Abstract
A growing body of evidence suggests that genes transcribed by RNA polymerase III exhibit multiple functions within a chromosome. While the predominant function of these genes is the synthesis of RNA molecules, certain RNA polymerase III genes also function as genomic landmarks. Transfer RNA genes are known to exhibit extra-transcriptional activities such as directing Ty element integration, pausing of replication forks, overriding nucleosome positioning sequences, repressing neighboring genes (tRNA position effect), and acting as a barrier to the spread of repressive chromatin. This study was designed to identify other tRNA loci that may act as barriers to chromatin-mediated repression, and focused on TRT2, a tRNAThr adjacent to the STE6 α2 operator. We show that TRT2 acts as a barrier to repression, protecting the upstream CBT1 gene from the influence of the STE6 α2 operator in MATα cells. Interestingly, deletion of TRT2 results in an increase in CBT1 mRNA levels in MATa cells, indicating a potential tRNA position effect. The transcription of TRT2 itself is unaffected by the presence of the α2 operator, suggesting a hierarchy that favors assembly of the RNA polymerase III complex versus assembly of adjacent α2 operator-mediated repressed chromatin structures. This proposed hierarchy could explain how tRNA genes function as barriers to the propagation of repressive chromatin.
INTRODUCTION
RNA polymerase III is predominantly responsible for the transcription of small cellular RNA molecules including tRNAs, 5S RNA, 7SL RNA and in Saccharomyces cerevisiae, the SNR6 gene encoding the spliceosome U6 RNA. Transcription of tRNA genes is mediated by the stepwise assembly of the TFIIIC transcription factor complex onto the box A and box B internal control region promoter elements, followed by recruitment of the TATA binding protein (TBP) containing complex TFIIIB. Once all transcription factors are in place, the RNA polymerase III enzymatic complex is recruited to initiate high level transcription of its target genes (1–3). These RNAs are extremely abundant in dividing cells, as tRNAs alone can account for as much as 15% of total RNA in log phase S.cerevisiae (4). This number suggests that tRNA genes are transcribed at an amazingly high rate during log phase growth (compared to RNA polymerase II genes), averaging approximately 104 transcription cycles/tRNA gene/generation, or roughly two cycles per second. This high rate of transcription can be explained in part by a facilitated recycling model in which an assembled RNA polymerase III complex is transferred from the termination site to the initiation site, remaining assembled on the tRNA gene through multiple rounds of transcription (5–7).
Such a persistently organized RNA polymerase III complex could also explain several observed ‘extra-transcriptional’ roles of tRNA genes within chromosomes. In S.cerevisiae, actively transcribed tRNA genes have been shown to direct Ty element integration (8–10), override nucleosome positioning signals (11), exert repressive position effects on neighboring RNA polymerase II promoters (12–15), act as replication fork pause sites (16), and act as a barrier to the propagation of heterochromatic repression, by blocking the spread of silent chromatin at the HMR locus (17). Of particular interest is the dichotomy that in certain cases a tRNA gene is capable of protecting a neighboring gene from repression (at HMR), while in other instances tRNA genes can directly repress or exert a negative influence on transcription of an adjacent RNA polymerase II gene, a process referred to as tRNA-mediated gene silencing (14) or tRNA position effect (15). While these types of effects have been observed in a limited number of cases (both natural and engineered), the genome-wide effects of the location of RNA polymerase III complex formation on neighboring chromosomal loci are largely unstudied.
We have previously described the heterochromatin barrier effect attributed to the HMR-tRNA (tRNAThr[AGU]C) on S.cerevisiae chromosome III. This tRNAThr gene prevents the spread of Sir protein-mediated gene silencing from the adjacent HMR locus in both reporter constructs and along the native chromosome (17). We asked if tRNAs adjacent to other repressed loci in S.cerevisiae could also function as barriers to repression of neighboring genes. TRT2 (coding for tRNAThr[CGU]K) is a single copy tRNAThr gene that lies just upstream of the α2 operator sequence that regulates the MATa cell-specific STE6 gene on S.cerevisiae chromosome XI. We specifically selected this locus for study as another example of a tRNA gene located adjacent to a repressed region of chromatin, and asked whether this tRNA gene might act as a barrier to the spread of repression. The α2 operator binds the Mcm1p/α2p complex, and initiates MATα cell-specific repression of MATa specific genes such as STE6 via multiple mechanisms, including nucleosome positioning (18,19), the recruitment of Ssn6p, Tup1p, and their associated histone deacetylases (20–23), and direct interaction with transcriptional machinery (24,25). This study asked whether TRT2 served as a barrier to α2 operator-mediated repression in MATα cells, and revealed that the same tRNA gene both protects the adjacent CBT1 gene from α2 operator repression in MATα cells, and exerts a potential negative tRNA position effect on CBT1 in MATa cells. This is the first example of a tRNA gene that displays multiple types of extra-transcriptional functions at the same locus.
MATERIALS AND METHODS
All yeast strains were derived from wild-type S.cerevisiae W303 (DDY2, DDY3 and DDY4, originally JRY4012, JRY4013 and JRY2334, obtained from Jasper Rine, University of California at Berkeley; genotypes of all yeast strains generated in this study are listed in Table 1). Since TRT2 is an essential single copy tRNA gene, a 0.32 kb fragment of TRT2 (SGD chromosome XI coordinates 46596–46919) was cloned by PCR into plasmids pRS414 and pRS415 (26) to cover deletions of the gene (plasmids pDD675 and pDD676, respectively). To construct the trt2–cbt1Δ::URA3 reporter strains described in Figure 1, a 2.1 kb segment of the TRT2 locus (coordinates 46162–48248) was amplified by PCR and cloned into pCR2.1-TOPO (Invitrogen) to make plasmid pDD689. The resulting plasmid was cut with Spe I and Xho I to remove TRT2 and CBT1, and was replaced with the Spe I-Xho I URA3 fragment from pDD588 (URA3 cloned into Bluescript SK+) to create plasmid pDD694, trt2–cbt1Δ::URA3. The modified locus was cut out of pDD694 and transformed into the diploid strain DDY2, and URA+ recombinants were selected and screened by PCR to verify proper integration. This diploid strain was then transformed with TRT2 plasmids pDD675 or pDD676 to cover the deletion, sporulated, and URA+ haploids were recovered. The cbt1Δ::URA3 control strains were made by direct PCR knockout of CBT1 with URA3, using pRS406 as template. Cells were grown on yeast minimal medium (YMD + 2% dextrose) lacking uracil to test for repression of the URA3 marker gene. Yeast Nitrogen Base was purchased from U.S. Biologicals, and YMD + all mix contained only those nutrients required for growth of W303 strains (adenine, histidine, leucine, lysine, tryptophan, and uracil).
Table 1. Genotypes of all yeast strains generated in this study.
| S.cerevisiae W303 strains | Source | |
|---|---|---|
| DDY2 | MATα/MATa ade2-1/ADE2 his3-11/his3-11 leu2-3, 112/leu2-3, 112 LYS2/lys2Δ trp1-1/trp1-1 ura3-1/ura3-1 | J. Rine |
| DDY3 | MATa ADE2 his3-11 leu2-3,112 lys2Δ trp1-1 ura3-1 | J. Rine |
| DDY4 | MATα ADE2 his3-11 leu2-3,112 lys2Δ trp1-1 ura3-1 | J. Rine |
| DDY889 | MATα ADE2 his3-11 leu2-3,112 LYS2 trp1-1 ura3-1 trt2-cbt1Δ::URA3 pTRT2:LEU2 | This study |
| DDY890 | MATa ade2-1 his3-11 leu2-3,112 LYS2 trp1-1 ura3-1 trt2-cbt1Δ::URA3 ppr1Δ::HIS3 pTRT2:LEU2 | This study |
| DDY891 | MATa ADE2 his3-11 leu2-3,112 LYS2 trp1-1 ura3-1 trt2-cbt1Δ::URA3 ppr1Δ::HIS3 pTRT2:TRP1 | This study |
| DDY902 | MATα ADE2 his3-11 leu2-3,112 lys2Δ trp1-1 ura3-1 trt2-cbt1Δ::URA3 ppr1Δ::HIS3 pTRT2:LEU2 | This study |
| DDY 903 | MATα ade2-1 his3-11 leu2-3,112 lys2Δ trp1-1 ura3-1 trt2-cbt1Δ::URA3 ppr1Δ::HIS3 pTRT2:LEU2 | This study |
| DDY974 | MATα ade2-1 his3-11 leu2-3,112 LYS2 trp1-1 ura3-1 cbt1Δ::URA3 ppr1Δ::HIS3 | This study |
| DDY975 | MATa ade2-1 his3-11 leu2-3,112 LYS2 trp1-1 ura3-1 cbt1Δ::URA3 ppr1Δ::HIS3 | This study |
| DDY1022 | MATa ADE2 his3-11 leu2-3,112 LYS2 trp1-1 ura3-1 trt2Δ ppr1Δ::HIS3 pTRT2:LEU2 | This study |
| DDY1024 | MATa ADE2 his3-11 leu2-3,112 LYS2 trp1-1 ura3-1 trt2Δ ppr1Δ::HIS3 pTRT2:LEU2 | This study |
| DDY1026 | MATα ADE2 his3-11 leu2-3,112 lys2Δ trp1-1 ura3-1 trt2Δ ppr1Δ::HIS3 pTRT2:LEU2 | This study |
| DDY1028 | MATα ADE2 his3-11 leu2-3,112 lys2Δ trp1-1 ura3-1 trt2Δ ppr1Δ::HIS3 pTRT2:LEU2 | This study |
| DDY1261 | MATα ADE2 his3-11 leu2-3,112 lys2Δ trp1-1 ura3-1 trt2Δ α2 operatorΔ ppr1Δ::HIS3 pTRT2:LEU2 | This study |
| DDY1262 | MATα ADE2 his3-11 leu2-3,112 lys2Δ trp1-1 ura3-1 trt2Δ α2 operatorΔ ppr1Δ::HIS3 pTRT2:LEU2 | This study |
| DDY1737 | MATα ADE2 his3-11 leu2-3,112 LYS2 trp1-1 ura3-1 α2 operatorΔ | This study |
| DDY1739 | MATa ADE2 his3-11 leu2-3,112 lys2Δ trp1-1 ura3-1 α2 operatorΔ | This study |
| DDY1740 | MATa ADE2 his3-11 leu2-3,112 LYS2 trp1-1 ura3-1 α2 operatorΔ | This study |
| DDY1742 | MATα ADE2 his3-11 leu2-3,112 lys2Δ trp1-1 ura3-1 α2 operatorΔ | This study |
| DDY1805 | MATα ade2-1 his3-11 leu2-3,112 LYS2 trp1-1 ura3-1 trt2Δ pTRT2:URA3 hos1::HIS3 hos2::TRP1 rpd3::LEU2 | This study |
| DDY1825 | MATα ADE21 his3-11 leu2-3,112 lys2Δ trp1-1 ura3-1 trt2Δ pTRT2:URA3 hos1::HIS3 hos2::TRP1 rpd3::LEU2 | This study |
| DDY1956 | MATα ADE2 his3-11 leu2-3,112 lys2Δ trp1-1 ura3-1 trt2Δ ppr1Δ::HIS3 pTRT2:LEU2 hda1Δ::KanMX | This study |
| DDY2021 | MATα ADE2 his3-11 leu2-3,112 lys2Δ trp1-1 ura3-1 trt2Δ ppr1Δ::HIS3 pTRT2:LEU2 hda1Δ::KanMX | This study |
Figure 1.
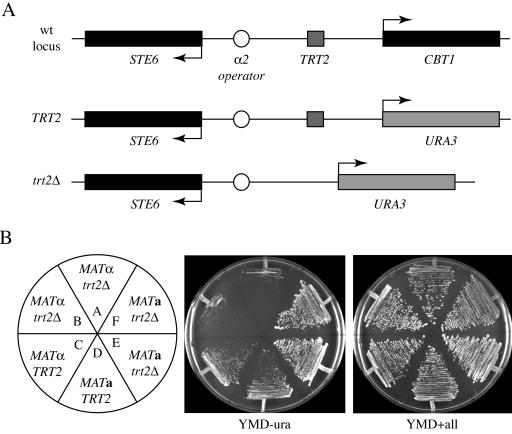
A URA3 marker gene is repressed when inserted upstream of the STE6 α2 operator site in S.cerevisiae chromosome XI. (A) The wild-type STE6–CBT1 region of chromosome XI is depicted on top. URA3 was inserted by homologous recombination upstream of the STE6 α2 operator to either delete the TRT2 tRNAThr gene (DDY890, DDY891, DDY902, and DDY903), or to retain the intervening TRT2 gene (DDY974 and DDY 975). (B) Each strain was streaked on YMD lacking uracil (YMD − ura) and incubated for 2 days. MATα strains lacking TRT2 showed inhibited growth on medium lacking uracil, while all strains grew equally on minimal YMD containing uracil (YMD + all).
To make the modified chromosomal loci, pDD689 was mutagenized using the Quik-change kit (Stratagene) to delete TRT2 (oligonucleotides DDO-96/97) from box A to the box B (chromosome XI coordinates 46747–46800). The α2 operator from coordinates 46472–46489 was deleted in the same way using oligonucleotides DDO-123/124. Plasmids containing deletions of TRT2 and/or the α2 operator were transformed into DDY889 (trt2–cbt1Δ::URA), selected on 5-FOA, and proper integration verified by PCR. Resulting strains containing modified STE6–CBT1 loci were backcrossed to trt2–cbt1Δ::URA3 strains to obtain sibling MATa and MATα versions.
For northern blot analysis, RNA was prepared as described in Iyer and Struhl (27). Northern blots contained 10 μg total RNA per lane, and were performed using Northern Max reagents (Ambion). CBT1 northern blots were run on 1.0% agarose gels, and the TRT2 blot in Figure 4 was run on a 1.2% agarose gel. Northern probes were generated from PCR products of the first 600 bp of each gene (except for TRT2, where the entire gene was amplified) that included a T7 RNA polymerase promoter attached to the downstream primer. These PCR products were used as templates to synthesize radiolabeled riboprobes using the Ambion Strip-EZ kit. All oligonucleotide sequences used for knockouts, PCR clonings, probe templates, and mutagenesis reactions are available on request.
Figure 4.
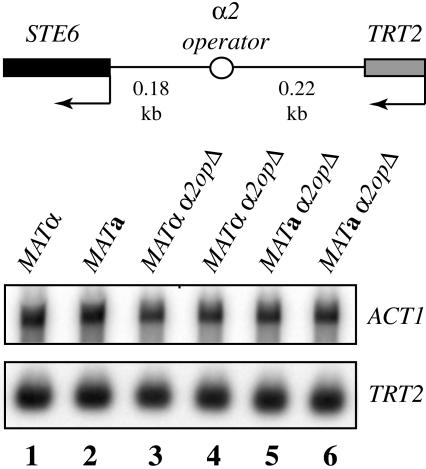
TRT2 expression is unaffected by the presence of an active α2 operator site. Northern blot analysis of TRT2 mRNA from wild-type MATα and MATa strains (DDY4 and DDY3, lanes 1 and 2), α2 operator deleted MATα strains (DDY1737 and DDY1742, lanes 3 and 4), and α2 operator deleted MATa strains (DDY1739 and DDY1740, lanes 5 and 6). After normalization to the ACT1 signal, TRT2 mRNA levels were identical in all strains.
HDA1 deletion in the trt2Δ strain was made by standard PCR knockout protocols using the plasmid pUG6 as a template (28). The hos1 hos2 rpd3 strains were made by crossing trt2Δ strains with strain DY6445 (MATα ade2 can1 his3 leu2 trp1 ura3 hos1::HIS3 hos2::TRP1 rpd3::LEU2), a gift from David Stillman (University of Utah).
Chromatin immunoprecipitation was performed as described in Kuo and Allis (29). Antibodies used were anti-acetyl-histone H3 and anti-acetyl-histone H4 from Upstate (catalog No. 06–599 and 06–866). An aliquot of 5 μl of a 1 : 10 dilution of DNA recovered from the immunoprecipitates was used to program PCR reactions (Taq polymerase purchased from Promega), and the same volume of a 1 : 40 dilution was used for the input controls. PCR conditions were 95°C for 2 min (initial denaturation), 95°C × 30 s, 55°C × 30 s, 72°C × 60 s (28 cycles).
RESULTS
TRT2 can protect an integrated URA3 marker gene from α2 operator repression
STE6 is a MATa cell-specific gene that is repressed in MATα cells by an upstream α2 operator sequence. Several α2 operator sequences, including this particular one, have been shown to be orientation independent in plasmid-based lacZ reporter gene assays (30), so we wished to determine if repression was also bi-directional in a chromosomal context. Also, since the TRT2 tRNAThr gene lies between this α2 operator and CBT1, the next RNA polymerase II transcribed gene upstream of STE6, we tested whether TRT2 acts as a barrier to repression of CBT1.
To test the hypothesis that repression spreads bi-directionally from a chromosomal α2 operator, and that the TRT2 gene acts as a barrier to α2 operator-mediated repression, we constructed yeast strains that contained URA3 integrated into chromosome XI in place of CBT1, upstream of the α2 operator site at the STE6 locus. Two sets of strains were constructed (Figure 1A), one that retained TRT2 between the α2 operator and URA3, and a second that replaced both CBT1 and TRT2 with URA3. Figure 1 shows the results when these strains were streaked on minimal media lacking uracil. MATα trt2–cbt1Δ::URA3 strains (Figure 1B, DDY 902 and DDY 903, wedges A and B) are considerably compromised for growth on YMD media lacking uracil compared to isogenic MATα (DDY974) or MATa (DDY975) strains containing TRT2 between the operator and URA3 (wedges C and D). URA3 is not completely repressed in these strains, as extended incubation eventually leads to formation of colonies. This delay in growth suggests that repression can spread from the α2 operator in both directions along chromosome XI and inhibit URA3 expression.
Interestingly, MATa trt2–cbt1Δ::URA3 strains grow slightly better on YMD lacking uracil than MATa strains containing TRT2, suggesting that in the absence of α2 operator-mediated repression in MATa cells, TRT2 may exert a repressive tRNA position effect on the URA3 reporter (compare DDY975, wedge D with DDY890 and 891, wedges E and F). These results prompted us to further investigate the effects of deleting TRT2 on the expression of CBT1, the gene naturally upstream of STE6 on chromosome XI, in both MATa and MATα cells.
Deletion of TRT2 from chromosome XI in MATα cells inhibits induction of CBT1 when cells are grown on acetate, and inhibition is dependent on the α2 operator
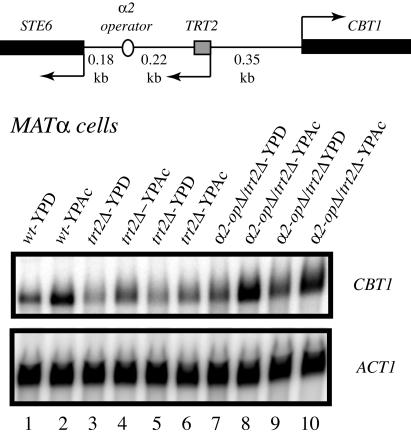
Cytochrome B termination (CBT1) is a gene required for proper maturation of cytochrome b mRNA in S.cerevisiae, and is essential for respiratory growth on non-fermentable carbon sources such as acetate and ethanol (31). CBT1 is located 862 bp upstream of STE6, placing it approximately 680 bp from the α2 operator. We observed that growth of wild-type MATα S.cerevisiae in media containing acetate as a sole carbon source (YPAc) resulted in a 3-fold induction of CBT1 mRNA compared to cells grown in dextrose (YPD, Figure 2, compare lanes 1 versus 2). We then asked whether CBT1 expression is affected by deletion of TRT2. Since TRT2 is an essential single copy tRNA gene, it was first deleted in a diploid strain, the deletion was covered with an episomal copy of TRT2 (pDD676, pRS415:TRT2:LEU2), and the resulting diploid strain was sporulated to obtain MATα trt2Δ:pTRT2:LEU2 cells. Deletion of TRT2 from chromosome XI in MATα cells reduced both the basal and induced levels of CBT1 expression to approximately 40% of normal levels as analyzed by northern blot analysis (Figure 2, lanes 3 and 4, 5 and 6 compared to lanes 1 and 2). This repression was dependent on the α2 operator, as deletion of both TRT2 and α2 operator sequences restored the normal levels of CBT1 mRNA induction (Figure 2, lanes 7–10). This result demonstrates that repression spreads along chromosome XI upstream of the α2 operator in the absence of TRT2, suggesting that TRT2 functions as a barrier to α2 operator-mediated repression of CBT1 in MATα cells.
Figure 2.
Deletion of TRT2 results in the repression of CBT1 transcription in MATα cells. Total RNA was isolated from strains containing a wild-type STE6–CBT1 locus (DDY4, lanes 1 and 2), a mutant locus deleted for TRT2 (DDY1026, lanes 3 and 4, DDY1028, lanes 5 and 6), and a mutant locus containing deletion of both TRT2 and the α2 operator (DDY1261, lanes 7 and 8, DDY1262, lanes 9 and 10). Odd numbered lanes contain RNA isolated from cells grown on dextrose as a carbon and energy source (YPD), and even numbered lanes from cells grown on acetate (YPAc), which induces CBT1 transcription. CBT1 mRNA levels were reduced approximately 3-fold in strains lacking only TRT2. Results from two independent isolates of each mutant strain are shown.
Deletion of TRT2 from chromosome XI in MATa cells results in an increase in expression of CBT1
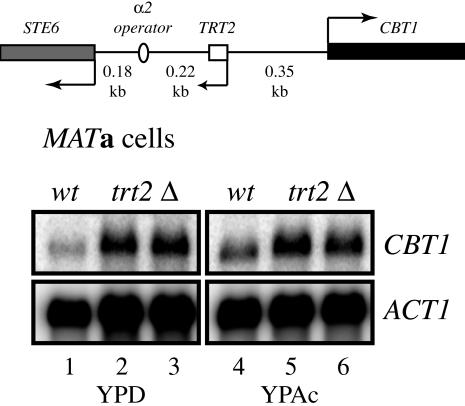
When CBT1 expression from a trt2Δ chromosome was analyzed in MATa cells, the opposite effect was observed. Figure 3 shows the results of northern blot analysis of wild-type and MATa trt2Δ strains probed for CBT1 message. Deletion of TRT2 in MATa cells leads to increased levels of CBT1 mRNA in either YPD or YPAc media, suggesting that in its native context in MATa cells, CBT1 may be subject to a tRNA position effect (Figure 3, compare lane 1 to lanes 2 and 3, lane 4 to lanes 5 and 6). The increased level of transcription of CBT1 in trt2Δ strains is consistent with observation of the strains analyzed in Figure 1, as MATa trt2–cbt1Δ::URA3 strains grew slightly better than MATa cbt1Δ::URA3 strains on YMD–uracil media.
Figure 3.
Deletion of TRT2 results in an increase in expression of CBT1 in MATa cells. Wild-type MATa S.cerevisiae (DDY3, lanes 1 and 4), and MATa trt2Δ (two independent isolates, DDY1022 lanes 2 and 5, and DDY1024 lanes 3 and 6), were grown on YPD (lanes 1–3) or on YPAc (lanes 4–6) and total RNA isolated. Northern blots were probed for CBT1 mRNA as in Figure 2.
Transcription of TRT2 is unaffected by α2 operator-mediated repression
Since TRT2 is a single copy tRNA gene, its expression level can be assayed directly by northern blotting. We next asked if the α2 operator affects expression of TRT2 itself. Figure 4 shows TRT2 expression levels in wild-type and α2 operator deleted MATa and MATα strains. After normalization to the ACT1 signal, no significant difference in the level of TRT2 RNA was seen in MATα versus MATa cells, therefore TRT2 is apparently unaffected by the presence of an adjacent active α2 operator (Figure 4, lanes 1 and 2). To further confirm that the TRT2 gene is refractory to α2 operator repression, the operator site was deleted in both MATα (Figure 4, lanes 3 and 4) and MATa (lanes 5 and 6) strains, and again no difference in TRT2 levels was seen. These results demonstrate that RNA polymerase III transcription of TRT2 is completely impervious to α2 operator-mediated repression.
Altered histone acetylation does not appear to be responsible for the spread of repression along a trt2Δ chromosome
The recent literature has described multiple yeast histone deacetylases as interacting with the Ssn6p/Tup1p complex to repress transcription. Increased histone H4 acetylation at the STE6 promoter is observed in class I histone deacetylase (HDAC) hos1 hos2 rpd3 triple mutant strains (23), however, loss of Rpd3p function affects both repression and activation of STE6 (32). Derepression of Ssn6–Tup1 regulated genes SUC2 and MFA2 is observed in triple hos1 hos2 rpd3 strains (23). Other Ssn6–Tup1 regulated genes, such as ENA1 appear to require the class II HDAC HDA1 for repression, and it has been reported that STE6 is partially derepressed in either hda1 or rpd3 strains (21). The Ssn6–Tup1 protein complex has been shown to physically interact with all of these HDACs in vitro (21–23).
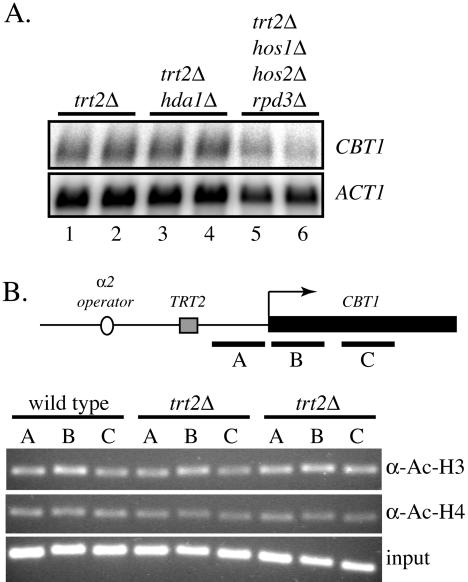
To assess whether HDAC recruitment by Ssn6–Tup1 at the α2 operator is responsible for CBT1 repression in the absence of TRT2, we performed northern blots in trt2Δ strains mutated for either hda1 or hos1 hos2 rpd3. Figure 5A shows that deletion of hda1 does not relieve repression of CBT1 in a trt2Δ background. The triple deletion of the class I HDACs results in even lower levels of CBT1, suggesting that, as for STE6 and other genes, RPD3 function is also required for normal activated expression (32). These results suggest that altered histone acetylation levels are not the major determinant in spreading of repression from the operator in the absence of TRT2.
Figure 5.
(A) Repression of CBT1 in trt2Δ strains is not relieved by mutation of histone deacetylases. Northern blot analysis of CBT1 mRNA from MATα trt2Δ cells containing HDAC mutations. Lanes 1 and 2, trt2Δ (DDY1026 and 1028); lanes 3 and 4, trt2Δ hda1Δ (DDY1956 and DDY2021); lanes 5 and 6, trt2Δ hos1Δ hos2Δ rpd3Δ (DDY1805 and DDY1825). (B) Chromatin immunoprecipitation of wild-type and trt2Δ strains using anti-acetylated histone H3 and H4 antibodies. MATα strains DDY4 (wild-type) and trt2Δ (DDY1026 and DDY1028) were grown and processed for chromatin immunoprecipitation. Primers sets for PCR analysis spanned the indicated regions (approximately 200 bp each PCR product) of the CBT1 gene. No significant difference in the level of CBT1 chromatin was seen in immunoprecipitates from wild-type versus trt2Δ strains.
In order to directly assess the histone acetylation state at CBT1 in wild-type and trt2Δ strains, we performed chromatin immunoprecipitation using antibodies against acetylated histone H3 or histone H4. DNA immunoprecipitated from wild-type or trt2Δ strains was probed by PCR with multiple primer sets spanning from −170 to +500 bp from the CBT1 start codon. The data in Figure 5B showed no significant difference in the amount of immunoprecipitated chromatin between wild-type and trt2Δ strains. These results also suggest that gross changes in histone acetylation are not the major determinant in repression of CBT1 in the trt2Δ background, and that other mechanisms of Ssn6–Tup1 repression, either nucleosome positioning or direct interaction with the transcriptional machinery, are responsible (see Discussion).
DISCUSSION
α2 operator-mediated repression is bi-directional at the STE6 locus
α2 operator sites mediate repression of transcription of MATa-cell-specific genes in MATα cells (20), and also regulate recombination enhancer activity in mating type switching (33,34). Transcriptional repression is mediated by binding of the α2/Mcm1p complex to the operator sites, which then recruit co-repressors such as the Ssn6p/Tup1p complex. Transcriptional repression by α2 operator sequences is mediated by the further recruitment of various histone deacetylases by Ssn6p/Tup1p (21,23), by the precise stable positioning of nucleosomes at the promoter region of the regulated gene (18,19,35), and possibly by direct interaction with transcriptional machinery (24,25). Despite a degree of asymmetry of natural α2 operator sites in Mcm1p/α2p regulated genes, cloned α2 operators in either orientation are able to repress transcription of plasmid-based reporter genes (30), suggesting that repression can spread bi-directionally from an α2 operator. This observation led us to analyze whether repression from the α2 operator upstream of the STE6 gene spreads bi-directionally on the native chromosome, and whether the TRT2 tRNAThr gene upstream acts as a barrier to such repression.
The results shown in Figures 1 and 2 show that the STE6 α2 operator can partially repress upstream genes specifically in MATα cells in a URA3 modified, or native chromosome XI lacking TRT2. The results from the northern blot analysis of CBT1 mRNA in trt2Δ strains shows a 3-fold repression compared to wild-type cells. This repression is clearly due to the operator sequence, as its deletion restores both the basal and induced levels of CBT1 transcription (Figure 2). One reason for the relatively mild repression (as compared to the complete repression of STE6 in MATα cells) could be due to the relative distance between the operator and the gene. The STE6 gene starts 182 bp from the operator, while the CBT1 gene is 650 bp away (598 bp in the trt2Δ strain). This increased distance may lead to weaker repression compared to that of STE6. The range of repression at this locus is limited to the CBT1 promoter, as deletion of TRT2 had no effect on expression of YKL207W, the next gene centromere proximal to CBT1 (D. Donze, unpublished data). Another possible reason for the relatively mild repression is the asymmetric nature of the STE6 α2 operator site, which could lead to differences in repression in each direction. A plasmid-based lacZ reporter gene was differentially repressed by opposite orientations of this operator, with the native orientation showing 1.5-fold higher repression then the reverse orientation (30). This asymmetry may lie in an asymmetry of direction of Hda1p activity from the operator, which has been proposed for the ENA1 promoter (21). Most likely, both distance and orientation are affecting the level of repression of CBT1 compared to STE6.
TRT2 acts as a barrier to repression
Since MATα cell-specific repression of CBT1 is observed only when the TRT2 gene is deleted (or contains only a box B point mutation; D. Donze, unpublished data), TRT2 is acting as a barrier to the spread of α2 operator-mediated repression. We have previously shown that the HMR-tRNA (tRNAThr[AGU] CR1) acts as a barrier to the spread of silencing at the HMR locus, as it blocks repression of a MATa1 reporter gene when juxtaposed between the gene and the silencer, and its deletion from the natural chromosome leads to a 60% reduction of expression of the downstream GIT1 gene (17). When tested alongside the HMR-tRNA in the MATa1 reporter gene assay, TRT2 showed a partial barrier activity to Sir protein-mediated silencing (17), while it appears to completely prevent the spread of α2 operator repression in this study. Therefore, different tRNA genes may vary in their ability to block repression, or may have evolved specificities for different types of repression.
The upstream spread of repression from the α2 operator into CBT1 does not appear to be mediated by major changes in histone acetylation, as suggested by the data in Figure 5. Deletion of HDACs known to be involved in Ssn6p–Tup1p-mediated repression do not result in derepression of CBT1 in trt2Δ strains, and chromatin immunoprecipitation with antibodies against acetylated histone H3 or H4 show no difference in the amount of CBT1 DNA immunoprecipitated in wild-type versus trt2Δ MATα strains. However, it may be that specific histone deacetylation events may be responsible, which would require a detailed analysis with antibodies specific for individual acetylated residues. Tup1p has been shown to utilize multiple mechanisms to repress transcription including recruitment of HDACs (21–23), inducing the stable positioning of nucleosomes (18,19,36), and also by direct interaction with the transcriptional machinery (24,25,37). The results presented here suggest that the latter two mechanisms of Tup1 transcriptional inhibition are most likely at work in the repression of CBT1 observed in the absence of TRT2. Since active tRNA genes have been demonstrated to directly override nucleosome positioning signals (11), we suggest that the barrier activity of TRT2 is at least in part due to an ability to block the spread of phased nucleosomes emanating from the α2 operator.
In the absence of repression, deletion of TRT2 results in elevated CBT1 mRNA levels
Transfer RNA genes in S.cerevisiae have been shown to exert a phenomenon referred to as either tRNA-mediated gene silencing or tRNA position effect. In the limited number of cases studied so far, a tRNA gene can exert a repressive effect on transcription from a nearby RNA polymerase II promoter, and this repression requires a transcriptionally active tRNA gene, or at least one competent to bind TFIIIC (13–15). The genome-wide extent of tRNA position effects is unknown, as it has previously only been observed at a single native chromosomal locus, PTR3. However, bioinformatic analysis suggests that tRNA position effects may exert a modest but general effect on nearby RNA polymerase II promoters at many loci, and has been suggested that position effects may regulate expression of genes that are derepressed when tRNA expression is downregulated (15). The results shown in Figure 3 demonstrate that deletion of TRT2 increases CBT1 expression in MATa cells, where α2 operator-mediated repression is absent. This provides a second example of a potential tRNA position effect on a native gene, supporting the bioinformatic predictions.
It should be noted that the mechanism of tRNA position effects has not been studied in detail on native chromosomal genes. Active tRNA genes have been shown to be localized to nucleoli in S.cerevisiae (38,39), and a mutation in the putative psuedouridine synthetase gene CBF5 disrupts both nucleolar localization of tRNA synthesis and suppresses tRNA-mediated gene silencing of a plasmid-based reporter gene (14). These studies have suggested that nucleolar localization may be responsible for both tRNA barrier function and tRNA position effects, however, other possibilities exist. One could speculate that inactivation of a tRNA gene could allow upstream activating sequences (UASs) from neighboring genes to inappropriately influence transcription of tRNA proximal genes, suggesting that a tRNA (or an engaged RNA polymerase III complex) might function somewhat as a classic metazoan insulator element, blocking the positive signal from the UAS.
TRT2 transcription is completely resistant to the presence of the α2 operator
Since the box B promoter element of TRT2 lies only 240 bp from the STE6 α2 operator, we wanted to ask if transcription of TRT2 itself was affected by its proximity to the repressive element. The results in Figure 4 show that TRT2 is unaffected by the presence of an active (MATα) or inactive (MATa) α2 operator, or by deletion of the operator in MATα cells. Therefore even in the presence of a nearby active operator site, a fully functional RNA polymerase III complex can form on the TRT2 gene and carry out normal levels of transcription. This suggests a hierarchy in the assembly of the RNA polymerase III complex onto a chromosome versus the assembly and propagation of repressive structures, and such a hierarchy may shed some light onto one aspect of the mechanism of the barrier activity of tRNA genes.
Working models of RNA polymerase III transcription depict the stepwise assembly of the TFIIIC transcription factor complex onto the box A and box B sites, followed by the recruitment of TFIIIB proteins Brf1p, Bdp1p, and TBP. Once assembled, this transcription factor platform is able to recruit the RNA polymerase III enzyme complex and initiate transcription (3), in a process that no longer requires TFIIIC. This sequence of events was determined largely from in vitro reconstitution experiments, but recent in vivo studies suggest a slightly different mechanism.
Chromatin immunoprecipitation studies of human cells progressing through mitosis show that as RNA polymerase III transcription decreases during mitosis, Bdp1p and polymerase subunits are mostly released from chromatin, but Brf1p and TBP remain associated with both tRNA and 5S genes (40). Studies in yeast cells during stationary phase or nutrient limited growth, conditions where RNA polymerase III transcription is markedly reduced, show that polymerase occupancy at a tRNA promoter is severely reduced, while TFIIIB subunit occupancy is only partially reduced (41,42). Interestingly, these studies show that the association of TFIIIC appears unchanged or even increased under conditions of reduced tRNA transcription. These results suggest a persistent association of at least part of the RNA polymerase III machinery with its target loci independent the transcriptional state of the gene. This partial association of RNA polymerase III transcription factors is also seen at ETC loci (extra TFIIIC), which appear to have TFIIIC constitutively bound in the absence of TFIIIB and polymerase (43). The persistent association of RNA polymerase III factors may in one sense serve as an ‘epigenetic mark’ of these loci for polymerase reassembly when changing conditions require the resumption of RNA polymerase III transcription. Such persistent ‘marking’ of RNA polymerase III promoters may also relate to their barrier function, as it might allow a preferential reassembly of the RNA polymerase III transcription complex after replication, even if the promoter lies adjacent to silencers or other repressive operator elements.
Another feature of RNA polymerase III that may contribute to barrier function is a process called facilitated recycling. Stably bound RNA polymerase III complexes are known to direct multiple rounds of transcription in vitro (44,45), and an individual enzyme complex appears to be able to recycle multiple times on an individual template without the need to reform a preinitiation complex (5–7). Although observed in vitro, this hyper-processive and persistent occupation of the RNA polymerase III complex is likely to occur in vivo to account for the transcription rate required to produce the large number of tRNA molecules per yeast cell. Such a persistent occupation of tRNA genes during all phases of the cell cycle could contribute to the barrier function of tRNA genes by again physically, and perhaps enzymatically (46) preventing the spread of repressive chromatin. With regard to the data in Figure 4, the level of TRT2 transcription from its single locus is identical with or without an active α2 operator, indicating that TRT2 is transcribed at normal levels by the RNA polymerase III machinery even when adjacent to repressive chromatin. This suggests that RNA polymerase III complex assembly, function and persistence at TRT2 are dominant over the encroachment of repressive chromatin structures.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the LSU genomics facility for DNA sequencing services, David Stillman for the HDAC mutants, and Giorgio Dieci and Mohamed Noor for critically reading the manuscript. This work in the Donze lab was supported by a Young Investigators Award from the Human Frontier Science Program, and a K22 Career Transition Award from the National Institute of Child Health and Human Development.
REFERENCES
- 1.Chedin S., Ferri,M.L., Peyroche,G., Andrau,J.C., Jourdain,S., Lefebvre,O., Werner,M., Carles,C. and Sentenac,A. (1998) The yeast RNA polymerase III transcription machinery: a paradigm for eukaryotic gene activation. Cold Spring Harb. Symp. Quant. Biol., 63, 381–389. [DOI] [PubMed] [Google Scholar]
- 2.Huang Y. and Maraia,R.J. (2001) Comparison of the RNA polymerase III transcription machinery in Schizosaccharomyces pombe, Saccharomyces cerevisiae and human. Nucleic Acids Res., 29, 2675–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paule M.R. and White,R.J. (2000) Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res., 28, 1283–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warner J.R. (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci., 24, 437–440. [DOI] [PubMed] [Google Scholar]
- 5.Dieci G., Giuliodori,S., Catellani,M., Percudani,R. and Ottonello,S. (2002) Intragenic promoter adaptation and facilitated RNA polymerase III recycling in the transcription of SCR1, the 7SL RNA gene of Saccharomyces cerevisiae. J. Biol. Chem., 277, 6903–6914. [DOI] [PubMed] [Google Scholar]
- 6.Dieci G. and Sentenac,A. (1996) Facilitated recycling pathway for RNA polymerase III. Cell, 84, 245–252. [DOI] [PubMed] [Google Scholar]
- 7.Ferrari R., Rivetti,C., Acker,J. and Dieci,G. (2004) Distinct roles of transcription factors TFIIIB and TFIIIC in RNA polymerase III transcription reinitiation. Proc. Natl Acad. Sci. USA, 101, 13442–13447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chalker D.L. and Sandmeyer,S.B. (1992) Ty3 integrates within the region of RNA polymerase III transcription initiation. Genes Dev., 6, 117–128. [DOI] [PubMed] [Google Scholar]
- 9.Kirchner J., Connolly,C.M. and Sandmeyer,S.B. (1995) Requirement of RNA polymerase III transcription factors for in vitro position-specific integration of a retrovirus like element. Science, 267, 1488–1491. [DOI] [PubMed] [Google Scholar]
- 10.Devine S.E. and Boeke,J.D. (1996) Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev., 10, 620–633. [DOI] [PubMed] [Google Scholar]
- 11.Morse R.H., Roth,S.Y. and Simpson,R.T. (1992) A transcriptionally active tRNA gene interferes with nucleosome positioning in vivo. Mol. Cell Biol., 12, 4015–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kinsey P.T. and Sandmeyer,S.B. (1991) Adjacent pol II and pol III promoters: transcription of the yeast retrotransposon Ty3 and a target tRNA gene. Nucleic Acids Res., 19, 1317–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hull M.W., Erickson,J., Johnston,M. and Engelke,D.R. (1994) tRNA genes as transcriptional repressor elements. Mol. Cell Biol., 14, 1266–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kendall A., Hull,M.W., Bertrand,E., Good,P.D., Singer,R.H. and Engelke,D.R. (2000) A CBF5 mutation that disrupts nucleolar localization of early tRNA biosynthesis in yeast also suppresses tRNA gene-mediated transcriptional silencing. Proc. Natl Acad. Sci. USA, 97, 13108–13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolton E.C. and Boeke,J.D. (2003) Transcriptional interactions between yeast tRNA genes, flanking genes and Ty elements: a genomic point of view. Genome Res., 13, 254–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deshpande A.M. and Newlon,C.S. (1996) DNA replication fork pause sites dependent on transcription. Science, 272, 1030–1033. [DOI] [PubMed] [Google Scholar]
- 17.Donze D. and Kamakaka,R.T. (2001) RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J., 20, 520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patterton H.G. and Simpson,R.T. (1994) Nucleosomal location of the STE6 TATA box and Mat alpha 2p-mediated repression. Mol. Cell Biol., 14, 4002–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimizu M., Roth,S.Y., Szent-Gyorgyi,C. and Simpson,R.T. (1991) Nucleosomes are positioned with base pair precision adjacent to the alpha 2 operator in Saccharomyces cerevisiae. EMBO J., 10, 3033–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith R.L. and Johnson,A.D. (2000) Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem. Sci., 25, 325–330. [DOI] [PubMed] [Google Scholar]
- 21.Wu J., Suka,N., Carlson,M. and Grunstein,M. (2001) TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol. Cell, 7, 117–126. [DOI] [PubMed] [Google Scholar]
- 22.Davie J.K., Edmondson,D.G., Coco,C.B. and Dent,S.Y. (2003) Tup1-Ssn6 interacts with multiple class I histone deacetylases in vivo. J. Biol. Chem., 278, 50158–50162. [DOI] [PubMed] [Google Scholar]
- 23.Watson A.D., Edmondson,D.G., Bone,J.R., Mukai,Y., Yu,Y., Du,W., Stillman,D.J. and Roth,S.Y. (2000) Ssn6-Tup1 interacts with class I histone deacetylases required for repression. Genes Dev., 14, 2737–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Redd M.J., Arnaud,M.B. and Johnson,A.D. (1997) A complex composed of tup1 and ssn6 represses transcription in vitro. J. Biol. Chem., 272, 11193–11197. [DOI] [PubMed] [Google Scholar]
- 25.Kuras L. and Struhl,K. (1999) Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature, 399, 609–613. [DOI] [PubMed] [Google Scholar]
- 26.Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iyer V. and Struhl,K. (1996) Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 93, 5208–5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guldener U., Heck,S., Fielder,T., Beinhauer,J. and Hegemann,J.H. (1996) A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res., 24, 2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuo M.H. and Allis,C.D. (1999) In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods, 19, 425–433. [DOI] [PubMed] [Google Scholar]
- 30.Zhong H. and Vershon,A.K. (1997) The yeast homeodomain protein MATalpha2 shows extended DNA binding specificity in complex with Mcm1. J. Biol. Chem., 272, 8402–8409. [DOI] [PubMed] [Google Scholar]
- 31.Rieger K.J., Aljinovic,G., Lazowska,J., Pohl,T.M. and Slonimski,P.P. (1997) A novel nuclear gene, CBT1, essential for mitochondrial cytochrome b formation: terminal processing of mRNA and intron dependence. Curr. Genet., 32, 163–174. [DOI] [PubMed] [Google Scholar]
- 32.Vidal M. and Gaber,R.F. (1991) RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol. Cell Biol., 11, 6317–6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu C., Weiss,K., Yang,C., Harris,M.A., Tye,B.K., Newlon,C.S., Simpson,R.T. and Haber,J.E. (1998) Mcm1 regulates donor preference controlled by the recombination enhancer in Saccharomyces mating-type switching. Genes Dev., 12, 1726–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szeto L., Fafalios,M.K., Zhong,H., Vershon,A.K. and Broach,J.R. (1997) Alpha2p controls donor preference during mating type interconversion in yeast by inactivating a recombinational enhancer of chromosome III. Genes Dev., 11, 1899–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roth S.Y., Shimizu,M., Johnson,L., Grunstein,M. and Simpson,R.T. (1992) Stable nucleosome positioning and complete repression by the yeast alpha 2 repressor are disrupted by amino-terminal mutations in histone H4. Genes Dev., 6, 411–425. [DOI] [PubMed] [Google Scholar]
- 36.Cooper J.P., Roth,S.Y. and Simpson,R.T. (1994) The global transcriptional regulators, SSN6 and TUP1, play distinct roles in the establishment of a repressive chromatin structure. Genes Dev., 8, 1400–1410. [DOI] [PubMed] [Google Scholar]
- 37.Gavin I.M., Kladde,M.P. and Simpson,R.T. (2000) Tup1p represses Mcm1p transcriptional activation and chromatin remodeling of an a-cell-specific gene. EMBO J., 19, 5875–5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bertrand E., Houser-Scott,F., Kendall,A., Singer,R.H. and Engelke,D.R. (1998) Nucleolar localization of early tRNA processing. Genes Dev., 12, 2463–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson M., Haeusler,R.A., Good,P.D. and Engelke,D.R. (2003) Nucleolar clustering of dispersed tRNA genes. Science, 302, 1399–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fairley J.A., Scott,P.H. and White,R.J. (2003) TFIIIB is phosphorylated, disrupted and selectively released from tRNA promoters during mitosis in vivo. EMBO J., 22, 5841–5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roberts D.N., Stewart,A.J., Huff,J.T. and Cairns,B.R. (2003) The RNA polymerase III transcriptome revealed by genome-wide localization and activity-occupancy relationships. Proc. Natl Acad. Sci. USA, 100, 14695–14700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harismendy O., Gendrel,C.G., Soularue,P., Gidrol,X., Sentenac,A., Werner,M. and Lefebvre,O. (2003) Genome-wide location of yeast RNA polymerase III transcription machinery. EMBO J., 22, 4738–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moqtaderi Z. and Struhl,K. (2004) Genome-wide occupancy profile of the RNA polymerase III machinery in Saccharomyces cerevisiae reveals loci with incomplete transcription complexes. Mol. Cell Biol., 24, 4118–4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lassar A.B., Martin,P.L. and Roeder,R.G. (1983) Transcription of class III genes: formation of preinitiation complexes. Science, 222, 740–748. [DOI] [PubMed] [Google Scholar]
- 45.Kassavetis G.A., Braun,B.R., Nguyen,L.H. and Geiduschek,E.P. (1990) S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell, 60, 235–245. [DOI] [PubMed] [Google Scholar]
- 46.Donze D. and Kamakaka,R.T. (2002) Braking the silence: how heterochromatic gene repression is stopped in its tracks. Bioessays, 24, 344–349. [DOI] [PubMed] [Google Scholar]