Abstract
Little evidence is available regarding the physiological effects of exposure to electronic cigarette (ECIG) aerosol. We sought to determine the molecular impact of ECIG aerosol exposure in human bronchial epithelial cells (HBECs). Gene-expression profiling was conducted in primary grown at air liquid interface and exposed to 1 of 4 different ECIG aerosols, traditional tobacco cigarette (TCIG) smoke, or clean air. Findings were validated experimentally with quantitative polymerase chain reaction and a reactive oxygen species immunoassay. Using gene set enrichment analysis, signatures of in vitro ECIG exposure were compared with those generated from bronchial epithelial brushings of current TCIG smokers and former TCIG smokers currently using ECIGs. We found 546 genes differentially expressed across the ECIG, TCIG, and air-exposed groups of HBECs (ANOVA; FDR q < .05; fold change > 1.5). A subset of these changes were shared between TCIG- and ECIG-exposed HBECs. ECIG exposure induced genes involved in oxidative and xenobiotic stress pathways and increased a marker of reactive oxygen species production in a dose-dependent manner. ECIG exposure decreased expression of genes involved in cilia assembly and movement. Furthermore, gene-expression differences observed in vitro were concordant with differences observed in airway epithelium collected from ECIG users (q < .01). In summary, our data suggest that ECIG aerosol can induce gene-expression changes in bronchial airway epithelium in vitro, some of which are shared with TCIG smoke. These changes were generally less pronounced than the effects of TCIG exposure and were more pronounced in ECIG products containing nicotine than those without nicotine. Our data further suggest that the gene-expression alterations seen with the in vitro exposure system reflects the physiological effects experienced in vivo by ECIG users.
Keywords: gene expression, tobacco, smoke, electronic cigarette.
The use of tobacco cigarettes (TCIGs) remains the leading preventable cause of death in the United States, responsible for more than 480 000 deaths per year (CDC, 2014). Smoking is the primary cause of lung cancer, chronic obstructive pulmonary disease (COPD), and emphysema and is also linked to heart disease and asthma (CDC, 2015).
Electronic cigarettes (ECIGs) are battery powered electronic nicotine delivery systems, which simulate tobacco smoking by delivering aerosolized nicotine. ECIG aerosols are generated by heating solutions of propylene glycol, vegetable glycerin, or similar mixtures together with nicotine and flavoring agents (Orellana-Barrios et al., 2015). ECIGs were first invented in 2003 and introduced to the United States in 2007, with sales surpassing 1.7 billion dollars in 2013 (Orellana-Barrios et al., 2015). Because ECIG aerosols should not contain high levels of the many toxic tobacco combustion products present in tobacco smoke, ECIGs are thought by many to be a safer alternative to traditional TCIG smoking (Grana and Ling, 2014; McNeill et al., 2015). In 2015, 12.6 percent of all adults and 47.6 percent of current smokers had used ECIGs (Schoenborn and Gindi, 2015). In 2015, ECIGs were the most commonly used tobacco product among middle and high school students, with 16 percent of high school students identifying as active users of ECIGs (up from 1.5 percent in 2011) (Singh et al., 2016). Increased advertising of these devices, the availability of attractive flavors, and the desire for nicotine alternatives in areas subject to indoor smoking bans have led to their popularity not only with current and former smokers but with nonsmokers as well (Farsalinos et al., 2014; Schoenborn and Gindi, 2015)
Despite the growing use of ECIGs in the United States, few studies have evaluated the potential physiological impact of this exposure. These efforts have been complicated by variations in chemical composition between brands, individual cartridges, and differences in labeled values (Cheng, 2014; Goniewicz et al., 2013; Trehy et al., 2011). Although there is some information on the physiological effects of ECIGs, the existing research has included indirect exposures (Romagna et al., 2013), exposure of nonlung cell types (Behar et al., 2014), or only evaluated the e-liquid itself (Kavvalakis et al., 2015) rather than the aerosol produced from the complete ECIG product. Those studies that have investigated the effect of ECIG exposure on airway epithelium have focused specifically on aspects of toxicology, cytotoxicity, and inflammation (Bahl et al., 2012; Lerner et al., 2015b; Neilson et al., 2015; Scheffler et al., 2015).
Previously, we have shown that airway epithelial gene expression is altered by TCIG smoking and can be used as a biomarker for smoking-associated lung disease, including early detection of lung cancer (Beane et al., 2007; Silvestri et al., 2015; Spira et al., 2007) and molecular subclasses of COPD (Christenson et al., 2015; Steiling et al., 2013; van den Berge et al., 2014). In this study, we aimed to determine the global gene-expression effects of ECIG exposure in human bronchial epithelial cells (HBECs) grown at the air liquid interface (ALI) (Mathis et al., 2013; Neilson et al., 2015) and compare it with the effect of TCIGs. Of the approximately 450 brands of ECIGs on the market (Scheffler et al., 2015), we studied the effects of Blu ECIGs given that Blu is currently the largest producer of ECIG products in the world, and among the most popular brands in the United States, occupying 45 percent of the market share (Scheffler et al., 2015; Thesing, 2014). Additionally, we compared our results with gene-expression differences between bronchial epithelial samples collected from former TCIG smokers and former TCIG smokers who currently use ECIGs.
MATERIALS AND METHODS
In vitro HBEC culture
Primary HBECs were isolated at MatTek Corp from the lungs of a 23-year-old Caucasian male nonsmoker donor with no history of respiratory disease obtained for research purposes with informed consent. The cells were grown using the EpiAirway ALI culture system as previously described (Mathis et al., 2013). This 21-day culture method allowed for the differentiation of bronchial epithelium cell types similar to those seen in vivo including ciliated cells, goblet cells, club cells, and basal cells. We performed standard EpiAirway quality control to ensure proper cell differentiation.
In vitro exposure system
Fully differentiated HBEC ALI cultures were exposed using a VitroCell Systems GmbH (Waldkirch, Germany) VC-1 smoking machine and 12/6 CF stainless-steel exposure module, as previously described (Neilson et al., 2015). Smoke/aerosol puffed from the cigarettes by the smoking machine was diluted with clean air at a rate of 0.5 l/min, before being drawn through the temperature controlled exposure chamber at a rate of 25 ml/min. Control cultures (sham treatment) were exposed to clean air only under the same conditions. Exposures were run in triplicate and included tobacco smoke generated from combustion of 3R4F reference cigarettes (University of Kentucky) that was drawn through the cigarette’s filter by the smoking machine similar to mainstream tobacco smoke. The ECIG exposures were generated from Blu-brand (Charlotte, North Carolina) disposables that were purchased from a retail source. These ECIGs were labeled as either menthol or tobacco-flavored and without or with nicotine (24 mg per cartridge). A drill was used to widen the opening of the smoking machine’s cigarette holder as the ECIG was slightly wider than the TCIG. Puff topographies were selected to mitigate toxicity and mimic physiologic exposure (Behar et al., 2015; Farsalinos et al., 2013). TCIG exposures were performed using the International Organization for Standardization (ISO) smoking regime (35 ml puffs with 1 min intervals) in accordance with ISO 3308:2012, using a 2 s puff draw, 8 s exhaust, and a bell-shaped smoking curve. TCIGs were smoked to 8 puffs/cig. E-cigarettes were puffed with an 80 ml puff drawn over 3 s, 8 s exhaust, with 30 s intervals, and using a square-wave puffing profile, to actuate the electronic device (Neilson et al., 2015). TCIG exposures were defined as 6 cigarettes (48 puffs), the maximum tolerable dose of TCIG exposure under the specific VC-1 dilution and vacuum conditions utilized in the study, while ECIG exposures included 400, 200, 100, and 50 puff exposures with the same VC-1 dilution and vacuum settings. Sham exposures, with only clean air infused into the climatic chamber, were equal to the longest exposure time for TCIG (48 min) or ECIG (200 min) exposures. Exposed cells were also compared with “incubator controls,” HBEC cells grown under the same ALI conditions but not placed into the smoking machine. Following exposure, ALI cultures were fed with fresh maintenance medium (MatTek Corp) and incubated under standard conditions (37°C, 5% CO2) for 22–24 h. Cultures were then fixed in RNALater (Ambion) and stored frozen at −80°C until processing for microarray and polymerase chain reaction (PCR) gene-expression assays.
Cytotoxicity assays
Airway epithelium viability was determined by measuring lactate dehydrogenase (LDH) release into the culture medium using a commercially available colorimetric (490/650 nm) assay kit following the manufacturer’s instructions (ClonTech, MK401). One hundred microliters of culture medium was utilized for the assay. The absorbance data for each tissue sample was normalized to 100% tissue death (ie, complete LDH release caused by tissue lysis with 0.2% Triton X-100 in PBS) and baseline LDH release (untreated incubator control tissue) using the following formula: corrected viability = 100 − [Abs(X) − Abs(Inc)]/[Abs(Triton) − Abs(Inc)]*100 where Abs(X) is the absorbance of the sample, Abs(Inc) is the absorbance of the incubator control, and Abs(Triton) is the absorbance of the sample incubated in 0.2% Triton X-100. Barrier integrity of the airway epithelium tight junctions was determined by measuring transepithelial electrical resistance (TEER) with an EVOM2 voltohmmeter and a 12 mm EndOhm electrode chamber (World Precision Instruments, Sarasota, Florida). Before TEER measurement, the apical surface of the tissues was rinsed 3 times with PBS. The background resistance without the epithelial barrier present was recorded and subtracted from all measurements. The raw resistance (after background subtraction) was multiplied by 1.12 (surface area of culture insert) resulting in final values with units of Ω • cm2. TEER measurements following exposure are presented as the percentage of the preexposure value.
Microarray data acquisition and data preprocessing
RNA was isolated using a standard Qiazol and Qiacube protocol from Qiagen (Valencia, California). RNA purity was assessed using a NanoDrop spectrophotometer and no samples were excluded from downstream analysis. One hundred nanograms of high molecular weight RNA was processed and hybridized to Affymetrix Human Gene 1.0 ST Arrays (Affymetrix, Santa Clara, California). Probeset normalization and summarization were performed using Robust Multi-array Analysis (RMA) (CDF v17.0.0). These data have been uploaded to the Gene Expression Omnibus under accession GSE82137.
Microarray gene-expression analysis
Data quality was assessed using relative log expression and normalized unscaled standard error metrics. Principal components analysis (PCA) was performed on the top 2000 genes by median absolute deviation. A 1-way analysis of variance (ANOVA) (q < .05, fold change > 1.5) was applied to detect major gene-expression differences across all exposure groups including air controls. Additionally, linear regression models were used to examine potential interaction effects between nicotine and flavoring additives in ECIGs. Differentially expressed genes were z-score normalized and hierarchically clustered using complete linkage clustering with a Euclidean distance metric. The gene dendrogram from the hierarchical clustering was cut using R’s cutree function to produce clusters. Cluster enrichment for biological pathways (P < .01) from the Gene Ontology biological terms taxonomy was performed using the gene list enrichment analysis tool EnrichR (Chen et al., 2013).
Real time PCR validation of select gene candidates
Quantitative Real-Time PCR (qRT-PCR) was used to validate the differential expression of select genes. Gene primers (GAPDH, CYP1A1, CYP1B1, FOXJ1, DNAH10) were obtained from SABiosciences (Valencia, California). Total RNA (500 ng) was reverse transcribed using SYBR Green reagents (Qiagen), and the resulting cDNA product was added to SYBR Green PCR Master Mix. All qPCR experiments were carried out in triplicate on each sample, and the CT values were averaged. The average CT value was then normalized to the housekeeping gene GAPDH, and fold change calculated relative to the average of the air controls.
Reactive oxygen species measurement
The production of reactive oxygen species (ROS) was determined by measuring levels of the ROS marker 8-isoprostane using an enzyme immunoassay from Cayman Chemical (Catalog No. 516351). This assay was performed using cell culture medium collected on the day following exposure. Media was frozen in 0.005% butylated hydroxytoluene at −80°C and thawed immediately before use. The assay was then performed following the manufacturer’s protocol, in triplicate for each sample; sample replicates were averaged and then normalized to the average of the air control values. Concentrations of 8-isoprostane in pg/ml are also included.
Patient population and in vivo sample processing
Bronchial airway epithelial cells were obtained from brushings of the right mainstem bronchus taken during fiberoptic bronchoscopy with an endoscopic cytobrush (Cellebrity Endoscopic Cytology Brush, Boston Scientific, Boston). Samples were collected from volunteer subjects at Boston Medical Center and University of California Los Angeles Medical Center between March 2014 and May 2015. Institutional review board approval was obtained at both institutions, and all subjects provided written informed consent. Volunteers were over the age of 21, not using marijuana, had no history of chronic lung disease and had no history of heart disease or other conditions that would increase the risk of undergoing bronchoscopy. Former smokers were required to have smoked at least 10 cigarettes a day for 2 years and to have abstained from TCIGs for at least 3 months prior to their visit. In addition, ECIG users were required to use an ECIG product at least 6 days a week, for at least 1 month. No current TCIG smokers were included in this study. Smoking cessation compliance was monitored by measuring exhaled carbon monoxide. Significant differences between the 2 groups were accessed using a Fishers Exact test for categorical variables and a T-test for continuous variables. RNA was isolated from bronchial brushings using the miRNeasy mini kit and Qiacube from Qiagen. RNA integrity was assessed using an Agilent 2100 Bioanalyzer, and RNA purity was assessed using a NanoDrop spectrophotometer. Samples from 2 volunteers were excluded due to a higher than expected exhaled carbon monoxide level at time of bronchoscopy. A total of 38 bronchial brushing samples passed quality control procedures and were used in downstream analysis.
In vivo bronchial epithelial gene-expression data generation and analysis
One hundred nanograms of high molecular weight RNA was processed and hybridized to Affymetrix Human Gene 1.0 ST Arrays. Samples were preprocessed using similar methods to in vitro arrays. A gene set representative of genes altered by ECIG exposure in vitro was derived by splitting the previously described ANOVA derived 546 gene signature into “up” and “down” regulated genes using a Student’s t test between ECIG versus air-exposed cells, and dividing by the subsequent t-statistic’s sign. To test whether these genes were concordantly changed among ECIG users, a LIMMA linear model (Ritchie et al., 2015; Smyth, 2004) was run for each gene on the platform, with coefficients for smoking status (ECIG, TCIG, or former smoker), subject age, and sample RNA integrity number (RIN). All genes were subsequently ranked by this model’s moderated t-statistic for the “ECIG versus Former” coefficient, and thus by their association with ECIG exposure in vivo. Gene set enrichment analysis (GSEA) was performed using this in vitro exposure gene set and in vivo derived ranked list using the JavaGSEA application (Subramanian et al., 2005, 2007).
Statistical analyses
All statistical analyses were performed with R 2.15.1 (available at https://www.r-project.org; last accessed October 7, 2016) and Bioconductor. Statistical significance of differential gene expression, 8-isoprostane concentrations, and LDH/TEER assays (with respect to air controls) was determined using the Mann-Whitney test.
RESULTS
Airway Epithelial Gene-Expression Changes Induced by ECIG Exposure In Vitro
Differentiated HBECs grown at an ALI were exposed to TCIG smoke (6 cigarettes), ECIG aerosol (50–400 puffs) from a tobacco-flavored Blu-brand ECIG labeled as containing 24 mg nicotine/cartridge or air controls. We first measured cytotoxicity via cell viability and TEER and found that while exposure of HBECs to TCIG smoke had a cytotoxic effect, there were no significant effects with up to 400 puffs ECIG exposure (Supplementary Figs. 1A and B). Moreover, we also failed to detect significant cytotoxicity when we exposed cells to 400 puffs of ECIG aerosol from a variety of Blu-branded products: tobacco-flavored without nicotine, tobacco-flavored with nicotine, menthol-flavored without nicotine, and menthol-flavored with nicotine (Supplementary Figure 1C). Based on these findings, we profiled gene expression in cells that had been exposed to either 6 cigarettes or 400 ECIG puffs (essentially the maximal dose of aerosol that could be extracted under normal usage from each Blu cartridge).
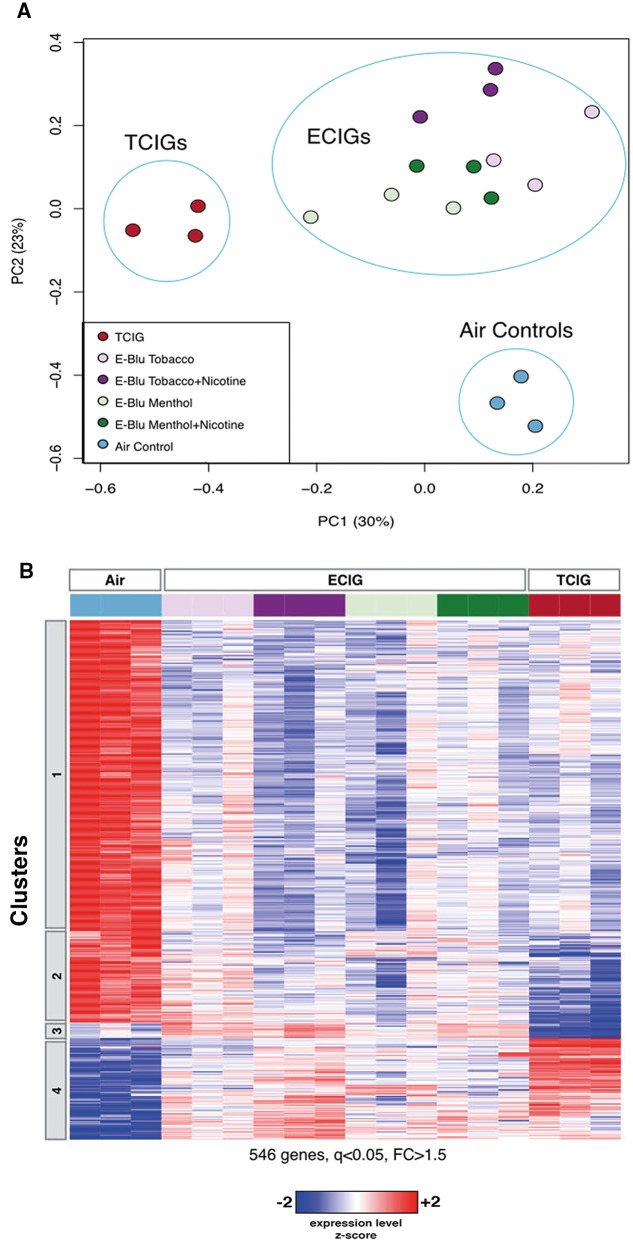
To examine the effect of these exposures on gene expression, we first performed a PCA of the gene-expression data and compared how TCIG, ECIG, and control samples organized relative to each other (Figure 1A). Interestingly, all 4 groups exposed to ECIG aerosol clustered together, separately from TCIG-exposed samples along the first principal component, and separately from air controls along the second principal component, indicating both similarities and differences between the effect of ECIGs and TCIGs on HBEC gene expression. To identify the genes associated with the effects of exposure, we performed an ANOVA and identified 546 genes that were significantly differentially expressed (False Discovery Rate (FDR) q < .05 and fold change > 1.5) between the 3 exposure groups (ECIG-exposed, TCIG-exposed, and air control) (Figure 1B).
FIG. 1.
Genes differentially expressed in human bronchial epithelial cells (HBECs) exposed to traditional tobacco cigarette (TCIG) smoke or electronic cigarette (ECIG) aerosol. A, Principal component analysis of top 2000 genes by median absolute deviation. HBECs exposed to TCIG smoke (red; n = 3), ECIG aerosol (purple/green; n =12), or Air (blue; n = 3). TCIG samples differ from air controls and ECIG samples along the first principal component (accounting for 30% of the variability in gene expression), while both ECIG and TCIG samples differ from air control along the second principal component (accounting for 23% of the gene-expression variability). B, Heatmap of the z-score normalized expression of genes that vary with exposure (546 genes, 1-way ANOVA q < .05 and fold change > 1.5). Genes were organized by hierarchical clustering and divided into 4 clusters.
Based on hierarchical gene clustering, these 546 genes resolved into 4 main clusters in which gene expression was similarly or differently altered by ECIG and TCIG exposure. Cluster 1 contains genes that are similarly expressed at lower levels following ECIG or TCIG exposure, while cluster 2 contains genes that are decreased in both exposure groups but more dramatically decreased following TCIG exposure than ECIG exposure. Cluster 4 consists of genes that are expressed at higher levels following either ECIG or TCIG exposure, whereas cluster 3 contains a relatively small number of genes that were more highly expressed specifically following ECIG exposure.
We performed pathway enrichment analysis to identify whether genes with roles in specific biological processes or pathways are significantly enriched in any of the clusters (Supplementary Tables 1–4). We found genes whose expression is downregulated by exposure to ECIG and TCIG (clusters 1 and 2) to be enriched for pathways related to cilium assembly and movement. Among the genes upregulated by exposure to ECIG or TCIG (cluster 4), we found enrichment for pathways related to apoptosis, xenobiotic stress, oxidative stress, and DNA damage. Among the genes expressed more highly specifically in response to ECIG exposure (cluster 3), we found enrichment for pathways related to cell cycle regulation and cell division, including nuclear division and cytokinesis.
Impact of ECIG Flavoring and Nicotine on Airway Gene Expression
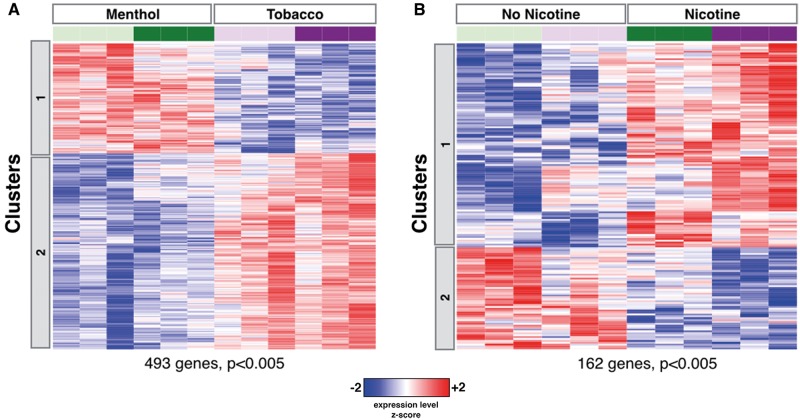
We next analyzed the gene-expression response to flavoring and nicotine components of ECIG (Figure 2). We identified 493 genes as differentially expressed (P < .005) between cells exposed to aerosol from ECIGs with menthol flavoring versus those exposed to aerosol from ECIGs with tobacco flavoring (Figure 2A). Our pathway analysis of this expression profile revealed an enrichment for cell adhesion and protein polymerization-related genes expressed at higher levels in the cells exposed to menthol-flavored ECIG aerosol; and cell cycle and superoxide response related genes expressed at higher levels in the cells exposed to tobacco-flavored aerosol (Supplementary Tables 5 and 6). A similar analysis looking at differences associated with exposure to aerosol from nicotine-containing ECIGs identified 162 genes with altered expression (P < .005) (Figure 2B). Genes upregulated after exposure to nicotine-containing ECIG aerosol were enriched for genes involved in ROS, epithelium differentiation, and the cytochrome P450 pathway, while downregulated genes were enriched for genes involved in the response to inorganic substances (Supplementary Tables 7 and 8).
FIG. 2.
Differential effects of ECIG flavoring and nicotine on HBEC gene expression. A, Heatmap of 493 genes (Student’s t test P < .005) differentially expressed after exposure to aerosol from ECIG products containing menthol flavoring versus tobacco flavoring. B, Heatmap of 162 genes (P < .005) differentially expressed after exposure to aerosol from ECIG products containing nicotine versus those without nicotine (24 mg nicotine per cartridge).
Validation of ECIG Impact on Airway Cilia Genes
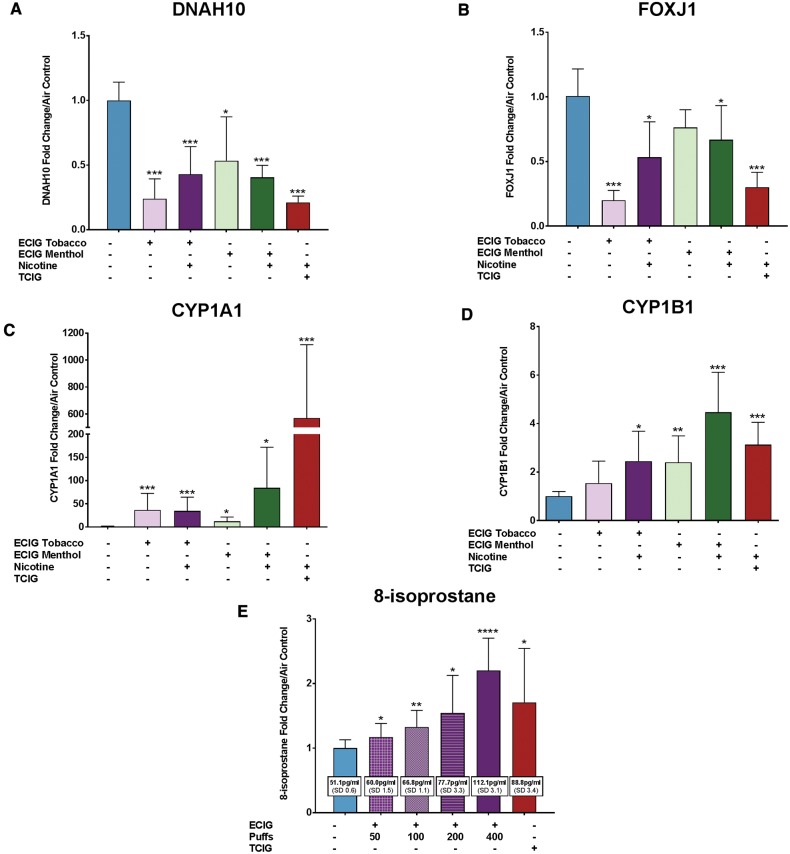
Given the well-established link between TCIG exposure and a reduction in ciliated cells in the airway epithelium, mucociliary clearance, and a shortening of airway cilia (Auerbach et al., 1961; Lam et al., 2013; Wolff, 1986), we sought to validate our findings suggesting that TCIGs and ECIGs both cause a decrease in cilia-related gene expression (Figure 1B, clusters 1 and 2). Using qRT-PCR, we validated the downregulation of the structural cilia dynein gene DNAH10 and ciliated cell marker FOXJ1 with TCIG and ECIG exposure (Figs. 3A and B). This data suggest that ECIGs might be similar to TCIGs in interfering with ciliated cells in the airway epithelium.
FIG. 3.
Increased cytochrome P450 and oxidative stress and reduced cilia gene expression in TCIG smoke and ECIG aerosol exposed HBECs. A–D, Exposures include 6 cigarettes (TCIG) and 400 puffs ECIG menthol, ECIG menthol with nicotine, ECIG tobacco, ECIG tobacco with nicotine and control (air). Samples are pooled from 2 experiments (n = 5 samples). A, Quantitative polymerase chain reaction (qPCR) of the dynein gene DNAH10. B, qPCR of the ciliated cell marker FOXJ1. C, qPCR of the cytochrome P450 gene CYP1A1. D, qPCR of the cytochrome P450 gene CYP1B1. E, Reactive oxygen species production measured via 8-isoprostane enzyme immunoassay. Exposures include air control, 6 cigarettes (TCIG), and either 50, 100, 200, or 400 puffs from a tobacco-flavored ECIG containing nicotine. Average 8-isoprostane concentrations in pg/ml are shown for each exposure in the white boxes. Samples were run in triplicate, with n = 3 experiments. For all panels, levels are shown as fold change relative to the mean of the air controls, and error bars represent the standard error. Statistical significance of the change relative to air control was assessed by Mann-Whitney test and is presented as * ≤ 0.05, ** ≤ 0.01, *** ≤ 0.001, **** ≤ 0.0001.
ECIG Exposure Induces AhR and Oxidative Stress Pathways in a Dose-Dependent Manner
We also identified biological responses that have previously been established as being important in the response to smoking (Ammous, 2008; Beane et al., 2007; Brody and Steiling, 2011; Mathis et al., 2013; Nagaraj et al., 2006; Zhang et al., 2008) among the genes that are upregulated in response to both ECIG and TCIG exposure. We found enrichment for genes related to the cytochrome P450 pathway, xenobiotic stress, and oxidative stress response induced with TCIG and ECIG exposure (Figure 1B, cluster 4; Supplementary Table 4). Genes involved in these pathways were further induced by nicotine-containing ECIG aerosol (Figure 2B, cluster 1; Supplementary Table 7). PCR validation of the aryl hydrocarbon receptor (AhR)-activated genes CYP1A1 and CYP1B1 revealed a significant induction with TCIG exposure and all 4 ECIG exposures (Figs. 3C and D). To determine whether the dose of ECIG (number of puffs of aerosol) influenced these gene-expression changes, we exposed ALI cultures to 400, 200, 100, and 50 puffs of an ECIG with nicotine and tobacco flavoring and measured CYP1A1 and CYP1B1 via PCR and oxidative stress via production of the ROS marker 8-isoprostane in the cell culture media. Although exposure to 400 puffs from an ECIG containing nicotine and tobacco flavoring highly induced both CYP1A1 and CYP1B1 relative to air controls, only CYP1A1 was significantly induced at lower doses (Supplementary Figure 3). CYP1A1 was also moderately induced in the air controls (as compared with incubator controls, cells which never entered the smoking chamber) (Supplementary Figure 2) emphasizing the importance of using air controls for isolating the specific effects of ECIG and TCIG exposure. Interestingly, 8-isoprostane levels increased with all doses of ECIG exposure as well as with TCIG exposure (Figure 3E).
In Vitro Exposure System Reflects the Airway Gene-Expression Alterations Experienced In Vivo by ECIG Users
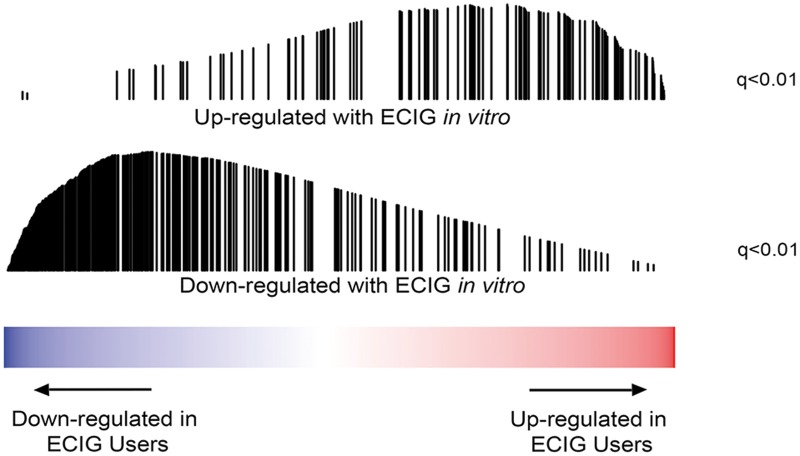
To evaluate the physiological relevance of the findings from our in vitro exposure studies, we compared the effects we observed in vitro to gene-expression profiles generated in bronchial epithelium samples from former cigarette smokers and former smokers who use ECIGs (Table 1). By GSEA, we found that the genes most upregulated by ECIG exposure in vitro were significantly enriched (q < .01) among the genes induced in ECIG users relative to former smokers while the genes most downregulated by ECIG exposure in vitro were significantly enriched (q < .01) among the repressed genes in ECIG users (Figure 4). These data suggest that the in vitro exposure system reflects some of the physiological effects of ECIGs experienced by ECIG users.
TABLE 1.
Patient Demographic Information for Bronchial Brush Samples Collected from Former Smokers Using ECIGs
| Former Smokers n = 21 | Former Smokers, ECIG Users n = 15 | P-value | |
|---|---|---|---|
| Age (years) | 43 (10.7) | 35.8 (10.4) | .05 |
| Sex | 11M/10F | 11M/4F | .30 |
| Race | Multiple: 2 | American Indian/Alaskan Native: 1 | .55 |
| Black/African American: 5 | Black/African American: 3 | ||
| White/Caucasian: 14 | White/Caucasian: 11 | ||
| Pack-years | 10.9 (10.5) | 13.8 (11.3) | .43 |
| Time since quit (months) | 67 (117.5) | 8.7 (4.4) | .06 |
| Carbon monoxide (ppm) | 2.3 (1.42) | 1.857 (1.51) [1 N/A] | .35 |
Samples were collected at Boston University and University of California Los Angeles. Mean values are shown, with standard deviations in parentheses. P-values are shown for the difference between the 2 groups.
FIG. 4.
Genes differentially expressed following in vitro ECIG exposure are enriched among genes differentially expressed in airway epithelial samples from ECIG users. (Top) Enrichment plot showing the rank of genes with increased expression in vitro in the list of genes ranked from most increased to most decreased by ECIG use in vivo. Genes upregulated with ECIG exposure in vitro are significantly enriched among the genes most induced in the airway of former smokers using ECIGs (n = 16) (q < .01). (Bottom) Enrichment plot showing the rank of genes with decreased expression in vitro in the list of genes ranked from most increased to most decreased by ECIG use in vivo. Genes downregulated with ECIG exposure in vitro are significantly enriched among the genes most downregulated in the airway of former smokers using ECIGs (n = 16) (q < .01).
DISCUSSION
We performed transcriptome profiling on differentiated bronchial epithelium, exposed directly to ECIG aerosol to identify genes whose expression is altered by ECIG exposure. These findings provide novel insights into how ECIGs might potentially alter airway biology. By including TCIG exposure as a comparator, we were able to determine which gene-expression changes induced by ECIG exposure are similar to the effects of TCIGs and which are distinct. We focused our studies on 4 ECIG products from a single manufacturer differing in flavoring and nicotine, allowing us to explore the effect of these ingredients. It remains to be determined if other ECIG products produce similar effects. Our finding of concordant gene-expression alterations in airway epithelial cells from individuals with real-world exposure to a variety of ECIG products mitigates this concern and more importantly, supports the physiological relevance of studying responses to ECIG exposures in vitro.
We found that exposure to TCIG smoke or ECIG aerosol each induced gene-expression changes in HBECs specifically related to xenobiotic metabolism, oxidative stress, DNA damage, and apoptosis. Relative to air controls, the magnitude of gene-expression changes was higher with TCIG exposures compared with ECIG exposures. In addition, our results indicate that these changes are more greatly induced by nicotine-containing ECIG aerosol than that of nonnicotine products and that this induction of CYP1A1, CYP1B1, and ROS production by ECIGs appears to be dose-dependent. Importantly, even in response to high dose ECIG exposure (400 puffs) the activation of these xenobiotic and oxidative stress pathways is substantially lower than that seen with lower-dose TCIG exposure (6 cigarettes). Activation of AhR and induction of the drug-metabolizing cytochrome P450 genes is associated with tobacco smoke-induced disease, including lung cancer (Lin et al., 2003; Tsay et al., 2013). This prompts further examination into the relationship between ECIG use and lung disease.
Oxidative stress and the increase in antioxidant genes in response to ROS and oxidative damage has also been well-established in smoking (Burke and FitzGerald, 2003; Crawford et al., 2000; Kosecik et al., 2005; Montuschi, 2004; van der Vaart et al., 2004) and recent publications have demonstrated the induction of oxidative stress by ECIG products in both cell culture and mouse models (Lerner et al., 2015a,b; Scheffler et al., 2015; Sussan et al., 2015). Consistent with these prior publications, we found that the production of 8-isoprostane increases in HBECs after exposure to TCIG smoke or ECIG aerosol, suggesting a corresponding increase in ROS. Interestingly, the production of 8-isoprostane appears at lower doses of ECIG aerosol than those required to induce gene-expression differences. Because ROS are important mediators of inflammation and have been implicated in the pathogenesis of diseases such as COPD and lung cancer, the production of ROS in response to ECIG aerosol exposure would suggest the potential for downstream adverse health effects of ECIGs (Lerner et al., 2015b). It should be emphasized that as 8-isoprostane is a biomarker of ROS production, it will be valuable in the future to confirm the increased production of ROS by directly assaying these species using mass spectrometry.
Additionally, genes with functions in cilium assembly and cilia movement pathways are downregulated after both TCIG and ECIG exposure. This decrease in gene expression may reflect impairment of cilia or a decrease in the number of ciliated cells and suggests a potential defect in airway clearance, and an increased susceptibility to respiratory infection (Lam et al., 2013).
Our results also suggest the presence of flavoring and nicotine specific effects. Nevertheless, our principal components analysis indicates that the primary differences in gene expression separate the TCIG-exposed HBECs from ECIG- and air-exposed HBECs. This suggests that a major effect of TCIGs on airway gene expression is largely distinct from the effect of ECIGs. However, the second principal component identifies a variability in gene expression that distinguishes the TCIG- and ECIG-exposed HBEC’s from the air-exposed HBECs and reflects a response to TCIG-exposure that is shared with ECIGs. Importantly, the effects of flavoring and nicotine, while detectable via linear modeling, are not apparent in the first 2 principal components suggesting that they are less dramatic sources of gene-expression heterogeneity. Similarly, by cluster analysis of the genes from our linear modeling analysis, we identified a small number of genes whose expression is specifically induced in response to ECIG exposure that includes genes involved in cell cycle and cell division pathways. The 16 genes upregulated only in ECIGs are the first identified to be expressed specifically in ECIGs, and further analysis of gene-expression effects unique to ECIGs is warranted. In particular, it will be important to determine which product component or components are responsible for inducing these responses.
Importantly, our analysis of in vivo bronchial epithelium samples from ECIG users indicated statistically significant similarities between the effects of ECIG exposure on gene expression in vitro and in vivo, suggesting that our in vitro findings are relevant to the in vivo effects of ECIG exposure. This connection, which was recovered despite the heterogeneity of products and users profiled in our in vivo dataset suggests that detrimental effects of ECIG use identified in vitro may be present in vivo.
There are a number of limitations to this study. HBECs were differentiated in an ALI system in order provide a more physiological system to measure the impact of ECIG aerosol. However, it is difficult to model the impact of chronic exposure that occurs in ECIG users using this relatively acute exposure system (< 24 h). Further, we profiled a relatively high dose of ECIG (400 puffs) in vitro over a relatively short period of time based on the amount of aerosol that could be extracted from a single ECIG cartridge. However, recent studies (Geiss et al., 2015) have indicated that based on nicotine content, 13 puffs e-liquid containing 18 mg/ml nicotine is equivalent to the smoke of 1 typical TCIG containing 0.5 mg nicotine. By these estimates, our 6 cigarette (48 puff) TCIG smoke exposure would be roughly equivalent to 6 × 13 = 78 puffs of ECIG aerosol. Seventy-eight puffs per 48 min is a rate of exposure roughly similar to our experimental ECIG dose of 100 puffs per 50 min, at which (and additionally at the 50 puff dose), we were still able to detect induction of both CYP1A1 and the ROS marker 8-isoprostane. The data from Geiss et al. (2015) suggests that our 400 puff ECIG dose are substantially greater than the 48 puff TCIG dose with regard to delivered nicotine. The generally more dramatic effect of TCIG exposure on bronchial epithelial gene expression relative to the 400 puff ECIG exposure therefore suggests that acute ECIG exposure is likely to have a less pronounced effect than acute TCIG exposure. In addition, while exposure of HBECs to TCIG smoke from 6 cigarettes significantly decreased cell viability, 400 puffs ECIG exposure did not have any significant effect on cell death. Importantly, despite using only 1 brand of ECIGs and cells from a single donor for our in vitro exposures, our finding of similar gene-expression changes in bronchial epithelial cells collected from users of diverse products supports both the physiological relevance of this system and the broad generalizability of the observed effects. Finally, our findings do not provide insight into the mechanisms by which ECIG alters airway gene expression, nor the downstream consequences of those transcriptomic changes as it relates to disease. These results have generated a number of specific biological hypotheses that can be tested in vitro and in vivo in future studies.
Overall, this work represents one of the first characterizations of the effects of ECIG exposure on airway epithelium gene expression. In addition, validation of our in vitro findings in in vivo samples suggests that the exposure of differentiated airway epithelium to ECIG aerosol will be a useful approach to understand the cellular effects of ECIG aerosol. Given the current lack of knowledge concerning the long-term health effects of ECIGs, we have focused our analysis on the aspects of the response to ECIG aerosol that are similar to the response to TCIG smoke as TCIG smoke exposure is known to have deleterious health effects. Importantly, we found that similar to TCIG smoke, ECIG aerosol exposure induces xenobiotic, oxidative, and additional stress pathways, and potentially impairs ciliated epithelium. However, we found that these effects were generally more severe in response to TCIG smoke exposure, indicating that ECIG exposure may be less harmful. Still, given that these responses to TCIGs are thought to be related to their long-term health effects, these findings raise concerns regarding the safety of ECIGs use, despite their relative dissimilarity to TCIGs. Further investigations into the dissimilarity of ECIGs and TCIGs should be pursued, as these studies may uncover additional physiological facets of ECIG exposure that can manifest through long-term ECIG use.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
ACKNOWLEDGMENTS
The authors acknowledge and thank Dr David Sherr, Dr Sarah Mazzilli, and Jessica Vick for their feedback on this article.
FUNDING
This work was supported by the National Cancer Institute at the National Institutes of Health (U01 CA152751).
REFERENCES
- Ammous Z. (2008). Variability in small airway epithelial gene expression among normal smokers. Chest 133, 1344–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach O., et al. (1961). Changes in bronchial epithelium in relation to cigarette smoking and in relation to lung cancer. N. Engl. J. Med. 265, 253–267. [DOI] [PubMed] [Google Scholar]
- Bahl V., et al. (2012). Comparison of electronic cigarette refill fluid cytotoxicity using embryonic and adult models. Reprod. Toxicol. 34, 529–537. [DOI] [PubMed] [Google Scholar]
- Beane J., et al. (2007). Reversible and permanent effects of tobacco smoke exposure on airway epithelial gene expression. Genome Biol. 8, R201.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar R. Z., et al. (2014). Identification of toxicants in cinnamon-flavored electronic cigarette refill fluids. Toxicol. In Vitro 28, 198–208. [DOI] [PubMed] [Google Scholar]
- Behar R. Z., et al. (2015). Puffing topography and nicotine intake of electronic cigarette users. PLoS One 10, e0117222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody J. S., Steiling K. (2011). Interaction of cigarette exposure and airway epithelial cell gene expression. Annu. Rev. Physiol. 73, 437–456. [DOI] [PubMed] [Google Scholar]
- Burke A., FitzGerald G. A. (2003). Oxidative stress and smoking-induced vascular injury. Prog. Cardiovasc. Dis. 46, 79–90. [DOI] [PubMed] [Google Scholar]
- CDC. (2016). Burden of Tobacco Use in the U.S. Centers for Disease Control and Prevention website. Available at: http://www.cdc.gov/tobacco/campaign/tips/resources/data/cigarette-smoking-in-united-states.html. Accessed October 7, 2016.
- CDC. (2015). Health Effects of Cigarette Smoking - Smoking & Tobacco Use. Centers for Disease Control and Prevention website. Available at: http://www.cdc.gov/tobacco/data_statistics/fact_sheets/health_effects/effects_cig_smoking. Accessed October 7, 2016.
- Chen E. Y., et al. (2013). Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T. (2014). Chemical evaluation of electronic cigarettes. Tob. Control 23(Suppl. 2), ii11–ii17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christenson S. A., et al. (2015). Asthma–COPD overlap. Clinical relevance of genomic signatures of type 2 inflammation in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 191, 758–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford E. L., Khuder S. A., Durham S. J., Frampton M., Utell M., Thilly W. G., Weaver D. A., Ferencak W. J., Jennings C. A., Hammersley J.R., et al. (2000). Normal bronchial epithelial cell expression of glutathione transferase P1, glutathione transferase M3, and glutathione peroxidase is low in subjects with bronchogenic carcinoma. Cancer Res. 60, 1609–1618. [PubMed] [Google Scholar]
- Farsalinos K. E., et al. (2013). Evaluation of electronic cigarette use (vaping) topography and estimation of liquid consumption: Implications for research protocol standards definition and for public health authorities’ regulation. Int. J. Environ. Res. Public Health 10, 2500–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farsalinos K. E., et al. (2014). Characteristics, perceived side effects and benefits of electronic cigarette use: A worldwide survey of more than 19,000 consumers. Int. J. Environ. Res. Public Health 11, 4356–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss O., et al. (2015). Characterisation of mainstream and passive vapours emitted by selected electronic cigarettes. Int. J. Hyg. Environ. Health 218, 169–180. [DOI] [PubMed] [Google Scholar]
- Goniewicz M. L., et al. (2013). Nicotine levels in electronic cigarettes. Nicotine Tob. Res. 15, 158–166. [DOI] [PubMed] [Google Scholar]
- Grana R. A., Ling P. M. (2014). ‘ Smoking revolution’ a content analysis of electronic cigarette retail websites. Am. J. Prev. Med. 46, 395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavvalakis M. P., et al. (2015). Multicomponent analysis of replacement liquids of electronic cigarettes using chromatographic techniques. J. Anal. Toxicol. 39(4), 262–269. [DOI] [PubMed] [Google Scholar]
- Kosecik M., et al. (2005). Increased oxidative stress in children exposed to passive smoking. Int. J. Cardiol. 100, 61–64. [DOI] [PubMed] [Google Scholar]
- Lam H.C., Cloonan S.M., Bhashyam A.R., Haspel J. A., Singh A., Sathirapongsasuti J. F., Cervo M., Yao H., Chung A. L., Mizumura K., et al. (2013). Histone deacetylase 6-mediated selective autophagy regulates COPD-associated cilia dysfunction. J. Clin. Invest. 123, 5212–5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner C. A., Sundar I. K., Watson R. M., Elder A., Jones R., Done D., Kurtzman R., Ossip D. J., Robinson R., McIntosh S., et al. (2015a). Environmental health hazards of e-cigarettes and their components: Oxidants and copper in e-cigarette aerosols. Environ. Pollut. 198, 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner C. A., Sundar I. K., Yao H., et al. (2015b). Vapors produced by electronic cigarettes and e-juices with flavorings induce toxicity, oxidative stress, and inflammatory response in lung epithelial cells and in mouse lung. PloS One 10, e0116732.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P., et al. (2003). Overexpression of aryl hydrocarbon receptor in human lung carcinomas. Toxicol. Pathol. 31, 22–30. [DOI] [PubMed] [Google Scholar]
- Mathis C., Poussin C., Weisensee D., Gebel S., Hengstermann A., Sewer A., Belcastro V., Xiang Y., Ansari S., Wagner S., et al. (2013). Human bronchial epithelial cells exposed in vitro to cigarette smoke at the air-liquid interface resemble bronchial epithelium from human smokers. Am. J. Physiol. Lung Cell. Mol. Physiol. 304, L489–L503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill A., et al. (2015). E-cigarettes: An evidence update. A report commissioned by Public Health England. Public Health England. Available at: https://www.gov.uk/government/publications/e-cigarettes-an-evidence-update. Accessed May 31, 2016. [Google Scholar]
- Montuschi P. (2004). Isoprostanes: Markers and mediators of oxidative stress. FASEB J. 18, 1791–1800. [DOI] [PubMed] [Google Scholar]
- Nagaraj N. S., et al. (2006). Cigarette smoke condensate induces cytochromes P450 and aldo-keto reductases in oral cancer cells. Toxicol. Lett. 165, 182–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilson L., et al. (2015). Development of an in vitro cytotoxicity model for aerosol exposure using 3D reconstructed human airway tissue; application for assessment of e-cigarette aerosol. Toxicol. In Vitro 29, 1952–1962. [DOI] [PubMed] [Google Scholar]
- Orellana-Barrios M. A., et al. (2015). Electronic cigarettes-a narrative review for clinicians. Am. J. Med. 28(7), 674–681. [DOI] [PubMed] [Google Scholar]
- Ritchie M. E., et al. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romagna G., et al. (2013). Cytotoxicity evaluation of electronic cigarette vapor extract on cultured mammalian fibroblasts (ClearStream-LIFE): Comparison with tobacco cigarette smoke extract. Inhal. Toxicol. 25, 354–361. [DOI] [PubMed] [Google Scholar]
- Scheffler S., et al. (2015). Evaluation of e-cigarette liquid vapor and mainstream cigarette smoke after direct exposure of primary human bronchial epithelial cells. Int. J. Environ. Res. Public Health 12, 3915–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenborn C. A., Gindi R. M. (2015). Electronic cigarette use among adults: United States, 2014 National Center for Health Statistics. Hyattsville, MD. [PubMed]
- Silvestri G. A., Vachani A., Whitney D., Elashoff M., Porta Smith K., Ferguson J. S., Parsons E., Mitra N., Brody J., Lenburg M. E., et al. (2015). A bronchial genomic classifier for the diagnostic evaluation of lung cancer. N. Engl. J. Med. 373, 243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh T., et al. (2016). Tobacco use among middle and high school students — United States, 2011–2015. MMWR Morb. Mortal. Wkly. Rep. 65, 361–367. [DOI] [PubMed] [Google Scholar]
- Smyth G. K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article3. [DOI] [PubMed] [Google Scholar]
- Spira A., Beane J. E., Shah V., Steiling K., Liu G., Schembri F., Gilman S., Dumas Y. M., Calner P., Sebastiani P., et al. (2007). Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat. Med. 13, 361–366. [DOI] [PubMed] [Google Scholar]
- Steiling K., et al. (2013). A dynamic bronchial airway gene expression signature of chronic obstructive pulmonary disease and lung function impairment. Am. J. Respir. Crit. Care Med. 187, 933–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., et al. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., et al. (2007). GSEA-P: A desktop application for gene set enrichment analysis. Bioinformatics 23, 3251–3253. [DOI] [PubMed] [Google Scholar]
- Sussan T. E., Gajghate S., Thimmulappa R. K., Ma J., Kim J., Sudini K., Consolini N., Cormier S. A., Lomnicki S., Hasan E., et al. (2015). Exposure to electronic cigarettes impairs pulmonary anti-bacterial and anti-viral defenses in a mouse model. PloS One 10, e0116861.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thesing G. (2014). Imperial tobacco’s Cooper adds blu to take e-cigarette leap. Bloom. Bus. Available at: http://www.bloomberg.com/news/articles/2014-07-15/imperial-tobacco-ceo-cooper-adds-blu-to-take-e-cigarette-leap. Accessed February 21, 2016. [Google Scholar]
- Trehy M. L., et al. (2011). Analysis of electronic cigarette cartridges, refill solutions, and smoke for nicotine and nicotine related impurities. J. Liq. Chromatogr. Relat. Technol. 34, 1442–1458. [Google Scholar]
- Tsay J. J., et al. (2013). Aryl hydrocarbon receptor and lung cancer. Anticancer Res. 33, 1247–1256. [PMC free article] [PubMed] [Google Scholar]
- van den Berge M., Steiling K., Timens W., Hiemstra P. S., Sterk P. J., Heijink I. H., Liu G., Alekseyev Y. O., Lenburg M. E., Spira A., et al. (2014). Airway gene expression in COPD is dynamic with inhaled corticosteroid treatment and reflects biological pathways associated with disease activity. Thorax 69, 14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berge M., Steiling K., Timens W., Hiemstra P. S., Sterk P. J., Heijink I. H., Liu G., Alekseyev Y. O., Lenburg M. E., Spira A., et al. (2004). Acute effects of cigarette smoke on inflammation and oxidative stress: A review. Thorax 59, 713–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff R. K. (1986). Effects of airborne pollutants on mucociliary clearance. Environ. Health Perspect. 66, 223–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., et al. (2008). Impact of smoking cessation on global gene expression in the bronchial epithelium of chronic smokers. Cancer Prev. Res. 1, 112–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.