Abstract
Multiple kelch13 alleles conferring artemisinin resistance (ART-R) are currently spreading through Southeast Asian malaria parasite populations, providing a unique opportunity to observe an ongoing soft selective sweep, investigate why resistance alleles have evolved multiple times and determine fundamental population genetic parameters for Plasmodium. We sequenced kelch13 (n = 1,876), genotyped 75 flanking SNPs, and measured clearance rate (n = 3,552) in parasite infections from Western Thailand (2001–2014). We describe 32 independent coding mutations including common mutations outside the kelch13 propeller associated with significant reductions in clearance rate. Mutations were first observed in 2003 and rose to 90% by 2014, consistent with a selection coefficient of ∼0.079. ART-R allele diversity rose until 2012 and then dropped as one allele (C580Y) spread to high frequency. The frequency with which adaptive alleles arise is determined by the rate of mutation and the population size. Two factors drive this soft sweep: (1) multiple kelch13 amino-acid mutations confer resistance providing a large mutational target—we estimate the target is 87–163 bp. (2) The population mutation parameter (Θ = 2Neμ) can be estimated from the frequency distribution of ART-R alleles and is ∼5.69, suggesting that short term effective population size is 88 thousand to 1.2 million. This is 52–705 times greater than Ne estimated from fluctuation in allele frequencies, suggesting that we have previously underestimated the capacity for adaptive evolution in Plasmodium. Our central conclusions are that retrospective studies may underestimate the complexity of selective events and the Ne relevant for adaptation for malaria is considerably higher than previously estimated.
Keywords: soft selective sweep, adaptation, effective population size, rapid evolution, drug resistance, plasmodium
Introduction
The number of times drug resistance alleles arise is a central question for disease intervention because it determines the useful life of drugs, the ease with which resistance arises, and the utility of diagnostic markers for tracking resistance. It is also a question of central interest for evolutionary biologists interested in adaptation, where the relative role of hard selective events, in which flanking genetic variation is purged from around the selected site, and soft selective events in which flanking genetic variation is maintained, is being actively debated (Jensen 2014; Schrider et al. 2015). The frequency with which beneficial alleles establish is expected to be determined by the rate (μ) at which mutations conferring resistance are introduced into the population (by mutation or migration) and the effective size (Ne) of the population during the time when selection is acting: these parameters are combined in the population mutation parameter (Θ = 2Neμ). Elegant theoretical work (Pennings and Hermisson 2006; Messer and Petrov 2013) has led to the prediction that when Θ < 0.01, mutations will be limiting and most selective events will involve single alleles generating hard selective sweeps, when Θ is between 0.01 and 1 both single and multiple origins of adaptive alleles are possible, and when Θ > 1, multiple origins of adaptive alleles are expected.
Most studies of selection in nature are retrospective. Typically, sequence data from present day populations is used to infer selective events that have occurred in the past. While valuable, these retrospective studies provide a simplified description of adaptive evolution, because only surviving allelic lineages are examined. In contrast, molecular description of ongoing adaptive evolution in nature provides a direct record of the dynamics of selective events, including documentation of allelic lineages that fail to survive. Studies of rapid evolution of microbial or insect populations in the face of ongoing selection with drugs of pesticides are particularly valuable in this respect (Karasov et al. 2010; Barroso-Batista et al. 2014; Pennings et al. 2014; Weetman et al. 2015).
Malaria parasites provide a powerful system for studying adaptation, because parasite populations have been exposed to a succession of antimalarials over the past 75 years, and selection is strong, with coefficients of 0.1–0.4 (Anderson et al. 2011). Parasite genomes are compact (23 Mb), blood stage parasites are haploid, and there is an obligate recombination in the mosquito, making this a useful system for understanding adaptation in other recombining eukaryotes. Retrospective molecular studies of resistance to chloroquine and pyrimethamine, in which variation at markers flanking resistance mutations were examined, have revealed remarkably few independent origins of high level resistance alleles (Wootton et al. 2002; Nair et al. 2003). Both high level resistance to chloroquine [involving the chloroquine resistance transporter (crt)] and pyrimethamine [involving mutations in dihyrofolate reductase (dhfr)] emerged once in SE Asia. This drove textbook “hard” selective sweeps, in which the spread of single resistant haplotypes purged genetic variation in the vicinity of these loci, and resistance haplotypes subsequently invaded sub-Saharan Africa (Wootton et al. 2002; Roper et al. 2004). While dhfr and crt alleles conferring high level resistance against both drugs also emerged independently in South American and Papua New Guinea (Fidock et al. 2000; Anderson and Roper 2005; Mita 2010), the rarity of independent resistance alleles is surprising as infected patient contains up to 1011 parasites, so each infected person will contain 10–100 parasites with mutations at each position in the genome assuming standard eukaryotic SNP mutation rates. However, the rarity of mutations is consistent with measures of short-term effective population size Ne, determined from temporal fluctuation in allele frequencies: these genetic-drift based Ne estimates range from <100 (Chang et al. 2012) to several 1000s (Nkhoma et al. 2013).
The recent discovery of a major ART-R gene, kelch13, in SE Asia, in which resistance alleles have multiple independent origins (Ashley et al. 2014; Takala-Harrison et al. 2015), supports a radically different model of resistance evolution, and provides a unique opportunity to examine an ongoing selective event in a recombining eukaryote and to estimate key population parameters for Plasmodium. ART-R was confirmed in SE Asia in 2009 (Dondorp et al. 2009). ART-R parasites show 100–1,000 fold slower clearance rate from the blood than sensitive parasites over a single 48 h parasite life-cycle, and increased treatment failure rates (Spring et al. 2015; Amaratunga et al. 2016; Phyo et al. 2016). Single amino acid edits in kelch13 increase resistance of cultured parasites (Straimer et al. 2015), showing that this gene is sufficient to cause ART-R. ART is thought to kill parasites through oxidative damage; kelch13 mutations are thought to reduce interaction with phosphatidylinositol-3-kinase (PfPI3K), enhancing the unfolded protein response (UPR) and reducing sensitivity of parasites to oxidative damage (Bozdech et al. 2015).
We describe the spread of ART-R alleles in parasites collected from the Thai-Myanmar border between 2001 and 2014. Thirty-two independent mutations have spread through this population generating an extraordinarily soft selective event. However, one particular mutation now dominates, so while this sweep is soft, the signature is expected to be much harder in the future. We use these data to estimate critical parameters such as the population mutation rate (Pennings and Hermisson 2006; Messer and Petrov 2013) and the short term Ne and to ask why there should be so many independent origins of ART-R.
Results
Dataset
We collected parasite genotypes, kelch13 sequence and/or parasite clearance data from 4,641 patients visiting four clinics on the Thailand-Myanmar border (2001–2014). These included 3,878 hyperparasitaemia patients, 691 patients from an ART-mefloquine efficacy study (Phyo et al. 2016), and 72 infections from uncomplicated malaria patients (supplementary fig. S1, Supplementary Material online). After removing clearance rate data that fitted poorly to a linear model (r2 ≥ 0.8) 3,552 remained. We excluded multiple clone infections by genotyping a panel of 93 variable SNPs (Phyo et al. 2012) in the hyperparasitemia samples. We determined sequence at the kelch13 locus in 1,876 infections. In addition, to examine the impact of kelch13 on flanking variation we genotyped 75 SNPs on chr. 13 in a subset of samples.
Change in Parasite Clearance Rate
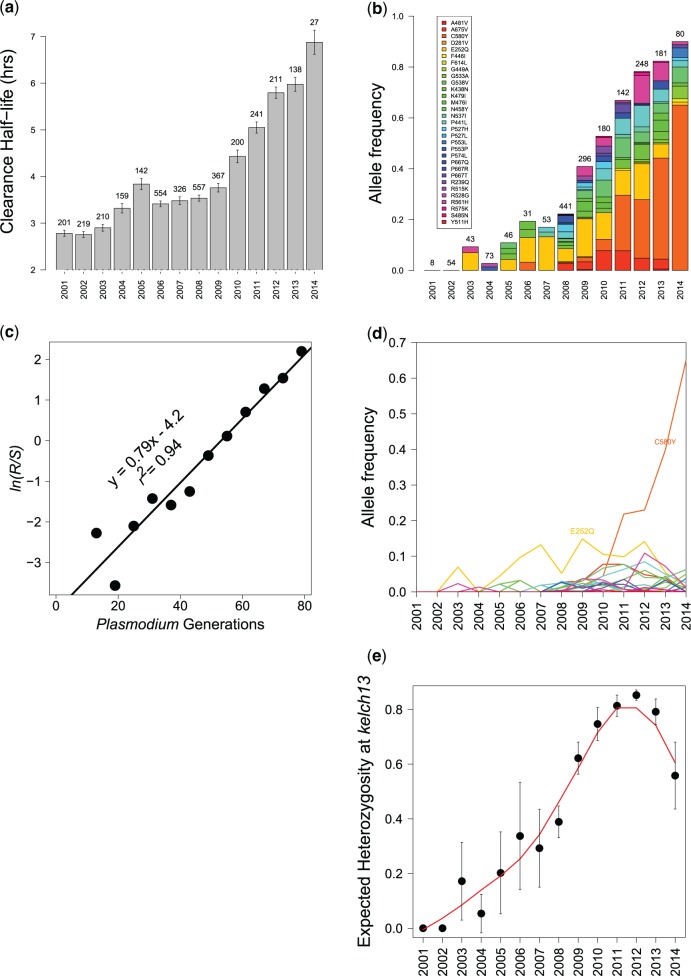
To determine clearance rate, we measured log linear decline in parasite density in blood samples collected at 6hr intervals following treatment. We used clearance rate half-life (T1/2P)—the time taken for parasite density to fall by half—as our comparison metric (Flegg et al. 2011). We previously described the change in T1/2P from 2001 to 2010 (Phyo et al. 2012). Mean T1/2P accelerates during 2011–2014, reaching over 6.0 hrs in 2014 (fig. 1A). The dynamics of T1/2P increase differ between locations (supplementary fig. S2, Supplementary Material online).
Fig. 1.
Evolution of ART-R on the Thailand–Myanmar border. (A) Change in clearance rate (T1/2P) between 2001 and 2014. (B) Frequency of different kelch13 mutations on the Thai–Myanmar border. Sample sizes are shown above the bars, and colors indicate the frequency of different alleles. The graph includes samples from both hyperparasitaemia (single genotype infections) and drug efficacy studies (all infections): these datasets are plotted independently in supplementary fig. S4, Supplementary Material online. (C) Selection coefficient (s) estimation for resistance alleles on the Thailand–Myanmar border. R and S are the frequency of kelch13 ART-S and ART-R alleles, and the slope (0.079) provides the estimation of s (Hartl and Dykhuizen 1981). Here, we assume a 6 generations per year. (D) Trajectory of different kelch13 alleles over time. (E) Expected heterozygosity (He) of kelch13 alleles over time.
Mutations in the kelch13 Locus
We observed 32 nonsynonymous SNPs in kelch13: 29 in the propeller region (1725980–1726940 bp, amino acids 419–707), and three outside the propeller (E252Q, D281V, and R239Q). There were multiple SNPs in several codons. For example P667→Q, R or T (CCA→CAA, CGA, or ACA), whereas at P527 consecutive mutations have generated H or L (CCT→CAT→CTT). kelch13 SNPs were first observed in 2003 and rose to 90% in 2014 (fig. 1B). The rise in kelch13 allele frequency tracks change in parasite T1/2P in the 4 clinics (fig. 1A and B and supplementary figs. S2 and S3, Supplementary Material online) and we see parallel changes in the hyperparasitemia and efficacy study data sets (supplementary fig. S4, Supplementary Material online). The most abundant allele before 2008 was E252Q (fig. 1B and D). C580Y allele was first observed in 2006, reached 4% frequency in 2010, and 65% by 2014 (fig. 1B and D) and was fixed or close to fixation in all clinics except MLA (supplementary fig. S3, Supplementary Material online). Only two other ART-R alleles reached >10% allele frequency (E252Q in 2007 and 2009–2012, and R561H in 2012) during the 14 year study.
Selection coefficients were 0.12, 0.079, and 0.059 respectively assuming 4, 6, or 8 parasite generations per year (fig. 1C). The analysis treats all alleles as being equivalent. The rapid rise of C580Y indicates much higher allele-specific selection coefficients: 0.24, 0.16, and 0.12 assuming 4, 6, or 8 parasite generations per year.
We measured expected heterozygosity (He) at kelch13 (the probability of drawing two different alleles). This rises from 0 in 2001 to 0.8 in 2011 and then drops to 0.5 as C580Y displaces other alleles (fig. 1E).
Genotype–Phenotype Associations
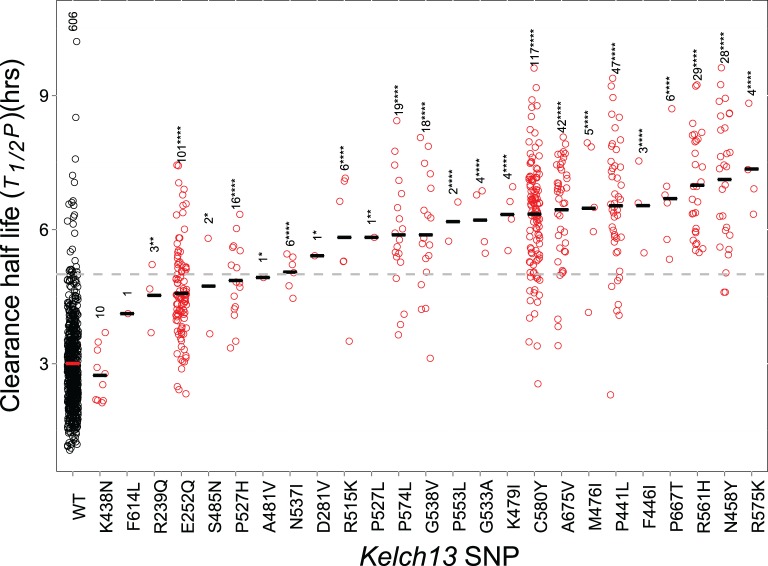
We compared T1/2P in infections carrying different kelch13 alleles (n = 1,044, fig. 2). WT parasites have low mean T1/2P, and were cleared from the bloodstream significantly faster than parasites with kelch13 mutations (t = −34.99, 3.77 × 10−165). The T1/2P distribution of WT parasites was broad (1.065–10.21 h, median = 2.86 h): however just 26/606 (4.2%) WT parasites had T1/2P ≥ 5 h. Kelch13 alleles varied significantly in T1/2P (fig. 2). Two nonsynonymous SNPs (K438N and F614L) did not show elevated T1/2P relative to WT. After removing these and WT alleles, we observed significant heterogeneity in T1/2P (F = 11.29, df = 22, P < 6.21 × 10−31). All three mutations outside the propeller region (R239Q, E252Q, and D281V) were associated with moderate T1/2P (4.53, 4.57, and 5.41 h, respectively): parasites with these mutations cleared significantly slower than WT (t = −14.44, 5.58 × 10−30), and significantly faster than parasites with C580Y (t = 11.77, 4.18 × 10−25). C580Y (T1/2P = 6.35) parasites cleared faster than several other common ART-R alleles [R561H (T1/2P = 7.00) and N458Y (T1/2P = 7.12)], although these differences were not significant after correction for multiple testing (supplementary fig. S5, Supplementary Material online).
Fig. 2.
Kelch13 alleles and parasite clearance phenotype A. Each circle represents T1/2P from a single patient. Sample sizes are shown for each mutation, and bars mark median values. The horizontal dotted line at T1/2P = 5 provides a cut-off for ART-R for categorizing clinical infections. Asterisks mark significant differences from WT (one-sided t-tests, *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
Whether additional loci in the genome impact T1/2P is an open question. We examined whether clonally indistinguishable groups of parasites isolated from different patients, but bearing the same mutation showed significant heterogeneity in T1/2P. Among the 612 WT parasites there were 88 groups of unique WT genotypes infecting 2–13 patients, but we observed no evidence for heterogeneity in T1/2P (F = 0.0373, df = 1, ns). Similarly for C580Y (n = 99, 15 groups infecting 2–6 patients, F = 0.031, df = 1, ns) and E252Q (n = 100, 18 groups infecting 2–10 patients, F = 1.1275, df = 1, ns) we found no evidence for genetic background effects on T1/2P (table 1).
Table 1.
No Evidence for Genetic Background Effects on T1/2P.
| Kelch13 genotype | n | n1 | No. MLGs | min | max | F-value | P-value |
|---|---|---|---|---|---|---|---|
| A675V | 40 | 16 | 7 | 2 | 4 | 0.08 | 0.79 |
| C580Y | 99 | 42 | 15 | 2 | 6 | 0.03 | 0.86 |
| E252Q | 100 | 53 | 18 | 2 | 10 | 1.20 | 0.28 |
| N458Y | 29 | 23 | 7 | 2 | 7 | 3.27 | 0.08 |
| P441L | 44 | 25 | 4 | 2 | 12 | 0.00 | 0.95 |
| R561H | 23 | 6 | 2 | 2 | 4 | 0.01 | 0.92 |
| WT | 613 | 244 | 86 | 2 | 13 | 0.00 | 0.97 |
n1—sample size after removal of singleton genotypes.
We compared clearance rates (T1/2P) among parasite multilocus genotypes (MLGs) carrying common kelch13 resistance mutations, represented >20 times in the dataset. For each allele, we used ANOVA to compare T1/2P among different multilocus genotypes. These analyses used parasite genotypes that were recovered from ≥2 patients. The number of MLGs (no. MLGs), the minimum (min.) and maximum (max.) number of parasites carrying each MLG, the F-value and associated P-value are shown. We found no evidence for an effect of genetic background on T1/2P.
Linkage Disequilibrium and Hitchhiking around kelch13
Soft sweeps are expected to have less impact on flanking diversity than hard sweeps, because multiple background haplotypes hitchhike with selected mutations. We measured SNP diversity (SNP-π) surrounding WT and ART-R alleles from 2008 to 2013. We observed no difference in SNP-π between parasites carrying WT and kelch13 ART-R alleles on any year examined (fig. 3A). However, when we examined SNPs surrounding individual ART-R SNPs we observed significant reduction in SNP-π relative to WT parasites for all common alleles except M461I and E252Q (fig. 3B). This was also reflected in increased linkage disequilibrium surrounding ART-R alleles (fig. 3C and D; supplementary fig. S6, Supplementary Material online). We inspected the length of haplotypes surrounding individual kelch13 alleles from the decay in expected haplotype homozygosity (EHH) (Sabeti et al. 2002). All common ART-R alleles showed elevated EHH (432–2,130 kb) relative to WT alleles (182 kb) (fig. 3D). Two alleles showed extremely long tracts of high EHH (G538V: 2446 kb; P441L: 2130 kb). E252Q has the shortest region of elevated EHH (432 kb). C580Y, which is rapidly increasing in frequency, showed moderate LD.
Fig. 3.
Genetic diversity and linkage disequilibrium around resistance alleles. (A) Mean diversity (SNP-π) for SNPs flanking kelch13 on WT (black bars) and kelch13 alleles carrying mutations (white bars). (B) Mean diversity (SNP-π) surrounding individual kelch13 alleles. (Bonferroni correct P-values: *P < 0.05, **P < 0.01, ***P < 0.001). (C) Expected haplotype homozygosity (EHH) across chr. 13. Black line shows EHH surrounding WT, whereas the red lines show EHH surrounding common kelch13 alleles. The markers are unevenly spaced on chr. 13 (supplementary fig. S6, Supplementary Material online). (D) Length (kb) of extended haplotypes surrounding kelch13 alleles. Bars show the distance at which EHH decays below 0.2 on either side of kelch13.
Multiple Origins of Individual Resistance Mutations
Different amino acid changes in ART-R alleles clearly demonstrate independent origins of ART-R. However, it is also possible that particular kelch13 mutations have arisen more than once: such independent alleles are expected to have different flanking SNP haplotypes. We used minimum spanning networks to examine haplotypes surrounding ART-R (fig. 4), focusing on 33 SNPs in a 472 kb region (1459805–1931812 bp) flanking kelch13 (supplementary fig. S6, Supplementary Material online). Three ART-R SNPs (C580Y, N458Y, and R561H) are found on divergent genotypes, suggesting 2–4 independent origins of each codon change. E252Q haplotypes are widely distributed across the minimum spanning networks. E252Q haplotypes also show limited LD (fig. 3A); for this ART-R mutation distinguishing between multiple origins and recombination is problematic.
Fig. 4.

Minimum spanning network of 332 kelch13 associated haplotypes. This analysis used 33 SNPs in a 472 kb region (1459805–1931812 bp) encompassing kelch13 to minimize impact of recombination and only common kelch13 alleles (represented ≥5 times in dataset) are included. Haplotypes are represented by filled circles, colored to show the associated kelch13 mutation. Circle size indicates number of samples represented (also marked). The grey lines linking circles indicate genetic relatedness: thin lines indicate ≥5 SNP differences, whereas thick lines indicate 1 SNP differences. C580Y, N458Y, and R561H associated haplotypes fall in differentiated haplotype groups consistent with ≥2 independent origins. E252Q alleles are also associated with multiple haplotypes—this ART-R mutation shows weak LD with surrounding SNPs (fig. 3) so we cannot exclude a single origin and subsequent recombination generating the pattern observed.
Estimation of the Mutational Target Size
The mutation target size (t) can be estimated from the number of ART-R mutations in kelch13. We observe 32 different nonsynonymous mutations in this study, of which 25 had associated T1/2P, and 23 showed slower clearance than WT parasites. The cumulative number observed in several published SE Asian studies is 75 (table 2). Note that this is a minimum estimate (t ≥ 75), because it includes only those sampled to date: rarefaction methods, based on the rate of sampling new mutations estimate t to be 107 (95% CI: 87–163) (supplementary fig. S7, Supplementary Material online).
Table 2.
Rarefaction Estimates of Total Numbers of Kelch Allele Present in SE Asian Populations.
| Study | Location | n | K13 Alleles (Obs) | K13 Alleles (Chao1 Est.) | Chao1 95% CI | Citation |
|---|---|---|---|---|---|---|
| Ashley | SE Asia | 874 | 27 | 31.19 | 27.78–49.31 | (Ashley et al. 2014) |
| Anderson | Thai-Myanmar border | 1876 | 32 | 38.99 | 33.45–66.73 | [This study] |
| Nyunt | Myanmar | 29 | 16 | 24.68 | 18.05–52.82 | (Nyunt et al. 2015) |
| Wang | China-Myanmar border | 191 | 18 | 22.64 | 18.94–40.72 | (Wang et al. 2015) |
| Tun | Myanmar | 940 | 29 | 40.98 | 31.65–83.14 | (Tun et al. 2015) |
| Takala-Harrison | SE Asia | 303 | 13 | 15.99 | 13.38–35.93 | (Takala-Harrison et al. 2015) |
| Amato | SE Asia | 1600 | 35 | 42.99 | 36–74.43 | (MalariaGEN Plasmodium falciparum Community Project 2016) |
| Huang | Southern China | 302 | 15 | 21.97 | 16.44–48.64 | (Huang et al. 2015) |
| All studies* | SE Asia | 6115 | 75 | 106.99 | 86.57–163.47 |
Note.—Survey of kelch13 mutations provide minimum estimates of the numbers of ART-R alleles present. We estimated actual numbers present from the rate of samples new mutations using rarefaction. This table shows results displayed graphically in supplementary fig. S4, Supplementary Material online. We used individual based analysis and sampling without replacement 100 times to estimate the number of different kelch13 alleles present in population from the observed data for this and several published studies. We used the Chao 1 estimator (the ACE and Chao1 estimates were identical) calculated using EstimateS v9.0.0 (http://viceroy.eeb.uconn.edu/estimates/) for these analyses. To estimate the total number of kelch13 alleles present in SE Asia from these population samples, we used the same software, and a sample based analysis. Observed and estimated numbers of alleles present in each population sample and in across all samples are shown in the summary table. The estimated numbers of alleles provides an estimate for the “mutation target size” (t) used for modeling emergence of ART-R (fig. 5 and table 4).
Chao1 Estimator used.
Estimation of Population Mutation Parameter and Short Term Ne
We estimated Θ (Pr Θ) given the observed numbers of independent ART-R alleles from 2008 to 2014 (fig. 5). Θ declines from 5.69 (95% CI: 3.32–10.61) in 2008 to 3.38 (95% CI: 1.76–7.34) in 2014 on the Thai–Myanmar border. Similar values are obtained from published studies of kelch13 allele frequencies in SE Asia (table 3). These are lower bounds, because we have not accounted for resistance alleles that have arisen more than once.
Fig. 5.
Population mutation parameter estimation. (A) Θ (=2Neµt) estimates from the number of different ART-R alleles observed in 2008. Dotted lines show the estimate (red text), and the 95% confidence errors. (B) Θ (and 95% C.I.) plotted between 2008 and 2014. Θ declines during this period. (C) Predicted evolution of kelch13 alleles on the Thai–Myanmar border based on Θ and s estimates (s = 0.079) from the current dataset. The parameter values are those listed in table 4. Time to establishment (Te) and fixation (Tfix) are estimated following (Messer and Petrov 2013). The colors indicate independent beneficial alleles and are illustrative only: the trajectories of individual alleles are not modeled here.
Table 3.
Estimates of the Population Mutation Parameter (Θ) from the Numbers of Alleles Observed within Population Subsamples.
| Study | Location | Theta Estimates |
|||
|---|---|---|---|---|---|
| n | k | theta | 95% CI | ||
| Ashley (Ashley et al. 2014)† | SE Asia | 100 | 17 | 5.63 | 3.29–10.49 |
| Anderson [This study] | Thai-Myanmar border | 98 | 17 | 5.69* | 3.32–10.61 |
| Nyunt (Nyunt et al. 2015) | Myanmar | 29 | 16 | 13.88 | 7.29–28.16 |
| Wang (Wang et al. 2015) | China-Myanmar border | 100 | 13 | 3.77 | 2.07–7.55 |
| Tun (Tun et al. 2015) | Myanmar | 100 | 17 | 5.63 | 3.29–10.49 |
| Takala-Harrison (Takala-Harrison et al. 2015) | SE Asia | 100 | 9 | 2.21 | 1.09–5 |
| (MalariaGEN Plasmodium falciparum Community Project 2016) † | SE Asia | 100 | 13 | 3.77 | 2.07–7.55 |
| Huang (Huang et al. 2015) | Southern China | 100 | 14 | 4.21 | 2.35–8.24 |
Note.—We estimated Θ (=2Neµt) from the number of different resistant alleles found in samples of parasites described in this and several published studies in SE Asia (see text and fig. 5 for details). Θ values range from 2.21 to 13.88. The high value calculated for the Nyunt et al. (2015) study in Myanmar may reflect low sample size.
Θ estimate from 2008: see fig. 5.
Includes some samples that are shared between these two publications.
We can estimate Ne during the time when selection is acting from the population mutation parameter, because Θ = 2Neμt and therefore Ne = Θ/2μt) (Karasov et al. 2010; Pennings et al. 2014). Using estimates of mutation rate (5.10 × 10−8 to 1.15 × 10−7 per nucleotide per generation) and target size (t = 87–163 bp), we determined short term Ne to be from 88,017 to 1,195,628 on the Thai–Myanmar border (table 4).
Table 4.
Summary of Parameters used for Estimation of Population Mutation Parameter (Θ) and Ne.
| Parameter | Estimate | Lower Bound | Upper Bound | Notes | Citation |
|---|---|---|---|---|---|
| Mutation rate/base/48 h asexual cycle | 3.39E−09 | 1.7E−09 | 3.82E−09 | Average values from 5 mutation accumulation experiments: upper and lower bounds are highest and lowest values recorded | (Bopp et al. 2013; Claessens et al. 2014) |
| Mutation rate/base/generation (μ) | 1.02E−07 | 5.10E−08 | 1.15E−07 | Assumes 6 generations/year (i.e. 2 month generation time from mosquito to mosquito). | (Bopp et al. 2013; Claessens et al. 2014) |
| Mutation target size (bp) (t) The number of base changes in kelch13 that result in ART-R | 106.99 | 86.57 | 163.47 | Estimates derived by rarefaction (table 2 and supplementary fig. S7, Supplementary Material online) using published datasets and this study | |
| Population mutation parameter (Θ) | 5.69 | 3.32 | 10.61 | Estimated following (Pennings and Hermisson 2006) from observed number of kelch13 alleles (k) in samples of size n from the Thai–Myanmar border (fig. 5). Value from 2008 shown | This study |
| Ne (estimated from Θ) | 2.61E+05 | 8.80E+04 | 1.20E+06 | Estimated from the parameters listed above: Ne = Θ/2μt | This study |
Discussion
Mutations Outside the Propeller Domain Are Associated with Slow Clearance
The current molecular screening approach for ART-R involves sequencing the kelch13 propeller (1,725,980–1,726,940 bp, residues 419–707): this is where nonsynonymous mutations associated with slow clearance were originally discovered in Cambodian parasites (Ariey et al. 2014). On the Thai–Myanmar border, the most common mutation prior to 2011 was E252Q. This mutation is 167 amino acids 5′ of the propeller and is associated with slower clearance than WT parasites, and elevated treatment failure rates (Phyo et al. 2016). Two other mutations (D281V and R239Q) close by are also associated with slow clearance, arguing that molecular surveillance for resistance should be expanded outside the kelch13 propeller.
Of 25 nonsynonymous mutations for which clearance rate data was available, 23/25 (92%) were associated with slower clearance than WT parasites: the vast majority of kelch13 SNPs influence phenotype. SNPs differed significantly in associated clearance rate. E252Q shows slower clearance than WT, but faster clearance than the majority of propeller mutations including C580Y. ART-R alleles also varied in their impact on treatment failure (Phyo et al. 2016), which should be directly linked to their capacity to spread. Phenotypic heterogeneity among ART-R alleles suggests that kelch13 mutations cause partial rather than complete loss of function. C580Y is of special interest as this allele is approaching fixation in this and other SE Asian parasite populations (Ariey et al. 2014; Ashley et al. 2014). The success of this allele may stem from the fact that C580Y disrupts of an internal disulphide bond, rather than changing the size or charge of amino acids exposed on the kelch13 surface.
One explanation for heterogeneity in clearance rate might be involvement of other loci in the genome (Miotto et al. 2015). We found no evidence for genetic background effects on clearance for common kelch13 alleles by comparing within and between clone variance in clearance rate. We emphasize that this does not rule out involvement of other loci in other aspects of resistance such as fitness costs. A longitudinal genome sequencing study on a subset of the parasites examined here identified several regions of the genome that show rapid changes in SNP frequency, identifying several putative genome regions that may be selected by ART (Cerqueira et al. 2015). Similarly, previous analyses have identified genome regions distant from kelch13 associated with ART-R (Cheeseman et al. 2012; Miotto et al. 2013; Takala-Harrison et al. 2013; Cheeseman et al. 2015; Miotto et al. 2015).
We observed 26/606 (4.2%) WT parasites had T1/2P ≥ 5 h. Inadequate dosing is unlikely to explain this because treatment was supervised. We cannot exclude defects in metabolism of artesunate, or alternative pathways to generate ART-R. We are currently analyzing a genetic cross (Vaughan et al. 2015) involving one such non-kelch13 ART-R parasite to investigate this further.
Retrospective Studies Underestimate Complexity of Selection Events
Kelch13 heterozygosity rises until 2012 then decreases as C580Y reaches high frequency (fig. 1D and E). Published datasets from Western Cambodia also support a model of diminishing variation at kelch13 over time. For example, kelch13 heterozygosity diminished in Pursat, Battambang, and Pailin in Western Cambodia between 2001 and 2012 (Ariey et al. 2014). As kelch13 resistance alleles reached high frequencies in western Cambodia several years before the Thailand–Myanmar border, they provide a snapshot of what we expect will occur on the Thai–Myanmar border.
Diminishing complexity of selective events at dihydrofolate reductase (dhfr) is also observed during evolution of pyrimethamine resistance in both Africa and Asia. On the Thai–Myanmar border, there were several independent origins of S108N mutation, the first step of resistance evolution (Nair et al. 2003). However, high level resistance involving N51I, C59R, and I164L was only associated with a single chr. 4 haplotype. Similarly, in Africa, there were multiple origins of parasites containing a single mutation (S108N), and three separate origins of parasites carrying double mutant (N51I/S108N or C59R/S108N), but these have now been replaced by triple mutant parasites originating in Asia (Anderson and Roper 2005). In this case, sequential sweeps involving several mutations on the same background result in a hard sweep.
Both kelch13 and dhfr datasets support a model in which selection events are soft initially, but rapidly become harder, as particular alleles dominate. Continuing surveillance of kelch13 on the Thai–Myanmar border will determine whether this trend continues. Most theoretical work assumes that beneficial alleles selected during soft selective sweeps are functionally equivalent, and whether particular beneficial mutations fix is dependent on genetic drift rather than selection (Pennings and Hermisson 2006): it would be useful to explore the impact of fitness heterogeneity on the trajectory of alleles in future theoretical investigations of soft sweeps. Demography alone can result in loss of adaptive alleles (Wilson et al. 2014). However, in the malaria examples, selective fixation of high fitness ART-R alleles is likely to be responsible for the hardening of the sweep. Kelch13 alleles are not functionally equivalent and differ in both clearance and ability to survive treatment (Phyo et al. 2016). That independent C580Y alleles have spread to high frequency in different countries in SE Asia also suggests that this allele has higher fitness than other ART-R alleles. More generally, we suggest that retrospective studies of extant populations are likely to underestimate the frequency and complexity of soft selective events.
Comparable evidence for sweeps that transition from soft to hard comes from laboratory selection with methylobactarium in which multiple adaptive alleles initally arose, but one high fitness allele eventually replaced the others (Lee and Marx 2013). If this pattern in common in nature, it has important implications for those interpreting retrospective studies. Sweeps, such as those observed in methylobactarium may eventually leave a hard signature but the bloom of independent adaptive alleles that initially arose clearly demonstrates that mutations were not limiting during adaptation.
An interesting feature of these data is that ART-R alleles with moderate fitness (e.g., E252Q) spread initially, but C580Y replaces other ART-R alleles after 2011. These data are consistent with both theoretical and empirical work showing that the distribution of fitness effects of beneficial mutations is exponential: whereas multiple mutations can generate phenotypes with modest increase in fitness, only a small subset show high fitness (Orr 2003; Kassen and Bataillon 2006). The waiting time for rare high fitness alleles to arise and establish is expected to be greater than for alleles of moderate fitness. We speculate that the rarity of high fitness ART-R alleles such as C580Y may explain why this allele appeared late, but subsequently swept through the population on the Thai–Myanmar border.
Population Mutation Parameter and Ne Estimates
Ne is the central parameter that determines the level of genetic drift in idealized populations, and the rate at which adaptive mutations are introduced into populations (Pennings and Hermisson 2006; Messer and Petrov 2013), but it is difficult to measure directly. Short term Ne can be inferred in natural populations by measuring variation in allele frequencies over time (i.e., by quantifying genetic drift). This approach gave average Ne estimate of 1965 on the Thai–Myanmar border (Nkhoma et al. 2013) and <100 in Senegal (Chang et al. 2012). We can also infer Θ from the numbers of independent adaptive alleles observed in our population sample and use this parameter to infer short-term Ne. This approach to Ne estimation from Θ follows work on Drosophila (Karasov et al. 2010) and HIV (Pennings et al. 2014) and utilizes the fact that Θ = 2Neμt (and therefore Ne = Θ/2μt). Θ ranges from 5.9 to 3.9 on the Thai–Myanmar border (2008–2014) and we obtain similar estimates from other SE Asian locations (table 3). Using estimates of mutation rate (5.10 × 10−8 to 1.15 × 10−7 per nucleotide per generation) and target size (87–163 bp), we determined short term Ne to be from 88,017 to 1,195,628 on the Thai–Myanmar border (table 4). These short term Ne estimates are 52–705 times greater than previous variance Ne estimates from the same parasite population (Nkhoma et al. 2013) and 9–120 times greater than long-term Ne (∼10,000) in SE Asia based on mtDNA nucleotide diversity (Joy et al. 2003). One caveat is that we may have underestimated the mutational target size (t). To provide a generous upper bound on the target size we can assume that mutations in all 1771 nonsynonymous sites within kelch13 can generate resistance alleles. Even with this obvious overestimate of t, Ne = 15,479 which is 9-fold greater than variance Ne values.
Two recent papers have challenged the application of classical population genetics models to malaria parasites (Chang et al. 2013; Chang and Hartl 2015). Classical population genetic models make Wright–Fisher assumptions that populations are constant in size, have nonoverlapping generations, and that each new generation is sampled from gametes produced by the previous generation. In contrast, the malaria parasite lifecycle consists of very large populations (≤1011) of asexually dividing blood stage forms (equivalent to gametes in a conventional diploid organism) within humans, from which small numbers of gametes (≤10) are sampled by mosquitoes each generation. Simulation studies demonstrate that genetic drift is amplified in the malaria lifecycle relative to the Wright–Fisher model because small numbers of gametes are sampled from infected people, whereas selection is amplified because there are large numbers of parasites within patients (Chang et al. 2013). Hence, drift-based measures of Ne, using fluctuation in allele frequencies, reflect most closely the numbers of parasites passing through the mosquito bottleneck, and underestimate Ne relevant for adaptation. We suggest that the decoupling of drift and selection may explain the discrepancy between Ne estimates for malaria, and that the Ne relevant for adaptation can best be estimated from Θ. Similar lifecycles are found in many pathogens and parasites: evaluation of the potential bias in applying classical population genetic tools to these organisms is urgently needed. Previous analyses have suggested that the availability of mutations limits drug resistance evolution in Plasmodium (Anderson and Roper 2005): the large short-term Ne values inferred here suggest we have underestimated the capacity for adaptive evolution in Plasmodium.
A Spectrum of Selective Events in Plasmodium
Evolutionary analyses of additional resistance loci in SE Asian parasite populations reveal a spectrum of sweeps ranging from hard to soft. We use estimates of Θ to better understand this spectrum. Both the chloroquine resistance transporter (crt) and dhfr provide examples of hard selective sweeps in SE Asia. A single allele (K76T) at crt spread through SE Asian malaria populations, purging genetic variation flanking crt, and this allele subsequently also spread across Africa (Wootton et al. 2002). In the case of dhfr, 2–4 mutations conferring resistance are found on the Thai–Myanmar border. However, all of these mutations share the same flanking haplotypes, consistent with a series of hard selective sweeps in which mutations arose on the same sweeping haplotype (Nair et al. 2003). Resistance at both crt and dhfr involves single SNP mutations, so the target size (t) is 1. As a consequence, Θ will be reduced ∼100-fold relative to kelch13 to ∼0.05. In this range of Θ values, multiple coexisting alleles are expected to be rare. Hence, hard sweeps at dhfr and crt are dependent on target size. Resistance alleles at the multidrug transporter (mdr1), GTP cyclohydrolase I (gch1) have multiple independent origins. Both mdr1 and gch1 involve independent origins of copy number variants (CNVs). There are 5–15 independent origins of CNVs containing mdr1 (Nair et al. 2007), and at least three independent origins of CNVs containing gch1 (Nair et al. 2008). The elevated mutation rate of CNV compared with SNPs most likely explains the soft sweeps at these adaptive CNVs. Mutation rates at CNVs are 5–7 orders of magnitude greater than SNPs in mammals (Inoue and Lupski 2002) and rates 10-fold greater than SNPs have been estimated at P. falciparum pfmdr1 (Preechapornkul et al. 2009).
Implications for Resistance Evolution and Management
Our re-estimation of Ne in SE Asia has several implications for resistance evolution and management. ART combination therapy was introduced on the Thai–Myanmar border in 1995, resistance was first suspected in 2008, and ART-R alleles have now almost fixed. When did resistance alleles first become established? The time to establishment (Te)—when resistance alleles first escape elimination by genetic drift—is: (Messer and Petrov 2013). Hence, Te is 2.23 generations or 4.66 months (assuming 6 generations/year) for this parasite population, suggesting that resistance alleles became established within a year of ART deployment. This is 8 years before ART-R alleles were first observed in this dataset (figs. 1 and 4), and 13 years before ART-R was suspected in SE Asia (Dondorp et al. 2009). The average number of ART-R mutations arising between 1995 and fixation is , where (Pennings and Hermisson 2006). Hence, with our estimated parameters (table 4) ART-R mutations will have arisen 765 times during this selective event.
Artemisinin-derivatives are used in combination with other drugs such as lumefantrine, mefloquine, and piperaquine to limit resistance evolution. This policy appears to have worked poorly in limiting ART-R evolution given that ART resistance has evolved multiple times. The use of partner drugs (e.g., mefloquine and lumefrantine) to which resistance has already emerged is one contributor to the failure of ACTs. Our results suggest that antimalarial combinations will need to contain more than two fully effective drugs to effectively constrain resistance evolution. This conclusion is similar to that reached by HIV researchers (Pennings 2013). Resistance breakthroughs occurred rapidly within patients treated with early HIV drug cocktails with multiple origins of resistance occurring within single infections. As HIV drug combinations increased in complexity, drug breakthroughs have become increasingly rare and drug resistance associated sweeps significantly harder (Feder et al. 2016). Triple antimalarial combinations (artesunate/mefloquine/piperaquine and artesunate/lumefantrine/amodiaquine) are now under clinical trials, but use of multidrug combinations is severely limited by the availability of effective drugs.
The evolution of ART-R demonstrates the importance of mutational target size for resistance evolution. Target size can be incorporated into evaluations of promising new antimalarials prior to deployment. Laboratory selection of cultured parasites using new antimalarials is now routinely carried out to evaluate the potential for resistance evolution (Flannery et al. 2013). Such studies can be used both to identify genes that underlie resistance, but also to determine how many nucleotides can generate resistance phenotype. An exciting approach to target size evaluation might be to use CRISPR/Cas9 approaches with degenerate homology regions to generate a population of mutations across a target gene, and then evaluate what fraction of these mutations responds to drug selection. In essence, this approach increases mutation rate in the target gene to allow thorough evaluation of mutational target size.
Finally, malaria parasites provide an empirical perspective on the current debate about the importance of soft versus hard sweeps in evolution (Jensen 2014). At a global scale all six known antimalarial resistance genes (dhfr, dihydopteroate synthase, gch1, pfcrt, mdr1, and kelch13) show multiple origins of resistance alleles (Fidock et al. 2000; Roper et al. 2003; Nair et al. 2007, 2008; Pearce et al. 2009). At a more local level, kelch13 mutations and alleles conferring low level dhfr resistance evolved multiple times in SE Asia, but the spread of high fitness alleles has lead to a hard sweep at dhfr (Nair et al. 2003; Roper et al. 2004) and hardening of the sweep at kelch13. Therefore, signatures of hard sweeps in extant populations do not necessarily suggest that sweeps were originally hard or that adaptation is limited by the availability of mutations. We suggest that retrospective inference of selective events from sequence variation may a weak approach to resolve this debate.
Materials and Methods
Study Site and Patients
We collected P. falciparum infected blood samples [blood spots or white-cell depleted venous blood samples (Venkatesan et al. 2012)] from patients visiting malaria clinics run by the Shoklo Malaria Research Unit (SMRU) from 2001 to 2014. The four clinics sampled, which span ∼100 km along the western border of Thailand, were Maela (refugee camp) and Wang Pha village to the north of Maesot, and Mae Kon Khen and Mawker Thai villages to the south of Maesot (Phyo et al. 2012). The majority of patients came from adjacent Myanmar. The parasite samples came from three different groups of patients: (a) uncomplicated hyperparasitaemia patients (≥4% red cells infected). These patients were treated with artesunate (ART) alone followed by combination therapy (Phyo et al. 2012). We then monitored parasite clearance at 6 h intervals until the patient was slide negative. (b) Uncomplicated malaria patients (<4% red cells infected) included in drug efficacy studies (2003–2013) (Phyo et al. 2016). Parasite clearance was not measured in these patients. (c) In 2014, we supplemented the numbers of samples by collecting blood spots from uncomplicated malaria patients admitted for treatment at SMRU clinics, because P. falciparum cases are declining rapidly in this region (Carrara et al. 2013; Nkhoma et al. 2013). Ethical approval was given by the Oxford Tropical Research Ethics Committee (OXTREC 562-15) and the Faculty of Tropical Medicine, Mahidol University (MUTM 2015-019-01).
Measurement of Parasite Clearance Half-Life
Slow parasite clearance from the blood of ART-treated patients is a hallmark of ART-R (Dondorp et al. 2010). We measured the parasite clearance in all hyperparasitaemia patients by plotting log parasite density against time (Flegg et al. 2011) using the online parasite clearance calculator (http://www.wwarn.org/tools-resources/toolkit/analyse/parasite-clearance-estimator-pce). We used the slope of the linear phase to determine the clearance half-life (T1/2P)—the time for parasitaemia to fall by half. We excluded clearance curves showing a poor fit (r2 < 0.8) to the log-linear model.
Genotyping of Parasites
We extracted DNA from blood spots using a two-step protocol: blood was eluted from six 3 mm punches (Gensolve kit, GenVault Corporation) and DNA extracted using QIAamp DNA Kits (Qiagen). Whole blood samples were extracted using the Gentra PureGene kit (QIAGEN).
We genotyped parasite infections from hyperparasitemic patients using 96-SNPs distributed across the P. falciparum genome using the Illumina Veracode platform. We considered infections to contain multiple clones if >5 SNPs showed heterozygous base calls. This threshold removes obvious multiple genotype infections whereas retaining samples with low numbers of heterozygous base calls due to genotyping error (Nkhoma et al. 2013). We used single clone infections for subsequent molecular analyses, because haplotypes can be unambiguously constructed in single clone infections with haploid parasites.
To further assess haplotype structure around kelch13, we genotyped an additional 163 SNPs using Illumina Veracode from a subset of samples (single clone infections from the hyperparasitemia study, and all samples from the treatment efficacy study). The 163 SNPs comprised 75 SNPs on chr 13 and 88 SNPs on other chromosomes. The SNPs were located in genome regions showing association with ART-R in association (Cheeseman et al. 2012) and pooled sequencing studies (Cheeseman et al. 2015), so are not randomly ascertained.
Sequencing the kelch13 Locus
We amplified three PCR fragments from the kelch13 locus and sequenced products in both directions on an ABI 3730 capillary sequencer (supplementary table S1, Supplementary Material online).
Analysis
Analyses were conducted using R version 3.2.3.
Selection Coefficients
We measured selection coefficients (s) for kelch13 mutations by plotting log(R/S) over time and measuring the slope. Time was measured in Plasmodium generations (the time taken for parasites to traverse both mosquito and human lifecycle stages). The length of a Plasmodium generation varies and is dependent on multiple factors such as gametocyte production and vector abundance. We used 4, 6, or 8 generations/year.
Modeling the Selective Sweep
Whether selective sweeps are soft or hard is dependent on the rate at which beneficial mutations are introduced into the population. This is determined by the population size during the period in which selection is acting, and the rate at which beneficial mutations are generated. We examined the fit of our data to theoretical predictions (Pennings and Hermisson 2006; Messer and Petrov 2013) for soft sweeps. The critical parameter predicting whether soft selective sweeps will occur is the population mutation parameter (Θ = 2Neμt), where Ne is the effective population size during drug selection (here between 1995, when ART treatment was introduced and the present where ART-R alleles are close to fixation), μ is the per nucleotide mutation rate per Plasmodium generation (the complete lifecycle including mosquito and human development), and t (the mutational “target” size) is the number of nucleotides at which mutations can generate a ART-R.
Θ can be determined directly from the number of independent origins of ART-R mutations in kelch13 in our population sample using Equation (12) in Pennings and Hermisson (Pennings and Hermisson 2006):
where is Stirling’s number of the first kind, n is the sample size and k is the number of haplotypes observed. We determined 95% confidence intervals from the probability distribution across a range of Θ values. Since Θ = 2Neμt, we estimated Ne using the mutation rate (μ) and target size (t): Ne = Θ/2μt.
The mutation rate (μ) has been measured in two mutation accumulation experiments (Bopp et al. 2013; Claessens et al. 2014). These estimates range from 1.7–3.2 × 10−9 (Bopp et al. 2013) to 3.82–4.28 × 10−10 per base/48 h asexual cycle (Claessens et al. 2014). The estimates are mutation rates for a single 48 h asexual parasite cycle. We use the mean mutation rate (3.39 × 10−9, range: 1.7–4.28 × 10−9) and converted per cycle rates to per generation mutation rates by multiplying by g/2, where g is the length in days of a malaria parasite generation (the duration of the complete parasite lifecycle, from mosquito to mosquito), and 2 days is the length of a single asexual cycle. We assumed 6 generations per year so g = ∼60 days. We assume that the cell division rate is the same in mosquito and human sections of the parasite lifecycle.
We estimated the mutational target size (t)—the number of bases at which mutations can generate ART-R—by counting the different mutations associated with ART-R in this study and in other papers describing sequence variation in kelch13. We then used rarefaction (Colwell 2013) to estimate the number of possible resistance alleles, from the rate at which new alleles are discovered.
Measurement of Parasite Clearance Half-Life
Slow parasite clearance from the bloodstream of infected patients is a hallmark of ART-R (Dondorp et al. 2010) and was determined in all hyperparasitaemia patients. We measured the rate of parasite clearance by plotting log parasite density against time using a standardized fitting method which separates the variable initial lag-phase from the subsequent log-linear decline (Flegg et al. 2011) using the online parasite clearance calculator (http://www.wwarn.org/tools-resources/toolkit/analyse/parasite-clearance-estimator-pce). We measured the slope of the linear phase and determined the parasite clearance half-life (T1/2P), defined as the time required for parasitaemia to fall by half during the log-linear decline. We excluded parasite clearance curves showing a poor fit (r2 < 0.8) to the log-linear model.
Genotype–Phenotype Associations
We compared clearance half-life (T1/2P) of parasites bearing kelch13 mutations with wild-type alleles using one-sided t-tests. We used uncorrected tests to avoid needless rejection of the null hypothesis (i.e., no difference in T1/2P) due to low sample size (8 mutations are found in ≤3 samples). In addition, we examined pair wise differences in T1/2P between different kelch13 alleles to determine allele specific variation in clearance: in this case Bonferroni correction was used.
SNP Variation on Chromosome 13
We measured extended haplotype homozygosity (EHH) using rehh (Gautier and Vitalis 2012) to examine hitchhiking associated with different kelch13 alleles. We computed SNP- π for each bi-allelic SNP: this is defined as the average proportion of pairwise differences at assayed SNP loci within a defined population (Neafsey et al. 2008). Average SNP-π across SNPs was then computed for SNPs flanking kelch13 to investigate changes in genetic variation over time. We used a single representative of each 93-locus genotype for this analysis, to avoid artificially increasing EHH. We measured expected heterozygosity (He) at the kelch13 locus by treating kelch13 as a single locus with multiple alleles. To evaluate evidence for independent origins of particular ART-R mutations, we used poppr (Kamvar et al. 2014) with default settings to construct minimum spanning networks comparing the relationships between different SNP haplotypes for the chr.13 region surrounding kelch13.
Supplementary Material
Supplementary figures S1–S7 and table S1 are available at Molecular Biology and Evolution online.
Acknowledgments
We thank the National Institutes for Health (R37 AI048071 (TJCA)) and the Bill and Melinda Gates Foundation for funding. Comments from Pleuni Pennings, Philipp Messer, and Dan Neafsey improved the manuscript. This work was conducted in facilities constructed with support from Research Facilities Improvement Program grant C06 RR013556 from the National Center for Research Resources. SMRU is part of the Mahidol Oxford University Research Unit supported by the Wellcome Trust of Great Britain.
References
- Amaratunga C, Lim P, Suon S, Sreng S, Mao S, Sopha C, Sam B, Dek D, Try V, Amato R, et al. 2016. Dihydroartemisinin-piperaquine resistance in Plasmodium falciparum malaria in Cambodia: a multisite prospective cohort study. Lancet Infect Dis. 16:357–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson T, Nkhoma S, Ecker A, Fidock D. 2011. How can we identify parasite genes that underlie antimalarial drug resistance? Pharmacogenomics 12:59–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson TJ, Roper C. 2005. The origins and spread of antimalarial drug resistance: lessons for policy makers. Acta Trop. 94:269–280. [DOI] [PubMed] [Google Scholar]
- Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, Kim S, Duru V, Bouchier C, Ma L, et al. 2014. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505:50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Mao S, Sam B, et al. 2014. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 371:411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso-Batista J, Sousa A, Lourenco M, Bergman ML, Sobral D, Demengeot J, Xavier KB, Gordo I. 2014. The first steps of adaptation of Escherichia coli to the gut are dominated by soft sweeps. PLoS Genet. 10:e1004182.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bopp SE, Manary MJ, Bright AT, Johnston GL, Dharia NV, Luna FL, McCormack S, Plouffe D, McNamara CW, Walker JR, et al. 2013. Mitotic evolution of Plasmodium falciparum shows a stable core genome but recombination in antigen families. PLoS Genet. 9:e1003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdech Z, Ferreira PE, Mok S. 2015. A crucial piece in the puzzle of the artemisinin resistance mechanism in Plasmodium falciparum. Trends Parasitol. 31:345–346. [DOI] [PubMed] [Google Scholar]
- Carrara VI, Lwin KM, Phyo AP, Ashley E, Wiladphaingern J, Sriprawat K, Rijken M, Boel M, McGready R, Proux S, et al. 2013. Malaria burden and artemisinin resistance in the mobile and migrant population on the Thai-Myanmar border, 1999-2011: an observational study. PLoS Med. 10:e1001398.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerqueira GC, Cheeseman IH, Nair S, McDew-White M, Melnikov A, Nosten F, Anderson TJ, Neafsey DE. 2015. Retrospective longitudinal genomic surveillance of Plasmodium falciparum malaria parasites documents the emergence of artemisinin resistance in Thailand. Abstract 583. October 25–29; Annual Meeting of the American Society of Tropical Medicine and Hygiene, Philadelphia.
- Chang HH, Hartl DL. 2015. Recurrent bottlenecks in the malaria life cycle obscure signals of positive selection. Parasitology 142 Suppl 1:S98–S107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HH, Moss EL, Park DJ, Ndiaye D, Mboup S, Volkman SK, Sabeti PC, Wirth DF, Neafsey DE, Hartl DL. 2013. Malaria life cycle intensifies both natural selection and random genetic drift. Proc Natl Acad Sci U S A. 110:20129–20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HH, Park DJ, Galinsky KJ, Schaffner SF, Ndiaye D, Ndir O, Mboup S, Wiegand RC, Volkman SK, Sabeti PC, et al. 2012. Genomic sequencing of Plasmodium falciparum malaria parasites from Senegal reveals the demographic history of the population. Mol Biol Evol. 29:3427–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IH, McDew-White M, Phyo AP, Sriprawat K, Nosten F, Anderson TJ. 2015. Pooled sequencing and rare variant association tests for identifying the determinants of emerging drug resistance in malaria parasites. Mol Biol Evol. 32:1080–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IH, Miller BA, Nair S, Nkhoma S, Tan A, Tan JC, Al Saai S, Phyo AP, Moo CL, Lwin KM, et al. 2012. A major genome region underlying artemisinin resistance in malaria. Science 336:79–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessens A, Hamilton WL, Kekre M, Otto TD, Faizullabhoy A, Rayner JC, Kwiatkowski D. 2014. Generation of antigenic diversity in Plasmodium falciparum by structured rearrangement of Var genes during mitosis. PLoS Genet. 10:e1004812.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell RK. 2013. EstimateS: statistical estimation of species richness and shared species from samples. Version 9. [cited 2013 Apr 12] Available from: http://purl.oclc.org/estimates.
- Dondorp AM, Yeung S, White L, Nguon C, Day NP, Socheat D, von Seidlein L. 2010. Artemisinin resistance: current status and scenarios for containment. Nat Rev Microbiol. 8:272–280. [DOI] [PubMed] [Google Scholar]
- Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, et al. 2009. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 361:455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder AF, Rhee SY, Holmes SP, Shafer RW, Petrov DA, Pennings PS. 2016. More effective drugs lead to harder selective sweeps in the evolution of drug resistance in HIV-1. Elife 5:10670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, Ursos LM, Sidhu AB, Naude B, Deitsch KW, et al. 2000. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 6:861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannery EL, Fidock DA, Winzeler EA. 2013. Using genetic methods to define the targets of compounds with antimalarial activity. J Med Chem. 56:7761–7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flegg JA, Guerin PJ, White NJ, Stepniewska K. 2011. Standardizing the measurement of parasite clearance in falciparum malaria: the parasite clearance estimator. Malar J. 10:339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier M, Vitalis R. 2012. rehh: an R package to detect footprints of selection in genome-wide SNP data from haplotype structure. Bioinformatics 28:1176–1177. [DOI] [PubMed] [Google Scholar]
- Hartl DL, Dykhuizen DE. 1981. Potential for selection among nearly neutral allozymes of 6-phosphogluconate dehydrogenase in Escherichia coli. Proc Natl Acad Sci U S A. 78:6344–6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Takala-Harrison S, Jacob CG, Liu H, Sun X, Yang H, Nyunt MM, Adams M, Zhou S, Xia Z, et al. 2015. A single mutation in K13 predominates in southern China and is associated with delayed clearance of Plasmodium falciparum following artemisinin treatment. J Infect Dis. 212:1629–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Lupski JR. 2002. Molecular mechanisms for genomic disorders. Annu Rev Genomics Hum Genet. 3:199–242. [DOI] [PubMed] [Google Scholar]
- Jensen JD. 2014. On the unfounded enthusiasm for soft selective sweeps. Nat Commun. 5:5281.. [DOI] [PubMed] [Google Scholar]
- Joy DA, Feng X, Mu J, Furuya T, Chotivanich K, Krettli AU, Ho M, Wang A, White NJ, Suh E, et al. 2003. Early origin and recent expansion of Plasmodium falciparum. Science 300:318–321. [DOI] [PubMed] [Google Scholar]
- Kamvar ZN, Tabima JF, Grunwald NJ. 2014. Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. Peer J. 2:e281.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasov T, Messer PW, Petrov DA. 2010. Evidence that adaptation in Drosophila is not limited by mutation at single sites. PLoS Genet. 6:e1000924.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassen R, Bataillon T. 2006. Distribution of fitness effects among beneficial mutations before selection in experimental populations of bacteria. Nat Genet. 38:484–488. [DOI] [PubMed] [Google Scholar]
- Lee MC, Marx CJ. 2013. Synchronous waves of failed soft sweeps in the laboratory: remarkably rampant clonal interference of alleles at a single locus. Genetics 193:943–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MalariaGEN Plasmodium falciparum Community Project 2016. Genomic epidemiology of artemisinin resistant malaria. Elife 5:e08714. 10.7554/eLife.08714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer PW, Petrov DA. 2013. Population genomics of rapid adaptation by soft selective sweeps. Trends Ecol Evol. 28:659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto O, Amato R, Ashley EA, MacInnis B, Almagro-Garcia J, Amaratunga C, Lim P, Mead D, Oyola SO, Dhorda M, et al. 2015. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat Genet. 47:226–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto O, Almagro-Garcia J, Manske M, Macinnis B, Campino S, Rockett KA, Amaratunga C, Lim P, Suon S, Sreng S, et al. 2013. Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat Genet. 45:648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita T. 2010. Origins and spread of pfdhfr mutant alleles in Plasmodium falciparum. Acta Trop. 114:166–170. [DOI] [PubMed] [Google Scholar]
- Nair S, Nash D, Sudimack D, Jaidee A, Barends M, Uhlemann AC, Krishna S, Nosten F, Anderson TJ. 2007. Recurrent gene amplification and soft selective sweeps during evolution of multidrug resistance in malaria parasites. Mol Biol Evol. 24:562–573. [DOI] [PubMed] [Google Scholar]
- Nair S, Miller B, Barends M, Jaidee A, Patel J, Mayxay M, Newton P, Nosten F, Ferdig MT, Anderson TJ. 2008. Adaptive copy number evolution in malaria parasites. PLoS Genet. 4:e1000243.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S, Williams JT, Brockman A, Paiphun L, Mayxay M, Newton PN, Guthmann JP, Smithuis FM, Hien TT, White NJ, et al. 2003. A selective sweep driven by pyrimethamine treatment in southeast Asian malaria parasites. Mol Biol Evol. 20:1526–1536. [DOI] [PubMed] [Google Scholar]
- Neafsey DE, Schaffner SF, Volkman SK, Park D, Montgomery P, Milner DA, Jr, Lukens A, Rosen D, Daniels R, Houde N, et al. 2008. Genome-wide SNP genotyping highlights the role of natural selection in Plasmodium falciparum population divergence. Genome Biol. 9:R171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nkhoma SC, Nair S, Al-Saai S, Ashley E, McGready R, Phyo AP, Nosten F, Anderson TJ. 2013. Population genetic correlates of declining transmission in a human pathogen. Mol Ecol. 22:273–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyunt MH, Hlaing T, Oo HW, Tin-Oo LL, Phway HP, Wang B, Zaw NN, Han SS, Tun T, San KK, et al. 2015. Molecular assessment of artemisinin resistance markers, polymorphisms in the k13 propeller, and a multidrug-resistance gene in the eastern and western border areas of Myanmar. Clin Infect Dis. 60:1208–1215. [DOI] [PubMed] [Google Scholar]
- Orr HA. 2003. The distribution of fitness effects among beneficial mutations. Genetics 163:1519–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce RJ, Pota H, Evehe MS, Ba e, Mombo-Ngoma G, Malisa AL, Ord R, Inojosa W, Matondo A, Diallo DA, et al. 2009. Multiple origins and regional dispersal of resistant DHPS in African Plasmodium falciparum malaria. PLoS Med. 6:e1000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennings PS. 2013. HIV drug resistance: problems and perspectives. Infect Dis Rep. 5:e5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennings PS, Hermisson J. 2006. Soft sweeps II–molecular population genetics of adaptation from recurrent mutation or migration. Mol Biol Evol. 23:1076–1084. [DOI] [PubMed] [Google Scholar]
- Pennings PS, Kryazhimskiy S, Wakeley J. 2014. Loss and recovery of genetic diversity in adapting populations of HIV. PLoS Genet. 10:e1004000.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phyo AP, Nkhoma S, Stepniewska K, Ashley EA, Nair S, McGready R, ler Moo C, Al-Saai S, Dondorp AM, Lwin KM, et al. 2012. Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. Lancet 379:1960–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phyo AP, Ashley EA, Anderson TJ, Bozdech Z, Carrara VI, Sriprawat K, Nair S, White MM, Dziekan J, Ling C, et al. 2016. Declining efficacy of artemisinin combination therapy against P. falciparum malaria on the Thai-Myanmar border (2003-2013): the role of parasite genetic factors. Clin Infect Dis. 63:784–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preechapornkul P, Imwong M, Chotivanich K, Pongtavornpinyo W, Dondorp AM, Day NP, White NJ, Pukrittayakamee S. 2009. Plasmodium falciparum pfmdr1 amplification, mefloquine resistance, and parasite fitness. Antimicrob Agents Chemother. 53:1509–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper C, Pearce R, Nair S, Sharp B, Nosten F, Anderson T. 2004. Intercontinental spread of pyrimethamine-resistant malaria. Science 305:1124.. [DOI] [PubMed] [Google Scholar]
- Roper C, Pearce R, Bredenkamp B, Gumede J, Drakeley C, Mosha F, Chandramohan D, Sharp B. 2003. Antifolate antimalarial resistance in southeast Africa: a population-based analysis. Lancet 361:1174–1181. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ, et al. 2002. Detecting recent positive selection in the human genome from haplotype structure. Nature 419:832–837. [DOI] [PubMed] [Google Scholar]
- Schrider DR, Mendes FK, Hahn MW, Kern AD. 2015. Soft shoulders ahead: spurious signatures of soft and partial selective sweeps result from linked hard sweeps. Genetics 200:267–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring MD, Lin JT, Manning JE, Vanachayangkul P, Somethy S, Bun R, Se Y, Chann S, Ittiverakul M, Sia-ngam P, et al. 2015. Dihydroartemisinin-piperaquine failure associated with a triple mutant including kelch13 C580Y in Cambodia: an observational cohort study. Lancet Infect Dis. 15:683–691. [DOI] [PubMed] [Google Scholar]
- Straimer J, Gnadig NF, Witkowski B, Amaratunga C, Duru V, Ramadani AP, Dacheux M, Khim N, Zhang L, Lam S, et al. 2015. Drug resistance. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science 347:428–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takala-Harrison S, Jacob CG, Arze C, Cummings MP, Silva JC, Dondorp AM, Fukuda MM, Hien TT, Mayxay M, Noedl H, et al. 2015. Independent emergence of artemisinin resistance mutations among Plasmodium falciparum in Southeast Asia. J Infect Dis. 211:670–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takala-Harrison S, Clark TG, Jacob CG, Cummings MP, Miotto O, Dondorp AM, Fukuda MM, Nosten F, Noedl H, Imwong M, et al. 2013. Genetic loci associated with delayed clearance of Plasmodium falciparum following artemisinin treatment in Southeast Asia. Proc Natl Acad Sci U S A. 110:240–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tun KM, Imwong M, Lwin KM, Win AA, Hlaing TM, Hlaing T, Lin K, Kyaw MP, Plewes K, Faiz MA, et al. 2015. Spread of artemisinin-resistant Plasmodium falciparum in Myanmar: a cross-sectional survey of the K13 molecular marker. Lancet Infect Dis. 15:415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan AM, Pinapati RS, Cheeseman IH, Camargo N, Fishbaugher M, Checkley LA, Nair S, Hutyra CA, Nosten FH, Anderson TJ, et al. 2015. Plasmodium falciparum genetic crosses in a humanized mouse model. Nat Methods 12:631–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan M, Amaratunga C, Campino S, Auburn S, Koch O, Lim P, Uk S, Socheat D, Kwiatkowski DP, Fairhurst RM, et al. 2012. Using CF11 cellulose columns to inexpensively and effectively remove human DNA from Plasmodium falciparum-infected whole blood samples. Malar J. 11:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wang Y, Cabrera M, Zhang Y, Gupta B, Wu Y, Kemirembe K, Hu Y, Liang X, Brashear A, et al. 2015. Artemisinin resistance at the China-Myanmar border and association with mutations in the K13 propeller gene. Antimicrob Agents Chemother. 59:6952–6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weetman D, Mitchell SN, Wilding CS, Birks DP, Yawson AE, Essandoh J, Mawejje HD, Djogbenou LS, Steen K, Rippon EJ, et al. 2015. Contemporary evolution of resistance at the major insecticide target site gene Ace-1 by mutation and copy number variation in the malaria mosquito Anopheles gambiae. Mol Ecol. 24:2656–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BA, Petrov DA, Messer PW. 2014. Soft selective sweeps in complex demographic scenarios. Genetics 198:669–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton JC, Feng X, Ferdig MT, Cooper RA, Mu J, Baruch DI, Magill AJ, Su XZ. 2002. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature 418:320–323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.