Abstract
The interaction of proteins with DNA results, in some cases, in DNA bending, and this might have functional importance. However, when the protein-induced bending of DNA is small, its measurement presents a problem. It is shown that the fluorescence resonance energy transfer between fluorophores placed on the ends of the specially designed U-shaped DNA, which contains the DNA-binding sites at its central part, can be successfully used for this purpose. The lever effect of the arms of such U-shaped DNA ensures that the distance between the fluorophores is very sensitive to bending of the central part. Using this technique, it was shown that (i) the AP-1 and ATF/CREB binding sites of GCN4 transcription factor are pre-bent to the same extent (∼12° toward the major groove) and (ii) binding of the GCN4 DNA-binding domain (GCN4-bZIP) results in additional bending of both these target sites but to a greater extent at the ATF/CREB site. In total, in the complex with GCN4-bZIP, the ATF/CREB site is bent by (25 ± 2)° and the AP-1 site by (20 ± 2)° toward the minor groove.
INTRODUCTION
The interaction of proteins with DNA results, in some cases, in significant bending of DNA, and this might have functional importance for transcriptional activation (1,2). According to crystal structures, association of the bZIP protein GCN4 with the ATF/CREB DNA-binding site results in a DNA bend of 20° toward the leucine zipper, while the association with the AP-1 site results in no distortion of the DNA (3–5). It was proposed that bending of the 10 bp palindromic ATF/CREB DNA occurs in order to accommodate the extra GC base pair in that site, relative to the 9 bp AP-1 site, and thereby conserves the protein–DNA interactions in these two complexes (4). One cannot exclude, however, that the difference in bending of these two sites is caused by the interactions between the molecules packed in the crystals. In order to reassess this issue, we turned to the fluorescence resonance energy transfer (FRET) technique, which has permitted us to show that GCN4-bZIP induces bending of both the ATF/CREB and AP-1 sites, although to differing extents.
The magnitude of FRET depends on the distance between donor and acceptor fluorophores. By placing them on each end of the DNA, one can use changes in the FRET effect (FE) which result from deformation of the DNA upon protein binding, in order to monitor DNA bending. In its classical form, FRET has proved very efficient for determining DNA bend angles when they are large (6,7). However, if the DNA bend is small, the distance between fluorophores does not change significantly, and the method becomes insensitive. If, however, bulges are placed in the arms of the DNA to create a U-shaped template with donor and acceptor at opposite ends, as shown in Figure 1, the lever effect ensures that the distance between the fluorophores is very sensitive to bending of the central tract between the bulges (8,9). By placing a protein-binding site in the central tract (‘base’ sequence), the induction of relatively small bends by protein binding is reflected in substantial changes in FE.
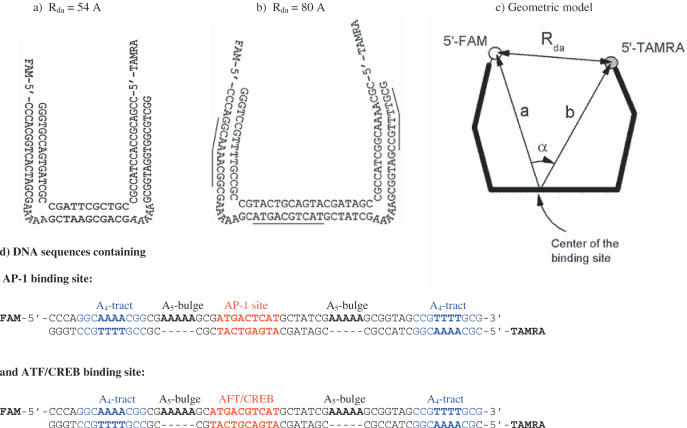
Figure 1.
Design of the double-labeled DNA construct for FRET analysis of binding and DNA bending by GCN4-bZIP protein. (a) Original DNA construction with DNA ‘base’ sequence between two A5-bulges and the arms, described in (9). The distance (Rda) between FAM (donor) and TAMRA (acceptor) attached to the 5′ ends was 54 Å. (b) Insertion of 10 bp DNA fragment (one full turn) containing target sequences, AP-1 or ATF/CREB, for binding of GCN4-bZIP protein increases the dye-to-dye distance (Rda) by 34 Å. Insertion of the A4-tracts into the arms decreases Rda to 80 Å. (c) Modeling of the DNA construct used for the quantitative calculation of the bend angle of the binding site from the measured Rda by Equation 1. (d) The sequences used for the construction of the U-shaped DNA.
The dissociation constant between bZIPs and DNA is rather low, in the nM range. To get well-resolved isotherm of this process, it must be studied at concentrations of a similar order of magnitude. However, at such a low concentration, the dimeric GCN4-bZIP dissociates (10,11). In that case, the observed association process of the bZIP with the DNA would not be a simple reaction but would proceed through a dimeric and monomeric pathway (12,13). To avoid this complication, we used a GCN4-bZIP modified with S-S cross-linked strands at the N-terminus; this does not change the ability of the bZIP to bind DNA but prevents its dissociation at the low concentrations used in the titration experiments (10,14).
MATERIALS AND METHODS
Design of the double-bulged DNA constructions
The original design of a double-bulged DNA (8,9) served as the basis for the DNA construct used in the present FRET experiments. This planar construct has two arms (16 and 17 bp), which are directed by A5-bulges to be almost perpendicular (∼84°) to the 9 bp base duplex at the center as shown in Figure 1a. The 5′ ends of the DNA are labeled with the fluorophores FAM (5,6-carboxyfluorescein) and TAMRA (5-carboxytetramethylrhodamine). The distance between these donor and acceptor fluorophores, Rda, determined by FRET, was found to be (54 ± 1) Å. This original construct required modifications to accommodate the bZIP dimer and its binding site between the arms.
Accommodation of the binding sites (6 and 7 bp for the AP-1 and ATF/CREB sites, respectively) in the central duplex required a significant extension. This can be carried out only by the insertion of a full turn of DNA (10 bp) so as to preserve the planarity of the construct. Since the bZIP binds to the major groove and induces DNA bending toward this groove (at least in the case of the GCN4:ATF/CREB complex), the target DNA sequence must be placed in such a way that the binding site faces toward the arms of the construct (see Figure 1b). If the inserted 10 bp fragment is straight, it should increase the distance between the ends of the arms by 34 Å, from 54 to 88 Å. This distance is too large for the FRET measurements. To decrease the distance between the ends of the arms, they were modified by inserting adenine-tracts (A4-tracts) that bend the arms inwards. According to the recent NMR analysis of a DNA duplex containing an A4-tract, 5′-GGCAAAACGG-3′, it is bent by 9° toward the minor groove (15). We inserted this fragment into both arms in such a way as to bend the arms in the plane of the construct. Insertion of these A4-tracts should decrease the Rda distance by 8 Å. With these modifications, the Rda distance is expected to be 80 Å, provided the inserted 10 bp elements in the base of the construct are straight. However, if the inserted binding site is bent toward the major groove, as shown for the ATF/CREB site (16), this distance should be smaller. As shown by our FRET experiments, this distance is indeed smaller for both the considered binding sites.
Simple geometric model for the calculation of a bend angle in the binding site
The double-labeled DNA molecule shown in Figure 1b was used to evaluate the relation between the measured dye-to-dye distance (Rda) and bend angle induced by binding of bZIP protein. The main features of the model are shown in Figure 1c. There is a simple geometric relation between Rda, the distance between the dyes and the center of binding site (a and b), and the angle (α) between them: (Rda)2 = a2 + b2 − 2abcos (α). Taking into account the known position of the dyes relative to the DNA [Hillisch et al. (8)], the length of the arms, the central duplex of DNA and the position of the binding site, we estimated that a ≅ b = 62 ± 3 Å, while the bend angle Δα can be expressed by the equation:
![]()
where (Rda)° = 80 Å is the modeled distance between the dyes (Figure 1c), and Rda is the measured dye-to-dye distance. This function, for the case when a = 62 Å, is plotted in Figure 3a as a solid line. The dashed lines show the range of uncertainty in the calculated bend angle. It is notable that this range is rather small. This functional dependence between the Rda and bend angle was used for determining the extent of DNA bending.
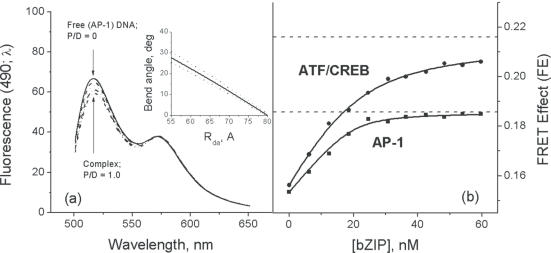
Figure 3.
FRET titration of the AP-1 and ATF/CREB binding sites located near the center of the U-shaped DNA with cross-linked bZIP. (a) Fluorescence spectra of the double-labeled DNA at different ratios bZIP/DNA (P/D) from 0 to 1.0 show the effect of energy migration from the donor (FAM, spectrum with maximum at 490 nm) to the acceptor (TAMRA, spectrum with maximum at 560 nm). The insert shows the calibration function derived from the geometric consideration of the DNA construct (see Figure 1) and plotted according to Equation 1. (b) FRET isotherm of binding bZIP to the AP-1 and ATF/CREB DNAs. The asymptotic values (AFE) of these isotherms (dotted lines), obtained by nonlinear least square fitting using Equation 4, show that the bZIP dimer bends the ATF/CREB DNA more significantly than the AP-1 DNA. The concentration of DNA was 20 nM.
DNA preparation
The oligonucleotides used for the construction of the double-bulged DNA are shown in Figure 1d. All oligonucleotides were purchased from Integrated DNA Technologies, Inc. They were purified using high-performance liquid chromatography and their homogeneity checked by mass spectrometry. The concentrations of single strands and duplexes were determined from the A260 of the nucleotides after complete digestion by phosphodiesterase I (Sigma) in 100 mM Tris–HCl, pH 8.0. For determination of the concentration of labeled single strands, the contribution of FAM and TAMRA absorption at 260 nm (E260 = 28 000 M−1 cm−1 and E260 = 29 000 M−1 cm−1, respectively) were taken into account.
The DNA duplexes were prepared by mixing the complementary oligonucleotides in equimolar amounts, heating to 70°C and cooling slowly to room temperature. The DNA duplexes obtained after annealing were analyzed by non-denaturing PAGE using the fluorescence of the attached FAM and TAMRA chromophores for visualization: this showed a single band, indicating the presence of only one structural form of the DNA (Figure 2). Solutions of DNA for the experiments were prepared by extensive dialysis against the solvent: 10 mM potassium phosphate buffer, pH 6.0, 100 mM KCl.
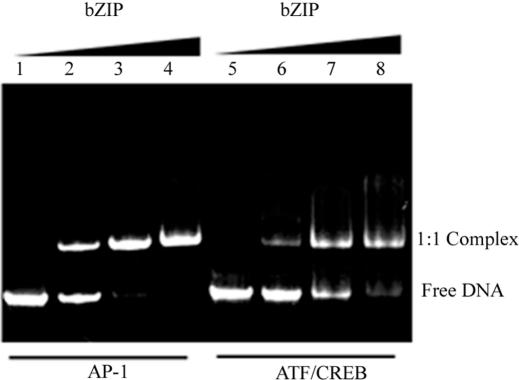
Figure 2.
The PAGE analysis of the constructed U-shaped DNAs and their complexes. Free U-shaped DNA with the AP-1 site (lane 1) and the ATF/CREB site (lane 5) show only one conformation. Lanes 2–4 and lanes 6–8 show binding of the cross-linked bZIP dimer to the DNA having the AP-1 and ATF/CREB sites, respectively. Binding mixtures in 20 μl contained 2.4 μg total DNA. The ratios of bZIP protein to DNA were: 0, 0.6, 1.2 and 1.8. Electrophoresis was on a gradient, 4–20%, non-denaturating polyacrylamide gel at 200 V for 40 min in 20 mM TBE buffer, pH 7.9. For visualization of the bands, the fluorescence of the attached FAM and TAMRA was used.
The S-S cross-linked GCN4-bZIP
The protein GCNK58 was produced from the pET3a-based plasmid pGCNK58 (17). A Y42W mutation was introduced into pGCNK58 to produce pGCNW58 using PCR mutagenesis techniques. Additionally, 4 amino acid residues (GSGC) were attached to the C-terminal end of GCNW58 to produce GCNW62. The plasmid pGCNW62 was constructed from pGCNW58 with PCR mutagenesis techniques. All plasmids were transformed into BL21(DE3)pLysS cells for protein expression. Induction of expression and purification of proteins were carried out with the same procedures as previously reported (17). Covalently cross-linked dimers of GCNW62 were formed by adding 25 mM Cu(II) (1, 10-phenanthroline)3 to a final concentration of 200 μM to a protein solution of 4 mg/ml in 30 mM NaPO4, pH 7.4 and 100 mM NaCl. The solution was left at room temperature for 30 min and was then dialyzed twice against 200 vol of buffer, each for ∼1.5 h (18). The extent of bZIP protein dimerization was determined by SDS–PAGE and mass spectroscopy, and it was shown that more than 95% of the protein was in the dimer cross-linked form:
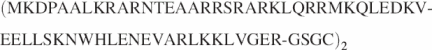
The homogeneity of the obtained S-S cross-linked dimeric GCN4-bZIP was checked by mass spectrometry and electrophoresis. The concentrations of the bZIP dimer were determined from the absorbance at 270 nm, using an extinction coefficient of 10 200 M−1 cm−1. All samples were studied in 100 mM KCl, 10 mM potassium phosphate, pH 6.0.
FRET measurements
Fluorescence measurements were carried out on a SPEX FluoroMax-3 spectrofluorimeter equipped with an accessory for steady-state fluorescence registration, a thermostated cell holder with a stirrer and a software controlled water bath. A 0.4 cm path length quartz Suprasil cells were used.
The methodology of FRET experiments for monitoring binding to DNA, together with analyses of the data obtained, was described in detail previously (6). FAM and TAMRA were excited at 490 and 560 nm, respectively. The FRET efficiency (E) was determined from the sensitization of the acceptor (TAMRA) fluorescence that normalizes the FRET signal for the quantum yield of TAMRA, for the concentration of the duplex molecule and for any error in the effectiveness of acceptor labeling (19). The fluorescence intensities of the emission spectra Fs(490), excited at 490 nm, where both donor (FAM) and acceptor (TAMRA) absorb light, were fitted to the weighted sum of the two spectral components: the standard spectrum of the donor (5′ FAM-DNA) excited at 490 nm, [Fd(490)] and the fluorescence spectrum of the acceptor (double-labeled 5′ FAM-DNA-5′ TAMRA) excited at 560 nm, Fa(560), (at this excitation wavelength only TAMRA absorbs),
![]()
A and FE are the fitted weighting factors of the two spectral components. The fit was made over the range 495–650 nm. The FRET effect [FE = Fa(490)/Fa(560)] is the TAMRA (acceptor) fluorescence component of the spectra, excited at 490 nm, normalized to the fluorescence spectra excited at 560 nm, Fa(560). It is linearly dependent on the efficiency of energy transfer, E.
![]()
The determined ratios ɛa(490)/ɛa(560) = 0.118 and ɛd(490)/ɛa(560) = 0.312 are in perfect agreement with those obtained previously (6) under the same conditions: 100 mM KCl, 10 mM potassium phosphate (pH 6.0). The FRET efficiency, E, varies as the sixth power of the separation between the donor (FAM) and acceptor (TAMRA), normalized to Ro, the characteristic Foster distance for 50% energy transfer efficiency: E = [1 + (Rda/Ro)6]−1. Accordingly, the dye-to-dye distance (Rda) was calculated as Rda = Ro[(1 − E)/E]1/6. For the FAM/TAMRA donor–acceptor pair, a Forster radius of Ro = 50 Å was used (6).
Analysis of bZIP binding to DNA
The dissociation constants were determined by direct fitting of the binding isotherms (see Figure 3b) using the following equation:
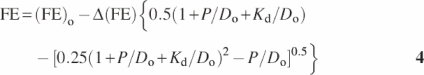
where FE is the observed FRET effect upon the addition of the protein to DNA; (FE)o is the FRET effect of free DNA; Δ(FE) = (AFE) − (FE)o is the maximum change in FE; (AFE) is the maximum (asymptotic) value of the FE obtained when all the DNA is complexed with protein; Kd is the observed dissociation constant; Do is the concentration of DNA and P is the concentration of the SS-cross-linked bZIP protein. Both Kd and Δ(FE) were treated as fitted parameters.
RESULTS AND DISCUSSION
Characterization of the free AP-1 and ATF/CREB sites
All FRET experiments with the double-bulged DNA constructs, one of which contained the AP-1 site and the other the ATF/CREB site, were carried out in 10 mM potassium phosphate buffer (pH 6.0), 100 mM KCl. For these two U-shaped DNA constructs, in the absence of bZIP proteins, the FE was found to be very similar. The FE for ATF/CREB was 0.1534 ± 0.0006, and the FE for the AP-1 site was 0.1562 ± 0.0006. Correspondingly, the calculated values of dye-to-dye distance (Rda) were 70.1 ± 0.5 and 69.1 ± 0.5 Å for the AP-1 and ATF/CREB sites, respectively (Table 1).
Table 1. FRET and bending parameters of the target DNAs upon binding of GCN4-bZIP dimer at 20°C in 100 mM KCl, 10 mM potassium phosphate (pH 6.0).
| U-shaped DNA | FE | FRET efficiency (E) | Rda (Å) | Bend angle (°) |
|---|---|---|---|---|
| Free AP-1 DNA | 0.1534 ± 0.0006 | 0.1163 ± 0.0005 | 70.1 ± 0.5 | 11 ± 1 |
| Complex:bZIP:AP-1 DNA | 0.1857 ± 0.0007a | 0.1295 ± 0.0006 | 61.8 ± 1.5 | 20 ± 2 |
| Free ATF/CREB DNA | 0.1562 ± 0.0006 | 0.1252 ± 0.0006 | 69.1 ± 0.5 | 12 ± 1 |
| Complex:bZIP:ATF/CREB DNA | 0.2140 ± 0.0007a | 0.3100 ± 0.0007 | 57.1 ± 1.5 | 25 ± 2 |
aAsymptotic values of FRET effect.
As explained above, for a totally planar construct, the calculated dye-to-dye distance is 80 Å, i.e. much greater than measured. This discrepancy cannot be explained by the deviation of the construct from planarity, e.g. due to a net residual twist in the 10 bp elements introduced into the base sequence or to the bending of the A-tracts in the arms being out-of-phase, since that would increase the dye-to-dye distance. It is very unlikely that the reduced end-to-end distance can be explained by larger bends in the A4-tracts, although earlier studies suggested that the A4-tracts bend DNA by a more substantial angle, up to 21° (20,21). The latest and most thorough investigation by NMR (15) clearly gives a bend of just 9° (see Materials and Methods). We therefore concluded that the lower Rda must result from an intrinsic bending of the 10 bp inserts that contain the binding sites. Calculation of the bend angles in the target sites using the derived Equation 1 and the measured Rda values gives very similar values of bend angle for the AP-1 and ATF/CREB sites: 11° and 12°, respectively (Table 1). Note that the functional dependence between Rda and the bend angle at the binding site is plotted in the insert to Figure 3a. The estimated bend angle is in agreement with that obtained in an NMR study of a 12 bp DNA duplex containing the ATF/CREB binding site, which showed that this DNA is bent by 13° toward the major groove, centered precisely at the bZIP recognition sequence (16). Any intrinsic bending in a DNA duplex containing the AP-1 site has not been investigated experimentally, but since we found that the Rda for constructs containing both the AP-1 and ATF/CREB sites are similar, we can conclude that the AP-1 site also has an intrinsic bend of ∼12°.
bZIP induced bending of the binding sites
Titration of the GCN4-bZIP into the double-bulged DNA constructs results in the formation of specific complexes (Figure 2). The FEs upon titration of the GCN4-bZIP into the double-bulged DNA constructs with the AP-1 and the ATF/CREB sites are presented in Figure 3. The dissociation constants, Kd, determined from these isotherms are 1.5 nM for the AP-1 site and 6.4 nM for the ATF/CREB site. These values are very close to those found by the fluorescence anisotropy titration (10), and a similar value for the Kd of an AP-1 complex with the S-S cross-linked bZIP was determined in (14). This correspondence with the Kd values obtained using short duplexes gives confidence that there are no additional constraints on binding introduced from the constructs used here. Figure 3 shows that these two isotherms start from about the same values of the FE and rise to asymptotically different levels (AFE), significantly higher for the ATF/CREB site than for the AP-1 site.
In the case of bZIP binding to AP-1 DNA, the FE changes from an initial value of 0.1534 to the asymptotic value of 0.1857. This corresponds to a decrease of the dye-to-dye distance by 8 Å, from 70 to 62 Å, for 100% complex, (Table 1). Binding of GCN4-bZIP to ATF/CREB DNA induces more pronounced changes in the FE, which increases from 0.1562 to 0.2140 at saturation, indicating a decrease of Rda by 11 Å (Table 1).
Assuming that upon binding of the bZIPs, the double-bulged DNA construct remains planar, then from Equation 1 the observed Rda values give an estimate for the additional bend of the DNA induced by binding of the bZIPs: ∼13° for the ATF/CREB site and ∼9° for the AP-1 site. Since both sites are pre-bent (Table 1), the overall bending of the ATF/CREB site should be about 25° and the AP-1 site about 20°. Although the difference in bending of these two sites is in qualitative correspondence with the values determined by crystallography (20° and 0° for the ATF/CREB and AP-1 sites, respectively), the absolute bending values differ considerably. This discrepancy might be caused either by interactions between the molecules in the crystal lattice or result from the assumptions used in the FRET analysis that all conformational changes in the double-bulged DNA construction are planar. However, the FRET experiments clearly demonstrate that the AP-1 and ATF/CREB sites contain an intrinsic bend of the same extent toward the major groove, and that bZIP-binding induces bending of the AP-1 site, although to a lesser extent than at the ATF/CREB site.
The experiments described above show that the measurements of the FE using a double-bulged U-shaped DNA is an efficient method for studying small DNA bends. The technique therefore permits detailed studies in solution of the DNA bending induced by bound transcription factors. Accumulation of such information for various protein–DNA complexes is required for understanding the biological relevance of DNA bending through small angles, a problem more discussed than studied.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Dr T. Ellenberger for the expression clones and protocol for pGCMK58 peptide expression and Dr C. Crane-Robinson for useful discussions. The financial support of NIH GM48036-12 is gratefully acknowledged.
REFERENCES
- 1.Falvo J.V., Thanos,D. and Maniatis,T. (1995) Reversal of intrinsic DNA bends in the IFN beta gene enhancer by transcription factors and the architectural protein HMG I(Y). Cell, 83, 1101–1111. [DOI] [PubMed] [Google Scholar]
- 2.Yie J., Merika,M., Munshi,N., Chen,G. and Thanos,D. (1999) The role of HMG I(Y) in the assembly and function of the IFN-beta enhanceosome. EMBO J., 18, 3074–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellenberger T.E. (1994) Getting a grip on DNA recognition: structures of the basic region leucine zipper, and the basic region helix–loop–helix DNA-binding domain. Curr. Opin. Stuct. Biol., 4, 12–21. [Google Scholar]
- 4.Keller W., Konig,P. and Richmond,T.J. (1995) Crystal structure of a bZIP/DNA complex at 2.2 A: determinants of DNA specific recognition. J. Mol. Biol., 254, 657–667. [DOI] [PubMed] [Google Scholar]
- 5.Konig P. and Richmond,T.J. (1993) The X-ray structure of the GCN4-bZIP bound to ATF/CREB site DNA shows the complex depends on DNA flexibility. J. Mol. Biol., 233, 139–154. [DOI] [PubMed] [Google Scholar]
- 6.Dragan A.I., Klass,J., Read,C., Churchill,M.E., Crane-Robinson,C. and Privalov,P.L. (2003) DNA binding of a non-sequence-specific HMG-D protein is entropy driven with a substantial non-electrostatic contribution. J. Mol. Biol., 331, 795–813. [DOI] [PubMed] [Google Scholar]
- 7.Dragan A.I., Read,C., Makeyeva,E.N., Milgotina,E.I., Churchill,M.E., Crane-Robinson,C. and Privalov,P.L. (2004) DNA binding and bending by HMG boxes: energetic determinants of specificity. J. Mol. Biol., in press. [DOI] [PubMed] [Google Scholar]
- 8.Hillisch A., Lorenz,M. and Diekmann,S. (2001) Recent advances in FRET: distance determination in protein–DNA complexes. Curr. Opin. Struct. Biol., 11, 201–207. [DOI] [PubMed] [Google Scholar]
- 9.Stuhmeier F., Hillisch,A., Clegg,R.M. and Diekman,S. (2000) Fluorescence energy transfer analysis of DNA structures containing several bulges and their interaction with CAP. J. Mol. Biol., 302, 1081–1100. [DOI] [PubMed] [Google Scholar]
- 10.Dragan A.I., Frank,L., Liu,Y., Makeyeva,E.N., Crane-Robinson,C. and Privalov,P.L. (2004) Thermodynamic signature of GCN4-bZIP binding to DNA indicates the role of water in discriminating between the AP-1 and ATF/CREB sites. J. Mol. Biol., in press. [DOI] [PubMed] [Google Scholar]
- 11.Wendt H., Berger,C., Baici,A., Thomas,R.M. and Bosshard,H.R. (1995) Kinetics of folding of leucine zipper domains. Biochemistry, 34, 4097–4107. [DOI] [PubMed] [Google Scholar]
- 12.Cranz S., Berger,C., Baici,A., Jelesarov,I. and Bosshard,H.R. (2004) Monomeric and dimeric bZIP transcription factor GCN4 bind at the same rate to their target DNA site. Biochemistry, 43, 718–727. [DOI] [PubMed] [Google Scholar]
- 13.Kohler J.J. and Schepartz,A. (2001) Kinetic studies of Fos.Jun.DNA complex formation: DNA binding prior to dimerization. Biochemistry, 40, 130–142. [DOI] [PubMed] [Google Scholar]
- 14.Berger C., Piubelli,L., Haditsch,U. and Bosshard,H.R. (1998) Diffusion-controlled DNA recognition by an unfolded, monomeric bZIP transcription factor. FEBS Lett., 425, 14–18. [DOI] [PubMed] [Google Scholar]
- 15.Barbic A., Zimmer,D.P. and Crothers,D.M. (2003) Structural origins of adenine-tract bending. Proc. Natl Acad. Sci. USA, 100, 2369–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khandelwal P., Panchal,S.C., Radha,P.K. and Hosur,R.V. (2001) Solution structure and dynamics of GCN4 cognate DNA: NMR investigations. Nucleic Acids Res., 29, 499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellenberger T.E., Brandl,C.J., Struhl,K. and Harrison,S.C. (1992) The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein–DNA complex. Cell, 71, 1223–1237. [DOI] [PubMed] [Google Scholar]
- 18.Tiebel B., Aung-Hilbrich,L.M., Schnappinger,D. and Hillen,W. (1998) Conformational changes necessary for gene regulation by Tet repressor assayed by reversible disulfide bond formation. EMBO J., 17, 5112–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clegg R.M. (1992) Fluorescence resonance energy-transfer and nucleic-acids. Methods Enzymol., 211, 353–388. [DOI] [PubMed] [Google Scholar]
- 20.Bolshoy A., McNamara,P., Harrington,R.E. and Trifonov,E.N. (1991) Curved DNA without A-A: experimental estimation of all 16 DNA wedge angles. Proc. Natl Acad. Sci. USA, 88, 2312–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Santis P., Palleschi,A., Savino,M. and Scipioni,A. (1990) Validity of the nearest-neighbor approximation in the evaluation of the electrophoretic manifestations of DNA curvature. Biochemistry, 29, 9269–9273. [DOI] [PubMed] [Google Scholar]