Abstract
Balance between the redox pair of nicotinamide adenine dinucleotides (oxidized NAD+ and reduced NADH), reflects the oxidative state of cells and the ability of biological systems to carry out energy production. A growing body of evidence suggests that an “immuno-oxidative” pathway including oxidative stress, mitochondrial dysfunction, neuroinflammation, and cell-mediated immune response may contribute to disruptions in brain activity in schizophrenia (SZ). The aim of this study is to assess possible redox imbalance in SZ patients by using a novel in vivo 31P MRS technique. The participants included 40 healthy controls, 21 chronic SZ, 13 first-episode (FE) SZ, and 18 FE bipolar disorder (BD) patients (as a psychiatric control group). All participants initially underwent structural imaging at a 3 Tesla (3 T) and 31P MRS measurements were performed on a 4 T MR scanner. NAD+ and NADH components were determined by nonlinear least-square fitting of the model simulated spectra; these incorporated prior chemical shift and coupling constant information to in vivo resonances obtained from 31P MRS experiments. We found a significant reduction in the NAD+/NADH ratio in chronically ill SZ patients compared to a matched healthy control group, and in FE SZ patients compared to both a matched FE BD patient group and a matched healthy control group. These findings provide evidence for redox imbalance in the brain in all phases of SZ, potentially reflecting oxidative stress.
Key words: redox state, NAD+ and NADH, schizophrenia, 31P MRS, oxidative stress
Introduction
Despite its public health impact and a century of biological research, the pathophysiology of schizophrenia (SZ) remains poorly understood. There is an urgent need to develop better insights into the neurobiology of SZ and to develop novel treatments. Several lines of evidence suggest SZ is associated with a range of biochemical brain abnormalities, including mitochondrial dysfunction,1–3 impaired bioenergetics,4 neuroinflammation,5–8 and oxidative stress.9–12
Nicotinamide adenine dinucleotide (NAD), which exists in oxidized (NAD+) and reduced form (NADH) has long been implicated in energy metabolism, reductive biosynthesis, and antioxidant activity. While the major biological function of NAD+ is to modulate cellular energy metabolism, mounting evidence indicates that NAD+ is also involved in biological activities such as cell death,13,14 calcium homeostasis,15,16 gene expression,17,18 aging, carcinogenesis, and immunological functions.15,19 The redox balance indicated by the ratio of concentrations of the redox pair NAD+ and NADH (Redox Ratio [RR] = [NAD+]/[NADH]) is critical to our understanding of oxidative stress for several reasons20: First, the redox pair of NAD+ and NADH are involved in critical roles for numerous oxidation-reduction reactions, and their ratio reflects cellular reducing potential. A more negative redox potential corresponds to a lower RR, indicating lower oxidation power and higher oxidative stress. Second, NAD+ kinases (NADKs) are the sole enzymes that convert NAD+ to nicotinamide adenine dinucleotide phosphate (NADP+), which is the precursor for NADPH (ie, reduced form of NADP). While NADKs can be inhibited by NADH and NADPH, oxidative stress can activate NADKs. Third, NADH may have direct antioxidant effects.21,22 Lastly, NAD+ can inhibit the generation of reactive oxygen species (ROS) from alpha-ketoglutarate dehydrogenase and pyruvate dehydrogenase.23
Despite the crucial roles of NAD+ and NADH (together termed NAD metabolites), their noninvasive in vivo detection is extremely challenging. Only 2 general approaches have been used thus far to investigate cellular NAD metabolism: the first is based on biochemical analyses such as high-performance liquid chromatography,24 capillary electrophoresis,25 or enzymatic cycling assays.26 These involve invasive techniques which preclude longitudinal assessment of NAD content in living organs. Furthermore, NAD+ and/or NADH content might be altered during sample preparation. The other approach is autofluorescence.27 The drawback of this method is low detection sensitivity of NADH signal from deep tissues due to limited penetration depth. Moreover NAD+ does not fluoresce.27 Thus, one cannot calculate RR in vivo using this approach. Given the importance of NAD metabolism in living organs, noninvasive in vivo detection of NAD+ and NADH signals would be a critical advance.
In vivo 31P MRS provides a critical window into membrane phospholipid metabolism and high-energy phosphate molecule dynamics. In addition to commonly observed signals such as phosphocreatine (PCr), inorganic phosphate (Pi), and the 3 adenosine triphosphate (ATP) resonances, NAD signals have been identified. However, because of low concentration of NAD metabolites (below 1mM), these have previously been considered below the in vivo MRS detection limit. Recently, a 31P MRS-based NAD quantification method has been developed to directly quantify NAD+ and NADH concentrations in vivo at ultra-high field MR scanners (7.0 and 9.4 Tesla).28,29 This method applied at a 4 Tesla (4 T) scanner was recently reported.30 This approach is based on a theoretical NMR model with prior knowledge (ie, chemical shift and J-coupling constants) to predict phosphorus NAD+ and NADH resonance signals. Thus, both NAD+ and NADH signals were quantified simultaneously by fitting the model simulated spectra to in vivo resonances.
Here we implemented the 31P MRS-based NAD quantification method at our 4 T scanner, then applied this novel method in a clinical study. We investigate the brain’s RR in SZ patients with chronic, well-established illness and first-episode (FE) SZ patients who have not been exposed to the confounding effects of chronic medication treatment and chronic psychosis. We also present data from a FE bipolar disorder (BD) cohort, as a psychiatric control group. Based on the prior literature we hypothesized that we would find a more negative redox potential (ie, lower RR) in the brains of SZ patients as direct evidence of oxidative stress. We did not have a hypothesis on findings in FE vs chronic SZ patients because the literature does not provide guidance on whether oxidative stress would progress with disease stage, or whether it may be more active in FE SZ patients and abate with chronic illness.
Methods
Participants
The participants included 40 healthy controls, 21 chronic SZ, 13 FE SZ, and 18 FE BD patients. For the test–retest study, 6 of these healthy participants were scanned twice within a 1-week period. Informed consent was obtained from all participants at study entry. The details of our IRB-approved human participants procedures were provided in supplementary material. Details for participant demographics are listed in supplementary table S1.
Magnetic Resonance Imaging and in vivo 31P MRS Experiments
Each participant underwent structural imaging at a 3 T Siemens scanner to rule out structural abnormalities. All 31P MRS measurements were performed on a 4 T whole-body scanner interfaced with Varian INOVA console and 31P spectra were acquired from frontal lobe of brain (supplementary figure S1). The data for chronic SZ and matched healthy-controls were from spectra collected in our previous study of creatine kinase (CK) and ATP synthesis reaction rates4 but now analyzed for an independent unrelated characteristic (RR). Additional data (ie, FE SZ and BD patients and age-matched healthy-controls) were also acquired. Detailed time frame for data acquisition is provided in supplementary material. For the unrelated purposes of the CK study, all 31P MRS data were acquired with a magnetization transfer strategy (31P-MT-MRS), where we collected a series of 31P spectra (every 26 signal averages) in the absence (control) and presence (6 series of spectra with varying saturation times) of a saturating pulse train centered on the γ-ATP frequency. Since the saturation pulse train (bandwidth 180 Hz) has no effect on the spectral region containing NAD+ and NADH, which is ~400 Hz upfield of the γ-ATP resonance (supplementary figure S2), this data collection strategy (31P-MT-MRS) could be used for NAD measurements without loss of signal due to saturation pulses. Details of the experiment have been published previously4 and more information was presented in supplementary material.
Voxel Segmentation
The volume fractions of different tissue types (gray/white matter and cerebrospinal fluid) were calculated by segmenting T1-weighted images acquired at 3 T; the percentage of different tissue types was extracted from the MRS voxel by incorporating voxel information (voxel position, size, orientation).
Spectral Processing, and NAD+ and NADH Quantification
The raw free induction decay (FID) of 31P MRS were zero-filled to increase spectral resolution and 8-Hz line broadening was applied before Fourier transformation to increase signal-to-noise ratio (SNR). After frequency drifts correction, we summed all series of spectra (control + saturated, total 7 series of spectra) to achieve sufficient SNR. Then, the spectra were phase and baseline corrected to ensure a symmetric α-ATP peak. The SNRs of each resonance (PCr, α-ATP, and NAD) were calculated by dividing the peak intensity by the standard deviation (SD) of the noise. The chemical shift assignment was done after setting PCr resonance at −2.5ppm as chemical shift reference. All spectral processing was conducted using in-house written MATLAB software.
For NAD+ and NADH quantification, we assumed that the half line width (HLW) of NAD+ and NADH resonances were the same. Fitting was performed on frequency domain and the unknown parameters (ie, signal intensities and HLW of α-ATP, NAD+, and NADH) were determined using a nonlinear least-square fitting of the model output to in vivo 31P resonances of NAD+, NADH and α-ATP. More detailed descriptions can be found in supplementary material. The fitting errors were calculated by dividing SD of residual signal by mean value of fitted signal. Intracellular RR was calculated from the fitted peak area ratio of NAD+ and NADH. In order to remain consistent with and compare to prior reports, the concentrations of NAD+ and NADH were determined using α-ATP signal as an internal reference, in which its brain concentration was set to 2.8mM.28,29
For the validity of our measurement, we summed all spectra of individual participant in group of chronic SZ and matched controls and the summed spectra were fitted again. In this case, uridine diphosphate glucose (UDPG) was also included in the fitting routine with published values of chemical shift and coupling constant31 to indentify the potential contaminations of UDPG to NAD content. However, for the fitting of individual spectra, we neglected UDPG model because of the limited SNR of this metabolite. The MRS data and voxel segmentation were processed blind to diagnosis.
Statistical Analysis
RR was our primary outcome measure. We first carried out an ANOVA comparing chronic SZ patients with matched controls. Since visual inspection revealed an unusual reduction in RR among the younger participants, we then carried out an ANOVA comparing the FE SZ patients to an age-matched group of healthy controls and a group of FE BD patients (post hoc Dunnett, using FE SZ as the group of interest). Finally, we compared the FE SZ group to the group of chronic SZ patients in an ANOVA. All ANOVAs used age as covariate. We considered 2-tailed P values lower than .0167 (=0.05/3 for 3 ANOVAs) as statistically significant. Furthermore, we compared RR in a small subgroup of patients not taking antipsychotic medication (N = 6) with RR in patients taking these medications (supplementary table S1).
We next computed correlation coefficients in order to assess the relationship between demographic/clinical parameters and RR. These included the whole sample for, body mass index (BMI) and, education; and the SZ patient sample for Positive and Negative Syndrome Scale (PANSS) and other symptom scales, Chlorpromazine (CPZ) equivalents, age at onset, illness duration (details in supplementary table S1). A large number of correlation analyses were performed on an exploratory basis; we considered a Pearson’s R value greater than 0.300 uncorrected for multiple comparisons as statistically and clinically significant.
Results
Validation of NAD Measures
Given NAD+ is structurally similar to NADH (supplementary figure S3a) with similar 31P NMR properties, we carried out a series of studies characterizing the technical approach: demonstration of excellent agreement between simulated and experimentally measured in vitro 31P spectra to validate model accuracy (supplementary figures S3b and S3c); Monte Carlo simulations with different SNR of α-ATP to demonstrate excellent fitting accuracy for our chosen threshold of SNR > 40 for processing in vivo data (supplementary figures S4a and S4b) and to demonstrate discrimination of patient and healthy control RR values (supplementary figure S4c); demonstration that HLW of α-ATP lower than 18 Hz in our in vivo data (after 8 Hz line broadening) provides excellent fitting accuracy without severely overlapping resonances with NAD (supplementary figure S5); the absence of impact of line broadening on in vivo data during post-processing (carried out to enhance SNR) (supplementary figure S6); fitting of spectra with and without NADH in the model along with α-ATP and NAD+ to demonstrate the validity of the NADH fit (supplementary figure S7); and demonstration of excellent reliability in vivo through a test–retest study of 6 healthy participants scanned twice within 1 week (coefficient of variation for RR = 1.7%). The details are provided in supplementary material.
In vivo NAD+ and NADH Quantification in SZ Patients at 4 T
The demographic and clinical characteristics of all participants are provided in supplementary table S1. The FE SZ group is not matched with controls for sex; we therefore tested for but found no sex effect on RR among all healthy participants (t(39) = −0.366, P = .717).
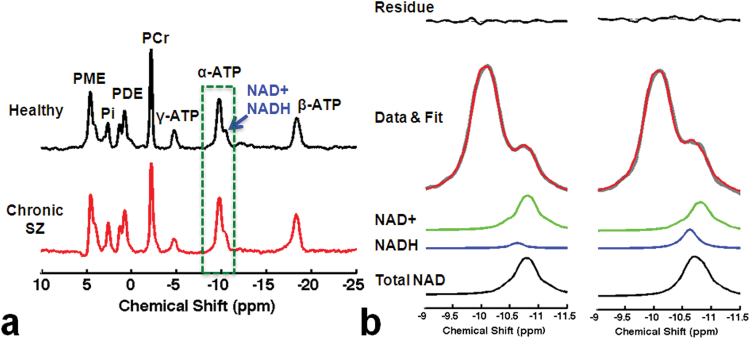
Figure 1a presents 31P spectra acquired from a healthy control (top) and a chronic SZ patient (bottom). The average SNR of PCr, α-ATP, and total NAD resonances (NAD+ and NADH), and HLW of PCr and α-ATP obtained from fitting procedures were not significantly different among the groups (P > .05; detailed parameters are listed in supplementary table S2). Figure 1b shows a magnified version of the relevant spectral region (stippled green box in figure 1a) with NAD+ and NADH fitting results and residual signal from the same participants. Overall fitting error was less than 5% and comparable between groups.
Fig. 1.
(a) In vivo summed 31P spectra from 7 series of spectra with/without magnetization transfer obtained from a representative healthy participant (top) and a chronic schizophrenia (SZ) patient (bottom). γ-ATP resonance is smaller compared with the other 2 ATP resonances due to the direct RF saturation effect as part of the 31P-MT-MRS experiment. (b) Fitting results from the healthy (left) and SZ participant (right).
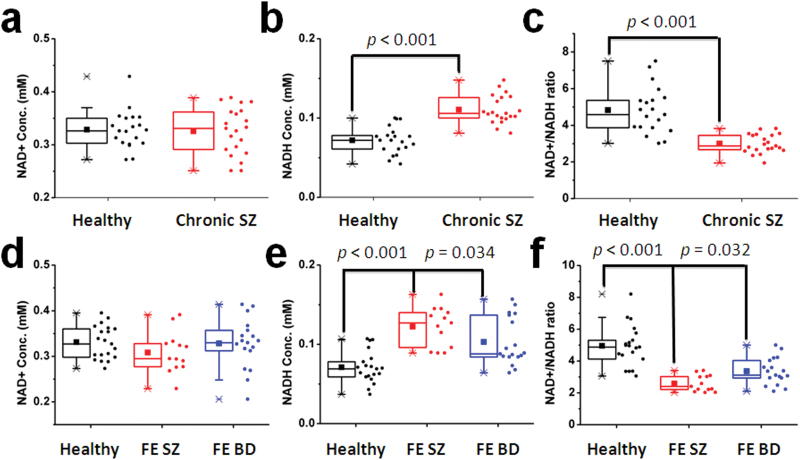
To test our a priori hypothesis, we first analyzed data from chronic SZ patients and matched healthy controls. Figure 2 (top panel) shows RR was significantly reduced by 38% in chronic SZ (F(1,40) = 42.7, P < .001). This finding was driven by a 53% NADH elevation in chronic SZ with no significant change in NAD+ (figure 2 and supplementary table S3). To investigate whether this significant abnormality is found in other phases of the illness, we collected data from FE SZ patients (N = 13) and age-matched (ie, young) controls (N = 20), as well as FE BD patients (N = 18; a psychiatric control group which receives similar treatments as SZ). Figure 2 (bottom panel) displays in vivo NAD+, NADH, and RR data in these 3 groups. Analysis revealed a significant difference in RR (F(2,48) = 25.5, P < .001). In post hoc testing, FE SZ patients had a highly significant 48% reduction in RR compared with healthy controls (Dunnett P < .001), as well as a more modest 23% reduction compared with FE BD patients (Dunnett P = .032). Again this pattern was driven by the elevated NADH without abnormalities in NAD+ (supplementary table S3). The FE SZ group also had 15% lower RR compared with chronic SZ, adjusting for age (F(2,31) = 21.9, P < .001). We found no correlations between RR and demographic (other than age) and clinical variables, which included CPZ equivalents (measure of antipsychotic dose) and tobacco smoking.
Fig. 2.
Box and whisker plot showing NAD+ (a and d), NADH (b and e), and RR (c and f) in the control and chronic schizophrenia (SZ) groups as well as in FE SZ, FE BD patients, and their age-matched control group. Individual data are also shown to the right of box plots. Bottom and top boundaries of each box indicate 25th and 75th percentiles, lower and upper whiskers indicate first and 99th percentiles. A horizontal line and filled square inside each box indicate median and mean values, respectively.
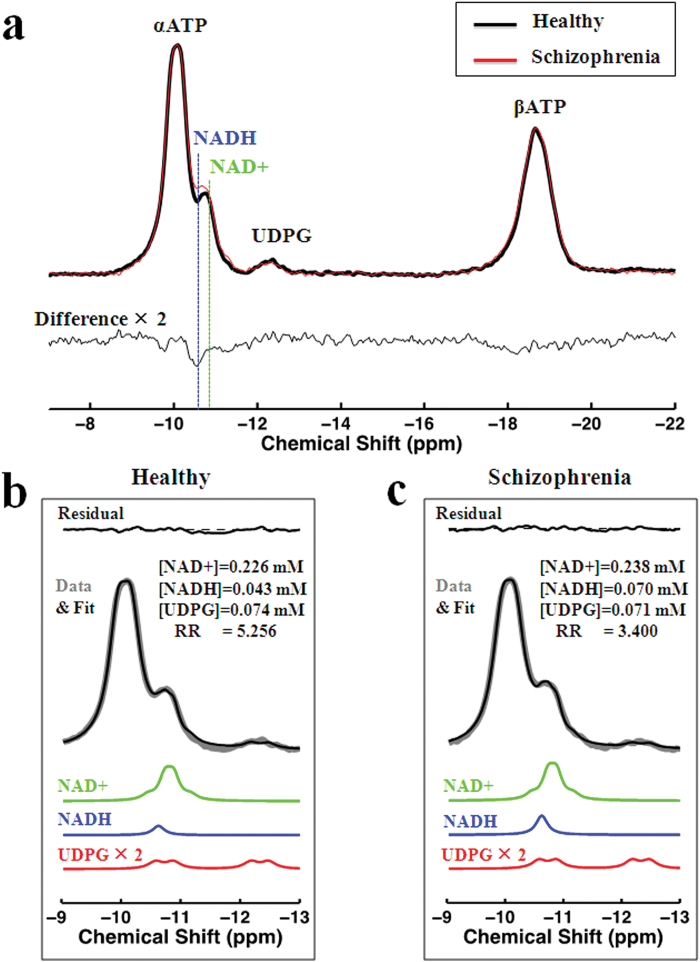
To provide a demonstration of NAD abnormalities independent of the fitting procedure and without other metabolite contributions (eg, UDPG), we summed individual participant spectra for the group of chronic SZ and matched controls, respectively, after normalization of peak intensity to α-ATP (internal reference) and spectral frequency alignment using a previously described algorithm (icoshift).32 Figure 3 shows summed spectra in range of −7ppm and −22ppm and the difference between them (figure 3a) as well as the fit results including UDPG resonances (figures 3b and 3c), clearly demonstrating an elevated NADH signal at −10.63ppm in chronic SZ compared to controls, without an apparent difference in the resonances of NAD+ and UDPG.
Fig. 3.
The summed spectra for healthy control and chronic schizophrenia (SZ) groups and the difference between them (a) as well as the fit results (b and c). The difference spectra and fitted Uridine diphosphate glucose (UDPG) signal are scaled by factor of 2.
Subgroup Analysis of Medication Effects
We found that there were no significant differences in RR between patients taking antipsychotic medications or lithium and those who were not (P > .05). Details are provided in supplementary material.
Voxel Segmentation
The voxel segmentation results revealed that there were no significant differences in voxel composition (ie, gray/white matter, CSF, muscle, and bone) across the groups. The detailed voxel compositions in each group are provided in supplementary table S3.
Age-Dependent Intracellular Redox State Changes
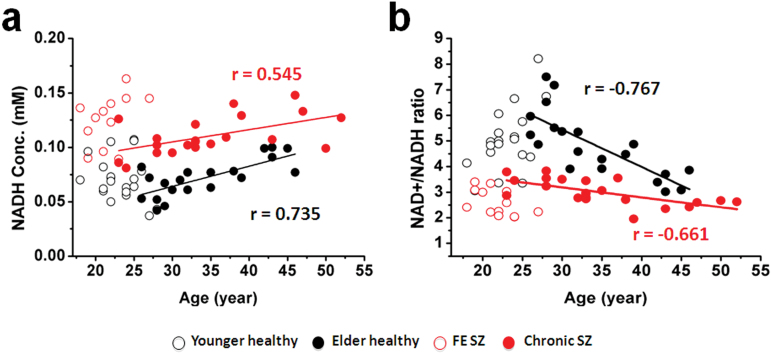
NAD measures and RR show strong age-dependence in healthy individuals.29 NADH and RR for SZ and healthy participants are plotted against age in figures 4a and 4b, demonstrating replication of RR decline in healthy participants29 and an extension of this finding to SZ patients. In addition, the younger participants have lower RR than older ones, apparently reversing the age-dependent trend. This finding requires further investigation in children and adolescents.
Fig. 4.
Age dependence of intracellular NADH level (a) and RR (b) in chronic schizophrenia (SZ) (red filled circles) and matched controls (black filled circles). R-values = Pearson correlation coefficients. In order to visualize the whole trend of redox state age-dependence, data from FE SZ (red open circles) and matched controls (black open circles) are presented. The older healthy group was recruited age/sex-matched for the chronic SZ group, and the younger healthy group matched for the FE SZ group. Some of the chronic SZ patients are relatively young, close in age to some FE SZ patients, and likewise for older and younger healthy participants. In fact, a correlation with age that includes all healthy participants together (since they are all comparable) is nonsignificant (R = 0.268, P = .095 for NADH and R = 9.297, P = .062 for Redox Ratio [RR]). This finding requires further investigation in children and adolescents.
Discussion
In this study, we carried out a substantial amount of work to demonstrate the feasibility of a 31P MRS-based in vivo intracellular redox state quantification technique at 4 T. Detailed discussion about our methods can be found in supplementary material. We then applied this novel method to investigate redox state in the frontal lobe of chronic and FE SZ, as well as FE BD patients. We found evidence for striking RR reductions in SZ, with every chronic SZ patient showing an RR of at least 1 SD below the control mean. RR reduction was of even greater magnitude among FE SZ patients. This study illustrates the power of examining in vivo brain redox dysregulation (measured as RR) in psychiatric disorders. Consistent with evidence from other studies, our findings suggest brain oxidative stress and altered energy production in SZ.9–11,33–35 Since RR can affect numerous critical enzymatic reactions, our findings for reduced RR in both FE and chronic SZ patients provide an important new insight into the pathophysiology of this major psychiatric condition.
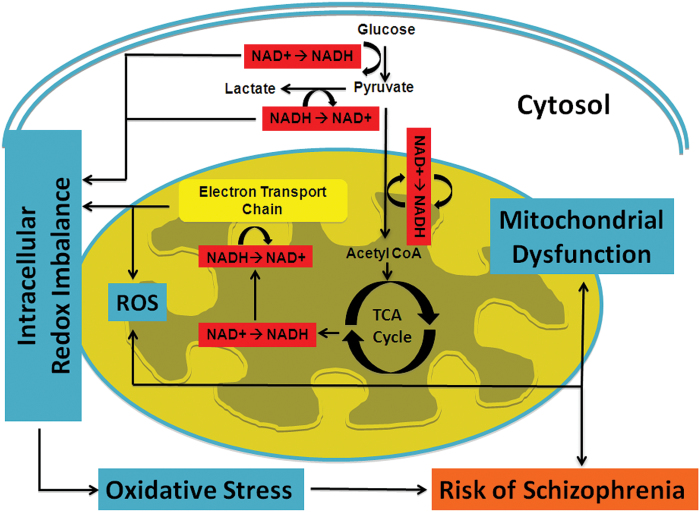
The redox pair of NAD+/NADH lies at the intersection of multiple critical biochemical pathways within cells. Figure 5 shows the metabolic pathways involving the NAD+/NADH redox pair in the cytosol and mitochondria. In the brain, most NADH molecules are produced from NAD+ during glycolysis in the cytosol (2 NADH per glucose), pyruvate decarboxylation (2 NADH per glucose), and the tricarboxylic acid (TCA) cycle (6 NADH per glucose) in mitochondria. The NADH molecules are converted back to NAD+ in the mitochondrial electron transport chain (ETC). Thus, RR is closely linked with brain carbohydrate metabolism and the balance between glycolysis and oxidative phosphorylation.29 Mitochondrial dysfunction can also contribute to abnormal RR independent of glycolysis. The regeneration of NAD+ in the ETC takes place in particular at the mitochondrial complex I. Expression of this complex is significantly reduced in SZ.36,37 Importantly, the ETC is also the site of generation of ROS in cells due to escape of unpaired electrons during oxidation reactions. Therefore, inefficient ETC activity would cause both NADH build-up and oxidative stress. Taken together, reduced RR in SZ can arise from a global shift from oxidative phosphorylation to glycolysis as the main source of ATP generation, or from abnormalities specifically in oxidative phosphorylation in mitochondria, or most likely both. These processes would lead to reduction in the number of ATP molecules and elevation in the number of ROSs generated per each molecule of glucose that is oxidized.
Fig. 5.
Diagram of metabolic pathways involving NAD+ and NADH redox reactions. Oxidative stress can induce redox imbalance in either cytosolic and/or mitochondria compartment, leading to mitochondrial dysfunction. The elevated NADH levels in patients with schizophrenia (SZ) can arise from either abnormal elevation in glycolysis in the cytosol, or disruption/inefficiency in the electron transport chain (ETC) of mitochondria, or both.
The significance of NAD metabolites (including NAD+ and NADH) goes beyond carbohydrate metabolism, however, since they are fundamental mediators for antioxidant defenses and therefore generation of oxidative stress. All major metabolic pathways including glycolysis, TCA cycle, respiratory chain, fatty acid oxidation as well as ketone body metabolism rely on redox reactions for which NAD+ is the most important co-substrate. Thus, the NAD+/NADH redox state is likely to impact flux through these metabolic pathways and vice versa. Sirtuins, a class of proteins involved in aging and stress damage, are NAD+ dependent, further highlighting the NAD metabolites’ critical role in cells.38 Interestingly, the brain is a major target for oxidative reactions, especially due to high amounts of oxidizable substrates, high oxygen tension, and relatively low antioxidant capacity.39,40 Our finding of reduced brain RR in SZ therefore suggests that the fragile balance of oxidation-reduction balance is disrupted in favor of oxidation in this condition.
There are additional points that should be noted. First, the redox state of cells is established by NADP+/NADPH and reduced glutathione (GSH)/oxidized glutathione (GSSG) in addition to NAD+/NADH and all 3 redox pairs are related.20,41,42 For instance, NAD+ is the substrate for the synthesis of NADP+ and NADP+ can be hydrolyzed to NAD+.43 Thus, alterations of the redox state of one of these redox pairs will also impact the others. RR reduction could reflect upstream perturbances in other metabolites.44,45 The intracellular concentrations of NADP+ and NADPH are low (~10%) compared with NAD+ and NADH, suggesting that our observed 31P signal from NAD groups is mainly attributed to NAD+ and NADH.46 It is generally accepted that NAD+/NADH is the predominant index of redox state.46 Therefore, RR still represents a useful index capturing oxidative stress in the brain in SZ even if it reflects abnormalities in other redox pairs. Given that neuroinflammation is associated with oxidative stress and downstream effects on antioxidants such as glutathione (GSH), it would be helpful to measure peripheral inflammation and/or in vivo GSH levels in order to better understand redox abnormality observed in this study. Second, we cannot rule out medication effects on RR because most patients who participated in this study were taking some medication. RR reduction was greater in the FE SZ patients than in the chronic SZ group even though the former had 60% lower CPZ equivalents. In addition, patients who were taking antipsychotic medications or lithium showed no significant difference in RR compared to those who were not, suggesting medication effects do not account for our findings. Lastly, figures 3b and 3c show that even including UDPG in spectral fitting, we can clearly identify increased NADH concentration and decreased RR and no significant differences in the concentrations of NAD+ and UDPG. All of these observations are consistent with our results derived from individual subject measurements indicating that the contamination from UDPG signal may not impact our findings. Of note, reported NAD+ and NADH concentrations differ somewhat depending on whether UDPG is included in the fitting (figures 3b and 3c and supplementary table S3).
In summary, using a noninvasive MR-based in vivo NAD assay, we provide direct evidence of redox abnormalities in a common and severe psychiatric disorder. We also find that FE SZ patients had more severe redox abnormalities compared with chronic patients, suggesting an active process in early stages of illness. This is partially improved in chronic illness, perhaps as a result of medication treatment. RR is influenced by multiple cellular signaling events and may constitute a metabolic integrator for local metabolic status within brain cells. The redox state is a key parameter in biological systems and oxidative stress may have widespread downstream effects on the brain, including on synaptic function and plasticity, as well as energy homeostasis. Therefore, our work provides new insights into the pathophysiology of SZ, as well as a biomarker for tracking the impact of treatment interventions.
Supplementary Material
Supplementary material is available at http://schizophreniabulletin.oxfordjournals.org.
Funding
MH094594 (D.O.); MH104449 (D.O.); NARSAD (D.O.); NARSAD (F.D.); MH092704 (F.D.); Shervert Frazier Research Institute (B.M.C.).
Supplementary Material
Acknowledgments
We thank X. Fan, J. Goldbach, and P. Hyunh for their assistance in acquiring clinical information and experimental help. The authors have no financial or other potential conflicts of interest to declare.
References
- 1. Gonçalves VF, Andreazza AC, Kennedy JL. Mitochondrial dysfunction in schizophrenia: an evolutionary perspective. Hum Genet. 2015;134:13–21. [DOI] [PubMed] [Google Scholar]
- 2. Prabakaran S, Swatton JE, Ryan MM, et al. Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol Psychiatry. 2004;9:684–697, 643. [DOI] [PubMed] [Google Scholar]
- 3. Rajasekaran A, Venkatasubramanian G, Berk M, Debnath M. Mitochondrial dysfunction in schizophrenia: pathways, mechanisms and implications. Neurosci Biobehav Rev. 2015;48:10–21. [DOI] [PubMed] [Google Scholar]
- 4. Du F, Cooper AJ, Thida T, et al. In vivo evidence for cerebral bioenergetic abnormalities in schizophrenia measured using 31P magnetization transfer spectroscopy. JAMA Psychiatry. 2014;71:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kenk M, Selvanathan T, Rao N, et al. Imaging neuroinflammation in gray and white matter in schizophrenia: an in-vivo PET study with [18F]-FEPPA. Schizophr Bull. 2015;41:85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Meyer U. Developmental neuroinflammation and schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2013;42:20–34. [DOI] [PubMed] [Google Scholar]
- 7. Monji A, Kato TA, Mizoguchi Y, et al. Neuroinflammation in schizophrenia especially focused on the role of microglia. Prog Neuropsychopharmacol Biol Psychiatry. 2013;42:115–121. [DOI] [PubMed] [Google Scholar]
- 8. Najjar S, Pearlman DM. Neuroinflammation and white matter pathology in schizophrenia: systematic review. Schizophr Res. 2015;161:102–112. [DOI] [PubMed] [Google Scholar]
- 9. Nagano T, Mizuno M, Morita K, Nawa H. Pathological Implications of Oxidative Stress in Patients and Animal Models with Schizophrenia: the Role of Epidermal Growth Factor Receptor Signaling. Curr Top Behav Neurosci. 2016;29:429–446. [DOI] [PubMed] [Google Scholar]
- 10. Sertan Copoglu U, Virit O, Hanifi Kokacya M, et al. Increased oxidative stress and oxidative DNA damage in non-remission schizophrenia patients. Psychiatry Res. 2015;229:200–205. [DOI] [PubMed] [Google Scholar]
- 11. Tuncel OK, Sarisoy G, Bilgici B, et al. Oxidative stress in bipolar and schizophrenia patients. Psychiatry Res. 2015;228:688–694. [DOI] [PubMed] [Google Scholar]
- 12. Yao JK, Keshavan MS. Antioxidants, redox signaling, and pathophysiology in schizophrenia: an integrative view. Antioxid Redox Signal. 2011;15:2011–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alano CC, Ying W, Swanson RA. Poly(ADP-ribose) polymerase-1-mediated cell death in astrocytes requires NAD+ depletion and mitochondrial permeability transition. J Biol Chem. 2004;279:18895–18902. [DOI] [PubMed] [Google Scholar]
- 14. Ying W, Garnier P, Swanson RA. NAD+ repletion prevents PARP-1-induced glycolytic blockade and cell death in cultured mouse astrocytes. Biochem Biophys Res Commun. 2003;308:809–813. [DOI] [PubMed] [Google Scholar]
- 15. Berger F, Ramirez-Hernandez MH, Ziegler M. The new life of a centenarian: signalling functions of NAD(P). Trends Biochem Sci. 2004;29:111–118. [DOI] [PubMed] [Google Scholar]
- 16. Ziegler M. New functions of a long-known molecule. Emerging roles of NAD in cellular signaling. Eur J Biochem. 2000;267:1550–1564. [DOI] [PubMed] [Google Scholar]
- 17. Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293:510–514. [DOI] [PubMed] [Google Scholar]
- 18. Zhang Q, Piston DW, Goodman RH. Regulation of corepressor function by nuclear NADH. Science. 2002;295:1895–1897. [DOI] [PubMed] [Google Scholar]
- 19. Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. [DOI] [PubMed] [Google Scholar]
- 20. Ying W. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxid Redox Signal. 2008;10:179–206. [DOI] [PubMed] [Google Scholar]
- 21. Kirsch M, De Groot H. NAD(P)H, a directly operating antioxidant? FASEB J. 2001;15:1569–1574. [DOI] [PubMed] [Google Scholar]
- 22. Olek RA, Ziolkowski W, Kaczor JJ, Greci L, Popinigis J, Antosiewicz J. Antioxidant activity of NADH and its analogue–an in vitro study. J Biochem Mol Biol. 2004;37:416–421. [DOI] [PubMed] [Google Scholar]
- 23. Starkov AA, Fiskum G, Chinopoulos C, et al. Mitochondrial alpha-ketoglutarate dehydrogenase complex generates reactive oxygen species. J Neurosci. 2004;24:7779–7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang H, Yang T, Baur JA, et al. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xie W, Xu A, Yeung ES. Determination of NAD(+) and NADH in a single cell under hydrogen peroxide stress by capillary electrophoresis. Anal Chem. 2009;81:1280–1284. [DOI] [PubMed] [Google Scholar]
- 26. Zhang Q, Wang SY, Nottke AC, Rocheleau JV, Piston DW, Goodman RH. Redox sensor CtBP mediates hypoxia-induced tumor cell migration. Proc Natl Acad Sci U S A. 2006;103:9029–9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Skala MC, Riching KM, Gendron-Fitzpatrick A, et al. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc Natl Acad Sci U S A. 2007;104:19494–19499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lu M, Zhu XH, Zhang Y, Chen W. Intracellular redox state revealed by in vivo (31) P MRS measurement of NAD(+) and NADH contents in brains. Magn Reson Med. 2014;71:1959–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhu XH, Lu M, Lee BY, Ugurbil K, Chen W. In vivo NAD assay reveals the intracellular NAD contents and redox state in healthy human brain and their age dependences. Proc Natl Acad Sci U S A. 2015;112:2876–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lu M, Zhu XH, Chen W. In vivo (31) P MRS assessment of intracellular NAD metabolites and NAD(+) /NADH redox state in human brain at 4 T. NMR Biomed. 2016;29:1010–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wehrli SL, Palmieri MJ, Berry GT, Kirkman HN, Segal S. 31P NMR analysis of red blood cell UDPGlucose and UDPGalactose: comparison with HPLC and enzymatic methods. Anal Biochem. 1992;202:105–110. [DOI] [PubMed] [Google Scholar]
- 32. Savorani F, Tomasi G, Engelsen SB. icoshift: a versatile tool for the rapid alignment of 1D NMR spectra. J Magn Reson. 2010;202:190–202. [DOI] [PubMed] [Google Scholar]
- 33. Cabungcal JH, Steullet P, Kraftsik R, Cuenod M, Do KQ. Early-life insults impair parvalbumin interneurons via oxidative stress: reversal by N-acetylcysteine. Biol Psychiatry. 2013;73:574–582. [DOI] [PubMed] [Google Scholar]
- 34. Flatow J, Buckley P, Miller BJ. Meta-analysis of oxidative stress in schizophrenia. Biol Psychiatry. 2013;74:400–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fournier M, Ferrari C, Baumann PS, et al. Impaired metabolic reactivity to oxidative stress in early psychosis patients. Schizophr Bull. 2014;40:973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Andreazza AC. Combining redox-proteomics and epigenomics to explain the involvement of oxidative stress in psychiatric disorders. Mol Biosyst. 2012;8:2503–2512. [DOI] [PubMed] [Google Scholar]
- 37. Karry R, Klein E, Ben Shachar D. Mitochondrial complex I subunits expression is altered in schizophrenia: a postmortem study. Biol Psychiatry. 2004;55:676–684. [DOI] [PubMed] [Google Scholar]
- 38. Donmez G, Outeiro TF. SIRT1 and SIRT2: emerging targets in neurodegeneration. EMBO Mol Med. 2013;5:344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bitanihirwe BK, Woo TU. Oxidative stress in schizophrenia: an integrated approach. Neurosci Biobehav Rev. 2011;35:878–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Erecinska M, Silver IA. Tissue oxygen tension and brain sensitivity to hypoxia. Respir Physiol. 2001;128:263–276. [DOI] [PubMed] [Google Scholar]
- 41. Dringen R. Metabolism and functions of glutathione in brain. Prog Neurobiol. 2000;62:649–671. [DOI] [PubMed] [Google Scholar]
- 42. Gu F, Chauhan V, Chauhan A. Glutathione redox imbalance in brain disorders. Curr Opin Clin Nutr Metab Care. 2015;18:89–95. [DOI] [PubMed] [Google Scholar]
- 43. Magni G, Orsomando G, Raffelli N, Ruggieri S. Enzymology of mammalian NAD metabolism in health and disease. Front Biosci. 2008;13:6135–6154. [DOI] [PubMed] [Google Scholar]
- 44. Sullivan EM, O’Donnell P. Inhibitory interneurons, oxidative stress, and schizophrenia. Schizophr Bull. 2012;38:373–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yarlagadda A. Role of calcium regulation in pathophysiology model of schizophrenia and possible interventions. Med Hypotheses. 2002;58:182–186. [DOI] [PubMed] [Google Scholar]
- 46. Blacker TS, Mann ZF, Gale JE, et al. Separating NADH and NADPH fluorescence in live cells and tissues using FLIM. Nat Commun. 2014;5:3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.