 68.16 (1.69) | 0.93 (0.01) | 0.74 (0.02) | |
| P-value | .662 | .537 | .585 | .662 | .634 | ||
| N6AMT1 Haplotype 3 + As3MT Haplotype 2 | 1/2 + 1/2 | 25 | 8.89 (1.39) | 23.84 (2.48) | 67.27 (2.73) | 0.91 (0.01) | 0.74 (0.03) |
| 1/2 + 0 | 36 | 8.53 (1.17) | 27.82 (2.08) | 63.65 (2.30) | 0.92 (0.01) | 0.70 (0.02) | |
| 0 + 1/2 | 106 | 6.30 (0.67) | 25.33 (1.20) | 68.37 (1.32) | 0.94 (0.01) | 0.73 (0.01) | |
| 0 + 0 | 122 | 8.37 (0.63) | 26.68 (1.11) | 64.96 (1.23) | 0.92 (0.01) | 0.71 (0.01) | |
| P-value | .085 | .539 | .172 | .085 | .436 | ||
| N6AMT1 Haplotype 3 + As3MT Haplotype 3 | 1/2 + 1/2 | 11 | 9.48 (2.09) | 25.43 (3.74) | 65.09 (4.13) | 0.91 (0.02) | 0.72 (0.04) |
| 1/2 + 0 | 50 | 8.46 (0.98) | 26.32 (1.76) | 65.23 (1.94) | 0.92 (0.01) | 0.71 (0.02) | |
| 0 + 1/2 | 51 | 9.15 (0.99) | 26.95 (1.77) | 63.91 (1.96) | 0.91 (0.01) | 0.70 (0.02) | |
| 0 + 0 | 177 | 6.92 (0.52) | 25.81 (0.93) | 67.28 (1.03) | 0.93 (0.01) | 0.72 (0.01) | |
| P-value | .132 | .946 | .429 | .132 | .808 |
Mean (Standard Error). ANCOVA were used to adjust for potential confounding variables at the following values: Gender = 1.4014, Age = 50.529, BMI = 24.616, Smoker = 0.3668, Drinker = 0.3080.
Comparison was done among any 2 groups, and those marked with same symbols (*,†) were significantly different (P < .05).
DISCUSSION
Chronic exposure to arsenic through drinking water has been shown in association with both cancerous and non-cancerous effects (Chen et al., 1988; Chen and Ahsan, 2004; Jomova et al. 2011; Mumford et al., 2007; Smith et al., 2006; Tseng et al., 1968). While the underlying mechanisms for arsenic-caused adverse effects are not clearly elucidated, it is widely accepted that arsenic metabolism plays a critical role in its toxic and carcinogenic activity (Drobna et al., 2005; Ferrario et al., 2008; Li et al., 2005; Thomas et al., 2007). Our study of a Chinese population suggests that genetic polymorphisms of N6AMT1 were capable of altering urinary arsenic profiles, which could also be affected by the haplotype combinations between N6AMT1 and As3MT.
In spite of the widely recognized fact that As3MT contributes substantially to arsenic metabolism and variability in urinary metabolites pattern among different populations (Drobna et al., 2005; Engström et al., 2011; Ferrario et al., 2008; Li et al., 2005; Thomas et al., 2007), evidence suggests there may exist other methyltransferases capable of arsenic metabolism. Retaining the capacity of arsenic methylation was observed in the As3MT eliminated human cells model (Drobna et al., 2006) and animal model (Chen et al., 2011; Drobna et al., 2009). In a previous study, we demonstrated that N6AMT1 was probably a promising methyltransferase participating in human arsenic metabolism, specifically converting high toxic MMAIII to the less toxic DMAV (Ren et al., 2011). This observation was further confirmed in our recent in vitro experiment, even though its effect was relatively minor and limited in contrast with the influence of As3MT (Zhang et al., 2015). A recent study showed a significant association between a genetic variation of N6AMT1 and urinary % MMA in Andean women (Harari et al., 2013). However, the arsenic metabolism pattern impacted by N6AMT1 or joint influence with As3MT has not been evaluated in other ethnically different populations.
In the present study of Chinese population, we observed that gender and smoking history had significant influence on arsenic metabolites profiles and methylation capacity (Table 2). Women demonstrated better arsenic methylation capacity than men, which was featured by remarkably lower % iAs and % MMA and higher % DMA, PMI and SMI. This finding was largely consistent with previous studies (Gamble et al., 2005; Lindberg et al., 2008; Tseng, 2005). Although the exact reason is unknown, it has been suggested that estrogen is likely play an important role, which involved in one-carbon metabolism, thus facilitating arsenic methylation by generating adequate supply of methyl group donor (Hall and Gamble, 2012). Similarly, non-smokers showed significantly better arsenic methylation capacity than current or former tobacco smokers, which is likely explained by the effect of some cigarettes chemicals competing for related enzymes or co-factors participating in the arsenic metabolism processes (Tseng, 2009). Ages, BMI and alcohol drinking have also been suggested to affect arsenic metabolism (Gomez-Rubio et al., 2011; Hopenhayn-Rich et al., 1996; Tseng, 2005, 2009). In our study, age, BMI or alcohol drinking had no effects in arsenic metabolites profiles and methylation indexes. However, study participants who were characterized by younger age, higher BMI or non-drinkers showed better arsenic metabolism capacity (relatively lower % MMA and higher % DMA and SMI).
In this Chinese population, we found that the genetic polymorphisms of N6AMT1 (rs1003671, rs7282257, rs2065266, rs2738966, rs2248501) were associated with the arsenic metabolites profiles and the methylation capacities (Table 3), particularly the amounts of urinary iAs. Rs1003671 with a genotype GA was showed to have a significant impact on all arsenic metabolites (the lowest % iAs and % MMA, and the highest % DMA) and methylation capacity indexes (highest PMI and SMI), indicating that genetic variance of rs1003671 could potentially contribute to the differential susceptibility of individuals to the toxic and carcinogenic activity of arsenic. Different from the finding by Harari et al. (2013) that variation of % MMA was associated with genetic polymorphism in an allele-dose dependent manner, we found, in our study of this Chinese population, that the allele-dose relation with urinary iAs was not existed, which may be due to the inherent dominance pattern of alleles on phenotypes (Miko, 2008). The people carrying N6AMT1 haplotype 2_GGCCAT had a significant impact on % iAs and PMI (Table 4), while the people carrying As3MT haplotype 2_GCAC showed significant effect on % DMA. When estimating the joint effect of N6AMT1 and As3MT haplotypes according to the combined copies number (Table 5), the association between some combinations and % iAs as well as PMI was observed at significant levels in the combination groups involving the N6AMT1 haplotype 2. For example, study participants with 1 or 2 copies of both N6AMT1 haplotype 2 and As3MT haplotype 2 had significantly lower % iAs, relatively lower % MMA, higher % DMA, PMI and SMI, which was consistent with the influence of each haplotype individually. This observation suggested that specific haplotype combinations of these 2 genes demonstrated an additive impact on the metabolism process, contributing to a strengthened effect in the ultimate metabolites pattern. The joint additive effects found in this study probably implied the importance of investigating the arsenic metabolism outcomes at gene–gene interaction level, and provided further explanation for differential susceptibility of individuals to the toxic and carcinogenic activity of arsenic. The % iAs was the main arsenic metabolite showing significant association with specific SNPs and haplotype combinations. The % iAs is negatively associated with PMI. PMI as (MMA + DMA)/tAs and SMI as DMA/(MMA + DMA) were calculated to reflect the different stages of arsenic bio-methylation process (Sun et al., 2007). Thus, individuals with lower urinary % iAs had a better methylation capacity by converting iAs to MMA. Previous studies suggested that higher urinary % iAs and % MMA, and lower % DMA and SMI, were an indication of incomplete and less-efficient arsenic metabolism (Chung et al., 2009; Lindberg et al., 2008). On the contrary, individuals expressing the opposite trend of these metabolites would associate with a relatively more efficient capacity for arsenic detoxification. We showed here that the study participants carrying the genotype or haplotypes mentioned above have a significant effect on urinary % iAs, suggesting that genetic variations of N6AMT1 are capable of modifying the activity of N6AMT1.
A number of studies have suggested that genetic variations may account for a large part of the altered arsenic metabolism among different population groups (Agusa et al., 2011; Engström et al., 2011; Fu et al., 2014). However, there were some inconsistences between genotypes and metabolism pattern in human from diverse ethnical backgrounds (Agusa et al., 2011). Though the As3MT SNPs investigated in this study were previously reported to have association with an arsenic metabolites profiles (Agusa et al., 2009; Engström et al., 2011; Schläwicke Engström et al., 2007), no such statistical significance was noted in our studied population, probably due to the limited sample power or other unknown environmental or biological factors. Arsenic methylation is a complicated process with the involvement of multiple enzymes functioning in a network pattern besides As3MT and N6AMT1, such as GSTO1 (Takeshita et al., 2009), GSTO2 (Janasik et al., 2015; Schmuck et al., 2005; Zakharyan et al., 2001), DNMT1a and DNMT3b (Engström et al., 2011), which have been shown to be related to inter-individual variations in arsenic metabolism outcomes at the genetic variation level or protein level. In addition, a number of studies suggested that demographic characteristics such as age, gender, body mass index (BMI), personal lifestyle (smoking history or alcohol consumption status) could also contribute to the varied efficiency in arsenic metabolism (Lindberg et al., 2010; Sun et al., 2007; Tseng, 2009; Zhang et al., 2014). Additional studies with larger human cohort are warrant to help clarify the genetic determinants of As3MT in arsenic biomethylation in this Chinese population with a focus on its interactive effect with N6AMT1 on arsenic metabolism.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
ACKNOWLEDGMENTS
We thank Prof Allan H. Smith (School of Public Health, University of California at Berkeley) for comments and preparation of the final manuscript. We gratefully acknowledge the assistance and cooperation of the Center for Disease Control and Prevention in Wuyuan, Inner Mongolia and thank all of the study participants. We acknowledge the NSF Major Research Instrumentation Program (CHE 0959565) for the acquisition of the HPLC-ICP-MS system used in this study. SNP genotyping was performed by the Genomics Shared Resource supported by Roswell Park Cancer Institute and National Cancer Institute (NCI) grant P30CA016056.
FUNDING
These studies were supported by National Institutes of Health (NIH) grant ES022329 and ES022629 (to X.R.), and by Science and Technology Department of Zhejiang Province (2013C33169), and in part by National Natural Science Foundation of China (81260413).
REFERENCES
- Abernathy C. O., Liu Y. P., Longfellow D., Aposhian H. V., Beck B., Fowler B., Goyer R., Menzer R., Rossman T., Thompson C., et al. (1999). Arsenic: health effects, mechanisms of actions, and research issues. Environ. Health Perspect. 107, 593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agusa T., Fujihara J., Takeshita H., Iwata H. (2011). Individual variations in inorganic arsenic metabolism associated with AS3MT genetic polymorphisms. Int. J. Mol. Sci. 12, 2351–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agusa T., Iwata H., Fujihara J., Kunito T., Takeshita H., Minh T. B., Trang P. T., Viet P. H., Tanabe S. (2009). Genetic polymorphisms in AS3MT and arsenic metabolism in residents of the Red River Delta, Vietnam. Toxicol. Appl. Pharmacol. 236, 131–141. [DOI] [PubMed] [Google Scholar]
- Barrett J. C. (2009). Haploview: visualization and analysis of SNP genotype data. Cold Spring HarbProtoc doi:10.1101/pdb.ip71. [DOI] [PubMed] [Google Scholar]
- Barrett J. C., Fry B., Maller J., Daly M. J. (2005). Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265. [DOI] [PubMed] [Google Scholar]
- Chen B., Arnold L. L., Cohen S. M., Thomas D. J., Le X. C. (2011). Mouse arsenic (+3 oxidation state) methyltransferase genotype affects metabolism and tissue dosimetry of arsenicals after arsenite administration in drinking water. Toxicol. Sci. 124, 320–326. [DOI] [PubMed] [Google Scholar]
- Chen C. J., Kuo T. L., Wu M. M. (1988). Arsenic and cancers. Lancet 1, 414–415. [DOI] [PubMed] [Google Scholar]
- Chen Y., Ahsan H. (2004). Cancer burden from arsenic in drinking water in Bangladesh. Am. J. Public Health 94, 741–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C. J., Hsueh Y. M., Bai C. H., Huang Y. K., Huang Y. L., Yang M. H., Chen C. J. (2009). Polymorphisms in arsenic metabolism genes, urinary arsenic methylation profile and cancer. Cancer Causes Control 20, 1653–1661. [DOI] [PubMed] [Google Scholar]
- de Bakker P. I., Yelensky R., Pe’er I., Gabriel S. B., Daly M. J., Altshuler D. (2005). Efficiency and power in genetic association studies. Nat. Genet. 37, 1217–1223. [DOI] [PubMed] [Google Scholar]
- Drobna Z., Naranmandura H., Kubachka K. M., Edwards B. C., Herbin-Davis K., Styblo M., Le X. C., Creed J. T., Maeda N., Hughes M. F., et al. (2009). Disruption of the arsenic (+3 oxidation state) methyltransferase gene in the mouse alters the phenotype for methylation of arsenic and affects distribution and retention of orally administered arsenate. Chem. Res. Toxicol. 22, 1713–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobna Z., Waters S. B., Devesa V., Harmon A. W., Thomas D. J., Styblo M. (2005). Metabolism and toxicity of arsenic in human urothelial cells expressing rat arsenic (+3 oxidation state) methyltransferase. Toxicol. Appl. Pharmacol. 207, 147–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobna Z., Waters S. B., Walton F. S., LeCluyse E. L., Thomas D. J., Styblo M. (2004). Interindividual variation in the metabolism of arsenic in cultured primary human hepatocytes. Toxicol. Appl. Pharmacol. 201, 166–177. [DOI] [PubMed] [Google Scholar]
- Drobna Z., Xing W., Thomas D. J., Styblo M. (2006). shRNA silencing of AS3MT expression minimizes arsenic methylation capacity of HepG2 cells. Chem. Res. Toxicol. 19, 894–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström K., Vahter M., Mlakar S. J., Concha G., Nermell B., Raqib R., Cardozo A., Broberg K. (2011). Polymorphisms in arsenic(+III oxidation state) methyltransferase (AS3MT) predict gene expression of AS3MT as well as arsenic metabolism. Environ. Health Perspect. 119, 182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrario D., Croera C., Brustio R., Collotta A., Bowe G., Vahter M., Gribaldo L. (2008). Toxicity of inorganic arsenic and its metabolites on haematopoietic progenitors “in vitro”: Comparison between species and sexes. Toxicology 249, 102–108. [DOI] [PubMed] [Google Scholar]
- Fu S., Wu J., Li Y., Liu Y., Gao Y., Yao F., Qiu C., Song L., Wu Y., Liao Y., et al. (2014). Urinary arsenic metabolism in a Western Chinese population exposed to high-dose inorganic arsenic in drinking water: Influence of ethnicity and genetic polymorphisms. Toxicol. Appl. Pharmacol. 274, 117–123. [DOI] [PubMed] [Google Scholar]
- Fujino Y., Guo X., Liu J., You L., Miyatake M., Yoshimura T. (2004). Mental health burden amongst inhabitants of an arsenic-affected area in Inner Mongolia, China. Soc. Sci. Med. 59, 1969–1973. [DOI] [PubMed] [Google Scholar]
- Fujino Y., Guo X., Shirane K., Liu J., Wu K., Miyatake M., Tanabe K., Kusuda T., Yoshimura T. (2006). Arsenic in drinking water and peripheral nerve conduction velocity among residents of a chronically arsenic-affected area in Inner Mongolia. J. Epidemiol. 16, 207–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble M. V., Liu X., Ahsan H., Pilsner R., Ilievski V., Slavkovich V., Parvez F., Levy D., Factor-Litvak P., Graziano J. H. (2005). Folate, homocysteine, and arsenic metabolism in arsenic-exposed individuals in Bangladesh. Environ. Health Perspect. 113, 1683–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner R. M., Engström K., Bottai M., Hoque W. A., Raqib R., Broberg K., Vahter M. (2012). Pregnancy and the methyltransferase genotype independently influence the arsenic methylation phenotype. Pharmacogenet. Genomics 22, 508–516. [DOI] [PubMed] [Google Scholar]
- Gomez-Rubio P., Klimecki W., Meza-Montenegro M., Cantu-Soto E. (2010). Genetic association between intronic variants in AS3MT and arsenic methylation efficiency is focused on a large linkage disequilibrium cluster in chromosome 10. J. Appl. Toxicol. 30, 260–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Rubio P., Roberge J., Arendell L., Harris R. B., O'Rourke M. K., Chen Z., Cantu-Soto E., Meza-Montenegro M. M., Billheimer D., Lu Z., et al. (2011). Association between body mass index and arsenic methylation efficiency in adult women from southwest U.S. and northwest Mexico. Toxicol. Appl. Pharmacol. 252, 176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. X., Hu L., Yand P. Z., Tanabe K., Miyatalre M., Chen Y. (2007). Chronic arsenic poisoning in drinking water in Inner Mongolia and its associated health effects. J. Environ. Sci. Health 42, 1853–1858. [DOI] [PubMed] [Google Scholar]
- Guo X., Fujino Y., Chai J., Wu K., Xia Y., Li Y., Lv J., Sun Z., Yoshimura T. (2003). The prevalence of subjective symptoms after exposure to arsenic in drinking water in Inner Mongolia, China. J. Epidemiol. 13, 211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Fujino Y., Kaneko S., Wu K., Xia Y., Yoshimura T. (2001). Arsenic contamination of groundwater and prevalence of arsenical dermatosis in the hetao plain area, Inner Mongolia, China. Mol. Cell. Biochem. 222, 137–140. [PubMed] [Google Scholar]
- Hall M. N., Gamble M. V. (2012). Nutritional manipulation of one-carbon metabolism: effects on arsenic methylation and toxicity. J. Toxicol. 2012, 595307.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harari F., Engström K., Concha G., Colque G., Vahter M., Broberg K. (2013). N-6-adenine-specific DNA methyltransferase 1 (N6AMT1) polymorphisms and arsenic methylation in Andean women. Environ. Health Perspect. 121, 797–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández A., Xamena N., Surralles J., Sekaran C., Tokunaga H., Quinteros D., Creus A., Marcos R. (2008). Role of the Met287Thr polymorphism in the AS3MT gene on the metabolic arsenic profile. Mutat. Res. 637, 80–92. [DOI] [PubMed] [Google Scholar]
- Hopenhayn-Rich C., Biggs M. L., Smith A. H., Kalman D. A., Moore L. E. (1996). Methylation study of a population environmentally exposed to arsenic in drinking water. Environ. Health Perspect. 104, 620–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janasik B., Reszka E., Stanislawska M., Wieczorek E., Fendler W., Wasowicz W. (2015). Biological monitoring and the influence of genetic polymorphism of As3MT and GSTs on distribution of urinary arsenic species in occupational exposure workers. Int. Arch. Occup. Environ. Health 88, 807–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jomova K., Jenisova Z., Feszterova M., Baros S., Liska J., Hudecova D., Rhodes C. J., Valko M. (2011). Arsenic: Toxicity, oxidative stress and human disease. J. Appl. Toxicol. 31, 95–107. [DOI] [PubMed] [Google Scholar]
- Kalman D. A., Dills R. L., Steinmaus C., Yunus M., Khan A. F., Prodhan M. M., Yuan Y., Smith A. H. (2014). Occurrence of trivalent monomethyl arsenic and other urinary arsenic species in a highly exposed juvenile population in Bangladesh. J. Expos. Sci. Environ. Epidemiol. 24, 113–120. [DOI] [PubMed] [Google Scholar]
- Li J., Waters S. B., Drobna Z., Devesa V., Styblo M., Thomas D. J. (2005). Arsenic (+3 oxidation state) methyltransferase and the inorganic arsenic methylation phenotype. Toxicol. Appl. Pharmacol. 204, 164–169. [DOI] [PubMed] [Google Scholar]
- Lindberg A. L., Rahman M., Persson L. A., Vahter M. (2008). The risk of arsenic induced skin lesions in Bangladeshi men and women is affected by arsenic metabolism and the age at first exposure. Toxicol. Appl. Pharmacol. 230, 9–16. [DOI] [PubMed] [Google Scholar]
- Lindberg A. L., Sohel N., Rahman M., Persson L. A., Vahter M. (2010). Impact of smoking and chewing tobacco on arsenic-induced skin lesions. Environ. Health Perspect. 118, 533–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C. W., Zhang C. Y., Sun Z. M., Yang Z. P., Bai F. M., Ding Y. S. (1996). The investigation of endemic arsenic poulting in Wuyuan County, Inner Mongolia. J. Endemic Dis. Contam. Treat. 2, 49–51. [Google Scholar]
- Mao G., Guo X., Kang R., Ren C., Yang Z., Sun Y., Zhang C., Zhang X., Zhang H., Yang W. (2010). Prevalence of disability in an arsenic exposure area in Inner Mongolia, China. Chemosphere 80, 978–981. [DOI] [PubMed] [Google Scholar]
- Marapakala K., Qin J., Rosen B. P. (2012). Identification of catalytic residues in the As(III) S-adenosylmethionine methyltransferase. Biochemistry 51, 944–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miko I. (2008). Genetic dominance: genotype-phenotype relationships. Nat. Educ. 1, 140. [Google Scholar]
- Mumford J. L., Wu K., Xia Y., Kwok R., Yang Z., Foster J., Sanders W. E. (2007). Chronic arsenic exposure and cardiac repolarization abnormalities with QT interval prolongation in a population-based study. Environ. Health Perspect. 115, 690–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Aleshin M., Jo W. J., Dills R., Kalman D. A., Vulpe C. D., Smith M. T., Zhang L. (2011). Involvement of N-6 adenine-specific DNA methyltransferase 1 (N6AMT1) in arsenic biomethylation and its role in arsenic-induced toxicity. Environ. Health Perspect. 119, 771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schläwicke Engström K., Broberg K., Concha G., Nermell B., Warholm M., Vahter M. (2007). Genetic polymorphisms influencing arsenic metabolism: Evidence from Argentina. Environ. Health Perspect. 115, 599–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schläwicke Engström K., Nermell B., Concha G., Stromberg U., Vahter M., Broberg K. (2009). Arsenic metabolism is influenced by polymorphisms in genes involved in one-carbon metabolism and reduction reactions. Mutat. Res. 667, 4–14. [DOI] [PubMed] [Google Scholar]
- Schmuck E. M., Board P. G., Whitbread A. K., Tetlow N., Cavanaugh J. A., Blackburn A. C., Masoumi A. (2005). Characterization of the monomethylarsonate reductase and dehydroascorbate reductase activities of Omega class glutathione transferase variants: implications for arsenic metabolism and the age-at-onset of Alzheimer's and Parkinson's diseases. Pharmacogenet. Genomics 15, 493–501. [DOI] [PubMed] [Google Scholar]
- Smith A. H., Marshall G., Yuan Y., Ferreccio C., Liaw J., von Ehrenstein O., Steinmaus C., Bates M. N., Selvin S. (2006). Increased mortality from lung cancer and bronchiectasis in young adults after exposure to arsenic in utero and in early childhood. Environ. Health Perspect. 114, 1293–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M., Donnelly P. (2003). A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 73, 1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G., Xu Y., Li X., Jin Y., Li B., Sun X. (2007). Urinary arsenic metabolites in children and adults exposed to arsenic in drinking water in Inner Mongolia, China. Environ. Health Perspect. 115, 648–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita H., Fujihara J., Takatsuka H., Agusa T., Yasuda T., Kunito T. (2009). Diversity of glutathione s-transferase omega 1 (a140d) and 2 (n142d) gene polymorphism in worldwide populations. Clin. Exp. Pharmacol. Physiol. 36, 283–286. [DOI] [PubMed] [Google Scholar]
- Tapio S., Grosche B. (2006). Arsenic in the aetiology of cancer. Mutat. Res. 612, 215–246. [DOI] [PubMed] [Google Scholar]
- Thomas D. J., Li J., Waters S. B., Xing W., Adair B. M., Drobna Z., Devesa V., Styblo M. (2007). Arsenic (+3 oxidation state) methyltransferase and the methylation of arsenicals. Exp. Biol. Med. 232, 3–13. [PMC free article] [PubMed] [Google Scholar]
- Tseng C. H. (2005). Blackfoot disease and arsenic: a never-ending story. J. Environ. Sci. Health C. Environ. Carcinog. Ecotoxicol. Rev. 23, 55–74. [DOI] [PubMed] [Google Scholar]
- Tseng C. H. (2009). A review on environmental factors regulating arsenic methylation in humans. Toxicol. Appl. Pharmacol. 235, 338–350. [DOI] [PubMed] [Google Scholar]
- Tseng W. P., Chu H. M., How S. W., Fong J. M., Lin C. S., Yeh S. (1968). Prevalence of skin cancer in an endemic area of chronic arsenicism in Taiwan. J. Natl. Cancer Inst. 40, 453–463. [PubMed] [Google Scholar]
- Valenzuela O. L., Drobna Z., Hernandez-Castellanos E., Sanchez-Pena L. C., Garcia-Vargas G. G., Borja-Aburto V. H., Styblo M., Del Razo L. M. (2009). Association of AS3MT polymorphisms and the risk of premalignant arsenic skin lesions. Toxicol. Appl. Pharmacol. 239, 200–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitbread A. K., Tetlow N., Eyre H. J., Sutherland G. R., Board P. G. (2003). Characterization of the human Omega class glutathione transferase genes and associated polymorphisms. Pharmacogenetics 13, 131–144. [DOI] [PubMed] [Google Scholar]
- Wood T. C., Salavagionne O. E., Mukherjee B., Wang L., Klumpp A. F., Thomae B. A., Eckloff B. W., Schaid D. J., Wieben E. D., Weinshilboum R. M. (2006). Human arsenic methyltransferase (AS3MT) pharmacogenetics: gene resequencing and functional genomics studies. J. Biol. Chem. 11, 7364–7373. [DOI] [PubMed] [Google Scholar]
- Zakharyan R. A., Sampayo-Reyes A., Healy S. M., Tsaprailis G., Board P. G., Liebler D. C., Aposhian H. V. (2001). Human monomethylarsonic acid (MMA(V)) reductase is a member of the glutathione-S-transferase superfamily. Chem. Res. Toxicol. 14, 1051–1057. [DOI] [PubMed] [Google Scholar]
- Zhang H., Ge Y., He P., Chen X., Carina A., Qiu Y., Aga D. S., Ren X. (2015). Interactive effects of N6AMT1 and As3MT in arsenic biomethylation. Toxicol. Sci. 146, 354–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Li Y., Liu J., Wang D., Zheng Q., Sun G. (2014). Differences of urinary arsenic metabolites and methylation capacity between individuals with and without skin lesions in Inner Mongolia, Northern China. Int. J. Environ. Res. Public Health 11, 7319–7332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.