Abstract
Mitochondrial biogenesis is a complex process requiring coordinated expression of nuclear and mitochondrial genomes. The peroxisome proliferator-activated receptor gamma co-activator 1-alpha (PGC-1α) is a key regulator of mitochondrial biogenesis, and it controls mitochondrial DNA (mtDNA) replication within diverse tissues, including muscle tissue. The aim of this study was to investigate the effects of eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) on mtDNA copy number and PGC-1α promoter activity in C2C12 muscle cells. mtDNA copy number and mRNA levels of genes related to mitochondrial biogenesis such as PGC-1α, nuclear respiratory factor 1 (NRF1) and mitochondrial transcription factor A (Tfam) were assayed by quantitative real-time PCR. The PGC-1α promoter from −970 to +412 bp was subcloned into the pGL3-basic vector, which includes a luciferase reporter gene. Both EPA and DHA significantly increased mtDNA copy number, dose and time dependently, and up-regulated mRNA levels of PGC-1α, NRF1, and Tfam. Furthermore, EPA and DHA stimulated PGC-1α promoter activity in a dose-dependent manner. These results suggest that EPA and DHA may modulate mitochondrial biogenesis, which was partially associated with increased mtDNA replication and PGC-1α gene expression in C2C12 muscle cells.
Keywords: EPA, DHA, mtDNA, PGC-1α, muscle cells
INTRODUCTION
Skeletal muscle plays a vital role in whole body energy balance as a major site of mitochondrial oxidative metabolism. To promote mitochondrial number and function in skeletal muscle, it is important to activate specific signal transduction mechanisms that stimulate peroxisome proliferator-activated receptor gamma co-activator 1-alpha (PGC-1α). Overexpressing PGC-1α in skeletal muscle of mice was reported to increase the amount of mitochondria (1). PGC-1α is a primary regulator of mitochondrial biogenesis by virtue of its ability to co-activate and augment the expression and activity of several transcription factors (2,3). Activated PGC-1α increases expression of nuclear respiratory factors 1 (NRF1) and mitochondrial transcription factor A (Tfam). NRF1 subsequently up-regulates Tfam to stimulate mitochondrial DNA (mtDNA) transcription and replication (4–6).
Omega-3 fatty acids are mainly found in the foods, such as fish oil, some plants and nut oils (7). Dietary omega-3 polyunsaturated fatty acids (ω-3 PUFA), such as eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) are essential nutrients for human health. In humans, EPA and DHA promote improved insulin sensitivity (8), lipid oxidation (9), and reduce body weight (10,11). The ω-3 PUFA have been shown to increase the expression of genes involved in mitochondrial biogenesis in white fat of C57BL/6J mice (12). Furthermore, a previous study showed that EPA induced mitochondrial biogenesis as consequence of transcriptional activity of PGC-1α and Tfam in C6 glioma cells (13). However, whether EPA and DHA directly regulate the mtDNA replication and PGC-1α activation in skeletal muscle cells in vitro remains unresolved.
Therefore, we investigated the direct effects of EPA and DHA on the expression of genes involved in mitochondrial biogenesis, mtDNA copy number, and PGC-1α promoter activity in C2C12 muscle cells.
MATERIALS AND METHODS
Materials and reagents
EPA, DHA, palmitate (PA), butylated hydroxytoluene (BHT), α-tocopherol, and bovine serum albumin (BSA) were purchased from Sigma-Aldrich Co. (St. Louis, MO, USA). The C2C12 mouse muscle cell line was obtained from American Type Culture Collection (Manassas, VA, USA). Dulbecco’s modified Eagle’s medium (DMEM), glutamine, penicillin-streptomycin, fetal bovine serum (FBS), and TRIzol reagent were obtained from Invitrogen (Carlsbad, CA, USA). Moloney murine leukemia virus (M-MLV) reverse transcriptase, pGEM® T easy vector, pGL3 basic vector, and luciferase reporter assay system kit were purchased from Promega (Madison, WI, USA). pCMV-β galactosidase was obtained from Clontech Laboratories, Inc. (Palo Alto, CA, USA). Puregene DNA isolation kit, Universal SYBR Green PCR Master Mix, and Superfect reagent were purchased from Qiagen (Chatsworth, CA, USA). Mlu I and Xho I were obtained from Takara (Tokyo, Japan).
Preparation of EPA, DHA, and PA
Non-esterified EPA (C20:5, ω-3), DHA (C22:6, ω-3), and PA (C16:0) were dissolved in 95% ethanol. EPA, DHA, and PA were combined with 7.5% BSA by stirring for 1 h at 37°C. Fatty acid/BSA complexes were added 0.1% BHT and 20 μM α-tocopherol to minimize oxidation. The saturated fatty acid, PA, was used to compare the relative change to unsaturated fatty acids (EPA and DHA).
Cell culture
Mouse C2C12 myoblasts were cultured in DMEM supplemented with 10% FBS and 1 U penicillin-streptomycin at 37°C in a 5% CO2 atmosphere. When C2C12 cells reached 90% confluence, differentiation was induced by incubation for 5 days with differentiation medium containing 2% horse serum. For the gene expression assay at mRNA level, differentiated C2C12 muscle cells were treated without (control) or with 50 μM PA, EPA, or DHA in serum-free medium for 24 h. For promoter activity assay, cells were cultured in serum-free media for 40 h with 0 (control), 1, 10, or 50 μM EPA and DHA. Control cells were treated with 0.1% BHT and 20 μM α-tocopherol without fatty acids. All measurements were performed in triplicate.
mtDNA copy number
The genomic DNA was extracted from muscle with a Puregene DNA isolation kit according to the manufacturer’s instructions. mtDNA copy number was calculated by real-time quantitative polymerase chain reaction (PCR) by measuring a mitochondrial gene [cytochrome oxidase subunit 1 (Cox1) vs. a nuclear gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH)].
Real-time quantitative PCR
Total RNA was extracted from cells with TRIzol reagent. The corresponding cDNA was synthesized from 4 μg of RNA with M-MLV reverse transcriptase. After cDNA synthesis, real-time quantitative PCR was performed by using Universal SYBR Green PCR Master Mix on a fluorometric thermal cycler (Corbett Research, Mortlake, Australia). Primers were designed by the program Primer3 (14). Sequences of the sense and antisense primers are shown in Table 1. The ΔΔCt method was used for relative quantification. The ΔΔCt value for each sample was determined by calculating the difference between the Ct value of the target gene and the Ct value of β-actin as a reference gene. The normalized expression level of each target gene was calculated as 2−ΔΔCt. Values are expressed as fold of the control.
Table 1.
Primers used for quantitative real-time polymerase chain reaction (PCR)
| Gene | GeneBank No. | Primer sequence (5′-3′) | |
|---|---|---|---|
| β-actin | NM_007393 | Forward | GGACCTGACAGACTACCTCA |
| Reverse | GTTGCCAATAGTGATGACCT | ||
| NRF1 | NM_010938 | Forward | AAGTATTCCACAGGTCGGGG |
| Reverse | TGGTGGCCTGAGTTTGTGTT | ||
| PGC-1α | NM_008904 | Forward | GGGCCAAACAGAGAGAGAGG |
| Reverse | GTTTCGTTCGACCTGCGTAA | ||
| Tfam | NM_009360 | Forward | GAGGCCAGTGTGAACCAGTG |
| Reverse | GTAGTGCCTGCTGCTCCTGA |
NRF1, nuclear respiratory factor 1; PGC-1α, peroxisome proliferative activated receptor gamma coactivator 1 alpha; Tfam, mitochondrial transcription factor A.
Construction of PGC-1α reporter gene
The human PGC-1α gene promoter was generated by PCR of human genomic DNA, using primers recognizing exon 1 of the PGC-1α gene (970 bp upstream to 412 bp downstream). The sequence information was deposited with GenBank (Accession no. AF108193). The primers of the PGC-1α gene promoter were designed such that the amplified promoter fragment was flanked by two restriction sites (Mlu I and Xho I). The 5′-primer, bearing a Mlu I site, was 5′-TGT ACG CGT CCC TCA GTT CAC AGA CAT TCT-3′, and the 3′-primer, bearing a Xho I site, was 5′-TCT CTC GAG ACA GTG CCA AAG TCA CAT GGA-3′. PGC-1α promoter amplification consisted of 95°C for 15 min followed by 30 cycles of 95°C for 1 min, 64°C for 1 min and 70°C for 2 min. The PGC-1α promoter fragment (−970/+412) was subcloned into the pGEM-T easy vector. It was then inserted into the pGL3 basic vector, which includes a luciferase reporter gene.
Transfection and luciferase assay
Transfection experiments were carried out with Superfect reagent according to the manufacturer’s instructions. The plasmids used were 2 μg of PGC-1α/luc reporter gene and 1 μg of pCMV-β-galactosidase as an internal standard to adjust for transfection efficiency. The pGL3-basic vector was used as a vector control. Three hours after transfection, cells were treated with 0 (control), 1, 10, or 50 μM EPA or DHA in 1% BSA serum-free medium for 40 h.
For the luciferase assay, cells were washed with PBS and harvested with lysis buffer. PGC-1α promoter activity was measured with the luciferase reporter assay system and a TD 20/20 luminometer (Turner Designs, Sunnyvale, CA, USA). β-Galactosidase activity was assayed enzymatically by using o-nitrophenyl-β-D-galactopyranoside as a substrate. Luciferase activity was calculated in relative light units and normalized to β-galactosidase activity.
Statistical analysis
Data are expressed as mean±standard deviation (SD). Statistical analyses were performed using SPSS software version 19 (IBM Corporation, Armonk, NY, USA). Significant differences among the treatment groups were assessed by one-way analysis of variance (ANOVA) and Tukey’s multiple comparison tests. P<0.05 was considered to indicate a statistically significant difference.
RESULTS
Effects of EPA and DHA on mtDNA copy number
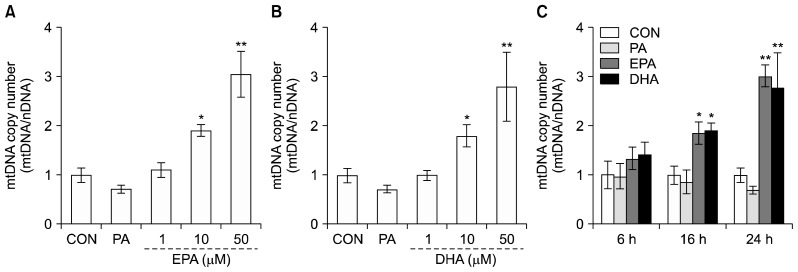
To investigate the effect of EPA and DHA on mtDNA copy number, we determined the ratio of mtDNA to nuclear DNA (nDNA). Differentiated C2C12 muscle cells were incubated in the presence of fatty acids with different degrees of saturation, including 50 μM PA (saturated) and various concentrations (0, 1, 10, or 50 μM) of EPA or DHA (unsaturated) for 24 h. The mtDNA/nDNA ratio increased by 1.4- and 1.9-fold in the presence of 10 and 50 μM EPA compared to the untreated control (Fig. 1A). Also, 50 μM DHA increased the mtDNA copy number by 1.8-fold compared to control cells (Fig. 1B). In addition, the mtDNA/nDNA ratio was increased time-dependently, reaching at 24 h after 50 μM EPA or DHA treatments (Fig. 1C). However, there were no significant differences for mtDNA copy number in the presence of 50 μM PA.
Fig. 1.
Effects of eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) on mtDNA copy number in C2C12 muscle cells. Differentiated C2C12 muscle cells were treated with 1% bovine serum albumin serum-free medium alone (control), 50 μM palmitate (PA) or 1, 10, and 50 μM EPA (A) or DHA (B) for 24 h. Cells were time-dependently exposed to 50 μM PA, EPA, or DHA for 6, 16, and 24 h (C). mtDNA copy number was measured by real-time quantitative PCR. Values are expressed as mean±SD (n=3) of three independent experiments. *P<0.05 and **P<0.01 compared to control.
Effects of EPA and DHA on expression of genes involved in mitochondrial biogenesis
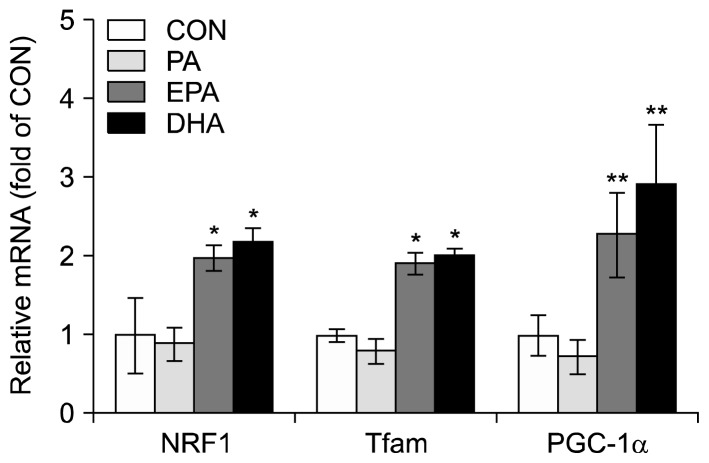
To determine the effect of EPA and DHA on expression of genes involved in mitochondrial biogenesis, the mRNA levels of PGC-1α, NRF1, and Tfam in C2C12 muscle cells were measured by real-time quantitative PCR. Cells were incubated without (control) or with 50 μM of PA, EPA, or DHA for 24 h. The mRNA levels of PGC-1α, NRF1, and Tfam increased by 2.3-, 2.0-, and 1.9-fold, respectively, in the presence of EPA compared with controls (Fig. 2). Furthermore, DHA induced 2.9-, 2.2-, and 2.0-fold increases, respectively, in mRNA levels of PGC-1α, NRF1, and Tfam compared to control (Fig. 2). In contrast, PA did not significantly increase in mRNA levels of PGC-1α, NRF1, and Tfam.
Fig. 2.
Effects of eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) on the mRNA levels of genes involved in mitochondrial biogenesis in C2C12 muscle cells. Differentiated C2C12 muscle cells were treated in 1% bovine serum albumin serum-free medium with 50 μM of palmitate (PA), EPA, or DHA for 24 h. The mRNA levels were measured by quantitative real-time RT-PCR. Values are means±SD (n=3) of three independent experiments. *P<0.05 and **P<0.01 compared to control.
Effects of EPA and DHA on PGC-1α promoter activity
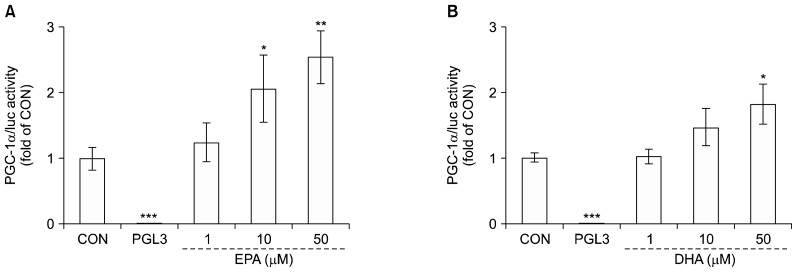
To further examine the up-regulated PGC-1α gene expression by EPA and DHA, we assayed the promoter activity in C2C12 muscle cells. Cells were treated with 0, 1, 10, or 50 μM EPA or DHA for 40 h. The PGC-1α promoter activity in the presence of 10 and 50 μM EPA increased by 2.0- and 2.5-fold, respectively, compared to control. Also, 50 μM DHA increased PGC-1α promoter activity by 1.8-fold (Fig. 3). Cotransfection with the control vector (pGL3-basic) had a negligible effect on luciferase activity.
Fig. 3.
Effects of eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) on peroxisome proliferator-activated receptor gamma co-activator 1-alpha (PGC-1α) promoter activity in C2C12 muscle cells. Differentiated cells were transfected with the PGC-1α (−1790/+52 bp)/luc reporter gene and pCMV-β galactosidase and were incubated 1% bovine serum albumin serum-free medium with 0 (control) to 50 μM EPA (A) and DHA (B) for 40 h. Promoter activities measured by luciferase activity were calculated in relative light units (RLU) and normalized to β-galactosidase activity. Values are expressed as mean±SD (n=3) of three independent experiments. *P<0.05, **P<0.01, and ***P<0.001 compared to control.
DISCUSSION
Mitochondrial biogenesis is a noticeable response of skeletal muscle to a variety of physiological conditions (15,16). There are several food components that have reported to induce mitochondrial biogenesis such as resveratrol (17), green tea polyphenols (18), and curcumin (19). Specifically, resveratrol increased the mitochondrial mass and mitochondrial DNA content, and the mRNA expression of NRF1, Tfam, and PGC-1α in endothelial cells. Also, green tea polyphenols increased mtDNA copy number and the mRNA expression of PGC-1α and Tfam in renal of rats. Curcumin has been shown to increase the mitochondrial DNA copy number and PGC-1α deacetylation in skeletal muscle during endurance training of rats.
EPA and DHA intake from marine sources has been reported to have diverse health benefits that include hypolipidemic effects, improved insulin sensitivity and fatty acid oxidation (20–22). While evidence supports a role for ω-3 PUFA in stimulating the expression of genes encoding regulatory factors for mitochondrial biogenesis and oxidative metabolism in adipose tissue (12), there is limited evidence evaluating the effects of EPA and DHA on muscle mitochondrial biogenesis.
To investigate the effects of EPA and DHA on mtDNA replication in C2C12 muscle cells, the mtDNA copy number was measured by real-time quantitative PCR. The EPA and DHA doses used in this study (1~50 μM) were non-cytotoxic for C2C12 cells, as previously demonstrated (23). EPA and DHA treatment significantly increased mtDNA copy number in skeletal muscle cells. These results agree with a previous study showing that EPA promoted elevated mtDNA copy number in inguinal and brown adipocytes (24). Thus, it can be postulated that EPA and DHA may enhance mitochondrial replication in skeletal muscle cells.
To understand the underlying mechanism of mitochondrial biogenesis by EPA and DHA, we investigated the mRNA levels of genes involved in mitochondrial biogenesis such as PGC-1α, NRF1, and Tfam in C2C12 cells. We found that both EPA and DHA significantly elevated mRNA levels of PGC-1α, NRF1, and Tfam compared to control and PA. PGC-1α serves a major integrative role in the transcriptional regulatory cascade upstream of mitochondrial biogenesis (25). A previous study showed that DHA up-regulated PGC-1α gene expression in 3T3-L1 cells (12). In addition, Gerhart-Hines et al. (26) reported that fasting induced PGC-1α deacetylation in skeletal muscle and that SIRT1 deacetylation of PGC-1α is required to activate mitochondrial fatty acid oxidation genes. Activated PGC-1α enhances NRF1 and NRF2 expression and increases Tfam expression. This result indicates a link between the nucleus and mitochondria by directly regulating mtDNA replication and transcription (4–6). Previously it was reported that conjugated linoleic acid isomers up-regulated PGC-1α, NRF1, and Tfam expression in C2C12 cells (27). Also, a recent study showed that ω-3 PUFA from cod fish oil induced the activation of PGC-1α and -β in rat skeletal muscles (28). In our study, EPA and DHA increased the expression of PGC-1α, NRF1, and Tfam genes in C2C12 cells. It is assumed that EPA and DHA directly modulate the expression of genes involved in muscle mitochondrial biogenesis.
Enhancement of the PGC-1α mRNA level may be derived from increased of transcription and/or promotion of mRNA stability. To distinguish between these possibilities, the effects of EPA and DHA on PGC-1α promoter activity were examined in C2C12 cells. PGC-1α promoter activity was elevated by both EPA and DHA treatment in a dose-dependent manner, in parallel with the altered mRNA expression. In a previous study, we showed that EPA and DHA increased expression of mitochondrial uncoupling protein 3 (UCP3) mRNA and enhanced UCP3 promoter activity in C2C12 cells (23). In addition, Siculella et al. (29) reported that PUFA down-regulated expression of the mitochondrial tricarboxylate carrier gene both transcriptionally and post-transcriptionally in rat liver. In our study, EPA and DHA increased PGC-1α promoter activity in parallel with mRNA expression. EPA and DHA may play positive roles in PGC-1α transcriptional regulation of mitochondrial biogenesis in C2C12 cells.
This study investigated the effects of EPA and DHA on mitochondrial biogenesis in C2C12 cells. The results showed that both EPA and DHA increased mtDNA copy number and up-regulated mRNA expression of genes involved in mitochondrial biogenesis, such as PGC-1α, NRF1, and Tfam in C2C12 cells. Moreover, both EPA and DHA increased PGC-1α promoter activity in parallel with mRNA expression. These findings suggest that EPA and DHA might regulate muscle mitochondrial biogenesis, potentially through mtDNA replication and PGC-1α promoter activation in C2C12 cells.
ACKNOWLEDGEMENTS
This study was supported by the National Research Foundation of Korea (NRF) funded by the Korean Government (No. 2013R1A1A2009522 and 2016 R1A2B4011021).
Footnotes
AUTHOR DISCLOSURE STATEMENT
The authors declare no conflict of interest.
REFERENCES
- 1.Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, Lowell BB, Bassel-Duby R, Spiegelman BM. Transcriptional co-activator PGC-1α drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 2.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 3.Handschin C, Spiegelman BM. Hypothesis article the role of exercise and PGC1α in inflammation and chronic disease. Nature. 2008;454:463–469. doi: 10.1038/nature07206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 5.Virbasius JV, Scarpulla RC. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc Natl Acad Sci USA. 1994;91:1309–1313. doi: 10.1073/pnas.91.4.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parisi MA, Clayton DA. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 1991;252:965–969. doi: 10.1126/science.2035027. [DOI] [PubMed] [Google Scholar]
- 7.Kris-Etherton PM, Taylor DS, Yu-Poth S, Huth P, Moriarty K, Fishell V, Hargrove RL, Zhao G, Etherton TD. Polyunsaturated fatty acids in the food chain in the United States. Am J Clin Nutr. 2000;71:179S–188S. doi: 10.1093/ajcn/71.1.179S. [DOI] [PubMed] [Google Scholar]
- 8.Bouchard-Mercier A, Rudkowska I, Lemieux S, Couture P, Pérusse L, Vohl MC. SREBF1 gene variations modulate insulin sensitivity in response to a fish oil supplementation. Lipids Health Dis. 2014;13:152. doi: 10.1186/1476-511X-13-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson EJ, Thayne KA, Harris M, Shaikh SR, Darden TM, Lark DS, Williams JM, Chitwood WR, Kypson AP, Rodriguez E. Do fish oil omega-3 fatty acids enhance antioxidant capacity and mitochondrial fatty acid oxidation in human atrial myocardium via PPARγ activation? Antioxid Redox Signal. 2014;21:1156–1163. doi: 10.1089/ars.2014.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thorsdottir I, Tomasson H, Gunnarsdottir I, Gisladottir E, Kiely M, Parra MD, Bandarra NM, Schaafsma G, Martinéz JA. Randomized trial of weight-loss-diets for young adults varying in fish and fish oil content. Int J Obes. 2007;31:1560–1566. doi: 10.1038/sj.ijo.0803643. [DOI] [PubMed] [Google Scholar]
- 11.Ramel A, Parra D, Martinéz JA, Kiely M, Thorsdottir I. Effects of seafood consumption and weight loss on fasting leptin and ghrelin concentrations in overweight and obese European young adults. Eur J Nutr. 2009;48:107–114. doi: 10.1007/s00394-008-0769-9. [DOI] [PubMed] [Google Scholar]
- 12.Flachs P, Horakova O, Brauner P, Rossmeisl M, Pecina P, Franssen-van Hal N, Ruzickova J, Sponarova J, Drahota Z, Vlcek C, Keijer J, Houstek J, Kopecky J. Polyunsaturated fatty acids of marine origin upregulate mitochondrial biogenesis and induce β-oxidation in white fat. Diabetologia. 2005;48:2365–2375. doi: 10.1007/s00125-005-1944-7. [DOI] [PubMed] [Google Scholar]
- 13.Jeng JY, Lee WH, Tsai YH, Chen CY, Chao SY, Hsieh RH. Functional modulation of mitochondria by eicosapentaenoic acid provides protection against ceramide toxicity to C6 glioma cells. J Agric Food Chem. 2009;57:11455–11462. doi: 10.1021/jf902021h. [DOI] [PubMed] [Google Scholar]
- 14.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 15.Freyssenet D, Berthon P, Denis C. Mitochondrial biogenesis in skeletal muscle in response to endurance exercises. Arch Physiol Biochem. 1996;104:129–141. doi: 10.1076/apab.104.2.129.12878. [DOI] [PubMed] [Google Scholar]
- 16.Hood DA, Takahashi M, Connor MK, Freyssenet D. Assembly of the cellular powerhouse: current issues in muscle mitochondrial biogenesis. Exerc Sport Sci Rev. 2000;28:68–73. [PubMed] [Google Scholar]
- 17.Csiszar A, Labinskyy N, Pinto JT, Ballabh P, Zhang H, Losonczy G, Pearson K, de Cabo R, Pacher P, Zhang C, Ungvari Z. Resveratrol induces mitochondrial biogenesis in endothelial cells. Am J Physiol Heart Circ Physiol. 2009;297:H13–H20. doi: 10.1152/ajpheart.00368.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rehman H, Krishnasamy Y, Haque K, Thurman RG, Lemasters JJ, Schnellmann RG, Zhong Z. Green tea polyphenols stimulate mitochondrial biogenesis and improve renal function after chronic cyclosporin A treatment in rats. PLoS One. 2013;8:e65029. doi: 10.1371/journal.pone.0065029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ray Hamidie RD, Yamada T, Ishizawa R, Saito Y, Masuda K. Curcumin treatment enhances the effect of exercise on mitochondrial biogenesis in skeletal muscle by increasing cAMP levels. Metabolism. 2015;64:1334–1347. doi: 10.1016/j.metabol.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 20.Elvevoll EO, Eilertsen KE, Brox J, Dragnes BT, Falkenberg P, Olsen JO, Kirkhus B, Lamglait A, Østerud B. Seafood diets: hypolipidemic and antiatherogenic effects of taurine and n-3 fatty acids. Atherosclerosis. 2008;200:396–402. doi: 10.1016/j.atherosclerosis.2007.12.021. [DOI] [PubMed] [Google Scholar]
- 21.Lombardo YB, Hein G, Chicco A. Metabolic syndrome: effects of n-3 pufas on a model of dyslipidemia, insulin resistance and adiposity. Lipids. 2007;42:427–437. doi: 10.1007/s11745-007-3039-3. [DOI] [PubMed] [Google Scholar]
- 22.Madsen L, Rustan AC, Vaagenes H, Berge K, Dyrøy E, Berge RK. Eicosapentaenoic and docosahexaenoic acid affect mitochondrial and peroxisomal fatty acid oxidation in relation to substrate preference. Lipids. 1999;34:951–963. doi: 10.1007/s11745-999-0445-x. [DOI] [PubMed] [Google Scholar]
- 23.Lee MS, Kim IH, Kim Y. Effects of eicosapentaenoic acid and docosahexaenoic acid on uncoupling protein 3 gene expression in C2C12 muscle cells. Nutrients. 2013;5:1660–1671. doi: 10.3390/nu5051660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao M, Chen X. Eicosapentaenoic acid promotes thermogenic and fatty acid storage capacity in mouse subcutaneous adipocytes. Biochem Biophys Res Commun. 2014;450:1446–1451. doi: 10.1016/j.bbrc.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 25.Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor γ coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1α. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim Y, Park Y. Conjugated linoleic acid (CLA) stimulates mitochondrial biogenesis signaling by the upregulation of PPARγ coactivator 1α (PGC-1α) in C2C12 cells. Lipids. 2015;50:329–338. doi: 10.1007/s11745-015-4000-5. [DOI] [PubMed] [Google Scholar]
- 28.Cavaliere G, Trinchese G, Bergamo P, De Filippo C, Mattace Raso G, Gifuni G, Putti R, Moni BH, Canani RB, Meli R, Mollica MP. Polyunsaturated fatty acids attenuate diet induced obesity and insulin resistance, modulating mitochondrial respiratory uncoupling in rat skeletal muscle. PLoS One. 2016;11:e0149033. doi: 10.1371/journal.pone.0149033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siculella L, Sabetta S, Damiano F, Giudetti AM, Gnoni GV. Different dietary fatty acids have dissimilar effects on activity and gene expression of mitochondrial tricarboxylate carrier in rat liver. FEBS Lett. 2004;578:280–284. doi: 10.1016/j.febslet.2004.11.014. [DOI] [PubMed] [Google Scholar]