Abstract
Disruption of protein quality control can be detrimental, having toxic effects on single cell organisms and contributing to neurodegenerative diseases such as Alzheimer’s, Parkinson’s and Huntington’s in humans. Here, we examined the effects of polyglutamine (polyQ) aggregation in a major fungal pathogen of humans, Candida albicans, with the goal of identifying new approaches to disable this fungus. However, we discovered that expression of polyQ stretches up to 230Q had no effect on C. albicans ability to grow and withstand proteotoxic stress. Bioinformatics analysis demonstrates that C. albicans has a similarly glutamine-rich proteome to the unicellular fungus Saccharomyces cerevisiae, which exhibits polyQ toxicity with as few as 72Q. Surprisingly, global transcriptional profiles indicated no significant change upon induction of up to 230Q. Proteomic analysis highlighted two key interactors of 230Q, Sis1 and Sgt2; however, loss of either protein had no additional effect on C. albicans toxicity. Our data suggest that C. albicans has evolved powerful mechanisms to overcome the toxicity associated with aggregation-prone proteins, providing a unique model for studying polyQ-associated diseases.
Keywords: Candida albicans, PolyQ, Aggregation, Toxicity, Saccharomyces cerevisiae
All cells use elaborate strategies to maintain cellular protein homeostasis and cope with environmental challenges. During stress, protein homeostasis is often compromised, eliciting protein unfolding, misfolding, and aggregation. Major cellular pathways controlling proteostasis in eukaryotic cells include the heat shock response (HSR) and the unfolded protein response (UPR), which involve molecular chaperones that aid in proper protein folding and prevent protein misfolding (Frydman 2001; Leach and Cowen 2014b). Both the HSR and UPR are rapidly activated upon stress to restore homeostasis and thereby prevent the toxic consequences of protein misfolding. Numerous neurodegenerative diseases are characterized by the accumulation of toxic misfolded proteins and a malfunctioning protein quality control system (Ross and Poirier 2004; Thomas et al. 1995). For example, genetic mutations resulting in aberrant expansions of polyQ repeats cause nine neurodegenerative disorders, including Huntington’s disease and spinocerebellar ataxias (Fan et al. 2014). In all these diseases, CAG trinucleotide repeat expansions in the translated region of specific genes lead to polyQ length-dependent aggregation, toxicity, and eventually neurodegeneration (Duyao et al. 1993).
PolyQ expansion proteins misfold into β sheet-rich, aggregating structures (Wickner et al. 2007), which mediate abnormal interactions with other proteins via exposed hydrophobic surfaces, thus leading to functional impairment and cellular toxicity (Chiti and Dobson 2006; Bolognesi et al. 2010; Olzscha et al. 2011). The presence of coiled-coil domains in polyQ peptides, which regulate polyQ aggregation and insolubility, are thought to enhance the pathogenesis of polyQ expansion diseases (Fiumara et al. 2010). PolyQ aggregation is thought to interfere with cellular protein quality control, including protein degradation, molecular chaperone-mediated protein folding, and the activity of the HSR and the UPR, thereby propagating protein misfolding within cells and entire tissues with toxic consequences (Bence et al. 2001; Gidalevitz et al. 2006).
Much of our understanding of polyQ-mediated toxicity and its interactions with cellular protein quality control comes from experiments expressing amino-terminal fragments of huntingtin (Htt)-containing polyQ expansions in the yeast Saccharomyces cerevisiae (Duennwald et al. 2006a,b; Krobitsch and Lindquist 2000). While only a small fraction of human proteins contain polyQ domains (defined as more than 10 consecutive glutamines in a protein), the yeast proteome is comparatively Q-rich (Michelitsch and Weissman 2000), with more than 50 polyQ proteins (Krobitsch and Lindquist 2000). Another organism that harbors unusually high numbers of polyQ proteins is the social amoeba Dictyostelium discoideum (Malinovska et al. 2015), which has an exceptionally high level of resilience to polyQ aggregation and toxicity in the absence of cellular stress (Malinovska et al. 2015; Santarriaga et al. 2015). As a consequence, D. discoideum was thought to exemplify the evolution of robust protein quality control machinery to cope with a proteome enriched in aggregation-prone proteins.
We aimed to understand how the major fungal pathogen of humans, Candida albicans, responds to polyQ proteins. C. albicans is a natural commensal of the human mucosal microbiota; however, in immunocompromised patients it can disseminate, accounting for over 400,000 life threatening infections world-wide every year (Horn et al. 2009). With limited drugs available and antifungal drug resistance on the rise (Pfaller 2012), we aimed to explore cellular protein quality control in C. albicans and its potential as a new therapeutic target. To this end, we expressed polyQ expansion proteins in C. albicans and assessed polyQ aggregation and toxicity. Our experiments document that C. albicans, whose proteome has a similar polyQ content to that of S. cerevisiae, is remarkably resistant to both polyQ aggregation and toxicity. Indeed, expression of expanded polyQ regions up to 230Qs had no effect on global transcription, resistance to stress, or general fitness of C. albicans cells. Heat shock and other proteotoxic stress conditions induced weak polyQ aggregation, without any toxic consequences. Our results, together with studies in S. cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster, Caenorhabditis elegans, and other model organisms, reveal that the prevalence of aggregation-prone proteins in an organism’s proteome does not automatically determine the capacity of the accompanying cellular protein quality control machinery to modulate protein aggregation and toxicity.
Materials and Methods
Strains and growth conditions
All strains are listed in Table 1. C. albicans strains were grown in YPD (1% yeast extract, 2% bactopeptone, and 2% glucose) (Sherman 1991). To induce expression of Htt polyglutamine expansions, strains grown overnight in YPD at 30° were diluted to OD600 0.2 in the absence or presence of 50 µg/ml doxycycline (BD Biosciences) for 24 hr. Cells were again diluted to OD600 0.05 under the same conditions and grown to midlog phase at the temperature indicated (∼6 hr) prior to microscopy, RNA extraction, spotting assays, Western blots, or semidenaturating detergent agarose gel electrophoresis (SDD-AGE) (Halfmann and Lindquist 2008). S. cerevisiae strains were grown in selective media containing 2% glucose overnight before being diluted to OD600 0.2 in selective media containing 2% glucose or 2% galactose to midlog phase (∼7 hr) to induce expression of the Htt polyglutamine expansions. A 30–42°, heat shock was imposed as described previously (Leach et al. 2012c). All drugs were added at the concentrations stated.
Table 1. C. albicans strains.
| Strain | Genotype | Source |
|---|---|---|
| SN95 | arg4∆/arg4∆ his1∆/his1∆ URA3/ura3:: λimm434 IRO1/iro1::λimm434 | Noble and Johnson (2005) |
| CaLC3067 HSP104/HSP104-GFP | arg4∆/arg4∆ his1∆/his1∆ URA3/ura3:: λimm434 IRO1/iro1::λimm434 HSP104/HSP104-GFP | This study |
| CaLC3069 25Q-1 | arg4∆/arg4∆ his1∆/his1∆ URA3/ura3:: λimm434 IRO1/iro1::λimm434 HSP104/HSP104-GFP, ADH1/adh1::Ptet-FLAG-25Q-RFP | This study |
| CaLC3070 25Q-2 | arg4∆/arg4∆ his1∆/his1∆ URA3/ura3:: λimm434 IRO1/iro1::λimm434 HSP104/HSP104-GFP, ADH1/adh1::Ptet-FLAG-25Q-RFP | This study |
| CaLC3069 72Q-1 | arg4∆/arg4∆ his1∆/his1∆ URA3/ura3:: λimm434 IRO1/iro1::λimm434 HSP104/HSP104-GFP, ADH1/adh1::Ptet-FLAG-72Q-RFP | This study |
| CaLC3070 72Q-2 | arg4∆/arg4∆ his1∆/his1∆ URA3/ura3:: λimm434 IRO1/iro1::λimm434 HSP104/HSP104-GFP, ADH1/adh1::Ptet-FLAG-72Q-RFP | This study |
| CaLC3252 103Q-1 | arg4∆/arg4∆ his1∆/his1∆ URA3/ura3:: λimm434 IRO1/iro1::λimm434 ADH1/adh1::Ptet-FLAG-103Q-RFP | This study |
| CaLC3253 103Q-2 | arg4∆/arg4∆ his1∆/his1∆ URA3/ura3:: λimm434 IRO1/iro1::λimm434 ADH1/adh1::Ptet-FLAG-103Q-RFP | This study |
| CaLC3256 230Q-1 | arg4∆/arg4∆ his1∆/his1∆ URA3/ura3:: λimm434 IRO1/iro1::λimm434 ADH1/adh1::Ptet-FLAG-230Q-RFP | This study |
| CaLC3257 230Q-2 | arg4∆/arg4∆ his1∆/his1∆ URA3/ura3:: λimm434 IRO1/iro1::λimm434 ADH1/adh1::Ptet-FLAG-230Q-RFP | This study |
| W303a | a can1-100, his3-11,15, leu2-3,112, trp1-1, ura3-1, ade2-1 | Duennwald et al. (2006b) |
| 25Q | As W303a GAL-FLAG-25Q-CFP | Duennwald et al. (2006b) |
| 72Q | As W303a GAL-FLAG-72Q-CFP | Duennwald et al. (2006b) |
| 103Q rnqΔ | As W303a GAL-FLAG-103Q-CFP rnqΔ | Duennwald et al. (2006b) |
| 103Q RNQ | As W303a GAL-FLAG-103Q-CFP RNQ | Duennwald et al. (2006b) |
| CaLC4012 hsp104Δ/hsp104Δ | arg4∆/arg4∆ his1∆/his1∆ URA3/ura3:: λimm434 IRO1/iro1::λimm434 hsp104Δ/hsp104Δ::FRT | This study |
| CaLC4088 hsp104Δ/hsp104Δ +103Q-1 | arg4∆/arg4∆ his1∆/his1∆ URA3/ura3:: λimm434 IRO1/iro1::λimm434 hsp104Δ/hsp104Δ::FRT, ADH1/adh1::Ptet-FLAG-103Q-RFP | This study |
| CaLC4082 hsp104Δ/hsp104Δ +103Q-2 | arg4∆/arg4∆ his1∆/his1∆ URA3/ura3:: λimm434 IRO1/iro1::λimm434 hsp104Δ/hsp104Δ::FRT, ADH1/adh1::Ptet-FLAG-103Q-RFP | This study |
| CaLC4083 hsp104Δ/hsp104Δ +230Q-1 | arg4∆/arg4∆ his1∆/his1∆ URA3/ura3:: λimm434 IRO1/iro1::λimm434 hsp104Δ/hsp104Δ::FRT, ADH1/adh1::Ptet-FLAG-230Q-RFP | This study |
| CaLC4084 hsp104Δ/hsp104Δ +230Q-2 | arg4∆/arg4∆ his1∆/his1∆ URA3/ura3:: λimm434 IRO1/iro1::λimm434 hsp104Δ/hsp104Δ::FRT, ADH1/adh1::Ptet-FLAG-230Q-RFP | This study |
| CaLC4385 sgt2Δ/sgt2Δ | arg4∆/arg4∆ his1∆/his1∆ URA3/ura3::imm434 IRO1/iro1::imm434 sgt2Δ/sgt2Δ::FRT | This study |
| CaLC4397 sgt2Δ/sgt2Δ+103Q-1 | arg4∆/arg4∆ his1∆/his1∆ URA3/ura3::imm434 IRO1/iro1::imm434 sgt2Δ/sgt2Δ::FRT, ADH1/adh1::Ptet-FLAG-103Q-RFP | This study |
| CaLC4398 sgt2Δ/sgt2Δ+103Q-2 | arg4∆/arg4∆ his1∆/his1∆ URA3/ura3::imm434 IRO1/iro1::imm434 sgt2Δ/sgt2Δ::FRT, ADH1/adh1::Ptet-FLAG-103Q-RFP | This study |
| CaLC4399 sgt2Δ/sgt2Δ+230Q-1 | arg4∆/arg4∆ his1∆/his1∆ URA3/ura3::imm434 IRO1/iro1::imm434 sgt2Δ/sgt2Δ::FRT, ADH1/adh1::Ptet-FLAG-230Q-RFP | This study |
| CaLC4400 sgt2Δ/sgt2Δ+230Q-2 | arg4∆/arg4∆ his1∆/his1∆ URA3/ura3::imm434 IRO1/iro1::imm434 sgt2Δ/sgt2Δ::FRT, ADH1/adh1::Ptet-FLAG-230Q-RFP | This study |
| CaLC4450 sis1Δ/sis1Δ | arg4∆/arg4∆ his1∆/his1∆ URA3/ura3::imm434 IRO1/iro1::imm434 sis1Δ/sis1Δ::FRT | This study |
| CaLC4462 sis1Δ/sis1Δ+103Q-1 | arg4∆/arg4∆ his1∆/his1∆ URA3/ura3::imm434 IRO1/iro1::imm434 sis1Δ/sis1Δ::FRT, ADH1/adh1::Ptet-FLAG-103Q-RFP | This study |
| CaLC4463 sis1Δ/sis1Δ+103Q-2 | arg4∆/arg4∆ his1∆/his1∆ URA3/ura3::imm434 IRO1/iro1::imm434 sis1Δ/sis1Δ::FRT, ADH1/adh1::Ptet-FLAG-103Q-RFP | This study |
| CaLC4464 sis1Δ/sis1Δ+230Q-1 | arg4∆/arg4∆ his1∆/his1∆ URA3/ura3::imm434 IRO1/iro1::imm434 sis1Δ/sis1Δ::FRT, ADH1/adh1::Ptet-FLAG-230Q-RFP | This study |
| CaLC4465 sis1Δ/sis1Δ+230Q-2 | arg4∆/arg4∆ his1∆/his1∆ URA3/ura3::imm434 IRO1/iro1::imm434 sis1Δ/sis1Δ::FRT, ADH1/adh1::Ptet-FLAG-230Q-RFP | This study |
| CaLC3491 BWP17 + CIp30 | ura3Δ::imm434/ura3Δ::imm434 arg4Δ::hisG/arg4Δ::hisG his1Δ::hisG/his1Δ::hisG, CIp30 (ARG4, HIS1, URA3) | This study |
| CaLC4483 BWP17+103Q-1 | ura3Δ::imm434/ura3Δ::imm434 arg4Δ::hisG/arg4Δ::hisG his1Δ::hisG/his1Δ::hisG, CIp30 (ARG4, HIS1, URA3), ADH1/adh1::Ptet-FLAG-103Q-RFP | This study |
| CaLC4484 BWP17+103Q-2 | ura3Δ::imm434/ura3Δ::imm434 arg4Δ::hisG/arg4Δ::hisG his1Δ::hisG/his1Δ::hisG, CIp30 (ARG4, HIS1, URA3), ADH1/adh1::Ptet-FLAG-103Q-RFP | This study |
| CaLC4485 BWP17+230Q-1 | ura3Δ::imm434/ura3Δ::imm434 arg4Δ::hisG/arg4Δ::hisG his1Δ::hisG/his1Δ::hisG, CIp30 (ARG4, HIS1, URA3), ADH1/adh1::Ptet-FLAG-230Q-RFP | This study |
| CaLC4486 BWP17+230Q-2 | ura3Δ::imm434/ura3Δ::imm434 arg4Δ::hisG/arg4Δ::hisG his1Δ::hisG/his1Δ::hisG, CIp30 (ARG4, HIS1, URA3), ADH1/adh1::Ptet-FLAG-230Q-RFP | This study |
| CaLC2369 ubi4Δ/ubi4Δ | ura3Δ::imm434/ura3Δ::imm434 arg4Δ::hisG/arg4Δ::hisG his1Δ::hisG/his1Δ::hisG, UBI4/loxP-HIS1-loxP::ubi4Δ/loxP-URA-loxP::ubi4Δ, NRG1-3xHA-ARG4 | This study |
| CaLC4487 ubi4Δ/ubi4Δ +103Q-1 | ura3Δ::imm434/ura3Δ::imm434 arg4Δ::hisG/arg4Δ::hisG his1Δ::hisG/his1Δ::hisG, UBI4/loxP-HIS1-loxP::ubi4Δ/loxP-URA-loxP::ubi4Δ, NRG1-3xHA-ARG4, ADH1/adh1::Ptet-FLAG-103Q-RFP | This study |
| CaLC4488 ubi4Δ/ubi4Δ +103Q-2 | ura3Δ::imm434/ura3Δ::imm434 arg4Δ::hisG/arg4Δ::hisG his1Δ::hisG/his1Δ::hisG, UBI4/loxP-HIS1-loxP::ubi4Δ/ loxP-URA-loxP::ubi4Δ, NRG1-3xHA-ARG4, ADH1/adh1::Ptet-FLAG-103Q-RFP | This study |
| CaLC4489 ubi4Δ/ubi4Δ +230Q-1 | ura3Δ::imm434/ura3Δ::imm434 arg4Δ::hisG/arg4Δ::hisG his1Δ::hisG/his1Δ::hisG, UBI4/loxP-HIS1-loxP::ubi4Δ/ loxP-URA-loxP::ubi4Δ, NRG1-3xHA-ARG4, ADH1/adh1::Ptet-FLAG-230Q-RFP | This study |
| CaLC4490 ubi4Δ/ubi4Δ +230Q-2 | ura3Δ::imm434/ura3Δ::imm434 arg4Δ::hisG/arg4Δ::hisG his1Δ::hisG/his1Δ::hisG, UBI4/loxP-HIS1-loxP::ubi4Δ/ loxP-URA-loxP::ubi4Δ, NRG1-3xHA-ARG4, ADH1/adh1::Ptet-FLAG-230Q-RFP | This study |
Plasmid construction
To generate C. albicans strains expressing various lengths of Htt-PolyQ repeats, two approaches were taken based on the size of the repeats. FLAG-HTT-25PolyQ and FLAG-HTT-72PolyQ were PCR amplified from Met-FLAG-25PolyQ-CFP and Met-FLAG-72PolyQ-CFP (Duennwald et al. 2006b), using oligos oLC3001/3035. oLC3001 contained three mutations for CUG codon optimization of the Htt gene for C. albicans. In addition, RFP was PCR amplified from pLC435 (Keppler-Ross et al. 2008), using primers oLC3011/3036. A fusion PCR was then performed with oLC3001/3011 to fuse FLAG-HTT-25PolyQ or 72PolyQ with RFP. The product, along with the plasmid pNIM1, which contains the tetracycline ON promoter (Park and Morschhauser 2005), was digested with SalI and BglII. Digested product and vector were ligated using T4 DNA ligase (NEB) and transformed into TOP10 cells (Invitrogen). Correct integration was tested for by PCR using oligos oLC3012/3013 (350 bp for 25PolyQ and 491 bp for 72PolyQ), and oLC3014/3015 (345 bp). This generated plasmids pLC774, tetON-FLAG-25PolyQ-RFP and pLC775, tetON-FLAG-72PolyQ-RFP. Plasmids were sequenced using oLC3012/3015.
Due to the nature of PolyQ repeats in generating 103Q and 230Q repeats, the FLAG-HTT-103PolyQ-RFP and FLAG-HTT-230PolyQ-RFP were synthesized (Invitrogen). The constructs and plasmid pNIM1 were liberated by digestion with BglII and SalI, gel purified, ligated, and transformed into Stbl2 cells (Invitrogen). Upstream integration was tested by PCR using oligos oLC3012/3013, with an expected size of 583 bp for 103PolyQ and 964 bp for 230PolyQ. Downstream integration was tested by PCR using oligos oLC3014/3015 with an expected size of 345 bp. This generated plasmids pLC807, tetON-FLAG-103PolyQ-RFP and pLC814, tetON-FLAG-230PolyQ-RFP. Plasmids were sequence verified using oLC3012/3036/3014. Primers are listed in Supplemental Material, Table S1.
Strain construction
C. albicans strains expressing 25Q and 72Q were generated by digesting pLC774 (25Q) or pLC775 (72Q) with ApaI and SacII to liberate the cassette. Cassettes were transformed into CaLC3067. NAT-resistant transformants were PCR tested for correct integration at the ADH1 locus by amplifying across both junctions using primer pairs oLC452/453 and oLC454/455. This generated CaLC3069 (25Q-1), CaLC3070 (25Q-2), CaLC3071 (72Q-1), and CaLC3072 (72Q-2).
To generate strains expressing 103Q or 230Q, pLC807 (103Q) or pLC814 (230Q) were digested with ApaI and SacII to liberate the cassettes. Constructs were transformed into SN95 (CaLC239). NAT-resistant transformants were PCR tested for correct integration at the ADH1 locus by amplifying across both junctions using primer pairs oLC452/453 and oLC454/455. This generated CaLC3252 (103Q-1), CaLC3253 (103Q-2), CaLC3256 (230Q-1), and CaLC3257 (230Q-2).
To generate an hsp104Δ/hsp104Δ mutant, the first allele of HSP104 was deleted by PCR amplifying the NAT flipper cassette (pLC49) with oLC3967/3968 containing homology to sequence upstream and downstream of HSP104. The PCR product was transformed into SN95 (CaLC239) and NAT-resistant transformants were PCR tested with oLC275/3021 and oLC274/3969 to verify integration of the cassette. The NAT cassette was excised, and a second round of transformation was performed to delete the second allele, using the same PCR product as was used to delete the first allele. Verification of deletion was ascertained as described above, and loss of the wild-type allele was determined by PCR using oligos oLC782/785. The NAT cassette was then excised, generating CaLC4012. The 103Q and 230Q constructs were transformed into this background as described above, generating CaLC4088 (hsp104Δ/hsp104Δ + 103Q-1), CaLC4082 (hsp104Δ/hsp104Δ + 103Q-2), CaLC4083 (hsp104Δ/hsp104Δ + 230Q-1), and CaLC4084 (hsp104Δ/hsp104Δ + 230Q-2).
To generate an sgt2Δ/sgt2Δ mutant, the first allele of SGT2 was deleted by PCR amplifying the NAT flipper cassette (pLC49) with oLC4197/4198 containing sequence homology upstream and downstream of SGT2. The PCR product was transformed into SN95 (CaLC239), and NAT-resistant transformants were PCR tested with oLC275/4199 and oLC274/4200 to verify integration of the cassette. The NAT cassette was excised, and a second round of transformation was performed to delete the second allele, using the same PCR product as was used to delete the first allele. Verification of deletion was ascertained as described above, and loss of the wild-type allele was determined by PCR using oligos oLC4201/4202. The NAT cassette was then excised, generating CaLC4385. The 103Q and 230Q constructs were transformed into this background as described above, generating CaLC4397 (sgt2Δ/sgt2Δ + 103Q-1), CaLC4398 (sgt2Δ/sgt2Δ + 103Q-2), CaLC4399 (sgt2Δ/sgt2Δ + 230Q-1), and CaLC4400 (sgt2Δ/sgt2Δ + 230Q-2).
To generate a sis1Δ/sis1Δ mutant, the first allele of SIS1 was deleted by PCR amplifying the NAT flipper cassette (pLC49) with oLC4270/4271 containing sequence homology upstream and downstream of SIS1. The PCR product was transformed into SN95 (CaLC239) and NAT-resistant transformants were PCR tested with oLC275/4272 and oLC274/4273 to verify integration of the cassette. The NAT cassette was excised, and a second round of transformation was performed to delete the second allele, using the same PCR product as was used to delete the first allele. Verification of deletion was ascertained as previously described, and loss of the wild-type allele was determined using oligos oLC4378/4379. The NAT cassette was then excised, generating CaLC4450. The 103Q and 230Q constructs were transformed into this background as described above, generating CaLC4462 (sis1Δ/sis1Δ + 103Q-1), CaLC4463 (sis1Δ/sis1Δ + 103Q-2), CaLC4464 (sis1Δ/sis1Δ + 230Q-1) and CaLC4465 (sis1Δ/sis1Δ + 230Q-2).
A ubi4Δ/ubi4Δ mutant had already been generated in the BWP17 background (Leach et al. 2011). As such, the 103Q and 230Q constructs were transformed into this background as described above, generating CaLC4483 (BWP17 + 103Q-1), CaLC4484 (BWP17 + 103Q-2), CaLC4485 (BWP17 + 230Q-1) and CaLC4486 (BWP17 + 230Q-2), CaLC4487 (ubi4Δ/ubi4Δ + 103Q-1), CaLC4488 (ubi4Δ/ubi4Δ + 103Q-2), CaLC4489 (ubi4Δ/ubi4Δ + 230Q-1), and CaLC4490 (ubi4Δ/ubi4Δ + 230Q-2).
Spotting and liquid assays
The spotting assay was performed with two independent strains of induced cells, grown at the temperature indicated, by determining OD600, diluting cells to an equal concentration, and spotting 10-fold dilutions (from ∼1 × 106 cells/ml) onto YPD. Plates were imaged after 48 hr. All spotting assays were performed in duplicate on at least two separate occasions. The liquid assays were performed in flat bottom, 96-well microtiter plates (Sarstedt). Cell densities of overnight cultures were determined and dilutions were prepared such that ∼103 cells were inoculated into each well. Assays were set up in a total volume of 0.2 ml/well of YPD or YPD containing 50 µg/ml doxycycline with twofold dilutions of the following drugs: Azetidine-2-carboxylic acid (Sigma, A0760), guanidine hydrochloride (Sigma, G4505), tunicamycin (Sigma, T7765), MG132 (Sigma, C2211), or geldanamycin (LC Labs, 4500). Plates were incubated in the dark at 30 or 42° for 48 hr, at which point plates were sealed and resuspended by agitation. Absorbance was determined at 600 nm using a spectrophotometer (Molecular Devices) and was corrected for background from the corresponding medium. Assays were performed in duplicate on at least two occasions. MIC data were quantitatively displayed with color using the program Java TreeView 1.1.3 (http://jtreeview.sourceforge.net).
SDD-AGE
Strains were grown as described in the growth conditions section. Midlog phase cells were harvested, washed once with dH2O, and snap frozen in liquid nitrogen. Cells were lysed in 300 μl lysis buffer (100 mM Tris pH 7.5, 200 mM NaCl, 1 mM EDTA, 5% glycerol, 1 mM DTT, 8 mM PMSF, and protease inhibitor cocktail), and ∼200 mg of 0.5 mm acid-washed beads was added to each tube. Cells were mechanically disrupted on a Biospec Mini-Beadbeater for six 30 sec periods, with 1 min on ice between each cycle. A 26G syringe was used to puncture the bottom of the tube, and supernatant was collected by spinning for 1 min at 5000 rpm. Protein was quantified using a Bradford assay. Gel was prepared by dissolving 1.5% agarose in 1 × TAE and, once dissolved, SDS was added at 0.5%. Running buffer was 1 × TAE with 0.5% SDS. Samples were prepared in 4 × sample buffer (2 × TAE, 20% glycerol, 8% SDS, and bromophenol blue) at 1 × and boiled for 5 min at 100°. Samples were run at 50 V for ∼3.5 hr. Gels were rinsed in dH2O and then TBS (20 mM Tris pH 7.5 and 9 g NaCl up to a volume of 1 L in dH2O), and transferred onto PVDF membranes using Whatman: Turboblotter Rapid Downward Transfer Systems for 5 hr. Membranes were blocked and proteins detected as per the western blotting protocol.
Microscopy
Imaging was performed on a Zeiss Imager M1 upright microscope and AxioCam MRm with AxioVision 4.7 software (Carl Zeiss, Inc.). An X-Cite series120 light source with ET HQ tetramethylrhodamine isothiocyanate (TRITC)/DsRED filter sets from ChromaTechnology (Bellows Falls, VT) was used for fluorescence microscopy.
Dot blots
Dot blots were performed as described before (Cashikar et al. 2005). In brief, protein lysates were prepared using glass beads and lysis buffer [100 mM Tris PH 7.5, 200 mM NaCl, 1 mM EDTA, 5% glycerol, 1 mM DTT, 4 mM PMSF, and protease inhibitor tablet (Sigma)]. Lysates were normalized using BCA and diluted in phosphate-buffered saline (PBS) in threefold serial dilutions. All dilutions were loaded onto the BIO-DOT apparatus (Bio-Rad), and assembled with a 0.2 µm nitrocellulose membrane. A vacuum was applied to the manifold to transfer the lysate from the apparatus to the membrane. Membrane was then probed with a 1:5000 anti-FLAG antibody or 1:10,000 anti-PSTAIRE antibody and processed as a western blot.
Quantitative RT-PCR
Cells were grown as stated in the growth conditions section. At midlog phase, strains were subjected to a 10 min 30–42° heat shock as described previously (Leach and Cowen 2014a), or left untreated at 30°. Cells were harvested from each culture, centrifuged at 3000 rpm for 2 min at 4°, and washed once with dH2O, before being snap frozen in liquid nitrogen and stored at −80°. RNA was subsequently isolated using the QIAGEN RNeasy kit and cDNA synthesis was performed using the AffinityScript cDNA synthesis kit (Stratagene). PCR was carried out using the SYBR Green JumpStart Taq ReadyMix (Sigma) with the following cycle conditions: 95° for 3 min, 95° for 10 sec and 60° for 30 sec for 39 rounds, 95° for 10 sec, and 65° for 5 sec. Reactions were performed in triplicate using the following primer pairs: HSP104 (oLC1620/1621) and HSP90 (oLC756/757). Transcript levels were normalized to ACT1 (oLC2285/2286). Data were analyzed using the Bio-Rad CFX Manager software, version 3.1 (Bio-Rad).
Western blotting
Total soluble protein was extracted and subjected to western blotting using published protocols (Leach and Cowen 2014a). Briefly, midlog-phase cells were pelleted by centrifugation, washed with sterile water, and resuspended in lysis buffer [0.1 M Tris-HCl (pH 8.0), 10% glycerol, 1 mM DTT, 1 mM PMSF, and protease inhibitor cocktail]. An equal volume of 0.5 mm acid-washed beads was added to each tube and cells were mechanically disrupted on a Biospec Mini-Beadbeater for six 30 sec periods, with 1 min on ice between each cycle. The lysate was pelleted by high-speed centrifugation and the supernatant removed for analysis. Protein concentration was determined by a Bradford assay. Protein samples were mixed with one-sixth volume of 6 × sample buffer [0.35 M Tris-HCl, 10% (wt/wt) SDS, 36% glycerol, 5% β-mercaptoethanol, and 0.012% bromophenol blue], and 75 μg protein was loaded in wells of an 8% SDS-PAGE gel. Separated proteins were transferred to a PVDF membrane for 1 hr at 100 V at 4°. Membranes were blocked in 5% milk in PBS containing 0.1% Tween 20 (PBS-T) at room temperature for 1 hr and subsequently incubated in a 1:10,000 dilution of anti-FLAG HRP-conjugated antibody (Sigma, A8592) in PBS-T + 5% milk [PBS 0.1% Tween-20 and 5% (w/v) milk]. Primary antibody was left on the membrane for 1 hr at room temperature. Membranes were washed in PBS-T and signals detected using an ECL western blotting kit as per the manufacturer’s instructions (Pierce).
RNA-seq
Wild-type (CaLC239, SN95), 103Q (CaLC3252)-, and 230Q (CaLC3256)-expressing cells were grown in the absence or presence of 50 μg/ml doxycycline as described in the growth conditions section, and RNA was isolated using the Ambion RiboPure RNA purification yeast kit. Three biological replicates grown on three separate occasions were analyzed. RNA integrity was assayed on an Agilent Bioanalyser with freshly prepared gel-dye matrix according to the Agilent Bioanalyser RNA Pico protocol. Ribosomal RNA was removed using the Ribo-Zero Gold rRNA Removal Kit and TruSeq RNA-seq libraries were prepared according to the manufacturer’s instructions (Illumina), and sequenced on the Illumina HiSeq2000 platform. The sequencing data were assessed for quality using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). All 51 bases for all 18 samples (six samples per biological replicate) had a minimum median Phred quality score over 32 (around 99.94% base call accuracy). The number of reads for each sample ranged from 12.3 M reads to 17.0 M reads with a minimum mapping rate of 95.9%. Each read was mapped against a C. albicans reference genome (SC5314, A21) using Tophat2 (version 2.1.0) with maximum mismatch of 1 and a gene transfer file (-N 1) (Kim et al. 2013), then expression of each RNA transcript was quantified and compared using cufflinks and cuffdiff (version 2.2.1) with the default setting (Trapnell et al. 2013). Both the reference genome and transcript annotation were downloaded from the Candida Genome Database (Inglis et al. 2012). All sequencing data are deposited at the Sequence Read Archive (Study Accession: SRP073330).
Detection of polyQ interactors
Untagged wild-type (CaLC239, SN95) and 230Q (CaLC3256)-expressing cells were grown in the absence or presence of 50 μg/ml doxycycline, as described in the growth conditions section. Cells were harvested at an OD600 0.8 at 4000 rpm for 10 min at 4°, washed twice with ice-cold 1 × PBS, and snap frozen in liquid nitrogen. Cells were resuspended in 10 ml of lysis buffer [20 mM Tris pH 7.5, 100 mM KCl, 5 mM MgCl, and 20% glycerol, with one protease inhibitor cocktail per 50 ml (complete, EDTA-free tablet, Roche Diagnostics), 1 mM PMSF (EMD Chemicals), and 0.5% Tween 20]. Cells were disrupted by bead-beating twice for 4 min with a 7 min break on ice between cycles. Lysates were centrifuged at 1300 × g for two 5 min cycles, recovering the supernatant at each stage. The combined lysate was centrifuged twice at 21,000 × g, first for 10 min and second for 5 min at 4°, and protein concentrations determined using the Bradford assay. Protein A/G beads (Santa Cruz SC-2003) were prepared for preclearing as per the manufacturer’s instructions, and protein samples were precleared for 1.5 hr at 4° with end-to-end rotation.
Anti-FLAG immunoprecipitations were performed by diluting protein samples to 20 mg/ml in lysis buffer containing 1 × Triton X-100, and incubating in 50 µl anti-FLAG M2 affinity gel (Sigma A2220) at 4° overnight with end-to-end rotation, as per the manufacturer’s specifications. Protein samples were eluted by washing beads twice with lysis buffer with detergent followed by three times with lysis buffer without detergent. Fresh elution buffer (9.5 M NH4OH, pH 11.0–12.0 and 0.5 mM EDTA) was then added at three times the bead volume and samples incubated at 4° for 15 min with end-over-end rotation. This was repeated three times. Samples were digested in-solution as described previously (Gundry et al. 2009). Briefly, DTT was added to a final concentration of 10 mM and samples were incubated with end-over-end rotation at room temperature for 30 min. Trypsin (Promega, MS grade) was added and samples were incubated overnight at 37° with end-over-end rotation. Reactions were stopped by adding 5 μl of 1.0% TFA.
Samples were analyzed on an Orbitrap analyzer (Q-Exactive, Thermo Fisher Scientific, San Jose, CA) outfitted with a nanospray source and EASY-nLC nano-LC system (Thermo Fisher Scientific). Lyophilized peptide mixtures were dissolved in 11 μl of 0.1% formic acid and 5 μl were loaded onto a 75 μm × 50 cm PepMax RSLC EASY-Spray column filled with 2 μM C18 beads (Thermo Fisher Scientific) at a pressure of 800 Bar. Peptides were eluted over 60 min at a rate of 250 nl/min using a 0–35% acetonitrile gradient in 0.1% formic acid. Peptides were introduced by nano-electrospray into the Q-Exactive mass spectrometer (Thermo Fisher Scientific). The instrument method consisted of one MS full scan (400–1500 m/z) in the Orbitrap mass analyzer with an automatic gain control (AGC) target of 1e6, maximum ion injection time of 120 msec, and a resolution of 70,000 followed by 10 data-dependent MS/MS scans with a resolution of 17,500, an AGC target of 1e6, maximum ion time of 120 msec, and one microscan. The intensity threshold to trigger a MS/MS scan was set to 1.7e4. Fragmentation occurred in the HCD trap with normalized collision energy set to 27. The dynamic exclusion was applied using a setting of 10 sec.
Database searching
Tandem mass spectra were extracted, charge state deconvoluted, and deisotoped by Xcalibur version 2.2. All MS/MS samples were analyzed using Sequest (Thermo Fisher Scientific; version 1.4.1.14) and X! Tandem [The GPM, thegpm.org; version CYCLONE (2010.12.01.1)]. Sequest was set up to search Uniprot-C_albicans.fasta (downloaded Feb 28 2014, 9082 entries) assuming the digestion enzyme trypsin. X! Tandem was set up to search the Uniprot-C_albicans database (downloaded Feb 28 2014, 9082 entries) also assuming trypsin. Sequest and X! Tandem were searched with a fragment ion mass tolerance of 0.020 kDa and a parent ion tolerance of 10.0 PPM. Deamidated of asparagine and glutamine, oxidation of methionine, and carbamidomethyl of cysteine were specified in Sequest as variable modifications. Glu->pyro-Glu of the N-terminus, ammonia-loss of the N-terminus, Gln->pyro-Glu of the N-terminus, deamidated of asparagine and glutamine, oxidation of methionine, and carbamidomethyl of cysteine were specified in X! Tandem as variable modifications.
Protein identification
Scaffold (version Scaffold_4.5.0, Proteome Software Inc., Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at > 95.0% probability. Peptide Probabilities from Sequest were assigned by the Scaffold Local FDR algorithm. Peptide Probabilities from X! Tandem were assigned by the Peptide Prophet algorithm (Keller et al. 2002) with Scaffold Δ-mass correction. Protein identifications were accepted if they could be established at > 95.0% probability, contained at least one identified peptide, and were present in both biological replicates. Protein probabilities were assigned by the Protein Prophet algorithm (Nesvizhskii et al. 2003). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins sharing significant peptide evidence were grouped into clusters. The mass spectrometry data has been deposited into the Proteome Exchange PRIDE database (http://www.proteomexchange.org/) under the accession number PXD003916.
Data availability
All sequencing data are deposited at the Sequence Read Archive (Study Accession: SRP073330). The mass spectrometry data has been deposited into the Proteome Exchange PRIDE database (http://www.proteomexchange.org/) under the accession number PXD003916.
Results
Absence of polyQ aggregation and toxicity in C. albicans
We generated a series of constructs containing human Htt exon 1 with varying lengths (25Q, 72Q, 103Q, and 230Q) of polyQ tracts (CAG repeats) with an amino-terminal FLAG tag and a C-terminal Red Fluorescent Protein (RFP), under the control of the tetracycline ON promoter, integrated at the ADH1 locus in C. albicans (Figure S1A) (Park and Morschhauser 2005). Our initial studies focused on 25Q and 72Q, where we examined cells for aggregation and toxicity; 25Q represents a nondisease related, nontoxic, and nonaggregating polyQ length, and 72Q represents a highly aggregation-prone and toxic polyQ length in many different model organisms including S. cerevisiae (Figure S1B), D. melanogaster, C. elegans, cultured mammalian cells, and rodents (Burright et al. 1995; Morley et al. 2002; Waelter et al. 2001; Marsh et al. 2000).
First, we examined PolyQ aggregation by microscopy in cells that were grown to midlog phase in the presence of 50 μg/ml doxycycline to induce polyQ expression (Figure S1A). Addition of doxycycline caused cells containing the polyQ expansions to fluoresce, with the signal uniformly and evenly distributed throughout the cell for both 25Q and 72Q. The lack of polyQ aggregation for 72Q suggests that C. albicans, in contrast to S. cerevisiae and many other organisms, is resistant to polyQ aggregation.
Further, we examined polyQ toxicity in C. albicans. Wild-type, 25Q-, and 72Q- expressing cells were grown at 30 or 42° in the absence and presence of doxycycline to induce expression of the expanded polyQ regions, and spotted on YPD plates to assess toxicity (Figure S1C). No growth defect was observed for any of the strains. Next, we monitored growth in 96-well plates in the absence and presence of doxycycline with a gradient of the nucleoside antibiotic tunicamycin, which induces ER stress and the UPR, and increases polyQ toxicity in S. cerevisiae and mammalian cells (Wimalasena et al. 2008). Comparable growth was observed for two independent 25Q- and 72Q-expressing strains and the wild-type strain under all conditions (Figure S1D). Thus, expression of 72Q does not elicit polyQ toxicity even in the context of ER stress. This is in contrast to S. cerevisiae, for which induction of 72Q using galactose causes toxicity compared to wild-type or 25Q-expressing strains (Figure S1E) (Duennwald et al. 2006b).
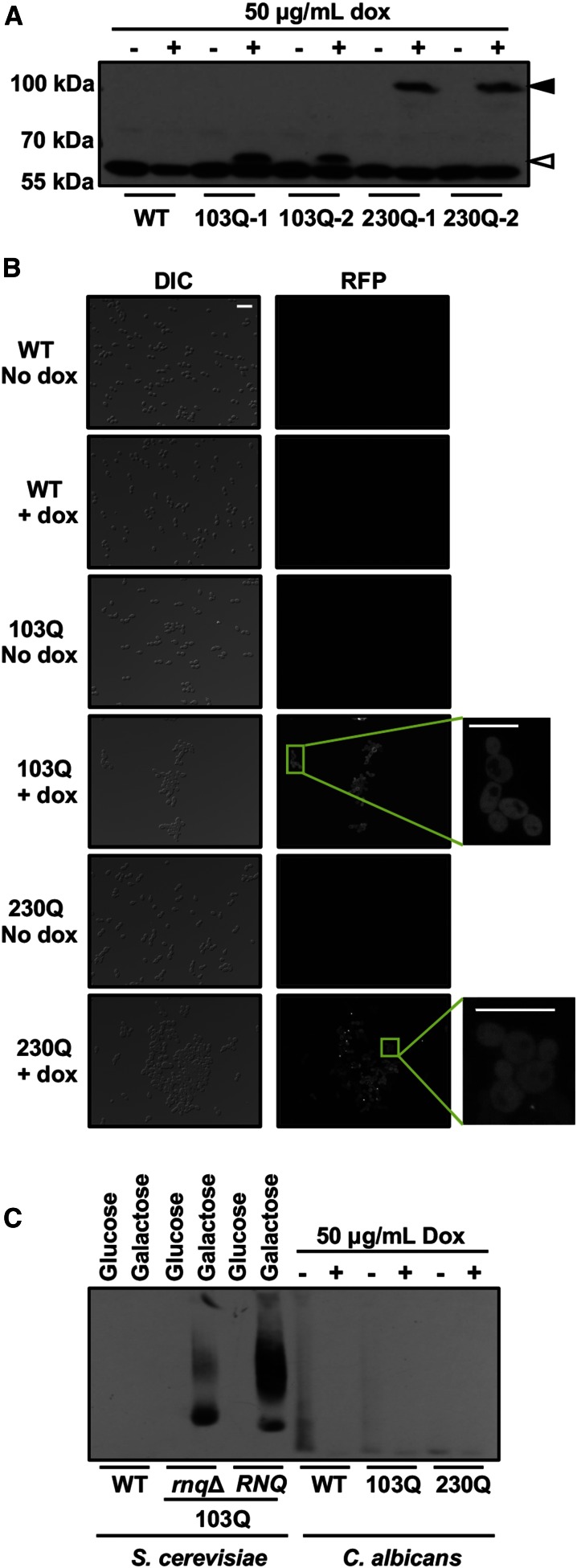
Next, we examined very long polyQ expansions, up to 230Q. We assessed polyQ expression by western blot in the absence (no induction) and presence (induction) of doxycycline. In two independent strains for 103Q and 230Q, we observed a clear signal in the presence of doxycycline (Figure 1A), confirming that the polyQ repeats are expressed. This also indicates solubility of the polyQ expansion proteins in C. albicans, in contrast to polyQ expression in S. cerevisiae and other organisms, for which polyQ expansions are unable to migrate in SDS-PAGE due to insolubility (Krobitsch and Lindquist 2000). To further study the aggregation of these very long polyQ expansions, we performed fluorescence microscopy to visualize RFP (Figure 1B). The fluorescence signal for the 103Q constructs was once again uniformly distributed within cells, with no obvious punctate structures visible (Figure 1B). Only 8% of cells expressing 230Q displayed some punctate structures, possibly indicating rare aggregation beginning in the few cells over the 30 hr time course of protein induction (Figure 1B).
Figure 1.
Expression of up to 230Q does not cause aggregation in C. albicans. (A) Western blot analysis using the FLAG epitope of wild-type (WT) and two independent strains harboring 103Q or 230Q expression constructs under the control of the tetracycline ON promoter. Protein expression was monitored after a total of 30 hr induction [−, no doxycycline (dox); +, 50 μg/ml dox]. White arrow = 103Q, black arrow = 230Q. (B) Live cell red fluorescent protein (RFP) (middle) and differential interference contrast (DIC) (left) microscopy of WT, and strains harboring 103Q or 230Q expression constructs in the absence (No dox) or presence (+ dox) of 50 μg/ml dox. Inset: magnification of a subset of cells representing polyQ expression. Scale bar, 20 μm. (C) Semidenaturating detergent agarose gel electrophoresis analysis of aggregated proteins expressed from S. cerevisiae WT, 103Q in rnqΔ, and 130Q in [RNQ+] grown in minimal medium with glucose (not inducing) or galactose (inducing) (lanes 1–6), C. albicans WT, and strains harboring 103Q or 230Q expression constructs grown in yeast peptone dextrose medium in the absence (−, not inducing) or presence (+, inducing) of 50 μg/ml dox. Detection via the N-terminal FLAG-tag.
In order to corroborate our microscopy results, we performed SDD-AGE assays to characterize polyQ aggregation biochemically in the 103Q- and 230Q-expressing strains. We included the rnqΔ and [RNQ+] 103Q polyglutamine-expanded Htt strains from S. cerevisiae, in which strains harboring as short a polyQ expansion as 46Q are known to be insoluble (Alberti et al. 2009; Chafekar et al. 2012). Rnq1 in its prion confirmation plays an essential role in polyQ aggregation, leading to toxicity (Meriin et al. 2002). Since C. albicans lacks prion proteins, we included the rnqΔ as a suitable control. In stark contrast to the insoluble protein aggregates observed in S. cerevisiae 103Q-expressing strains, the 103Q and even 230Q expansions remained completely soluble in C. albicans (Figure 1C). Thus, in C. albicans, polyQ expansions do not form aggregates detectable by microscopy or biochemical assays, even with extremely long polyQ stretches that would readily aggregate in many other eukaryotic cell types.
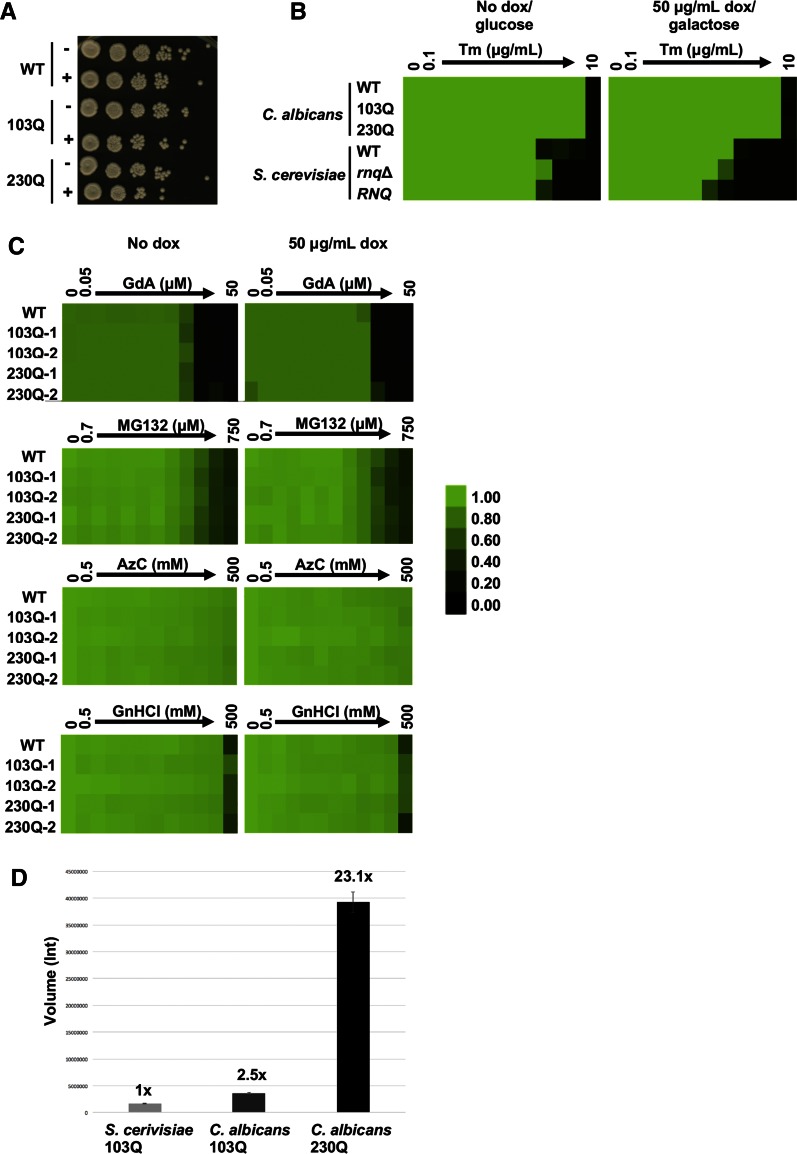
We next tested whether the expression of 103Q or 230Q causes polyQ toxicity, as evidenced by reduced growth in C. albicans. PolyQ expansions were induced by growing cells for 24 hr, followed by subculture and growth for an additional 6 hr before plating cells on YPD in serial dilutions to assess potential growth defects (Figure 2A). All strains grew at similar rates, suggesting that expression of polyQ proteins with long glutamine regions is not toxic to C. albicans (Figure 2A).
Figure 2.
Expression of up to 230Q in combination with proteotoxic stressors does not cause toxicity in C. albicans. (A) Wild-type (WT) and strains harboring 103Q or 230Q expression were monitored after a total of 30 hr induction (−, no doxycycline (dox); +, 50 μg/ml dox) before being serially diluted ten-fold and spotted onto YPD solid medium, grown at 30°, and imaged after 48 hr. (B) Toxicity of S. cerevisiae WT, 103Q in rnqΔ, and 103Q in [RNQ+] compared to C. albicans WT and strains harboring 103Q or 230Q expression constructs was assessed in the presence of a tunicamycin (Tm) gradient from 0 to 10 μg/ml, in twofold dilutions in YPD with (inducing) or without (not inducing) 50 μg/ml dox or minimal medium with glucose (not inducing) or galactose (inducing). Growth was measured after 48 hr static incubation by absorbance at 600 nm and normalized relative to the no Tm control. For each strain, optical densities were averaged for duplicate measurements and displayed quantitatively using Treeview, as shown in the color bar. Data are representative for three biological replicates. (C) Toxicity of the C. albicans strains harboring 103Q and 230Q was assessed in the presence of proteotoxic stressors. Gradients were performed in twofold dilutions, and strains set up in YPD in the absence (no dox) or presence of 50 μg/ml dox. Growth was measured after 48 hr static incubation at 30° by absorbance at 600 nm and normalized relative to the untreated control. AzC, azetidine-2-carboxylic acid; GdA, geldanamycin; GnHCl, guanidine hydrochloride. (D) Dot blots were performed to determine the expression levels of 103Q in S. cerevisiae, and 103Q and 230Q in C. albicans. Protein lysates were serially diluted in PBS and loaded on the dot blot apparatus assembled with a nitrocellulose membrane. Blot quantification was carried out using Bio-Rad Image Lab 5.2 software following the blot scan. PolyQ expression levels have been normalized to the corresponding loading control (PSTAIRE). S. cerevisiae 103Q was set to 1. 103Q expression in C. albicans is 2.5 times greater than in S. cerevisiae, and 230Q is 23 times greater. AzC, azetidine-2-carboxylic acid; GdA, geldanamycin; GnHCl, guanidine hydrochloride.
In an attempt to unmask polyQ toxicity, we expressed the polyQ expansion proteins in the presence of additional proteotoxic stresses. First, we set up a growth assay in 96-well plates in the presence of doxycycline to express the expanded polyQ regions with a concentration gradient of tunicamycin. We included strains in the absence of doxycycline and the S. cerevisiae wild-type, rnqΔ, and [RNQ+] 103Q polyglutamine-expanded Htt strains as controls. As expected, expression of the 103Q polyglutamine-expanded Htt in S. cerevisiae [RNQ+] strain impaired growth compared to wild-type cells lacking the polyQ expansion, with a minor effect seen in the rnqΔ strain (Figure 2B). In stark contrast, neither the C. albicans 103Q or 230Q strains exhibited any growth defect (Figure 2B). We further tested other compounds that perturb protein homeostasis, including the Hsp90 inhibitor geldanamycin, the proteasome inhibitor MG132, the protein unfolding inducer azetidine-2-carboxylic acid, and the Hsp104 inhibitor guanidine hydrochloride (Figure 2C). Even at very high concentrations, all except geldanamycin had no impact on the growth of wild-type cells. Expression of up to 230Q had no additional effect on cell growth, demonstrating that C. albicans does not display polyQ toxicity even in the presence of proteotoxic stress (Figure 2C). To ensure that polyQ Htt protein levels were similar between S. cerevisiae and C. albicans, we compared polyQ Htt proteins using dot blots. The 103Q polyglutamine-expanded protein levels in S. cerevisiae were comparable to the expression levels of 103Q in C. albicans. However, 230Q expression levels in C. albicans were more than 20-fold higher than 103Q in S. cerevisiae, ruling out the possibility that the lack of toxicity in C. albicans could be due to low levels of polyQ expression (Figure 2D).
High temperature induces polyQ aggregation independent of toxicity
When cells are subjected to a heat shock or continuous growth at high temperatures, many proteins unfold, which reduces growth and general fitness (Leach et al. 2012a). We asked whether polyQ expression combined with growth at high temperature would overwhelm the protein folding machinery, leading to increased aggregation and toxicity. First, we examined aggregation of our polyQ expansions by fluorescence microscopy. PolyQ expansions were induced by growing cells for 24 hr, followed by subculture and growth for an additional 6 hr at 42° before imaging. The fluorescence signal for the 103Q constructs was once again evenly distributed throughout the cell, with only minimal punctate structures visible (Figure S2A). In contrast, induction of 230Q produced extensive punctate structures, indicating that the longer polyQ expansions no longer remain soluble and aggregate.
Based on this result and previously published work, we postulated that expressing 103Q and 230Q at high temperature with a gradient of the unfolded protein stressor tunicamycin would unmask polyQ toxicity. To our surprise, once again, none of the polyQ expressing strains exhibited any growth defect (Figure S2B). We also tested the unfolded protein inducer azetidine-2-carboxylic acid and the Hsp104 inhibitor guanidine hydrochloride on cells incubated at 42°. Again, we observed no toxicity whatsoever in any of the polyQ expressing strains (Figure S2C).
Expression of polyglutamine-expanded Htt has no effect on the global transcriptional response
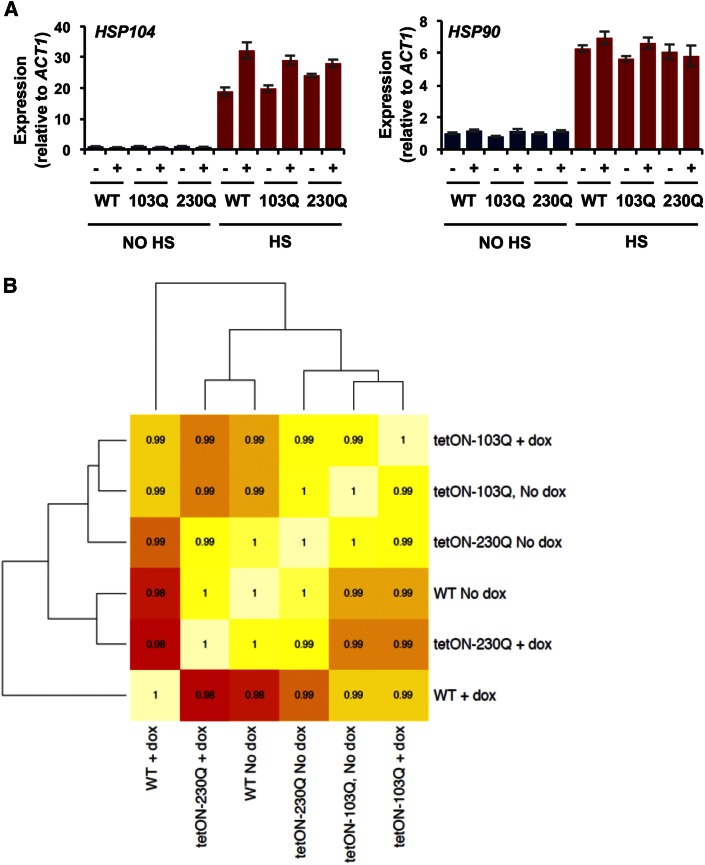
Studies in cultured mammalian cells, yeast, and mice, have documented a strong transcriptional response to the expression of polyQ expansion proteins (Hughes et al. 2001; Mohan et al. 2014). Given our findings of the exceptional resistance of C. albicans to polyQ aggregation and toxicity, we asked whether the cells effectively upregulate protective genes, such as molecular chaperones, to prevent polyQ aggregation and toxic consequences. Specifically, we tested whether polyQ expansion proteins activate the HSR, leading to upregulation of key molecular chaperones that could modulate polyQ proteins. To this end, we expressed 103Q and 230Q expansions by growing cells for 24 hr, followed by subculture and growth for an additional 6 hr before subjecting cells to a short 10 min 42° heat shock. Expression of HSP90, a key molecular chaperone known to aid in the folding of ∼10% of the proteome (Leach et al. 2012b), and HSP104, which functions as a potent protein disaggregase (Grimminger-Marquardt and Lashuel 2010), was then examined. In the absence of heat shock, there was no difference in HSP90 or HSP104 transcript levels between wild-type and polyQ-expressing strains (Figure 3A). Heat shock induced transcript levels of both genes, as expected, but once again no significant difference was observed between wild-type and polyQ expansion strains (Figure 3A). These experiments indicate that polyQ expression does not induce a HSR under normal growth conditions, and that C. albicans cells expressing polyQ proteins elicit a normal HSR, in contrast to S. cerevisiae and mammalian cells.
Figure 3.
Expression of polyQ does not affect the transcriptome of C. albicans. (A) C. albicans wild-type (WT) and strains harboring 103Q or 230Q expression constructs were grown at 30° (no heat shock [HS]) or subjected to a 10 min 30–42° HS, in the absence (−) and presence (+) of 50 μg/ml doxycycline (dox) to induce polyQ expression; transcript levels of target genes were measured and normalized to the ACT1 loading control. Data represent mean values ± SD from two independent biological replicates. (B) Heat map illustrating the Pearson correlation of RNA sequencing (RNA-seq) global gene expression between WT and strains harboring 103Q or 230Q expression constructs in the absence and presence of 50 μg/ml dox to induce polyQ expression.
In addition, we probed global transcriptional changes using RNA-seq to determine the response to 103Q and 230Q expression. PolyQ expression was induced for a total of 30 hr before RNA extraction and sequencing. Surprisingly, very few gene expression differences were observed between wild-type, 103Q and 230Q data sets (Figure 3B). Indeed, in each of three replicates performed, the correlation coefficient between any two samples remained above 0.88 (Figure S3), with few genes being up or downregulated in any of the conditions tested (File S1). The data document that expression of polyQ proteins has little or no effect on gene expression in C. albicans.
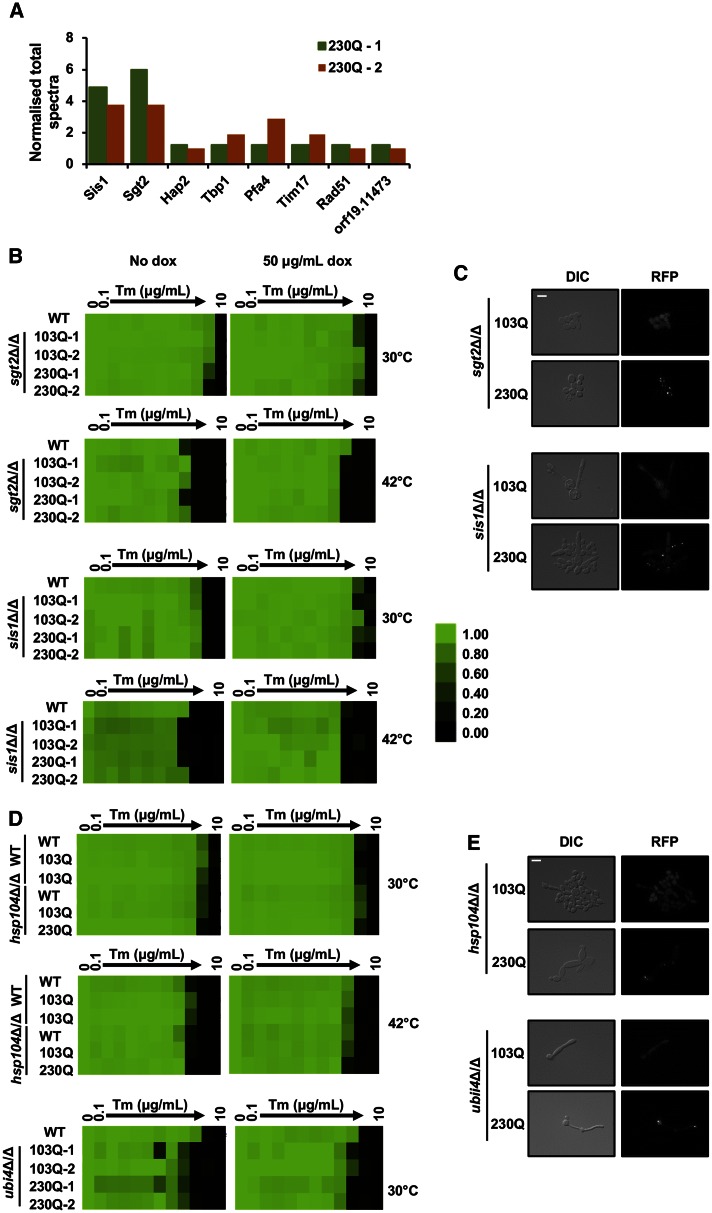
Sis1 and Sgt2 interact with polyglutamine-expanded Htt
The negligible transcriptional response to polyQ expansions suggested that C. albicans cells cope with high levels of polyQ proteins with minimal changes in gene expression programs. Therefore, we explored a mass spectrometry-based approach to determine which proteins interact with the polyQ proteins using 230Q as an example. IP-mass spectrometry showed that only eight proteins are enriched in the 230Q strain upon induction of the polyQ expansion, compared to the uninduced condition and the wild-type strain (Table 2). Interestingly, the molecular chaperones Sgt2 and Sis1 had among the strongest interactions with the 230Q expansion (Figure 4A). Sgt2 has previously been implicated as an amyloid sensor, regulating the targeting of chaperones to aggregation-prone proteins in S. cerevisiae (Kiktev et al. 2012). Furthermore, Sis1 has recently been found to be sequestered by polyQ aggregates, which in turn inhibits degradation of misfolded proteins via the ubiquitin–proteasome system (Park et al. 2013). To determine if these proteins prevent polyQ aggregation and toxicity, we generated deletion mutants for each of them in C. albicans, and expressed our 103Q and 230Q expansions. We then tested polyQ toxicity using a growth assay with an increasing concentration of tunicamycin at either 30 or 42° (Figure 4B). In both sis1 and sgt2 deletion mutants, no polyQ toxicity was observed compared to wild-type strains (Figure 4B). We also explored polyQ aggregation via fluorescence microscopy after growth at 42° (Figure 4C). We found that the 103Q expansion remains evenly distributed throughout the cell, indicative of a soluble polyQ protein. Yet when the 230Q expansion is expressed at higher temperatures, we observe a few punctate structures forming, indicative of aggregation, similar to our experiment in heat-shocked wild-type cells (Figure S2A).
Table 2. PolyQ interactors.
| Protein | Role | PolyQ | PolyN |
|---|---|---|---|
| Sis1 | Putative type II HSP40 cochaperone | N/A | N/A |
| Sgt2 | Putative small tetratricopeptide repeat (TPR)-containing protein | N/A | N/A |
| Hap2 | CCAAT-binding transcription factor; regulates low-iron induction of FRP1 | 13 | 9 |
| Tbp1 | Transcription initiation factor; binds TATA box sequence | N/A | N/A |
| Pfa4 | Palmitoyltransferase with autoacylation activity; required for palmitoylation of amino acid permeases | N/A | N/A |
| Tim17 | Predicted component of the Translocase of the Inner Mitochondrial membrane (TIM23 complex), involved in protein import into mitochondria | N/A | N/A |
| Rad51 | Protein involved in homologous recombination and DNA repair | N/A | N/A |
| Orf19.11473 | Ortholog(s) have DNA-directed RNA polymerase activity, RNA polymerase III activity and role in tRNA (transfer RNA) transcription from RNA polymerase III promoter, transcription initiation from RNA polymerase III promoter | N/A | N/A |
N/A, not applicable.
Figure 4.
Deletion of polyQ protein interactors does not increase toxicity or aggregation. (A) Protein interactors of 230Q. Histogram depicting the normalized total spectra count of proteins found to interact with 230Q in two biological replicates (230Q-1 and 230Q-2). (B) Toxicity in wild-type (WT) and independent sis1Δ/sis1Δ or sgt2Δ/sgt2Δ mutants, harboring 103Q or 230Q expression constructs, was assessed in the presence of a tunicamycin (Tm) gradient from 0 to 10 μg/ml, in twofold dilutions in YPD with or without 50 μg/ml doxycycline (dox) at 30 or 42°. Growth was measured after 48 hr of static incubation by absorbance at 600 nm and normalized relative to the no Tm control. For each strain, optical densities were averaged for duplicate measurements and displayed quantitatively using Treeview, as shown in the color bar. Data are representative for three biological replicates. (C) Live cell microscopy of 103Q and 230Q in sis1Δ/sis1Δ or sgt2Δ/sgt2Δ mutants grown at 42°. Scale bar, 10 μm. (D) Toxicity of WT and independent hsp104Δ/hsp104Δ or ubi4Δ/ubi4Δ mutants harboring 103Q or 230Q expression constructs was assessed in the presence of a Tm gradient from 0 to 10 μg/ml, in twofold dilutions in YPD with or without 50 μg/ml dox at 30 or 42°. Growth was measured after 48 hr of static incubation by absorbance at 600 nm and normalized relative to the no Tm control. For each strain, optical densities were averaged for duplicate measurements and displayed quantitatively using Treeview, as shown in the color bar. Data are representative for three biological replicates. (E) Live cell microscopy of 103Q and 230Q in hsp104Δ/hsp104Δ or ubi4Δ/ubi4Δ mutants grown at 42°. Scale bar, 10 μm. DIC, differential interference contrast; RFP, red fluorescent protein.
Next, we tested the impact of additional key regulators of protein homeostasis on polyQ toxicity and aggregation. Given the established role of Hsp104 in modulating polyQ aggregation and toxicity, we assessed the impact of HSP104 deletion in C. albicans on the aggregation and toxicity of 103Q and 230Q. In addition, we postulated that the ubiquitin-proteasome pathway may also play a role by targeting the expansions for efficient degradation, preventing any cellular toxicity. Thus, we also used a deletion of UBI4, the gene encoding polyubiquitin. Once again, we subjected the strains to ER stress via tunicamycin treatment and performed growth assays at 30 or 42°. Notably, the ubi4Δ/ubi4Δ is temperature-sensitive, and we could only test this mutant at 30° (Leach et al. 2011). As with SIS1 and SGT2, deletion of HSP104 or UBI4 did not exacerbate growth defects upon expression of 103Q and 230Q (Figure 4D). Again, we performed fluorescence microscopy to determine if any further aggregation was occurring in the mutants. Cells were grown at 42° for a short period of time to ensure that viability of the temperature-sensitive mutants was not affected, and then cells were imaged. Similar to our other mutants, 103Q remained evenly distributed across the cytosol and 230Q formed few punctate structures (Figure 4E).
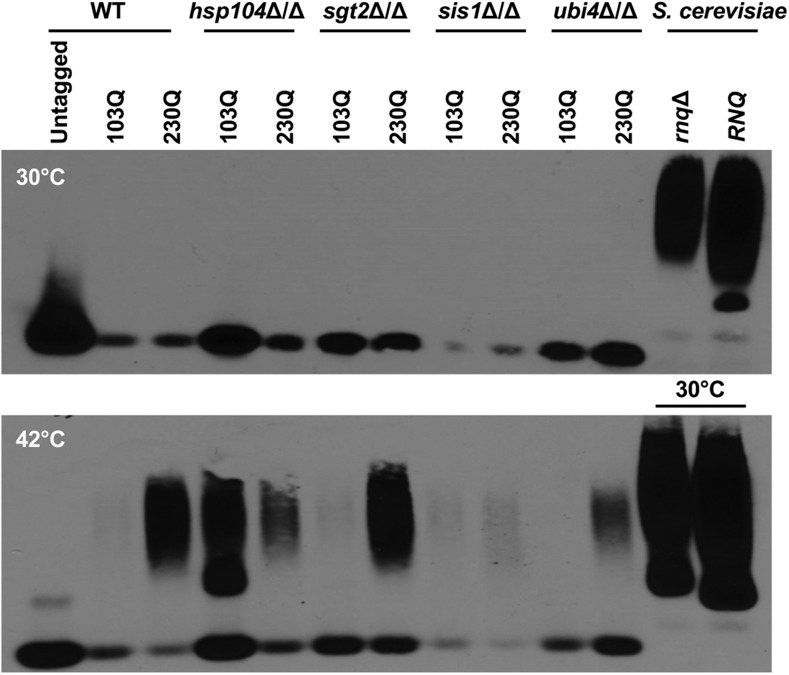
Finally, we corroborated these data by performing SDD-AGE assays in each of the deletion strains at 30 and 42°. PolyQ expansions were induced in strains grown for 24 hr, followed by subculturing for an additional 6 hr at 30 or 42°. At 30°, polyQ aggregates were observed in the S. cerevisiae 103Q expressing rnqΔ and [RNQ+] strains, but not in any of the C. albicans strains (Figure 5). However, there was considerable aggregation observed in most of our C. albicans mutants expressing 230Q grown at 42°, as well as in the hsp104Δ/hsp104Δ mutant expressing 103Q. Due to the temperature sensitivity of S. cerevisiae, aggregation of our C. albicans mutants at 42° was compared to our S. cerevisiae strains grown at 30°, whereby we still observed significantly lower levels of aggregation (Figure 5).
Figure 5.
Expression of up to 230Q in mutants involved in protein quality control does not cause aggregation in C. albicans. Semidenaturating detergent agarose gel electrophoresis analysis of aggregated proteins expressed from C. albicans wild type (WT) and different mutant backgrounds harboring 103Q or 230Q expression constructs at 30° (top) vs. 42° (bottom). S. cerevisiae 103Q in rnqΔ and [RNQ+] were grown at 30° only and were included as a control. Detection via the N-terminal FLAG-tag.
Discussion
PolyQ expansion proteins, their misfolding, aggregation, and toxicity are closely associated with at least nine neurodegenerative diseases (Fan et al. 2014). In addition, polyQ expansion proteins have successfully served as experimental paradigms to explore basic mechanisms by which cellular defense systems modulate the toxicity often associated with protein misfolding. Following this logic, we established a C. albicans model expressing polyQ expansion proteins to gain insights into the protein quality control system and stress response programs of this major human fungal pathogen. However, we found that the expression of amino-terminal fragments of a polyQ-expanded huntingtin protein (103Q and 230Q) in C. albicans does not share any of the previously established biochemical and cellular hallmarks of polyQ expansion proteins and their interactions with cellular protein control systems.
The amino acid sequence of a protein determines its propensity to misfold and aggregate, with sequences rich in asparagine and glutamine favoring aggregation (Alberti et al. 2009). Indeed, in S. cerevisiae, cultured mammalian cells, D. melanogaster, C. elegans, rodents, and many other models, expression of polyQ expansion proteins leads to considerable aggregation in the cytosol or the nucleus (Brignull et al. 2006; Bradford et al. 2010; Faber et al. 1999; Jackson et al. 1998; Duennwald et al. 2006b; Duennwald 2011), which is often accompanied by reduced cellular and organismal fitness due to impaired core cellular functions, particularly those involved in protein quality control. Strikingly, our findings clearly demonstrate that C. albicans cells expressing polyQ expansion proteins had no polyQ aggregation under basal physiological conditions, and there was negligible polyQ toxicity or transcriptional response, with the HSR also being unaffected. This result is striking given that the C. albicans polyQ constructs are expressed at higher levels than the corresponding polyQ constructs in S. cerevisiae (Figure 2D).
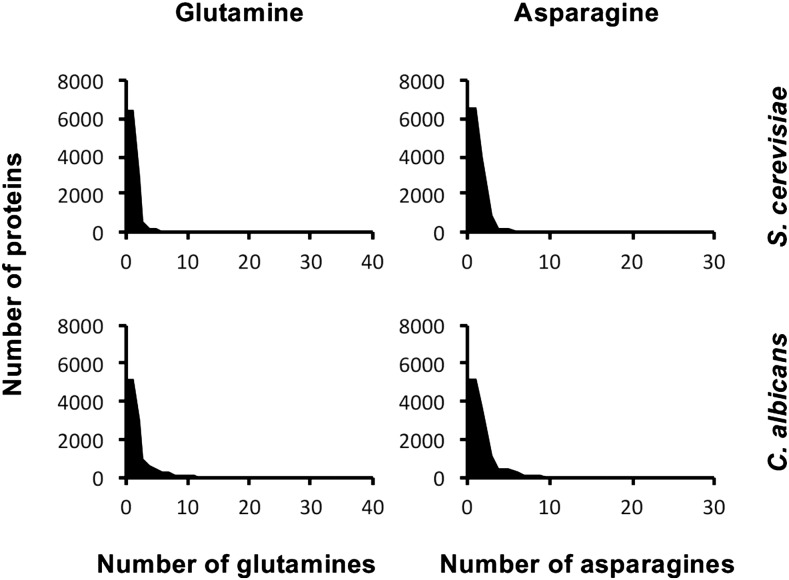
This lack of toxicity associated with polyQ expansions resonates with recent findings in D. discoideum, an organism with a proteome that is highly enriched in polyN and polyQ proteins (Malinovska et al. 2015; Santarriaga et al. 2015). In D. discoideum, polyQ aggregation could only be induced by exposure to strong proteotoxic stress conditions, such as a heat shock. This was interpreted as evidence that organisms with proteomes enriched in aggregation-prone proteins, such as polyQ expansion proteins, have evolved highly effective protein quality control systems to prevent the toxic consequences of protein misfolding. Notably, our results in C. albicans, along with studies performed in other organisms, such as S. pombe and D. melanogaster, do not fall in line with this correlation. Unlike D. discoideum, the proteome of C. albicans does not have a high proportion of aggregation-prone proteins (polyQ expansion and other proteins) (Figure 6), and S. pombe contains very few aggregation-prone proteins in its proteome (Schaefer et al. 2012). Yet both yeasts show little to no polyQ aggregation and polyQ toxicity. In fact, even when we challenge C. albicans protein quality control systems by genetic manipulation or exposure to proteotoxic stress conditions, C. albicans exhibits little polyQ aggregation and no polyQ toxicity. Further, the proteome of D. melanogaster, similar to the proteome of D. discoideum, is highly enriched in Q- and N-rich aggregation-prone proteins (Schaefer et al. 2012). Yet unlike D. discoideum, the expression of polyQ expansion proteins results in polyQ aggregation and toxicity in fly cells (Jackson et al. 1998). PolyQ-expanded proteins engage in many unusually stable protein–protein interactions and often sequester other proteins rich in N or Q repeats, which can impair their functions, resulting in cellular toxicity (Duennwald et al. 2006a; Kochneva-Pervukhova et al. 2012; Lessing and Bonini 2008; Kayatekin et al. 2014). However, the proteomes of C. albicans and S. cerevisiae do not differ significantly in the proportions of Q- or N-rich proteins in their proteasomes (Figure 6). In fact, our IP-mass spec experiments identified only eight proteins that interact with 230Q in C. albicans, only one of which is N- or Q-rich (Table 2). Some interactions may have been missed due to technical challenges associated with aggregation-prone proteins, however, the lack of polyQ aggregation and toxicity in C. albicans suggests minimal functional consequences. Clearly, there is no simple correlation between the proportion of aggregation-prone proteins in the proteome of a given organism and resistance to polyQ aggregation and toxicity.
Figure 6.
Bioinformatic analysis reveals similar features between S. cerevisiae and C. albicans Q/N-rich proteome. Length distribution of glutamine and asparagine runs in S. cerevisiae and C. albicans, with both species containing one protein with ≤40 glutamines in a row or ≤30 asparagines in a row.
We further explored the resistance of C. albicans to polyQ aggregation and toxicity by directly targeting major chaperone pathways with central roles in protein quality control. We found that the chaperones Sis1 and Sgt2 interact with 230Q in C. albicans. These chaperones are known to assist protein folding, prevent aggregation, and target misfolded proteins for proteolytic degradation (Verghese et al. 2012). Sgt2 and its mammalian homolog, SGTA, are recruited into polyglutamine aggregates in S. cerevisiae and in mammalian cells, respectively (Wear et al. 2015; Wang et al. 2007). Sgt2 may regulate prion propagation by modulating interactions of Hsp104 and Hsp70 proteins with prion polymers (Kiktev et al. 2012), and may positively regulate polyQ expansions in S. cerevisiae. Yet the deletion of SGT2 in C. albicans did not exacerbate aggregation or toxicity of polyQ expression (Figure 4, B and C). In S. cerevisiae, the Hsp40 chaperone Sis1 interacts with soluble polyQ species and transports polyQ aggregates, along with other misfolded proteins, to the nucleus for proteasomal degradation (Park et al. 2013). Depletion of SIS1 in S. cerevisiae causes protein misfolding and aggregation in the cytosol (Park et al. 2013). Yet again, we found that deletion of SIS1 coupled with polyQ expression had no effect on protein aggregation, solubility, or toxicity in C. albicans (Figure 4, B and C). Finally, even exposing C. albicans to geldanamycin to inhibit Hsp90 during expression of our polyQ-expanded Htt had no effect on toxicity (Figure 2C). Likewise, deletion of HSP104, a major fungal disaggregase, had no effect on polyQ aggregation, solubility, or toxicity (Figure 4, D and E and Figure 5).
Our results, combined with data from other model organisms, reveals a perplexing interplay between protein quality control systems, polyQ aggregation, and toxicity, and possibly more global aspects of protein misfolding. Our findings suggest that C. albicans has evolved powerful mechanisms to control protein aggregation during normal growth conditions and upon stress. Systematic analysis of aggregation and toxicity associated with polyQ expansion proteins, prions, and other proteins with a high propensity to misfold across different organisms and cell types will be central to elucidating fundamental mechanisms that have evolved across the tree of life to mitigate proteotoxicity.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.035675/-/DC1.
Acknowledgments
We thank the Donnelly Sequencing Centre for sequencing, and Jonathan Krieger at the SikKids Proteomics, Analytics, Robotics & Chemical Biology Centre at The Hospital for Sick Children for mass spectrometry analysis. M.D.L. is supported by a Sir Henry Wellcome Postdoctoral Fellowship (Wellcome Trust grant 096072), T.K. is supported by a Queen Elizabeth II Graduate Scholarship in Science and Technology (University of Toronto), M.L.D. is supported by a Canadian Institutes of Health Research (CIHR) Operating grant 325538, L.E.C. is supported by a Canada Research Chair in Microbial Genomics and Infectious Disease, by CIHR grants MOP-119520 and MOP-86452, and by the Natural Sciences and Engineering Research Council (NSERC) of Canada (grants 06261 and 462167).
Footnotes
Communicating editor: J. Berman
Literature Cited
- Alberti S., Halfmann R., King O., Kapila A., Lindquist S., 2009. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137(1): 146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bence N. F., Sampat R. M., Kopito R. R., 2001. Impairment of the ubiquitin-proteasome system by protein aggregation. Science 292(5521): 1552–1555. [DOI] [PubMed] [Google Scholar]
- Bolognesi B., Kumita J. R., Barros T. P., Esbjorner E. K., Luheshi L. M., et al. , 2010. ANS binding reveals common features of cytotoxic amyloid species. ACS Chem. Biol. 5(8): 735–740. [DOI] [PubMed] [Google Scholar]
- Bradford J. W., Li S., Li X.-J., 2010. Polyglutamine toxicity in non-neuronal cells. Cell Res. 20(4): 400–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brignull H. R., Morley J. F., Garcia S. M., Morimoto R. I., 2006. Modeling polyglutamine pathogenesis in C. elegans, pp. 256–282 in Methods in Enzymology. Academic Press, San Diego. [DOI] [PubMed] [Google Scholar]
- Burright E. N., Brent Clark H., Servadio A., Matilla T., Feddersen R. M., et al. , 1995. SCA1 transgenic mice: a model for neurodegeneration caused by an expanded CAG trinucleotide repeat. Cell 82(6): 937–948. [DOI] [PubMed] [Google Scholar]
- Cashikar A. G., Duennwald M., Lindquist S. L., 2005. A chaperone pathway in protein disaggregation. Hsp26 alters the nature of protein aggregates to facilitate reactivation by Hsp104. J. Biol. Chem. 280(25): 23869–23875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chafekar S. M., Wisén S., Thompson A. D., Echeverria A., Walter G. M., et al. , 2012. Pharmacological tuning of heat shock protein 70 modulates polyglutamine toxicity and aggregation. ACS Chem. Biol. 7(9): 1556–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F., Dobson C. M., 2006. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75(1): 333–366. [DOI] [PubMed] [Google Scholar]
- Duennwald M. L., 2011. Polyglutamine misfolding in yeast: toxic and protective aggregation. Prion 5(4): 285–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duennwald M. L., Jagadish S., Giorgini F., Muchowski P. J., Lindquist S., 2006a A network of protein interactions determines polyglutamine toxicity. Proc. Natl. Acad. Sci. USA 103(29): 11051–11056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duennwald M. L., Jagadish S., Muchowski P. J., Lindquist S., 2006b Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc. Natl. Acad. Sci. USA 103(29): 11045–11050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyao M., Ambrose C., Myers R., Novelletto A., Persichetti F., et al. , 1993. Trinucleotide repeat length instability and age of onset in Huntington’s disease. Nat. Genet. 4(4): 387–392. [DOI] [PubMed] [Google Scholar]
- Faber P. W., Alter J. R., MacDonald M. E., Hart A. C., 1999. Polyglutamine-mediated dysfunction and apoptotic death of a Caenorhabditis elegans sensory neuron. Proc. Natl. Acad. Sci. USA 96(1): 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H.-C., Ho L.-I., Chi C.-S., Chen S.-J., Peng G.-S., et al. , 2014. Polyglutamine (polyQ) diseases: genetics to treatments. Cell Transplant. 23(4–5): 441–458. [DOI] [PubMed] [Google Scholar]
- Fiumara F., Fioriti L., Kandel E. R., Hendrickson W. A., 2010. Essential role of coiled coils for aggregation and activity of Q/N-rich prions and polyQ proteins. Cell 143(7): 1121–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman J., 2001. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem. 70(1): 603–647. [DOI] [PubMed] [Google Scholar]
- Gidalevitz T., Ben-Zvi A., Ho K. H., Brignull H. R., Morimoto R. I., 2006. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science 311(5766): 1471–1474. [DOI] [PubMed] [Google Scholar]
- Grimminger-Marquardt V., Lashuel H. A., 2010. Structure and function of the molecular chaperone Hsp104 from yeast. Biopolymers 93(3): 252–276. [DOI] [PubMed] [Google Scholar]
- Gundry R.L., White M.Y., Murray C.I., Kane L.A., Fu Q., et al. , 2009. Preparation of proteins and peptides for mass spectrometry analysis in a bottom-up proteomics workflow. Curr. Protoc. Mol. Biol. 88: 10.25.1–10.25.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R., Lindquist S., 2008. Screening for amyloid aggregation by semi-denaturing detergent-agarose gel electrophoresis. J. Vis. Exp. 17: e838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn D. L., Neofytos D., Anaissie E. J., Fishman J. A., Steinbach W. J., et al. , 2009. Epidemiology and outcomes of candidemia in 2019 patients: data from the prospective antifungal therapy alliance registry. Clin. Infect. Dis. 48(12): 1695–1703. [DOI] [PubMed] [Google Scholar]
- Hughes R. E., Lo R. S., Davis C., Strand A. D., Neal C. L., et al. , 2001. Altered transcription in yeast expressing expanded polyglutamine. Proc. Natl. Acad. Sci. USA 98(23): 13201–13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis D. O., Arnaud M. B., Binkley J., Shah P., Skrzypek M. S., et al. , 2012. The Candida genome database incorporates multiple Candida species: multispecies search and analysis tools with curated gene and protein information for Candida albicans and Candida glabrata. Nucleic Acids Res. 40(Database issue): D667–D674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson G. R., Salecker I., Dong X., Yao X., Arnheim N., et al. , 1998. Polyglutamine-expanded human huntingtin transgenes induce degeneration of Drosophila photoreceptor neurons. Neuron 21(3): 633–642. [DOI] [PubMed] [Google Scholar]
- Kayatekin C., Matlack K. E. S., Hesse W. R., Guan Y., Chakrabortee S., et al. , 2014. Prion-like proteins sequester and suppress the toxicity of huntingtin exon 1. Proc. Natl. Acad. Sci. USA 111(33): 12085–12090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A., Nesvizhskii A. I., Kolker E., Aebersold R., 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74(20): 5383–5392. [DOI] [PubMed] [Google Scholar]
- Keppler-Ross S., Noffz C., Dean N., 2008. A new purple fluorescent color marker for genetic studies in Saccharomyces cerevisiae and Candida albicans. Genetics 179(1): 705–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiktev D. A., Patterson J. C., Müller S., Bariar B., Pan T., et al. , 2012. Regulation of chaperone effects on a yeast prion by cochaperone Sgt2. Mol. Cell. Biol. 32(24): 4960–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., et al. , 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14(4): R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochneva-Pervukhova N. V., Alexandrov A. I., Ter-Avanesyan M. D., 2012. Amyloid-mediated sequestration of essential proteins contributes to mutant huntingtin toxicity in yeast. PLoS One 7(1): e29832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krobitsch S., Lindquist S., 2000. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc. Natl. Acad. Sci. USA 97(4): 1589–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach M. D., Cowen L. E., 2014a Membrane fluidity and temperature sensing are coupled via circuitry comprised of Ole1, Rsp5, and Hsf1 in Candida albicans. Eukaryot. Cell 13(8): 1077–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach M. D., Cowen L. E., 2014b To sense or die: mechanisms of temperature sensing in fungal pathogens. Curr. Fungal Infect. Rep. 8(2): 185–191. [Google Scholar]
- Leach M. D., Stead D. A., Argo E., MacCallum D. M., Brown A. J., 2011. Molecular and proteomic analyses highlight the importance of ubiquitination for the stress resistance, metabolic adaptation, morphogenetic regulation and virulence of Candida albicans. Mol. Microbiol. 79(6): 1574–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach M. D., Budge S., Walker L., Munro C., Cowen L. E., et al. , 2012a Hsp90 orchestrates transcriptional regulation by Hsf1 and cell wall remodelling by MAPK signalling during thermal adaptation in a pathogenic yeast. PLoS Pathog. 8(12): e1003069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach M. D., Klipp E., Cowen L. E., Brown A. J., 2012b Fungal Hsp90: a biological transistor that tunes cellular outputs to thermal inputs. Nat. Rev. Microbiol. 10(10): 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach M. D., Tyc K. M., Brown A. J. P., Klipp E., 2012c Modelling the regulation of thermal adaptation in Candida albicans, a major fungal pathogen of humans. PLoS One 7(3): e32467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessing D., Bonini N. M., 2008. Polyglutamine genes interact to modulate the severity and progression of neurodegeneration in Drosophila. PLoS Biol. 6(2): e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovska L., Palm S., Gibson K., Verbavatz J.-M., Alberti S., 2015. Dictyostelium discoideum has a highly Q/N-rich proteome and shows an unusual resilience to protein aggregation. Proc. Natl. Acad. Sci. USA 112(20): E2620–E2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh J. L., Walker H., Theisen H., Zhu Y.-Z., Fielder T., et al. , 2000. Expanded polyglutamine peptides alone are intrinsically cytotoxic and cause neurodegeneration in Drosophila. Hum. Mol. Genet. 9(1): 13–25. [DOI] [PubMed] [Google Scholar]
- Meriin A. B., Zhang X., He X., Newnam G. P., Chernoff Y. O., et al. , 2002. Huntingtin toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J. Cell Biol. 157(6): 997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelitsch M. D., Weissman J. S., 2000. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc. Natl. Acad. Sci. USA 97(22): 11910–11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan R. D., Abmayr S. M., Workman J. L., 2014. The expanding role for chromatin and transcription in polyglutamine disease. Curr. Opin. Genet. Dev. 26: 96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley J. F., Brignull H. R., Weyers J. J., Morimoto R. I., 2002. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 99(16): 10417–10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii A. I., Keller A., Kolker E., Aebersold R., 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75(17): 4646–4658. [DOI] [PubMed] [Google Scholar]
- Noble S. M., Johnson A. D., 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell 4(2): 298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzscha H., Schermann S. M., Woerner A. C., Pinkert S., Hecht M. H., et al. , 2011. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144(1): 67–78. [DOI] [PubMed] [Google Scholar]
- Park S.-H., Kukushkin Y., Gupta R., Chen T., Konagai A., et al. , 2013. PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell 154(1): 134–145. [DOI] [PubMed] [Google Scholar]
- Park Y. N., Morschhauser J., 2005. Tetracycline-inducible gene expression and gene deletion in Candida albicans. Eukaryot. Cell 4(8): 1328–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller M. A., 2012. Antifungal drug resistance: mechanisms, epidemiology, and consequences for treatment. Am. J. Med. 125(1): S3–S13. [DOI] [PubMed] [Google Scholar]
- Ross C. A., Poirier M. A., 2004. Protein aggregation and neurodegenerative disease. Nat. Med. 10(Suppl.): S10–S17. [DOI] [PubMed] [Google Scholar]
- Santarriaga S., Petersen A., Ndukwe K., Brandt A., Gerges N., et al. , 2015. The social amoeba Dictyostelium discoideum is highly resistant to polyglutamine aggregation. J. Biol. Chem. 290(42): 25571–25578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M. H., Wanker E. E., Andrade-Navarro M. A., 2012. Evolution and function of CAG/polyglutamine repeats in protein–protein interaction networks. Nucleic Acids Res. 40: 4273–4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., 1991. Getting started with yeast. Methods Enzymol. 194: 3–21. [DOI] [PubMed] [Google Scholar]
- Thomas P. J., Qu B.-H., Pedersen P. L., 1995. Defective protein folding as a basis of human disease. Trends Biochem. Sci. 20(11): 456–459. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D. G., Sauvageau M., Goff L., Rinn J. L., et al. , 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31(1): 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verghese J., Abrams J., Wang Y., Morano K. A., 2012. Biology of the heat shock response and protein chaperones: budding yeast (Saccharomyces cerevisiae) as a model system. Microbiol. Mol. Biol. Rev. 76(2): 115–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waelter S., Boeddrich A., Lurz R., Scherzinger E., Lueder G., et al. , 2001. Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol. Biol. Cell 12(5): 1393–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Meriin A. B., Costello C. E., Sherman M. Y., 2007. Characterization of proteins associated with polyglutamine aggregates: a novel approach towards isolation of aggregates from protein conformation disorders. Prion 1(2): 128–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wear M. P., Kryndushkin D., O’Meally R., Sonnenberg J. L., Cole R. N., et al. , 2015. Proteins with intrinsically disordered domains are preferentially recruited to polyglutamine aggregates. PLoS One 10(8): e0136362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B., Edskes H. K., Shewmaker F., Nakayashiki T., 2007. Prions of fungi: inherited structures and biological roles. Nat. Rev. Microbiol. 5(8): 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimalasena T. T., Enjalbert B., Guillemette T., Plumridge A., Budge S., et al. , 2008. Impact of the unfolded protein response upon genome-wide expression patterns, and the role of Hac1 in the polarized growth, of Candida albicans. Fungal Genet. Biol. 45(9): 1235–1247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data are deposited at the Sequence Read Archive (Study Accession: SRP073330). The mass spectrometry data has been deposited into the Proteome Exchange PRIDE database (http://www.proteomexchange.org/) under the accession number PXD003916.