Abstract
The Virginia chicken lines have been divergently selected for juvenile body weight for more than 50 generations. Today, the high- and low-weight lines show a >12-fold difference for the selected trait, 56-d body weight. These lines provide unique opportunities to study the genetic architecture of long-term, single-trait selection. Previously, several quantitative trait loci (QTL) contributing to weight differences between the lines were mapped in an F2-cross between them, and these were later replicated and fine-mapped in a nine-generation advanced intercross of them. Here, we explore the possibility to further increase the fine-mapping resolution of these QTL via a pedigree-based imputation strategy that aims to better capture the genetic diversity in the divergently selected, but outbred, founder lines. The founders of the intercross were high-density genotyped, and then pedigree-based imputation was used to assign genotypes throughout the pedigree. Imputation increased the marker density 20-fold in the selected QTL, providing 6911 markers for the subsequent analysis. Both single-marker association and multi-marker backward-elimination analyses were used to explore regions associated with 56-d body weight. The approach revealed several statistically and population structure independent associations and increased the mapping resolution. Further, most QTL were also found to contain multiple independent associations to markers that were not fixed in the founder populations, implying a complex underlying architecture due to the combined effects of multiple, linked loci perhaps located on independent haplotypes that still segregate in the selected lines.
Keywords: imputation-based association, advanced intercross body weight line, Virginia chicken lines, QTL fine-mapping
Long-term selective breeding of animals and plants for extreme phenotypes has resulted in genetically distinct lines that are a valuable resource for dissecting the genetic architecture of complex traits (Hill 2005). Most traits of interest in animal breeding (e.g., production of eggs or meat, resistance to disease) are influenced by a combination of genetic and environmental factors. Due to their multi-factorial nature and despite the ability to obtain data on both genome-wide genetic markers and phenotypes from large numbers of individuals, it is challenging to disentangle their genetic architecture by analyzing data from the commercial populations. An alternate strategy is to make use of experimental populations resulting from long-term selection experiments, where the focus has been to develop divergent lines from a common base population using more coherent selection criteria. Such populations will display larger phenotypic differences than populations subjected to composite, commercial breeding programs and hence facilitate in-depth studies of the genetic basis underlying the selection response and general genetic architecture of these traits (Andersson and Georges 2004). Given that many of the agriculturally important traits are related to metabolism, feeding-behavior and growth, they also provide a good model for translational studies to decipher the genetic architecture of traits of interest in human medicine, including obesity, eating disorders, and diabetes.
Kemper et al. (2012) recently reviewed the literature on the genetic basis of body size and highlighted how complex the genetic architectures of body size are in species with contributions by many loci with large, intermediate, and small individual effects. Also within species, the genetic basis of variations in body size among strains of mice (Valdar et al. 2006), breeds of cattle (Saatchi et al. 2014), pigs (Yoo et al. 2014), and chickens (Van Goor et al. 2015) is often polygenic and due to polymorphisms with modest individual effects. Studies of experimental crosses from artificially selected populations with extreme body sizes in the mouse (Bevova et al. 2006; Parker et al. 2011) and chicken (Sheng et al. 2015) using, for example, chromosome substitution strains (Bevova et al. 2006) and advanced intercross lines (AILs) (Darvasi and Soller 1995; Besnier et al. 2011; Parker et al. 2011) have revealed that the responses to selection in these populations has resulted from selection on highly complex and polygenic genetic architectures.
The Virginia lines are experimental populations established in 1957 to study the genetic effects of long-term (>50 generations), divergent, single-trait selection for 56-d high (HWS) or low (LWS) body weight in chickens (Dunnington and Siegel 1996; Márquez et al. 2010; Dunnington et al. 2013). The lines originated from the same base population, composed by crossing seven partially inbred White Plymouth Rock chicken lines, and today display more than a 12-fold difference in body weight at 56 d of age (Márquez et al. 2010; Dunnington et al. 2013). In addition to the direct effects of selection on body weight, the selected lines also display correlated selection responses for a range of metabolic and behavioral traits including disrupted appetite, obesity, and antibody response (Dunnington et al. 2013).
The Virginia HWS and LWS lines have been used extensively for studying the genetic architecture of body weight and other metabolic traits. These studies have uncovered a number of loci with minor direct effects on body weight, metabolic traits, and body-stature traits by quantitative trait loci (QTL) mapping in an F2 intercross (Jacobsson et al. 2005; Park et al. 2006; Wahlberg et al. 2009). Also, a network of epistatic loci has been found to make a significant contribution to long-term selection response through the release of selection-induced additive variation (Carlborg et al. 2006; Le Rouzic et al. 2007; Le Rouzic and Carlborg 2008). Explorations of the genome-wide footprint of selection by selective-sweep mapping suggests that perhaps >100 loci throughout the genome have contributed to selection response (Johansson et al. 2010; Pettersson et al. 2013), and many of these contribute to 56 d body weight (Sheng et al. 2015).
To replicate and fine-map the body weight QTL inferred in the F2 intercross, we developed, genotyped and phenotyped for body weight at 56 d of age (BW56), a nine-generation AIL. This large AIL originated from the same founders as the F2 intercross, but was selectively genotyped at a higher resolution (∼1 marker/cM) in nine QTL (Besnier et al. 2011). In this population, most of the original minor (Besnier et al. 2011) and epistatic (Pettersson et al. 2011) QTL were replicated and fine-mapped. These earlier studies analyzed the data using a haplotype-based linkage-mapping approach in a variance-component based model framework to infer single-locus effects (Besnier et al. 2011) or a fixed-effect model framework assuming fixed alternative alleles in the two founder lines for detecting epistasis (Pettersson et al. 2011). The variance-component model was used in the replication study to avoid the assumption of allelic fixation in the founder lines. By implementing it in a Flexible Intercross Analysis modeling framework (Rönnegård et al. 2008), it was expected to improve power when the parental lines carry alleles with correlated effects (e.g., multiple alleles with similar effects).
Although the initial studies mapped QTL under the assumption of fixation, or an effect correlation, of divergent alleles in the parental lines, the results at the same time implied that multiple alleles might be segregating in several of the mapped regions. To this end, the first QTL replication study in the AIL population (Besnier et al. 2011) found a large within founder line heterogeneity in the allelic effects. Later the selective-sweep studies, which utilized data from multiple generations of divergently selected and relaxed lines, identified ongoing selection and multiple sweeps in many QTL (Johansson et al. 2010; Pettersson et al. 2013), as well as extensive allelic purging (Pettersson et al. 2013). This allelic heterogeneity challenges attempts to dissect the architecture of the selected trait via, e.g., QTL introgression (Ek et al. 2012). Alternative approaches are therefore needed to uncover multi-locus, multi-allelic genetic architectures in QTL and their contributions to the long-term response to directional selection.
In this study, we explore an imputation-based association-mapping strategy for further dissection of previously mapped and replicated QTL (Besnier et al. 2011; Pettersson et al. 2011). For this, we made use of available high-density (60K SNP-chip) genotypes for founders (Johansson et al. 2010; Pettersson et al. 2013) and intermediate-density SNP-genotypes in several QTL in the entire nine-generation AIL pedigree. By increasing the marker density in the QTL throughout the AIL by imputation, we aimed to better capture the effects of segregating haplotypes within and between the divergently selected founder populations than with the previously used markers. This aim can be achieved as the original markers genotyped in the AIL were selected to identify high- and low-line derived alleles, and not alleles that segregate within or across the founder lines. By testing for association between imputed markers and body weight, the fine-mapping analyses were less constrained by the original selection of markers and facilitated a more thorough exploration of the genetic architectures of the nine evaluated QTL. A single-marker association analysis was first used to identify regions with candidate associations. These were then simultaneously analyzed using a backward-elimination approach with bootstrapping to identify statistically independent signals that were robust to the effects of markers elsewhere in the genome and the pedigree-structure in the population. In regions where the signals were robust to the pedigree-structure, the results from the single-marker association analysis were used to fine-map the region. Our imputation-based approach replicated most QTL and also improved the resolution in the fine-mapping analyses by not only using the recombination events in the AIL, but also the historical recombinations in the pedigree. We found that several of the original QTL are likely due to the combined effects of multiple linked loci, several of which are segregating in the founder lines of the AIL.
Materials and Methods
Animals
The Virginia chicken lines are part of an ongoing selection experiment to study the genetics of long-term, single-trait selection (Márquez et al. 2010; Dunnington et al. 2013). It was initiated in 1957 from a base population, generated by intercrossing seven partially inbred lines of White Plymouth Rock chickens. From the offspring of the partially inbred lines, resulting from the intercrossing, the birds with the highest and lowest 56 d body weights (with some restrictions), respectively, were selected to produce the high- and low-weight selected lines (HWS and LWS) (Márquez et al. 2010; Dunnington et al. 2013). Since then, the lines have undergone divergent selection for increased and decreased body weights with one new generation hatched in March of every year.
An AIL was founded by reciprocal crosses of 29 HWS and 30 LWS founder birds from generation 40 (Besnier et al. 2011). The mean, sex-averaged 56 d body weights for HWS and LWS at this generation were 1522 g and 181 g, respectively. Repeated intercrossing of birds was used to develop a nine-generation AIL consisting of generations F0–F8. In each generation, ∼90 birds were bred by paired mating, genotyped, and weighed at 56 d of age (BW56). In total, the AIL population consisted of 1536 F0–F8 individuals with complete records on pedigree and genotypes (see Genotyping), and 1348 F2–F8 individuals with juvenile body weight (BW56) records.
Genotyping
The complete AIL pedigree (1536 birds) had earlier been genotyped in nine selected QTL for 304 SNP-markers that passed quality control as described in Besnier et al. (2011). Further, 40 of the founders for the pedigree (20 HWS and 20 LWS) had also earlier been genotyped using a whole genome 60K SNP-chip (Johansson et al. 2010; Pettersson et al. 2013). The 6607 markers from the SNP-chip that were informative and passed quality control in that study are located in the nine QTL regions targeted in this study. When merging the information from the 60K SNP-chip and the information from the 304 markers genotyped earlier, 55 markers in 40 founders were genotyped using both methods. Out of these 55 markers, 28 markers with genotype inconsistencies between the genotyping technologies were removed during quality control. In total, our analyses were based on 6888 markers, where 40 of the 59 AIL founders had genotypes for all markers, and the remaining individuals in the pedigree had genotypes for 281 markers. Table 1 shows how these markers are distributed across the nine QTL regions.
Table 1. Genotyped and imputed markers across the nine analyzed QTL.
| GGAa | QTLb | Startc (bp) | Endc (bp) | QTL Size (bp) | Markers AILd | Markers 60ke | Markers Totalf | Marker Densityg |
|---|---|---|---|---|---|---|---|---|
| 1 | Growth1 | 169 634 954 | 181 087 961 | 11 453 008 | 26 | 504 | 530 | 46 |
| 2 | Growth2 | 47 929 675 | 65 460 002 | 17 530 328 | 33 | 667 | 700 | 40 |
| 2 | Growth3 | 124 333 151 | 133 581 122 | 9 247 972 | 19 | 395 | 414 | 45 |
| 3 | Growth4 | 24 029 841 | 68 029 533 | 43 999 693 | 57 | 1885 | 1942 | 44 |
| 4 | Growth6 | 1 354 213 | 13 511 203 | 12 156 991 | 23 | 514 | 537 | 44 |
| 4 | Growth7 | 85 459 943 | 88 832 107 | 3 372 165 | 14 | 141 | 155 | 46 |
| 5 | Growth8 | 33 696 791 | 39 052 438 | 5 355 648 | 5 | 221 | 226 | 42 |
| 7 | Growth9 | 10 916 819 | 35 491 706 | 24 574 888 | 76 | 1397 | 1473 | 60 |
| 20 | Growth12 | 7 109 709 | 13 899 993 | 6 790 285 | 28 | 883 | 911 | 134 |
Gallus Gallus Autosome.
QTL names as in Jacobsson et al. (2005).
Base pair position according to Chicken genome assembly (galGal3) of May 2006.
Markers as in Besnier et al. (2011).
Markers as in Johansson et al. (2010).
Total markers in d and e.
Markers/Mb.
Phasing and imputation of markers
All genotyped markers in the QTL (Table 1) were first ordered according to their physical location in the chicken genome assembly of May 2006 (galGal3). In the ordered marker set, the SNP-chip markers were evenly distributed in the intervals between the sparser set of markers genotyped across the entire AIL.
Using the software ChromoPhase (Daetwyler et al. 2011), we phased and imputed genotypes for the complete set of 6888 markers across the entire AIL pedigree. ChromoPhase first phases large segments of chromosomes, in our case the QTL regions. It then imputes the missing genotypes in the AIL individuals genotyped with the sparse set of markers from the genotype information available in high-density genotyped founders utilizing the pedigree information. It thus predicts both phased haplotypes across the nine studied QTL and genotypes at markers that were only genotyped in a subset of the founder individuals in the pedigree.
A two-step fine-mapping approach accounting for population structure
Earlier studies have shown that the genetic architecture of body weight is highly polygenic in the Virginia lines (e.g., Siegel 1962a,b; Jacobsson et al. 2005; Wahlberg et al. 2009; Johansson et al. 2010; Besnier et al. 2011; Pettersson et al. 2011, 2013; Sheng et al. 2015). We therefore implemented a forward-selection/backward-elimination procedure with a termination criteria suitable for a polygenic trait in a bootstrap-based framework to correct for population structure in the AIL (Valdar et al. 2009; Sheng et al. 2015). As all markers with genotypes could not be included in a backward-elimination analysis due to the limited sample size, we first used a forward-selection based single-marker association analysis to identify a smaller set of statistically suggestive independent signals within each QTL region. The backward-elimination analysis (Valdar et al. 2009; Sheng et al. 2015) was then used to identify associations robust to possible influences of genetic dependencies (linkage or LD) between markers within the QTL or population structure in the AIL (Peirce et al. 2008; Cheng et al. 2010).
Single-marker association analyses:
The qtscore function in the GenABEL package (Aulchenko et al. 2007) was used to test for association between body weight at 56 d of age and, genotyped or imputed, individual genetic markers within the targeted QTL. The allelic effect of each marker, was estimated using a regression model (Model 1), where the genotype at each marker was coded in Z as 0 if homozygous for the major allele, 1 if heterozygous, and 2 if homozygous for the minor allele. Sex and generation were added as categorical covariates, with two different levels for sex and seven different levels for generation, defined for each individual in X. The phenotype, body weight at 56 d of age, is given in the numerical variable y.
ε was assumed to be iid and normally distributed around 0 with variance μ is the intercept, which in this model represented the mean body weight at 56 d of age for individual F2 females. The associations for the individual markers from this model were used for comparisons to results from earlier linkage-mapping analyses to fine-map the QTL in this pedigree that did not account for the possible effects of pedigree-structure (Model A in Besnier et al. 2011). Further, they were also used to evaluate the resolution of regions with associations robust to the pedigree-structure in the population (described in detail in Results).
Next, a forward-selection analysis was performed by scanning across all markers within each QTL using Model 1. If any of the markers were nominally significant (P < 0.05) in the scan, the marker with the strongest association was added as a covariate in the model. This procedure was repeated until no additional significant markers were detected. The markers from this analysis with an allele-frequency > 0.10 in the population were subjected to the full backward-elimination analysis described in the next section.
A multi-locus association analysis to identify regions with associations that are robust to the pedigree-structure in the population:
In short, we used a bootstrap-based backward-elimination model selection framework (Sheng et al. 2015) across the markers selected by forward-selection in the QTL. An adaptive model selection criterion controlling the False Discovery Rate (Abramovich et al. 2006; Gavrilov et al. 2009) was used during backward-elimination in a standard linear model framework, starting with a full model including the fixed effects of sex and generation, and the additive effects of all markers (Model 2):
where phenotype, sex, and generation were coded as described for Model 1 and where ε again is assumed to be iid and normally distributed around 0 with variance The intercept, represents the mean body weight at 56 d of age for female individuals from the F2 generation. In Model 2, genotypes were coded based on the line-origin of the alleles at each locus. Genotypes of individuals homozygous for the major allele in the AIL founders from the high-weight selection line were coded as 1 at that locus. If an individual was heterozygous, its genotype was coded as 0. Genotypes of individuals homozygous for the allele corresponding to the major allele in AIL founders from the low-weight selection line were coded as −1. By coding genotypes in a −1, 0, and 1 manner, the estimates of the marginal allele-substitution effects, from fitting Model 2 will be negative if the allele that is at highest frequency in the high-weight line decreases weight or if the allele with highest frequency in the low-weight line increases weight.
Convergence was based on a 20% False Discovery Rate (FDR) level. The analysis was performed using bootstrapping with 1000 resamples. Markers with an RMIP (Resample Model Inclusion Probability) > 0.46, as suggested for an AIL generation F18 (Valdar et al. 2009), were included in the final model. The FDR in the final model was confirmed using the original FDR procedure described in Benjamini and Hochberg (1995) as implemented in the p.adjust function in the R stats-package (R Development Core Team 2015). The additive genetic effect for each locus was estimated using the multi-locus genetic model described above (Model 2). The contribution of a set of n associated markers to the founder line difference was calculated as where is the allele-substitution effect for marker i, and are the frequencies of the major AIL allele at marker i in the HWS and LWS founders, respectively.
Data availability
Genotype, phenotype, and pedigree data are included in the supplemental files. Supplemental Material, File S1 contains detailed descriptions of all supplemental data files. File S2 contains the genotypes, File S3 the pedigree, and File S4 the phenotypes.
Results and Discussion
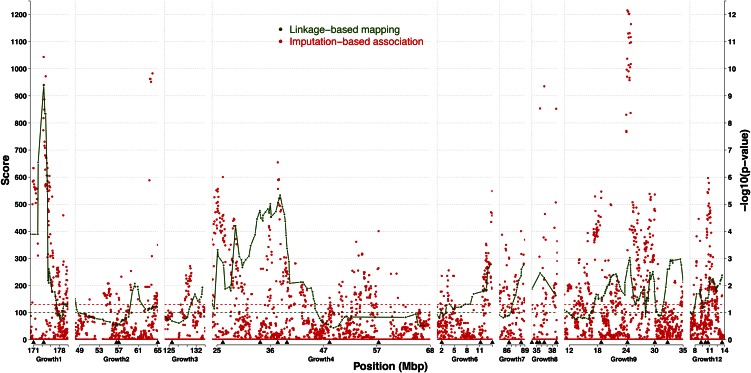
We compared the results of the imputation-based association analyses with the previously reported results from the linkage-based analysis of the same nine QTL in Besnier et al. (2011). Figure 1 shows the statistical support for association and linkage to BW56 across the QTL. The significances for all the genotyped and imputed markers from the single-marker associations are provided together with the results from model A in Besnier et al. (2011) that were also obtained without correction for population structure. Figure 1 also highlights those regions that contain associations robust to the pedigree-structure in the bootstrap-based forward-selection/backward-elimination analyses (Figure 1 and Table 2). Overall, the results from these three analyses overlap well. Together, they show that most regions with strong associations in the single-marker association were robust to the pedigree-structure and that the association analysis approach using imputed genotypes for SNPs suggests that several QTL were likely due to multiple linked loci. In the sections below, these results are described and discussed in more detail.
Figure 1.
Comparison between linkage- and association-based fine-mapping analyses of nine QTL in an AIL chicken population. Green lines show the statistical support curve (score statistics from Model A) for the linkage-based mapping study of (Besnier et al. 2011) and the red dots associations to each analyzed marker in the new imputation-based association analysis (this study). The green and red horizontal dotted lines indicate the experiment-wide significance threshold for earlier linkage-analysis and the nominal significance in the imputation-based association analysis, respectively. Arrowheads under the x-axis indicate the position of markers identified as experiment-wide significant (20% FDR) in the bootstrap-based backward-elimination procedure.
Table 2. Estimated additive effects and standard error for experiment-wide independent association signals, between body weight at 56 d of age and genotype, identified in a bootstrap-based approach implemented in a backward-elimination model selection framework across the markers in the genotyped QTL.
| GGAa | QTLb | Locationc (bp) | Markerd | ΔAFe | a ± SEf | Signg |
|---|---|---|---|---|---|---|
| 1 | Growth1 | 170 637 618 | rs13968052i | 0.28 | 16.3 ± 6.6 | 1.3 × 10−2 |
| 173 709 608 | rs14916997 | 0.71 | 19.3 ± 5.5 | 4.8 × 10−4 | ||
| 2 | Growth2 | 56 720 515 | rs14185295 | 0.32 | 13.2 ± 6.2 | 3.3 × 10−2 |
| 57 198 629 | rs14185836i | 0.16 | −12.1 ± 6.2 | 5.4 × 10−2 | ||
| 65 460 002 | rs14196021 | 0.32 | 14.0 ± 5.9 | 1.7 × 10−2 | ||
| 2 | Growth3 | 126 000 254 | rs16120360 | 0.36 | 12.6 ± 5.8 | 3.0 × 10−2 |
| 3 | Growth4 | 26 215 175 | rs14328509 | 0.53 | 19.8 ± 5.4 | 2.3 × 10−4 |
| 33 743 569 | rs314044798i | 0.46 | −21.9 ± 6.1 | 3.8 × 10−4 | ||
| 37 287 334 | rs316425755i | 0.12 | 27.8 ± 6.9 | 5.7 × 10−5 | ||
| 39 139 081 | rs15468467i | 0.23 | 27.0 ± 7.3 | 2.1 × 10−4 | ||
| 47 729 342 | rs316384373i | 0.30 | −13.1 ± 6.8 | 5.5 × 10−2 | ||
| 57 624 596 | rs14363139i | 0.64 | 18.2 ± 4.1 | 2.9 × 10−4 | ||
| 4 | Growth6 | 2 392 397 | rs14419462i | 0.30 | 19.7 ± 7.6 | 1.0 × 10−2 |
| 10 914 312 | rs14428120i | 0.77 | −14.5 ± 5.8 | 1.3 × 10−2 | ||
| 13 511 203 | rs15500313 | 0.78 | 17.7 ± 6.0 | 3.4 × 10−3 | ||
| 4 | Growth7 | 86 755 267 | rs14499758 | 0.32 | 18.1 ± 6.3 | 3.9 × 10−3 |
| 88 325 118 | rs15639000 | 0.06 | 13.4 ± 5.4 | 1.3 × 10−2 | ||
| 5 | Growth8 | 33 713 055 | rs14530756i | 0.46 | −13.5 ± 5.3 | 1.1 × 10−2 |
| 34 772 650 | rs16487762i | 0.53 | 15.0 ± 6.8 | 2.7 × 10−2 | ||
| 35 299 978 | rs16487933i | 0.40 | −18.6 ± 6.0 | 1.8 × 10−3 | ||
| 36 291 277 | rs13585490 | 0.15 | 14.1 ± 5.3 | 7.3 × 10−3 | ||
| 38 774 986 | rs315605733i | 0.23 | 23.3 ± 7.3 | 1.4 × 10−3 | ||
| 38 867 279 | rs314075508i | 0.37 | 16.4 ± 5.5 | 3.0 × 10−3 | ||
| 7 | Growth9 | 18 544 622 | rs14611566i | 0.00 | −20.1 ± 5.1 | 9.4 × 10−5 |
| 23 959 214 | rs16596357i | 0.17 | 18.9 ± 5.6 | 7.7 × 10−4 | ||
| 29 631 963 | rs10727581i | 0.32 | 15.9 ± 6.1 | 9.0 × 10−3 | ||
| 32 262 733 | rs317586448i | 0.33 | −13.2 ± 5.0 | 8.7 × 10−3 | ||
| 20 | Growth12 | 9 302 754 | rs14277526i | 0.34 | 21.8 ± 6.8 | 1.4 × 10−3 |
| 10 165 171 | rs14278292i | 0.37 | −14.7 ± 6.5 | 2.4 × 10−2 | ||
| 10 667 729 | rs16172598i | 0.56 | 14.3 ± 5.1 | 5.1 × 10−3 | ||
| 13 427 530 | rs16176151i | 0.13 | 8.5 ± 5.2 | 1.0 × 10−1 |
For a marker with a positive estimated additive effect, the effect on weight is caused by the allele with its origin in the line associated with the sign of the effect, i.e., an allele with its origin in high-line is associated with an increase in body weight and an allele with its origin in low-line is associated with a decrease in body weight. In cases where a weight-increasing allele has its origin in the low-line or a weight-decreasing allele has its origin in the high-line the sign of the estimated additive effect will be negative.
Gallus Gallus Autosome.
QTL name as in Jacobsson et al. (2005).
Base pair position according to Chicken genome assembly (galGal3) of May 2006.
SNP name as in NCBI dbSNP where imputed markers are labeled with i after the marker name.
Difference in allele-frequency between the HWS and LWS founder lines.
Additive effect ± SE calculated in a model including all loci in the table.
Significance of the estimated additive genetic effect in a model including all loci in the table.
Four statistically independent associated markers in the GGA7 QTL Growth9
The QTL Growth9 on GGA7 (Gallus Gallus Autosome 7) (Figure 1; 10.9–35.5 Mb) was the only QTL that reached genome-wide significance in the first F2 intercross between the HWS and LWS lines (Jacobsson et al. 2005). It was later identified as a central QTL in an epistatic network explaining a large part of the difference in weight between HWS and LWS lines (Carlborg et al. 2006). In the earlier fine-mapping analysis, the linkage signal covered most of the QTL region (from 15 to 35 Mb), but subsequent analyses showed that two independent loci were segregating in the region (Besnier et al. 2011). The signal in the imputation-based association analysis performed here is more focused, with a highly significant signal in a 2.8 Mb region between 23.7 and 26.4 Mb. This region overlaps with the strongest signal in the linkage-scan and is tagged by a single imputed marker (rs16596357i; Table 2) in the multi-locus analysis accounting for population structure. The major allele in the HWS line (P = 0.67) increases weight by 18.9 (SE 5.6) g, and it still segregates at an intermediary frequency (P = 0.50) in the LWS line. Previously, Ahsan et al. (2013) explored potential candidate mutations in the QTL and found two regulatory SNPs near the peak at 21 Mb (21.6 and 22.7 Mb) and a synonymous-coding SNP in a CpG island in an exon of the Insulin-like growth factor binding protein 2 (IGFBP2) gene in the middle of the major association peak at 24.8 Mb. In addition to the strong association around 24 Mb, the association analysis also highlights two additional regions (centered around 18 and 29 Mb). A single imputed marker (rs14611566i; Table 2) is retained in the first region in the multi-locus analysis accounting for population structure. This marker has an estimated allele-substitution effect of −20.1 (SE 5.1) g, but as it segregates at equal, intermediary frequencies in the HWS and LWS lineages (P = 0.50) it did not contribute to the founder line difference. Two linked imputed markers are kept in the third region (rs10727581i; rs317586448i; 2.6 Mb apart; Table 2). Here, the first marker is nearly fixed for one allele in the LWS line (P = 0.93), but segregates at an intermediary frequency in the HWS line (P = 0.61). At the second marker, the major allele in the HWS (P = 0.74) segregates at an intermediary frequency in the LWS (P = 0.41). Due to the close linkage between the associated markers, it is difficult to interpret their individual effects and disentangle whether the detected associations are due to the LD-pattern of multiple closely linked loci or a single locus with multiple segregating alleles. The peak in the F2 QTL overlaps the major 23.7–26.4 Mb association peak detected in this analysis (Wahlberg et al. 2009). Due to the low differences in allele frequencies between the associated markers in the three regions, their total contribution to the founder line difference is small (8 g) amounting to only about 10% of the original estimated F2 QTL effect of 86 g (Wahlberg et al. 2009).
Two statistically independent associated markers in the GGA1 QTL Growth1
The strongest association in the study by Besnier et al. (2011) was found on GGA1 in the QTL Growth1 (Figure 1; 169.6–181.1 Mb). Here, the second strongest association was detected in that QTL. The imputation-based association analysis highlights two significant associations separated by a region of very low association and both associations remained in the multi-locus analysis accounting for population structure (the imputed marker rs13968052i and the genotyped marker rs14916997 at 170.6 and 173.7 Mb, respectively). The strongest of these association-peaks was located near the peak detected using the earlier linkage-based analysis. Several of the significant associated markers were located in this region (173.6–175.3 Mb). A candidate gene for growth, Asparagine-linked glycosylation 11 homolog gene (ALG11), is located at 174.6 Mb and has a strong mutation in its regulatory region (Ahsan et al. 2013). The second association was found to a group of significant markers in a narrower region upstream from the main linkage-peak (170.3–171.7 Mb). The association analysis thus suggests that the original 10.6 Mb QTL region that has its peak between markers located at 175.2–177.7 Mb is due to the effects of two separate loci located in these confined 1.5 and 1.8 Mb regions. As the two associated markers are closely linked in this population, it is difficult to interpret their individual effects, but their total contribution to the founder line difference (37 g) is about 75% of that by the original Growth1 QTL as estimated in the F2 analysis (49 g; Wahlberg et al. 2009).
Four statistically independent associated markers in the GGA2 QTL Growth2 and Growth3
GGA2 contains two QTL, Growth2 (Figure 1; 47.9–65.5 Mb) and Growth3 (Figure 1; 124.3–133.6 Mb). The multi-locus analyses identified three significantly associated markers in Growth2, where the first two are clustered at 56.7 and 57.2 Mb (genotyped marker rs14185295 and imputed marker rs14185836i, respectively), with the last genotyped marker (rs14196021) located at 65.5 Mb. The distance between the markers, and the region of low association between them in the single-marker analysis (Figure 1), suggests that two linked loci contribute to the Growth2 QTL. In the earlier linkage-based analysis the strongest signal in Growth2 was located in between these markers at 60.6 Mb. The QTL-peak in the original F2 analysis (Jacobsson et al. 2005) is difficult to assess as nearest marker (MCW130) is not mapped to the chicken genome and no significant signal was found using a denser marker-map by Wahlberg et al. (2009). As the first two markers in the QTL are tightly linked, it is difficult to interpret the individual estimates of their effects; however, the third marker located 8 Mb upstream from them has a small independent effect. The estimated contribution by these loci to the founder line difference is small (14 g), which amounts to about 30% of the original contribution by the Growth2 QTL in the F2 analyses (Jacobsson et al. 2005). In Growth3, a single association was detected to a genotyped marker (rs16120360) in the multi-locus analysis and this peak was inside the original F2 QTL [101.6–131.9 Mb; (Jacobsson et al. 2005)] but shifted almost 4 Mb upstream from the top signal found in the earlier linkage-based analysis. The contribution by this marker to the line difference is small (9 g) and about 15% of that in the original F2 analysis (Jacobsson et al. 2005).
Six statistically independent associated markers in the GGA5 QTL Growth8
One of the strongest association signals was found on GGA5 in the QTL Growth8 (Figure 1; 33.7–39.1 Mb) and six markers were retained in this region after accounting for population structure in the multi-locus analysis (Table 2). The single-locus analysis suggests that these markers tag two loci – one from 33.7–36.3 Mb (three imputed and one genotyped marker; Table 2) and one around 38.8 Mb (two imputed markers). The markers were located between the markers flanking the original QTL (21.6–44.2 Mb) in Jacobsson et al. (2005) and overlaps with the earlier linkage signal. The association signal was, however, stronger than the linkage signal suggesting that the imputed markers tag the QTL better than the haplotypes inferred from the sparser set of genotyped markers. Although the linkage between the markers again makes it difficult to obtain stable estimates for the effects of individual markers in the two associated loci, their estimated contribution to the founder line difference (16 g) amounts to about one third of that estimated effect in the F2 (Jacobsson et al. 2005).
Six statistically independent associated markers in the GGA3 QTL Growth4
In the QTL Growth4 on GGA3 (Figure 1; 24.0–68.1 Mb), both the association and linkage analyses identify a broad region of association from 24 to 41 Mb. Although the statistical support curve in the linkage analysis contains multiple peaks, that analysis was unable to fine-map the region into multiple, independent signals. Here, the multi-locus analysis suggests that perhaps up to four independent regions contribute to this QTL, with one associated genotyped marker at 26.2 Mb, three imputed markers from 33.7–39.1 Mb, one imputed marker at 47.7 Mb, and one at 57.6 Mb. The single-locus association analysis highlights two particularly strong and distinct association-peaks located approximately between 24–27 and 33–37 Mb, respectively. A candidate mutation in Growth4 was found near the second association region at 33.6 Mb inside the regulatory region of Cysteine rich transmembrane BMP regulator 1 (CRIM1) (Ahsan et al. 2013). The associated region around 55–57 Mb displayed very low significance in the previous linkage analysis. The outmost markers (26.2 and 57.6 Mb) have allele-substitution effects of 19.8 (SE 5.4) and 18.2 (SE 4.1) g, respectively, and are rather diverged between the lines (∼50% difference between the lines). For the other two clusters of markers, it is difficult to obtain stable estimates of their individual effects. Their estimated joint contribution to the founder line difference (35 g) is about 65% of that in the original F2 analysis (Jacobsson et al. 2005).
Four statistically independent associated markers in the GGA20 QTL Growth12
The earlier linkage analysis replicated the QTL Growth12 on GGA20 (Figure 1; 7.1–13.9 Mb), with the strongest associated marker at 10.7 Mb, and the signal covered most of the region (8–13.9 Mb). Four markers were significant in the multi-locus analysis and three imputed markers were located in the main single-marker association peak covering the region from 9 to 11 Mb, while the fourth associated imputed marker was located about 4 Mb upstream (13.4 Mb). Again, it is difficult to interpret the individual effects of the tightly linked markers; however, their estimated contribution to the line difference (22 g) is about 75% of that estimated in the F2 analysis (Jacobsson et al. 2005).
Five statistically independent associated markers in the GGA4 QTL Growth6 and Growth7
In both Growth6 (Figure 1; 1.3–13.6 Mb) and Growth7 (Figure 1; 85.4–88.9 Mb) on GGA4, several markers were significant in the multi-marker analysis. The single-marker analysis illustrates that these markers tag association-peaks that were located very close to the main peaks in the earlier linkage-based analysis, suggesting that both analyses identified the same underlying loci. The association analysis identified two genotyped markers in a region in Growth7 with strong association around 86.5–88.5 Mb. It is difficult to interpret their individual effects due to the close linkage, but their estimated contribution to the founder line difference was small (13 g) amounting to about a fifth of that estimated in the F2 population (66 g; Jacobsson et al. 2005). In Growth6, the multi-marker analysis detected associations to two imputed and one genotyped marker representing one locus at 2.3 Mb and a second locus at 10.9–13.5 Mb. Here, the tight linkage in the second locus also makes interpretation of their individual effects difficult, whereas the allele frequencies are not that differentiated in the first locus. Their estimated contribution to the line difference is therefore small (17 g) amounting to about a fifth of the 92 g estimated in the original F2 analysis (Jacobsson et al. 2005).
General comments
Here we report the results from using an imputation-based association-mapping strategy to fine-map QTL in a nine-generation, outbred AIL. By combining high-density genotyping of the AIL founders with imputation throughout the rest of the pedigree utilizing a sparser genotyped marker backbone, we increased the marker density ∼20-fold in the studied regions. This subsequent association analysis had a comparable power for replication of QTL to the earlier used linkage-based strategy. In addition to this, the new analyses also detected multiple association-peaks in several of the QTL and narrowed the associated regions considerably compared to the regions detected previously (Besnier et al. 2011). Together, they suggest that this imputation-based association-mapping approach is a promising strategy for improving the resolution in fine-mapping studies in outbred pedigrees, where high-density marker genotypes are not available for all studied individuals. When interpreting the full results from the multi-locus backward-elimination analysis (Table 2), it should be noted that the results are reported at a 20% False Discovery Rate. Although a significant proportion of the markers could thus be false positive associations, as illustrated by the individual P-values for the additive effects of the associated markers, most of the peaks on the chromosomes also contain markers with more significant individual associations. We used this threshold because earlier mapping and replication studies confirmed that the QTL contain at least one small-effect locus contributing to 56 d weight, and that the major aim of this study was to provide an overall view of the most likely genetic architecture of the fine-mapped QTL, rather than high-confidence estimates for the individual regions.
In both Growth1 and Growth4 two strong, distinct association signals were identified. Also in the QTL Growth8 and Growth9 the new analysis identified strong association-peaks covering many markers. In these regions, the strongest linkage signals identified in the previous fine-mapping analysis (Besnier et al. 2011) overlap with the strongest signals in the current analyses. However, the association analysis also separates the signals into multiple peaks and highlights narrower regions. Hence, it provides more useful input for further analyses to identify candidate genes underlying the QTL. In most cases the associated regions are restricted to distinct 2–3 Mb regions, which as indicated by the findings from Ahsan et al. (2013), is useful for restricting the bioinformatics analyses to only the most promising candidate genes for further functional studies. In Growth6, Growth7, and Growth12, the association signals were not as significant as in the other QTL. Despite this, the multi-locus analyses suggest that the linkage signals in the earlier analyses were due to distinct loci with independent effects, mapped here into narrower association-peaks.
Overall, the location of the association signals in this study overlapped well with the top signals in the earlier linkage analyses. However, in two of the QTL (Growth2 and Growth3), the association-peaks are shifted when comparing results from both studies. Further work is needed to explore whether this reflects separate loci with distinct genetic architectures that could only be detected with the respective methods, or if they reflect a signal of the same underlying causal locus.
Here, we estimated the additive genetic effects of the fine-mapped regions using data from the F2–F8 generations of the AIL. To evaluate whether they were in general agreement with estimates obtained for the same regions in earlier studies, we compared them to the estimates obtained from our first large F2 population (Jacobsson et al. 2005; Wahlberg et al. 2009). The QTL effects were generally lower in the F2–F8 data than in the F2. Although this may be interpreted as the F2 estimates being inflated, several other factors could also be considered. First, the 56 d body weights were considerably lower for the F8 generation because younger dams were used to generate these (Besnier et al. 2011). In the analyses, a fixed effect of generation was used to account for the mean weight differences between generations. However, it did not account for the likely scenario that the QTL effects were smaller in these F8 birds due to their lower body weight. As about 30% of the birds in the pedigree are from this generation, this would bias the overall effects downward. Standardization of the phenotypes from different generations to the same mean and variance is a way to possibly account for this, but a caveat of that approach is an introduction of an upward bias of the effects if the QTL effects in the F8 are, in fact, not that much smaller. We therefore chose to report the more conservative estimates based on analyzing the nonstandardized phenotypes. Second, eight of the nine QTL contain fine-mapped regions with associations to several tightly linked markers. If these markers are located on the same haplotypes, it is not possible to disentangle their effects in this pedigree as too few recombination events have accumulated in the F2–F8 generations of the AIL, and due to such collinearities the estimates for the individual markers reported here would not properly describe the contributions of these haplotypes to the line difference. In several of the regions with multiple associated markers, the estimates of the additive effects were also negative for at least one of these markers. Although this could be interpreted as transgression being common in the population, we find them more likely to result from the collinearities among the closely linked markers. Further analyses utilizing, for example, later AIL generations, markers that specifically tag the haplotype-structure of the founder lines and methods that can account for multi-allelic genetic architectures will be needed to disentangle the genetic architectures of these loci and quantify their contributions to the founder line difference. Third, the F2 QTL estimates were obtained using a line-cross analysis where it is assumed that the founder lines are fixed for alternative QTL alleles (Jacobsson et al. 2005; Wahlberg et al. 2009). In the current association analysis, it is instead assumed that the alternative alleles at the tested markers tag nearby functional alleles. As none of the associated markers were fixed for alternative alleles in the founder lines (Table 2 and Table S1), the current fine-mapping analysis suggests that one, or both, founder lines segregate for multiple functional alleles in the QTL. To compare the estimates from the line-cross analysis in the F2 and the association analyses in the AIL F2–F8 generations, they need to be compared using a common reference. Here, we did this by estimating how much the associated markers in each QTL were expected to contribute to the founder line difference under the assumption that they act completely additively. Under this assumption, their contribution would equal twice the sum of the allele-substitution effects of the markers in a QTL weighted by their respective allele-frequency differences between the founder lines. That is, if the markers are fixed for alternative alleles in the founder lines they would contribute two allele-substitution effects to the founder line difference, whereas they would contribute nothing if the allele was present at equal frequencies in both founder lines. This estimate is conservative as, for example, dominance leads to an underestimation of the contribution of the locus. This as, in the presence of a dominant allele, one line will not need to be fixed to contribute most of its effect because this is also displayed in the heterozygotes for that allele. When comparing estimates this way, the combined effects of the associated markers in each of the QTL contribute from 10 to 75% of that estimated in the F2 by Jacobsson et al. (2005). In total, the QTL replicated here contributed 171 g to the founder line difference, compared to the 416 g in the F2. As discussed above, further analyses in other populations with more informative genetic markers using other statistical methods are, however, required to explore this further.
Our analyses suggest that there is extensive within-line segregation in the QTL regions. One possible explanation for the slow fixation in these loci could be that the beneficial alleles at the linked fine-mapped loci were located on different haplotypes at the onset of selection. Due to the low selection pressure on each QTL region resulting from the highly polygenic architecture of the selected trait, the close linkage between the loci contributing to the QTL, and the small effective breeding population, the probability that beneficial recombinant haplotypes are selected and increase in frequency in the population should be low. Another alternative explanation could be that the effects of the linked loci are dependent on the genetic background (epistasis) or dominance, which might have affected the selection pressure on individual contributing loci. As we did not explore the contributions by dominance or epistasis in this study, further work would be necessary to evaluate their contributions to the low fixation in the QTL.
A key for successful imputation of the high-density marker set throughout the AIL pedigree is that the haplotypes across these markers are correctly estimated in the founders. There are several properties of the Virginia lines that improve haplotype estimation from high-density genotypes. First, as the number of generations since the lines diverged is relatively few (40 generations), most new haplotypes will result from recombination of original haplotypes, rather than by new mutations. Second, the strong artificial selection imposed on the populations since they were founded is likely to have further reduced haplotype diversity across the genome. This is likely the reason that many selective-sweeps across long haplotypes have been found to be fixed, or nearly fixed, across the genome within and between the lineages (Johansson et al. 2010; Pettersson et al. 2013). This is reflected in a large average LD-block size (> 50 kb) across the genome (Marklund and Carlborg 2010). Given the density of the 60k SNP-chip genotyping used here, several markers will be present on each such LD-block and hence improve efficiency in haplotype estimation. Additional genotyping will, however, be necessary in subsequent generations to experimentally confirm the associations to imputed markers reported here.
Genotype data are available for all individuals in the AIL pedigree. The dense marker backbone (∼1 marker/cM) from the first genotyping of the AIL (Besnier et al. 2011), allow the relatively long haplotypes that are inherited as intact segments from parents to offspring to be efficiently phased, imputed, and traced throughout the pedigree for later association analyses.
The highly polygenic genetic architecture in this population is consistent with what has been revealed in other fine-mapping analyses in deep intercrosses (Parker et al. 2011) and chromosomal substitution strains (Bevova et al. 2006) involving intensively selected mouse populations. Recent work on a mouse population that has evolved to an extreme body size in nature has also uncovered a highly polygenic architecture of adaptation (Gray et al. 2015), illustrating that complex genetic architectures are likely to be involved in responses to both natural and experimental selection. Further, our detection of multiple associations to nearby markers in our AIL is also consistent with reports from other AIL-based fine-mapping studies in chickens from outbred base-populations (Van Goor et al. 2015) and association studies within and across cattle breeds (Saatchi et al. 2014). Subsequent studies will help to elucidate whether the underlying genetic architecture of associations detected to linked markers in this and other outbred populations are primarily due to the segregation of multiple haplotypes in the outbred founder populations and breeds or a reflection of several tightly linked functional polymorphisms.
Here, the association analysis was performed using a linear model including fixed effects of genotype, sex, and AIL generation. Sex and generation were included as both these environmental factors had significant effects on BW56 (Besnier et al. 2011). Implementing the model selection by backward-elimination in a bootstrap-based framework is a way to account for possible effects of population structure in the AIL that might increase the risk for reporting false positives. However, since the association signals in most cases overlap well with the final marker set resulting from the testing of experiment-wide significant associations, we do not find this to be any cause of great concern in this experiment.
Conclusions
In conclusion, this study shows that the proposed imputation-based association-mapping strategy, and further model selection by backward-elimination in a bootstrap-based framework, is useful for identifying independent association signals within and across the nine evaluated QTL. The association-peaks were narrower than those obtained in the earlier performed linkage analysis, often highlighting regions down to 2–3 Mb in length allowing the identification of multiple association signals in several QTL. This suggests that the association-based strategy has higher resolution, as well as provides an improved power to disentangle the effects of multiple linked loci inside QTL, compared to linkage-based fine-mapping. Combining traditional linkage-based approaches to analyze outbred advanced intercross populations with imputation-based association-mapping approaches might thus be an important and cost-effective approach to improve the efficiency in postassociation bioinformatics analyses and functional explorations aiming to identify candidate mutations. A previous candidate gene study based on the nine QTL fine-mapped here has already reported some interesting mutations in growth-related genes (Ahsan et al. 2013) overlapping with the association signals reported here. Further bioinformatics investigations of the regions fine-mapped here could potentially reveal new important genes and mutations affecting body weight in these chicken lines and provide new candidate genes for studying the genetic architecture of metabolic traits in other species, including humans.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.036012/-/DC1.
Acknowledgments
We thank Leif Andersson for initiating the AIL experiment with P.B.S. and sharing the data from the F2 intercross. Per Wahlberg and Francois Besnier are acknowledged for their valuable contributions during preparation and quality control work of the genotype and phenotype data from the AIL. Genotyping was performed by the SNP&SEQ Technology Platform in Uppsala, which is part of Science for Life Laboratory at Uppsala University and is supported as a national infrastructure by the Swedish Research Council (VR-RFI). Formas (grant 221-2013-450 to Ö.C.) and the Swedish Research Council (grant 621-2012-4634 to Ö.C.) are acknowledged for financial support.
Author contributions: Ö.C. and P.B.S. initiated the study. Ö.C. designed the project with M.A. and M.B.; P.B.S. developed the Virginia chicken lines; P.B.S. designed, planned, and bred the Virginia Advanced Intercross Line; P.B.S. and C.F.H. designed, planned, bred, bled, phenotyped, and extracted DNA from the Virginia Advanced Intercross Line population; Ö.C. designed the statistical analyses; M.A., M.B., and Ö.C. contributed analysis scripts; Ö.C., M.B., and M.A. performed the data analyses, summarized the results, and wrote the manuscript. All authors read and approved the final manuscript.
Footnotes
Communicating editor: D. J. de Koning
Literature Cited
- Abramovich F., Benjamini Y., Donoho D. L., Johnstone I. M., 2006. Special invited lecture: adapting to unknown sparsity by controlling the false discovery rate. Ann. Stat. 34: 584–653. [Google Scholar]
- Ahsan M., Li X., Lundberg A. E., Kierczak M., Siegel P. B., et al. , 2013. Identification of candidate genes and mutations in QTL regions for chicken growth using bioinformatic analysis of NGS and SNP-chip data. Front. Genet. 4: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson L., Georges M., 2004. Domestic-animal genomics: deciphering the genetics of complex traits. Nat. Rev. Genet. 5: 202–212. [DOI] [PubMed] [Google Scholar]
- Aulchenko Y. S., Ripke S., Isaacs A., van Duijn C. M., 2007. GenABEL: an R package for genome-wide association analysis. Bioinformatics 23: 1294–1296. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Besnier F., Wahlberg P., Rönnegård L., Ek W., Andersson L., et al. , 2011. Fine mapping and replication of QTL in outbred chicken advanced intercross lines. Genet. Sel. Evol. 43: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevova M. R., Aulchenko Y. S., Aksu S., Renne U., Brockmann G. A., 2006. Chromosome-wise dissection of the genome of the extremely big mouse line DU6i. Genetics 172: 401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlborg Ö., Jacobsson L., Åhgren P., Siegel P., Andersson L., 2006. Epistasis and the release of genetic variation during long-term selection. Nat. Genet. 38: 418–420. [DOI] [PubMed] [Google Scholar]
- Cheng R., Lim J. E., Samocha K. E., Sokoloff G., Abney M., et al. , 2010. Genome-wide association studies and the problem of relatedness among advanced intercross lines and other highly recombinant populations. Genetics 185: 1033–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler H. D., Wiggans G. R., Hayes B. J., Woolliams J. A., Goddard M. E., 2011. Imputation of missing genotypes from sparse to high density using long-range phasing. Genetics 189: 317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvasi A., Soller M., 1995. Advanced intercross lines, an experimental population for fine genetic mapping. Genetics 141: 1199–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnington E. A., Siegel P. B., 1996. Long-term divergent selection for eight-week body weight in White Plymouth Rock chickens. Poult. Sci. 75: 1168–1179. [DOI] [PubMed] [Google Scholar]
- Dunnington E. A., Honaker C. F., McGilliard M. L., Siegel P. B., 2013. Phenotypic responses of chickens to long-term, bidirectional selection for juvenile body weight–historical perspective. Poult. Sci. 92: 1724–1734. [DOI] [PubMed] [Google Scholar]
- Ek W., Marklund S., Ragavendran A., Siegel P., Muir W., et al. , 2012. Generation of a multi-locus chicken introgression line to study the effects of genetic interactions on metabolic phenotypes in chickens. Front. Genet. 3: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilov Y., Benjamini Y., Sarkar S. K., 2009. An adaptive step-down procedure with proven FDR control under independence. Ann. Stat. 37: 619–629. [Google Scholar]
- Gray M. M., Parmenter M. D., Hogan C. A., Ford I., Cuthbert R. J., et al. , 2015. Genetics of rapid and extreme size evolution in island mice. Genetics 201: 213–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. G., 2005. A century of corn selection. Science 307: 683–684. [DOI] [PubMed] [Google Scholar]
- Jacobsson L., Park H.-B., Wahlberg P., Fredriksson R., Perez-Enciso M., et al. , 2005. Many QTLs with minor additive effects are associated with a large difference in growth between two selection lines in chickens. Genet. Res. 86: 115–125. [DOI] [PubMed] [Google Scholar]
- Johansson A. M., Pettersson M. E., Siegel P. B., Carlborg Ö., 2010. Genome-wide effects of long-term divergent selection. PLoS Genet. 6: e1001188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper K. E., Visscher P. M., Goddard M. E., 2012. Genetic architecture of body size in mammals. Genome Biol. 13: 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Rouzic A., Carlborg Ö., 2008. Evolutionary potential of hidden genetic variation. Trends Ecol. Evol. 23: 33–37. [DOI] [PubMed] [Google Scholar]
- Le Rouzic A., Siegel P. B., Carlborg Ö., 2007. Phenotypic evolution from genetic polymorphisms in a radial network architecture. BMC Biol. 5: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marklund S., Carlborg Ö., 2010. SNP detection and prediction of variability between chicken lines using genome resequencing of DNA pools. BMC Genomics 11: 665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez G. C., Siegel P. B., Lewis R. M., 2010. Genetic diversity and population structure in lines of chickens divergently selected for high and low 8-week body weight. Poult. Sci. 89: 2580–2588. [DOI] [PubMed] [Google Scholar]
- Park H.-B., Jacobsson L., Wahlberg P., Siegel P. B., Andersson L., 2006. QTL analysis of body composition and metabolic traits in an intercross between chicken lines divergently selected for growth. Physiol. Genomics 25: 216–223. [DOI] [PubMed] [Google Scholar]
- Parker C. C., Cheng R., Sokoloff G., Lim J. E., Skol A. D., et al. , 2011. Fine-mapping alleles for body weight in LG/J × SM/J F2 and F34 advanced intercross lines. Mamm. Genome 22: 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirce J. L., Broman K. W., Lu L., Chesler E. J., Zhou G., et al. , 2008. Genome reshuffling for advanced intercross permutation (GRAIP): simulation and permutation for advanced intercross population analysis. PLoS One 3: e1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson M., Besnier F., Siegel P. B., Carlborg Ö., 2011. Replication and explorations of high-order epistasis using a large advanced intercross line pedigree. PLoS Genet. 7: e1002180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson M. E., Johansson A. M., Siegel P. B., Carlborg Ö., 2013. Dynamics of adaptive alleles in divergently selected body weight lines of chickens. G3 (Bethesda) 3: 2305–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team, 2015 R: A Language and Environment for Statistical Computing. https://www.r-project.org.
- Rönnegård L., Besnier F., Carlborg Ö., 2008. An improved method for quantitative trait loci detection and identification of within-line segregation in F2 intercross designs. Genetics 178: 2315–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saatchi M., Schnabel R. D., Taylor J. F., Garrick D. J., 2014. Large-effect pleiotropic or closely linked QTL segregate within and across ten US cattle breeds. BMC Genomics 15: 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Z., Pettersson M. E., Honaker C. F., Siegel P. B., Carlborg Ö., 2015. Standing genetic variation as a major contributor to adaptation in the Virginia chicken lines selection experiment. Genome Biol. 16: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel P. B., 1962a A double selection experiment for body weight and breast angle at eight weeks of age in chickens. Genetics 47: 1313–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel P. B., 1962b Selection for body weight at eight weeks of age. 1. Short term response and heritabilities. Poult. Sci. 41: 954–962. [Google Scholar]
- Valdar W., Solberg L. C., Gauguier D., Burnett S., Klenerman P., et al. , 2006. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat. Genet. 38: 879–887. [DOI] [PubMed] [Google Scholar]
- Valdar W., Holmes C. C., Mott R., Flint J., 2009. Mapping in structured populations by resample model averaging. Genetics 182: 1263–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Goor A., Bolek K. J., Ashwell C. M., Persia M. E., Rothschild M. F., et al. , 2015. Identification of quantitative trait loci for body temperature, body weight, breast yield, and digestibility in an advanced intercross line of chickens under heat stress. Genet. Sel. Evol. 47: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlberg P., Carlborg Ö., Foglio M., Tordoir X., Syvänen A.-C., et al. , 2009. Genetic analysis of an F(2) intercross between two chicken lines divergently selected for body-weight. BMC Genomics 10: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo C. K., Park H.-B., Lee J. B., Jung E. J., Kim B. M., et al. , 2014. QTL analysis of body weight and carcass body length traits in an F2 intercross between Landrace and Korean native pigs. Anim. Genet. 45: 589–592. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genotype, phenotype, and pedigree data are included in the supplemental files. Supplemental Material, File S1 contains detailed descriptions of all supplemental data files. File S2 contains the genotypes, File S3 the pedigree, and File S4 the phenotypes.