Abstract
Background
Accumulating evidence suggests the involvement of long non-coding RNAs (lncRNAs) as oncogenic or tumor suppressive regulators in the development of various cancers. In the present study, we aimed to identify a lncRNA signature based on RNA sequencing (RNA-seq) data to predict survival in esophageal cancer.
Material/Methods
The RNA-seq lncRNA expression data and clinical information were downloaded from The Cancer Genome Atlas (TCGA) database. Differentially expressed lncRNAs were screened out between esophageal cancer and normal tissues. Univariate and multivariate Cox regression analysis were performed to establish a lncRNA-related prognostic model. Receiver operating characteristic (ROC) analysis was conducted to test the sensitivity and specificity of the model. GO (gene ontology) functional and KEGG pathway enrichment analyses were performed for mRNAs co-expressed with the lncRNAs to explore the potential functions of the prognostic lncRNAs.
Results
A total of 265 differentially expressed lncRNAs were identified between esophageal cancer and normal tissues. After univariate and multivariate Cox regression analysis, eight lncRNAs (GS1-600G8.5, LINC00365, CTD-2357A8.3, RP11-705O24.1, LINC01554, RP1-90J4.1, RP11-327J17.1, and LINC00176) were finally screened out to establish a predictive model by which patients could be classified into high-risk and low-risk groups with significantly different overall survival. Further analysis indicated independent prognostic capability of the 8-lncRNA signature from other clinicopathological factors. ROC curve analysis demonstrated good performance of the 8-lncRNA signature. Functional enrichment analysis showed that the prognostic lncRNAs were mainly associated with esophageal cancer related biological processes such as regulation of glucose metabolic process and amino acid and lipids metabolism.
Conclusions
Our study developed a novel candidate model providing additional and more powerful prognostic information beyond conventional clinicopathological factors for survival prediction of esophageal cancer patients. Moreover, it also brings us new insights into the molecular mechanisms underlying esophageal cancer.
MeSH Keywords: Biological Markers; Esophageal Neoplasms; RNA, Long Noncoding; Survival Analysis
Background
Esophageal cancer is one of the most common digestive malignancies with the 5-year relative survival rate less than 20% [1], ranking the fourth leading cause of cancer death among both men and women in China [2]. Like most solid tumors, pathological tumor node metastasis (TNM) stage is still a main prognostic indicator of esophageal cancer patient survival. However, the molecular heterogeneity and complexity of esophageal cancer make clinical outcomes difficult to predict and even patients within the same stage present wide variations in survival [3]. Therefore, it is urgent to develop novel biomarkers or models for survival risk prediction in esophageal cancer which would provide patients more effective therapies.
Long non-coding RNAs (lncRNAs) are mostly defined as RNA transcripts exceeding 200 nucleotides (nt) in length apparently without protein coding capacity [4]. Accumulating evidence indicates that lncRNAs play critical roles in a spectrum of biological processes via transcriptional, post-transcriptional and epigenetic mechanisms [5]. Dysregulation of lncRNAs has been observed in various cancers, including breast cancer, colorectal cancer, lung cancer, prostate cancer as well as esophageal cancer [6–10]. It has been reported that dysregulated lncRNAs are associated with cancer pathogenesis and function as oncogenic or tumor suppressive regulators in cancer development [11]. They have been shown to affect cell proliferation, migration, and invasion through regulating the expression of genes involved in various tumorigenetic pathways [11,12]. One of the recent techniques used for transcriptome analyses is RNA sequencing (RNA-seq), a next-generation sequencing technique with high-sensitivity, high-throughput and the ability of detecting novel exons, splice sites, and transcripts [13,14]. However, RNA-seq based work focused on the prognostic power of lncRNA signatures for survival risk of esophageal cancer patients is quite rare.
In this study, we identified for the first time a RNA-seq based lncRNA signature as a predictor of survival risk of esophageal cancer patients using a cohort of more than 100 cases from The Cancer Genome Atlas (TCGA) database. Cox regression analysis and risk score model method were utilized to develop an 8-lncRNA signature which could distinguish patients with good and poor survival. A higher area under curve (AUC) of the receiver operating characteristic (ROC) curve confirmed good sensitivity and specificity of the prognostic model while multivariate Cox regression analysis and stratified analysis indicated the independence of predictive capacity of the 8-lncRNA prognostic signature from other clinicopathological factors. Furthermore, our functional enrichment analysis suggested that the eight predictive lncRNAs were probably involved in the progression of esophageal cancer through exerting their roles in esophageal cancer related biological processes and pathways, such as regulation of glucose metabolic process, positive regulation of MAPK cascade, and amino acid and lipids metabolism. Our results provided further insights into the predictive capacity of lncRNAs underlying esophageal cancer.
Material and Methods
The esophageal cancer patient dataset
The preprocessed level 3 RNA-seq data and corresponding clinical information of esophageal cancer patients were collected from The Cancer Genome Atlas (TCGA) database (http://cancergenome.nih.gov/) (as of September, 2016). The patients meeting the following criteria were included in the study: (1) patients with complete information of lncRNA expression profiles and clinical characteristics (including age, gender, race, stage, survival status and survival time) (2) the overall survival time was more than one month.
Differentially expressed lncRNAs screening between esophageal cancer and normal tissues
According to the inclusion criteria, a total of 122 esophageal cancer patients were enrolled in the study (Table 1). The gene expression profiling data of the 122 esophageal cancer samples and 11 normal samples were downloaded from the TCGA database. The package of edgeR [15] in R language was employed to identify the differentially expressed lncRNAs between esophageal cancer and normal tissues with the |log2FC| >2 and FDR <0.01 set as the threshold. Then the unsupervised hierarchical clustering was performed based on the expression of these altered lncRNAs by the pheatmap package in R (https://cran.r-project.org/web/packages/pheatmap/index.html, version 1.0.8) [16].
Table 1.
Summary of esophageal cancer patient clinical characteristics.
| Characteristic | Patients (N=122) | |
|---|---|---|
| n | % | |
| Age category | ||
| <60 y | 54 | 44.26 |
| ≥60 y | 68 | 55.74 |
| Gender | ||
| Male | 102 | 83.61 |
| Female | 20 | 16.39 |
| Race | ||
| White | 83 | 68.03 |
| Asian | 36 | 29.51 |
| Black or African American | 3 | 2.46 |
| Pathological Stage | ||
| Stage I | 14 | 11.47 |
| Stage II | 63 | 51.64 |
| Stage III | 39 | 31.97 |
| Stage IV | 6 | 4.92 |
| Vital Status | ||
| Alive | 85 | 69.67 |
| Dead | 37 | 30.33 |
Survival analysis and definition of lncRNA related prognostic model
The association between the expression of differentially expressed lncRNAs and patient overall survival was evaluated by univariate Cox proportional hazards regression analysis using the survival R package. Only those lncRNAs with p-value <0.05 were considered as candidate variables and entered into a stepwise multivariate Cox regression analysis tested by AIC (Akaike Information Criterion, assessing the goodness of fit of a statistical model) to identify the predictive model with the best explanatory and informative efficacy. Then, a lncRNA-related prognostic model was established to evaluate each patient’s survival risk as follows:
where k is the number of prognostic lncRNAs, Ci represents the coefficient of the ith lncRNA in the multivariate Cox regression analysis, Vi is the expression value of the ith lncRNA. The lncRNAs with Ci >0 were defined as high-risk signatures while those with Ci <0 were defined as protective lncRNAs.
Risk stratification and ROC curve
According to the predictive lncRNA signature model, the risk score of each of the 122 patients was calculated. The patients were then classified into high-risk or low-risk group using the median risk score as the cutoff value. Overall survival curves were generated using the Kaplan-Meier method, and two-sided log-rank tests were employed to compare the differences in overall survival time between the high-risk and low-risk patient groups. The sensitivity and specificity of the lncRNA prognostic model to predict clinical outcome were evaluated by calculating the area under curve (AUC) of the receiver operating characteristic (ROC) curve in the R package of “survival ROC” [17].
Independence of survival prediction by the prognostic lncRNA signature from other clinical variables
To determine whether predictive capacity of the lncRNA signature was independent of other clinical factors (including race, gender, stage, and age) of esophageal cancer patients, multivariate Cox regression analysis was carried out using overall survival as the dependent variable and lncRNA signature and other conventional clinical factors as independent variables. For clinical features with p-value <0.01 in Cox regression analysis, stratification analysis was further performed to determine whether the lncRNA signature exhibit prognostic value within the same clinical factor.
Functional enrichment analysis
To identify potential biological processes and pathways which the predictive lncRNAs were involved in, functional enrichment analysis was performed. First, the Pearson correlation coefficients between the expression profiles of the eight prognostic lncRNAs and protein-coding genes (PCGs) were calculated to determine the co-expression relationships of the lncRNAs and PCGs. The PCGs with |Pearson correlation coefficient| >0.40 were considered to be lncRNAs-related PCGs. Gene ontology (GO) biological process (BP) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were carried out for those PCGs using the Database for Annotation, Visualization, and Integrated Discovery (DAVID, https://david.ncifcrf.gov/, version 6.8) [18]. The p-value <0.05 was set as the cutoff criterion for both GO and KEGG functional analysis.
Results
Differentially expressed lncRNAs between esophageal cancer and normal tissues
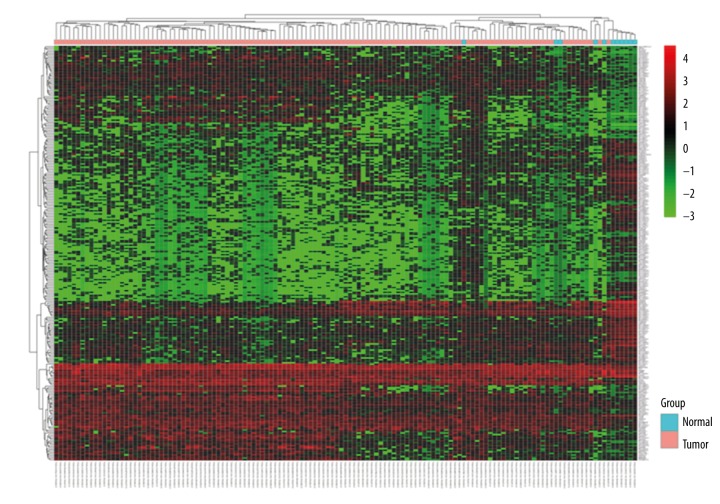
According to the cutoff criteria, a total of 265 differentially expressed (including 112 upregulated and 153 downregulated) lncRNAs were identified between esophageal cancer tissues and normal tissues. The results of unsupervised hierarchical cluster analysis in Figure 1 showed that the esophageal cancer samples could be clearly distinguished from the normal controls with the expression of differentially expressed lncRNAs.
Figure 1.
Unsupervised hierarchical clustering analysis of the differentially expressed lncRNAs between esophageal cancer and normal tissues.
Establishment of the 8-lncRNA signature associated with overall survival of esophageal cancer patients
To identify prognosis-related lncRNAs, we first used univariate Cox regression analysis to evaluate the associations between the expression level of each of the differentially expressed lncRNAs and patients’ overall survival, and found that 13 lncRNAs were significantly related to overall survival (p<0.05). Then a stepwise multivariate Cox regression analysis was performed and eight lncRNAs therein (as shown in Table 2) were finally screened out to establish a predictive model. As previously described, the predictive model was defined as the linear combination of the expression levels of the eight lncRNAs weighted by their relative coefficient in multivariate Cox regression as follows: survival risk score = (0.0398× expression value of GS1-600G8.5) + (0.9990× expression value of LINC00365) + (0.6216× expression value of CTD-2357A8.3) + (13.6225× expression value of RP11-705O24.1) + (1.7105× expression value of LINC01554) + (0.8297× expression value of RP1-90J4.1) + (−6.2336× expression value of RP11-327J17.1) + (−1.2226× expression value of LINC00176). Among these, GS1-600G8.5, LINC00365, CTD-2357A8.3, RP11-705O24.1, LINC01554, and RP1-90J4.1 showed positive coefficients in Cox regression analysis, indicating high-risk signatures for these six lncRNAs since their high expression signified a shorter overall survival of patients. For the remaining two lncRNAs, we observed negative coefficients in Cox regression analysis, implying that these lncRNAs could be regarded as protective lncRNAs since patients with higher expression levels of these lncRNAs tended to have longer overall survival compared with those with lower expression levels of these lncRNAs.
Table 2.
Overall information of 8 prognostic lncRNAs associated with OS of esophageal cancer patients.
| Ensembl ID | Gene symbol | Chromosome | Relative coefficient | P value* |
|---|---|---|---|---|
| ENSG00000235385 | GS1-600G8.5 | Chr X: 13,266,048–13,303,452 (−) | 0.0398 | 6.92E-04 |
| ENSG00000224511 | LINC00365 | Chr 13: 30,103,178–30,108,875 (−) | 0.999 | 1.89E-03 |
| ENSG00000267123 | CTD-2357A8.3 | Chr 17: 78,617,389–78,632,057 (−) | 0.6216 | 6.78E-03 |
| ENSG00000254119 | RP11-705O24.1 | Chr 8: 61,785,047–61,944,180 (+) | 13.6225 | 0.013 |
| ENSG00000236882 | LINC01554 | Chr 5: 95,852,232–95,860,133 (+) | 1.7105 | 0.016 |
| ENSG00000257906 | RP1-90J4.1 | Chr 12: 47,415,008–47,420,179 (+) | 0.8297 | 0.020 |
| ENSG00000259763 | RP11-327J17.1 | Chr 15: 96,235,785–96,236,703 (+) | −6.2336 | 0.036 |
| ENSG00000196421 | LINC00176 | Chr 20: 64,034,344–64,039,962 (+) | −1.2226 | 0.039 |
Derived from the univariate Cox regression analysis
Risk stratification and ROC curve indicate good performance of the 8-lncRNA signature in predicting the overall survival of esophageal cancer patients
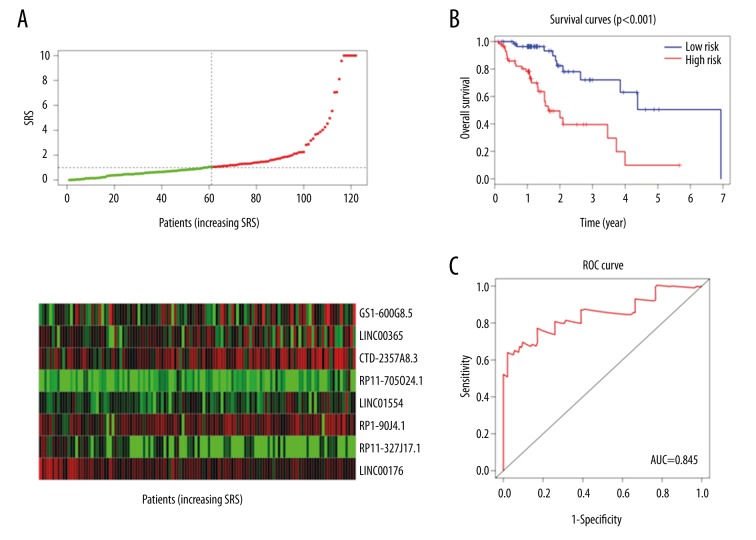
For each of the 122 patients in our study, we were able to calculate an 8-lncRNA expression-based survival risk score (referred to as “SRS”) and assigned them into a high-risk group or a low-risk group using the median risk score of 1.058 as the cutoff point. As a result, 61 patients were classified into the high-risk group since their SRSs were greater than the cutoff value, whereas the other 61 patients were assigned to the low-risk group with their SRSs less than the cutoff point (Figure 2A). The Kaplan-Meier overall survival curves of the two groups based on the eight lncRNAs were notably different (log-rank p=1.72 e-05 <0.001), showing overall survival in 9.89% and 50.5% at five years for patients with high-risk and low-risk SRS, respectively (Figure 2B). The prognostic power of the 8-lncRNA signature was evaluated by calculating the AUC of ROC curve. Higher AUC indicates better model performance and AUC more than 0.80 is considered good performance. In our study, the ROC curve analysis achieved AUC of 0.845, showing good sensitivity and specificity of the 8-lncRNA signature model in predicting esophageal cancer patient survival risk (Figure 2C).
Figure 2.
Prognostic evaluation of the 8-lncRNA signature in esophageal cancer patients. (A) The distribution of lncRNA-related SRS and the expression heatmap of 8 prognostic lncRNAs. (B) Kaplan-Meier survival curve analysis for overall survival of esophageal cancer patients using the 8-lncRNA signature. (C) ROC curve analysis of the 8-lncRNA signature.
Prognostic value of the 8-lncRNA signature is independent of conventional clinical factors
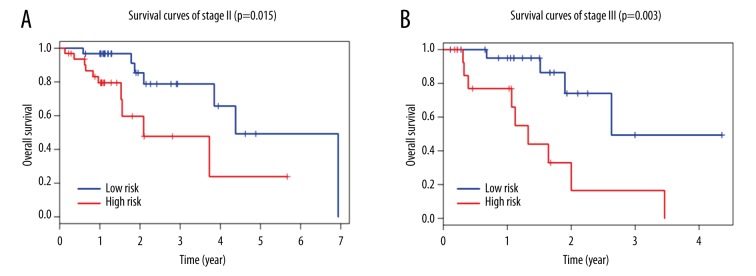
Multivariate Cox regression analysis demonstrated that the 8-lncRNA signature risk score maintained an independent predictive ability from other clinical factors (HR=5.951, 95% CI 2.577–13.741, p=2.95 e-05, shown in Table 3). Meanwhile, we also found that TNM stage was an independent predictor for overall survival of esophageal cancer patients. Therefore, stratification analysis was further carried out to examine whether the 8-lncRNA signature could provide predict value for patients within the same TNM stage. Because the sample sizes in stage I and IV were too small to draw any reliable conclusions (n=14 in stage I and n=6 in stage IV), stratification analysis was performed only in stage II and III patients. Log-rank test for patients in stage II demonstrated that the 8-lncRNA signature could distinguish patients with significantly different survival (p=0.015, Figure 3A). Similar predictive value of the 8-lncRNA signature was observed for stage III patients (p=0.003, Figure 3B). Altogether, these results indicated that the prognostic capability of the 8-lncRNA signature is independent of conventional clinical factors for survival prediction of esophageal cancer patients.
Table 3.
Multivariate Cox regression analysis of overall survival.
| Variables | HR | 95% CI of HR | P value |
|---|---|---|---|
| Age | 1.01 | 0.979–1.043 | 0.524 |
| Race | 0.903 | 0.553–1.474 | 0.683 |
| Gender (Male vs. Female) | 2.157 | 0.571–8.152 | 0.257 |
| Stage (III + IV vs. I+II) | 2.501 | 1.53–4.089 | 2.58E-04 |
| Eight-lncRNA risk score (High vs. Low) | 5.951 | 2.577–13.741 | 2.95E-05 |
HR – hazard ratio; CI – confidence interval.
Figure 3.
Survival prediction in stage II and III patients. (A) Kaplan-Meier survival curves of stage II patients with esophageal cancer classified into high-risk and low-risk groups by the 8-lncRNA signature (p=0.015). (B) Kaplan-Meier survival curves of stage III patients divided into high-risk and low-risk groups (p=0.003).
Identification of the 8-lncRNA signature related biological processes and pathways
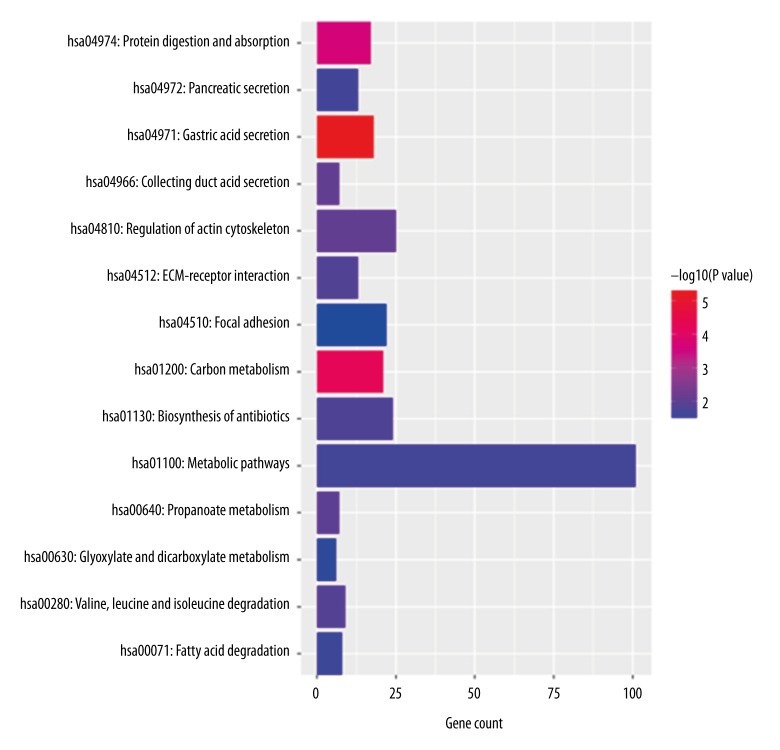
We performed GO and KEGG functional enrichment analysis for the PCGs co-expressed with the lncRNAs in the predictive signature to reveal the potential functions of the eight prognostic lncRNAs. The results showed that co-expressed PCGs were enriched in 73 GO BP terms, which mainly clustered in regulation of diverse biological processes (such as GO: 0010906~regulation of glucose metabolic process, GO: 0010628~positive regulation of gene expression, GO: 0043410~positive regulation of MAPK cascade), transport of various substances (including GO: 0071805~potassium ion transmembrane transport, GO: 0035879~plasma membrane lactate transport, GO: 0098719~sodium ion import across plasma membrane) and response to different stimulants (such as GO: 0042594~response to starvation, GO: 0043627~response to estrogen, GO: 0042493~response to drug) (The top 30 GO BP terms were shown in Table 4). Fourteen KEGG pathways were enriched which mainly focused on digestive functions (including hsa04971: Gastric acid secretion, hsa04974: Protein digestion and absorption, hsa04972: Pancreatic secretion) and basic substance metabolism (such as hsa01200: Carbon metabolism, hsa00280: Valine, leucine and isoleucine degradation, hsa01100: Metabolic pathways and hsa00071: Fatty acid degradation) (Figure 4).
Table 4.
Enrichment analysis of top 30 GO BP terms for lncRNA-related PCGs.
| Term | Count | P value |
|---|---|---|
| GO: 0071805~potassium ion transmembrane transport | 26 | 3.26E-07 |
| GO: 0007586~digestion | 18 | 3.78E-07 |
| GO: 0051453~regulation of intracellular pH | 12 | 1.40E-05 |
| GO: 0010906~regulation of glucose metabolic process | 9 | 5.23E-05 |
| GO: 0010107~potassium ion import | 9 | 3.55E-04 |
| GO: 0010628~positive regulation of gene expression | 33 | 4.76E-04 |
| GO: 0001696~gastric acid secretion | 5 | 5.84E-04 |
| GO: 0042391~regulation of membrane potential | 14 | 1.11E-03 |
| GO: 0035313~wound healing, spreading of epidermal cells | 5 | 2.98E-03 |
| GO: 0008152~metabolic process | 23 | 3.04E-03 |
| GO: 0043401~steroid hormone mediated signaling pathway | 11 | 4.07E-03 |
| GO: 0035879~plasma membrane lactate transport | 5 | 4.44E-03 |
| GO: 0098719~sodium ion import across plasma membrane | 5 | 4.44E-03 |
| GO: 0006811~ion transport | 18 | 5.77E-03 |
| GO: 0031581~hemidesmosome assembly | 5 | 6.31E-03 |
| GO: 0007565~female pregnancy | 15 | 6.43E-03 |
| GO: 0042594~response to starvation | 9 | 6.81E-03 |
| GO: 0009083~branched-chain amino acid catabolic process | 6 | 6.90E-03 |
| GO: 0055085~transmembrane transport | 28 | 7.00E-03 |
| GO: 0007605~sensory perception of sound | 18 | 7.80E-03 |
| GO: 0051056~regulation of small GTPase mediated signal transduction | 18 | 7.80E-03 |
| GO: 0022010~central nervous system myelination | 4 | 8.44E-03 |
| GO: 0043627~response to estrogen | 12 | 8.67E-03 |
| GO: 0006813~potassium ion transport | 13 | 8.70E-03 |
| GO: 0042493~response to drug | 36 | 0.0116 |
| GO: 0042853~L-alanine catabolic process | 3 | 0.0127 |
| GO: 0030036~actin cytoskeleton organization | 17 | 0.0127 |
| GO: 0001937~negative regulation of endothelial cell proliferation | 7 | 0.0129 |
| GO: 0034765~regulation of ion transmembrane transport | 15 | 0.0156 |
| GO: 0043410~positive regulation of MAPK cascade | 12 | 0.0199 |
GO – gene ontology; BP – biological process; PCGs – protein-coding genes.
Figure 4.
KEGG pathway for lncRNA-related PCGs.
Discussion
Esophageal cancer is a global health threat with high morbidity and mortality [19]. Owing to the heterogeneity, conventional prognostic systems such as TNM staging system often show insufficient prediction for risk stratification and clinical outcome estimations. Therefore, considerable efforts have been made in recent decades to develop novel prognostic signatures to promote the prediction of esophageal cancer patient survival [20–22].
As a new focused class of ncRNAs, lncRNAs were indicated to participate in multiple biological processes including X chromosome inactivation, genomic imprinting, and tumor related alterations [23]. Accumulating reports have demonstrated the dysregulation of lncRNAs and their potential as biomarkers in various cancers [24–26]. Li et al. have established a 3-lncRNA signature associated with the patient survival in esophageal squamous cell carcinoma using microarray analysis [27]. However, microarray technology has some undesired shortcomings, such as the bias due to the probe selection and the limitation of identifying only known transcripts [13,14]. Compared with microarrays, RNA-seq technique has been developed as an emerging sequencing technology with advantages reducing these defects and standing out especially with the ability of finding novel transcripts [14]. Nevertheless, research investigating the impact of lncRNAs in esophageal cancer patient survival, which were based on RNA-seq technology, are quite deficient.
In this study, we developed an 8-lncRNA signature which was able to predict the clinical outcome of esophageal cancer. To the best of our knowledge, this is the first lncRNA-related predictive model based on RNA-seq technology using a cohort of more than 100 cases in esophageal cancer. The differentially expressed lncRNAs were first screened out between esophageal cancer and normal tissues with the data downloaded from TCGA database. Then the expression profiles of these lncRNAs of 122 esophageal cancer patients were analyzed by univariate and stepwise multiple Cox proportional hazards regression analysis. Then eight lncRNAs were finally identified to establish a predictive model based on the linear combination of these lncRNAs. A distinctive separation was observed in survival curves between patient groups with high-risk and low-risk scores using the predictive model. And ROC analysis achieved an AUC of 0.845 which demonstrated high sensitivity and specificity of the lncRNA signature model. When taking other clinical factors together, multivariate Cox regression analysis revealed that the 8-lncRNA signature was independent of these conventional clinicopathological factors including race, gender, age, and tumor stage. Further stratification analysis indicated favorable discrimination of the 8-lncRNA signature in predicting different survival of patients in the same TNM stage. This might provide more references for clinical doctors to select better individualized and effective treatment for patients with different survival risk.
The carcinogenesis of esophageal cancer is a multi-step process hallmarked by a series of genetic alterations [28]. Although growing attention has begun to focus on the study of lncRNAs, the functions of most lncRNAs are still unknown. Computational annotation of lncRNA functions through their co-expressed mRNAs has been proven to be effective [29]. In the present study, we performed GO and KEGG enrichment analysis for the co-expressed mRNAs of the eight lncRNAs to explore the functions of the predictive lncRNAs. The results showed that the prognostic lncRNAs were involved in significant biological processes such as regulation of glucose metabolic process, positive regulation of gene expression, positive regulation of MAPK cascade and enriched in KEGG pathways including gastric acid secretion, amino acid metabolism (valine, leucine and isoleucine degradation), and lipids metabolism (fatty acid degradation). Studies have identified aberrant glucose metabolic processes in esophageal cancer, including high plasma and urine glucose levels [30], excessive glucose uptake and accumulation [31] and alterations of glucose metabolism associated genes [32]. MAPK signaling cascade play significant roles in converting external stimuli into broad cellular responses [33]. Inhibition of MAPK pathways suppresses proliferation and induces apoptosis of esophageal cancer cells [34]. Higher gastric acid secretion has been observed in gastroesophageal junction adenocarcinoma, including Barrett esophageal cancer compared to normal subjects [35,36]. Amino acid and lipids metabolism disorders, such as increasing leucine level [37] and highly activated fatty acids metabolism [38], were also found to be responsible for the development of esophageal cancer. Therefore, it is plausible to infer that the eight prognostic lncRNAs participate in the progression of esophageal cancer through interacting with PCGs in these esophageal cancer-related biological pathways. However, further experimental studies are needed to confirm the functions of these lncRNAs.
Conclusions
This study identified a RNA-seq based 8-lncRNA signature which could predict the survival risk of esophageal cancer patients. The signature displayed independent prognostic capacity of conventional clinicopathological factors and could robustly predict survival outcomes of esophageal cancer patients within the same TNM stage. It could be used to identify patients with high-risk scores who will benefit from more effective and individualized therapy. It could not only serve as a novel potential biomarker for esophageal cancer patient survival risk stratification, but also provide us a better understanding of molecular mechanisms involved in the development of esophageal cancer. However, further clinical studies validating the predictive efficacy of the signature and experimental research investigating the functions of the prognostic lncRNAs need to be conducted.
Footnotes
Conflicts of interest
None.
Source of support: Departmental sources
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Peters CJ, Rees JR, Hardwick RH, et al. A 4-gene signature predicts survival of patients with resected adenocarcinoma of the esophagus, junction, and gastric cardia. Gastroenterology. 2010;139(6):1995–2004.e15. doi: 10.1053/j.gastro.2010.05.080. [DOI] [PubMed] [Google Scholar]
- 4.Young RS, Ponting CP. Identification and function of long non-coding RNAs. Essays Biochem. 2013;54:113–26. doi: 10.1042/bse0540113. [DOI] [PubMed] [Google Scholar]
- 5.Quan M, Chen J, Zhang D. Exploring the secrets of long noncoding RNAs. Int J Mol Sci. 2015;16(3):5467–96. doi: 10.3390/ijms16035467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cerk S, Schwarzenbacher D, Adiprasito JB, et al. Current status of long non-coding RNAs in human breast cancer. Int J Mol Sci. 2016;17(9) doi: 10.3390/ijms17091485. pii: E1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saus E, Brunet-Vega A, Iraola-Guzmán S, et al. Long non-coding RNAs as potential novel prognostic biomarkers in colorectal cancer. Front Genet. 2016;7:54. doi: 10.3389/fgene.2016.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng Z, Zhang C, Duan C. Functions and mechanisms of long noncoding RNAs in lung cancer. Onco Targets Ther. 2016;9:4411–24. doi: 10.2147/OTT.S109549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mouraviev V, Lee B, Patel V, et al. Clinical prospects of long noncoding RNAs as novel biomarkers and therapeutic targets in prostate cancer. Prostate Cancer Prostatic Dis. 2016;19(1):14–20. doi: 10.1038/pcan.2015.48. [DOI] [PubMed] [Google Scholar]
- 10.Tang WW, Wu Q, Li SQ, et al. Implication of lncRNAs in pathogenesis of esophageal cancer. Onco Targets Ther. 2015;8:3219–26. doi: 10.2147/OTT.S87856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H, Chen Z, Wang X, et al. Long non-coding RNA: A new player in cancer. J Hematol Oncol. 2013;6:37. doi: 10.1186/1756-8722-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ling H, Vincent K, Pichler M, et al. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene. 2015;34(39):5003–11. doi: 10.1038/onc.2014.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han Y, Gao S, Muegge K, et al. Advanced applications of RNA sequencing and challenges. Bioinform Biol Insights. 2015;9(Suppl 1):29–46. doi: 10.4137/BBI.S28991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farkas MH, Au ED, Sousa ME, Pierce EA. RNA-Seq: Improving our understanding of retinal biology and disease. Cold Spring Harb Perspect Med. 2015;5(9):a017152. doi: 10.1101/cshperspect.a017152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolde R. Pheatmap: Pretty heatmaps. R package version 1.0.8. 2015. [Google Scholar]
- 17.Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56(2):337–44. doi: 10.1111/j.0006-341x.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 18.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 19.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 20.Feber A, Xi L, Pennathur A, et al. MicroRNA prognostic signature for nodal metastases and survival in esophageal adenocarcinoma. Ann Thorac Surg. 2011;91(5):1523–30. doi: 10.1016/j.athoracsur.2011.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao JY, Wang F, Li Y, et al. Five miRNAs considered as molecular targets for predicting esophageal cancer. Med Sci Monit. 2015;21:3222–30. doi: 10.12659/MSM.895001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu P, Qiao J, He W, et al. Genome-wide gene expression profile analyses identify CTTN as a potential prognostic marker in esophageal cancer. PLoS One. 2014;9(2):e88918. doi: 10.1371/journal.pone.0088918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: Past, present, and future. Genetics. 2013;193(3):651–69. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ren W, Zhang J, Li W, et al. A Tumor-specific prognostic long non-coding RNA signature in gastric cancer. Med Sci Monit. 2016;22:3647–57. doi: 10.12659/MSM.901190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu Y, Chen HY, Yu CY, et al. A long non-coding RNA signature to improve prognosis prediction of colorectal cancer. Oncotarget. 2014;5(8):2230–42. doi: 10.18632/oncotarget.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sørensen KP, Thomassen M, Tan Q, et al. Long non-coding RNA expression profiles predict metastasis in lymph node-negative breast cancer independently of traditional prognostic markers. Breast Cancer Res. 2015;17:55. doi: 10.1186/s13058-015-0557-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Chen Z, Tian L, et al. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut. 2014;63(11):1700–10. doi: 10.1136/gutjnl-2013-305806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denlinger CE, Thompson RK. Molecular basis of esophageal cancer development and progression. Surg Clin North Am. 2012;92(5):1089–103. doi: 10.1016/j.suc.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 29.Liao Q, Liu C, Yuan X, et al. Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucleic Acids Res. 2011;39(9):3864–78. doi: 10.1093/nar/gkq1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hasim A, Ma H, Mamtimin B, et al. Revealing the metabonomic variation of EC using 1H-NMR spectroscopy and its association with the clinicopathological characteristics. Mol Biol Rep. 2012;39(9):8955–64. doi: 10.1007/s11033-012-1764-z. [DOI] [PubMed] [Google Scholar]
- 31.Jadvar H, Henderson RW, Conti PS. 2-deoxy-2-[F-18]fluoro-D-glucose- positron emission tomography/computed tomography imaging evaluation of esophageal cancer. Mol Imaging Biol. 2006;8(3):193–200. doi: 10.1007/s11307-006-0036-5. [DOI] [PubMed] [Google Scholar]
- 32.Hochwald JS, Zhang J. Glucose oncometabolism of esophageal cancer. Anticancer Agents Med Chem. 2016 doi: 10.2174/1871520616666160627092716. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 33.Wagner EF, Nebreda ÁR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9(8):537–49. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 34.Wu K, Yang Y, Liu D, et al. Activation of PPARγ suppresses proliferation and induces apoptosis of esophageal cancer cells by inhibiting TLR4-dependent MAPK pathway. Oncotarget. 2016;7(28):44572–82. doi: 10.18632/oncotarget.10067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inomata Y, Koike T, Ohara S, et al. Presevation of gastric acid secretion may be important for the development of gastroesophageal junction adenocarcinoma in Japanese people, irrespective of the H. pylori infection status. Am J Gastroenterol. 2006;101(5):926–33. doi: 10.1111/j.1572-0241.2006.00497.x. [DOI] [PubMed] [Google Scholar]
- 36.Koike T, Ohara S, Inomata Y, et al. The prevalence of Helicobacter pylori infection and the status of gastric acid secretion in patients with gastroesophageal junction adenocarcinoma in Japan. Inflammopharmacology. 2007;15(2):61–64. doi: 10.1007/s10787-006-1549-x. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X, Xu L, Shen J, et al. Metabolic signatures of esophageal cancer: NMR-based metabolomics and UHPLC-based focused metabolomics of blood serum. Biochim Biophys Acta. 2013;1832(8):1207–16. doi: 10.1016/j.bbadis.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 38.Wang L, Chen J, Chen L, et al. 1H-NMR based metabonomic profiling of human esophageal cancer tissue. Mol Cancer. 2013;12:25. doi: 10.1186/1476-4598-12-25. [DOI] [PMC free article] [PubMed] [Google Scholar]