Abstract
Brassica napus introgression lines (ILs), having B-genome segments from B. carinata, were assessed genetically for extent of introgression and phenotypically for siliqua shatter resistance. Introgression lines had 7–9% higher DNA content, were meiotically stable, and had almost normal pollen fertility/seed set. Segment introgressions were confirmed by fluorescent genomic in situ hybridization (fl-GISH), SSR analyses, and SNP studies. Genotyping with 48 B-genome specific SSRs detected substitutions from B3, B4, B6, and B7 chromosomes on 39 of the 69 ILs whereas SNP genotyping detected a total of 23 B-segments (≥3 Mb) from B4, B6, and B7 introgressed into 10 of the 19 (C1, C2, C3, C5, C6, C8, C9, A3, A9, A10) chromosomes in 17 ILs. The size of substitutions varied from 3.0 Mb on chromosome A9 (IL59) to 42.44 Mb on chromosome C2 (IL54), ranging from 7 to 83% of the recipient chromosome. Average siliqua strength in ILs was observed to be higher than that of B. napus parents (2.2–6.0 vs. 1.9–4.0 mJ) while siliqua strength in some of the lines was almost equal to that of the donor parent B. carinata (6.0 vs.7.2 mJ). These ILs, with large chunks of substituted B-genome, can prove to be a useful prebreeding resource for germplasm enhancement in B. napus, especially for siliqua shatter resistance.
Keywords: oilseed rape, siliqua shatter resistance, fl-GISH, graphical genotyping, SNP genotyping
A relatively young polyploid, Brassica napus originated through several hybridization events between its diploid progenitors B. rapa (AA; 2n = 20) and B. oleracea (CC; 2n = 18) (Allender and King 2010). The geographic center of origin of B. napus is unknown, as records of this crop over the last 500 yr do not predate its origin (Gómez-Campo and Prakash 1999). Although feral populations are common in Europe, there is no evidence of truly wild B. napus populations (Prakash et al. 2011). Due to its relatively recent origin, probably as an agricultural hybrid, it has low allelic diversity as compared to that of its progenitors. Genetic studies based on DNA polymorphisms show a narrow genetic base (Diers et al. 1996; Lombard et al. 2000; Cartea et al. 2005; Zhou et al. 2006; Chen et al. 2008; Wang et al. 2009; Gyawali et al. 2013) with germplasm tending to cluster by growth habit (Qian et al. 2006; Zhou et al. 2006; Chen et al. 2008) or geographic origin (Hu et al. 2007). The inherent problem of low genetic diversity in B. napus has been further intensified by aggressive plant breeding efforts toward improving oil quality and facilitating adaptation to restricted photoperiod and temperature regimes. Plant breeders are now confronted with diminishing selection gains for yield, oil content, and adaptability to climatic challenges.
Loss of genetic diversity in the modern cultivars of B. napus is well documented in Australia (Cowling 2007; Wang et al. 2009), Canada (Fu and Gugel 2010), and Germany (Abbadi and Leckband 2011). There is evidence that the C-genome of B. napus possesses a relatively narrow genetic base compared to the A-genome (Bus et al. 2011). Efforts to broaden genetic diversity in B. napus through interspecific hybridization with related species have also benefited the A-genome more than the C-genome (Rahman 2013).
Acquiring new genetic diversity in breeding programs is crucial for continuous germplasm enhancement. Introgression of novel genes from progenitor species through direct resynthesis has shown excellent potential in enhancing heterosis of resynthesized rapeseed genotypes (Girke et al. 1999; Gehringer et al. 2007). However, a serious limitation to this approach is the association of undesirable linkages in terms of poor seed quality traits and unacceptable agronomic characteristics (Girke et al. 1999). In spite of the pervasive limitation of linkage drag, the introgression of exotic alleles from progenitor diploids, related allopolyploids, and wild crucifers is crucial for a continuous pipeline of canola germplasm improvement, both for qualitative and quantitative traits such as pest resistance and productivity.
Development of genetically more diverse B. napus lines through substitution of the B. napus A-genome with the A-genome of B. rapa and substitution of the B. napus C-genome with the C-genome of B. carinata has also been attempted. This method involved development of trigenomic hexaploids (AABBCC), followed by subsequent backcrossing to natural B. napus (Li et al. 2004, 2006; Xiao et al. 2010). The resulting B. napus lines carried >75% of the genome from B. rapa (A-genome) and B. carinata (C-genome). Introgression lines (ILs) can provide a promising avenue to effectively exploit the genetic potential of related crop allopolyploids and wild species. B. carinata (BBCC; 2n = 34) is a known source of tolerance to many biotic (Navabi et al. 2010) and abiotic stresses. However, attempts to introgress genetic variation from the B-genome of B. carinata have largely been unsuccessful; B-genome chromosomes were either eliminated in the early generations (Li et al. 2004; Schelfhout et al. 2006) or retained as intact (B1, B3, B6, B7 and B8) or broken chromosomes in advanced progenies of interspecific crosses (Navabi et al. 2010, 2011; Fredua-Agyeman et al. 2014).
In this communication, we report the cytogenetic and molecular characterization of 69 B-genome ILs of B. napus. These ILs were synthesized with the aim of introgressing useful gene(s), especially siliqua shatter resistance from B-genome of B. carinata, as siliqua shattering is a major commercial cultivation bottleneck in B. napus.
Materials and Methods
Synthesis of ILs
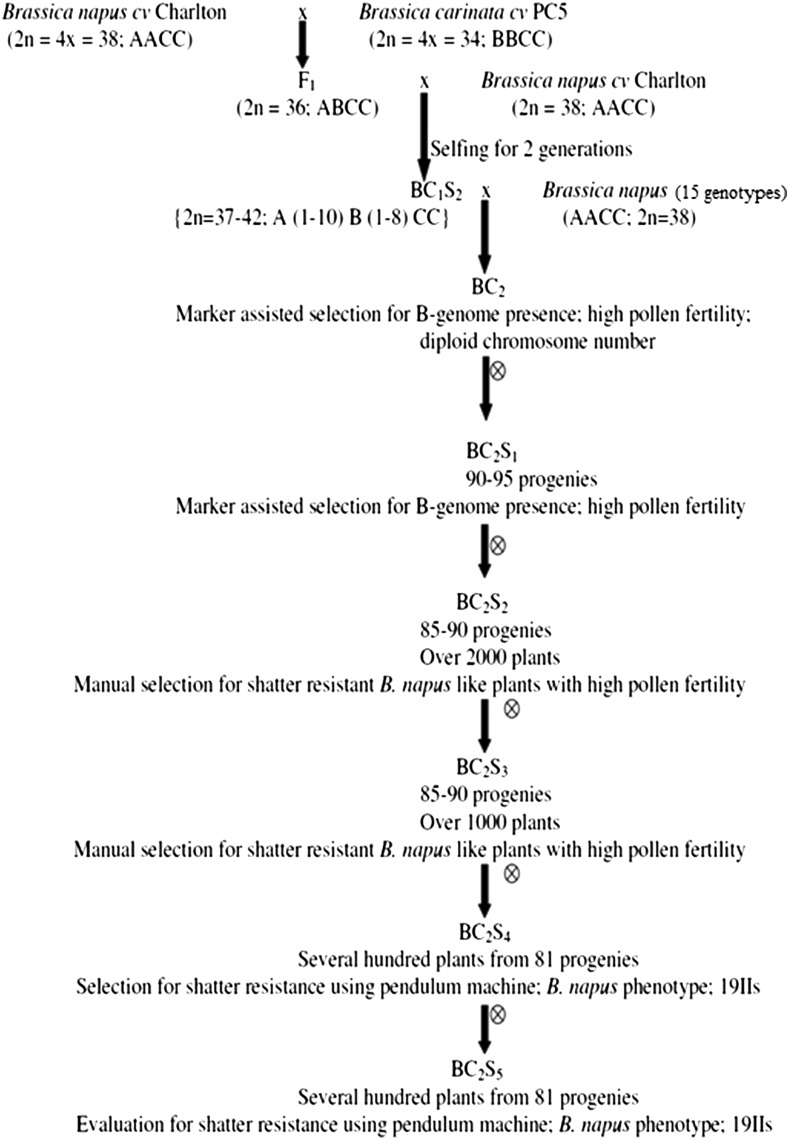
Continuous attempts have been made over the past 10 years to introgress genes associated with pod shatter resistance from B. carinata cv. PC5 into the genetic background of a group of 15 B. napus genotypes. The procedure followed for the synthesis of ILs is depicted in Figure 1. The crossing scheme was designed to promote homologous chromosome pairing between the C-genome chromosomes of B. napus and B. carinata, but homeologous exchanges were also expected to take place between combining B-/A- and B-/C-genomes especially during backcross generations. A large plant population (over 200 plants) was raised in each backcross or selfed generation. Every plant was assayed for pollen grain stainability and also probed using B-genome-specific chromatin marker pBNBH35 (Gupta et al. 1992; Schelfhout et al. 2004) to identify putative plants with B-genome introgressions. A strong phenotypic selection pressure for siliqua shatter resistance was applied between the BC2S1 and BC2S5 generations. Plants with high pollen grain stainability and hard-to-shatter siliquae only were retained for further generations of selfing and backcrossing. A total of 81 ILs were retained at the end of BC2S5.
Figure 1.
Breeding strategy followed for developing B. napus–B. carinata introgression lines. B. napus cv. Charlton was crossed with B. carinata cv. PC5 and the resultant F1 was backcrossed to Charlton. BC1 was selfed for two generations to get BC1S2 which was crossed with 15 diverse B. napus pollen parents. Resulting 15 BC2 progenies were subjected to intense selection and thereafter carried upto BC2S5 as depicted above.
Phenotyping of ILs for pod shatter resistance
The set of 81 B. napus ILs was raised along with 15 B. napus recipient genotypes under two environments during 2012–2013 and one environment during 2013–2014 in an α lattice design with two replications. Paired rows each of B. carinata cv. PC5 (donor for siliqua shatter resistance) and B. nigra cv. UP (diploid B-genome parent of B. carinata) were raised separately, to be used as controls for estimation of siliqua shatter energy as well as for molecular analysis. Since high-quality DNA, required for SNP genotyping, could only be obtained for 69 ILs, five B. napus parents, B. carinata cv. PC 5, and B. nigra cv. UP, this study reports results for molecular analysis on the basis of these lines only.
Estimation of nuclear DNA content
Self-pollinated progeny from the introgressed material was analyzed in the BC1S5 generation to determine variation in genome size and ploidy level using a standard flow cytometry DNA content estimation protocol (Doležel and Bartoš 2005). The PartecCyStain UV precise P reagent kit was used for nuclei extraction and DNA staining. Approximately 1–2 mg of young leaf tissue was finely chopped with a sharp razor blade in 400 μl of extraction buffer. The sample was filtered into a test tube containing 1.6 ml staining solution through a 50 μm CellTrics filter. This solution was incubated at room temperature for 30–60 sec and then ∼10,000 nuclei per sample were analyzed using a Partec CyFlow Ploidy Analyzer (Partec, Münster, Germany). Before analyzing samples on a particular day, the flow cytometer was calibrated first using fixed chicken red blood cells, having a known 2C DNA content of 2.33–2.5 pg, and then using an international DNA reference standard Lycopersicon esculentum var stupicke polni tyckoverane, which has a known 2C DNA content value of 1.96 pg (Doležel et al. 1992). B. carinata was used as an internal standard, with its DNA content determined in relation to the Lycopersicon sample. The absolute DNA amount of a sample was computed using the formula:
For determining the genome size, the 1C content of the sample was multiplied by 978 Mb since 1 pg has been reported to be equal to 978 Mb (Doležel et al. 2003).
Cytogenetic studies
Pollen grain stainability was determined from squash preparations of five freshly dehisced anthers in 1% acetocarmine solution. A total of five plants from each genotype were sampled for the purpose. For meiotic studies, squash preparations of anthers at the appropriate stage were made in 2% acetocarmine, and at least 50 PMCs (pollen mother cells) per introgression line with well spread metaphase-I/diakinesis/anaphase-I were observed under an Olympus BX61TRF microscope.
Fluorescent genomic in situ hybridization (fl-GISH)
The GISH protocol as proposed by Schwarzacher and Heslop-Harrison (2000) was followed with minor modifications, to study B-genome introgressions.
Preparation of chromosome spreads:
Flower buds were fixed directly in Farmer’s solution (three parts ethanol: one part acetic acid) in the early morning hours. To remove the fixative, anthers of appropriate size were washed in a citrate buffer for 30 min and subsequently incubated in an enzyme mixture containing 2% (v/w) cellulase and 20% (v/v) pectinase in 4 mmol/l citrate buffer (pH 4.8) for 1 hr at 37°. These anthers were agitated in a microcentrifuge tube to release PMCs, centrifuged for 3 min at 600–800 × g, and then treated for 45 min in 150 mmol/l KCl. The PMCs were washed three times at 800 × g for 3 min in freshly prepared fixative to clear the cytoplasm. One drop of 7 µl PMC suspension was released on acid-cleaned chilled slides for proper spread. Slides were air-dried in a desiccator before further use.
Preparation of probes:
Purified DNA of B. nigra cv UP, extracted using the DNeasy plant mini kit (Qiagen), was used for preparing the probe. DNA was sheared in an autoclave for 2 min and then allowed to cool slowly for reannealing. Sheared DNA of the desired fragment size (500–1000 bp) was labeled with Tetramethyl-Rhodamine-5-dUTP (Sigma Aldrich) dye using a nick-translation kit. The DNA of B. napus (200–500 bp) was autoclaved and was used as a blocker at 200 times the probe concentration to prevent nonspecific intergenomic cross-hybridization.
In situ hybridization:
Forty microliters of hybridization mixture containing 50% formamide, 2× SSC (saline sodium citrate buffer), 10% dextran sulfate, 0.025 µg salmon sperm DNA, 1.25 mM EDTA, 0.1255% acetic acid, 200 ng of labeled probe, and 200-fold blocking DNA was applied on slides with good chromosome spreads, followed by incubation at 80° for 4 min in a thermocycler for simultaneous denaturation of probe and chromosomes. These slides were kept for hybridization overnight at 37° and then washed at 42° for 2 min in 2× SSC and twice for 5 min in 0.1× SSC. Slides were dehydrated through an ethanol series (70, 90, and 96% for 2 min each) and air-dried. Chromosomes were counterstained with DAPI (4′, 6-diamidino-2-phenylindole) by incubating slides with 100 µl DAPI solution (4 µg/ml in McIlvaine’s buffer) at room temperature for 10–30 min in the dark. Slides were rinsed in detection buffer and two drops of antifade was added. Fluorescence was visualized using a Carl Zeiss microscope (Imager Z2AX10). At least 25 cells were photographed using a computer-assisted cooled charge-coupled device camera and images were merged with Image-I software.
DNA extraction
Fresh leaves of individual ILs were collected at the juvenile plant stage and ground in liquid nitrogen. DNA was extracted from the ground tissues using the CTAB (cetyltrimethylammonium bromide) method (Doyle and Doyle 1990). The DNA was lyophilized and sent to the Centre for Integrative Legume Research (CILR) Laboratory, The University of Queensland, Australia for SNP genotyping.
B-genome-specific SSR marker allele presence in ILs
The 69 ILs were analyzed using 48 SSR markers (at six markers per chromosome) across the B-genome (Dr. Isobel Parkin, personal communication available under a materials transfer agreement from Agriculture and AgriFood Canada). Known approximate map positions were also supplied for these markers. The 10.3 µl of PCR master mix contained 5 µl of 5 ng/µl template DNA, 1 µl of 10× PCR buffer with 15 mM MgCl2 (Sigma Aldrich), 1 µl of 10 µM each primer, 2 µl of 1 mM dNTPS (Sigma Aldrich), and 0.3 µl of 3 U/µl Taq DNA polymerase (Sigma Aldrich). PCR analysis was carried out in 96-well PCR plates using an Eppendorf AG 6325 thermocycler on the following profile: initial cycle of denaturation at 94° for 5 min; 35 amplification cycles each of denaturation at 94° for 30 sec, optimal annealing temperature for 30 sec, elongation at 72° for 1 min, and a final extension cycle at 72° for 7 min. The amplified DNA product was resolved using 6% nondenaturing polyacrylamide gel and visualized using a Syngene Gene Genius gel documentation system. The size of PCR product was recorded with reference to a 100-bp ladder. B. rapa (AA), B. oleracea (CC), B. nigra (BB), B. carinata (BBCC), B. juncea (AABB), and B. napus (AACC) were used as controls. Any marker that showed amplification in B. rapa (AA), B. oleracea (CC), or B. napus (AACC) was rejected. Markers which showed expected size amplification in all three B-genome-bearing species, i.e., B. nigra (BB), B. juncea (AABB) or B. carinata (BBCC), were retained to detect the presence of B-chromosomes in the ILs. Thus, 22 markers representing all the eight B-genome chromosomes amplified on the ILs. Further, introgression was only considered if two or more consecutive markers from a single chromosome were present in an IL. Consequently, 11 markers (SJ7046, SB1752, SB2141AI, SB1935A, SJ1505, SJ0338, SJ0502, SJ7104, SB31138, SJ39119I, SJ13133) representing four chromosomes were such that met these very stringent criteria. Software Graphical GenoTypes v.2.0 (vanBerloo 2008) was used for graphical representation of the polymorphism data thus generated to confirm the presence of B-genome alleles in each IL (Supplemental Material, Figure S1).
SNP genotyping
An Illumina Infinium 60k SNP array developed for B. napus (http://www.illumina.com), carrying 52,157 SNPs across the 19 A- and C-genome chromosomes, was used for genotyping. Hybridization protocols were run as per manufacturer’s specifications for all samples in the population and controls (parent species). The chips were scanned using Illumina HiScanSQ. Genotypic data were visualized using Genome Studio v.2011.1 (Illumina, Inc., San Diego, CA). Single nucleotide polymorphisms were filtered in Genome Studio to retain genome-specic SNPs. Polymorphic SNP markers were distributed evenly across all 19 B. napus chromosomes. SNPs that did not show three clear genotype clusters (AA, AB, BB) in the population were removed. As the available SNP chip was designed for A- and C-genomes it could not be used directly to score B-genome chromatin substitutions. All SNPs giving >20% hetero calls (AB), >10% no calls (NC), or cross-amplifying the B-genome of B. nigra/B. carinata were removed. Finally, 1458 A-genome and 1936 C-genome SNPs (a total of 3394) were retained. NCs were recorded for many SNPs in several regions of ILs. Strings of NCs ≥3 Mb were color coded and treated as B-genome introgressions. All the preexisting A–C translocations in recipient B. napus genotypes were not included in the introgression count.
Phenotyping for resistance to siliqua shatter
The set of 81 ILs and 15 B. napus parents grown in the replicated trial under three environments were compared with hard to thresh donor parent B. carinata cv. PC5 for introgression of shatter resistance. Five plants were randomly tagged from the two center rows of each plot in each replicate in each environment. Five siliquae from the center of the main raceme were carefully detached at physiological maturity and stored in coarse silica gel blue self-indicating granules to equilibrate them to constant moisture content at room temperature. The siliquae were oven dried at 70° for 24 hr immediately before assessing siliqua strength. The relative resistance to siliqua shatter was measured in terms of rupture energy using an improvised pendulum apparatus (Liu et al. 1994; Kadkol 2009) wherein the pendulum strikes the siliqua with a known force and records the energy absorbed to split it open in millijoules.
Data availability
All data for confirming the conclusions in this research paper are represented in the figures and tables. Detailed data will be made available on request.
Results
Genome size variation, meiotic configuration, and pollen fertility in ILs
The 1C genome size in the ILs ranged from 1246 to 1336 Mb (Table 1), which is 7–9% higher than the reported genome size range of 1165–1222 Mb for B. napus (http://www.brassica.info/info/reference/genome-sizes.php). Analysis of the meiotic configurations revealed a mode of 19 bivalents at metaphase-I with regular anaphase chromosome separation (Table 1) in all evaluated ILs. Only a few PMCs showed rare univalent (<0.12 per cell) or multivalent (<0.04 per cell) formations. Pollen grain stainability varied from 60% (IL43) to 97.5% (IL11, IL12, IL24, IL46). Seed set on self-pollination was normal, suggesting high female fertility.
Table 1. Pollen fertility, meiotic configuration, genome size variation, and B-genome presence in B. napus introgression lines.
| Sr. No. | Introgression Line | 1C Genome Size (Mb) | 2C DNA Content (pg) | Meiotic Configuration | Pollen Fertility (%) | Presence of B-Genome-Specific Marker “pBNBH35” | |
|---|---|---|---|---|---|---|---|
| Metaphase | Anaphase | ||||||
| (2n = 38) | (n = 19) | ||||||
| 1 | IL1 | 1280 | 2.62 | ─ | ─ | 85.0 | + |
| 2 | IL3 | 1257 | 2.57 | ─ | ─ | 92.5 | + |
| 3 | IL4 | ─ | ─ | 19 IIs | 19 Is | 97.0 | + |
| 4 | IL5 | 1336 | 2.73 | ─ | ─ | 82.5 | + |
| 5 | IL6 | ─ | ─ | ─ | ─ | 90.0 | + |
| 6 | IL7 | ─ | ─ | ─ | ─ | 90.0 | + |
| 7 | IL9 | 1246 | 2.55 | 19 IIs | 19 Is | 92.5 | + |
| 8 | IL10 | 1336 | 2.73 | ─ | ─ | 90.0 | + |
| 9 | IL11 | 1308 | 2.67 | ─ | ─ | 97.5 | + |
| 10 | IL12 | 1285 | 2.63 | 19 IIs | 19 Is | 97.5 | + |
| 11 | IL13 | ─ | ─ | ─ | ─ | 87.5 | + |
| 12 | IL14 | 1302 | 2.66 | 19 IIs | 19 Is | 90.0 | + |
| 13 | IL16 | 1285 | 2.63 | 19 IIs | 19 Is | 90.0 | + |
| 14 | IL17 | ─ | ─ | ─ | ─ | 90.0 | + |
| 15 | IL18 | 1274 | 2.61 | ─ | ─ | 87.5 | + |
| 16 | IL19 | ─ | ─ | ─ | ─ | 90.0 | + |
| 17 | IL20 | 1285 | 2.63 | ─ | ─ | 90.0 | − |
| 18 | IL21 | 1274 | 2.61 | ─ | ─ | 77.5 | + |
| 19 | IL22 | 1325 | 2.71 | ─ | ─ | 97.0 | − |
| 20 | IL23 | 1285 | 2.63 | 19 IIs | 19 Is | 82.5 | + |
| 21 | IL24 | 1313 | 2.69 | ─ | ─ | 97.5 | + |
| 22 | IL26 | ─ | ─ | ─ | ─ | 92.5 | + |
| 23 | IL27 | 1302 | 2.66 | ─ | ─ | 85.0 | + |
| 24 | IL28 | ─ | ─ | 19 IIs | 19 Is | 67.5 | + |
| 25 | IL30 | ─ | ─ | ─ | ─ | 87.5 | − |
| 26 | IL31 | ─ | ─ | ─ | ─ | 72.5 | + |
| 27 | IL32 | 1302 | 2.66 | 19 IIs | 19 Is | 90.0 | + |
| 28 | IL33 | ─ | ─ | ─ | ─ | 77.5 | + |
| 29 | IL34 | 1319 | 2.7 | 19 IIs | 19 Is | 87.5 | + |
| 30 | IL37 | ─ | ─ | 19 IIs | 19 Is | 67.5 | + |
| 31 | IL38 | 1280 | 2.62 | ─ | ─ | 72.5 | + |
| 32 | IL39 | ─ | ─ | 19 IIs | 19 Is | 62.5 | + |
| 33 | IL40 | 1268 | 2.59 | 19 IIs | 19 Is | 65.0 | + |
| 34 | IL41 | ─ | ─ | 19 IIs | 19 Is | 87.5 | + |
| 35 | IL42 | ─ | ─ | 19 IIs | 19 Is | 85.0 | + |
| 36 | IL43 | 1280 | 2.62 | ─ | ─ | 60.0 | + |
| 37 | IL44 | ─ | ─ | ─ | ─ | 75.0 | + |
| 38 | IL45 | 1319 | 2.7 | ─ | ─ | 77.5 | + |
| 39 | IL46 | 1330 | 2.72 | ─ | ─ | 97.5 | + |
| 40 | IL47 | 1280 | 2.62 | 19 IIs | 19 Is | 92.5 | + |
| 41 | IL48 | ─ | ─ | 19 IIs | 19 Is | 95.0 | − |
| 42 | IL49 | ─ | ─ | ─ | ─ | 85.0 | + |
| 43 | IL50 | 1308 | 2.67 | ─ | ─ | 75.0 | + |
| 44 | IL51 | ─ | ─ | ─ | ─ | 72.5 | + |
| 45 | IL52 | 1296 | 2.65 | 19 IIs | 19 Is | 82.5 | − |
| 46 | IL53 | ─ | ─ | ─ | ─ | 77.5 | − |
| 47 | IL54 | ─ | ─ | ─ | ─ | 82.5 | + |
| 48 | IL56 | ─ | ─ | ─ | ─ | 87.5 | + |
| 49 | IL57 | ─ | ─ | ─ | ─ | 62.5 | − |
| 50 | IL59 | ─ | ─ | ─ | ─ | 77.5 | + |
| 51 | IL61 | ─ | ─ | ─ | ─ | 77.5 | + |
| 52 | IL62 | 1297 | 2.65 | ─ | ─ | 90.0 | + |
| 53 | IL63 | ─ | ─ | ─ | ─ | 75.0 | − |
| 54 | IL64 | ─ | ─ | ─ | ─ | 67.5 | + |
| 55 | IL65 | 1285 | 2.63 | ─ | ─ | 77.5 | + |
| 56 | IL66 | 1274 | 2.61 | ─ | ─ | 90.0 | − |
| 57 | IL67 | 1274 | 2.61 | ─ | ─ | 62.5 | + |
| 58 | IL68 | ─ | ─ | 19 IIs | 19 Is | 92.5 | − |
| 59 | IL69 | ─ | ─ | ─ | ─ | 87.5 | + |
| 60 | IL70 | 1325 | 2.71 | ─ | ─ | 92.5 | + |
| 61 | IL71 | ─ | ─ | ─ | ─ | 75.0 | + |
| 62 | IL72 | ─ | ─ | 19 IIs | 19 Is | 80.0 | + |
| 63 | IL73 | 1325 | 2.71 | ─ | ─ | 90.0 | + |
| 64 | IL74 | ─ | ─ | 19 IIs | 19 Is | 60.0 | + |
| 65 | IL75 | ─ | ─ | ─ | ─ | 85.0 | + |
| 66 | IL76 | ─ | ─ | ─ | ─ | 85.0 | + |
| 67 | IL78 | ─ | ─ | ─ | ─ | 85.0 | + |
| 68 | IL79 | 1330 | 2.72 | ─ | ─ | 85.0 | + |
| 69 | IL80 | ─ | ─ | 19 IIs | 19 Is | 80.0 | + |
Fluorescent genomic in situ hybridization
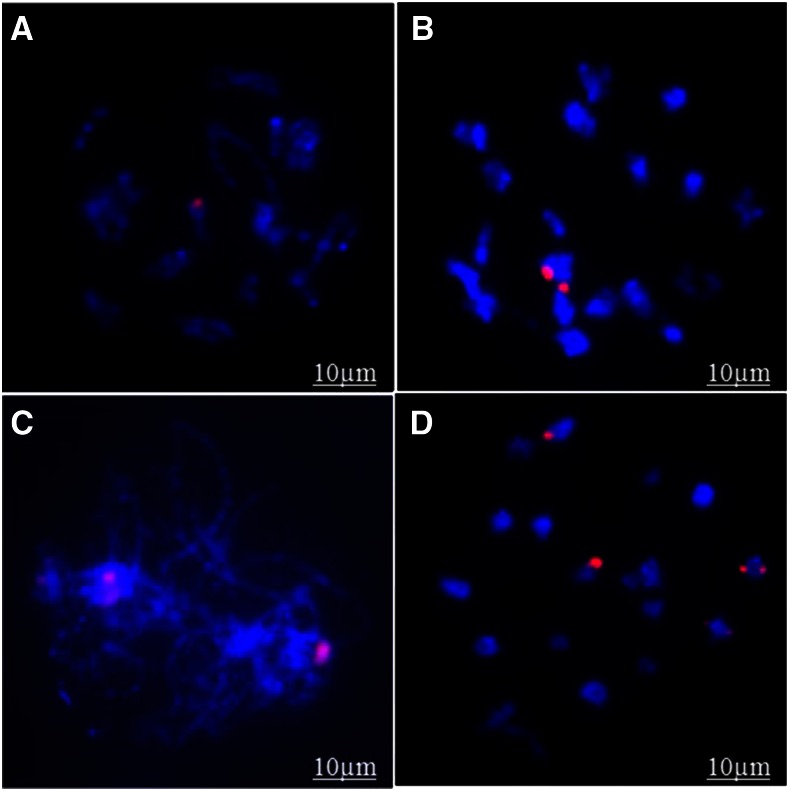
GISH with a B. nigra chromatin probe did not show any B-genome signal in any of the B. napus controls whereas 1–4 bivalents having B-genome fragment substitutions (stained red) were observed in the ILs (Figure 2). The remaining 18-15 A-/C-genome bivalents counter-stained blue with DAPI and had no detectable (≥10 kbp) fragment substitution.
Figure 2.
GISH signals showing the substituted B-genome fragment(s) in B. napus introgression lines, (A) IL16 at metaphase, (B) IL75 at metaphase, (C) IL69 at pachytene, (D) IL59 at metaphase. All 19 A-/C-genome chromosomes are characterized by blue color due to staining with DAPI. B-genome chromosome introgressions are expressed in red color due to labeling with B-genome specific probe.
Genotyping with B-genome-specific SSR primers
Molecular genotyping with chromosome-specific SSR markers allowed us to identify B-genome chromosomes involved in the introgressions (Figure 3, Figure S1, and Table 2). Out of the evaluated 48 SSR markers from eight B-genome chromosomes (at six per chromosome), only 22 markers representing all the eight chromosomes got amplified. Further, this number was reduced to only 11 markers (SJ7046, SB1752, SB2141AI, SB1935A, SJ1505, SJ0338, SJ0502, SJ7104, SB31138, SJ39119I, SJ13133) from chromosomes B3, B4, B6, and B7, respectively, due to the stringent criterion followed for marking an introgression as positive, i.e., the amplification of two or more consecutive markers on a single chromosome in an inclusive introgression. On this basis, 39 of the 69 evaluated ILs had introgressed B-genome fragments. Number of fragments introgressed per IL varied from one to five; 13 ILs had 1, 11 had 2, 9 had 3, 5 had 4, and 1 had 5 introgressed fragments. An interstitial marker SJ7046 and a terminal marker SB1752 of chromosome B3 showed amplification in five ILs. Two markers, SB2141AI and SB1935A from chromosome B4 showed amplification in eight ILs (IL4, IL6, IL9, IL31, IL39, IL73, IL79, IL80). Out of the six B6 markers used, five (SB31138, SJ7104, SJ0338, SJ1505, SJ0502) showed amplification in 28 ILs: consecutive markers SB31138 and SJ7104 amplified in 3 ILs (IL27, IL59, IL61); SJ7104 and SJ0338 amplified in 26 ILs and SJ1505 and SJ0502 amplified in 4 ILs. Interestingly all the five polymorphic B6 markers amplified in IL59. Interstitial markers SJ39119I and SJ13133 confirmed the presence of substituted B-genome fragments from chromosome B7 in 14 ILs.
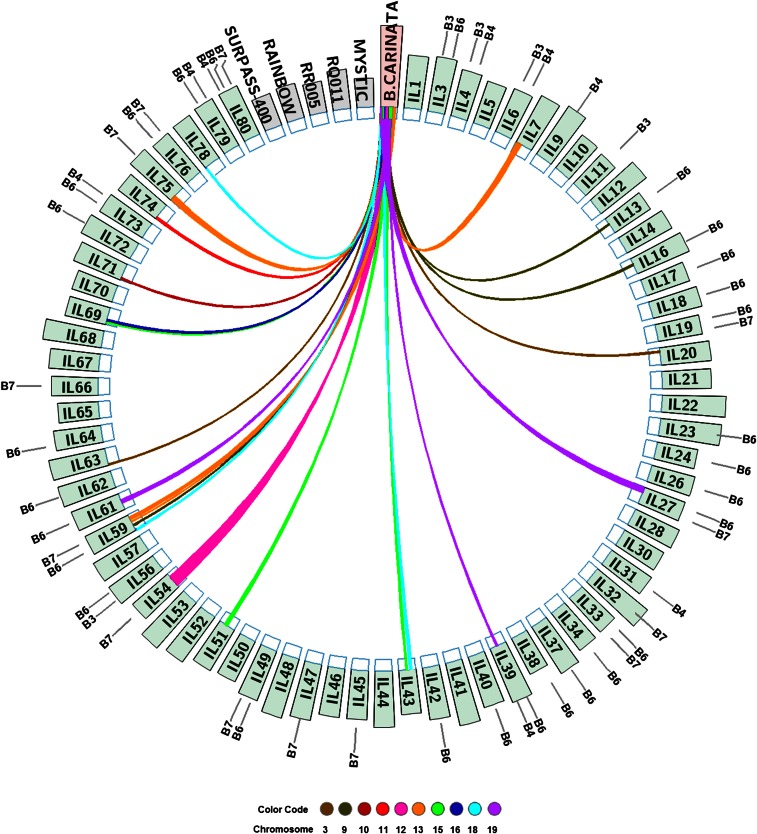
Figure 3.
CIRCOS diagram depicting ILs having B-genome fragment introgression(s) > 3 Mb in specific A-/C- chromosomes based on SNP analysis (inner circle), comparative siliqua shatter energy (outer circle) and B-genome chromosomes involved in introgression in different ILs based on SSR analysis (outermost notations).
Table 2. B-genome chromosome segment substitutions as per SNP assays in the A- and C-genome chromosomes of B. napus introgression lines.
| Introgression Line | Number of Introgressed B-Fragments | Recipient Chromosome (A/C) | Size of the Recipient (A/C) Chromosome (Mb) | Size of the Introgressed B-Fragment (Mb) | Relative Size of the Introgressed Fragment (%) | Putative B-Chromosome Associateda |
|---|---|---|---|---|---|---|
| IL7 | 1 | C3 | 67.05 | 18.73 | 27.93 | — |
| IL13 | 1 | A9 | 37.98 | 4.25 | 11.18 | B6 |
| IL16 | 1 | A9 | 37.98 | 4.52 | 11.90 | B6 |
| IL20 | 1 | A3 | 35.76 | 3.44 | 9.62 | — |
| IL27 | 1 | C9 | 52.92 | 20.53 | 38.79 | B6/B7 |
| IL39 | 1 | C9 | 52.92 | 3.99 | 7.54 | B4/B6 |
| IL43 | 2 | C5 | 46.89 | 4.36 | 9.30 | — |
| — | — | C8 | 42.97 | 6.70 | 15.60 | — |
| IL51 | 1 | C5 | 46.89 | 6.87 | 14.65 | — |
| IL54 | 1 | C2 | 51.34 | 42.44 | 82.66 | B7 |
| IL59 | 4 | A9 | 37.98 | 3.00 | 7.90 | B6/B7 |
| — | — | C3b | 67.05 | 20.96 | 31.26 | B6/B7 |
| — | — | C8 | 42.97 | 3.11 | 7.23 | B6/B7 |
| IL61 | 1 | C9 | 52.92 | 23.65 | 44.70 | B6 |
| IL63 | 1 | A3 | 35.76 | 3.55 | 9.93 | — |
| IL69 | 3 | C2 | 51.34 | 12.33 | 24.01 | — |
| — | — | C5 | 46.89 | 4.97 | 10.60 | — |
| — | — | C6 | 40.57 | 4.35 | 10.72 | — |
| IL71 | 1 | A10 | 19.66 | 4.25 | 21.63 | — |
| IL74 | 1 | C1 | 43.24 | 7.20 | 16.66 | — |
| IL75 | 1 | C3 | 67.05 | 18.88 | 28.16 | B7 |
| IL78 | 1 | C8 | 42.97 | 3.78 | 8.81 | — |
As per B-genome-specific SSRs.
Chromosome C3 has two introgressed B-fragments of the collective size of 20.96 Mb.
SNP genotyping to delineate introgressed segments
A stringent criterion (size ≥3.0 Mb) followed to classify a translocation allowed the detection of 23 substituted segments into 17 ILs. These substituted chromosome segments are enumerated in Table 2 and depicted through the circular genome data visualization software (CIRCOS) (Krzywinski et al. 2009) in Figure 3. Seven C-genome chromosomes (C1, C2, C3, C5, C6, C8, and C9) and three A-genome chromosomes (A3, A9, and A10) showed substituted B-chromosome fragments in different ILs. The majority of the ILs (14 of the 17) had only one B-fragment (≥3.0 Mb) introgression; two ILs (IL43, IL69) had two chromosomes showing introgressions while only one IL (IL59) had three different chromosomes with B-fragments. The size of the substituted B-genome fragments varied from 3.0 Mb (A9, IL59) to 42.44 Mb (C2, IL54). Of all the 10 chromosomes showing introgressed B-fragments, only one IL (IL59) was identified to be carrying more than one fragment of ≥3.0 Mb size, on chromosome C3. In terms of proportions, the substituted segments ranged from 7.23% (C8, IL59) to almost 83% (C2, IL54) of the actual size of the recipient A/C chromosomes. Only eight (IL13, IL16, IL27, IL39, IL54, IL59, IL61, and IL75) of the 17 ILs showing the presence of B-genome fragments on the basis of SNP assay were common to SSR analysis. In the remaining nine ILs, the absence of such correspondence may be due to the very stringent criterion followed for marking an introgression on the basis of SSR markers (i.e., amplification of two or more consecutive markers on the same chromosome).
Variation for siliqua shatter resistance
There was significant variation for siliqua strength in the 81 phenotyped ILs. The range varied from 2.2 to 6.0 mJ (timely sown 2012–2013), from 2.7 to 5.5 mJ (late sown 2012–2013), and from 3.1 to 5.0 mJ (timely sown 2013–2014), respectively, in the three experiments. In comparison, the range estimates for the 15 B. napus parents varied from 1.8 to 4.0 mJ, from 1.9 to 3.8 mJ, and from 1.8 to 3.9 mJ, respectively. The shatter energy range for the donor parent B. carinata was observed to be between 5.6 and 7.2 mJ for the three test environments. Average shatter energy over the environments for the 69 ILs is depicted as histograms on the CIRCOS (Figure 3). Siliqua strength in some of the ILs, namely, IL7 (5.7 mJ), IL8 (5.6 mJ), IL9 (6.0 mJ), IL12 (5.8 mJ), IL16 (5.5 mJ), IL18 (5.9 mJ), IL22 (5.5 mJ), IL23 (6.0 mJ), IL32 (5.7 mJ), IL41 (6.0 mJ), IL48 (5.8 mJ), IL52 (6.0 mJ), IL53 (6.0 mJ), IL63 (5.5 mJ), IL68 (6.0 mJ), IL72 (5.5 mJ), IL75 (6.0 mJ), and IL76 (5.5 mJ), was almost equal to that of the donor parent B. carinata.
Discussion
Crop Brassica diploids are ancient polyploids which evolved from a paleo-genome through genome duplication, rearrangements, and fractionation (Warwick and Black 1991; Lagercrantz 1998; Lysak et al. 2005). Of the three Brassica genomes, the A- and C- genomes are very closely related while the B-genome is phylogenetically distant. Although Panjabi et al. (2008) have demonstrated some degree of homeology between the B- and A-/C-genomes; homeologous pairing between B-genome chromosomes with either of the A- and C-genomes is rare (Mason et al. 2010, 2011). This poses a challenge to attempts at transfer of desirable traits from the three B-genome-containing species to B. napus. Some studies have nevertheless achieved transfer of desirable traits. Chèvre et al. (1997) stably introgressed blackleg resistance encoded by genes located on the B-genome of B. juncea into B. napus. Plieske et al. (1998) found that B-genome genes for resistance to blackleg from B. nigra, B. juncea, and B. carinata were introgressed at the same location on the B. napus genome. It was argued that this occurred due to the three species sharing the same copy of the resistance gene with sufficient colinearity of the genomic region containing the resistance gene between the B-genome and B. napus to allow recombination to occur. In contrast, Fredua-Agyeman et al. (2014) could not achieve B-genome introgression from B. carinata into B. napus. They found intact or broken B-genome chromosomes inherited together with the B. napus chromosomes over several generations. This was broadly confirmatory of an earlier study by Navabi et al. (2011) who found no evidence of B-genome chromosome introgression into the A- or C- chromosomes of B. napus. They also showed that B-genome chromosomes were inherited as a whole linkage group, with the occasional loss of terminal segments, and several of the B-genome chromosomes were retained across generations. Although Chèvre et al. (1991) have shown excellent transmission frequency (50–100%) of B-genome chromosomes in B. napus–B. nigra disomic chromosome addition lines, low transmission of translocated chromosomes during early stages of the backcross program may be the reasons for failure to introgress B-genome fragments in previous studies. Lack of selection pressure for retention of B-chromosomes during initial backcross generations and small sample size of backcross or selfed progenies may have been the other limiting factors.
A strategy of parallel backcrossing, selfing, and phenotypic selection supplemented with marker assisted selection was followed for developing the ILs. Retention of a higher proportion of the donor B-genome was favored by limiting the cycles of backcrossing to two. This was followed by five cycles of selfing and phenotypic selection for hard to thresh siliquae. A very large population base in each selfing cycle seemed crucial for this scheme. Employing a large set of B. napus parents was opted for making BC2 crosses to facilitate introgression while importing more genetic diversity from the B. napus genome. Preselection in backcross generations based either on genotypic or phenotypic selection was followed which has also been proved successful in other crops (Eshed and Zamir 1994; Eduardo et al. 2005).
Despite selection for high pollen fertility in each recombination cycle, pollen grain stainability in the 69 ILs varied from 60 to 97.5%. High pollen fertility is normally considered as a reflection of stable meiosis, which was confirmed from meiotic analysis wherein 19 normal bivalents with regular anaphase chromosome separation were observed in all the ILs. It is likely that selection pressure for improved pollen grain fertility helped to stabilize translocations as translocation homozygotes. GISH confirmed the presence of large (≥3.0 Mb) B-genome fragments for up to four B. napus chromosomes. Amplification with B-genome SSR markers suggested fragment substitutions in 39 ILs showing introgressed fragments from only four (B3, B4, B6, and B7) of the eight B-genome chromosomes. There appeared to be no retention of intact B-genome chromosomes. In a few cases, the substituted B-genome fragments were from terminal regions of the B-genome chromosomes. B-genome markers of two B-chromosomes from interstitial and terminal regions were present in IL59. These emphasized the possibility of large and multiple translocations resulting from chromosome breakage and reunions. Their presence also indicates that chromatin substitutions must have occurred in BC1 generation, as different recipient B. napus parents were used for producing the BC2. ILs carrying fragments from chromosome B3 may be of special interest for canola breeders due to reported association of B. carinata chromosomes B3 and B8 with cotyledon stage resistance against blackleg (Fredua-Agyeman et al. 2014).
B-genome segment substitution was detected in 7 of the 9 C-genome chromosomes (C1, C2, C3, C5, C6, C8, and C9) and in 3 of the 10 A-genome chromosomes (A3, A9, and A10). The size of the substituted B-genome fragments varied from 3.00 Mb (A9, IL59) to 42.44 Mb (C2, IL54). This translates into 7.23% to almost 83% of the recipient A/C chromosomes, depending on the size of the recipient chromosome. Due to the stringent criterion followed to classify a translocation (minimum 3.0 Mb size of substituted fragment), SNP genotyping provided a conservative estimate of B-genome presence. The introgressed fragments were scattered throughout the recipient chromosome. This may have resulted from preferential chromosome recombination events between areas of high homeology, plus selection pressure for the target trait during development of ILs. Several mapping studies in the past have demonstrated homologous regions that the B-genome chromosomes shared with the A- and C-genomes (Lagercrantz and Lydiate 1996; Panjabi et al. 2008). Comparative mapping of different Brassicaceae lineages has illustrated the presence of conserved genome blocks in B. napus (Parkin et al. 2005; Schranz et al. 2006). Molecular characterization of B. napus, resynthesized through B. juncea × B. carinata hybridizations, had also revealed introgressed B-genome segments (Chatterjee et al. 2016) from all the B-chromosomes but more so from B6. More recently, Gupta et al. (2016) have reported a sizeable number of C-genome chromosome substitution lines in the progenies of derived B. juncea, suggesting that the substituting C-genome chromosomes were likely to have replaced the B-genome chromosomes.
Many ILs revealed a very high rupture energy requirement, which was almost as good (6.0 mJ) as that recorded for B. carinata (7.2 mJ), the donor parent. Special mention may be made of IL9, IL23, IL41, IL52, IL53, IL63, and IL68. Transferability of introgressed shatter resistance was also confirmed in the progenies of crosses involving ILs and several genotypes of euploid B. napus.
Introgression lines constitute permanent diversity conduits for crop improvement. Apart from their use as a plant breeding resource, these have been utilized for identifying genes (Eshed and Zamir 1996), analyzing pleiotropic effects, differentiating between pseudo overdominance and true-dominance (Yamamoto et al. 1998), and map-based QTL cloning. Phenotypic variation and presence of a significant proportion of the donor genome in the ILs is critical for their optimal utilization. Frey et al. (1983) were the first to recognize the productivity-increasing potential of alien genes in oats. This was followed by the synergistic association of QTL mapping and alien gene introgression conceived by Tanksley et al. (1996), who also suggested the advanced backcross QTL strategy. The ILs reported in our study can be used to construct a new platform for genetic and functional genomics analysis in B. napus. These contain substituted B-genome segments on the C-/A-genome chromosomes and may carry QTL for key productivity or defense-related traits. However, linkage drag, distal location of the translocated segment, interaction with recipient chromosome(s), transmission frequency of translocated chromosome, and pleiotropy may limit their utility. It may still be possible to develop fine ILs carrying a single and small introgressed segment for future use in advanced backcrossing and marker-assisted programs. Developing secondary F2 or DH populations through hybridization between ILs and recipient parents may be another option to fine map introgressed QTL (Tian et al. 2006). The ILs show significant differences for many yield-related traits, especially siliqua shatter resistance, and will be used in future to map the genes/QTL controlling productivity and siliqua shattering. Further investigations may involve complete genome or transcriptome sequencing to rapidly identify possible candidate genes in the tagged QTL regions.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.036442/-/DC1.
Acknowledgments
Research was carried out with financial assistance received from the Indian Council of Agricultural Research (ICAR). The Australian Centre for International Agricultural Research and Grains Research & Development Corporation (GRDC), Australia collectively funded the research project “Oilseed Brassica improvement in China, India, and Australia” until 2009 and subsequently under the GRDC-funded research project “Expanding the Brassica germplasm base through collaboration with China and India.” S.S.B. acknowledges salary support from ICAR for the latter part of the project. J.B. is supported by an Australian Research Council Future Fellowship (FT130100604). A.S.M. acknowledges salary support from an Australian Research Council Discovery Early Career Researcher Award (DE120100668) and Deutsche Forschungsgemeinschaft Emmy Noether award. Financial assistance available under Indo Australia biotechnology fund (AISRF06520) during the latter stage of the studies is also acknowledged. I.D. acknowledges salary support from Punjab Agricultural University during her doctorate studies.
Footnotes
Communicating editor: J. A. Birchler
Literature Cited
- Abbadi A., Leckband G., 2011. Rapeseed breeding for oil content, quality and sustainability. Eur. J. Lipid Sci. Technol. 113: 1198–1206. [Google Scholar]
- Allender C. J., King G. J., 2010. Origins of the amphiploid species Brassica napus L. investigated by chloroplast and nuclear molecular markers. BMC Plant Biol. 10: 54. .10.1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bus A., Kӧrber N., Snowdon R. J., Stich B., 2011. Patterns of molecular variation in a species-wide germplasm set of Brassica napus. Theor. Appl. Genet. 123: 1413–1423. [DOI] [PubMed] [Google Scholar]
- Cartea M. E., Soengas P., Picoaga A., Ordas A., 2005. Relationships among Brassica napus (L.) germplasm from Spain and Great Britain as determined by RAPD markers. Genet. Resour. Crop Evol. 52: 655–662. [Google Scholar]
- Chatterjee D., Banga S., Gupta M., Bharti S., Salisbury P. A., et al. , 2016. Resynthesis of Brassica napus through hybridization between B. juncea and B. carinata. Theor. Appl. Genet. 129: 977–990. [DOI] [PubMed] [Google Scholar]
- Chen S., Nelson M. N., Ghamkhar K., Fu T., Cowling W. A., 2008. Divergent patterns of allelic diversity from similar origins: the case of oilseed rape (Brassica napus L.) in China and Australia. Genome 51: 1–10. [DOI] [PubMed] [Google Scholar]
- Chèvre A. M., This P., Eber F., Deschamps M., Renard M., et al. , 1991. Characterization of disomic addition lines of Brassica napus-Brassica nigra by isozyme, fatty acid and RFLP markers. Theor. Appl. Genet. 81: 43–49. [DOI] [PubMed] [Google Scholar]
- Chèvre A. M., Barret P., Eber F., Dupuy P., Brun H., et al. , 1997. Selection of stable Brassica napus-B. juncea recombinant lines resistant to blackleg (Leptosphaeria masculans). 1: identification of molecular markers, chromosomal and genomic origin of the introgression. Theor. Appl. Genet. 95: 1104–1111. [Google Scholar]
- Cowling W. A., 2007. Genetic diversity in Australian canola and implications for crop breeding for changing future environments. Field Crops Res. 104: 103–111. [Google Scholar]
- Diers B. W., McVetty P. B. E., Osborn T. C., 1996. Relationship between heterosis and genetic distance based on restriction fragment length polymorphism markers in oilseed rape (Brassica napus L.). Crop Sci. 36: 79–83. [Google Scholar]
- Doležel J., Bartoš J., 2005. Plant DNA flow cytometry and estimation of nuclear genome size. Ann. Bot. (Lond.) 95: 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doležel J., Sgorbati S., Lucretti S., 1992. Comparison of three DNA fluorochromes for flow cytometric estimation of nuclear DNA content in plants. Physiol. Plant. 85: 625–631. [Google Scholar]
- Doležel J., Bartoš J., Voglmayr H., Greilhuber J., 2003. Nuclear DNA content and genome size of trout and human. Cytometry 51: 127–128. [DOI] [PubMed] [Google Scholar]
- Doyle J. J., Doyle J. L., 1990. Isolation of plant DNA from fresh tissue. Focus 12: 13–15. [Google Scholar]
- Eduardo I., Arús P., Monforte A. J., 2005. Development of a genomic library of near isogenic lines (NILs) in melon (Cucumismelo L.) from the exotic accession PI161375. Theor. Appl. Genet. 112: 139–148. [DOI] [PubMed] [Google Scholar]
- Eshed Y., Zamir D., 1994. Introgressions from Lycopersicon pennellii can improve the soluble-solids yield of tomato hybrids. Theor. Appl. Genet. 88: 891–897. [DOI] [PubMed] [Google Scholar]
- Eshed Y., Zamir D., 1996. Less-than-additive epistatic interactions of quantitative trait loci in tomato. Genetics 143: 1807–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredua-Agyeman R., Coriton O., Huteau V., Parkin I. A. P., Chèvre A. M., et al. , 2014. Molecular cytogenetic identification of B-genome chromosomes linked to blackleg disease resistance in Brassica napus × B. carinata interspecific hybrids. Theor. Appl. Genet. 127: 1305–1317. [DOI] [PubMed] [Google Scholar]
- Frey K. J., Cox T. S., Rodgers D. M., Bramel-Cox P., 1983. Increasing cereal yields with genes from wild and weedy species, pp. 51–68 in Proceedings of the 15th International Genetics Congress, edited by Swaminathan M. S. Oxford and IBH Publishing Company, New Delhi, India. [Google Scholar]
- Fu Y. B., Gugel R. K., 2010. Genetic diversity of Canadian elite summer rape (B. napus) cultivars from the pre- to post-canola quality era. Can. J. Plant Sci. 90: 23–33. [Google Scholar]
- Gehringer A., Snowdon R., Spiller T., Basunanda P., Friedt W., 2007. New oilseed rape (Brassica napus L.) hybrids with high levels of heterosis for seed under nutrient-poor conditions. Breed. Sci. 57: 315–320. [Google Scholar]
- Girke A., Becker H. C., Enqvist G. M., 1999. Resynthesized rapeseed as a new gene pool for hybrid breeding. Proceedings of 10th International Rapeseed Congress, Canberra, Australia, September 26–29. [Google Scholar]
- Gómez-Campo C., Prakash S., 1999. Origin and domestication, pp. 33–58 in Biology of Brassica Coenospecies, Vol. 4, edited by Gómez-Campo C. Elsevier, Amsterdam, Netherlands. [Google Scholar]
- Gupta M., Mason A., Batley J., Bharti S., Banga S. S., 2016. Molecular-cytogenetic characterization of C-genome chromosome substitution lines in Brassica juncea (L.) Czern and Coss. Theor. Appl. Genet. 129: 1153–1166. [DOI] [PubMed] [Google Scholar]
- Gupta V., Lakshmisita G., Shaila M. S., Jagannathan V., Lakshmikumaran M. S., 1992. Characterization of species-specific repeated DNA sequences from B. nigra. Theor. Appl. Genet. 84: 397–402. [DOI] [PubMed] [Google Scholar]
- Gyawali S., Hegedus D. D., Parkin I. A. P., Poon J., Higgins E., et al. , 2013. Genetic diversity and population structure in a world collection of Brassica napus accessions with emphasis on South Korea, Japan and Pakistan. Crop Sci. 53: 1537–1545. [Google Scholar]
- Hu S., Yu C., Zhao H., Sun G., Zhao S., et al. , 2007. Genetic diversity of Brassica napus L. germplasm from China and Europe assessed by some agronomically important characters. Euphytica 154: 9–16. [Google Scholar]
- Kadkol G., 2009. Brassica shatter-resistance research update, pp. 1–6 in Proceedings of 16th Australian Research Assembly on Brassicas, Ballarat, VIC, Australia. [Google Scholar]
- Krzywinski M. I., Schein J. E., Birol I., Connors J., Gascoyne R., et al. , 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagercrantz U., 1998. Comparative mapping between Arabidopsis thaliana and Brassica nigra indicates that Brassica genomes have evolved through extensive genome replication accompanied by chromosome fusions and frequent rearrangements. Genetics 150: 1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagercrantz U., Lydiate D., 1996. Comparative genome mapping in Brassica. Genetics 144: 1903–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Qian W., Meng J., Li Z., 2004. Construction of novel Brassica napus genotypes through chromosomal substitution and elimination using interploid species hybridization. Chromosome Res. 12: 417–426. [DOI] [PubMed] [Google Scholar]
- Li M., Chen X., Meng J., 2006. Inter-sub-genomic heterosis in rapeseed production with a partial new-typed Brassica napus containing subgenome Ar from Brassica rapa and Cc from Brassica carinata. Crop Sci. 46: 234–242. [Google Scholar]
- Liu X.-Y., Macmillan R. H., Burrow R. P., Kadkol G. P., Halloran G. M., 1994. Pendulum test for evaluation of rupture strength of seed siliquae. J. Texture Stud. 25: 179–189. [Google Scholar]
- Lombard V., Baril C. P., Dubreuil P., Blouet F., Zhang D., 2000. Genetic relationships and fingerprinting of rapeseed cultivars using AFLP: consequences for varietal registration. Crop Sci. 40: 1417–1425. [Google Scholar]
- Lysak M. A., Koch M. A., Pecinka A., Schubert I., 2005. Chromosome triplication found across the tribe Brassiceae. Genome Res. 15: 516–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason A. S., Huteau V., Eber F., Coriton O., Yan G., et al. , 2010. Genome structure affects the rate of autosyndesis and allosyndesis in AABC, BBAC and CCAB Brassica interspecific hybrids. Chromosome Res. 18: 655–666. [DOI] [PubMed] [Google Scholar]
- Mason A. S., Nelson M. N., Castello M.-C., Yan G., Cowling W. A., 2011. Genotypic effects on the frequency of homoeologous and homologous recombination in Brassica napus × B. carinata hybrids. Theor. Appl. Genet. 122: 543–553. [DOI] [PubMed] [Google Scholar]
- Navabi Z. K., Parkin I. A. P., Pires J. C., Xiong Z., Thiagarajah M. R., 2010. Introgression of B-genome chromosomes in a doubled haploid population of Brassica napus × B. carinata. Genome 53: 619–629. [DOI] [PubMed] [Google Scholar]
- Navabi Z. K., Stead K. E., Pires C. J., Xiong Z., Sharpe A. G., et al. , 2011. Analysis of B-genome chromosome introgression in interspecific hybrids of Brassica napus × B. carinata. Genetics 187: 659–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panjabi P., Jagannath A., Bisht N. C., Padmaja K. L., Sharma S., et al. , 2008. Comparative mapping of Brassica juncea and Arabidopsis thaliana using Intron Polymorphism (IP) markers: homologous relationships, diversification and evolution of the A, B and C Brassica genomes. BMC Genomics 9: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin I., Gulden S. M., Sharpe A., Lukens L., Trick M., et al. , 2005. Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171: 765–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plieske J., Struss D., Rӧbbelen G., 1998. Inheritance of resistance derived from the B-genome of Brassica against Phoma lingam in rapeseed and the development of molecular markers. Theor. Appl. Genet. 97: 929–936. [Google Scholar]
- Prakash S., Wu X. M., Bhat S. R., 2011. History, evolution, and domestication of Brassica crops. Plant Breed. Rev. 35: 19–84. [Google Scholar]
- Qian W., Meng J., Li M., Frauen M., Sass O., et al. , 2006. Introgression of genomic components from Chinese Brassica rapa contributes to widening the genetic diversity in rapeseed (B. napus L.), with emphasis on the evolution of Chinese rapeseed. Theor. Appl. Genet. 113: 49–54. [DOI] [PubMed] [Google Scholar]
- Rahman H., 2013. Breeding spring canola (Brassica napus L.) by the use of exotic germplasm. Can. J. Plant Sci. 93: 363–373. [Google Scholar]
- Schelfhout C. J., Snowdon R., Cowling W. A., Wroth J. M., 2004. A PCR based B-genome specific marker in Brassica species. Theor. Appl. Genet. 109: 917–921. [DOI] [PubMed] [Google Scholar]
- Schelfhout C. J., Snowdon R., Cowling W. A., Wroth J. M., 2006. Tracing B-genome chromatin in Brassica napus x Brassica juncea interspecific progeny. Genome 49: 1490–1497. [DOI] [PubMed] [Google Scholar]
- Schranz M. E., Lysak M. A., Mitchell-Olds T., 2006. The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 11: 535–542. [DOI] [PubMed] [Google Scholar]
- Schwarzacher T., Heslop-Harrison P., 2000. Practical In situ Hybridization. BIOS Scientific Publishers, Oxford. [Google Scholar]
- Tanksley S. D., Grandillo S., Fulton T. M., Zamir D., Eshed Y., et al. , 1996. Advanced backcross QTL analysis in a cross between an elite processing line of tomato and its wild relative L. pimpinellifolium. Theor. Appl. Genet. 92: 213–224. [DOI] [PubMed] [Google Scholar]
- Tian F., Li D. J., Fu Q., Zhu Z. F., Fu Y. C., et al. , 2006. Construction of introgression lines carrying wild rice (Oryza rufipogon Griff.) segments in cultivated rice (Oryza sativa L.) background and characterization of introgressed segments associated with yield-related traits. Theor. Appl. Genet. 112: 570–580. [DOI] [PubMed] [Google Scholar]
- vanBerloo R., 2008. GGT 2.0: versatile software for visualization and analysis of genetic data. J. Hered. 99: 232–236. [DOI] [PubMed] [Google Scholar]
- Wang A., Yu Z., Ding Y., 2009. Genetic diversity analysis of wild close relatives of barley from Tibet and the Middle East by ISSR and SSR markers. C. R. Biol. 332: 393–403. [DOI] [PubMed] [Google Scholar]
- Warwick S. I., Black L. D., 1991. Molecular systematics of Brassica and allied genera (Subtribe Brassicinae, Brassiceae): chloroplast genome and cytodeme congruence. Theor. Appl. Genet. 82: 81–92. [DOI] [PubMed] [Google Scholar]
- Xiao Y., Chen L., Zou J., Tian E., Xia W., et al. , 2010. Development of a population for substantial new type Brassica napus diversified at both A/C genomes. Theor. Appl. Genet. 121: 1141–1150. [DOI] [PubMed] [Google Scholar]
- Yamamoto T., Kuboki Y., Lin S. Y., Sasaki T., Yano M., 1998. Fine mapping of quantitative trait loci Hd-1, Hd-2 and Hd-3 controlling heading date of rice, as single Mendelian factor. Theor. Appl. Genet. 97: 37–44. [Google Scholar]
- Zhou W. J., Zhang G. Q., Tuvesson S., Dayteg C., Gertsson B., 2006. Genetic survey of Chinese and Swedish oilseed rape (Brassica napus L.) by simple sequence repeats (SSRs). Genet. Resour. Crop Evol. 53: 443–447. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data for confirming the conclusions in this research paper are represented in the figures and tables. Detailed data will be made available on request.