Abstract
The developmental origins of obesity hypothesis posits a multifaceted contribution of factors to the fetal origins of obesity and metabolic disease. Adipocyte hyperplasia in gestation and early childhood may result in predisposition for obesity later in life. Rodent in vitro and in vivo studies indicate that some chemicals may directly affect adipose progenitor cell differentiation, but the human relevance of these findings is unclear. The nuclear receptor peroxisome proliferator-activated receptor gamma (PPARG) is the master regulator of adipogenesis. Human adipose-derived stem cells (hASC) isolated from adipose tissue express endogenous isoforms of PPARG and represent a biologically relevant cell-type for evaluating activity of PPARG ligands. Here, a multi-endpoint approach based on a phenotypic adipogenesis assay was applied to screen a set of 60 chemical compounds identified in ToxCast Phase I as PPARG active (49) or inactive (11). Chemicals showing activity in the adipogenesis screen were further evaluated in a series of 4 orthogonal assays representing 7 different key events in PPARG-dependent adipogenesis, including gene transcription, protein expression, and adipokine secretion. An siRNA screen was also used to evaluate PPARG-dependence of the adipogenesis phenotype. A universal concentration-response design enabled inter-assay comparability and implementation of a weight-of-evidence approach for bioactivity classification. Collectively, a total of 14/49 (29%) prioritized chemicals were identified with moderate-to-strong activity for human adipogenesis. These results provide the first integrated screening approach of prioritized ToxCast chemicals in a human stem cell model of adipogenesis and provide insight into the capacity of PPARG-activating chemicals to modulate early life programming of adipose tissue.
Keywords: adipose-derived stem cell, adipogenesis, PPARG, ToxCast, endocrine disrupting chemicals, obesogens
The increased prevalence of obesity in the United States continues to be a serious health concern. Data from the National Health and Nutrition Examination Survey reports obesity levels at 35–37% of the population, independent of race, gender, or age (Yang and Colditz, 2015). Given the rise in obesity rates, efforts are underway to identify contributions of genetic and environmental risk factors. Together with genotype, nutrition, and activity levels, chemical exposures may contribute to adiposity through altered adipocyte development. The notion that chemicals may alter prenatal or early life adipose tissue development has been coined the “obesogen hypothesis” (Grun and Blumberg, 2006) and was reviewed in a National Toxicology Program workshop (Thayer et al., 2012). Metabolic set points established at critical windows in early life may significantly influence later-life disposition to obesity and metabolic disease (Boekelheide et al., 2012; Hatch et al., 2010; Heindel et al., 2015; Janesick and Blumberg, 2012; La Merrill and Birnbaum, 2011; Legler et al., 2011; Regnier and Sargis, 2014; Thayer et al., 2012).
Adipose tissue is derived from the mesoderm germ layer during embryonic development, with overall fat mass composition determined by adipocyte hypertrophy (cell size) and hyperplasia (cell proliferation and/or maturation). The second trimester of gestation is a critical window for adipose tissue hyperplasia in humans (Poissonnet et al., 1983, 1984). Tissue cellularity continues to develop through childhood, reaching a terminal set point at approximately 18 years of age (Spalding et al., 2008). Once this set point is established, turnover of adipocytes continues at a steady rate for the duration of adult life without any significant alteration to total cell number (Spalding et al., 2008). Individuals with a greater number of adipocytes are far more likely to have an overweight/obese phenotype (Knittle et al., 1977, 1979; Spalding et al., 2008). In this manner, chemicals that induce a higher number of adipocytes in adipose tissue might affect weight gain by pre-disposing individuals to other dietary and lifestyle risk factors more commonly associated with obesity.
Human adipose-derived stem cells (hASC) are progenitor cells within the adipose stromal mesenchyme that are recruited for lineage specification to form fully functional adipocytes. The essential regulator of early adipocyte differentiation is the nuclear receptor peroxisome proliferator-activated receptor gamma, PPARG (Rosen et al., 1999). Expression and activation of this ligand-inducible transcription factor is the key molecular initiating event driving the transcriptional program that upregulates lipid metabolism and supports adipokine secretion in mature adipocytes.
The EPA’s ToxCast (Dix et al., 2007) and NIH’s Tox21 programs include PPARG as a target endpoint for chemical screening and prioritization (Judson et al., 2010; Kavlock et al., 2012). These programs aim to modernize toxicity testing by implementing high-throughput in vitro assays to characterize chemical–biological activity. ToxCast generated data sets are a valuable resource for prioritizing chemicals for further testing in assays specifically designed to recapitulate key cellular events in an Adverse Outcome Pathway (AOP). Such prioritization efforts are being used by the Endocrine Disruption Screening Program (EDSP), where Phase I and II ToxCast data are evaluated to reduce the number of chemicals moving on to more traditional animal-based tests for disruption of the estrogen, androgen, and thyroid endocrine pathways. A similar approach has also been recommended for other pathways, including those related to the glucocorticoid and PPAR signaling networks (Filer et al., 2014b; Reif et al., 2010). To date, ToxCast prioritized chemical sets have yet to be screened in biologically relevant assays for evaluating human adipogenesis.
A number of compounds have been linked to PPARG-dependent adipogenesis or increased fat accumulation in rodent in vivo models or in cell lines (Feige et al., 2007; Hao et al., 2012; Inadera and Shimomura, 2005; Kanayama et al., 2005; Li et al., 2012; Pereira-Fernandes et al., 2013a). However, there is no primary human cell model to assess the PPARG pathway and adipogenesis. Building on a medium-throughput assay we had developed (Foley et al., 2015), this study focused more explicitly on the PPARG pathway in hASC differentiation. A multi-endpoint approach to assay design was implemented with 5 orthogonal assays spanning neutral lipid accumulation, PPARG-dependence, gene and protein expression, and adipokine secretion endpoints. The assay was multiplexed with acute and chronic cytotoxicity endpoints to identify appropriate viability ranges for each test compound. Each assay endpoint was evaluated with a universal concentration-response design to simplify inter-assay comparison and provide greater weight-of-evidence for bioactivity classification (See flowchart in Figure 1). Our results demonstrate the first integrated screening approach of prioritized ToxCast Phase I chemicals in a human stem cell model of adipogenesis and provide insight into the capacity of PPARG candidate chemicals to modulate early life programming of adipose tissue.
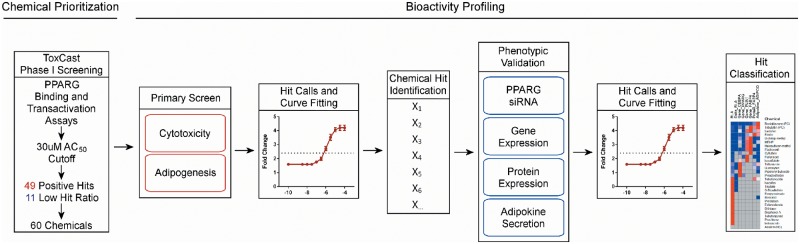
FIG. 1.
Overview of the human adipogenesis study design. Chemical selection from ToxCast Phase 1 screening consisted of 49 chemicals identified in 3 independent PPARG binding and transactivation assays. 11 “low active” chemicals were selected as experimental negatives based on hit ratios across all ToxCast assays. A primary screen with 60 chemicals was run in a functional adipogenesis assay to identify viability ranges for each chemical, as well as activity for the differentiation endpoint. Assay hits were then evaluated across 4 additional orthologous assays spanning 7 endpoints that are specific for molecular and phenotypic features of human adipocytes. All data was analyzed in concentration-response format using a universal assay platform. Chemical hits identified in each assay were scored and classified according to hit frequency, efficacy, and potency.
MATERIALS AND METHODS
Cell culture
Human subcutaneous adipose-derived stem cells obtained at passage 3 were from a pooled donor super lot (SL0048, Zen-Bio, Research Triangle Park, North Carolina) and used for all experiments. The donor pool was derived from a range of anatomical sites of 8 Caucasian female donors with a mean age of 44 [range: 29–51] and mean BMI of 26.3 [range: 25.1–29.2] (Table 1). Cells were maintained in Dulbecco’s Modified Eagle’s Medium (Thermo Scientific, Waltham, Massachusetts) supplemented with 10% mesenchymal stem cell-fetal bovine serum (LifeTech, Carlsbad, California), l-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin, and 0.25 µg/ml Amphotericin B at 5% CO2 in a 37 °C humidified chamber. Cells did not exceed passage 4 for any experiments.
TABLE 1.
Human Adipose-Derived Stem Cell Donor Demographics.
| Lot # | Donors | Gender | Age (mean) | BMI (mean) | Diabetic | Ethnicity | Smoker |
|---|---|---|---|---|---|---|---|
| SL0048 | 8 | Female | 44 [Range: 29–51] | 26.3 [Range: 25.1–29.2] | No | Caucasian | No |
Lot and donor number, gender, age, body mass index (BMI), diabetes status, ethnicity, and smoking status are reported for the donor lot used for all experiments.
Chemicals
Reference chemicals for PPARG-dependent adipogenesis assay development and validation were rosiglitazone (Cayman Chemical, Ann Arbor, Michigan) and tributyltin chloride (Sigma-Aldrich, St. Louis, Missouri) (Supplementary Table 1). Prioritized candidate chemicals for PPARG specific activity were selected using 3 ToxCast Phase I assays: NVS_NR_hPPARg, ATG_PPARg_TRANS, and NCGC_PPARg_Agonist (public release January 2010; www3.epa.gov/research/COMPTOX/previously_published.html; last accessed 27 September 2016). The NovaScreen radioligand binding assay, the Attagene Trans-Factorial assay, and NCGC GeneBlazer assay were selected based on specificity for the nuclear receptor PPARG and were restricted to an AC50 cutoff of 30 µM, occurring in at least 1 of 3 assays. No chemical was identified in more than 2 assays. Based on these parameters, 49 test chemicals were selected for screening (Supplementary Table 2). To identify suitable negative controls, all 309 chemicals from Phase I screening were rank-ordered by hit ratio to identify chemicals that exhibited the lowest activity across all assay platforms, and were negative for all 3 PPARG assays. Using these criteria, 11 chemicals had a hit ratio ≤0.0038 and were incorporated as “low-active” negative controls into the primary screen (Supplementary Table 3). A total of 60 test chemicals were run in the primary adipogenesis assay.
Primary adipocyte differentiation assay
A quantitative adipogenesis assay multiplexed with cytotoxicity endpoints was performed as previously described (Foley et al., 2015) with minor modifications. Adipose-derived stem cells were expanded for 1 passage, seeded at a density of 46,875 cells/cm2 in 96-well clear bottom, black-walled ViewPlates (Perkin Elmer, Waltham, Massachusetts), and cultured in maintenance medium for 48 h. For differentiation, a positive control cocktail was composed of maintenance medium with 10 µg/ml human insulin (Sigma-Aldrich, St. Louis, Missouri), 200 µM isobutylmethylxanthine (Sigma-Aldrich), 20 nM dexamethasone (Selleckchem, Houston, Texas), and 1µM rosiglitazone. To establish a medium for targeting PPARG activity, rosiglitazone was omitted from the cocktail formulation. To demonstrate PPARG-dependent activity in this medium formulation, 1 µM rosiglitazone or 100 nM tributyltin chloride were used as chemical reference compounds. To initiate the assay, medium was added with positive control (rosiglitazone or tributyltin) or treatment compound at 0, 48, 96, and 144 h with termination of the assay at the end of 192 h; a total of 4 treatments over an 8 day incubation period. Compound plates were prepared from DMSO master plates as 4X stock in treatment medium using a Viaflo Assist (Integra Biosciences, Hudson, New Hampshire) to automate chemical dilutions. Treatments were performed using a semi-automated Viaflo 384 liquid handling system with 96-channel head (Integra Biosciences). Samples were fixed in freshly prepared 4% paraformaldehyde for 30 min, washed 2 times in PBS, and subsequently stained with the neutral lipid stain AdipoRed (Lonza, Basel, Switzerland) and the nuclear dye Hoechst 33342 (Sigma-Aldrich) for an additional 30 min. After washing 2 times in PBS, fluorescence of AdipoRed (Ex485/Em572) and Hoechst (Ex330/Em470) was collected on a Spectramax M3 microplate reader (Molecular Devices, Sunnyvale, California). Relative lipid accumulation (RLA) was calculated by determining the ratio of lipid intensity to Hoechst intensity values. RLA values were normalized to plate-based non-differentiated hASC controls to determine fold-change values across multiple test plates.
Two cytotoxicity assays for viability and cell counts were multiplexed with the differentiation assay. Following the initial 48-h treatment window, medium was collected for viability determination using the Cytotoxicity Detection Kit PLUS (LDH) (Roche, Indianapolis, Indiana). Protocol was used according to manufacturer’s instructions. For cell counts, Hoechst stained cell nuclei were used to acquire single channel images on an ArrayScan XTI (Cellomics, Pittsburgh, Pennsylvania) using a dry 10X objective (0.3 NA) following fixation. The Target Activation algorithm was used to apply segmentation masking and calculate total object count across 10 fields. Values were plotted as fold-change to vehicle control.
PPARG siRNA assay
A loss-of-function PPARG siRNA assay for assessment of transcript knockdown was performed as previously described (Foley et al., 2015). Cells were seeded at 24,000 cells/cm2 in 96-well plates and transfected with 25nM SMARTpool siRNA duplexes (Dharmacon/Thermo Fisher Scientific, Waltham, Massachusetts) using Lipofectamine RNAiMax (LifeTech, Carlsbad, California). Seventy-two hours post-transfection, cells were initiated to differentiate according to the adipogenesis assay protocol. The following siRNA oligo duplexes were used: ON-TARGETplus Non-targeting Pool (Thermo; D-001810-10-05) and ON-TARGETplus SMARTpool Human PPARG (Thermo; L-003436-00-0005). For screening, adipogenesis assays were conducted for test compounds using matched plates of non-targeting and PPARG siRNA, respectively. RLA fold-change values were determined relative to plate-based vehicle controls and a ΔRLA metric calculated to determine the relative magnitude of PPARG knockdown on test chemical induced differentiation. For each concentration point, [1+ (RLA non-targeting siRNA − RLA PPARG siRNA)] was calculated and plotted as concentration-response. Three independent replicates of the assay were performed.
QuantiGene Plex assay
A multiplexed quantitative mRNA assay was developed using the Luminex bead (xMap) technology in the QuantiGene Plex system (eBioscience, San Diego, California). Four phenotypic marker genes for human adipogenesis were selected: CCAAT/enhancer binding protein alpha (CEBPA) (NM_004364), Peroxisome proliferator activated receptor gamma (PPARG) (NM_005037), Perilipin 1 (PLIN1) (NM_002666), and Fatty acid binding protein 4 (FABP4) (NM_001442). Two internal reference standards were selected for expression normalization: Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (NM_002046.5) and Beta-2 microglobulin (B2M) (NM_004048.2). Samples were harvested at the end of the adipogenesis assay in 100 µl of proprietary lysis mixture and hybridized to probe sets according to the manufacturer’s protocol. The dynamic range of the assay was established from control samples treated with the reference compounds. Signal data was acquired on a BioPlex 200 system and analyzed using the Bio-Plex Manager Software v4.0 (Bio-Rad, Hercules, CA). The raw intensities for each gene were normalized to the geometric mean of GAPDH and B2M at each concentration point to determine the relative gene expression level (RGEL). The RGEL ratio to matched plate-based non-differentiated negative control was calculated, Log2 transformed, and plotted as fold-change. Two independent replicates of the assay were performed.
Protein simple quantitative protein expression assay
A duplexed quantitative protein expression assay was developed using capillary Western technology on a Wes instrument (Protein Simple, San Jose, California). Phenotypic marker FABP4 was combined with an internal GAPDH reference standard to screen chemicals in a concentration-responsive manner. Samples were harvested after completion of the adipogenesis assay in 50 µl of Mammalian Protein Extraction Reagent (Thermo Fisher Scientific) supplemented with 1X HALT Protease Inhibitor cocktail (Thermo Fisher Scientific). Primary antibodies were goat anti-A-FABP (C-15) (Santa Cruz Biotechnology, Dallas, Texas; sc-18661), and goat anti-GAPDH (AbCam, Cambridge, UK; ab9483). Secondary antibody was donkey anti-goat IgG-HRP (Santa Cruz Biotechnology; sc-2020). Samples were separated by molecular weight according to manufacturer’s protocol on 25-lane, 12–230 kDa resolution cartridges (Protein Simple; PS-MK01), substituting the included anti-rabbit secondary antibody with the anti-goat HRP. The dynamic range for the assay was established from control samples treated with reference compounds rosiglitazone and tributyltin. Raw image files were acquired and analyzed using Compass v2.6.6 software (Protein Simple) to determine relative intensities for each protein. FABP4 intensity values were normalized to GAPDH at each concentration point to calculate relative protein expression levels (RPEL). RPELs were normalized as a percentage of the mean for each plate-based positive reference standard (1 µM rosiglitazone). Two independent replicates of the assay were performed.
Meso scale discovery (MSD) adipokine secretion assay
A singleplex electrochemiluminescent ELISA (Meso Scale Discovery, Gaithersburg, Maryland) was used for detecting PPARG-dependent adipokine secretion from the adipogenesis assay. The human adiponectin kit (K151BXC), designed for analysis of human serum and plasma samples, was adapted for cell culture applications. The assay was performed as described in the manufacturer’s protocol with 3 modifications to improve small-volume sensitivity for cell culture samples. (1) Undiluted sample volumes were doubled to a maximum capacity of 20 µl; (2) primary incubation periods with test sample were increased to 16 h at 4 °C; and (3) the linear range of the calibrator standard curve was extended down to 0.00256 ng/ml to improve the lower limit of detection. The dynamic range for the assay was established from control samples treated with reference compounds. The MSD Discovery Workbench software was used to extrapolate raw sample concentrations (ng/ml) from the standard curve using a 4-parameter logistic model with 1/Y2 weighting function to enhance low end curve-fitting. Sample values were normalized to plate-based vehicle controls and plotted as fold-change. Three independent replicates of the assay were performed.
Curve fitting and data analysis
Parameters for hit calls were similar to those used for the ToxCast Analysis Pipeline (Filer et al., 2015a) with modifications. Threshold cutoffs were determined as a multiple of the noise-band for each assay using all of the vehicle control values for each specific endpoint. Initial outlier removal was performed by excluding values outside of the mean ± 3 standard deviations of the vehicle data set. The outlier adjusted median was used to calculate a baseline median absolute deviation (BMAD) based on the following equation:
where b = 1.4826 (assuming normality of the data), is each data point is the n original observations. Based on this value, the following cutoffs were used for each assay endpoint: 3*BMAD (LDH release, cell counts, gene expression), and 5*BMAD (RLA, protein expression, adiponectin secretion). Due to small variability in the vehicle controls, a default 20% cutoff was applied for the ΔRLA endpoint.
Any point across the concentration series with a mean value greater than the cutoff threshold was considered a positive hit. For chemical hits, a non-linear regression curve was fit to the data using Prism 6 (GraphPad Software Inc, La Jolla, California). The viability range for each chemical was established with a Toxicity Concentration Cutoff (TCC) derived from the hits identified in the multiplexed cytotoxicity endpoints (LDH release, cell counts) of the primary adipogenesis screen. The lowest concentration point exceeding the cutoff threshold for either cytotoxicity assay was deemed the TCC, and thus the preceding concentration established the top of the viability range. If no significant cytotoxicity was observed, the TCC was set at 1000 µM. Downstream curves were fit only within the viability range for hits identified in all of the phenotypic assay endpoints. Summary values derived from the curves were maximum fold-change, half-maximal activity concentration (AC50) for potency, and total peak area under the concentration-response curve (AUC) for efficacy. All data was plotted, visualized, and analyzed using Prism 6. Heat maps were generated using GENE-E software v3.0.204 (Broad Institute, Cambridge, Massachusetts; www.broadinstitute.org/cancer/software/GENE-E/).
RESULTS
Optimization of an In Vitro Model of Human Adipogenesis for PPARG Function
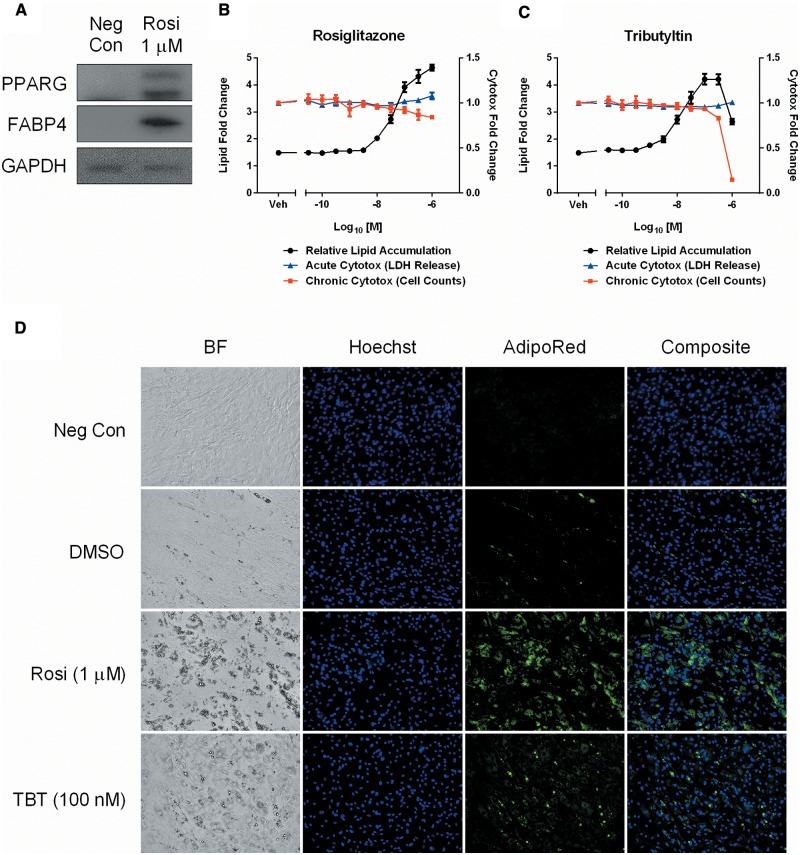
We previously developed an 8 day, repeat-dosing, multiplexed assay for differentiation steps within the adipocyte lineage. To identify a suitable donor pool for screening, several lots were evaluated using a positive control hormone cocktail (10 µg/ml human insulin, 20 nM dexamethasone, 200 µM IBMX, and 1 µM rosiglitazone) to assess differentiation potential. Western blot analysis for the nuclear receptor PPARG and the terminal differentiation marker FABP4 confirmed a suitable donor pool that expressed both isoforms of PPARG, in addition to showing increases in FABP4 when terminally differentiated at 8 days (Figure 2A). Both the intra-assay Z′-factor (0.85) and signal/noise (5.1-fold change) validated the suitability for screening and dynamic range of the chosen lot, respectively (data not shown).
FIG. 2.
Optimization of a PPARG-dependent human adipogenesis assay. A donor pool of non-differentiated adipose-derived stem cells was tested for adipogenic capacity and expression of endogenous PPARG isoforms at the end of the 8 day assay protocol. A Western blot of non-differentiated (Neg Con) and differentiated (Rosi 1 µM) cells was probed for PPARG, FABP4, and GAPDH expression (A). A modified medium formulation illustrates concentration-responsive differentiation of rosiglitazone (B) and tributyltin (C) treated cells, multiplexed with acute (LDH release) and chronic (cell counts) cytotoxicity endpoints. Concentrations are mean ± SEM of 3 experimental replicates. Micrographs taken with a 20× objective demonstrate brightfield (BF), nuclear (Hoechst), neutral lipid (AdipoRed), and composite images of the 4 control samples used in the adipogenesis screen (D). Neg Con = non-differentiated hASC, DMSO= vehicle, Rosi= rosiglitazone, TBT= tributyltin.
The adipogenic hormone cocktail used to differentiate hASCs induced coordinated activation of several key signaling pathways essential for lineage progression. Dexamethasone is a synthetic glucocorticoid receptor (GR) agonist that promotes GR-dependent gene transcription, IBMX is a cyclic nucleotide phosphodiesterase inhibitor that promotes accumulation of cAMP, insulin stimulates the PI3K/AKT pathway to modulate glucose and lipid metabolism, and rosiglitazone is a high-affinity PPARG agonist that promotes gene transactivation. Modification of the cocktail formulation was evaluated to determine the potential for specifically probing PPARG-dependent activation. In the absence of a PPARG agonist, hASCs did not differentiate efficiently within the 8-day time frame, showing negligible neutral lipid accumulation with the DMSO vehicle (Figure 2B and D). However, a robust relative lipid accumulation (RLA) concentration-response was observed when adding rosiglitazone back to the base hormone cocktail up to the positive control concentration of 1 µM. Rosiglitazone caused differentiation to an early adipocyte phenotype showing multiple perinuclear lipid droplets and a more rounded morphology (Figure 2B and D). The multiplexed acute and chronic cytotoxicity endpoints demonstrated that no significant toxicity was observed across the concentration ranges tested. To incorporate a chemical-specific reference compound into the assay, tributyltin chloride was dosed in the same base cocktail (without rosiglitazone) at the prescribed 48 h intervals. A similar RLA concentration-response was observed with tributyltin and rosiglitazone (Figure 2C). The maximum observed effect level, without any significant toxicity, was 1 µM for rosiglitazone and 100 nM for tributyltin (Figure 2D). Treatment with these concentrations served as positive controls, in conjunction with vehicle (DMSO) and non-differentiated negative controls (Figure 2D).
Primary Adipogenesis Screen
Chemicals prioritized from Phase I of the ToxCast program were screened in the adipogenesis assay, including 49 PPARG “active” chemicals and 11 “low-active” negative controls. Chemicals were dosed in a 10-point concentration series (3 nM–100 µM) to evaluate activity on human adipogenesis. Each experiment was run in plate triplicate and performed 3 times over independent weeks. Inter-experimental performance metrics were robust for RLA, LDH release, and cell counts, signifying a reliable screening system (Supplementary Figure 1). Initial analysis of the cytotoxicity data (LDH release and cell counts) was conducted to identify toxicity concentration cutoffs (TCC). Similar to a range-finding experiment in a rodent bioassay, this pre-filtering step was taken to identify the appropriate viability range of each chemical compound. Of the 49 compounds identified as PPARG agonists in the ToxCast dataset, 15 (31%) produced significant activity in the 2 day LDH release assay, resulting in identification of a TCC (Supplementary Figure 2A and B). The 8-day cell count assay had 22 (45%) significant hits, with similar TCC determinations (Supplementary Figure 3A and B). There was no significant toxicity associated with any of the low-active compounds in either assay. The viability range was defined by the lowest concentration point in the series, up to the point preceding any respective TCC. All curve-fitting for chemical hits was restricted to the viability range for each chemical in all subsequent assay endpoints.
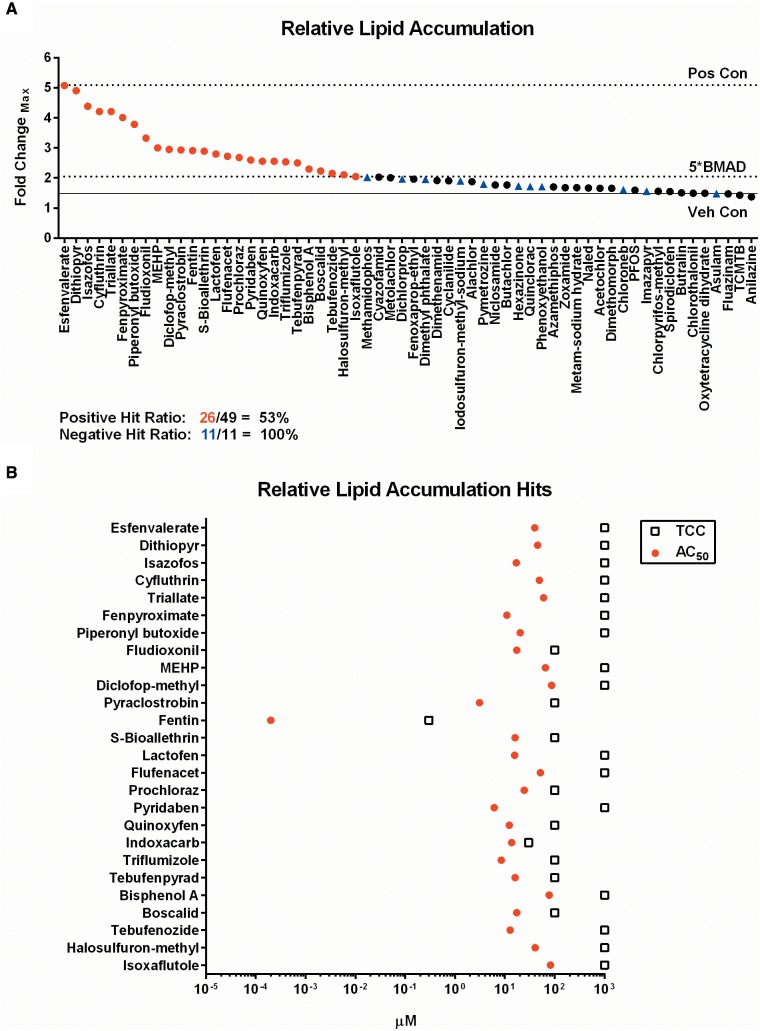
Of the compounds screened in the RLA assay, 26/49 (53%) of ToxCast PPARG agonists demonstrated lipid accumulation exceeding the 5*BMAD cutoff, whereas 11/11 (100%) low-active controls exhibited no activity. Efficacy of activity is illustrated by the maximum fold-change observed for each chemical (Figure 3A). The potency of the response was determined by plotting the rank-ordered hits by efficacy (top-to-bottom) and plotting the derived AC50 values with the corresponding TCC (Figure 3B). Hit concordance was 3/9 (33%) with the ToxCast NovaScreen assay (NVS_NR_hPPARg), 26/45 (58%) for the Attagene assay (ATG_PPARg_TRANS), and 1/4 (25%) for the NIH Chemical Genomics Center GeneBlazer assay (NCGC_PPARg_Agonist) (Supplementary Figure 9D). The AC50 concordance for these assays was deemed non-significant for the NovaScreen assay (r = 0.321, P = .792), significant for the Attagene assay (r = 0.452, P = .021), and could not be determined for the NCGC assay (Supplementary Figure 9A–C). Individual concentration-response graphs for the 3 assay endpoints in the primary screen are located in the supplementary graphs (Appendix 1, p2–16). The 26 chemical hits from the adipogenesis assay, and 1 negative control (asulam), were selected for additional phenotypic characterization.
FIG. 3.
Primary adipogenesis screening results. Sixty chemicals screened in the adipogenesis assay were scored for significance and plotted as rank-ordered (left-to-right) maximum fold-change observed for any concentration point in the viable range of each chemical (A). The corresponding AC50 values and cytotoxicity derived TCC are plotted for each hit identified (B). Red circles are PPARG candidate chemical hits, black circles are non-significant PPARG candidate chemical hits, and blue triangles are negative controls. Pos Con = rosiglitazone positive control, Veh Con = DMSO vehicle control, 5*BMAD = significance threshold cutoff, TCC = toxicity concentration cutoff.
Loss-of-Function PPARG siRNA Assay
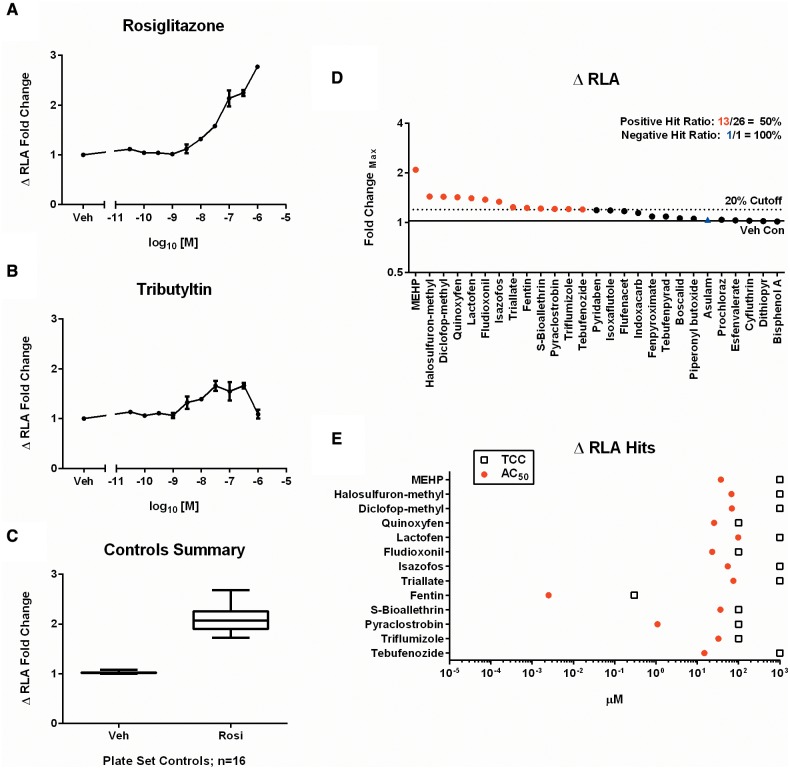
Despite the qualitative and quantitative advantages to using a functional phenotype like neutral lipid accumulation to define PPARG-dependent adipogenesis, an increase in lipid content does not discriminate chemically directed transcriptional reprogramming of adipogenesis from direct modulation of lipid metabolism. We previously validated a loss-of-function siRNA knockdown protocol for the adipogenesis assay (Foley et al., 2015). To develop an assay that could be scored quantitatively, a ΔRLA metric was developed which calculates the difference between control (non-targeting siRNA) and knockdown (PPARG siRNA) phenotypes, enabling plots for gain-of-function concentration-response. This siRNA assay assists in discriminating true positive hits that are dependent on PPARG for promoting neutral lipid accumulation from false positives that could either result from an assay artifact or enhanced lipid accumulation via mechanisms other than PPARG-mediated transcription (Supplementary Figure 4). A concentration-dependent increase in ΔRLA was validated with both rosiglitazone (Figure 4A) and tributyltin (Figure 4B). The shape of the curves was consistent with the original adipogenesis assay, demonstrating consistency of the RLA phenotype and dependence on PPARG. Plate-matched sets of non-targeting and PPARG siRNA were run for all 27 chemicals in experimental triplicate. Individual concentration-response graphs for the ΔRLA endpoint are located in the supplementary graphs (Appendix 1, p17–19). The mean dynamic range for rosiglitazone controls was 2.09 ± 0.26 fold-change (Figure 4C). 13/26 (50%) of the PPARG candidates were significant in the assay. The other 13 chemicals with activity in the primary screen showed a similar response in the wild-type and knockdown cells, resulting in insignificant ΔRLA (Figure 4D). The potency was also determined for each compound, with the organotin fentin hydroxide yielding the lowest AC50 value (2.5 nM) (Figure 4E).
FIG. 4.
Loss-of-function evaluation in the PPARG siRNA assay. Plate sets of non-targeting and PPARG siRNA were evaluated in concentration-response format for loss of RLA. ΔRLA fold-change plotted for rosiglitazone (A) and tributyltin (B) demonstrate a reduction in lipid accumulation. Concentrations are mean ± SEM of 3 experimental replicates. The performance of control values from all plate sets run in the assay (n = 16) are shown (C). Each box and whisker plot displays the median, first and third quartiles (boxes), and the 2.5–97.5 percentile (whiskers). Results from the chemicals screened in the ΔRLA assay were scored for significance and plotted as rank-ordered (left-to-right) maximum fold-change observed for any concentration point in the viable range determined for each chemical (D). The corresponding AC50 values and TCC are plotted for each hit identified (E).
Gene Expression Assay
Initial lineage commitment and progression is encompassed by several core genes that drive the adipogenic program. One of the earliest key molecular initiating events is activation of CEBPA, which directly promotes induction of PPARG expression. Ligand-dependent binding to PPARG promotes a downstream cascade of gene activation that includes induction of lipid metabolism genes associated with mature adipocytes. Two of these genes, PLIN1 and FABP4, are associated with lipid droplet formation and fatty acid transport, respectively.
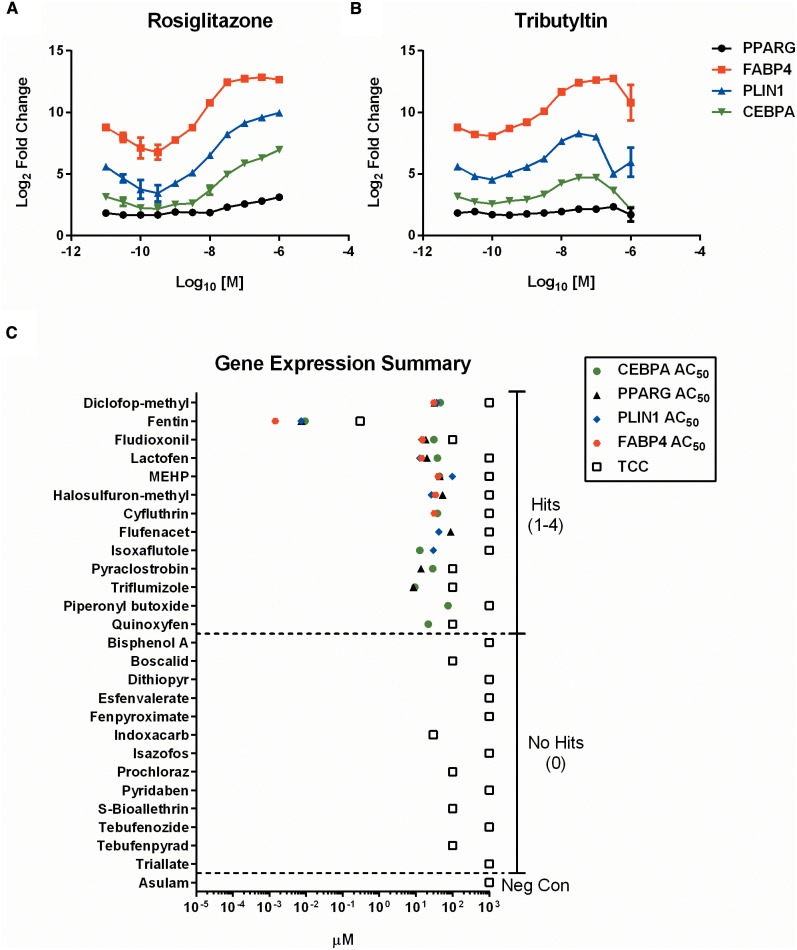
Using a multiplexed approach, transcript levels of the 4 phenotypic marker genes were measured simultaneously from lysates harvested from the adipogenesis assay. Validation of the selected probe sets with rosiglitazone demonstrated a consistent concentration-response that coincided with expected relative gene expression levels (RGEL). The maximum observed Log2 RGELs for CEBPA, PPARG, and PLIN1 were 7.0, 3.1, and 10.0, respectively, at the 1 µM dose. FABP4 RGEL peaked earlier with a 12.8-fold change at the 300 nM dose (Figure 5A). The plateau observed for FABP4 indicated likely saturation of the probe signal in the multiplexed format, but was deemed suitable for screening. Similar expected results were seen with tributyltin where RGELs for CEBPA, PPARG, PLIN1, and FABP4 were 4.7, 2.1, 8.0, and 12.6, respectively, at the 100 nM concentration (Figure 5B).
FIG. 5.
Multiplexed gene expression results. CEBPA, PPARG, PLIN1, and FABP4 mRNA expression are plotted as Log2 ratios of treated versus DMSO vehicle controls for rosiglitazone (A) and tributyltin (B). Each gene endpoint was scored independently and comprehensive AC50 and TCC values plotted for each chemical (C). Dashed lines separate regions of hit frequency (0–4), with activity of negative control (asulam) shown.
The 27 chemicals selected from the primary adipogenesis screen were then run in the gene expression assay. Each chemical was scored independently for each of the 4 gene endpoints. Individual concentration-response graphs for each endpoint are located in the supplementary graphs (Appendix 1, p20–31). Asulam, the negative control compound, was negative for all adipogenic markers. 11/26 (42%) compounds were positive for CEBPA with a maximum RGEL of 7.0 observed for lactofen (Supplementary Figure 5A) and a maximum potency (ie, lowest AC50) for fentin hydroxide (Supplementary Figure 5B). 9/26 (35%) of compounds were positive for PPARG expression, where fentin had both the maximum expression (2.7-fold), and the greatest potency (AC50 of 7.4 nM) (Supplementary Figure 6A and B). Fewer genes were positive for PLIN1, with 8/26 (31%) demonstrating some activity. Lactofen had the greatest efficacy with an RGEL of 10.8, and fentin hydroxide consistently the most potent, with an AC50 of 7.4 nM (Supplementary Figure 7A and B). Finally, FABP4 had the fewest hits, 7/26 (27%). Fentin hydroxide had the greatest effect on FABP4, with an RGEL of 13.6 and an AC50 of 1.5 nM (Supplementary Figure 8). Overall, there were at least 1–4 gene expression hits for 13/26 (50%) PPARG candidate chemicals (Figure 5C). Diclofop-methyl, fentin, fludioxonil, lactofen, and MEHP (4 hits), halosulfuron-methyl (3 hits), cyfluthrin, flufenacet, isoxaflutole, pyraclostrobin, and triflumizole (2 hits), piperonyl butoxide and quinoxyfen (1 hit). In general, the range of AC50 values was within the same order of magnitude for all chemical-gene associated hits and below the TCC for each respective compound.
Protein Expression Assay
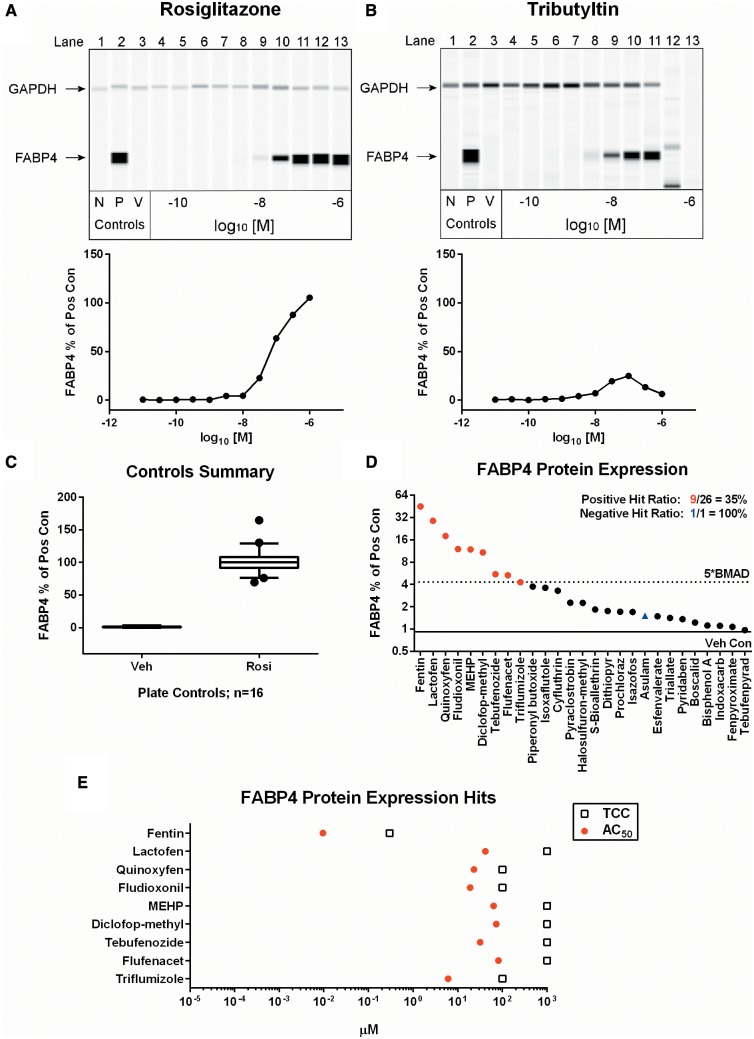
Target protein expression remains a valuable indicator for phenotypic characterization, but traditional Western blot methods are limited in throughput, variable on blotting consistency, require a large volume of starting material, and are restricted in the ability to reliably quantify relative expression levels. To create a quantitative assay for concentration-response assessments of protein expression from the 96-well adipogenesis assay, a capillary Western immunoblotting system was employed. The target protein selected was FABP4, which is highly expressed in both early and fully mature adipocytes. GAPDH expression is relatively consistent between hASC and mature adipocytes, so it was used as an internal reference standard for normalization. Validation of the technology with rosiglitazone demonstrated the expected qualitative and quantitative concentration-response for FABP4 relative protein expression levels (RPEL) at the 8-day termination point of the assay (Figure 6A). Tributyltin demonstrated a mean peak RPEL of 25%, with a subsequent loss of signal at the 300 nM and 1 µM concentrations, consistent with the TCC for this compound (Figure 6B). As with the previous endpoints, each of the 26 RLA hits, and the negative control, were screened for FABP4 protein expression in experimental duplicate. Individual concentration-response graphs for FABP4 expression are in the supplementary graphs (Appendix 1, p32–34). The assay proved to have a fair amount of variability between runs, as demonstrated by the fluctuation in vehicle (range 0–3.3%) and rosiglitazone control RPELs (range 70–165%) (Figure 6C). The number of chemicals exceeding the 5*BMAD cutoff was 9/26 (35%), with no activity observed for the negative control (Figure 6D). Fentin hydroxide yielded the highest efficacy at 45% FABP4 expression relative to rosiglitazone, and was the most potent (AC50 = 9.5 nM) of the tested compounds (Figure 6E).
FIG. 6.
FABP4 protein expression results. Qualitative and quantitative FABP4 and GAPDH protein expression levels are shown in concentration-response format for rosiglitazone (A) and tributyltin (B). Values are representative of 3 experimental replicates. N = non-differentiated controls, P = rosiglitazone positive control, V = DMSO vehicle control. The performance of control values from all plates run in the assay (n = 16) are shown (C). Each box and whisker plot displays the median, first and third quartiles (boxes), and the 2.5–97.5 percentile (whiskers). Results from the chemicals screened in the FABP4 assay were scored for significance and plotted as rank-ordered (left-to-right) maximum percentage of positive control observed for any concentration point in the viable range of each chemical (D). The corresponding AC50 values and TCC are plotted for each hit identified (E).
Adiponectin Secretion Assay
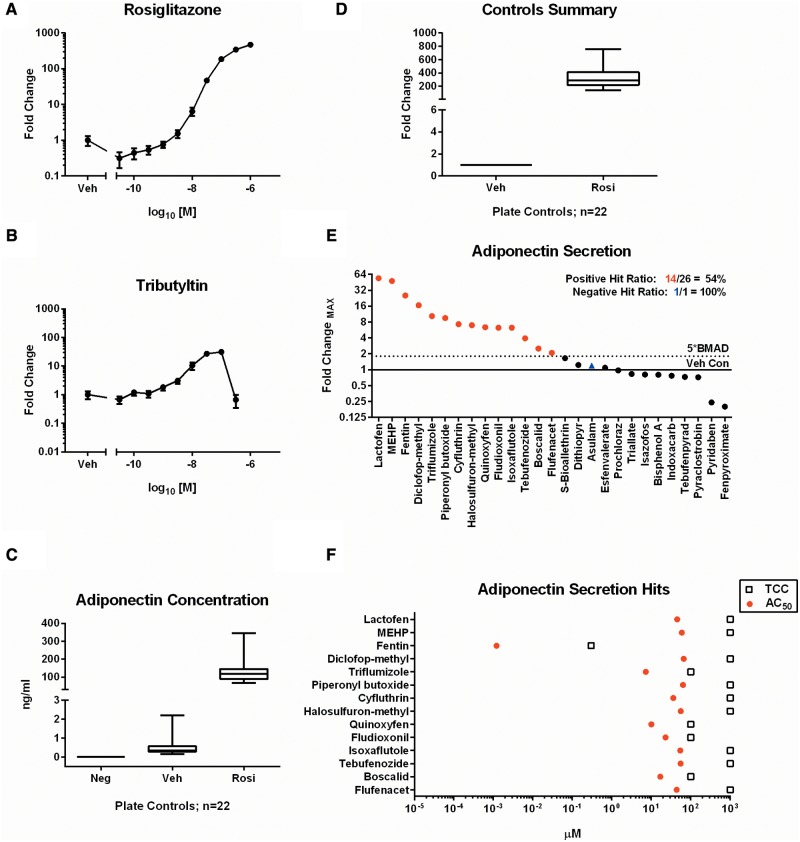
Fully mature adipocytes secrete cytokine proteins, so-called adipokines, such as leptin, adiponectin, and IL-6 that influence both local and peripheral tissue functions. Adiponectin (ADIPOQ) expression is enhanced by PPARG activation and increases insulin sensitivity, and modulates glucose and fatty acid metabolism (Combs et al., 2002). An electrochemiluminescent ELISA was employed to measure adiponectin protein levels in supernatant derived on the final day of treatment in the adipogenesis assay. Initial validation with rosiglitazone exhibited a robust concentration-response with peak activity of 469 fold-change over vehicle treated controls (Figure 7A). Tributyltin also promoted increased accumulation with a peak 31-fold change at 100 nM (Figure 2B). Samples were collected from 3 independent replicates of the 26 compounds, and negative control, screened in the previous assays. Individual concentration-response graphs for ADIPOQ secretion are located in the supplementary graphs (Appendix 1, p35–37). Median concentrations of ADIPOQ for the composite non-differentiated, vehicle, and rosiglitazone controls were 0, 0.3, and 117.9 ng/ml, respectively (Figure 7C), corresponding to a 286.5-fold change for rosiglitazone controls (Figure 7D). The results confirmed the specificity and range of adiponectin secretion in response to PPARG activation. 14/26 (54%) of compounds had significant secretion, whereas the negative control produced no effect. Lactofen exhibited the highest efficacy that was 54.1-fold over vehicle treated cells (Figure 7E). The most potent chemical was fentin hydroxide (AC50 = 1.2 nM) (Figure 7F).
FIG. 7.
Adiponectin secretion results. Secretion of adiponectin into the assay medium was assessed in concentration-response format for rosiglitazone (A) and tributyltin (B). Values are mean ± SEM of 3 independent replicates. The performance of control values from all plates run in the assay (n = 22) demonstrate concentration (C) and dynamic range (D) of the assay. Each box and whisker plot displays the median, first and third quartiles (boxes), and the 2.5–97.5 percentile (whiskers). Results from the chemicals screened in the adiponectin assay were scored for significance and plotted as rank-ordered (left-to-right) maximum fold-change observed for any concentration point in the viable range of each chemical (D). The corresponding AC50 values and TCC are plotted for each hit identified (E).
Summary Analysis
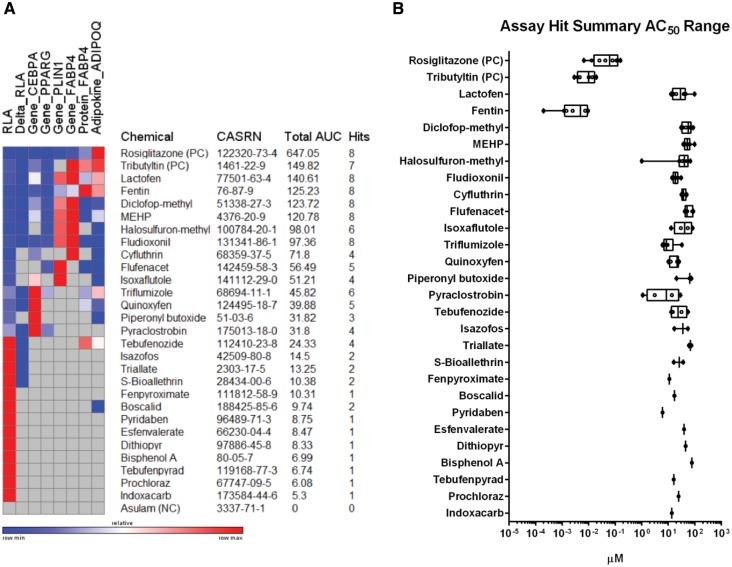
To profile the bioactivity of chemicals identified in the primary screen, key summary data for hit frequency, total peak area under the concentration-response curve (AUC) for efficacy, and AC50 for potency were determined for the 27 chemicals carried through all 8 phenotypic assay endpoints. Given the common concentration-response design, an approach was taken to calculate the AUC (concentration × response) for the prescribed viability range of each identified hit to determine the overall efficacy for each test compound across the endpoints. Values were plotted on a heat map where relative AUC could be visualized row-wise for each independent chemical across the respective endpoint hits (Figure 8A). Chemicals were rank-ordered by AUC sum values (Figure 8A). Positive (rosiglitazone and tributyltin) and negative (asulam) controls were included in the ranking to provide context. Each chemical was classified according to hit frequency as a strong (6–8 hits), moderate (3–5 hits), weak (1–2 hits), or inactive (0 hits) adipogenic compound (Table 2). The highest hit frequency was observed for lactofen, fentin, diclofop-methyl, MEHP, and fludioxonil which exhibited activity in all 8 assay endpoints. Apart from the reference compounds, these same chemicals also occupied the top 5/6 spots for total AUC, providing context for the magnitude of overall bioactivity observed across the assay endpoints. The 12 chemicals identified as weak activators were hits derived from the primary screen that exhibited minimal activity across the other assay endpoints, and are therefore likely to be false positives in the absence of additional phenotypic validation.
FIG. 8.
Efficacy, hit frequency, and potency summary values for adipogenesis hits. The total peak area under the curve (AUC) of significant hits across 8 assay endpoints was calculated and plotted on a heat map for each of the 26 chemical hits identified in the primary adipogenesis screen. Rosiglitazone and tributyltin positive controls (PC) and asulam negative control (NC) are included for context. Compounds were rank-ordered (top-to-bottom) by total AUC and include CASRN and hit frequency (A). The range of AC50 values for each endpoint hit is displayed for each compound as in part A (B). Each box and whisker plot displays the median, first and third quartiles (boxes), and the min–max (whiskers).
TABLE 2.
Classification of Hit Frequency for Chemicals Run in All Adipogenesis Assays
| Hit Classification |
||||||||
|---|---|---|---|---|---|---|---|---|
| Chemical | CASRN | Total AUC | Median AC50 (μM) | # Hits | Strong (6–8) | Moderate (3–5) | Weak (1–2) | Inactive (0) |
| Rosiglitazone | 122320-73-4 | 647.05 | 0.0610 | 8 | * | |||
| Tributyltin Chloride | 1461-22-9 | 149.82 | 0.0100 | 7 | * | |||
| Lactofen | 77501-63-4 | 140.61 | 29.21 | 8 | * | |||
| Fentin | 76-87-9 | 125.23 | 0.0049 | 8 | * | |||
| Diclofop-methyl | 51338-27-3 | 123.72 | 56.47 | 8 | * | |||
| MEHP | 4376-20-9 | 120.78 | 52.46 | 8 | * | |||
| Halosulfuron-methyl | 100784-20-1 | 98.01 | 40.65 | 6 | * | |||
| Fludioxonil | 131341-86-1 | 97.36 | 18.77 | 8 | * | |||
| Cyfluthrin | 68359-37-5 | 71.80 | 37.38 | 4 | * | |||
| Flufenacet | 142459-58-3 | 56.49 | 51.87 | 5 | * | |||
| Isoxaflutole | 141112-29-0 | 51.21 | 42.09 | 4 | * | |||
| Triflumizole | 68694-11-1 | 45.82 | 8.46 | 6 | * | |||
| Quinoxyfen | 124495-18-7 | 39.88 | 21.76 | 5 | * | |||
| Piperonyl butoxide | 51-03-6 | 31.82 | 63.81 | 3 | * | |||
| Pyraclostrobin | 175013-18-0 | 31.80 | 8.41 | 4 | * | |||
| Tebufenozide | 112410-23-8 | 24.33 | 23.27 | 4 | * | |||
| Isazofos | 42509-80-8 | 14.50 | 35.87 | 2 | * | |||
| Triallate | 2303-17-5 | 13.25 | 67.51 | 2 | * | |||
| S-Bioallethrin | 28434-00-6 | 10.38 | 26.18 | 2 | * | |||
| Fenpyroximate | 111812-58-9 | 10.31 | 11.00 | 1 | * | |||
| Boscalid | 188425-85-6 | 9.74 | 17.14 | 2 | * | |||
| Pyridaben | 96489-71-3 | 8.75 | 6.10 | 1 | * | |||
| Esfenvalerate | 66230-04-4 | 8.47 | 39.74 | 1 | * | |||
| Dithiopyr | 97886-45-8 | 8.33 | 45.44 | 1 | * | |||
| Bisphenol A | 80-05-7 | 6.99 | 77.66 | 1 | * | |||
| Tebufenpyrad | 119168-77-3 | 6.74 | 16.10 | 1 | * | |||
| Prochloraz | 67747-09-5 | 6.08 | 24.49 | 1 | * | |||
| Indoxacarb | 173584-44-6 | 5.30 | 13.72 | 1 | * | |||
| Asulam | 3337-71-1 | 0.00 | NA | 0 | * | |||
Summary values for chemical efficacy (AUC) and potency (AC50) are shown for the 26 chemical hits identified from the adipogenesis screen, as well as rosiglitazone and tributyltin positive controls (PC), and asulam negative control (NC). Hit frequency is reported across the 8 orthologous adipocyte endpoints, and are stratified into strong (6–8), moderate (3–5), weak (1–2), and inactive (0) hits.
The AC50 ranges for all assay hits were primarily restricted to µM concentrations, with the exception of the organotin fentin hydroxide that was in the low nM range (Figure 8B). In addition, the clustering of AC50 values for each individual test compound fell within 1–2 orders of magnitude. Of the original 60 chemicals screened, 26 demonstrated functional modulation of lipid accumulation in the differentiation assay. All 26 were derived from the PPARG ToxCast prioritized set. From this pool, additional characterization identified 14/49 (29%) that had moderate-to-strong PPARG function in our human stem cell model of adipogenesis. Strong hit classification (6–8 hits) encompassed 7 chemicals: lactofen, fentin hydroxide, diclofop-methyl, MEHP, halosulfuron-methyl, fludioxonil, and triflumizole. The other 7 chemicals were classified as moderately active (3–5 hits): cyfluthrin, flufenacet, isoxaflutole, quinoxyfen, piperonyl butoxide, pyraclostrobin, and tebufenozide.
DISCUSSION
Perhaps the most common cell model employed for investigating chemical perturbation of adipogenesis is the mouse 3T3-L1 cell line (Janesick et al., 2016; Pereira-Fernandes et al., 2013a); a committed adipocyte progenitor that has been invaluable in defining the cellular pathways involved in adipogenesis. An additional transgene assay in Xenopus (Punzon et al., 2013) or whole-organism zebrafish assay (Tingaud-Sequeira et al., 2011) provide additional understanding of PPARG signaling. Despite orthologous PPARG genes, these alternative model systems are limited in human relevance due to the absence of a cellular environment that includes physiological expression of human nuclear receptor and a complement of co-factors, as well as temporal activation of a gene program that drives terminal differentiation from a multipotent progenitor. The ToxCast PPARG binding and transactivation reporter assays used for chemical prioritization have similar limitations. While certainly useful as a first tier effort for high-volume screening, they have less utility in defining potential for phenotypic outcomes in the intact human. For this, assays fit for the purpose of addressing functional toxicological effects are necessary. By manipulating the classic hormonal cocktail that stimulates adipogenesis, we identified a method to initiate lineage commitment in a multipotent hASC population, and enable activation of PPARG by reference agonists to promote chemical-dependent differentiation. This approach provides a means for directly addressing activity of endogenous PPARG activity in a native cellular environment to track lineage progression of the stem cells. The model more closely resembles the true in vivo biological process and should provide more physiologically relevant insight into chemical activity.
Of the 49 PPARG candidate chemicals screened in the primary adipogenesis assay, 26 of them exhibited some differentiation activity. Of those 26, 14 were considered moderate or strong hits based on hit frequency. Only 5 chemicals (lactofen, fentin hydroxide, diclofop-methyl, MEHP, and fludioxonil) demonstrated activity in every assay endpoint measured. The organotin, fentin hydroxide (aka triphenyltin hydroxide), was the most potent, with a median AC50 of 4.90 nM. This predicted activity is consistent with other chemicals in its class, including the reference compound tributylin chloride. These organotins are direct ligands for PPARG and RXRA, and stimulate 3T3-L1 adipogenesis and associated gene expression (Kanayama et al., 2005). Several other studies have reported mechanistic functions for tributyltin in promoting adipogenesis, lipid accumulation, and epigenetic modification (Chamorro-Garcia et al., 2013; Grun et al., 2006; Kirchner et al., 2010; Li et al., 2011; Pereira-Fernandes et al., 2013b; Yanik et al., 2011). We previously demonstrated that tributyltin, triphenyltin, dibutyltin, and tetrabutyltin all induced hASC differentiation (Foley et al., 2015). These findings lend further support for organotins as PPARG activators with bioactivity in human adipogenesis.
Several of the highest ranking active compounds—lactofen, diclofop-methyl, and MEHP—are hepatic peroxisome proliferators. Lactofen, a diphenyl ether herbicide, causes peroxisome proliferation and liver tumors in mice (Butler et al., 1988). Diclofop-methyl, another diphenyl ether with specificity for PPARA in transient reporter assays, increases hepatic Cyp4A, a PPARA target gene, in mice (Takeuchi et al., 2006). MEHP activates both PPARA and PPARG in reporter assays (Maloney and Waxman, 1999), causes peroxisome proliferation and liver tumors in rodents, and induces adipogenesis (Feige et al., 2007; Hao et al., 2012; Pereira-Fernandes et al., 2013a; Watt and Schlezinger, 2015). As a structurally related member of the PPAR nuclear receptor super family, PPARA is predominantly expressed in liver, heart, and kidney, with lower expression in muscle and brown adipose tissues. PPARG on the other hand, particularly the short isoform (PPARG1), is predominantly expressed in white adipose tissue and the large intestine (Auboeuf et al., 1997). Both PPARA and PPARG subtypes can induce adipogenesis when overexpressed in non-adipocyte progenitor NIH-3T3 cells (Brun et al., 1996). However, PPARA is expressed at very low levels in normal hASC (Auboeuf et al., 1997) and is unlikely to play a prominent role in differentiation in vivo. Here, the effects of PPARA activators on PPARG-dependent adipogenesis may be due to considerable cross reactivity among the receptor subtypes and warrants broader testing of PPARA prioritized compounds for adipogenic potential.
Triflumizole and halosulfuron-methyl each had 6 hits. Triflumizole is a fungicide that has been previously reported as a PPARG agonist with adipogenic potential in both mouse 3T3-L1 cells and human mesenchymal stem cells (Li et al., 2012). The sulfonylurea herbicide halosulfuron-methyl, a chemical pulled from later releases of the ToxCast datasets, is a novel hit in the data series that has no prior evidence for adipogenic potential. Likewise, the 6/7 moderate actives—cyfluthrin, flufenacet, isoxaflutole, piperonyl-butoxide, pyraclostrobin, and tebufenozide—have no published references to an ability to promote adipogenesis. Updates in the ToxCast processing pipeline could have an impact on chemical selection, so later releases of the ToxCast datasets may yield a different set of active compounds in this assay.
A recent report investigating the utility of ToxCast and ToxPi for evaluating adipogenic potential of novel chemicals identified, among others, triphenytlin, quinoxyfen, and fludioxonil; moderate and strong activators in our data series (Janesick et al., 2016). Discordance among the Janesick study results include, but were not limited to, pyraclostrobin and tebufenozide (moderate actives in the hASC assays, inactive in mPPARG reporter assay), as well as zoxamide and spirodiclofen (negative in the hASC screen, positive in mPPARG reporter assays). Whereas some difference in outcomes may be explained by species specificity for chemical activity, other variables include cell sourcing, assay design, duration of differentiation, analytical parameters, and technical execution. As demonstrated in our study, even a universal assay platform using an identical cell population, dosing scheme, and termination point resulted in disparate responses across the chemical test set. Using the weight-of-evidence for all assay endpoints, a more complete picture of likely “true positive” chemical effects emerged, highlighting the value in relying on a multi-parameter approach to evaluate the phenotype; an approach whose value has been demonstrated for other endocrine pathways with ToxCast data (Browne et al., 2015; Rotroff et al., 2013).
A clear advantage of this primary cell system is the ability to evaluate susceptibility directly in human populations of interest. However, as with most primary cell sourcing, heterogeneity of the genetic background may affect differentiation potential (Sen et al., 2001). To mitigate phenotypic bias from single donor sourcing, we used a pooled lot of 8 donors with cells derived from a range of anatomical sites. Whereas all our donors were adult Caucasian females, other potential confounders, such as age, race and gender, have not been addressed in this initial screen. Future experiments could focus on additional populations, particularly younger donors, to evaluate effects of the identified compounds where relevance to early life development is of particular concern.
Many of the moderate-to-strong acting chemicals identified in this study are food-use pesticide-active ingredients. Exposure estimates and toxicokinetic data have already been collected with these compounds to support estimation of oral equivalent doses (OED) by in vitro-to-in vivo extrapolations (Rotroff et al., 2010; Wetmore et al., 2012, 2015). Lipophilic compounds have preferentially higher concentrations in fat depots, the target tissue for adipogenic responses, than in other tissues. The current kinetic models used for absorption, clearance, and plasma binding may also need to consider the role of fat partitioning in altering response patterns to compounds with adipogenic potential.
CONCLUSION
The transition to high-throughput in vitro chemical testing for bioactivity profiling has led to questions regarding the relevance of these tests to human safety assessments. Second tier testing in assays fit for the purpose of evaluating responses in human cells have been proposed as a means of confirming activity and improving measures of dose response for relevant endpoints. The current study used a multi-endpoint approach to evaluate the potential of a prioritized set of chemicals to promote adipogenesis in human adipose-derived stem cells. Assessment of key adipocyte phenotypic characteristics including neutral lipid formation, gene and protein expression, PPARG dependence, and adipokine secretion provided a comprehensive picture of the potential for candidate chemicals to promote differentiation. Inclusion of cytotoxicity endpoints supported range-finding for each chemical, thereby restricting consideration of bioactivity to appropriate viability ranges. Evaluation of the weight-of-evidence across endpoints enabled classification of chemicals according to hit frequency, efficacy, and potency. This approach demonstrates a process of prioritizing compounds from ToxCast high-throughput screens and moving them into a mechanistic in vitro model to identify a mode-of-action in context with a phenotypic outcome. Specifically, this assay provides a biologically sound weight-of-evidence for ranking the potential of environmental compounds to activate PPARG and induce human adipocyte differentiation.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
This work was supported by the Long-Range Research Initiative (LRI) of the American Chemistry Council.
Supplementary Material
Appendix
Appendix 1. Individual graphs for chemicals tested across all the adipogenesis assay endpoints.
REFERENCES
- Auboeuf D., Rieusset J., Fajas L., Vallier P., Frering V., Riou J. P., Staels B., Auwerx J., Laville M., Vidal H. (1997). Tissue distribution and quantification of the expression of mRNAs of peroxisome proliferator-activated receptors and liver X receptor-alpha in humans: no alteration in adipose tissue of obese and NIDDM patients. Diabetes 46, 1319–1327. [DOI] [PubMed] [Google Scholar]
- Boekelheide K., Blumberg B., Chapin R. E., Cote I., Graziano J. H., Janesick A., Lane R., Lillycrop K., Myatt L., States J. C., et al. (2012). Predicting later-life outcomes of early-life exposures. Environ. Health Perspect. 120, 1353–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne P., Judson R. S., Casey W. M., Kleinstreuer N. C., Thomas R. S. (2015). Screening chemicals for estrogen receptor bioactivity using a computational model. Environ. Sci. Technol. 49, 8804–8814. [DOI] [PubMed] [Google Scholar]
- Brun R. P., Tontonoz P., Forman B. M., Ellis R., Chen J., Evans R. M., Spiegelman B. M. (1996). Differential activation of adipogenesis by multiple PPAR isoforms. Genes Dev. 10, 974–984. [DOI] [PubMed] [Google Scholar]
- Butler E. G., Tanaka T., Ichida T., Maruyama H., Leber A. P., Williams G. M. (1988). Induction of hepatic peroxisome proliferation in mice by lactofen, a diphenyl ether herbicide. Toxicol. Appl. Pharmacol. 93, 72–80. [DOI] [PubMed] [Google Scholar]
- Chamorro-Garcia R., Sahu M., Abbey R. J., Laude J., Pham N., Blumberg B. (2013). Transgenerational inheritance of increased fat depot size, stem cell reprogramming, and hepatic steatosis elicited by prenatal exposure to the obesogen tributyltin in mice. Environ. Health Perspect. 121, 359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combs T. P., Wagner J. A., Berger J., Doebber T., Wang W. J., Zhang B. B., Tanen M., Berg A. H., O'Rahilly S., Savage D. B., et al. (2002). Induction of adipocyte complement-related protein of 30 kilodaltons by PPARgamma agonists: a potential mechanism of insulin sensitization. Endocrinology 143, 998–1007. [DOI] [PubMed] [Google Scholar]
- Dix D. J., Houck K. A., Martin M. T., Richard A. M., Setzer R. W., Kavlock R. J. (2007). The ToxCast program for prioritizing toxicity testing of environmental chemicals. Toxicol. Sci. 95, 5–12. [DOI] [PubMed] [Google Scholar]
- Feige J. N., Gelman L., Rossi D., Zoete V., Metivier R., Tudor C., Anghel S. I., Grosdidier A., Lathion C., Engelborghs Y. (2007). The endocrine disruptor mono-ethyl-hexyl-phthalate is a selective PPARγ modulator which promotes adipogenesis . J. Biol. Chem. 282, 19152–19166. [DOI] [PubMed] [Google Scholar]
- Filer D. L., Kothiya P., Setzer W. R., Judson R. S., Martin M. T. (2015a). The ToxCast analysis pipeline: an R package for processing and modeling chemical screening data. US Environmental Protection Agency. Available from: https://www.epa.gov/sites/production/files/2015-08/documents/pipeline_overview.pdf.
- Filer D., Patisaul H. B., Schug T., Reif D., Thayer K. (2014b). Test driving ToxCast: endocrine profiling for 1858 chemicals included in phase II. Curr. Opin. Pharmacol. 19, 145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley B., Clewell R., Deisenroth C. (2015). Development of a human adipose-derived stem cell model for characterization of chemical modulation of adipogenesis. Appl. In Vitro Toxicol. 1, 66–78. [Google Scholar]
- Grun F., Blumberg B. (2006). Environmental obesogens: organotins and endocrine disruption via nuclear receptor signaling. Endocrinology 147, S50–S55. [DOI] [PubMed] [Google Scholar]
- Grun F., Watanabe H., Zamanian Z., Maeda L., Arima K., Cubacha R., Gardiner D. M., Kanno J., Iguchi T., Blumberg B. (2006). Endocrine-disrupting organotin compounds are potent inducers of adipogenesis in vertebrates. Mol. Endocrinol. 20, 2141–2155. [DOI] [PubMed] [Google Scholar]
- Hao C., Cheng X., Xia H., Ma X. (2012). The endocrine disruptor mono-(2-ethylhexyl) phthalate promotes adipocyte differentiation and induces obesity in mice. Biosci. Rep. 32, 619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch E. E., Nelson J. W., Stahlhut R. W., Webster T. F. (2010). Association of endocrine disruptors and obesity: perspectives from epidemiological studies. Int. J. Androl. 33, 324–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heindel J. J., Newbold R., Schug T. T. (2015). Endocrine disruptors and obesity. Nat. Rev. Endocrinol. 11, 653–661. [DOI] [PubMed] [Google Scholar]
- Inadera H., Shimomura A. (2005). Environmental chemical tributyltin augments adipocyte differentiation. Toxicol. Lett. 159, 226–234. [DOI] [PubMed] [Google Scholar]
- Janesick A., Blumberg B. (2012). Obesogens, stem cells and the developmental programming of obesity. Int. J. Androl. 35, 437–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janesick A. S., Dimastrogiovanni G., Vanek L., Boulos C., Chamorro-Garcia R., Tang W., Blumberg B. (2016). On the utility of ToxCast and ToxPi as methods for identifying new obesogens. Environ. Health Perspect. 124, 1214–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson R. S., Houck K. A., Kavlock R. J., Knudsen T. B., Martin M. T., Mortensen H. M., Reif D. M., Rotroff D. M., Shah I., Richard A. M., et al. (2010). In vitro screening of environmental chemicals for targeted testing prioritization: the ToxCast project. Environ. Health Perspect. 118, 485–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanayama T., Kobayashi N., Mamiya S., Nakanishi T., Nishikawa J. (2005). Organotin compounds promote adipocyte differentiation as agonists of the peroxisome proliferator-activated receptor gamma/retinoid X receptor pathway. Mol. Pharmacol. 67, 766–774. [DOI] [PubMed] [Google Scholar]
- Kavlock R., Chandler K., Houck K., Hunter S., Judson R., Kleinstreuer N., Knudsen T., Martin M., Padilla S., Reif D., et al. (2012). Update on EPA's ToxCast program: providing high throughput decision support tools for chemical risk management. Chem. Res. Toxicol. 25, 1287–1302. [DOI] [PubMed] [Google Scholar]
- Kirchner S., Kieu T., Chow C., Casey S., Blumberg B. (2010). Prenatal exposure to the environmental obesogen tributyltin predisposes multipotent stem cells to become adipocytes. Mol. Endocrinol. 24, 526–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knittle J. L., Ginsberg-Fellner F., Brown R. E. (1977). Adipose tissue development in man. Am. J. Clin. Nutr. 30, 762–766. [DOI] [PubMed] [Google Scholar]
- Knittle J. L., Timmers K., Ginsberg-Fellner F., Brown R. E., Katz D. P. (1979). The growth of adipose tissue in children and adolescents. Cross-sectional and longitudinal studies of adipose cell number and size. J. Clin. Invest. 63, 239–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Merrill M., Birnbaum L. S. (2011). Childhood obesity and environmental chemicals. Mt. Sinai J. Med. 78, 22–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legler J., Hamers T., van Eck van der Sluijs-van de Bor M., Schoeters G., van der Ven L., Eggesbo M., Koppe J., Feinberg M., Trnovec T. (2011). The OBELIX project: early life exposure to endocrine disruptors and obesity. Am. J. Clin. Nutr. 94, 1933S–1938S. [DOI] [PubMed] [Google Scholar]
- Li X., Pham H. T., Janesick A. S., Blumberg B. (2012). Triflumizole is an obesogen in mice that acts through peroxisome proliferator activated receptor gamma (PPARgamma). Environ. Health Perspect. 120, 1720–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Ycaza J., Blumberg B. (2011). The environmental obesogen tributyltin chloride acts via peroxisome proliferator activated receptor gamma to induce adipogenesis in murine 3T3-L1 preadipocytes. J. Steroid. Biochem. Mol. Biol. 127, 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney E. K., Waxman D. J. (1999). trans-Activation of PPARalpha and PPARgamma by structurally diverse environmental chemicals. Toxicol. Appl. Pharmacol. 161, 209–218. [DOI] [PubMed] [Google Scholar]
- Pereira-Fernandes A., Demaegdt H., Vandermeiren K., Hectors T. L., Jorens P. G., Blust R., Vanparys C. (2013a). Evaluation of a screening system for obesogenic compounds: screening of endocrine disrupting compounds and evaluation of the PPAR dependency of the effect. PLoS One 8, e77481.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Fernandes A., Vanparys C., Hectors T. L., Vergauwen L., Knapen D., Jorens P. G., Blust R. (2013b). Unraveling the mode of action of an obesogen: mechanistic analysis of the model obesogen tributyltin in the 3T3-L1 cell line. Mol. Cell Endocrinol. 370, 52–64. [DOI] [PubMed] [Google Scholar]
- Poissonnet C. M., Burdi A. R., Bookstein F. L. (1983). Growth and development of human adipose tissue during early gestation. Early Hum. Dev. 8, 1–11. [DOI] [PubMed] [Google Scholar]
- Poissonnet C. M., Burdi A. R., Garn S. M. (1984). The chronology of adipose tissue appearance and distribution in the human fetus. Early Hum. Dev. 10, 1–11. [DOI] [PubMed] [Google Scholar]
- Punzon I., Latapie V., Le Mevel S., Hagneau A., Jolivet P., Palmier K., Fini J. B., Demeneix B. A. (2013). Towards a humanized PPARgamma reporter system for in vivo screening of obesogens. Mol. Cell Endocrinol. [DOI] [PubMed] [Google Scholar]
- Regnier S. M., Sargis R. M. (2014). Adipocytes under assault: Environmental disruption of adipose physiology. Biochim. Biophys. Acta (BBA) –Mol. Basis Dis. 1842, 520–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif D. M., Martin M. T., Tan S. W., Houck K. A., Judson R. S., Richard A. M., Knudsen T. B., Dix D. J., Kavlock R. J. (2010). Endocrine profiling and prioritization of environmental chemicals using ToxCast data. Environ. Health Perspect. 118, 1714–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen E. D., Sarraf P., Troy A. E., Bradwin G., Moore K., Milstone D. S., Spiegelman B. M., Mortensen R. M. (1999). PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell 4, 611–617. [DOI] [PubMed] [Google Scholar]
- Rotroff D. M., Dix D. J., Houck K. A., Knudsen T. B., Martin M. T., McLaurin K. W., Reif D. M., Crofton K. M., Singh A. V., Xia M., et al. (2013). Using in vitro high throughput screening assays to identify potential endocrine-disrupting chemicals. Environ. Health Perspect. 121, 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotroff D. M., Wetmore B. A., Dix D. J., Ferguson S. S., Clewell H. J., Houck K. A., Lecluyse E. L., Andersen M. E., Judson R. S., Smith C. M., et al. (2010). Incorporating human dosimetry and exposure into high-throughput in vitro toxicity screening. Toxicol. Sci. 117, 348–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A., Lea-Currie Y. R., Sujkowska D., Franklin D. M., Wilkison W. O., Halvorsen Y. D., Gimble J. M. (2001). Adipogenic potential of human adipose derived stromal cells from multiple donors is heterogeneous. J. Cell Biochem. 81, 312–319. [DOI] [PubMed] [Google Scholar]
- Spalding K. L., Arner E., Westermark P. O., Bernard S., Buchholz B. A., Bergmann O., Blomqvist L., Hoffstedt J., Naslund E., Britton T., et al. (2008). Dynamics of fat cell turnover in humans. Nature 453, 783–787. [DOI] [PubMed] [Google Scholar]
- Takeuchi S., Matsuda T., Kobayashi S., Takahashi T., Kojima H. (2006). In vitro screening of 200 pesticides for agonistic activity via mouse peroxisome proliferator-activated receptor (PPAR)alpha and PPARgamma and quantitative analysis of in vivo induction pathway. Toxicol. Appl. Pharmacol. 217, 235–244. [DOI] [PubMed] [Google Scholar]
- Thayer K. A., Heindel J. J., Bucher J. R., Gallo M. A. (2012). Role of environmental chemicals in diabetes and obesity: a National Toxicology Program workshop review. Environ. Health Perspect. 120, 779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tingaud-Sequeira A., Ouadah N., Babin P. J. (2011). Zebrafish obesogenic test: a tool for screening molecules that target adiposity. J. Lipid Res. 52, 1765–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt J., Schlezinger J. J. (2015). Structurally-diverse, PPARgamma-activating environmental toxicants induce adipogenesis and suppress osteogenesis in bone marrow mesenchymal stromal cells. Toxicology 331, 66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmore B. A., Wambaugh J. F., Allen B., Ferguson S. S., Sochaski M. A., Setzer R. W., Houck K. A., Strope C. L., Cantwell K., Judson R. S., et al. (2015). Incorporating high-throughput exposure predictions with dosimetry-adjusted in vitro bioactivity to inform chemical toxicity testing. Toxicol. Sci. 148, 121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmore B. A., Wambaugh J. F., Ferguson S. S., Sochaski M. A., Rotroff D. M., Freeman K., Clewell H. J., 3rd, Dix D. J., Andersen M. E., Houck K. A., et al. (2012). Integration of dosimetry, exposure, and high-throughput screening data in chemical toxicity assessment. Toxicol. Sci. 125, 157–174. [DOI] [PubMed] [Google Scholar]
- Yang L., Colditz G. A. (2015). Prevalence of overweight and obesity in the United States, 2007-2012. JAMA Intern. Med. 175, 1412–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanik S. C., Baker A. H., Mann K. K., Schlezinger J. J. (2011). Organotins are potent activators of PPARgamma and adipocyte differentiation in bone marrow multipotent mesenchymal stromal cells. Toxicol. Sci. 122, 476–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.