Abstract
PCR amplification of limited amounts of DNA template carries an increased risk of product redundancy and contamination. We use molecular barcoding to label each genomic DNA template with an individual sequence tag prior to PCR amplification. In addition, we include molecular ‘batch-stamps’ that effectively label each genomic template with a sample ID and analysis date. This highly sensitive method identifies redundant and contaminant sequences and serves as a reliable method for positive identification of desired sequences; we can therefore capture accurately the genomic template diversity in the sample analyzed. Although our application described here involves the use of hairpin-bisulfite PCR for amplification of double-stranded DNA, the method can readily be adapted to single-strand PCR. Useful applications will include analyses of limited template DNA for biomedical, ancient DNA and forensic purposes.
INTRODUCTION
The polymerase chain reaction (PCR) allows multiple copies of selected DNA sequences to be copied from limited amounts of DNA template (1). Reactions with limited template, however, increase the risk of amplifying contaminant DNA and can also result in a skewed yield of PCR products such that there is a high degree of redundancy for a small portion of the original genomic sequences (2). Redundancy can either be useful, e.g. in tracking mutations arising during the PCR amplification of individual molecules, or unwelcome, for example, when the goal is to compare and quantify sequences from different cells represented in the same DNA sample, as in bisulfite methylation analysis (3). The frequent observation of multiple amplified sequences derived from a single original molecule was also noted by Millar et al. (4) in the context of bisulfite genomic sequencing, a method increasingly used in epigenetic research.
In response to the challenges of PCR redundancy and contamination associated with PCR amplification of limited amounts of DNA template, we have labeled genomic DNA fragments with molecular sequence barcodes and ‘batch-stamps’ prior to PCR amplification. This was accomplished by including these molecular labels in the hairpin linker sequence that we use in hairpin-bisulfite PCR (5). This encoded information enables us to track the genomic origin of each sequence obtained from PCR and subsequent bacterial cloning. Each genomic fragment is marked prior to amplification, allowing us to identify contaminant and redundant sequences and to quantify accurately the proportion of cells carrying a particular sequence variant by counting only distinctly tagged sequences. This highly sensitive method offers confirmation of the independent genomic origin of all sequences in final data sets derived from PCR amplification.
MATERIALS AND METHODS
Conditions for hairpin-bisulfite PCR of human genomic FMR1 sequences (5) were as follows: 5 μg of genomic DNA was cleaved by 10 U each of restriction endonucleases DraIII and AluI for 1 h at 37°C, followed by enzyme inactivation at 65°C for 20 min. The use of a second restriction endonuclease, in this case AluI, removed the CG-rich sequence distal to the region analyzed. Ligation of the hairpin linker (5′P-AGCGATGCDDDDDDDGCATCGCT-TGA, with variations in the non-random nucleotides for batch-stamps) to DraIII-cleaved genomic DNA was for 15 min at 20°C, using 400 U of T4 ligase in 20 μl with 1× ligation buffer (New England Biolabs), followed by enzyme inactivation at 65°C for 20 min.
The bisulfite conversion followed the protocol of Laird et al. (5) with additional thermal denaturation steps. Hairpin-ligated DNA was denatured in 0.3 M NaOH for 20 min, then heated to 100°C for 1 min before addition of sodium bisulfite and hydroquinone to 3.4 M and 1 mM, respectively. The reaction mixture was incubated for 6 h at 55°C, with additional thermal denaturation steps (99°C for 90 s, 10 times over the 6 h), and then incubated for an additional 6 h at 55°C. This was followed by a purification step using QIAquick PCR purification columns (Qiagen), subsequent treament with NaOH (final concentration 0.3 M) at 37°C for 20 min, and another purification using Microspin S-200 HR columns (Amersham Pharmacia Biosciences). PCR conditions were Hotstar Master Mix (Qiagen), with denaturation at 95°C for 15 min, followed by 38 cycles of denaturing at 95°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 45 s; this was followed by a final extension at 72°C for 5 min. Primers used were (i) first primer, 5′-CCTCTCTCTTCAAATAACCTAAAAAC-3′ and (ii) second primer, 5′-GTTGYGGGTGTAAATATTGAAATTA-3′.
All PCR products were analyzed by agarose gel electrophoresis; further cloning and sequencing of appropriately sized products was with TOPO TA Cloning Kits (Invitrogen Life Technologies); sequencing reactions were carried out with fluorescent dideoxy nucleotides (BIGDYE Terminator 3.1, Applied Biosystems), at either the DNA Sequencing Facility, Department of Biochemistry, or the Comparative Genomics Center, Department of Biology, University of Washington. Each sequence was proofread against the sequence trace; errant base calling was corrected manually before being presented here. For purposes of analysis and presentation, the output sequence was folded, using word-processing software, into a hairpin conformation so that both strands aligned.
RESULTS
The challenge of amplifying limited amounts of DNA template can result from trace amounts of initial DNA sample, or from laboratory analyses that include substantial DNA degradation as a necessary side effect of processing, as in bisulfite genomic sequencing (6). One of the major problems encountered in these analyses is to capture accurately the genomic template diversity following the steps of PCR and bacterial cloning. Hairpin-bisulfite PCR involves the ligation of a synthetic hairpin linker to the ends of a double-stranded genomic DNA fragment prior to bisulfite conversion and PCR amplification (5). While the primary purpose of the hairpin linker is to maintain attachment of complementary strands, it can also be used to encode each ligated genomic fragment with information that distinguishes it from other sequences within a sample, allowing us to evaluate cloned sequences for redundancy and contamination. To accomplish this, we replaced the 6 nt loop of our hairpin linker (5) with 7 nt randomly selected from A, G and T. Cytosine was not used because its identity would be ambiguous after bisulfite conversion. With a random 7 nt barcode, the number of possible codes is 2187; in selecting 15 cloned PCR products from one DNA sample, the probability that two of these will be different genomic fragments labeled with identical 7 nt barcodes is 0.047 (see Supplementary Material). Some applications will require a larger pool of random-sequence barcodes if more independently derived sequences are required. We have used linkers with up to 13 nt in the hairpin loop with no observable detriments to sequence recovery. A 13 nt barcode gives ∼1.6 × 106 different codes; even for a selection of 100 cloned PCR products, the probability that two of these would be different genomic fragments labeled with identical barcodes is only 0.0031 (see Supplementary Material).
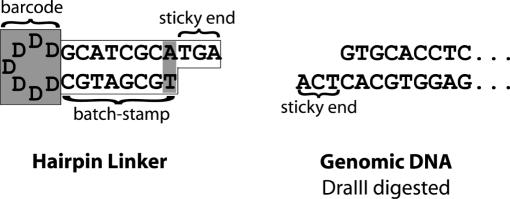
In addition to adding the random barcode, we ‘batch-stamped’ molecules by encoding the hairpin linker with information that would designate the sample analyzed and the date of analysis. We designed multiple variants of the hairpin linker by changing nucleotides in the stem of the linker. These stem changes represented different batches of linkers, each of which we used for the analysis of a different sample. Thus, the resulting sequences each bear a consistent ‘batch-stamp’ encoded in the stem, and a randomly variable barcode encoded in the loop (Figure 1).
Figure 1.
Schematic of barcoded and batch-stamped hairpin linker, designed for ligation to DraIII-cut genomic DNA of FMR1. The letter D represents a nucleotide randomly selected from A, G and T.
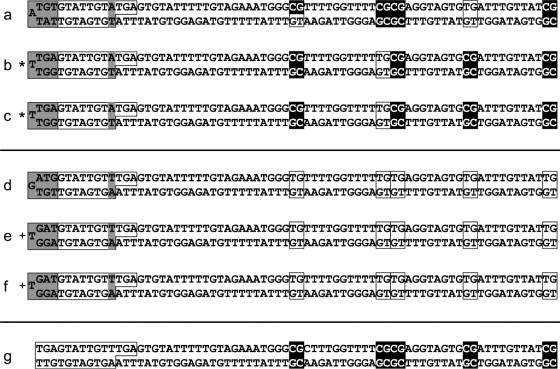
We applied our enhanced hairpin-bisulfite PCR method to the FMR1 promoter region in the DNA of males with fragile X syndrome. The classes of sequences recovered included hypermethylated sequences with distinctive barcodes and patterns of methylation (Figure 2a), redundant hypermethylated sequences with identical barcodes and methylation patterns (Figure 2b–c), hypomethylated sequences with distinctive barcodes (Figure 2d), redundant hypomethylated sequences with identical barcodes (Figure 2e–f), and contaminant sequences with our original linker that predates the barcoding (Figure 2g). The number of sequences cloned influenced the observed proportion of redundancy among the recovered sequences; the observed proportions of both redundancy and contamination appeared to depend on the initial amount of DNA used and the quality of the bisulfite conversion. Among eight different DNA samples analyzed, the proportion of sequences that were redundant ranged from 7 to 51%, and the proportion of sequences that were contaminants ranged from 0 to 14%. Occasionally, contaminant sequences were cloned from PCR reactions in which control reactions (those without template DNA) showed no DNA bands on ethidium–bromide-stained agarose gels. In these contexts, barcoding serves as a highly accurate method for positive identification of desired sequences.
Figure 2.
FMR1 promoter sequences, with inferred methylation states of CpG sites, recovered from male fragile X patients using hairpin-bisulfite PCR with linker barcoding and batch-stamping. Methods are as described in the text. Unconverted (methylated) CpG dyads are black, and converted (unmethylated) CpG dyads are boxed. Within the 26 nt linker (boxed region at left), the randomized 7 nt variable barcodes are shaded at far left; the designated variable batch-stamps (A:T or T:A) are shaded at right. All sequences show 100% conversion of non-CpG cytosines. (a) A distinctive hypermethylated sequence. (b and c) Redundant hypermethylated sequences recovered from independent bacterial colonies, with identical barcodes and methylation patterns. (d) A hypomethylated sequence with a distinctive barcode. (e and f) Redundant hypomethylated sequences with identical barcodes recovered from independent bacterial colonies. These are distinguishable as redundant and as different from the hypomethylated sequence ‘d’ only because of barcoding. (g) A contaminant sequence bearing our original hairpin linker that predates the addition of the barcode and batch-stamp. This sequence was recovered during analysis of the same sample that generated sequences ‘a–c’. Sequences ‘a–c’ carry a different batch-stamp than sequences ‘d–f’, with the inversion of the A–T base pair, confirming that these sequence sets came from different DNA samples. Redundant hypermethylated sequences are denoted with asterisks (*), and redundant hypomethylated sequences with plus signs (+).
Within 142 barcodes recovered from multiple reactions with FMR1, the average nucleotide composition was 54% T, 26% G and 19% A. This bias is similar to that previously reported for the influence of loop nucleotides on the stability of DNA hairpin structures (7).
DISCUSSION
The concept of molecular barcoding has previously been used in signature-tagged mutagenesis (8,9), to track the origins of expressed sequence tags (10), and to label objects for identification and authentication (11,12). Here, we apply this concept to the labeling of individual genomic fragments with distinct sequence tags. The ability to barcode and ‘batch-stamp’ genomic DNA sequences from individual alleles is useful in situations where template DNA is limited, thus identifying contaminants and redundant sequences arising from template re-cloning. We have identified contaminant sequences even when multiple control (no DNA) PCR samples were negative. Barcoding allows for quantification of the relative abundance of genomic methylation patterns or polymorphic sequences by correcting for skewing that can arise from PCR amplification or the cloning of the products. The barcoding method thus provides a definitive solution to the problem identified by both Taylor et al. (2) and Millar et al. (4), in which multiple amplified sequences are derived from a single original molecule when template DNA is limited or of poor quality. The method also allows for the analysis of mutations arising during PCR amplification. Although our application described here involves the use of hairpin-bisulfite PCR for amplification of double-stranded DNA, the method can readily be adapted to single-strand PCR. Useful applications will include analyses of limited template DNA for biomedical, ancient DNA and forensic purposes.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Stanley Gartler, Diane Genereux and Carl Bergstrom for helpful discussions and suggestions. Support was provided by National Institutes of Health Grants GM 53805, HD 02274 and HD 16659.
REFERENCES
- 1.Saiki R.K., Scharf,S., Faloona,F., Mullis,K.B., Horn,G.T., Erlich,H.A. and Arnheim,N. (1985) Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle-cell anemia. Science, 230, 1350–1354. [DOI] [PubMed] [Google Scholar]
- 2.Taylor J.M., Spagnolo,D.V. and Kay,P.H. (1997) B-cell target DNA quantity is a critical factor in the interpretation of B-cell clonality by PCR. Pathology, 29, 309–312. [DOI] [PubMed] [Google Scholar]
- 3.Stöger R., Kajimura,T.M., Brown,W.T. and Laird,C.D. (1997) Epigenetic variation illustrated by DNA methylation patterns of the fragile-X gene FMR1. Hum. Mol. Genet., 6, 1791–1801. [DOI] [PubMed] [Google Scholar]
- 4.Millar D.S., Warnecke,P.M., Melki,J.R. and Clark,S.J. (2002) Methylation sequencing from limiting DNA: embryonic, fixed, and microdissected cells. Methods, 27, 108–113. [DOI] [PubMed] [Google Scholar]
- 5.Laird C.D., Pleasant,N.D., Clark,A.D., Sneeden,J.L., Hassan,K.M., Manley,N.C., Vary,J.C., Jr., Morgan,T., Hansen,R.S. and Stöger,R. (2004) Hairpin-bisulfite PCR: assessing epigenetic methylation patterns on complementary strands of individual DNA molecules. Proc. Natl Acad. Sci. USA, 101, 204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grunau C., Clark,S.J. and Rosenthal,A. (2001) Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res., 29, e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Senior M.M., Jones,R.A. and Breslauer,K.J. (1988) Influence of loop residues on the relative stabilities of DNA hairpin structures. Proc. Natl Acad. Sci. USA, 85, 6242–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hensel M., Shea,J.E., Gleeson,C., Jones,M.D., Dalton,E. and Holden,D.W. (1995) Simultaneous identification of bacterial virulence genes by negative selection. Science, 269, 400–403. [DOI] [PubMed] [Google Scholar]
- 9.Shoemaker D.D., Lashkari,D.A., Morris,D., Mittmann,M. and Davis,R.W. (1996) Quantitative phenotypic analysis of yeast deletion mutants using a highly parallel molecular bar-coding strategy. Nature Genet., 14, 450–456. [DOI] [PubMed] [Google Scholar]
- 10.Qiu F., Guo,L., Wen,T.J., Liu,F., Ashlock,D.A. and Schnable,P.S. (2003) DNA sequence-based ‘bar codes’ for tracking the origins of expressed sequence tags from a maize cDNA library constructed using multiple mRNA sources. Plant Physiol., 133, 475–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cook L.J. and Cox,J.P.L. (2003) Methylated DNA labels for marking objects. Biotechnol. Lett., 25, 89–94. [DOI] [PubMed] [Google Scholar]
- 12.Cox J.P. (2001) Bar coding objects with DNA. Analyst, 126, 545–547. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.