Summary
Infections due to multidrug‐resistant bacteria represent a major global health challenge. To combat this problem, new antibiotics are urgently needed and some plant‐associated bacteria are a promising source. The rhizobacterium Serratia plymuthica A153 produces several bioactive secondary metabolites, including the anti‐oomycete and antifungal haterumalide, oocydin A and the broad spectrum polyamine antibiotic, zeamine. In this study, we show that A153 produces a second broad spectrum antibiotic, andrimid. Using genome sequencing, comparative genomics and mutagenesis, we defined new genes involved in andrimid (adm) biosynthesis. Both the expression of the adm gene cluster and regulation of andrimid synthesis were investigated. The biosynthetic cluster is operonic and its expression is modulated by various environmental cues, including temperature and carbon source. Analysis of the genome context of the adm operon revealed a gene encoding a predicted LysR‐type regulator, AdmX, apparently unique to Serratia strains. Mutagenesis and gene expression assays demonstrated that AdmX is a transcriptional activator of the adm gene cluster. At the post‐transcriptional level, the expression of the adm cluster is positively regulated by the RNA chaperone, Hfq, in an RpoS‐independent manner. Our results highlight the complexity of andrimid biosynthesis – an antibiotic with potential clinical and agricultural utility.
Introduction
The discovery of antibiotics is one of the main milestones in the history of medicine. However, excessive overuse of antibiotics has encouraged the emergence of multidrug‐resistant bacteria, leading to a global increase in the spectrum of untreatable infections, which are currently responsible for around 50,000 annual deaths in Europe and the United States (Woodford et al., 2011; Blair et al., 2015). There is therefore an urgent need to identify new antibiotics, but efforts focussed on discovery and development of new antibiotics have met with only limited success (Lewis, 2013; Pidot et al., 2014). New platforms for antibiotic discovery include the generation of synthetic antimicrobials and development of species‐specific antibiotics (Fischbach and Walsh, 2009; Lewis, 2013; Liu et al., 2013; Pidot et al., 2014). It has been estimated that 90% of microbial natural products, and more than 99% of the total number of secondary metabolites, remain to be discovered. Consequently, recent approaches to antibiotic discovery include screening of microbes from new ecological niches and attempts at exploitation of previously uncultured microbes (Fischbach and Walsh, 2009).
Natural products (and their synthetic derivatives) comprise most of the antibiotics used clinically (Newman and Cragg, 2007), many of which are based on non‐ribosomal peptides and/or polyketides. Both families of secondary metabolites are synthesised by multifunctional enzymes, known as non‐ribosomal peptide synthetases (NRPSs) and polyketide synthases (PKSs), through sequential rounds of condensation of amino acids and acyl‐CoA building units, respectively (Sattely et al., 2008; Hertweck, 2009). The great structural diversity of non‐ribosomal peptides and polyketides results from the number of condensed building units and a range of pre‐ and post‐assembly processing reactions (Sattely et al., 2008; Hertweck, 2009). This chemical diversity is consequently reflected in a broad spectrum of biological activities (Sattely et al., 2008; Fischbach and Walsh, 2009; Hertweck, 2009; Pidot et al., 2014; Mousa and Raizada, 2015).
Some bacteria can devote up to 10% of their genomes to secondary metabolism (Udwary et al., 2007; Nett et al., 2009; Chowdhury et al., 2015). The biological synthesis of such metabolites can be energetically costly and so production is generally highly regulated (Coulthurst et al., 2005; Williamson et al., 2006; Liu et al., 2013). Biosynthetic gene clusters are often linked to their own regulatory genes (Chen et al., 2006; Zhao et al., 2010; Gurney and Thomas, 2011; Liu et al., 2013), the products of which are involved in sensing factors such as physiological state, population density and diverse environmental cues. As a result, the synthesis of the cognate secondary metabolite can be modulated appropriately. Quorum sensing regulatory circuits (Coulthurst et al., 2005; Williamson et al., 2006; Müller et al., 2009; Matilla et al., 2015), two‐component systems (Sola‐Landa et al., 2003; Haas and Défago, 2005; Williamson et al., 2006), orphan transcriptional regulators (Williamson et al., 2006; Lu et al., 2011; Klaponski et al., 2014) and post‐transcriptional regulators (Vogel and Luisi, 2011; Romeo et al., 2013) can all be involved in the regulatory complexity of bacterial secondary metabolite control.
The rhizosphere is one of the most complex environments on earth, with many organisms interacting and competing for nutrients and space (Lugtenberg and Kamilova, 2009; Mendes et al., 2013). Many rhizosphere microbes have evolved the capacity to synthesize bioactive secondary metabolites that allow them to efficiently antagonize diverse niche competitors (Berg et al., 2002; De Vleesschauwer and Höfte, 2007; Raaijmakers et al., 2009; Pidot et al., 2014; Mousa and Raizada, 2015). Consequently, this defines the rhizosphere as a habitat with great potential for exploitation as a source of new natural products with pharmacological, chemotherapeutic and agricultural applications.
Serratia plymuthica strains are near‐ubiquitous in nature but have been commonly isolated from soil and the rhizosphere of many economically important crops (De Vleesschauwer and Höfte, 2007). Serratia plymuthica strains possess great potential as biocontrol agents by antagonizing the growth of plant‐pathogens through the production of diverse bioactive secondary metabolites, siderophores and lytic enzymes (Alström, 2001; De Vleesschauwer and Höfte, 2007; Matilla et al., 2015). The strain used in this study, Serratia plymuthica A153, was isolated from the rhizosphere of wheat (Åstrom and Gerhardson, 1988) and it has been shown to possess bioactivity against fungi, oomycetes, bacteria and nematodes (Thaning et al., 2001; Matilla et al., 2012; Hellberg et al., 2015). These activities are mainly due to the synthesis of NRPS‐ and PKS‐based secondary metabolites, such as the haterumalide, oocydin A (Thaning et al., 2001), and the polyamine antibiotic, zeamine (Hellberg et al., 2015).
Our previous work showed that the strain A153, in addition to zeamine, produces an unidentified second antibacterial compound (Hellberg et al., 2015). In this study, we employed genome sequencing, comparative genomics and mutagenesis approaches to identify the genes involved in the biosynthesis of the unknown secondary metabolite. The regulation of the production of the antibacterial compound was also investigated and the results showed that the expression of the biosynthetic genes is tightly regulated at transcriptional and post‐transcriptional levels. Different environmental cues controlling the transcription of the biosynthetic genes were also identified.
Results
Serratia plymuthica A153 produces the hybrid non‐ribosomal peptide‐polyketide antibiotic, andrimid
Characterization of the biocontrol rhizobacterium, S. plymuthica A153, showed that this strain possesses a strong bioactivity against Bacillus subtilis (Fig. 1A). The observed antibacterial activity was not associated with the production of other known bioactive secondary metabolites produced by A153, namely oocydin A (Matilla et al., 2012) or zeamine (Hellberg et al., 2015).
Figure 1.
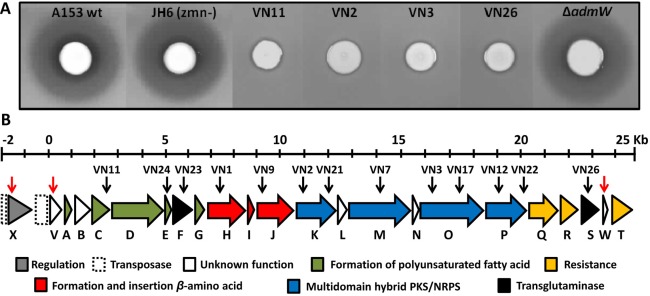
Identification and characterization of the andrimid gene cluster in Serratia plymuthica A153.
A. Antibacterial activities against Bacillus subtilis of Serratia plymuthica A153, and derivative strains with mutations in the zeamine (zmn) and andrimid (adm) biosynthetic gene clusters.
B. Genetic organization of the adm gene cluster in S. plymuthica A153. The same genetic organization was found in S. marcescens MSU97 and S. marcescens 90‐166 (Fig. S3). Location of the Tn‐KRCPN1 transposon insertions and in‐frame deletion mutants are indicated by black and red arrows, respectively. Colour code representing the functional category of each gene of the gene cluster is given where possible, based on the biosynthetic pathway for andrimid proposed by Jin et al. (2006). Genes admV, admW and admX were not previously associated with the regulation or biosynthesis of andrimid.
During the in silico analysis of the A153 genome sequence (Matilla et al., 2016) we identified at least five candidate biosynthetic PKS and NRPS gene clusters which could be responsible for the synthesis of the unknown antibacterial compound. To identify the genes responsible for this bioactivity, a random transposon insertion strain library was constructed and screened for mutants defective in antibacterial activity against Bacillus subtilis. Several transposon insertion mutants showing loss of antibacterial properties were isolated and all the insertions were transduced back into the wild type genetic background using the transducing phage ϕMAM1 (Matilla and Salmond, 2014) to confirm the link between transposon insertions and mutant phenotype. Random primed PCR confirmed that most of the transposons were located in a hybrid PKS/NRPS gene cluster described previously as responsible for the biosynthesis of the broad‐spectrum antibiotic, andrimid (Figs. 1 and Supporting information Fig. S1) (Jin et al., 2006).
We had access to another plant‐associated oocydin A producing strain, Serratia marcescens MSU97. This strain also showed strong antibacterial activity towards B. subtilis (Supporting information Fig. S2) and sequencing of its genome (Matilla and Salmond, unpubl. data) revealed that the andrimid (adm) gene cluster is also present in this plant epiphytic bacterium (Supporting information Fig. S3). We reported previously that MSU97 is recalcitrant to various genetic tools (Matilla et al., 2012) and attempts at isolating mutants defective in the adm gene cluster of this strain were unsuccessful.
Comparative analyses of sequenced andrimid gene clusters
The biosynthesis of andrimid has been demonstrated in a broad range of bacteria (Fredenhagen et al., 1987; Long et al., 2005; Jin et al., 2006; Wietz et al., 2011; Sánchez et al., 2013). However, few adm gene clusters have been sequenced, including those of the marine bacteria, Vibrionales SWAT‐3 (PATRIC Genome ID 391574.12) and Vibrio coralliilyticus S2052 (Machado et al, 2015), and the plant‐associated enterobacterium Pantoea agglomerans Eh335 (Jin et al., 2006; GenBankTM Accession No. AY192157.1). Additionally, our genomic analyses revealed that the adm gene cluster is also present in the recently sequenced rhizobacterium Serratia marcescens 90‐166 (Supporting information Figs. S3 and S4) (Jeong et al., 2015).
Comparative analyses showed that the genomic context of the sequenced adm gene clusters in A153, MSU97, Eh335, S2052 and SWAT‐3 is completely different and, consequently, the upstream and downstream predicted ends of the biosynthetic clusters were assigned based only on their homologies (Supporting information Figs. S3 and S4). These analyses allowed the identification of a gene, designated admV, located immediately upstream of admA. AdmV was not previously associated with andrimid biosynthesis (Jin et al., 2006) and in silico analyses did not shed light on its putative function. However, we found that the gene admV is conserved in all adm gene clusters (Supporting information Figs. S3 and S4). To further investigate its role in the synthesis of andrimid, we constructed an in frame deletion mutant defective in admV, thereby avoiding polar effects in the expression of the downstream adm genes. The resulting mutant strain no longer exhibited antibacterial activity and the bioactivity could be complemented by the in‐trans expression of AdmV (Fig. 2).
Figure 2.

Role of the hypothetical protein AdmV in the biosynthesis of andrimid. Bioactivities against Bacillus subtilis of an in‐frame admV deletion mutant of Serratia plymuthica A153. Induction of the expression of the wild type proteins was done by addition of 1 mM of IPTG. The bioassays were repeated at least three times, and representative results are shown. Pictures were taken after 48 h of incubation at 25 °C.
The adm gene clusters of A153, MSU97, 90‐166, Eh335, SWAT‐3 and S2052 are 25.1, 24.9, 24.8, 24.7, 25.6 and 25.6 kb, respectively, and they are between 70.1% and 99.0% identical at the DNA level (Supporting information Fig. S4 and Table S1) suggesting that adm biosynthetic clusters may have been moved horizontally between the producing strains. In accordance with this hypothesis, the overall genomic G + C content of A153 (56.0%), MSU97 (58.9%), 90‐166 (59.1%), SWAT‐3 (44.5%) and S2052 (45.7%) is considerably different from the G + C content of their respective adm biosynthetic clusters, which are 45.8%, 47.6%, 46.27, 51.1% and 51.2%, respectively. Furthermore, remnant sequences of transposable genetic elements were found flanking the gene clusters of all the producing strains, although located in different regions in these loci (Supporting information Fig. S3).
In contrast, some remarkable differences were found between the six biosynthetic clusters. First, the intergenic regions of several adm contiguous genes are not conserved between the adm gene clusters (Supporting information Fig. S4) which could suggest differential regulation in the expression of the biosynthetic clusters. Second, the intergenic region between admS and admT is considerably larger in A153, MSU97 and 90‐166, and we identified a putative ORF, admW, in these three Serratia strains (Supporting information Figs. S3 and S4). However, the in frame deletion of admW did not alter the antibacterial activity of A153 (Fig. 1A). Finally, a gene encoding a LysR‐type transcriptional regulator was found upstream of admT in the adm gene clusters of Vibrionales SWAT‐3 and Vibrio coralliilyticus S2052 (Supporting information Fig. S3).
The andrimid gene cluster consists of a large polycistronic unit
The adm gene cluster in Serratia consists of 22 predicted ORFs and its genetic organization, together with the small intergenic distances between contiguous genes, suggests the presence of a single transcriptional unit (Figs. 3A and Supporting information Fig. S3). To further understand the biosynthesis of andrimid, we investigated its transcriptional regulation. With primers spanning the 3′ end of the upstream gene and the 5′ end of the contiguous downstream gene (Fig. 3A), we used reverse transcription‐PCR (RT‐PCR) to assess co‐transcription of the adm genes in S. plymuthica A153. PCR products were obtained across all intergenic regions covering the complete adm biosynthetic cluster, indicating the presence of one long polycistronic transcript (Fig. 3B) – although the possibility of internal promoters cannot be discarded.
Figure 3.
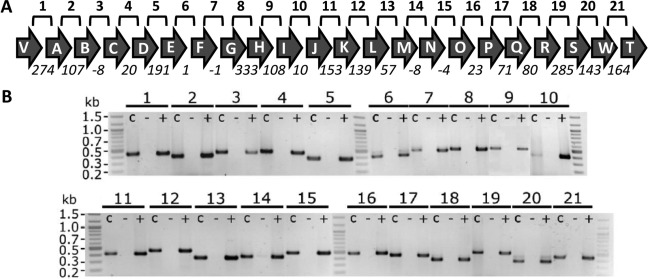
The andrimid gene cluster is organized as a polycistronic transcriptional unit.
A. Schematic representation of the adm gene cluster in Serratia plymuthica A153. Lines labelled 1‐21 above the gene cluster represent the regions amplified by RT‐PCR and shown in B. Numbers below the arrows represent the intergenic distance in base pairs, and negative numbers indicate overlapping genes.
B. Transcript analysis by RT‐PCR using primers designed to span the intergenic region between two adjacent genes. For each region, three PCR analyses were carried out: +, RT‐PCR on cDNA; −, negative control with no reverse transcriptase; c, positive control with genomic DNA as template. Culture samples for RNA isolation were taken at early stationary phase (Fig. 4).
The transcription of the adm biosynthetic cluster is growth phase dependent
We identified several mutants in which the Tn‐KRCPN1 transposon generated β‐galactosidase transcriptional fusions (Table 1). Because our RT‐PCR analyses demonstrated the presence of an operonic adm biosynthetic cluster, we investigated transcription throughout growth in a Lac‐ derivative of A153. Using a chromosomal fusion located in the first multidomain PKS/NRPS‐encoding gene of the biosynthetic cluster (admK) our β‐galactosidase assays showed that the transcription of the adm operon started in late‐logarithmic phase of growth (Figs. 4A and 5B). Expression of the biosynthetic cluster correlated perfectly with the presence of andrimid in cell‐free supernatants (Fig. 4B).
Table 1.
Bacteria and phages used in this study.
| Bacteria/phage | Genotype or relevant characteristic a | Reference or source |
|---|---|---|
| Escherichia coli DH5α | supE44 lacU169(Ø80lacZΔ M15) hsdR17 ( ) recA1 endA1 gyrA96 thi‐1 relA1 | Woodcock et al. (1989) |
| E. coli CC118λpir | araD, Δ(ara, leu), ΔlacZ74, phoA20, galK, thi‐1, rspE, rpoB, argE, recA1, λpir | Herrero et al. (1990) |
| E. coli HH26 | Mobilizing strain for conjugal transfer | Kaniga et al. (1991) |
| E. coli β2163 | F‐ RP4‐2‐Tc::Mu ΔdapA::(erm‐pir); KmR EmR | Demarre et al. (2005) |
| Serratia plymuthica A153 | Wild type, rhizosphere isolate | Hökeberg et al. (1997) |
| LacZ | A153 ΔlacZ (1470 bp Δ) | Matilla et al. (2015) |
| VN1 | A153 transposon mutant admH::Tn‐KRCPN1lacZ; KmR | This study |
| VN2 | A153 transposon mutant admK::Tn‐KRCPN1lacZ; KmR | This study |
| VN3 | A153 transposon mutant admO::Tn‐KRCPN1lacZ; KmR | This study |
| VN7 | A153 transposon mutant admM::Tn‐KRCPN1; KmR | This study |
| VN9 | A153 transposon mutant admJ::Tn‐KRCPN1; KmR | This study |
| VN11 | A153 transposon mutant admC::Tn‐KRCPN1lacZ; KmR | This study |
| VN12 | A153 transposon mutant admP::Tn‐KRCPN1; KmR | This study |
| VN17 | A153 transposon mutant admO::Tn‐KRCPN1; KmR | This study |
| VN21 | A153 transposon mutant admK::Tn‐KRCPN1; KmR | This study |
| VN22 | A153 transposon mutant admP::Tn‐KRCPN1lacZ; KmR | This study |
| VN23 | A153 transposon mutant admF::Tn‐KRCPN1; KmR | This study |
| VN24 | A153 transposon mutant admE::Tn‐KRCPN1lacZ; KmR | This study |
| VN26 | A153 transposon mutant admS::Tn‐KRCPN1lacZ; KmR | This study |
| A153JH6 | A153 ΔlacZ, zmn13::Tn‐KRCPN1; Zeamine‐; KmR | Hellberg et al. (2015) |
| ANDV | A153 ΔadmV (336 bp Δ) | This study |
| ANDW | A153 ΔadmW (150 bp Δ) | This study |
| ANDX | A153 ΔadmX (789 bp Δ) | This study |
| XJH6 | A153 ΔadmX, zmn13::Tn‐KRCPN1; andrimid‐, zeamine‐; KmR | This study |
| ARpoS | A153 rpoS::Km; KmR | Matilla et al. (2015) |
| AHfq | A153 Δhfq::Km (252 bp Δ); KmR | Matilla et al. (2015) |
| A153H | A153 Δhfq (252 bp Δ) | This study |
| A153HL | A153 ΔlacZ, Δhfq | This study |
| LVN2 | A153 ΔlacZ, admK::Tn‐KRCPN1lacZ; generated by transduction using ϕMAM1; KmR | This study |
| HLVN2 | A153 ΔlacZ, Δhfq, admK::Tn‐KRCPN1lacZ; generated by transduction using ϕMAM1; KmR | This study |
| XLVN2 | A153 ΔlacZ, ΔadmX, admK::Tn‐KRCPN1lacZ; generated by transduction using ϕMAM1; KmR | This study |
| ASptI | A153 in‐frame sptI (483 bp Δ) | Matilla and Salmond (unpubl. data) |
| ASptR | A153 sptR::Km; KmR | Matilla and Salmond (unpubl. data) |
| ASplR | A153 splR::Km; KmR | Matilla and Salmond (unpubl. data) |
| ASpsR | A153 spsR::Km; KmR | Matilla and Salmond (unpubl. data) |
| A153C | A153 in‐frame csrB (267 bp Δ) | Matilla and Salmond (unpubl. data) |
| Serratia marcescens MSU97 | Wild type, plant epiphyte, pigmented | Strobel et al. (1999) |
| Bacillus subtilis JH642 | pheA1 trpC2 | J.A. Hoch |
| Dickeya solani MK10 | Wild type, plant pathogen | Pritchard et al. (2013) |
| Xanthomonas campestris pv. campestris | Wild type, plant pathogen | R. Penyalver |
| Phages | ||
| ϕMAM1 | Generalized transducing phage for S. plymuthica A153 | Matilla and Salmond (2014) |
a. The following abbreviations are used: Km, kanamycin; Tc, tetracycline; Em, erythromycin.
Figure 4.
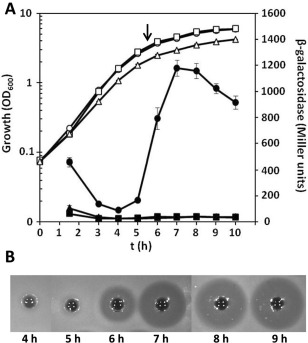
AdmX and Hfq regulate andrimid production by activating the expression of the adm biosynthetic gene cluster.
A. β‐galactosidase activity (filled symbols) throughout growth measured from a chromosomal fusion admK::lacZ in Serratia plymuthica A153 LacZ (circles), and its ΔadmX (squares) and Δhfq (triangles) derivative strains in LB medium at 25 °C. Open symbols represent bacterial growth. Data are the mean and standard deviation of three biological replicates. Arrow, time point when samples for RT‐PCR and qPCR were taken (Figs. 3 and S6).
B. Andrimid production by S. plymuthica A153 strain JH6 (zeamine negative) throughout growth in LB medium at 25 °C. For the assays, a Bacillus subtilis top agar lawn was prepared and 300 µl of filter‐sterilized supernatants were added to holes punched in the Bacillus bioassay plates.
Figure 5.
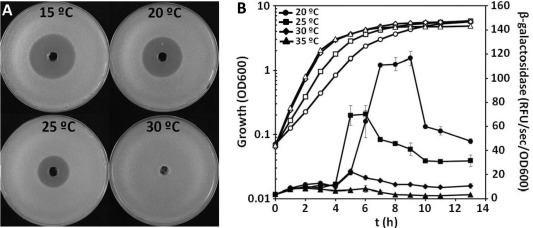
The production of andrimid in Serratia plymuthica A153 is temperature‐dependent and correlates with the expression of the adm gene cluster.
A. Halos of antibiosis against Bacillus subtilis of filter‐sterilized supernatants of A153 strain JH6 (zeamine negative) grown in LB at different temperatures. The bioassays were repeated at least three times, and representative results are shown.
B. β‐Galactosidase activity (filled symbols) throughout growth measured from the chromosomal fusion admK::lacZ in Serratia plymuthica A153 LacZ. Open symbols represent bacterial growth. Data are the mean and standard deviation of three biological replicates. Doubling times at 20, 25, 30 and 35 °C were 66.8 ± 0.4, 47.3 ± 0.7, 40.7 ± 0.4 and 40.5 ± 0.2 minutes respectively.
Temperature and carbon source regulate the transcription of the adm operon
The biosynthesis of secondary metabolites can be energetically costly and other secondary metabolites produced by Serratia strains have been shown to be highly regulated by various environmental cues (Williamson et al., 2006; Coulthurst et al., 2005). To shed light on the regulation of andrimid production in A153, we investigated the impact of different environmental parameters in the transcription of the adm operon.
At the optimal growth temperature (30 °C) for A153, andrimid production was abolished (Fig. 5A). However, as the temperature decreased, a gradual increase in the production of andrimid was observed, with higher production levels at 15 °C than at 25 °C (Fig. 5A). To examine whether the increase in andrimid production was reflected in the expression of the adm gene cluster, we evaluated the impact of temperature on the transcription of the biosynthetic cluster. β‐galactosidase assays showed that the transcription was also thermoregulated (Fig. 5B). In accordance with this, no adm expression was observed at temperatures above 30 °C, confirming the tight correlation between andrimid production and transcription of the biosynthetic operon (Fig. 5B).
Serratia plymuthica A153 was originally isolated from the rhizosphere (Astrom and Gerhardson, 1988), a soil environment which is under the direct influence of plant‐root exudates. The composition of root exudates is chemically complex but quantitatively it consists mainly of carbon‐based compounds, primarily organic acids and sugars (Uren, 2007; Badri and Vivanco, 2009; Suzuki et al., 2009). Thus, we investigated the production of andrimid in response to different carbon sources found in plant root exudates. Our results showed that andrimid is differentially produced depending on carbon source. Maximal antibacterial activity was observed in the presence of citrate, gluconate or glycerol whereas no activity was detected in the presence of arabinose or succinate as sole carbon sources (Fig. 6A). Although the growth rates of A153 varied between the tested carbon sources (Supporting information Fig. S5), the observed antibacterial activity was not correlated with the growth rate per se or the final optical density reached in each of the culture media. Consistent with these observations, higher transcriptional levels of the adm operon were observed in media containing citrate or gluconate, whereas no transcription was seen in arabinose media (Fig. 6B), confirming that both transcriptional activity and andrimid production, were carbon source‐dependent.
Figure 6.
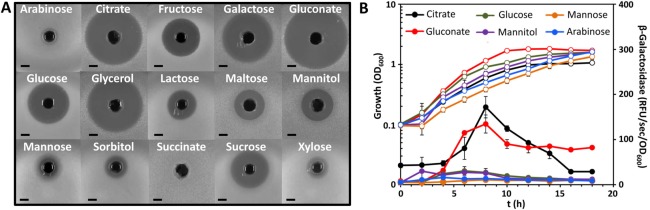
Effect of carbon source on the production of andrimid and expression of the adm gene cluster in Serratia plymuthica A153.
A. Halos of antibiosis against Bacillus subtilis of filter‐sterilized supernatants of A153 strain JH6 (zeamine negative) grown in minimal medium with different carbon sources. The bioassays were repeated at least three times, and representative results are shown. All the carbon sources were used at a final concentration of 15 mM. With the exception of lactose, all carbon sources used are frequently found in plant root exudates. Bars, 5 mm.
B. β‐galactosidase activity (filled symbols) throughout growth measured from the chromosomal fusion admK::lacZ in Serratia plymuthica A153 LacZ in minimal medium with different carbon sources. Open symbols represent bacterial growth. Data are the mean and standard deviation of three biological replicates. Growth and doubling times of A153 in all the carbon sources used are shown in Fig. S5.
Finally, we also evaluated the production of andrimid at different pH, aeration conditions and NaCl concentrations, but no impacts on antibacterial synthesis were observed (not shown).
The LysR‐type regulator, AdmX, activates andrimid production
Analysis of the genomic context of the adm gene clusters in A153, MSU97, 90‐166, Eh335, S2052 and SWAT‐3 revealed a 2 kb region upstream of admV which was highly conserved in the Serratia strains (Supporting information Figs. S3 and S4). This region contained a gene encoding a putative LysR‐type transcriptional regulator (LTTR; Supporting information Figs. S3 and S4). The family of LTTRs comprises one of the largest classes of transcriptional regulators in bacteria, functioning either as repressors or activators of the transcription of their target genes (Maddocks and Oyston, 2008). To investigate the potential role of the identified LTTR in andrimid biosynthesis, we engineered an in frame deletion mutant in S. plymuthica A153. The resulting mutant did not inhibit growth of B. subtilis but antibacterial activity was fully restored by in trans expression of the LTTR encoding gene (Fig. 7A). These results suggested that the regulator, designated AdmX, is responsible for activating transcription of the adm gene cluster and β‐galactosidase assays confirmed that expression of the biosynthetic cluster was abolished in an admX‐deficient strain (Fig. 4A). These results were supported by qPCR analyses showing decreased adm transcript levels in the A153 ΔadmX strain (Supporting information Fig. S6).
Figure 7.
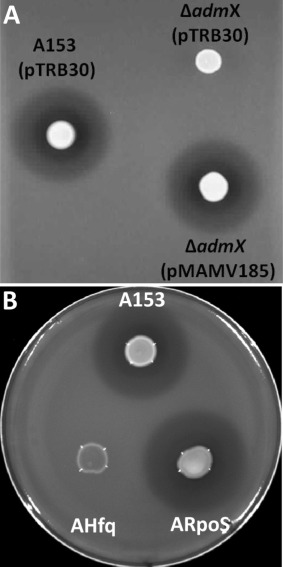
The LysR‐type regulator AdmX, and the RNA chaperone Hfq positively regulate the biosynthesis of andrimid. Bioactivities against Bacillus subtillis of S. plymuthica A153 and derivative strains are shown (A, B). In frame deletion of admX was functionally complemented by the in trans expression of AdmX using the pQE80L‐based vector, pMAMV185 (A). Induction of the expression of the AdmX was done by addition of 0.1 mM of IPTG. Complementation of the Δhfq strain was done using a pQE‐80L‐based vector (Fig. S8). The bioassays were repeated at least three times, and representative results are shown. Pictures were taken after 48 h of incubation at 25 °C.
To assess the transcription of admX throughout growth, we constructed a transcriptional fusion of the admX promoter to the reporter gene lacZ. β‐galactosidase assays showed that the transcription of admX started in late‐logarithmic phase of growth, correlating with transcription of the adm biosynthetic cluster (Supporting information Fig. S7). We explored the effect of AdmX on expression of its cognate gene. However, the transcription of admX remained unaltered in an admX mutant strain (Supporting information Fig. S7). Finally, we also investigated whether andrimid thermoregulation is mediated by AdmX. Previous studies showed that the expression of another LTTR (PecT), and consequently the expression of its target genes, is thermoregulated (Hérault et al., 2014). However, no differences in transcripts levels of admX were observed (using qPCR analysis) at 25, 30 and 37 °C (data not shown).
The chaperone Hfq regulates the expression of the adm gene cluster
The biosynthesis of secondary metabolites is frequently regulated at post‐transcriptional level and, in gammaproteobacteria, this is mainly mediated by the Csr/Rsm (Romeo et al., 2013) and Hfq (Vogel and Luisi, 2011) systems. To assess a potential role of these post‐transcriptional regulatory systems, we analysed the production of andrimid in different genetic backgrounds in S. plymuthica A153. A mutant with a deletion in the non‐coding sRNA, csrB, showed the same antibacterial activity as the wild type strain (not shown). However, in frame deletion of hfq led to complete loss of andrimid production (Fig. 7B) and this phenotype was partially complemented by the in trans expression of hfq (Supporting information Fig. S8).
β‐galactosidase assays were performed to assess the expression of the adm gene cluster throughout growth in A153 Δhfq. The results showed that transcription of the andrimid operon was abolished in a hfq‐deficient strain, indicating that the chaperone Hfq positively regulates expression of the adm biosynthetic cluster (Fig. 4A). qPCR analyses confirmed that the transcriptional levels of the adm gene cluster were reduced by 99.9% in an hfq‐deficient background (Supporting information Fig. S6). To investigate whether Hfq regulates admX expression, qPCR experiments were also performed. However, admX transcript levels were unaltered in the Δhfq mutant (Supporting information Fig. S6).
The expression of the adm gene cluster starts in late‐logarithmic phase of growth (Fig. 4) and so we investigated whether quorum sensing (QS) could be regulating andrimid production. A153 carries sptI, sptR, splR and spsR genes encoding LuxI‐ and LuxR‐type proteins very similar to those of the three QS systems identified in the taxonomically related rhizobacterium, S. plymuthica G3 (Liu et al., 2011; Duan et al., 2012). However mutations in each of these four genes in A153 had no observable impacts on antibacterial activity (not shown). Similarly, the role of the stationary phase sigma factor, RpoS, in the production of andrimid was also investigated, but the antibacterial activity of an rpoS mutant was indistinguishable from that of the wild type strain (Fig. 7B).
Discussion
Since the golden age of antimicrobial discovery in the 1940–1960s, there has been a diminution in the rate of discovery of new antibiotics and this problem has been exacerbated by the progressive emergence worldwide of antibiotic‐resistant bacteria (Lewis, 2013; Blair et al., 2015). However, there are many antibiotics described in the literature that, although not currently exploited, could prove to be lead molecules for drug discovery programmes leading to clinical or agricultural utility. The hybrid NRPS/PKS antibacterial compound, andrimid, was first isolated in the late 1980s from a brown planthopper intracellular symbiont belonging to the Enterobacter genus (Fredenhagen et al., 1987). After its discovery, three related antibacterial compounds, moiramides A‐C (Supporting information Fig. S1), were also isolated from Pseudomonas fluorescens (Needham et al., 1994).
Andrimid is a nanomolar inhibitor of the bacterial acetyl‐CoA carboxylase (Freiberg et al., 2004), an enzyme responsible for the conversion of acetyl‐CoA to malonyl‐CoA in the first committed step for the synthesis of fatty acids (Harwood, 2007). The discovery of the andrimid target, together with its unusual chemistry, has stimulated interest in the biochemistry and genetics of the antibiotic. The andrimid gene cluster was first described almost a decade ago and a model for its biosynthesis proposed (Jin et al., 2006). In this model, the function for most of the Adm proteins was predicted and, in some cases, later demonstrated biochemically (Fortin et al., 2007; Magarvey et al., 2008; Ratnayake et al., 2011). However, the roles of the hypothetical proteins AdmB, AdmL and AdmN in the synthesis of andrimid remain unknown. In this study, we redefined the length of the adm gene cluster by identifying a new hypothetical protein encoding gene, admV, located immediately upstream of admA and forming part of the adm operon (Figs. 3 and Supporting information Fig. S3). Using mutagenesis and complementation analyses we demonstrated that AdmV is required for the biosynthesis of andrimid. Our bioinformatic analyses did not categorically clarify the function of AdmV. However, in silico studies predicted three alpha helices with a high content of positively charged amino acids at the C‐terminal end, suggesting that AdmV may be a DNA binding protein.
Andrimid has been shown to be a broad‐spectrum antibacterial compound that acts on Gram‐positive and Gram‐negative bacteria. It was first isolated after it was shown to have potent activity against the different pathovars of the phytopathogen, Xanthomonas campestris (Fredenhagen et al., 1987) but further research showed that it is also active against bacterial pathogens belonging to Bacillus, Enterococcus, Escherichia, Salmonella, Staphylococcus, Vibrio and Yersinia genera (Needham et al., 1994; Singh et al. 1997; Long et al., 2005; Wietz et al., 2010; Sánchez et al., 2013). Resistance to andrimid is dependent on multidrug efflux pumps (Freiberg et al., 2004; Jin et al., 2006) or the presence of andrimid‐resistant acetyl‐CoA carboxyltransferases (ACC) hypothesized to show decreased affinity for the inhibitor (Liu et al., 2008). Interestingly, we showed that the plant‐pathogen Agrobacterium tumefaciens and multiple rhizobacteria strains in the family Enterobacteriaceae (i.e., Kluyvera, Pantoea, Serratia, Weeksella, Xenorhabdus, and the emerging phytopathogen, Dickeya solani) (Supporting information Figs. S9 and S10) are sensitive to andrimid. However, the differential sensitivities to the antibiotic of these plant‐associated strains could also indicate the presence of weaker intrinsic resistance mechanism(s) of different efficiencies.
NRPS and PKS constitute the main enzymatic source of secondary metabolites in bacteria; 36% of which are generated by hybrid NRPS/PKS gene clusters (Wang et al., 2014). Secondary metabolites have been associated with important roles in the physiology and development of the bacterial host, but are also implicated in overcoming competitors in the same nutritional niche (Liu et al., 2013; Pidot et al., 2014; Wang et al., 2014; Mousa and Raizada, 2015). The synthesis in situ of andrimid has been demonstrated in a sponge‐associated bacterium (Oclarit et al., 1994; Long et al., 2005) and its production is favoured over that of other metabolites in conditions mimicking natural environments ‐ suggesting an important ecophysiological role of andrimid (Wietz et al., 2011). Consistent with this view, even sub‐lethal concentrations of andrimid can promote a negative chemotactic response (in sensitive competitors) away from the antibiotic, thereby enhancing competitiveness of the producers (Graff et al., 2013). In S. plymuthica A153, the synthesis of andrimid is carbon source‐dependent and induced in the presence of sugars and organic acids commonly found in plant root exudates (Fig. 6). Carbon source has been shown to regulate antibiotic synthesis at both molecular and physiological levels and, in general, rapidly metabolized carbon sources are involved in repression of bacterial secondary metabolites (Sánchez et al., 2010). However, our results did not show a correlation between preferred carbon sources and andrimid production (Figs. 6 and S5). Importantly, in A153, the production of the antibiotic zeamine is high in the presence of carbon sources which show low andrimid production (i.e., arabinose, sorbitol and succinate) whereas the synthesis of zeamine is repressed by carbon sources that stimulate andrimid production (i.e., citrate, gluconic acid and glycerol) (Fig. 6; Hellberg et al., 2015). This “mirror image” metabolic regulation of andrimid and zeamine production when presented with different environmental carbon sources could be beneficial in maintaining the capacity of A153 to overcome bacterial competitors in its natural niche, the rhizosphere, where specific carbon source availability is likely to fluctuate.
Soil bacteria are subjected to daily and seasonal abiotic variations and temperature is considered a key abiotic factor influencing bacterial metabolic activity – which may therefore modulate inter‐ and intra‐specific interactions between microbes. The results reported here show that andrimid synthesis in A153 is thermoregulated, with enhanced antibiotic production at lower temperature. Previous studies also showed that andrimid production in Vibrio (Long et al., 2005; Wietz et al., 2011) and Serratia strains (Sánchez et al., 2013) is also thermosensitive. In this study, we demonstrate that the observed thermoregulation is exerted at the transcriptional level (Fig. 5) and in a AdmX‐independent fashion. During the analysis of the bioactive properties of A153, we showed that the production of the haterumalide, oocydin A, is also thermosensitive (Matilla and Salmond, unpubl. data) and this may indicate that a common control pathway is modulating the thermoregulation of the biosynthesis of secondary metabolites in A153. The ecological role of this thermosensitivity is unknown, although it could be related to competitiveness and efficient colonization of specific niches by the producing rhizobacterium, S. plymuthica A153.
To date, most of the published research on andrimid has focussed on its biochemistry, but the regulation of its biosynthesis has attracted little attention. The first adm gene cluster was identified in Pantoea agglomerans but no regulatory proteins were found in this biosynthetic cluster (Jin et al., 2006). However, our analysis of the genomic context of the andrimid gene cluster in A153, MSU97 and 90‐166, revealed a LysR‐type regulator‐encoding gene, admX, which was highly conserved in these producing strains (Supporting information Figs. S3 and S4). Mutagenesis, complementation and gene expression analyses demonstrated that AdmX is a positive regulator of andrimid biosynthesis (Figs. 4 and 7A). AdmX is a 305 amino acid protein composed of a helix‐turn‐helix motif‐containing DNA‐binding domain and a probable effector binding domain at the N‐terminal and C‐terminal regions, respectively (Supporting information Fig. S11). By analogy with other LTTR structures (Ezezika et al., 2007; Monferrer et al., 2010; Devesse et al., 2011), AdmX is proposed to possess a potential effector binding site located in between the two lobes of the effector binding domain (Supporting information Fig. S11). Inspection of a homology model shows that this site is primarily composed of amino acids with hydrophobic side chains (Supporting information Fig. S11) and further investigations are necessary to identify candidate effector molecules.
In general, LysR transcriptional regulators activate the expression of their target genes while negatively autoregulating their own transcription (Maddocks and Oyston, 2008). LTTRs have been shown to regulate genes involved in metabolism, virulence, motility, chemotaxis, quorum sensing and biofilm formation (Maddocks and Oyston, 2008). However, only a limited number of LTTRs have been found to regulate the biosynthesis of secondary metabolites – such as actinorhodin (Mao et al., 2013), phenazines (Klaponski et al., 2014), pyoluteorin (Li et al., 2012), ralfuranones (Kai et al., 2014) and undecylprodigiosin (Mao et al., 2013). LTTRs are broadly distributed within the prokaryotic kingdom, suggesting dissemination and acquisition by horizontal gene transfer (HGT; Maddocks and Oyston, 2008). In accordance with this notion, sequences reminiscent of transposases were found flanking admX (Supporting information Figs. S3 and S4) and the G + C content of the admX genes in A153, MSU97 and 90‐166 is considerably higher than that of their respective adm gene clusters. Although AdmX seems to be restricted to the adm biosynthetic clusters present in Serratia strains, BLAST analyses showed that AdmX is up to 84% identical (90% similar) to an orphan LysR‐type regulator highly conserved within Enterobacter and Klebsiella genera, again suggesting that this LTTR‐encoding gene may have been acquired by HGT. Interestingly, we found an LTTR‐encoding gene immediately upstream of admT in the adm gene clusters of Vibrio coralliilyticus S2052 and Vibrionales SWAT‐3 (Supporting information Fig. S3), perhaps indicating an important regulatory role in the expression of the adm gene clusters in these strains. It has been proposed that the fragmented andrimid biosynthetic pathway reflects a recent evolutionary origin (Magarvey et al., 2008) and so the insertion of the LTTR‐encoding genes in the adm gene clusters of Vibrio and Serratia may represent a step forward in the evolution of these biosynthetic clusters.
The post‐transcriptional regulation of the adm gene cluster in A153 was also investigated and our results showed that the RNA binding protein, Hfq, positively regulates the expression of the andrimid operon. Previous studies of Serratia strains showed that the synthesis of secondary metabolites such as a carbapenem antibiotic (Wilf et al., 2011), prodigiosin (Wilf et al., 2011) and pyrrolnitrin (Zhou et al., 2012) is regulated by Hfq, and we recently showed that Hfq positively regulates the expression of the oocydin A (Matilla et al., 2015) and zeamine (Hellberg et al., 2015) gene clusters in S. plymuthica A153. It is known that Hfq also stimulates rpoS translation (Vogel and Luisi, 2011) and the deletion of hfq results in reduced rpoS transcripts levels in A153 (Matilla et al., 2015). However, contrary to our observations on regulation of oocydin A in A153 (Matilla et al., 2015), Hfq‐mediated regulation of andrimid is independent of RpoS (Fig. 7B), as observed in other Serratia strains for the biosynthesis of zeamine (Hellberg et al., 2015), carbapenem (Wilf and Salmond, 2012) and prodigiosin (Wilf and Salmond, 2012) antibiotics.
In summary, we have shown that the plant‐associated bacterium, Serratia plymuthica A153, produces the broad spectrum antibacterial compound, andrimid. Comparative genomics, molecular genetics and transcriptional approaches led us to re‐define the borders of the adm gene clusters and identify new genes involved in the biosynthesis and regulation of andrimid. Further, in vivo assays expanded the spectrum of bacterial strains which are sensitive to andrimid. For the first time, the regulation of the biosynthesis of andrimid was investigated at transcriptional and post‐transcriptional levels. Future research will provide information about the effectors recognized by AdmX and such knowledge, coupled with the work described in this study, may encourage the use of plant‐associated andrimid‐producing strains as biocontrol agents in sustainable agriculture strategies.
Experimental procedures
Bacterial strains, culture media and growth conditions
Bacterial strains used in this study are listed in Table 1 and Supporting Information Table S2. Serratia, Bacillus, Agrobacterium, Dickeya, Kluyvera, Pantoea, Weeksella, Xanthomonas, Xenorhabdus, Yersinia and their derivative strains were grown routinely at 30 °C, unless otherwise indicated, in Luria Broth (LB; 5 g yeast extract l−1,10 g Bacto tryptone l−1 and 5 g NaCl l−1) or minimal medium (0.1%, w/v, (NH4)2SO4, 0.41 mM MgSO4, 40 mM K2HPO4, 14.7 mM KH2PO4, pH 6.9–7.1) with glucose (0.2%; w/v) as carbon source, unless otherwise indicated. Escherichia coli strains were grown at 37 °C in LB. Escherichia coli DH5α was used as a host for gene cloning. Media for propagation of E. coli β2163 were supplemented with 300 µM 2,6‐diaminopimelic acid. When appropriate, antibiotics were used at the following final concentrations (in µg ml−1): ampicillin, 100; chloramphenicol, 25; kanamycin, 25 (E. coli strains) and 50 (Serratia strains); streptomycin, 50; tetracycline, 10. Sucrose was added to a final concentration of 10% (w/v) when required to select derivatives that had undergone a second crossover event during marker‐exchange mutagenesis. Bacterial growth (OD600 nm) was measured on a Unicam Heλios spectrophotometer at 600 nm, 1 cm path length.
In vitro nucleic acid techniques and bioinformatic analyses
Plasmid DNA was isolated using the Anachem Keyprep plasmid kit. For DNA digestion, the manufacturer's instructions were followed (New England Biolabs, Roche and Fermentas). Separated DNA fragments were recovered from agarose gels using the Anachem gel recovery kit. Ligation reactions and total DNA extraction were performed as previously described (Sambrook et al., 1989). Competent cells were prepared using calcium chloride and transformations were performed by standard protocols (Sambrook et al., 1989). Phusion® high fidelity DNA polymerase (New England Biolabs) was used in the amplification of PCR fragments for cloning. PCR reactions were purified using the Anachem PCR Clean‐up kit. PCR fragments were verified by DNA sequencing that was carried out at the University of Cambridge DNA Sequencing Facility (Cambridge, UK) or at the Institute of Parasitology and Biomedicine Lopez‐Neyra (CSIC; Granada, Spain). Sequence comparison analyses were performed employing the wgVISTA online tool (Frazer et al., 2004). Open reading frames (ORFs) in the andrimid gene clusters were predicted using Glimmer 3.0 (Delcher et al., 1999). Blast analyses were used for the functional gene assignment. Protein domain organization was identified using the NCBI conserved domains database. Multiple sequence alignments were carried out with ClustalW2 (European Bioinformatics Institute). Artemis software (Wellcome Trust Sanger Institute) was used to visualize genomic sequences.
Random transposon mutagenesis
Random transposon mutagenesis of S. plymuthica A153 using Tn‐KRCPN1 was performed by biparental conjugation mating using Escherichia coli β2163, as described previously (Matilla et al., 2012). In total, three thousand kanamycin‐resistant insertion mutants were screened for their antibacterial activity against Bacillus subtilis using dual drop culture bioassays. Auxotrophic mutants were discarded and insertion mutations were transduced into the wild type strain A153 using phage ϕMAM1 (Matilla and Salmond, 2014). The insertion site of transposon Tn‐KRCPN1 in mutants of interest was determined using random primed PCR following the method described previously (Matilla et al., 2012) and using primers described in Supporting Information Table S3.
Antibacterial assays
Antibiotic activity was tested using agar lawn assays. Briefly, indicator plates for andrimid production contained a 0.8% LB agar (LBA) top lawn containing 200 µl of an overnight culture of the bacterial strain to test. Five microliters of overnight cultures of the andrimid‐producing strains were spotted on the surface of the indicator agar lawn and incubated for 48 h at 25 °C, unless otherwise indicated. To determine andrimid levels in bacterial supernatants, culture samples were taken, bacterial cells were pelleted by centrifugation (10,000×g, 10 min), and the supernatant was filtered (0.2 µm). Three hundred microliters of the filter‐sterilized supernatant were added to wells cut into the LBA plate and incubated at 25 °C for 24 h. All experiments were repeated at least three times.
Construction of strains and plasmids
Chromosomal mutants of Serratia plymuthica strains were constructed by homologous recombination using derivative plasmids of the suicide vector pKNG101. These plasmids, which are listed in Supporting Information Table S4, were confirmed by DNA sequencing and they carried mutant in‐frame deletions for the replacement of wild type genes in the chromosome. Primers used in this study are listed in Supporting Information Table S3. In all cases, plasmids for mutagenesis were transferred to S. plymuthica strains by triparental conjugation using E. coli CC118λpir and E. coli HH26 (pNJ500) as helper. The plasmids for the construction of the in‐frame deletion mutants were generated by amplifying the up‐ and downstream flanking regions of the gene, or domain to be deleted. The resulting PCR products were digested with the enzymes specified in Supporting Information Table S4 and ligated in a three‐way ligation into pUC18Not, previously cloned into the marker exchange vector pKNG101. The in‐frame deletion mutant strains ANDX, ANDV and ANDW were generated using plasmids pMAMV175, pMAMV191 and pMAMV192, respectively. Mutant strains defective in hfq, A153H and A153HL, were constructed using plasmid pMAMV193. All relevant mutations were confirmed by PCR and sequencing.
For the construction of the complementing plasmids, the genes were amplified using primers described in Supporting Information Table S3 and cloned into pTRB30. All the inserts were confirmed by PCR and sequencing. Complementing plasmids were used to transform A153 by electroporation.
Genetic complementation assays
Complementation of mutations was carried out by the introduction of a wild type copy of the corresponding mutated gene in trans on plasmid pTRB30. For the complementation assays, LBA containing the appropriate antibiotic (to maintain the plasmid) and isopropyl‐β‐D‐thiogalactopyranoside (IPTG) at 0.1 or 1 mM were added to holes punched in Bacillus subtilis bioassay plates. Then, 5 μl of overnight cultures of the selected strains were spotted on the surface of the LBA containing the antibiotic and IPTG and were incubated at 25 °C for 2 days.
Generalized transduction
The generalized transducing viunalikevirus, ϕMAM1, was used for transduction of chromosomal mutations, as described previously (Matilla and Salmond, 2014).
β‐Galactosidase assays
Expression of the lacZ reporter gene was performed using the fluorogenic substrate 4‐methylumbelliferyl β‐D‐galactoside (Melford Cat No. M1095) at a final concentration of 0.125 mg ml−1, as described previously (Ramsay, 2013). Samples were measured in a SpectraMax Gemini XPS fluorescence microplate reader (Molecular Devices) using the following settings: excitation 360 nm, emission 450 nm, cut‐off 435 nm, reading every 30 s for 20 min at 37 °C. β‐Galactosidase activity was expressed as relative fluorescent units s−1 and normalize to the OD600 nm of the corresponding sample. Alternatively, β‐galactosidase activity was measured as described previously (Miller, 1972) using 2‐Nitrophenyl β‐D‐galactopyranoside (ONPG; Sigma–Aldrich Cat No. N1127) as substrate. All the transcriptional fusion assays were carried out using S. plymuthica A153 LacZ (control) or derived mutants.
RNA extraction, cDNA synthesis, reverse transcription‐PCR (RT‐PCR) and quantitative real time PCR analyses
RNA was extracted from early stationary phase cultures grown in LB medium using an RNeasy mini kit (Qiagen) according to the manufacturer's instructions. RNA concentration was determined spectrophotometrically and RNA integrity was assessed by agarose gel electrophoresis. Genomic DNA contamination was eliminated by treating total RNA with Turbo DNA‐free (Ambion). The synthesis of cDNA was performed using random hexamers (GE Healthcare) and SuperScript II reverse transcriptase (Invitrogen) in a 30 µl reaction with 2 µg of total RNA and incubation at 42 °C for 2 h. As negative control the reaction was performed omitting the reverse transcriptase. For the RT‐PCR analysis, the equivalent of 50 ng of total RNA was subjected to PCR amplification using primers to amplify across the junctions (Supporting Information Table S3). Positive and negative control PCR reactions were performed using genomic DNA and no‐RT cDNA samples, respectively, as templates. PCR conditions consisted of 30 cycles of denaturation for 1 min at 94 °C, annealing for 1 min at 62 °C, and extension for 40 s at 72 °C. qPCRs were performed as described previously (Burr et al., 2006) using primers specific for admX and admV (Supporting Information Table S3). qPCR amplifications were performed using an MyiQTM2 Two‐Color Real‐Time PCR Detection System (Bio‐Rad). To confirm the absence of contaminating genomic DNA, control PCRs were carried out using no RT cDNA samples as templates. Melting curve analyses were conducted to ensure amplification of a single product. The relative gene expression was calculated using the critical threshold (ΔΔCt) method (Pfaffl, 2001) and using 16S rRNA as the internal control to normalize the data.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's web‐site:
Fig. S1. Structures of andrimid and moiramide B. The structure of andrimid consists of an unsaturated fatty acid chain, a pyrrolidinedione ring, a valine and glycine derived β‐ketoamide and the amino acid β‐phenylalanine. Chemical synthesis studies showed that the fatty acid chain and β‐phenylalanine are involved in bacterial cell penetration of the antibiotic, whereas the pyrrolidinedione head and the β‐ ketoamide moiety are responsible for the antibacterial activity (Pohlmann et al., 2005; Freiberg et al., 2006).
Fig. S2. Antibacterial activity of Serratia marcescens MSU97 against Bacillus subtilis.
Fig. S3. Schematic representation of the andrimid gene clusters of Serratia marcescens MSU97, Serratia marcescens 90‐166, Pantoea agglomerans Eh335, Vibrio coralliilyticus S2052 and Vibrionales bacterium SWAT‐3. Numbers below the arrows represent the intergenic distance between contiguous genes, with negative numbers indicate overlapping genes
Fig. S4. DNA homology between the andrimid gene cluster of Serratia plymuthica A153 and the andrimid gene clusters of the other producing strains. A, Schematic representation of the adm gene cluster in Serratia strains. B‐F, Alignments representing the percentage of DNA homology between the adm gene cluster of A153 and those of S. marcescens MSU97 (B), S. marcescens 90‐166 (C), Pantoea agglomerans Eh335 (D) Vibrio coralliilyticus S2052 (E) and Vibrionales SWAT‐3 (F). Alignments were performed using wgVISTA (Frazer et al., 2004).
Fig. S5. Growth of Serratia plymuthica A153 in minimal medium with different carbon sources. Growth curves showing the doubling time in sorbitol (181.2 ± 1 min), mannitol (153 ± 1 min), fructose (145.8 ± 2 min), galactose (115.2 ± 1 min), mannose (181.6 ± 2 min), lactose (413.4 ± 6 min), xylose (208.2 ± 2 min), succinic acid (142.8 ± 1 min), maltose (145.2 ± 1 min), sucrose (106.2 ± 1 min), glucose (115.2 ± 1 min), glycerol (121 ± 1 min), gluconic acid (96.6 ± 1 min), arabinose (235.2 ± 2 min) and citrate (158.7 ± 3 min) as sole carbon source. Data are the mean and standard deviation of three biological replicates. The assays were done at 25 ºC with shaking at 200 rpm.
Fig S6. Impact of Hfq (A) and AdmX (B) on the expression of admV and admX. Quantitative real‐time PCR was used to measure transcript levels of admV (grey bars) and admX (white bars) in Serratia plymuthica A153, and derivative strains. The values showed the average expression relative to wild type expression. The arrow in Fig. 4A indicates the time point when the samples for qPCR were taken. The data are the mean and standard deviation of three biological replicates.
Fig. S7. AdmX transcription correlates with the expression of the andrimid gene cluster. Transcription of the admX (PadmX::lacZ; pMAMV244) promoter fusion throughout growth in Serratia plymuthica A153 strains. β‐Galactosidase activity (filled symbols) and growth curves (open symbols) were determined in LacZ (red) and ΔadmX (blue) in LB medium at 25 °C. A153 wt harbouring the empty reporter plasmid (black) was used as negative control in the assays. Data are the mean and standard deviation of three biological replicates.
Fig. S8. Genetic complementation of Serratia plymuthica A153 strain A153H. Expression of hfq in trans in A153 Δhfq restored andrimid production and therefore the antibacterial activity against Bacillus subtilis. Induction of Hfq expression was done by addition of 0.1 mM of IPTG. The bioassays were repeated at least three times, and a representative figure is shown. Pictures were taken after 48 h of incubation at 25 °C.
Fig. S9. Sensitivities of different bacterial strains to the antibiotic andrimid. Bioactivities of Serratia plymuthica A153 and the non‐andrimid producing mutant of A153, VN2, against ecologically different bacterial strains. For the assays, an indicator top agar lawn was prepared as described in “Experimental procedures,” and 5 μl overnight cultures of the A153 strains were spotted on the surface of the bacterial indicator agar lawns. The bioassays were repeated three times, and representative results are shown. Pictures were taken after 48 h of incubation at 25 °C. The strains used are described in Table 1 and supplementary Table S2.
Fig. S10. Andrimid shows antibacterial activity against Bacillus subtilis, Dickeya solani and Xanthomonas campestris pv. campestris. Recovery of viable Bacillus, Dickeya and Xanthomonas cells grown in the presence of A153 JH6 (andrimid positive, zeamine negative) and A153 XJH6 (andrimid and zeamine negative) supernatants. The values showed the percentage of viable cells in the presence of JH6 supernatants relative to the number of viable cells in the presence of XJH6 supernatants. For the assays, overnight bacterial cultures of Bacillus, Dickeya and Xanthomonas were adjusted to an optical density at 600 nm (OD600) of 0.1 and grown at 30 ºC with orbital shaking (225 rpm). At an OD600 of 0.4, 10 mL of the bacterial culture was removed and pelleted by centrifugation at 4,000 x g for 10 min at room temperature. The pellet was resuspended in 5 ml of 2X LB and 5 ml supernatants of an overnight culture of A153 JH6 or A153 XJH6 were added to the bacterial culture. Samples were taken after 5 and 10 h of incubation and the number of colony forming units (CFU) were determined. Data are the mean and standard deviation of three biological replicates.
Fig. S11. Homology model of AdmX. The model was generated by the Geno3D modeling algorithm (Combet et al., 2000) and the structure of the BenM transcriptional regulator (PDB ID 3K1N) as template. The site for the binding of potential effector molecules is indicated.
Table S1. Identity at DNA level of the andrimid gene clusters between producing strains.
Table S2. Additional bacterial strains used in this study.
Table S3. Oligonucleotides used in this study.
Table S4. Plasmids used in this study.
Acknowledgements
We thank Kornelia Smalla and Ian Toth for the generous donation of bacterial strains. Work in the Salmond laboratory is supported by funding through the Biotechnology and Biological Sciences Research Council (UK). M.A.M. was supported by the EU Marie‐Curie Intra‐European Fellowship for Career Development (FP7‐PEOPLE‐2011‐IEF) Grant No. 298003 and the Spanish Ministry of Economy and Competitiveness Postdoctoral Research Program, Juan de la Cierva (JCI‐2012‐11815). The Krell laboratory is supported by FEDER funds and Fondo Social Europeo through grants from the Junta de Andalucía (Grant CVI‐7335) and the Spanish Ministry for Economy and Competitiveness (Grants BIO2013‐42297 and RTC‐2014‐1777‐3).
Contributor Information
Miguel A. Matilla, Email: miguel.matilla@eez.csic.es
George P. C. Salmond, Email: gpcs2@cam.ac.uk
References
- Alström, S. (2001) Characteristics of bacteria from oilseed rape in relation to their biocontrol activity against Verticillium dahliae . J Phytopathol 149: 57–64. [Google Scholar]
- Åström, B. , and Gerhardson, B. (1988). Differential reactions of wheat and pea genotypes to root inoculation with growth‐affecting rhizosphere bacteria. Plant Soil 109: 263–269. [Google Scholar]
- Badri, D.V. , and Vivanco, J.M. (2009) Regulation and function of root exudates. Plant Cell Environ 32: 666–681. [DOI] [PubMed] [Google Scholar]
- Berg, G. , Roskot, N. , Steidle, A. , Eberl, L. , Zock, A. , and Smalla, K. (2002) Plant‐dependent genotypic and phenotypic diversity of antagonistic rhizobacteria isolated from different Verticillium host plants. Appl Environ Microbiol 68: 3328–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair, J.M. , Webber, M.A. , Baylay, A.J. , Ogbolu, D.O. , and Piddock, L.J. (2015) Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol 13: 42–51. [DOI] [PubMed] [Google Scholar]
- Burr, T. , Barnard, A.M. , Corbett, M.J. , Pemberton, C.L. , Simpson, N.J. , and Salmond, G.P.C. (2006) Identification of the central quorum sensing regulator of virulence in the enteric phytopathogen, Erwinia carotovora: the VirR repressor. Mol Microbiol 59: 113–125. [DOI] [PubMed] [Google Scholar]
- Chen, X.H. , Vater, J. , Piel, J. , Franke, P. , Scholz, R. , Schneider, K. , et al (2006) Structural and functional characterization of three polyketide synthase gene clusters in Bacillus amyloliquefaciens FZB42. J Bacteriol 188: 4024–4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury, S.P. , Hartmann, A. , Gao, X. , and Borriss, R. (2015) Biocontrol mechanism by root‐associated Bacillus amyloliquefaciens FZB42 – a review. Front Microbiol 6: 780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulthurst, S.J. , Barnard, A.M. , and Salmond, G.P.C. (2005) Regulation and biosynthesis of carbapenem antibiotics in bacteria. Nat Rev Microbiol 3: 295–306. [DOI] [PubMed] [Google Scholar]
- Delcher, A.L. , Harmon, D. , Kasif, S. , White, O. , and Salzberg, S.L. (1999) Improved microbial gene identification with GLIMMER. Nucl. Acids Res 27: 4636–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarre, G. , Guérout, A.M. , Matsumoto‐Mashimo, C. , Rowe‐Magnus, D.A. , Marlière, P. , and Mazel, D. (2005) A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPalpha) conjugative machineries and their cognate Escherichia coli host strains. Res Microbiol 156: 245–255. [DOI] [PubMed] [Google Scholar]
- Devesse, L. , Smirnova, I. , Lönneborg, R. , Kapp, U. , Brzezinski, P. , Leonard, G.A. , and Dian, C. (2011) Crystal structures of DntR inducer binding domains in complex with salicylate offer insights into the activation of LysR‐type transcriptional regulators. Mol Microbiol 81: 354–367. [DOI] [PubMed] [Google Scholar]
- De Vleesschauwer, D. , and Hofte, M. (2007) Using Serratia plymuthica to control fungal pathogens of plants. CAB Rev Perspect Agric Vet Sci Nutr Nat Resour 2: 1–12. [Google Scholar]
- Duan, Y. , Zhou, M. , Guo, S. , Ding, L. , Gao, K. , and Liu, X. (2012) Cloning and preliminary identification of SptR, a LuxR‐like regulator from Serratia plymuthica . Afr J Microbiol Res 6: 6333–6339. [Google Scholar]
- Ezezika, O.C. , Haddad, S. , Clark, T.J. , Neidle, E.L. , and Momany, C. (2007) Distinct effector‐binding sites enable synergistic transcriptional activation by BenM, a LysR‐type regulator. J Mol Biol 367: 616–629. [DOI] [PubMed] [Google Scholar]
- Fischbach, M.A. , and Walsh, C.T. (2009) Antibiotics for emerging pathogens. Science 325: 1089–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer, K.A. , Pachter, L. , Poliakov, A. , Rubin, E.M. , and Dubchak, I. (2004) VISTA. Computational tools for comparative genomics. Nucl Acids Res 32: W273–W279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredenhagen, A. , Tamure, S.Y. , Kenny, P.T.M. , Komura, H. , Naya, Y. , and Nakanishi, K. (1987) Andrimid, a new peptide antibiotic produced by an intracellular bacterial symbiont isolated from a brown planthopper. J Am Chem Soc 109: 4409–4411. [Google Scholar]
- Freiberg, C. , Brunner, N.A. , Schiffer, G. , Lampe, T. , Pohlmann, J. , Brands, M. , et al (2004) Identification and characterization of the first class of potent bacterial acetyl‐CoA carboxylase inhibitors with antibacterial activity. J Biol Chem 279: 26066–26073. [DOI] [PubMed] [Google Scholar]
- Fortin, P.D. , Walsh, C.T. , and Magarvey, N.A. (2007) A transglutaminase homologue as a condensation catalyst in antibiotic assembly lines. Nature 448: 824–827. [DOI] [PubMed] [Google Scholar]
- Graff, J.R. , Forschner‐Dancause, S.R. , Menden‐Deuer, S. , Long, R.A. , and Rowley, D.C. (2013) Vibrio cholerae exploits sub‐lethal concentrations of a competitor‐produced antibiotic to avoid toxic interactions. Front Microbiol 4: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney, R. , and Thomas, C.M. (2011) Mupirocin: biosynthesis, special features and applications of an antibiotic from a gram‐negative bacterium. Appl Microbiol Biotechnol 90: 11–21. [DOI] [PubMed] [Google Scholar]
- Haas, D. , and Défago, G. (2005) Biological control of soil‐borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol 3: 307–319. [DOI] [PubMed] [Google Scholar]
- Harwood, J.L. (2007) Lipid metabolism In The Lipid Handbook, 3rd ed Harwood J.L., Gunstone F.D., and Dijkstra A.J. (eds.). New York: CRC Press, pp. 637–702. [Google Scholar]
- Hellberg, J.E. , Matilla, M.A. , and Salmond, G.P. (2015) The broad‐spectrum antibiotic, zeamine, kills the nematode worm Caenorhabditis elegans . Front Microbiol 6: 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hérault, E. , Reverchon, S. , and Nasser, W. (2014) Role of the LysR‐type transcriptional regulator PecT and DNA supercoiling in the thermoregulation of pel genes, the major virulence factors in Dickeya dadantii . Environ Microbiol 16: 734–745. [DOI] [PubMed] [Google Scholar]
- Herrero, M. , de Lorenzo, V. , and Timmis, K.N. (1990) Transposon vectors containing nonantibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in Gram‐negative bacteria. J Bacteriol 172: 6557–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertweck, C. (2009) The biosynthetic logic of polyketide diversity. Angew Chem Int Ed Engl 48: 4688–4716. [DOI] [PubMed] [Google Scholar]
- Hökeberg, M. , Gerhardson, B. , and Johnsson, L. (1997) Biological control of cereal seed‐borne diseases by seed bacterization with greenhouse‐selected bacteria. Eur J Plant Pathol 103: 25–33. [Google Scholar]
- Jeong, H. , Kloepper, J.W. , and Ryu, C.M. (2015) Genome sequence of rhizobacterium Serratia marcescens strain 90‐166, which triggers induced systemic resistance and plant growth promotion. Genome Announc 3: e00667‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, M. , Fischbach, M.A. , and Clardy, J. (2006) A biosynthetic gene cluster for the acetyl‐CoA carboxylase inhibitor andrimid. J Am Chem Soc 128: 10660–10661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai, K. , Ohnishi, H. , Mori, Y. , Kiba, A. , Ohnishi, K. , and Hikichi, Y. (2014) Involvement of ralfuranone production in the virulence of Ralstonia solanacearum OE1‐1. ChemBioChem 15: 2590–2597. [DOI] [PubMed] [Google Scholar]
- Kaniga, K. , Delor, I. , and Cornelis, G.R. (1991) A wide‐host range suicide vector for improving reverse genetics in Gram‐negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica . Gene 109: 137–141. [DOI] [PubMed] [Google Scholar]
- Klaponski, N. , Selin, C. , Duke, K. , Spicer, V. , Fernando, D.W. , Belmonte, M.F. , and de Kievit, T.R. (2014) The requirement for the LysR‐type regulator PtrA for Pseudomonas chlororaphis PA23 biocontrol revealed through proteomic and phenotypic analysis. BMC Microbiol 14: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, K. (2013) Platforms for antibiotic discovery. Nat Rev Drug Discov 12: 371–387. [DOI] [PubMed] [Google Scholar]
- Li, S. , Huang, X. , Wang, G. , and Xu, Y. (2012) Transcriptional activation of pyoluteorin operon mediated by the LysR‐type regulator PltR bound at a 22 bp lys box in Pseudomonas aeruginosa M18. PLoS One 7: e39538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, G. , Chater, K.F. , Chandra, G. , Niu, G. , and Tan, H. (2013) Molecular regulation of antibiotic biosynthesis in Streptomyces . Microbiol Mol Biol Rev 77: 112–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Fortin, P.D. , and Walsh, C.T. (2008) Andrimid producers encode an acetyl‐CoA carboxyltransferase subunit resistant to the action of the antibiotic. Proc Natl Acad Sci U S A 105: 13321–13326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Jia, J. , Popat, R. , Ortori, C.A. , Li, J. , Diggle, S.P. , Gao, K. , and Cámara, M. (2011) Characterisation of two quorum sensing systems in the endophytic Serratia plymuthica strain G3: differential control of motility and biofilm formation according to life‐style. BMC Microbiol 11: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, R.A. , Rowley, D.C. , Zamora, E. , Liu, J. , Bartlett, D.H. , and Azam, F. (2005) Antagonistic interactions among marine bacteria impede the proliferation of Vibrio cholerae . Appl Environ Microbiol 71: 8531–8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Y. , He, J. , Zhu, H. , Yu, Z. , Wang, R. , Chen, Y. , et al (2011) An orphan histidine kinase, OhkA, regulates both secondary metabolism and morphological differentiation in Streptomyces coelicolor . J Bacteriol 193: 3020–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg, B. , and Kamilova, F. (2009) Plant‐growth‐promoting rhizobacteria. Annu Rev Microbiol 63: 541–556. [DOI] [PubMed] [Google Scholar]
- Machado, H. , Sonnenschein, E.C. , Melchiorsen, J. , and Gram, L. (2015) Genome mining reveals unlocked bioactive potential of marine Gram‐negative bacteria. BMC Genomics 16: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks, S.E. , and Oyston, P.C. (2008) Structure and function of the LysR‐type transcriptional regulator (LTTR) family proteins. Microbiology 154: 3609–3623. [DOI] [PubMed] [Google Scholar]
- Magarvey, N.A. , Fortin, P.D. , Thomas, P.M. , Kelleher, N.L. , and Walsh, C.T. (2008) Gatekeeping versus promiscuity in the early stages of the andrimid biosynthetic assembly line. ACS Chem Biol 3: 542–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, X.M. , Sun, Z.H. , Liang, B.R. , Wang, Z.B. , Feng, W.H. , Huang, F.L. , and Li, Y.Q. (2013) Positive feedback regulation of stgR expression for secondary metabolism in Streptomyces coelicolor . J Bacteriol 195: 2072–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla, M.A. , Stöckmann, H. , Leeper, F.J. , and Salmond, G.P.C. (2012) Bacterial biosynthetic gene clusters encoding the anti‐cancer haterumalide class of molecules: biogenesis of the broad spectrum antifungal and antioomycete compound, oocydin A. J Biol Chem 287: 39125–39138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla, M.A. , and Salmond, G.P.C. (2014) The Viunalikevirus, bacteriophage φMAM1, is a broad host range, high efficiency generalised transducing phage that infects environmental and clinical isolates of the enterobacteria, Serratia and Kluyvera . Appl Environ Microbiol 80: 6446–6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla, M.A. , Leeper, F.J. , and Salmond, G.P. (2015) Biosynthesis of the antifungal haterumalide, oocydin A, in Serratia, and its regulation by quorum sensing, RpoS and Hfq. Environ Microbiol 17: 2993–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla, M.A. , Drew, A. , Udaondo, Z. , Krell, T. , and Salmond, G.P. (2016) Genome sequence of Serratia plymuthica A153, a model rhizobacterium for the investigation of the synthesis and regulation of haterumalides, zeamine and andrimid. Genome Announcements. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes, R. , Garbeva, P. , and Raaijmakers, J.M. (2013) The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev 37: 634–663. [DOI] [PubMed] [Google Scholar]
- Monferrer, D. , Tralau, T. , Kertesz, M.A. , Dix, I. , Solà, M. , and Usón, I. (2010) Structural studies on the full‐length LysR‐type regulator TsaR from Comamonas testosteroni T‐2 reveal a novel open conformation of the tetrameric LTTR fold. Mol Microbiol 75: 1199–1214. [DOI] [PubMed] [Google Scholar]
- Mousa, W.K. , and Raizada, M.N. (2015) Biodiversity of genes encoding anti‐microbial traits within plant associated microbes. Front Plant Sci 6: 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, H. , Westendorf, C. , Leitner, E. , Chernin, L. , Riedel, K. , Schmidt, S. , Eberl, L. , and Berg, G. (2009) Quorum‐sensing effects in the antagonistic rhizosphere bacterium Serratia plymuthica HRO‐C48. FEMS Microbiol Ecol 67: 468–78. [DOI] [PubMed] [Google Scholar]
- Needham, J. , Kelly, M.Y. , Ishige, M. , and Andersen, R.J. (1994) Andrimid and moiramides A‐C, metabolites produced in culture by a marine isolate of the bacterium Pseudomonas Fluorescens – structure elucidation and biosynthesis. J Org Chem 59: 2058–2063. [Google Scholar]
- Nett, M. , Ikeda, H. , and Moore, B.S. (2009) Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep 26: 1362–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, D.J. , and Cragg, G.M. (2007) Natural products as sources of new drugs over the last 25 years. J Nat Prod 70: 461–477. [DOI] [PubMed] [Google Scholar]
- Oclarit, J.M. , Okada, H. , Ohta, S. , Kaminura, K. , Yamaoka, Y. , Iizuka, T. , Miyashiro, S. , and Ikegami, S. (1994) Anti‐Bacillus substance in the marine sponge, Hyatella species, produced by an associated Vibrio species bacterium. Microbios 78: 7–16. [PubMed] [Google Scholar]
- Pfaffl, M.W. (2001) A new mathematical model for relative quantification in real‐time RT‐PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidot, S.J. , Coyne, S. , Kloss, F. , and Hertweck, C. (2014) Antibiotics from neglected bacterial sources. Int J Med Microbiol 304: 14–22. [DOI] [PubMed] [Google Scholar]
- Pritchard, L. , Humphris, S. , Baeyen, S. , Maes, M. , Van Vaerenbergh, J. , Elphinstone, et al (2013a). Draft genome sequences of four Dickeya dianthicola and four Dickeya solani strains. Genome Announc 1: e00087‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers, J.M. , Paulitz, T.C. , Steinberg, C. , Alabouvette, C. , and Moënne‐Loccoz, Y. (2009) The rhizosphere. A playground for soil‐borne pathogen and beneficial microorganisms. Plant Soil 321: 341–361. [Google Scholar]
- Ramsay, J. (2013) High‐throughput β‐galactosidase and β‐glucuronidase assays using fluorogenic substrates. Bio‐protocol 3: e827. [Google Scholar]
- Ratnayake, N.D. , Wanninayake, U. , Geiger, J.H. , and Walker, K.D. (2011) Stereochemistry and mechanism of a microbial phenylalanine aminomutase. J Am Chem Soc 133: 8531–8533. [DOI] [PubMed] [Google Scholar]
- Romeo, T. , Vakulskas, C.A. , and Babitzke, P. (2013) Post‐transcriptional regulation on a global scale: form and function of Csr/Rsm systems. Environ Microbiol 15: 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch, E.F. , and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd ed New York, NY, USA: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Sánchez, S. , Chávez, A. , Forero, A. , García‐Huante, Y. , Romero, A. , Sánchez, M. , et al (2010) Carbon source regulation of antibiotic production. J Antibiot 63: 442–459. [DOI] [PubMed] [Google Scholar]
- Sánchez, L.A. , Sierra, M.G. , Siñeriz, F. , and Delgado, O. (2013) Andrimid production at low temperature by a psychrotolerant Serratia proteamaculans strain. World J Microbiol Biotechnol 29: 1773–1781. [DOI] [PubMed] [Google Scholar]
- Sattely, E.S. , Fischbach, M.A. , and Walsh, C.T. (2008) Total biosynthesis: in vitro reconstitution of polyketide and nonribosomal peptide pathways. Nat Prod Rep 25: 757–793. [DOI] [PubMed] [Google Scholar]
- Singh, M.P. , Mroczenski‐Wildey, M.J. , Steinberg, D.A. , Andersen, R.J. , Maiese, W.M. , and Greenstein, M. (1997) Biological activity and mechanistic studies of andrimid. J Antibiot 50: 270–273. [PubMed] [Google Scholar]
- Sola‐Landa, A. , Moura, R.S. , and Martín, J.F. (2003) The two‐component PhoR‐PhoP system controls both primary metabolism and secondary metabolite biosynthesis in Streptomyces lividans . Proc Natl Acad Sci U S A 100: 6133–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel, G. , Li, J.Y. , Sugawara, F. , Koshino, H. , Harper, J. , and Hess, W.M. (1999) Oocydin A, a chlorinated macrocyclic lactone with potent anti‐oomycete activity from Serratia marcescens . Microbiology 145: 3557–3564. [DOI] [PubMed] [Google Scholar]
- Suzuki, K , Okazaki, K , Tawaraya, K. , Osaki, M. , and Shinana, T. (2009) Gas chromatography–mass spectrometry associated global analysis of rice root exudates under aseptical conditions. Soil Sci Plant Nutr 55: 505–513. [Google Scholar]
- Thaning, C. , Welch, C.J. , Borowicz, J.J. , Hedman, R. , and Gerhardson, B. (2001) Suppression of Sclerotinia sclerotiorum apothecial formation by the soil bacterium Serratia plymuthica: identification of a chlorinated macrolide as one of the causal agents. Soil Biol Biochem 33: 1817–1826. [Google Scholar]
- Udwary, D.W. , Zeigler, L. , Asolkar, R.N. , Singan, V. , Lapidus, A. , Fenical, et al (2007) Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica . Proc Natl Acad Sci U S A 104: 10376–10381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren, N.C. (2007) Types, amounts, and possible functions of compounds released into rhizosphere by soil‐grown plants In The Rhizosphere: Biochemistry and Organic Substances at the Soil‐Plant Interface. Pinton R., Varanini Z. and Nannipieri P. (eds.). New York: CRC Press, pp. 1–21. [Google Scholar]
- Vogel, J. , and Luisi, B.F. (2011) Hfq and its constellation of RNA. Nat Rev Microbiol 9: 578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Fewer, D.P. , Holm, L. , Rouhiainen, L. , and Sivonen, K. (2014) Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc Natl Acad Sci U S A 111: 9259–9264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wietz, M. , Mansson, M. , Gotfredsen, C.H. , Larsen, T.O. , and Gram, L. (2010) Antibacterial compounds from marine Vibrionaceae isolated on a global expedition. Mar Drugs 8: 2946–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wietz, M. , Månsson, M. , and Gram, L. (2011) Chitin stimulates production of the antibiotic andrimid in a Vibrio coralliilyticus strain. Environ Microbiol Rep 3: 559–564. [DOI] [PubMed] [Google Scholar]
- Wilf, N.M. , and Salmond, G.P.C. (2012) The stationary phase sigma factor, RpoS, regulates the production of a carbapenem antibiotic, a bioactive prodigiosin and virulence in the enterobacterial pathogen Serratia sp. ATCC 39006. Microbiology 158: 648–658. [DOI] [PubMed] [Google Scholar]
- Wilf, N.M. , Williamson, N.R. , Ramsay, J.P. , Poulter, S. , Bandyra, K.J. , and Salmond, G.P.C. (2011) The RNA chaperone, Hfq, controls two luxR‐type regulators and plays a key role in pathogenesis and production of antibiotics in Serratia sp. ATCC 39006. Environ Microbiol 13: 2649–2666. [DOI] [PubMed] [Google Scholar]
- Williamson, N.R. , Fineran, P.C. , Leeper, F.J. , and Salmond, G.P.C. (2006) The biosynthesis and regulation of bacterial prodiginines. Nat Rev Microbiol 4: 887–899. [DOI] [PubMed] [Google Scholar]
- Woodcock, D.M. , Crowther, P.J. , Doherty, J. , Jefferson, S. , DeCruz, E. , Noyer‐ Weidner, M. , et al (1989) Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucl Acids Res 17: 3469–3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodford, N. , Turton, J.F. , and Livermore, D.M. (2011) Multiresistant Gram‐negative bacteria: the role of high‐risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev 35: 736–755. [DOI] [PubMed] [Google Scholar]
- Zhao, C. , Coughlin, J.M. , Ju, J. , Zhu, D. , Wendt‐Pienkowski, E. , Zhou, X. , et al (2010) Oxazolomycin biosynthesis in Streptomyces albus JA3453 featuring an ‘acyltransferase‐less’ type I polyketide synthase that incorporates two distinct extender units. J Biol Chem 285: 20097–20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, M. , Gao, K. , Zeng, J. , Yu, X. , Wu, Y. , Ge, J. , et al (2012) Role of the RNA‐binding protein Hfq in Serratia plymuthica . Front Biosci 4: 1263–1275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's web‐site:
Fig. S1. Structures of andrimid and moiramide B. The structure of andrimid consists of an unsaturated fatty acid chain, a pyrrolidinedione ring, a valine and glycine derived β‐ketoamide and the amino acid β‐phenylalanine. Chemical synthesis studies showed that the fatty acid chain and β‐phenylalanine are involved in bacterial cell penetration of the antibiotic, whereas the pyrrolidinedione head and the β‐ ketoamide moiety are responsible for the antibacterial activity (Pohlmann et al., 2005; Freiberg et al., 2006).
Fig. S2. Antibacterial activity of Serratia marcescens MSU97 against Bacillus subtilis.
Fig. S3. Schematic representation of the andrimid gene clusters of Serratia marcescens MSU97, Serratia marcescens 90‐166, Pantoea agglomerans Eh335, Vibrio coralliilyticus S2052 and Vibrionales bacterium SWAT‐3. Numbers below the arrows represent the intergenic distance between contiguous genes, with negative numbers indicate overlapping genes
Fig. S4. DNA homology between the andrimid gene cluster of Serratia plymuthica A153 and the andrimid gene clusters of the other producing strains. A, Schematic representation of the adm gene cluster in Serratia strains. B‐F, Alignments representing the percentage of DNA homology between the adm gene cluster of A153 and those of S. marcescens MSU97 (B), S. marcescens 90‐166 (C), Pantoea agglomerans Eh335 (D) Vibrio coralliilyticus S2052 (E) and Vibrionales SWAT‐3 (F). Alignments were performed using wgVISTA (Frazer et al., 2004).
Fig. S5. Growth of Serratia plymuthica A153 in minimal medium with different carbon sources. Growth curves showing the doubling time in sorbitol (181.2 ± 1 min), mannitol (153 ± 1 min), fructose (145.8 ± 2 min), galactose (115.2 ± 1 min), mannose (181.6 ± 2 min), lactose (413.4 ± 6 min), xylose (208.2 ± 2 min), succinic acid (142.8 ± 1 min), maltose (145.2 ± 1 min), sucrose (106.2 ± 1 min), glucose (115.2 ± 1 min), glycerol (121 ± 1 min), gluconic acid (96.6 ± 1 min), arabinose (235.2 ± 2 min) and citrate (158.7 ± 3 min) as sole carbon source. Data are the mean and standard deviation of three biological replicates. The assays were done at 25 ºC with shaking at 200 rpm.
Fig S6. Impact of Hfq (A) and AdmX (B) on the expression of admV and admX. Quantitative real‐time PCR was used to measure transcript levels of admV (grey bars) and admX (white bars) in Serratia plymuthica A153, and derivative strains. The values showed the average expression relative to wild type expression. The arrow in Fig. 4A indicates the time point when the samples for qPCR were taken. The data are the mean and standard deviation of three biological replicates.
Fig. S7. AdmX transcription correlates with the expression of the andrimid gene cluster. Transcription of the admX (PadmX::lacZ; pMAMV244) promoter fusion throughout growth in Serratia plymuthica A153 strains. β‐Galactosidase activity (filled symbols) and growth curves (open symbols) were determined in LacZ (red) and ΔadmX (blue) in LB medium at 25 °C. A153 wt harbouring the empty reporter plasmid (black) was used as negative control in the assays. Data are the mean and standard deviation of three biological replicates.
Fig. S8. Genetic complementation of Serratia plymuthica A153 strain A153H. Expression of hfq in trans in A153 Δhfq restored andrimid production and therefore the antibacterial activity against Bacillus subtilis. Induction of Hfq expression was done by addition of 0.1 mM of IPTG. The bioassays were repeated at least three times, and a representative figure is shown. Pictures were taken after 48 h of incubation at 25 °C.
Fig. S9. Sensitivities of different bacterial strains to the antibiotic andrimid. Bioactivities of Serratia plymuthica A153 and the non‐andrimid producing mutant of A153, VN2, against ecologically different bacterial strains. For the assays, an indicator top agar lawn was prepared as described in “Experimental procedures,” and 5 μl overnight cultures of the A153 strains were spotted on the surface of the bacterial indicator agar lawns. The bioassays were repeated three times, and representative results are shown. Pictures were taken after 48 h of incubation at 25 °C. The strains used are described in Table 1 and supplementary Table S2.
Fig. S10. Andrimid shows antibacterial activity against Bacillus subtilis, Dickeya solani and Xanthomonas campestris pv. campestris. Recovery of viable Bacillus, Dickeya and Xanthomonas cells grown in the presence of A153 JH6 (andrimid positive, zeamine negative) and A153 XJH6 (andrimid and zeamine negative) supernatants. The values showed the percentage of viable cells in the presence of JH6 supernatants relative to the number of viable cells in the presence of XJH6 supernatants. For the assays, overnight bacterial cultures of Bacillus, Dickeya and Xanthomonas were adjusted to an optical density at 600 nm (OD600) of 0.1 and grown at 30 ºC with orbital shaking (225 rpm). At an OD600 of 0.4, 10 mL of the bacterial culture was removed and pelleted by centrifugation at 4,000 x g for 10 min at room temperature. The pellet was resuspended in 5 ml of 2X LB and 5 ml supernatants of an overnight culture of A153 JH6 or A153 XJH6 were added to the bacterial culture. Samples were taken after 5 and 10 h of incubation and the number of colony forming units (CFU) were determined. Data are the mean and standard deviation of three biological replicates.
Fig. S11. Homology model of AdmX. The model was generated by the Geno3D modeling algorithm (Combet et al., 2000) and the structure of the BenM transcriptional regulator (PDB ID 3K1N) as template. The site for the binding of potential effector molecules is indicated.
Table S1. Identity at DNA level of the andrimid gene clusters between producing strains.
Table S2. Additional bacterial strains used in this study.
Table S3. Oligonucleotides used in this study.
Table S4. Plasmids used in this study.


