Summary
Mycobacterium tuberculosis produces several bacterial effectors impacting the colonization of phagocytes. Here, we report that the putative lipoprotein LppM hinders phagocytosis by macrophages in a toll‐like receptor 2‐dependent manner. Moreover, recombinant LppM is able to functionally complement the phenotype of the mutant, when exogenously added during macrophage infection. LppM is also implicated in the phagosomal maturation, as a lppM deletion mutant is more easily addressed towards the acidified compartments of the macrophage than its isogenic parental strain. In addition, this mutant was affected in its ability to induce the secretion of pro‐inflammatory chemokines, interferon‐gamma‐inducible protein‐10, monocyte chemoattractant protein‐1 and macrophage inflammatory protein‐1α. Thus, our results describe a new mycobacterial protein involved in the early trafficking of the tubercle bacillus and its manipulation of the host immune response.
Introduction
Tuberculosis (TB) remains a leading infectious disease around the world with 1.5 million deaths and 9.6 million new cases per year. Approximately one‐third of the world population is considered to have latent TB, and although they show no clinical symptoms, they are at risk to progress to active disease (Barry et al., 2009; Young et al., 2009). The treatment of active TB requires chemotherapy to be taken for at least 6 months, which often leads to a poor compliance because of the toxicity of the treatment and thereby to the emergence of multi‐drug‐resistant or extremely‐drug‐resistant strains of Mycobacterium tuberculosis. The emergence of totally drug‐resistant strains of M. tuberculosis has become a major global threat (Klopper et al., 2013), urging the need to better understand TB pathogenesis in order to develop new and more potent drugs. TB pathogenesis is linked to a tight interplay between the host immune response and M. tuberculosis, which produces factors that contribute to its successful colonization and its persistence within host cells. Tubercle bacilli are able to infect macrophages and to actively replicate within their phagosomes. After interaction with several cell‐host receptors and subsequent phagocytosis, the bacilli are able to block phagosomal maturation, to escape from the phagosome and to subvert the host immune response (for reviews (Killick et al., 2013; Stanley and Cox, 2013)).
The cell colonization and intracellular trafficking of M. tuberculosis involve several distinct bacterial effectors, and deciphering the contribution of each of the different actors at the host‐pathogen interface is essential to understand the intracellular persistence of M. tuberculosis. The bacterial virulence factors that have been extensively studied to impact on the invasion process are the ESX type VII secretion system, the serine protein kinase PknG, the lipid phosphatase SapM and components of the mycobacterial cell wall (Walburger et al., 2004; Vergne et al., 2005; Brodin et al., 2006; Cambier et al., 2014). Using a non‐biased global screening approach on an 11 000 M. tuberculosis transposon mutant library, we identified additional mycobacterial genes whose expression impairment impacts the intracellular trafficking (Brodin et al., 2010). Among these genes, rv2171 (lppM) was shown to affect colocalization of M. tuberculosis Beijing GC1237 with LysoTracker, a marker of the acidic lysosomal compartment, suggesting that LppM is involved in preventing early phagosomal maturation in infected macrophages (Brodin et al., 2010). Interestingly, lppM expression has been shown to be induced during macrophage infection (Dubnau et al., 2002), and it was recently selected by bioinformatic analysis as a potential M. tuberculosis effector involved in host–pathogen interaction (Li et al., 2015).
In this study, we demonstrate that LppM limits the early step of M. tuberculosis entry into the macrophage. This phenotype can be reversed by the exogenous addition of recombinant LppM and is dependent on toll‐like receptor 2 (TLR2). We further characterized the involvement of lppM in M. tuberculosis macrophage colonization by showing its implication in the blocking of the phagosomal maturation using different cellular markers: LysoTracker, lysosomal‐associated membrane protein‐1 (LAMP‐1) and cathepsin D. Furthermore, we show that the lppM deletion mutant is affected in its ability to induce the secretion of the pro‐inflammatory chemokines interferon‐gamma‐inducible protein‐10 (IP‐10) (CXCL10), monocyte chemoattractant protein‐1 (MCP‐1) (CCL2) and macrophage inflammatory protein‐1α (MIP‐1α) (CCL3). Altogether, these data suggest that LppM is an additional bacterial effector implicated in the early steps of M. tuberculosis infection.
Results
LppM hinders mycobacterial uptake into macrophages
To investigate the contribution of LppM in the colonization of macrophages, a lppM deletion mutant (MtbH37RvΔlppM) was constructed in M. tuberculosis H37Rv by substituting the lppM gene with a cassette coding for hygromycin resistance (Fig. S1A). Allelic exchange was confirmed by polymerase chain reaction (PCR) with the amplification of a 2.9 kb fragment (Fig. S1B) and checked by sequencing to confirm the insertion of the hygromycin‐resistance cassette at the expected locus (data not shown). In axenic cultures of M. tuberculosis H37Rv, LppM is constitutively expressed, and its apparent migration weight around 28 kDa after SDS‐PAGE is above its predicted molecular weight (Fig. S1C). As expected, the production of LppM was totally abolished in M. tuberculosis H37RvΔlppM (Fig. S1C). The deletion mutant was complemented with a copy of lppM under the control of its own promoter (promlppM::lppM) or the strong hsp60 promoter (promhsp60::lppM) in the integrative vector pMV306 (Fig. S1D).
DsRed‐expressing M. tuberculosis H37Rv and M. tuberculosis H37RvΔlppM were used to infect murine bone marrow‐derived macrophages (BMDM) at multiplicities of infection (MOI) of 5 and 10, and uptake of fluorescent mycobacteria was determined 2 h post‐infection. The percentage of BMDM containing bacteria was largely increased for M. tuberculosis H37RvΔlppM in comparison with M. tuberculosis H37Rv (Fig. 1A). As the use of a third antibiotic marker was detrimental to the normal growth of M. tuberculosis H37RvΔlppM, preventing complementation by an additional plasmid, we checked complementation by covalently staining the bacteria with the red fluorescent dye CypHer5E. Although labelling and detection were not as strong as DsRed‐labelled bacteria, complementation of CypHer5E‐labelled M. tuberculosis H37RvΔlppM with promlppM::lppM vector restored the infectivity to levels similar to those of M. tuberculosis H37Rv (Fig. 1B). To discard any impact of clumping bacteria, we assessed the median of the bacterial area in infected BMDM. In these conditions, there was no difference between M. tuberculosis H37Rv and M. tuberculosis H37RvΔlppM, indicating that the deletion of lppM did not increase clumping of the culture (Fig. S2A). On the contrary, heat‐killed M. tuberculosis H37Rv and M. tuberculosis H37RvΔlppM displayed a significantly higher bacterial area reflecting their tendency to form bigger aggregates (Fig. S2A).
Figure 1.
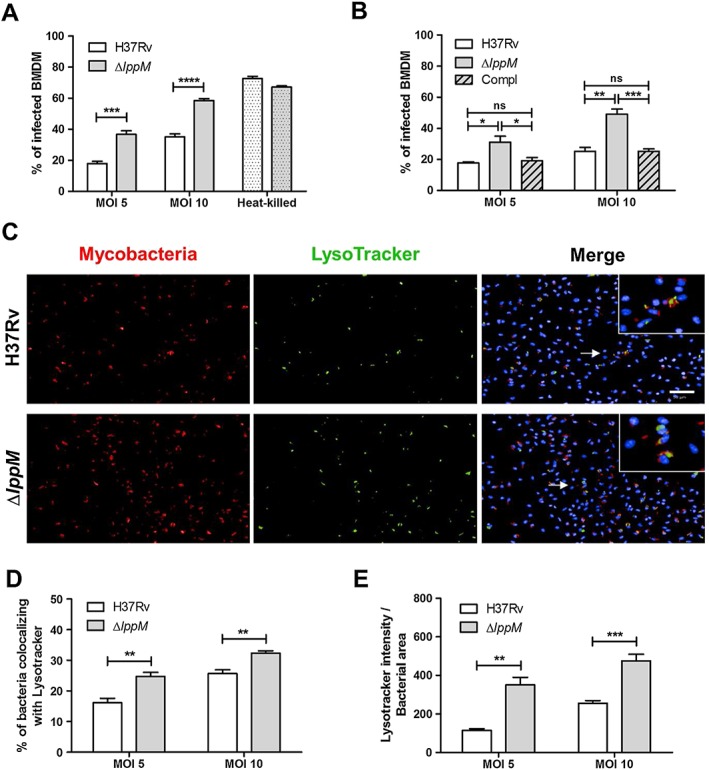
Mycobacterium tuberculosis H37RvΔlppM is more prone to phagocytosis by macrophages than M. tuberculosis H37Rv and is impaired in blocking phagosomal maturation.
A. Percentage of infected BMDM 2 h post‐infection, using M. tuberculosis H37Rv or M. tuberculosis H37RvΔlppM producing fluorescent DsRed, or their respective heat‐killed bacteria [multiplicities of infection (MOI) 10] as a control.
B. Percentage of infected bone marrow‐derived macrophages (BMDM) 2 h post‐infection, using M. tuberculosis H37Rv, M. tuberculosis H37RvΔlppM or the complemented strain (promlppM::lppM) labelled with CypHer5E.
C. DsRed‐producing bacteria (red), LysoTracker staining (green) and Hoechst staining (blue) of BMDM infected with M. tuberculosis H37Rv or M. tuberculosis H37RvΔlppM. The bar corresponds to 50 µm. Zoomed‐in inset (right panel) corresponds to the area indicated by the white arrow.
D. Percentage of M. tuberculosis H37Rv or M. tuberculosis H37RvΔlppM colocalizing with LysoTracker staining in BMDM.
E. Standardization of LysoTracker intensity by bacterial area among the population of M. tuberculosis H37Rv‐infected or M. tuberculosis H37RvΔlppM‐infected BMDM. All graphs are representative of at least three independent experiments.
To obtain insight into the fate of M. tuberculosis H37RvΔlppM after macrophage uptake, we assessed the bacterial area at different time points (from Days 0 to 4) after BMDM infection, using a lower MOI of 2. As M. tuberculosis H37RvΔlppM uptake was higher in comparison with M. tuberculosis H37Rv, each bacterial area was standardized by the corresponding bacterial area obtained at Day 0. In these conditions, M. tuberculosis H37RvΔlppM growth in the macrophage was slower than that of M. tuberculosis H37Rv during the first 3 days of infection. However, M. tuberculosis H37RvΔlppM growth was identical to that of the wild‐type at Day 4 (Fig. S2B). These data illustrate the importance of LppM for the early steps of macrophage colonization and less so at later time points.
LppM hinders phagosomal maturation
To determine whether M. tuberculosis H37RvΔlppM is affected in a subsequent early trafficking process, we investigated the effect of the lppM deletion on phagosome maturation. DsRed‐labelled M. tuberculosis H37Rv‐infected or M. tuberculosis H37RvΔlppM‐infected BMDM were stained with LysoTracker, a fluorescent marker of acidified cellular compartments and analysed by confocal imaging using a dedicated multiparameter automated image analysis system (Brodin et al., 2010). This analysis showed that the percentage of bacteria colocalizing with LysoTracker was higher for M. tuberculosis H37RvΔlppM than for M. tuberculosis H37Rv, indicating increased targeting of the mutant to the acidified compartment (Fig. 1C and D). The percentage of LysoTracker‐positive cells was higher for M. tuberculosis H37RvΔlppM than for M. tuberculosis H37Rv (Fig. S2C), in agreement with our previous observation on a ΔlppM transposon mutant in the M. tuberculosis Beijing GC1237 background (Brodin et al., 2010). Complementation by lppM under the dependence of its own promoter restored the wild‐type phenotype (Fig. S2C). However, as CypHer5E‐staining is pH sensitive, we used DsRed‐expressing bacteria to standardize the LysoTracker intensity to quantify the bacteria (bacterial area) present in the macrophages. This ratio highlights the increase of Lysotracker intensity in the case of M. tuberculosis H37RvΔlppM compared with M. tuberculosis H37Rv, regardless of the number of intracellular bacteria, which reflects an enhanced acidification of the vacuoles containing M. tuberculosis H37RvΔlppM (Fig. 1E). These results indicate that lppM partly contributes to the blocking of the phagosomal maturation by the bacilli.
This was further characterized by using other cellular markers: the LAMP‐1 and the endosomal protease, cathepsin D. Similarly to LysoTracker staining, LAMP‐1 and cathepsin D colocalized at a significantly higher extent with M. tuberculosis H37RvΔlppM compared with M. tuberculosis H37Rv (Fig. 2A–D), confirming the role of LppM in the early steps of macrophage infection.
Figure 2.
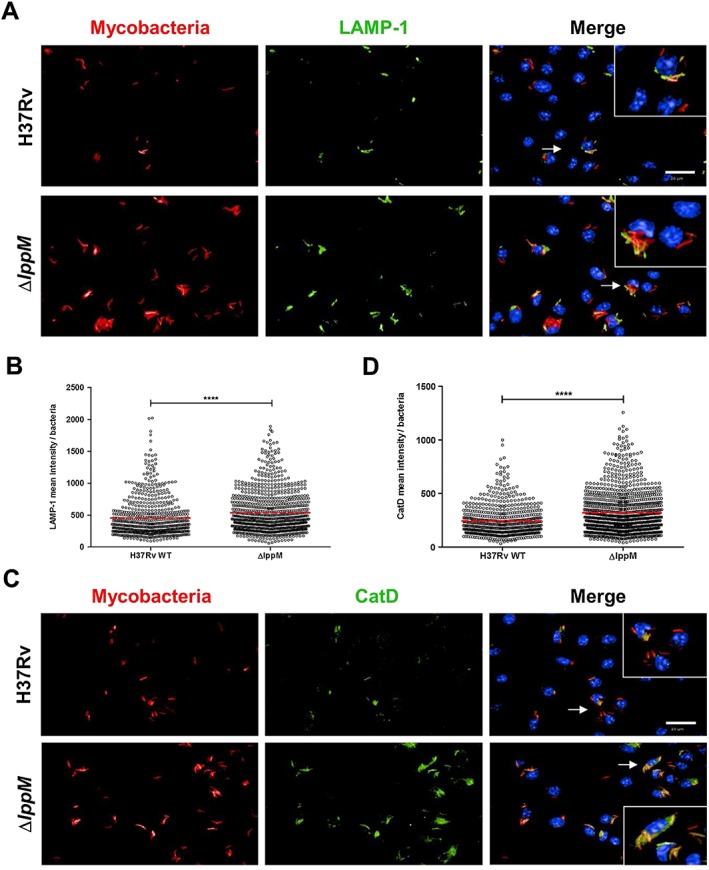
Mycobacterium tuberculosis H37RvΔlppM is more easily addressed towards the degradative compartments of macrophages than M. tuberculosis H37Rv.
A. DsRed‐producing bacteria (red), lysosomal‐associated membrane protein‐1 (LAMP‐1) staining (green) and Hoechst staining (blue) of bone marrow‐derived macrophages (BMDM) infected with M. tuberculosis H37Rv or M. tuberculosis H37RvΔlppM. The bar corresponds to 20 µm. Zoomed‐in inset (right panel) corresponds to the area indicated by the white arrow.
B. LAMP‐1 mean intensity per bacteria after infection of BMDM with M. tuberculosis H37Rv or M. tuberculosis H37RvΔlppM.
C. DsRed‐producing bacteria (red), Cathepsin D (CatD) staining (green) and Hoechst staining (blue) of BMDM infected with M. tuberculosis H37Rv or M. tuberculosis H37RvΔlppM. The bar corresponds to 20 µm. Zoomed‐in inset (right panel) corresponds to the area indicated by the white arrow.
D. Cathepsin D mean intensity per bacteria after infection of BMDM with M. tuberculosis H37Rv or M. tuberculosis H37RvΔlppM.
Recombinant 6His‐LppM26–185 limits phagocytosis
Recombinant soluble 6His‐LppM26–185, starting right after the residue Cys25 and devoid of the putative C‐terminal transmembrane domain (Barthe et al., unpublished), was produced in Escherichia coli (Fig. 3A). When 6His‐LppM26–185 was added to the medium of M. tuberculosis H37RvΔlppM, the phagocytosis level of the mutant strain was reduced to that of M. tuberculosis H37Rv (Fig. 3B). This complementation of the uptake defect did not depend on LppM acylation, as Cys25 was not present in 6His‐LppM26–185, inferring that LppM action is mediated by another structural element. However, the addition of 6His‐LppM26–185 did not complement the M. tuberculosis H37RvΔlppM phenotype regarding the bacterial colocalization with acidified compartments (Fig. 3C) or LysoTracker intensity (Fig. 3D).
Figure 3.
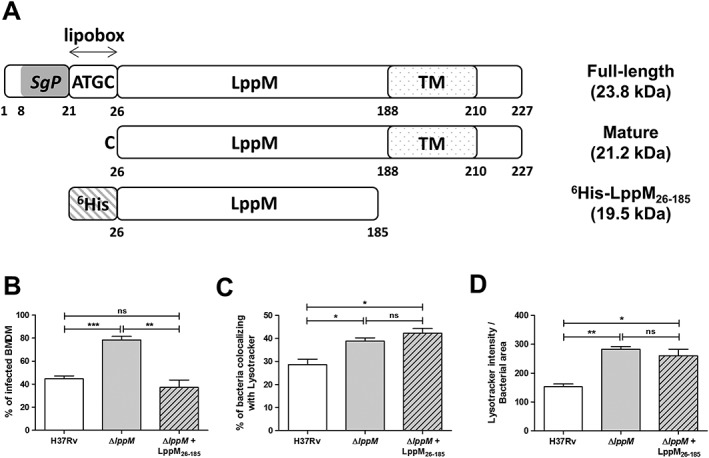
LppM impacts phagocytosis by bone marrow‐derived macrophages (BMDM).
A. Schematic representation of the different forms of LppM with their molecular weight. The “full‐length” LppM corresponds to the entire protein containing a signal peptide (SgP, grey box), a putative lipobox motif (22ATGC25) and a transmembrane sssdomain (TM, dotted box). The mature LppM corresponds to the form after cleavage of its signal peptide, starting at Cys25. Soluble domain of LppM corresponds to the N‐terminal 6His‐tagged (stripped box) construction lacking the initial cysteine (Cys25) and its C‐terminal transmembrane domain.
B. Complementation of the Mycobacterium tuberculosis H37RvΔlppM phagocytosis phenotype by adding 10 μM of the recombinant LppM (6His‐LppM26–185) purified from Escherichia coli.
C. Percentage of M. tuberculosis H37Rv and M. tuberculosis H37RvΔlppM, without or with addition of 6His‐LppM26–185, colocalizing with LysoTracker staining in BMDM.
D. Standardization of LysoTracker intensity by bacterial area among the population of M. tuberculosis H37Rv‐infected and M. tuberculosis H37RvΔlppM‐infected BMDM, without or with addition of 6His‐LppM26–185. All graphs are representative of at least two independent experiments.
Phenotype of Mycobacterium tuberculosis H37RvΔlppM is mediated by toll‐like receptor 2‐dependent signalling
Toll‐like receptor 2 operates as a primary sensor of a large variety of mycobacterial components and thereby triggers innate immune responses (Underhill et al., 1999; Harding and Boom, 2010). Therefore, we examined the effect of the lppM deletion on murine TLR2−/− BMDM. Interestingly, no difference was observed between M. tuberculosis H37Rv and M. tuberculosis H37RvΔlppM in bacterial uptake by TLR2−/− BMDM (Fig. 4A). Bacterial colocalization in the acidified compartments of TLR2−/− BMDM was also similar between M. tuberculosis H37Rv and M. tuberculosis H37RvΔlppM (Fig. 4B and C). In addition, there was no significant difference in Lysotracker intensity when TLR2−/− BMDM were infected with M. tuberculosis H37Rv or M. tuberculosis H37RvΔlppM (Fig. 4D). These results indicate that LppM modulates bacterial uptake through TLR2 signalling.
Figure 4.
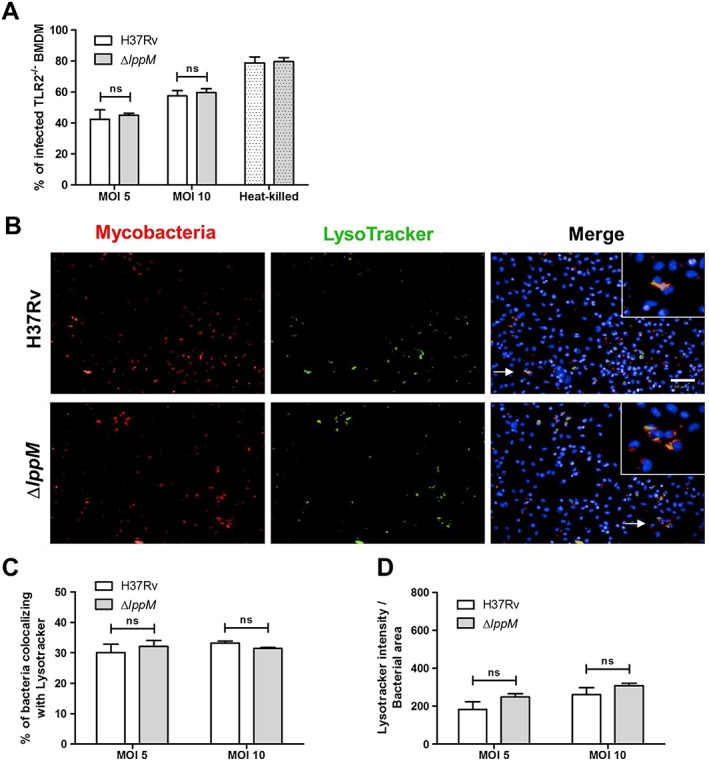
Mycobacterium tuberculosis H37RvΔlppM mutant phenotype depends on toll‐like receptor 2 (TLR2)‐signalling.
A. Percentage of infected TLR2−/− BMDM 2 h post‐infection, using M. tuberculosis H37Rv or M. tuberculosis H37RvΔlppM producing DsRed and respective heat‐killed bacteria [multiplicities of infection (MOI) 10] as a control.
B. DsRed‐producing bacteria (red), LysoTracker staining (green) and Hoechst staining (blue) of TLR2−/− BMDM infected with M. tuberculosis H37Rv or M. tuberculosis H37RvΔlppM. The bar corresponds to 50 µm.
C. Percentage of M. tuberculosis H37Rv or M. tuberculosis H37RvΔlppM colocalizing with LysoTracker staining in TLR2−/− BMDM.
D. Standardization of LysoTracker intensity by bacterial area among the population of M. tuberculosis H37Rv‐infected or M. tuberculosis H37RvΔlppM‐infected TLR2−/− BMDM. All graphs are representative of at least two independent experiments.
Mycobacterium tuberculosis H37RvΔlppM induces reduced secretion of pro‐inflammatory chemokines over Mycobacterium tuberculosis H37Rv
We used the Luminex technology to further characterize the cytokine signatures induced by M. tuberculosis H37RvΔlppM. M. tuberculosis H37RvΔlppM was affected in its ability to induce the secretion of the pro‐inflammatory chemokines IP‐10 (Fig. 5A), MCP‐1 (Fig. 5B) and MIP‐1α (Fig. 5C), and this phenotype was (partially) reversed with the complemented strain. In contrast to the phagocytosis phenotype, the defect in chemokine secretion was not mediated by TLR2, as the profile of chemokines was almost identical between wild‐type and TLR2−/− BMDM infected by M. tuberculosis H37RvΔlppM (Fig. 5A–C). As LppM is implicated in cytokine production in a TLR2‐independent manner, it appears that LppM displays its action through a mechanism different from that of other mycobacterial lipoproteins.
Figure 5.
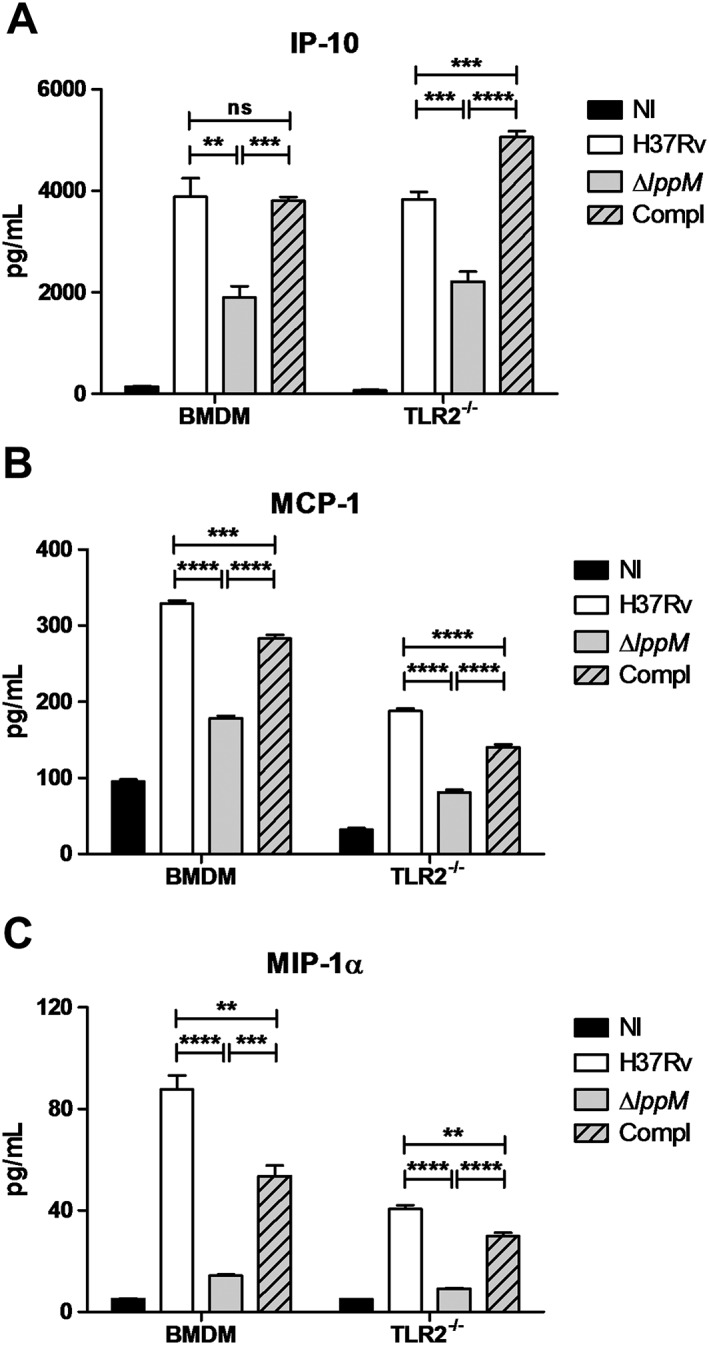
Mycobacterium tuberculosis H37RvΔlppM fails to fully induce the secretion of the pro‐inflammatory chemokines. Secretion of (A) interferon‐gamma‐inducible protein‐10 (IP‐10), (B) monocyte chemoattractant protein‐1 (MCP‐1) and (C) macrophage inflammatory protein‐1α (MIP‐1α) in supernatant of non‐infected (NI) and infected wild‐type or toll‐like receptor 2 (TLR2)−/− bone marrow‐derived macrophages (BMDM) with M. tuberculosis H37Rv, M. tuberculosis H37RvΔlppM or complemented strain (Compl).
Discussion
In this study, we show that LppM limits M. tuberculosis uptake by macrophages and phagosomal maturation. 6His‐LppM26–185 is able to complement the phenotype of M. tuberculosis H37RvΔlppM with respect of phagocytosis, arguing for a direct involvement of LppM in macrophage uptake, regardless of LppM acylation. However, the addition of 6His‐LppM26–185 to M. tuberculosis H37RvΔlppM did not complement the phagosomal maturation phenotype, suggesting that the exogenous protein is not phagocytosed to the same extend as the live bacilli that is unstable once internalized by macrophage or that additional partners of LppM are required for its intracellular complementation. It has been previously reported that LprG, another mycobacterial lipoprotein, is able to control phagolysosomal fusion and that its action is mediated by the binding of LipoArabinoMannan (LAM) in its hydrophobic pocket (Gaur et al., 2014; Shukla et al., 2014), a lipoglycan known to be involved in this process. Therefore, it is tempting to hypothesize that the presence of ligands is required to fully complement the intracellular phenotype of M. tuberculosis H37RvΔlppM, similarly to LprG with LAM. Consistent with this contention, we found that 6His‐LppM26–185 produced by Mycobacterium smegmatis is able to bind PhosphatidylInositol Mannosides (PIM) derivatives, such as PI, PIM2Ac2 and PIM6Ac2 (Barthe et al., unpublished), suggesting a role of these PIM derivatives in the LppM‐mediated phagosomal maturation defect. Therefore, a defect in PIM presentation by M. tuberculosis H37RvΔlppM could be directly responsible for the increased phagocytosis phenotype. Alternatively, the absence of LppM may have perturbed the general architecture of the mycobacterial cell wall and may have favoured the recognition of some other mycobacterial ligands by macrophages.
Several mycobacterial lipoproteins have been shown to be TLR2 agonists (Brightbill et al., 1999; Gehring et al., 2004; Pecora et al., 2006; Drage et al., 2009; Drage et al., 2010; Lancioni et al., 2011). Here, we show that LppM inhibits macrophage uptake of TB bacilli through a TLR2‐dependent signalling, as M. tuberculosis H37Rv and M. tuberculosis H37RvΔlppM show similar colocalization with acidified compartments within TLR2−/− BMDM. There is previous evidence that phagosomal maturation may be controlled by TLR‐dependent signals, as E. coli‐containing or Staphylococcus aureus‐containing phagosomes can fuse with lysosomes in wild‐type macrophages, whereas the phagosomes of MyD88−/− or TLR2x4−/− macrophages cannot (Blander and Medzhitov, 2004). TLR2 has also been shown to be expressed on the surface of phagosomes (Underhill et al., 1999). Therefore, LppM would be able to trigger a TLR2 signalling while the bacilli reside in the extracellular environment and during its prolonged stay in the phagosomal compartment.
Mycobacterium tuberculosis H37RvΔlppM induces lower levels of pro‐inflammatory chemokines IP‐10, MCP‐1 and MIP‐1α than M. tuberculosis H37Rv, reflecting a potential attenuation of the mutant on the very first hours of the infection. Indeed, IP‐10 is expressed in very high amounts in patients with active TB and is one of the most promising alternative markers to interferon‐gamma to detect patients with active TB (Ruhwald et al., 2007). Furthermore, high levels of MCP‐1 are correlated with the disease severity in TB patients (Hasan et al., 2009). Moreover, infection of macrophages results in the induction of MIP‐1α, which is required for the inhibition of M. tuberculosis growth (Saukkonen et al., 2002). Interestingly, it was shown that non‐capped lipoarabinomannan plays a direct role in inflammation by inducing the expression of MCP‐1 and MIP‐1α (Moller et al., 2003). This could be in direct correlation with the binding of PIM derivatives by LppM in M. smegmatis mc2155 (Barthe et al., unpublished), considering that PIM are precursors of LAM. Consequently, for M. tuberculosis H37RvΔlppM, a defect in the presentation of PIM derivatives by LppM or in the recognition of other mycobacterial ligands may potentially drive a decreased expression of both MCP‐1 and MIP‐1α.
It appears that mycobacterial lipoproteins display a large panel of cellular functions, ranging from phagosome–lysosome fusion inhibition in the case of LprG to the induction of macrophage apoptosis in the case of LpqH (Sanchez et al., 2012). Here, we propose that LppM is specialized in the initial steps of macrophage infection by M. tuberculosis. Increased macrophage uptake, impairment of the phagosomal maturation blocking and decreased pro‐inflammatory chemokine production induced by M. tuberculosis H37RvΔlppM are the hallmarks of a defect in efficient macrophage colonization, and these features highlight the importance of LppM for the initial steps of M. tuberculosis infection.
Experimental procedures
Bacterial strains
Escherichia coli TOP10 (Invitrogen) and E. coli BL21(DE3) (Stratagene) were used for cloning and expression of recombinant proteins (Table S1). All strains were grown in LB medium (Difco) at 37°C supplemented with ampicillin (100 µg ml−1), hygromycin (100 µg ml−1) or kanamycin (25 µg ml−1), when required. M. tuberculosis H37Rv, M. tuberculosis H37RvΔlppM and complemented strains were grown in Middlebrook 7H9 plus 0.05% Tween80 or on Middlebrook 7H11 agar plates, with OADC enrichment (Difco) supplemented with hygromycin (50 µg ml−1) or kanamycin (25 µg ml−1), when required. (Table S1).
Generation of a lppM deletion mutant in Mycobacterium tuberculosis H37Rv
To generate M. tuberculosis H37RvΔlppM, we followed the protocol previously described by van Kessel et al. (van Kessel et al., 2008). Briefly, upstream and downstream regions of lppM were amplified from M. tuberculosis H37Rv genomic DNA using [Up_lppM_dir and Up_lppM_rev] and [Down_lppM_dir and Down_lppM_rev] as primers (Table S2). Both fragments were inserted at either side of a hygromycin resistance cassette. After linearization, the resulting construction was electroporated in competent M. tuberculosis H37Rv containing pJV53 (Table S1) after acetamide induction. Several clones were selected and tested for the insertion of the hygromycin resistance cassette by PCR using Seq_lppM_dir and Seq_lppM_rev (Fig. S1A and Fig. S1B, Table S2). The resulting PCR fragment was further checked by sequencing to confirm the insertion at the expected M. tuberculosis locus. In addition, M. tuberculosis H37Rv and M. tuberculosis H37RvΔlppM were transformed by pMRF1 (Table S1), a derivative of pMV261 carrying a kanamycin resistance cassette, which allows the expression of the red fluorescent protein DsRed.
Plasmid constructions
Fragments of lppM were amplified by PCR using M. tuberculosis H37Rv genomic DNA as a template and [pET15b_lppM 26‐227_dir and pET15b_lppM 26‐227_rev] or [pET15b_lppM 26‐227_dir and pET15b_lppM 26‐185_rev] (Table S2), containing NdeI and NheI sites, as primers. The resulting fragments were digested with NdeI and NheI and ligated into pETPhos, generating pETPhos::lppM 26‐227 and pETPhos::lppM 26‐185 (Table S1). The same approach was undertaken to insert lppM into pMV306 and pVV16, using the following pairs of primers [pMV306_lppM_dir and pMV306_lppM_rev] and [pVV16_lppM_dir and pVV16_lppM_rev] (Table S2), together with the relevant restriction enzymes, to obtain pMV306::promlppM::lppM and pVV16::lppM (Table S1). Finally, promhsp60::lppM was subcloned from pVV16::lppM into pMV306 using XbaI and ClaI to yield pMV306::promhsp60::lppM (Table S1). All plasmids were checked by sequencing.
Purification of recombinant 6His‐LppM
Escherichia coli BL21(DE3) strains containing pETPhos::lppM 26‐227 or pETPhos::lppM 26‐185 were used to inoculate 1 l of LB medium supplemented with ampicillin (100 µg ml−1) and cultured at 37°C with shaking until the A600 reached ~0.5. Then, 1 mm (final concentration) of isopropyl 1‐thio‐β‐d‐galactopyranoside was added, and growth was continued for 3 h at 37°C. Soluble 6His‐LppM26–227 and 6His‐LppM26–185 were purified using Ni‐NTA agarose beads (Qiagen) as previously described (Veyron‐Churlet et al., 2010).
Generation of anti‐LppM antibodies and Western‐blot analysis
Recombinant 6His‐LppM26–227 purified from E. coli was used to generate specific rabbit antibodies against LppM according to the Speedy protocol (Eurogentec). Proteins were resolved by SDS‐PAGE using 4–15% acrylamide gels (BioRad) and then transferred onto a polyvinylidene difluoride membrane. The membrane was saturated with TBS‐Tween‐5% milk and probed overnight with anti‐LppM antibodies diluted 1:1000 with TBS‐Tween‐3% milk. The membrane was then incubated with anti‐rabbit horseradish peroxidase‐conjugated secondary antibodies (Jackson Immunoresearch) for 1 h at room temperature, followed by detection using Immobilon kit (Millipore). Chemiluminescence was determined using the ImageQuant LAS 4000 (GE Healthcare).
Cell culture
Wild‐type and TLR2−/− bone marrow progenitors were obtained from tibias and femur bones of 7 to 11 week‐old C57BL/6 mice and frozen at −80°C. The BMDM were obtained by seeding 107 frozen bone marrow cells in 75 cm2 flasks in RPMI 1640 Glutamax medium (Gibco) supplemented with 10% heat‐inactivated fetal bovine serum (FBS) (Gibco) and 10% L929‐conditioned medium containing macrophage colony‐stimulating factor. After 7 days of incubation at 37°C in 5% CO2, BMDM were rinsed with Dulbecco's phosphate‐buffered saline (D‐PBS) and harvested with Versene (Gibco) for further use.
Mycobacterial preparation and labelling
Bacteria were harvested, washed three times and resuspended in PBS. The bacteria were allowed to stand for 30 min to allow residual aggregates to settle. The bacterial suspensions were then aliquoted and frozen at −80°C for further use. When needed, M. tuberculosis H37Rv, M. tuberculosis H37RvΔlppM and the complemented strain (promlppM::lppM) were covalently labelled with CypHer5 mono ester dye, which is a red‐excitable cyanine dye derivative, available as an N‐Hydroxysuccinimide ester that can be directly coupled to a broad range of cell surface receptors (Sigma Aldrich, Saint‐Louis, MO). 108 bacteria were diluted in 300 µl of 0.1 mg ml−1 CypHer5E in 0.1 m sodium carbonate buffer (pH 9). The bacterial suspensions were incubated at 30°C under stirring at 600 r.p.m. and washed three times with PBS before use.
Infection for intracellular growth and LysoTracker assays
The bacteria were quantified by measuring the optical density (OD600) and/or DsRed‐fluorescence on a Victor Multilabel Counter (Perkin Elmer). The heat‐killed bacteria were used as a positive control for infection in LysoTracker assays. Twenty microliter of bacterial suspensions and LppM26‐185‐coated beads was seeded in 384‐well plates (Greiner) at MOI ranging from 2 to 10 bacteria per cell. Then, 2.104 cells in a 30 µl volume were seeded per well. The plates were incubated at 37°C in 5% CO2 for 2 h. To follow the mycobacterial replication in the BMDM, the cells were washed and treated for 1 h with 50 ng ml Amikacin (Sigma‐Aldrich) to remove extracellular bacteria. Cells were finally incubated at 37°C with 5% CO2 in complete culture medium containing a non‐cytotoxic concentration of Hoechst (Sigma‐Aldrich), corresponding to 300 ng ml−1, to stain the cell nuclei. Image acquisition was performed daily. To study the phagosomal maturation/acidification, 50 µl of RPMI containing 1 µm of LysoTracker green DND‐26 (Invitrogen) was added per well, and cells were incubated at 37°C with 5% CO2 for 1.5 h. Cell nuclei were stained using 10 µg ml−1 Hoechst for 30 min at 37°C with 5% CO2. Finally, cells were fixed with 10% neutral buffered Formalin solution (Sigma‐Aldrich), containing paraformaldehyde at 4%, for 30 min at room temperature, washed in PBS and kept at 4°C in D‐PBS 1% FBS until image acquisition.
Immunofluorescence assays
Macrophage infection was performed as described earlier. After 2 h of infection, cell monolayers were fixed with Formalin and permeabilized with D‐PBS 0.2% Triton X100 for 5 min at room temperature. Cells were incubated for 30 min with blocking buffer, corresponding to 10% Donkey serum in D‐PBS, prior to overnight incubation at 4°C with mouse anti‐LAMP‐1 [H4A3] (Abcam, Cambridge, UK) or goat anti‐Cathepsin D (C‐20) (Santa Cruz Biotechnology, Texas, USA) antibodies. Then, cells were incubated for 1 h at room temperature with Alexa‐Fluor 488‐conjugated goat anti‐mouse and donkey anti‐goat (Life technologies) secondary antibodies respectively. Cell nuclei were fluorescently labelled using 5 µg ml DAPI (Sigma‐Aldrich) for 10 min. Cell monolayers were washed in PBS and kept in D‐PBS 1% FBS until further use.
Image acquisition by automated confocal microscopy
Images were acquired using an automated fluorescent confocal microscope OPERA (PerkinElmer), with a 20X (Numeric Aperture 0.7) or 63X (NA 1.2) water immersion lens. The microscope was equipped with 405, 488, 561 and 640 nm excitation lasers. The emitted fluorescence was captured using three cameras associated with a set of filters covering a detection wavelength ranging from 450 to 690 nm. Nucleus staining with Hoechst was detected using the 405 nm laser (blue channel) with a 450/50 emission filter. LysoTracker‐positive, LAMP‐1‐positive and cathepsin D‐positive compartments (green channel) and mycobacteria (red channel) were detected on 488 and 561 nm laser with a 540/75 and 600/40 emission filters respectively. CypHer5E dye was detected with laser and filter that match the 644 nm absorbance and 663 nm emission wavelengths respectively.
Image‐based analysis
For bacteria replication and acidic compartment colocalization, images were analysed using a multi‐parameter script developed with Columbus system (version 2.3.1; PerkinElmer). Cells were detected and counted using internal methods provided by the software. The cell number and the bacterial area were determined by well. Intracellular bacterial growth was daily quantified by the ratio of bacterial area per cell divided by the bacterial area at Day 0. Acidic compartments stained by LysoTracker and bacteria were individually localized by a manual threshold method, using non‐infected wells to determine the minimum threshold required to discard any background signals. The bacterial area was then stretched from 2 pixels to define the bacterial ring region. LysoTracker fluorescence area was detected in the red channel by keeping only pixels with a higher intensity than a manually defined threshold. Cells were split into four groups based on the presence (infected) or the absence (non‐infected) of an intracellular bacterial signal and a LysoTracker positive or negative signal. Results are expressed as the average of all images in four different wells in at least three independent experiments. The percentage of infection and bacteria/LysoTracker colocalization was calculated as follows:
Lysosomal‐associated membrane protein‐1‐positive and cathepsin D‐positive compartments were identified according to an arbitrary threshold of fluorescence intensity, determined on non‐infected cells, associated with anti‐LAMP‐1 or cathepsin D immunolabelling. The same thresholds were then applied to infected cells. The LAMP‐1 and cathepsin D mean intensities were measured for each bacterium.
Infection for the determination of the cytokine profile
Murine macrophages were seeded at a density of 3.8 × 105 cells per well in 24‐well plates. After 24 h, at 37°C in 5% CO2, supernatants were removed, and cells were infected with 950 µl of M. tuberculosis H37Rv, M. tuberculosis H37RvΔlppM or complemented strain (promlppM::lppM) suspensions to reach MOI of 1, 2 and 3. The plates were incubated at 37°C in 5% CO2 for 5 or 24 h. The supernatants were then filtered using a 0.22 µm PVDF filter, sampled and stored at −80°C until analysis. Mouse cytokine concentrations were quantified using Cytokine Mouse Magnetic 20‐Plex Panel kit (Life Technologies) and Luminex® platform, accordingly to the manufacturer's protocol.
Statistics
For bacteria replication and acidic compartment colocalization using the LysoTracker probe, the statistical significance (P‐value) was tested with a two‐tailed unpaired t‐test (GraphPad Prism version 5.04), and P‐values ≤ 0.05 were considered to show significant differences. For the comparison of the LAMP‐1 and cathepsin D mean intensities obtained for wild‐type and mutant bacteria, the statistical significant value was obtained using a Wilcoxon unpaired test (or Mann–Whitney U‐test).
Supporting information
Fig. S1. Generation of M. tuberculosis H37RvΔlppM. (A) Genomic organization of the lppM locus in M. tuberculosis H37Rv and schematic representation of the insertion of the hygromycin cassette resistance. Primers 1 and 2 used for PCR are symbolized by arrows. (B) PCR using primers 1 and 2 with genomic DNA extracted from M. tuberculosis H37Rv or M. tuberculosis H37RvΔlppM. (C) Western‐blot analysis on crude lysates of M. tuberculosis H37Rv and M. tuberculosis H37RvΔlppM cultures using polyclonal antibodies against LppM and anti‐Hsp65 monoclonal antibodies, used as a control. (D) Western‐blot analysis using anti‐LppM antibodies on crude lysates of M. tuberculosis H37RvΔlppM transformed by integrative empty vector (pMV306), vector containing lppM under the control of its own promoter (promlppM::lppM), vector containing lppM under the control of the strong hsp60 promoter (promhsp60::lppM) and the recombinant mature 6His‐LppM26‐227 purified from E. coli.
Fig. S2. Analysis of M. tuberculosis H37RvΔlppM phenotype after infection of BMDM. (A) Median of the bacterial area after infection of BMDM by M. tuberculosis H37Rv, M. tuberculosis H37RvΔlppM or respective heat‐killed bacteria (MOI 10) as a control. (B) Fold change of the bacterial area after infection of BMDM by M. tuberculosis H37Rv or M. tuberculosis H37RvΔlppM (using a MOI of 2) from Day 0 to Day 4. (C) Percentage of Lysotracker‐positive BMDM 2 h post‐infection, using M. tuberculosis H37Rv, M. tuberculosis H37RvΔlppM or the complemented strain (promlppM::lppM) labelled with CypHer5E.
Table S1. List of strains and plasmids used in this study.
Table S2. List of primers used in this study.
Supporting info item
Acknowledgements
We thank Dr Sang Hyun Cho (Institute for Tuberculosis Research, Chicago, USA) for kindly providing pMRF1. RVC acknowledges support from the Région Nord‐Pas‐de‐Calais. Financial support for this work was provided by the European Community (ERC‐STG INTRACELLTB Grant no 260901, MM4TB Grant no 260872), the Agence Nationale de la Recherche (ANR‐10‐EQPX‐04‐01, ANR‐14‐CE14‐0024‐03), the Projet Transversal de Recherche de l'Institut Pasteur (PTR441), the Feder (12001407 (D‐AL) Equipex Imaginex BioMed) and the Région Nord‐Pas‐de‐Calais (convention no 12000080).
Deboosère N., Iantomasi R., Queval C. J., Song O.‐R., Deloison G., Jouny S., Debrie A.‐S., Chamaillard M., Nigou J., Cohen‐Gonsaud M., Locht C., Brodin P., and Veyron‐Churlet R. (2017) LppM impact on the colonization of macrophages by Mycobacterium tuberculosis , Cellular Microbiology, 19, e12619. doi: 10.1111/cmi.12619.
Contributor Information
Priscille Brodin, Email: priscille.brodin@inserm.fr.
Romain Veyron‐Churlet, Email: romain_veyron@yahoo.fr.
References
- Barry, C.E., 3rd , Boshoff, H.I. , Dartois, V. , Dick, T. , Ehrt, S. , Flynn, J. , et al. (2009) The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat Rev Microbiol 7: 845–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander, J.M. , and Medzhitov, R. (2004) Regulation of phagosome maturation by signals from toll‐like receptors. Science 304: 1014–1018. [DOI] [PubMed] [Google Scholar]
- Brightbill, H.D. , Libraty, D.H. , Krutzik, S.R. , Yang, R.B. , Belisle, J.T. , Bleharski, J.R. , et al. (1999) Host defense mechanisms triggered by microbial lipoproteins through toll‐like receptors. Science 285: 732–736. [DOI] [PubMed] [Google Scholar]
- Brodin, P. , Majlessi, L. , Marsollier, L. , de Jonge, M.I. , Bottai, D. , Demangel, C. , et al. (2006) Dissection of ESAT‐6 system 1 of Mycobacterium tuberculosis and impact on immunogenicity and virulence. Infect Immun 74: 88–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin, P. , Poquet, Y. , Levillain, F. , Peguillet, I. , Larrouy‐Maumus, G. , Gilleron, M. , et al. (2010) High content phenotypic cell‐based visual screen identifies Mycobacterium tuberculosis acyltrehalose‐containing glycolipids involved in phagosome remodeling. PLoS Pathog: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambier, C.J. , Takaki, K.K. , Larson, R.P. , Hernandez, R.E. , Tobin, D.M. , Urdahl, K.B. , et al. (2014) Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature 505: 218–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drage, M.G. , Pecora, N.D. , Hise, A.G. , Febbraio, M. , Silverstein, R.L. , Golenbock, D.T. , et al. (2009) TLR2 and its co‐receptors determine responses of macrophages and dendritic cells to lipoproteins of Mycobacterium tuberculosis . Cell Immunol 258: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drage, M.G. , Tsai, H.C. , Pecora, N.D. , Cheng, T.Y. , Arida, A.R. , Shukla, S. , et al. (2010) Mycobacterium tuberculosis lipoprotein LprG (Rv1411c) binds triacylated glycolipid agonists of Toll‐like receptor 2. Nat Struct Mol Biol 17: 1088–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau, E. , Fontan, P. , Manganelli, R. , Soares‐Appel, S. , and Smith, I. (2002) Mycobacterium tuberculosis genes induced during infection of human macrophages. Infect Immun 70: 2787–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur, R.L. , Ren, K. , Blumenthal, A. , Bhamidi, S. , Gibbs, S. , Jackson, M. , et al. (2014) LprG‐mediated surface expression of lipoarabinomannan is essential for virulence of Mycobacterium tuberculosis . PLoS Pathog 10 e1004376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring, A.J. , Dobos, K.M. , Belisle, J.T. , Harding, C.V. , and Boom, W.H. (2004) Mycobacterium tuberculosis LprG (Rv1411c): a novel TLR‐2 ligand that inhibits human macrophage class II MHC antigen processing. J Immunol 173: 2660–2668. [DOI] [PubMed] [Google Scholar]
- Harding, C.V. , and Boom, W.H. (2010) Regulation of antigen presentation by Mycobacterium tuberculosis: a role for Toll‐like receptors. Nat Rev Microbiol 8: 296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan, Z. , Cliff, J.M. , Dockrell, H.M. , Jamil, B. , Irfan, M. , Ashraf, M. , and Hussain, R. (2009) CCL2 responses to Mycobacterium tuberculosis are associated with disease severity in tuberculosis. PLoS One 4 e8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killick, K.E. , Ni Cheallaigh, C. , O'Farrelly, C. , Hokamp, K. , MacHugh, D.E. , and Harris, J. (2013) Receptor‐mediated recognition of mycobacterial pathogens. Cell Microbiol 15: 1484–1495. [DOI] [PubMed] [Google Scholar]
- Klopper, M. , Warren, R.M. , Hayes, C. , Gey van Pittius, N.C. , Streicher, E.M. , Muller, B. , et al. (2013) Emergence and spread of extensively and totally drug‐resistant tuberculosis, South Africa. Emerg Infect Dis 19: 449–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancioni, C.L. , Li, Q. , Thomas, J.J. , Ding, X. , Thiel, B. , Drage, M.G. , et al. (2011) Mycobacterium tuberculosis lipoproteins directly regulate human memory CD4(+) T cell activation via Toll‐like receptors 1 and 2. Infect Immun 79: 663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Fan, X. , Long, Q. , Xie, L. , and Xie, J. (2015) Mycobacterium tuberculosis effectors involved in host‐pathogen interaction revealed by a multiple scales integrative pipeline In Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases, Vol. 32, pp. 1–11. [DOI] [PubMed] [Google Scholar]
- Moller, A.S. , Ovstebo, R. , Westvik, A.B. , Joo, G.B. , Haug, K.B. , and Kierulf, P. (2003) Effects of bacterial cell wall components (PAMPs) on the expression of monocyte chemoattractant protein‐1 (MCP‐1), macrophage inflammatory protein‐1alpha (MIP‐1alpha) and the chemokine receptor CCR2 by purified human blood monocytes. J Endotoxin Res 9: 349–360. [DOI] [PubMed] [Google Scholar]
- Pecora, N.D. , Gehring, A.J. , Canaday, D.H. , Boom, W.H. , and Harding, C.V. (2006) Mycobacterium tuberculosis LprA is a lipoprotein agonist of TLR2 that regulates innate immunity and APC function. J Immunol 177: 422–429. [DOI] [PubMed] [Google Scholar]
- Ruhwald, M. , Bjerregaard‐Andersen, M. , Rabna, P. , Kofoed, K. , Eugen‐Olsen, J. , and Ravn, P. (2007) CXCL10/IP‐10 release is induced by incubation of whole blood from tuberculosis patients with ESAT‐6, CFP10 and TB7.7 In Microbes and infection/Institut Pasteur, Vol. 9, pp. 806–812. [DOI] [PubMed] [Google Scholar]
- Sanchez, A. , Espinosa, P. , Garcia, T. , and Mancilla, R. (2012) The 19 kDa Mycobacterium tuberculosis lipoprotein (LpqH) induces macrophage apoptosis through extrinsic and intrinsic pathways: a role for the mitochondrial apoptosis‐inducing factor. Clin Dev Immunol 2012: 950503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saukkonen, J.J. , Bazydlo, B. , Thomas, M. , Strieter, R.M. , Keane, J. , and Kornfeld, H. (2002) Beta‐chemokines are induced by Mycobacterium tuberculosis and inhibit its growth. Infect Immun 70: 1684–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla, S. , Richardson, E.T. , Athman, J.J. , Shi, L. , Wearsch, P.A. , McDonald, D. , et al. (2014) Mycobacterium tuberculosis lipoprotein LprG binds lipoarabinomannan and determines its cell envelope localization to control phagolysosomal fusion. PLoS Pathog 10 e1004471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley, S.A. , and Cox, J.S. (2013) Host–pathogen interactions during Mycobacterium tuberculosis infections. Curr Top Microbiol Immunol 374: 211–241. [DOI] [PubMed] [Google Scholar]
- Underhill, D.M. , Ozinsky, A. , Smith, K.D. , and Aderem, A. (1999) Toll‐like receptor‐2 mediates mycobacteria‐induced proinflammatory signaling in macrophages. Proc Natl Acad Sci U S A 96: 14459–14463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kessel, J.C. , Marinelli, L.J. , and Hatfull, G.F. (2008) Recombineering mycobacteria and their phages. Nat Rev Microbiol 6: 851–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergne, I. , Chua, J. , Lee, H.H. , Lucas, M. , Belisle, J. , and Deretic, V. (2005) Mechanism of phagolysosome biogenesis block by viable Mycobacterium tuberculosis . Proc Natl Acad Sci U S A 102: 4033–4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyron‐Churlet, R. , Zanella‐Cleon, I. , Cohen‐Gonsaud, M. , Molle, V. , and Kremer, L. (2010) Phosphorylation of the Mycobacterium tuberculosis beta‐ketoacyl‐acyl carrier protein reductase MabA regulates mycolic acid biosynthesis. J Biol Chem 285: 12714–12725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walburger, A. , Koul, A. , Ferrari, G. , Nguyen, L. , Prescianotto‐Baschong, C. , Huygen, K. , et al. (2004) Protein kinase G from pathogenic mycobacteria promotes survival within macrophages. Science 304: 1800–1804. [DOI] [PubMed] [Google Scholar]
- Young, D.B. , Gideon, H.P. , and Wilkinson, R.J. (2009) Eliminating latent tuberculosis. Trends Microbiol 17: 183–188. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Generation of M. tuberculosis H37RvΔlppM. (A) Genomic organization of the lppM locus in M. tuberculosis H37Rv and schematic representation of the insertion of the hygromycin cassette resistance. Primers 1 and 2 used for PCR are symbolized by arrows. (B) PCR using primers 1 and 2 with genomic DNA extracted from M. tuberculosis H37Rv or M. tuberculosis H37RvΔlppM. (C) Western‐blot analysis on crude lysates of M. tuberculosis H37Rv and M. tuberculosis H37RvΔlppM cultures using polyclonal antibodies against LppM and anti‐Hsp65 monoclonal antibodies, used as a control. (D) Western‐blot analysis using anti‐LppM antibodies on crude lysates of M. tuberculosis H37RvΔlppM transformed by integrative empty vector (pMV306), vector containing lppM under the control of its own promoter (promlppM::lppM), vector containing lppM under the control of the strong hsp60 promoter (promhsp60::lppM) and the recombinant mature 6His‐LppM26‐227 purified from E. coli.
Fig. S2. Analysis of M. tuberculosis H37RvΔlppM phenotype after infection of BMDM. (A) Median of the bacterial area after infection of BMDM by M. tuberculosis H37Rv, M. tuberculosis H37RvΔlppM or respective heat‐killed bacteria (MOI 10) as a control. (B) Fold change of the bacterial area after infection of BMDM by M. tuberculosis H37Rv or M. tuberculosis H37RvΔlppM (using a MOI of 2) from Day 0 to Day 4. (C) Percentage of Lysotracker‐positive BMDM 2 h post‐infection, using M. tuberculosis H37Rv, M. tuberculosis H37RvΔlppM or the complemented strain (promlppM::lppM) labelled with CypHer5E.
Table S1. List of strains and plasmids used in this study.
Table S2. List of primers used in this study.
Supporting info item


