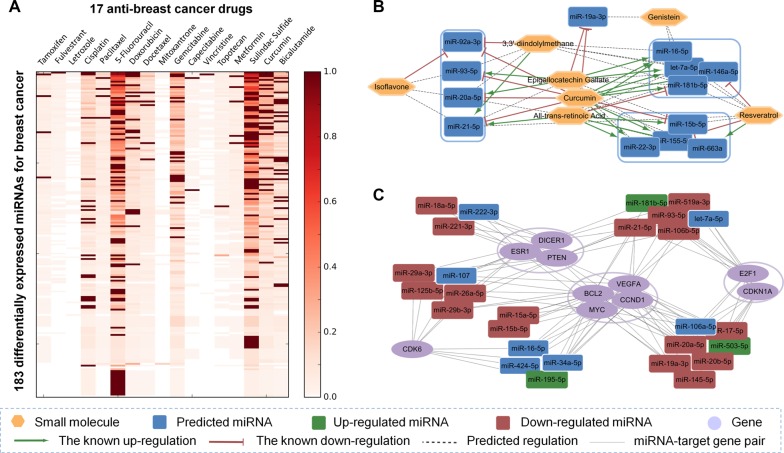
Figure 4. Identification of potential pharmacogenomic biomarkers for breast cancer, natural products and non-steroidal anti-inflammatory drugs (NSAIDs) via the SMiR-NBI model.
(A) The heatmap showed the predicted scores (color keys) of the SMiR-NBI model for 183 differentially expressed miRNAs in breast cancer from The Cancer Genome Atlas (see Methods) and 17 anti-breast cancer drugs. (B) The representative miRNA pharmacogenomic pathway for natural products included 13 common miRNAs regulated by at least 3 different natural products. (C) The representative miRNA pharmacogenomic pathway for NSAIDs contained 10 hub genes with the highest degrees in NSAIDs-regulating miRNA-gene subnetwork and 27 miRNAs targeting at least 3 hub genes. Regulations between NSAIDs and miRNAs were denoted by different colors.