Abstract
Lotus (Nelumbo nucifera Gaertn.) seed proteome has been the focus of our studies, and we have recently established the first proteome dataset for its mature seed endosperm. The current study unravels the immature endosperm, as well as the embryo proteome, to provide a comprehensive dataset of the lotus seed proteins and a comparison between the mature and immature endosperm tissues across the seed’s development. One-dimensional gel electrophoresis (SDS-PAGE) linked with tandem mass spectrometry provided a protein inventory of the immature endosperm (122 non-redundant proteins) and embryo (141 non-redundant proteins) tissues. Comparing with the previous mature endosperm dataset (66 non-redundant proteins), a total of 206 non-redundant proteins were identified across all three tissues of the lotus seed. Results revealed some significant differences in proteome composition between the three lotus seed tissues, most notably between the mature endosperm and its immature developmental stage shifting the proteins from nutrient production to nutrient storage.
Keywords: 1-DGE, LC-MS/MS, lotus, seed, proteome analysis, plant proteomics
1. Introduction
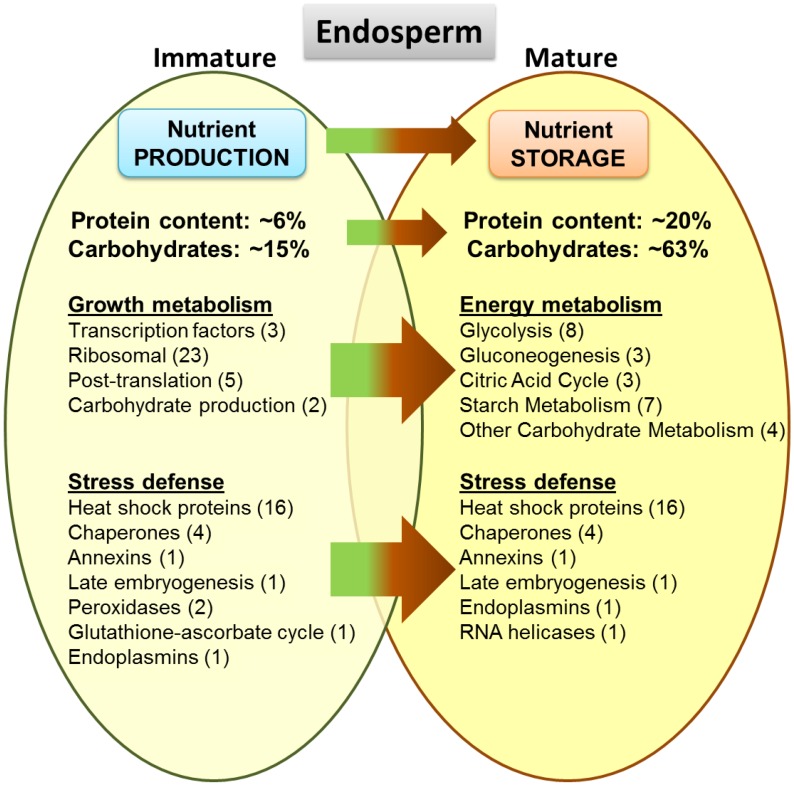
Nelumbo nucifera (Gaertn.) is an aquatic perennial belonging to the family of Nelumbonaceae, whose most used common name is the lotus. The lotus typically grows in shallow ponds, with its rhizomes under the mud and its large leaves rising on stalks 1–2 m above the water surface. Flowers are white to rosy, sweet-scented, solitary, hermaphrodite and 10–25 cm in diameter, while its fruits are ovoid having nut like achenes. Seeds are black, hard and ovoid [1]. In its immature form, the lotus seed is initially of a yellowish color (early stages) and becomes green as it grows and matures. In its late immature stages, the seed is a 1.2–1.5 cm long ovoid covered in a soft green husk containing a moist and soft endosperm and the developing embryo. When the seed reaches maturity, the husk turns dark brown and hardens, and both the endosperm and embryo become considerably dry. The lotus embryo, or germ, is a small, stalk-like tissue at the core of the lotus seed. The embryo is green and yellow in color. In the mature seed, the embryo tissue is dry, and while inside an intact seed, it can remain viable for germination for more than a thousand years, making it the most durable seed known [2,3,4,5]. The immature seed, which is composed largely of the endosperm, has a water content of 77.5%, as opposed to the 13.1% water content of the mature seed. The immature seed also has lower protein and carbohydrate content, 5.9% and14.9%, respectively, compared to 19.1% and 62.6% for the mature seed [6].
The lotus seeds and rhizome are extensively consumed as food in China and Japan and regarded as a health food [7,8,9], and the plant is also utilized as a source of traditional medicine in India and China [1,10]. Furthermore, extracts from the lotus leaves, rhizomes, and seeds have been shown possess multiple health benefits and a diverse amount of secondary metabolites (more details are given in our review [11] and references therein). The genome of the lotus has only recently been sequenced [12], and a few targeted genome and transcriptome-level works have led to the identification of some functional proteins, as well as their successful cloning and transgenic expression [13,14,15,16]. Considering its documented health benefits and several desirable characteristics for nutritional, agricultural and scientific uses, such as its protein content, ability to be cultivated in flooded areas, growth and germination vigor, and extreme seed durability, the lotus plant would consist of an excellent candidate as a crop, source of recombinant genes, or even as potential model organism. However, despite these characteristics, proteome analysis of the plant is still at the initial stages of research. Figure 1 depicts the lotus fruit and seed, its importance and proteomic study goals.
Figure 1.
Overview of the significance and goals of the proteomic research of the lotus. The fruit (seedpod) with seeds from a lotus plant growing in Ibaraki University pond, and the open seed with endosperm and embryo is shown.
Aiming to develop a proteome catalogue of the lotus plant—starting with its seed, the nutrient rich food source—the first study by our research group has unraveled the mature endosperm proteome of the lotus seed, which included the establishment of protocols for protein extraction and analyses by one-dimensional gel electrophoresis (1-DGE) and by two-dimensional gel electrophoresis (2-DGE) in conjunction with mass spectrometry [17]. In the present work, we advance our study of the lotus seed by further analyzing the endosperm of the lotus seed in its immature stage and the embryo, the other prominent component of the mature seed, by utilizing 1-DGE linked with tandem mass spectrometry proteomic approach. The resulting proteome from each tissue (immature endosperm and embryo) is compared with the mature endosperm proteins in hope to bring to light any notable differences in protein content between the different tissue locations and developmental stages.
2. Experimental Section
2.1. Plant Material and Tissues (Immature Endosperm and Embryo of Lotus Seed) Preparation
Lotus seeds, both mature and immature, were obtained from a small cultivation pond in the Ibaraki University’s College of Agriculture campus in Ami town, Ibaraki, Japan [17]. The immature seed endosperm was collected from seeds extracted from the lotus seedpod in their post-pollination late immature stage. At the point of collection, the seeds were approximately 1.3 cm long, and the external husk was still green and soft. The seeds were washed and stored whole at −80 °C until tissue extraction. The seeds were cut open and the soft and white core was removed whole and then cut across its length. The translucent sheet around the core, any discernible embryo tissue, as well as the central portion of the core immediately around the embryo was removed. The remaining soft endosperm fragments were ground under liquid nitrogen and the resulting powder was stored in sterile BD Falcon tubes at −80 °C until extraction of protein. For embryo tissue sample preparation, the mature seeds (stored at room temperature) were cracked open in a clean environment and the endosperm and embryo portions were cleanly separated and stored in sterile BD Falcon tubes at −80 °C. The embryo fragments were ground into a fine powder in liquid nitrogen, with a pre-chilled mortar and pestle. Resulting powder was stored in sterile 2.0 mL microfuge tubes at −80 °C until further analysis.
2.2. Extraction of the Lotus Seed Immature Endosperm and Embryo Proteins
Proteins were extracted from the powdered samples using the Tris-buffered saline (TBS) extraction method described in a previous study [17]. Briefly, a 3:1 mixture of TBS-20 buffer [10 mM Tris-HCl, 150 mM NaCl, pH 7.4, 0.1% (v/v) Tween-20, plus one tablet of EDTA-free proteinase inhibitor (cOmplete Mini, Roche) per 50 mL] and SDS (sodium dodecyl sulfate) reducing buffer [62 mM Tris (pH 6.8), 10% (v/v) glycerol, 2.5% (w/v) SDS, 5% (v/v) 2-mercaptoethanol] was used to extract the powdered samples at 2 mL/100 mg. The sample/buffer mixtures were also subjected to several 30 s ultrasonic bath cycles and at 95 °C heating for 5 min to help extraction. The extract was separated by centrifugation, and its proteins precipitated and purified using the ProteoExtract kit (Calbiochem). The dry protein pellets obtained were either resolubilized in LB-TT (7 M urea, 2 M thiourea, 4% (w/v) CHAPS, 18 mM Tris-HCl (pH 8.0), 14 mM Trizma base, 0.2% (v/v) Triton X-100 and 50 mM dithiothreitol) for immediate use or stored at −80 °C. Prior to use, protein content of the resolubilized extracts was measured by Bradford assay [18].
2.3. Extraction of the Lotus Seed Immature Endosperm and Embryo Proteins
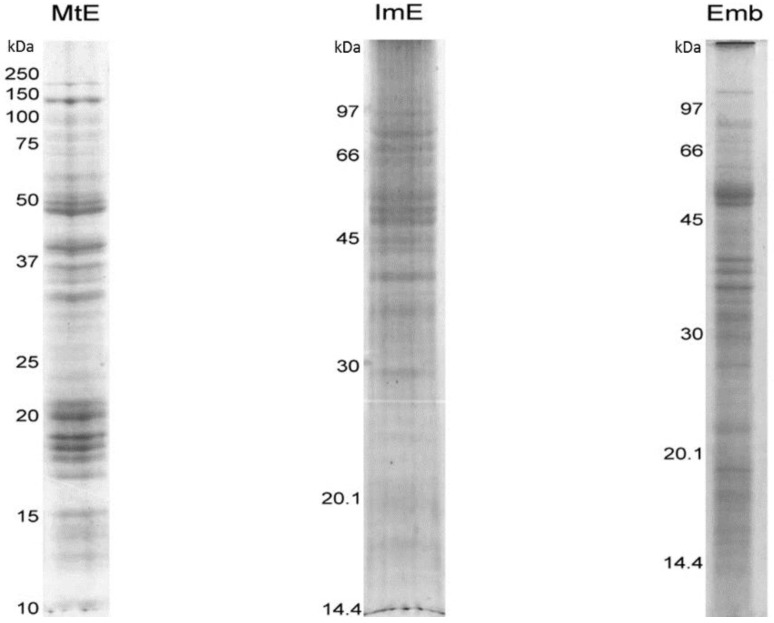
Protein samples from both tissues were subjected to 1-DGE (SDS-PAGE, 12.5%), both for visualization of protein profiles (Figure 2) using Coomassie Brilliant Blue [19] staining, and prior to analysis by 1DGE-MS.
Figure 2.
1D SDS-PAGE of protein extracts from lotus seed mature endosperm (MtE), immature endosperm (ImE), and embryo (Emb). SDS-PAGE, 12.5%; Coomassie brilliant blue stained. Molecular weight markers are shown on left-hand side of each gel image.
The 1DGE-MS analyses followed the same methodology as with the previous lotus seed analyses [17]. The extracts were initially separated using SDS-PAGE. The resulting vertical protein lanes were sliced into eight pieces of equal length (regardless of apparent protein concentration) giving fraction 1: <120 kDa, fraction 2: 120–60 kDa, fraction 3: 60–40 kDa, fraction 4: 40–30 kDa, fraction 5: 30–22 kDa, fraction 6: 22–17 kDa, fraction 7: 17–14 kDa, and fraction 8: 14–10 kDa. Each fraction was digested with 1 µg of trypsin at 37 °C for 16 h [17,18,19,20]. Digested peptides were recovered twice with 20 µL of 5% (v/v) formic acid in 50% (v/v) acetonitrile. Extracted peptides were combined and then evaporated in a vacuum concentrator until liquid was dry. Dried peptides were dissolved into 20 µL of 5% acetonitrile/0.1% formic acid and then filtrated by the Ultrafree-MC Centrifugal Filters (Millipore, PVDF 0.45 µm, Darmstadt, Germany). Liquid chromatography–tandem mass spectrometry (MS/MS) analysis was performed using the LTQ-Orbitrap XL-HTC-PAL system (Thermo, Waltham, MA, USA). Trypsin digests were loaded on the column (100 µm internal diameter, 15 cm length, l-Column, CERI) using the Paradigm MS4 HPLC pump (Michrom BioResources, Auburn, AL, USA) and HTC-PAL Autosampler (CTC Analytics, Zwingen, Switzerland), and were eluted by a gradient of 5%–45% (v/v) acetonitrile in 0.1% (v/v) formic acid for 26 min. The eluted peptides were introduced directly into an LTQ-Orbitrap with a flow rate of 500 nL/min, and a spray voltage of 2.0 kV. The range of MS scan was m/z 450–1500. The top three peaks were subjected to MS/MS analysis. MS/MS spectra were analyzed by Mascot server (version 2.4.1, Matrix Science, Boston, MA, USA) in house (http://www.matrixscience.com/) and compared against proteins registered in the SwissProt (SwissProt_2012_03) database (total sequences: 428650; sequences after taxonomy filter (Viridiplantae): 27008; date: 26 July 2013). The Mascot search parameters were set as follows: threshold of the ion score cutoff, 0.05, peptide tolerance, 10 ppm, MS/MS tolerance, 0.5 Da, and peptide charge, 2+ or 3+. The search was also set to allow one missed cleavage by trypsin, a carboxymethylation modification of Cys residues, and a variable oxidation modification of Met residues. Gene ontology analysis on the data was performed using the Uniprot (www.uniprot.org) and the EMBL-EBI (www.ebi.ac.uk) databases.
3. Results and Discussion
3.1. Protein Content of the Immature Endosperm and Embryo Tissues
Protein extracts from the lotus immature seed endosperm presented very low protein yield (ca. 1.5% in the TBS method), requiring larger amounts of tissue to be extracted in order to obtain a suitable amount of protein. The reason for low protein yield lies in the high water content of the immature seed compared to its mature form. The lotus seed embryo showed a similar total protein yield to the endosperm extract [17] when extracted by the TBS/clean-up method (ca. 9%, compared to ca. 11% for the mature endosperm).
A comparison of the of the 1-D band profile on the SDS-PAGE of the embryo extract with the endosperm one showed many similarities, but also some noticeable differences, such as an absence of strongly stained bands at ca. 20 kDa and 40 kDa, and more numerous bands at low-molecular weights, under 30 and 20 kDa (see above, Figure 2). In the case of the immature endosperm, the 1-D profile is more similar to the mature endosperm than the embryo, but still was found to be different from both tissues profiles. Compared with the mature endosperm extract, the immature endosperm extract most notably does not present a high amount of protein bands around the 20 kDa range. The cluster of bands around 50 kDa is similar to that in both the endosperm and embryo, and the immature endosperms profile of bands in the 60–90 kDa range seems more similar to the mature endosperm than the embryo.
3.2. Lotus Immature Endosperm Proteins Identified by 1-DGE and MS/MS Analyses
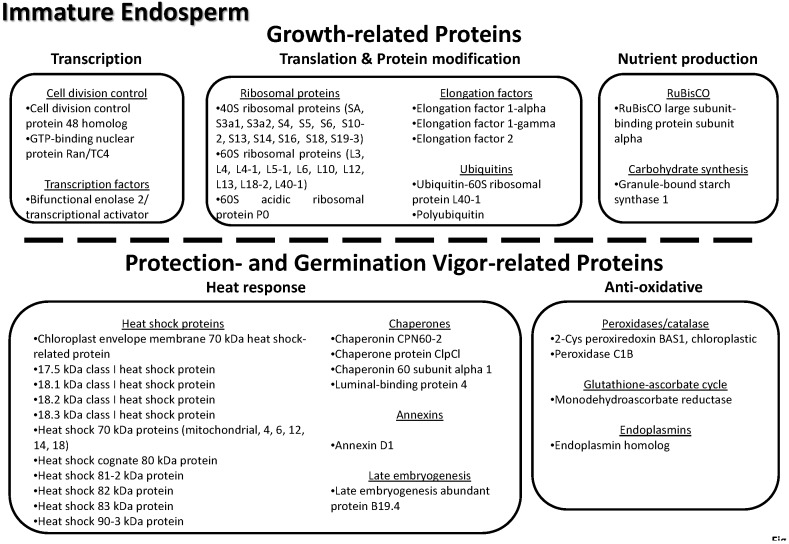
The 1-DGE separation (SDS-PAGE) of proteins in an extract, followed by MS/MS analysis is part of the so-called “bottom-up” approach to proteomics, a methodology in which proteins are proteolytically digested into peptides prior to mass spectrometric analysis, and the ensuing peptide masses and sequences are used to identify corresponding proteins. This simple approach is a useful method for performing large-scale analyses of complex samples [21]. For the sample consisting of a purified extract of lotus immature endosperm proteins, after separation by SDS-PAGE, the sample was divided into eight fractions, analyzed by LC-MS/MS, and matched against a green plant database, as detailed in the Experimental Section. Results revealed more than 500 protein matches with at least two confirmed peptide fragment matches were identified amongst all fractions, and from these 333 unique protein matches were identified. Different database matches that were likely to refer to the same protein in the sample, such as two or more matches for the same protein but from different database organisms, were grouped together based on taxonomical proximity and similarity of identified peptide sequences. Finally, 122 non-redundant (nr) protein matches were listed, along with the number of repeated matches found for each one (Table 1), with the protein match listed being the one with the highest score amongst its group of similar proteins.
Table 1.
List of top-scored non-redundant (nr) protein matches of the lotus immature endosperm 1-D shotgun mass spectroscopy results, as matched to Green Plant proteome database (SwissProt 57.0, http://www.uniprot.org/statistics/UniProtKB%2015).
| Fractions 1 | Protein Accession | Protein Description | Similar 2 | Score 3 | Cover (%) | PEPTIDE Sequences | Sig. Peptide Number | Func. Cat. 4 |
|---|---|---|---|---|---|---|---|---|
| 6,5,8,(7,4,3,1) | ENO1_HEVBR | Enolase 1 OS = Hevea brasiliensis | 11 | 1471 | 43.4 | TAIAK, YNQLLR, LTSEIGEK, ACNALLLK, DGGSDYLGK, AGWGVMASHR, EKACNALLLK, MGAEVYHHLK, RAGWGVMASHR, LGANAILAVSLAVCK, VQIVGDDLLVTNPK, AAVPSGASTGIYEALELR, LAMQEFMILPVGASSFK, SGETEDTFIADLSVGLATGQIK, YGQDATNVGDEGGFAPNIQENK, KYGQDATNVGDEGGFAPNIQENK, YGQDATNVGDEGGFAPNIQENKEGLELLK | 17 | II |
| 7,8,6,5,1,4,2 | G3PC_ANTMA | Glyceraldehyde-3-phosphate dehydrogenase, cytosolic OS = Antirrhinum majus | 23 | 1242 | 43.6 | AAAHLK, KATYEQIK, AAIKEESEGK, AGIALNDNFVK, DAPMFVVGVNEK, AASFNIIPSSTGAAK, VPTVDVSVVDLTVR, DAPMFVVGVNEKEYK, VPTVDVSVVDLTVRLEK, FGIVEGLMTTVHSITATQK, GILGYTEDDVVSTDFVGDSR, LTGMSFRVPTVDVSVVDLTVR, LKGILGYTEDDVVSTDFVGDSR, VINDRFGIVEGLMTTVHSITATQK | 14 | II |
| 4,8,6,7,5 | HSP7D_ARATH | Heat shock 70 kDa protein 4 OS = Arabidopsis thaliana | 10 | 625 | 23.8 | IEEVD, LSKEEIEK, ITITNDKGR, DAGVISGLNVMR, NALENYAYNMR, MVNHFVQEFKR, TTPSYVAFTDSER, IINEPTAAAIAYGLDK, ATAGDTHLGGEDFDNR, NAVVTVPAYFNDSQR, IINEPTAAAIAYGLDKK, EQIFSTYSDNQPGVLIQVYEGER | 12 | IX |
| 4 | HSP7E_SPIOL | Chloroplast envelope membrane 70 kDa heat shock-related protein OS = Spinacia oleracea | 1 | 580 | 21.7 | LSKEEIEK, DAGVISGLNVMR, EIAEAYLGSTVK, NALENYAYNMR, TTPSYVAFTDSER, IINEPTAAAIAYGLDK, ATAGDTHLGGEDFDNR, NAVVTVPAYFNDSQR, IINEPTAAAIAYGLDKK, EQVFSTYSDNQPGVLIQVYEGER | 10 | IX |
| 4 | BIP4_TOBAC | Luminal-binding protein 4 OS = Nicotiana tabacum | 5 | 542 | 21.6 | VQQLLK, NTVIPTKK, IMEYFIK, LSQEEIER, ITITNDKGR, DYFDGKEPNK, FEELNNDLFR, EAEEFAEEDKK, IVNKDGKPYIQVK, ARFEELNNDLFR, NGHVEIIANDQGNR, IINEPTAAAIAYGLDK, IINEPTAAAIAYGLDKK, IKDAVVTVPAYFNDAQR | 14 | IX |
| 4,5,6,7,8 | METE_ARATH | 5-methyltetrahydropteroyltriglutamate—homocysteine methyltransferase OS = Arabidopsis thaliana | 4 | 516 | 19.2 | AAAALK, VVEVNALAK, SWLAFAAQK, AVNEYKEAK, YLFAGVVDGR, SDEKLLSVFR, FALESFWDGK, GNASVPAMEMTK, YGAGIGPGVYDIHSPR, GMLTGPVTILNWSFVR | 10 | I |
| 6,1,5,(7,8,3,2,4) | EF1A_TOBAC | Elongation factor 1-alpha OS = Nicotiana tabacum | 8 | 484 | 34.7 | YDEIVK, GFVASNSK, QTVAVGVIK, EVSSYLKK, LPLQDVYK, ARYDEIVK, IGGIGTVPVGR, STNLDWYK, STTTGHLIYK, EHALLAFTLGVK, GFVASNSKDDPAK, YYCTVIDAPGHR, MIPTKPMVVETFSEYPPLGR, NMITGTSQADCAVLIIDSTTGGFEAGISK | 14 | V |
| 4 | HSP7L_ARATH | Heat shock 70 kDa protein 12 OS = Arabidopsis thaliana | 1 | 479 | 16.5 | VQQLLK, NTVIPTKK, IMEYFIK, FDLTGVPPAPR, FEELNNDLFR, EAEEFAEEDKK, ARFEELNNDLFR, NGHVEIIANDQGNR, IINEPTAAAIAYGLDK, IINEPTAAAIAYGLDKK, IKDAVVTVPAYFNDAQR | 11 | IX |
| 4,8,6 | HSP7N_ARATH | Heat shock 70 kDa protein 18 OS = Arabidopsis thaliana | 1 | 474 | 18.5 | ITITNDKGR, EIAEAYLGSSIK, MVNHFVQEFKR, TTPSYVAFTDSER, IINEPTAAAIAYGLDK, ATAGDTHLGGEDFDNR, NAVVTVPAYFNDSQR, IINEPTAAAIAYGLDKK | 8 | IX |
| 7 | MDHM_CITLA | Malate dehydrogenase, mitochondrial OS = Citrullus lanatus | 5 | 459 | 18.2 | TFYAGK, LFGVTTLDVVR, TQDGGTEVVEAK, DDLFNINAGIVK, KLFGVTTLDVVR, RTQDGGTEVVEAK, VAVLGAAGGIGQPLALLMK, KVAVLGAAGGIGQPLALLMK | 8 | II |
| 4,5 | HSP80_SOLLC | Heat shock cognate protein 80 OS = Solanum lycopersicum | 1 | 427 | 20.9 | AVENSPFLEK, LGIHEDSQNR, ADLVNNLGTIAR, KAVENSPFLEK, HFSVEGQLEFK, GIVDSEDLPLNISR, SLTNDWEEHLAVK, SGDEMTSLKDYVTR, KPEEITKEEYAAFYK, MKEGQNDIYYITGESK | 10 | IX |
| 5 | CH62_MAIZE | Chaperonin CPN60-2, mitochondrial OS = Zea mays | 6 | 422 | 23.3 | VTDALNATK, GVEELADAVK, IGGASEAEVGEK, SVAAGMNAMDLR, IGGASEAEVGEKK, NVVIEQSFGAPK, AAVEEGIVPGGGVALLYASK, TPVHTIASNAGVEGAVVVGK, QRPLLIVAEDVESEALGTLIINK | 9 | IX |
| 7,8,6,1 | ACT_GOSHI | Actin OS = Gossypium hirsutum | 19 | 414 | 32.6 | AGFAGDDAPR, GYSFTTTAER, EITALAPSSMK, DAYVGDEAQSK, AVFPSIVGRPR, DAYVGDEAQSKR, SYELPDGQVITIGAER, VAPEEHPVLLTEAPLNPK, TTGIVLDSGDGVSHTVPIYEGYALPHAILR | 9 | VII |
| 5,8,7 | ENO2_ARATH | Bifunctional enolase 2/transcriptional activator OS = Arabidopsis thaliana | 1 | 411 | 34.5 | YNQLLR, DGGSDYLGK, ISGDALKDLYK, LGANAILAVSLAVCK, VNQIGSVTESIEAVK, TYDLNFKEENNNGSQK, SGETEDTFIADLAVGLSTGQIK, YGQDATNVGDEGGFAPNIQENK, YGQDATNVGDEGGFAPNIQENKEGLELLK | 9 | IV |
| 4,5,6 | HSP83_IPONI | Heat shock protein 83 OS = Ipomoea nil | 1 | 405 | 23 | VIVTTK, VVVSDR, AILFVPK, DVDGEQLGR, APFDLFDTR, AVENSPFLER, LGIHEDSQNR, LDAQPELFIR, RAPFDLFDTR, ADLVNNLGTIAR, ELISNASDALDK, HFSVEGQLEFK, GVVDSDDLPLNISR, ELISNASDALDKIR, ITLFLKEDQLEYLEER | 14 | IX |
| 7,6,1,2 | ADH1_SOLTU | Alcohol dehydrogenase 1 OS = Solanum tuberosum | 7 | 390 | 23.7 | ELELEK, SDIPSVVEK, FGVTEFVNPK, GTFFGNYKPR, THPMNLLNER, KFGVTEFVNPK, YMNKELELEK, TLKGTFFGNYKPR, GSSVAIFGLGAVGLAAAEGAR | 9 | II |
| 4,5,7,8 | ENPL_CATRO | Endoplasmin homolog OS = Catharanthus roseus | 3 | 387 | 10.5 | FWNEFGK, ESFKELTK, YGWSSNMER, ELISNASDALDK, IMQSQTLSDASK, GLVDSDTLPLNVSR, ELISNASDALDKIR, VFISDEFDELLPK, RVFISDEFDELLPK | 9 | IX |
| 7,(8,6) | RBL_MAIZE | Ribulose bisphosphate carboxylase large chain OS = Zea mays | 52 | 382 | 27.5 | AMHAVIDR, AQAETGEIK, DTDILAAFR, DDFIEKDR, VALEACVQAR, EITLGFVDLLR, LTYYTPEYETK, MSGGDHIHSGTVVGK, YGRPLLGCTIKPK, GGLDFTKDDENVNSQPFMR | 10 | I |
| 6 | SAHH_MEDSA | Adenosylhomocysteinase OS = Medicago sativa | 3 | 369 | 15.1 | ATDVMIAGK, HSLPDGLMR, ITIKPQTDR, TEFGPSQPFK, VAVVCGYGDVGK, SKFDNLYGCR, IVGVSEETTTGVK, IVGVSEETTTGVKR | 8 | I |
| 4,(5,6) | HSP82_ORYSJ | Heat shock protein 81-2 OS = Oryza sativa subsp. Japonica | 2 | 368 | 21.5 | VVVSDR, IAELLR, AILFVPK, APFDLFDTR, AVENSPFLEK, RAPFDLFDTR, KAVENSPFLEK, SDLVNNLGTIAR, HFSVEGQLEFK, GIVDSEDLPLNISR, SLTNDWEEHLAVK, HSEFISYPISLWTEK, KPEEITKEEYAAFYK | 12 | IX |
| 4,6,7 | HSP70_DAUCA | Heat shock 70 kDa protein OS = Daucus carota | 1 | 335 | 15.7 | IEEVD, NALENYAYNMR, NQVAMNPSNTVFDAK, NQVAMNPSNTVFDAKR, SINPDEAVAYGAAVQAAILSGEGNER, EQIFSTYSDNQPGVLIQVYEGER | 6 | IX |
| 5 | CPNA1_ARATH | Chaperonin 60 subunit alpha 1, chloroplastic OS = Arabidopsis thaliana | 1 | 331 | 16.4 | KVTISK, VVNDGVTIAR, NVVLDEFGSPK, VGAATETELEDR, GYISPQFVTNPEK, TNDSAGDGTTTASILAR | 6 | IX |
| 4,5 | HSP82_MAIZE | Heat shock protein 82 OS = Zea mays | 2 | 297 | 14.4 | APFDLFDTR, AVENSPFLER, LGIHEDSQNR, RAPFDLFDTR, SDLVNNLGTIAR, ELISNASDALDK, HFSVEGQLEFK, GVVDSDDLPLNISR, ELISNASDALDKIR | 9 | IX |
| 8,7 | ALF_CICAR | Fructose-bisphosphate aldolase, cytoplasmic isozyme OS = Cicer arietinum | 2 | 289 | 14.2 | ANSEATLGTYK, GILAADESTGTIGK, GILAADESTGTIGKR, YHDELIANAAYIGTPGK | 4 | II |
| 4,(3,5) | CD48A_ARATH | Cell division control protein 48 homolog A OS = Arabidopsis thaliana | 3 | 285 | 14 | TLLAK, KGDLFLVR, ELVELPLR, LAEDVDLER, LAGESESNLR, GILLYGPPGSGK, IVSQLLTLMDGLK, ELVELPLRHPQLFK, NAPSIIFIDEIDSIAPK | 9 | III/IV |
| 7,8,6,1 | PGKH_TOBAC | Phosphoglycerate kinase, chloroplastic OS = Nicotiana tabacum | 6 | 284 | 15.4 | AAVPTIK, AHASTEGVTK, FAVGTEAIAK, VILSSHLGRPK, GVTTIIGGGDSVAAVEK, LASLADLYVNDAFGTAHR, KLASLADLYVNDAFGTAHR | 7 | II |
| 5,(4) | PGMC_POPTN | Phosphoglucomutase, cytoplasmic OS = Populus tremula | 1 | 281 | 12.2 | YLFEDGSR, FFEVPTGWK, LSGTGSEGATIR, SMPTSAALDVVAK, YDYENVDAGAAK, VETTPFGDQKPGTSGLR | 6 | II |
| 4,(5) | HS903_ARATH | Heat shock protein 90-3 OS = Arabidopsis thaliana | 3 | 269 | 20.2 | IAELLR, AILFVPK, AVENSPFLEK, LGIHEDSQNR, ADLVNNLGTIAR, KAVENSPFLEK, HFSVEGQLEFK, GIVDSEDLPLNISR, HSEFISYPISLWIEK | 9 | IX |
| 5,6 | PMG2_ARATH | Probable 2,3-bisphosphoglycerate-independent phosphoglycerate mutase 2 OS = Arabidopsis thaliana | 1 | 257 | 13.6 | VHILTDGR, ARDAILSGK, LVDLALASGK, TFACSETVK, MKALEIAEK, GWDAQVLGEAPHK, RGWDAQVLGEAPHK, AVGPIVDGDAVVTFNFR | 8 | II |
| 5,6 | PMG1_ARATH | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase 1 OS = Arabidopsis thaliana | 5 | 224 | 10.2 | VHILTDGR, ARDAILSGK, LDQLQLLIK, GWDAQVLGEAPHK, RGWDAQVLGEAPHK, AVGPIVDGDAVVTFNFR | 6 | II |
| 5 | SSG1_HORVU | Granule-bound starch synthase 1, chloroplastic/amyloplastic OS = Hordeum vulgare | 5 | 214 | 7.5 | FFHCYK, EALQAEVGLPVDR, FSLLCQAALEAPR, VAFCIHNISYQGR | 4 | I |
| 6 | RL4_PRUAR | 60S ribosomal protein L4 OS = Prunus armeniaca | 3 | 200 | 20.8 | AGQGAFGNMCR, AGHQTSAESWGTGR, YAVVSAIAASAVPSLVLAR, AWYQTMISDSDYTEFDNFTK | 4 | V |
| 8 | H2B_GOSHI | Histone H2B OS = Gossypium hirsutum | 5 | 200 | 49 | IYIFK, LVLPGELAK, AMGIMNSFINDIFEK | 3 | VII |
| 5 | RUBA_RICCO | RuBisCO large subunit-binding protein subunit alpha (Fragment) OS = Ricinus communis | 2 | 200 | 16 | NVVLDEFGSPK VGAATETELEDR, GYISPQFVTNPEK, LGLLSVTSGANPVSIK | 4 | I |
| 8 | H2B1_MEDTR | Probable histone H2B.1 OS = Medicago truncatula | 2 | 199 | 45.3 | IYIFK, LVLPGELAK, AMGIMNSFINDIFEK | 3 | VII |
| 8 | RL182_ARATH | 60S ribosomal protein L18-2 OS = Arabidopsis thaliana | 1 | 185 | 13.4 | APLGQNTVLLR, AGGECLTFDQLALR | 2 | V |
| 5 | CALR_BERST | Calreticulin OS = Berberis stolonifera | 3 | 175 | 9.1 | LAEETWGK, LLSGDVDQK, KLAEETWGK, TLVFQFSVK, LLSGDVDQKK, YVGIELWQVK | 6 | V |
| 8 | 1433E_TOBAC | 14-3-3-like protein E OS = Nicotiana tabacum | 5 | 172 | 26.8 | NVIGAR, NLLSVAYK, DSTLIMQLLR, TVDVEELTVEER, IISSIEQKEESR, SAQDIALAELAPTHPIR | 6 | VIII |
| 8 | H4_ARATH | Histone H4 OS = Arabidopsis thaliana | 1 | 160 | 45.6 | TLYGFGG, IFLENVIR, DAVTYTEHAR, ISGLIYEETR, DNIQGITKPAIR | 5 | VII |
| 6,5,8 | KPYC_SOYBN | Pyruvate kinase, cytosolic isozyme OS = Glycine max | 2 | 157 | 9.2 | KGSDLVNVR, GDLGMEIPVEK, VENQEGVLNFDEILR | 3 | II |
| 8 | RS6_ASPOF | 40S ribosomal protein S6 OS = Asparagus officinalis | 2 | 155 | 11.6 | LVTPLTLQR, ISQEVSGDALGEEFK, ISQEVSGDALGEEFKGYVFK | 3 | V |
| 5 | TCPA_ARATH | T-complex protein 1 subunit alpha OS = Arabidopsis thaliana | 1 | 153 | 6.4 | YFVEAGAIAVR, VLVELAELQDR, NKIHPTSIISGYR | 3 | V |
| 1 | ADT1_GOSHI | ADP, ATP carrier protein 1, mitochondrial OS = Gossypium hirsutum | 1 | 143 | 5.7 | SSLDAFSQILK, LLIQNQDEMIK | 2 | VI |
| 4 | HSP7S_SPIOL | Stromal 70 kDa heat shock-related protein, chloroplastic (Fragment) OS = Spinacia oleracea | 2 | 142 | 7.2 | QFAAEEISAQVLR, AVVTVPAYFNDSQR, IINEPTAASLAYGFEK | 3 | IX |
| 7,8 | GCST_PEA | Aminomethyltransferase, mitochondrial OS = Pisum sativum | 2 | 140 | 14.7 | LYFGEFR, GGAIDDSVITK, SLLALQGPLAAPVLQHLTK, TGYTGEDGFEISVPSEHGVELAK | 4 | I |
| 3,2 | HSP7O_ARATH | Heat shock 70 kDa protein 14 OS = Arabidopsis thaliana | 1 | 139 | 7.7 | ILSHAFDR, AVLDAATIAGLHPLR, AVEKEFEMALQDR, RAVLDAATIAGLHPLR | 4 | IX |
| 4 | HSP7F_ARATH | Heat shock 70 kDa protein 6, chloroplastic OS = Arabidopsis thaliana | 1 | 139 | 7.5 | TTPSVVAYTK, QFAAEEISAQVLR, QAVVNPENTFFSVK, LSFKDIDEVILVGGSTR | 4 | IX |
| 8 | RS4_GOSHI | 40S ribosomal protein S4 OS = Gossypium hirsutum | 2 | 135 | 20.6 | LSIIEEAR, LGNVFTIGK, FDVGNVVMVTGGR, LGGAFAPKPSSGPHK | 4 | V |
| 3,4 | CLPA_BRANA | ATP-dependent Clp protease ATP-binding subunit clpA homolog, chloroplastic (Fragment) OS = Brassica napus | 1 | 135 | 5.8 | VIGQDEAVK, TAIAEGLAQR, YRGEFEER, VLELSLEEAR | 4 | I |
| 4,5,8 | EF2_BETVU | Elongation factor 2 OS = Beta vulgaris | 1 | 131 | 5.5 | GGGQIIPTAR, EGALAEENMR, RVFYASQLTAKPR, LWGENFFDPATKK | 4 | V |
| 8 | RL12_PRUAR | 60S ribosomal protein L12 OS = Prunus armeniaca | 1 | 129 | 22.3 | VSVVPSAAALVIK, VTGGEVGAASSLAPK | 2 | V |
| 6,(7,8) | ATPBM_NICPL | ATP synthase subunit beta, mitochondrial OS = Nicotiana plumbaginifolia | 4 | 129 | 12.5 | VLNTGSPITVPVGR, TVLIMELINNVAK, IPSAVGYQPTLATDLGGLQER | 3 | I |
| 4,(7) | PHSH_SOLTU | Alpha-glucan phosphorylase, H isozyme OS = Solanum tuberosum | 6 | 125 | 5.6 | AFATYTNAK, QLLNILGVIYR, HMEIIEEIDKR, TIAYTNHTVLPEALEK | 4 | II |
| 6 | RL3_ORYSJ | 60S ribosomal protein L3 OS = Oryza sativa subsp. Japonica | 2 | 124 | 6.9 | VIAHTQIR, HGSLGFLPR, GKGYEGVVTR | 3 | V |
| 8 | TPIS_MAIZE | Triosephosphate isomerase, cytosolic OS = Zea mays | 2 | 124 | 11.9 | FFVGGNWK, VAYALSQGLK, VIACVGETLEQR | 3 | VII |
| 8 | LE194_HORVU | Late embryogenesis abundant protein B19.4 OS = Hordeum vulgare | 1 | 121 | 9.2 | GGLSTMNESGGER, KGGLSTMNESGGER | 2 | IX |
| 1,(2) | AVP_VIGRR | Pyrophosphate-energized vacuolar membrane proton pump OS = Vigna radiata var. Radiata | 2 | 117 | 3.3 | AADVGADLVGK, YIEAGASEHAR, AADVGADLVGKVER | 3 | VI |
| 8 | RL6_MESCR | 60S ribosomal protein L6 OS = Mesembryanthemum crystallinum | 1 | 115 | 10.7 | VDISGVNVEK, ASITPGTVLIILAGR | 2 | V |
| 5 | SSG1_ARATH | Probable granule-bound starch synthase 1, chloroplastic/amyloplastic OS = Arabidopsis thaliana | 1 | 115 | 3.9 | FFHCYK, YGTVPIVASTGGLVDTVK | 2 | I |
| 8 | RL10_VITRI | 60S ribosomal protein L10 OS = Vitis riparia | 1 | 114 | 10.5 | VSIGQVLLSVR, ENVSSEALEAAR | 2 | V |
| 8 | RS18_ARATH | 40S ribosomal protein S18 OS = Arabidopsis thaliana | 1 | 111 | 21.7 | LRDDLER, VLNTNVDGK, IMFALTSIK, IPDWFLNR | 4 | V |
| 7 | AATM_LUPAN | Aspartate aminotransferase P2, mitochondrial (Fragment) OS = Lupinus angustifolius | 1 | 111 | 6.8 | IADVIQEK, LNLGVGAYR, VATVQGLSGTGSLR | 3 | I |
| 8 | GBLPA_ORYSJ | Guanine nucleotide-binding protein subunit beta-like protein A OS = Oryza sativa subsp. Japonica | 1 | 110 | 9 | DGVTLLWDLAEGK, FSPNTFQPTIVSGSWDR | 2 | VIII |
| 8 | H2AX_CICAR | Histone H2AX OS = Cicer arietinum | 1 | 108 | 15.1 | AGLQFPVGR, GKGEIGSASQEF | 2 | VII |
| 4 | VATA_GOSHI | V-type proton ATPase catalytic subunit A OS = Gossypium hirsutum | 2 | 108 | 5 | LAADTPLLTGQR, LVSQKFEDPAEGEEALVAK | 2 | VI |
| 8 | PARP3_SOYBN | Poly [ADP-ribose] polymerase 3 OS = Glycine max | 1 | 107 | 4.2 | VLCSQEIYK, LEPLVANFMK, LFEEITGNEFEPWER | 3 | III |
| 3,4,(5) | CLPC1_ARATH | Chaperone protein ClpC1, chloroplastic OS = Arabidopsis thaliana | 5 | 106 | 5.4 | TAIAEGLAQR, YRGEFEER, VLELSLEEAR | 3 | IX |
| 8 | NDK1_ARATH | Nucleoside diphosphate kinase 1 OS = Arabidopsis thaliana | 2 | 103 | 9.4 | NVIHGSDSVESAR, NVIHGSDSVESARK | 2 | I |
| 8 | RL13_TOBAC | 60S ribosomal protein L13 OS = Nicotiana tabacum | 1 | 98 | 16.3 | SLEGLQTNVQR, KLAPTIGIAVDHR | 2 | V |
| 8 | RS5_CICAR | 40S ribosomal protein S5 (Fragment) OS = Cicer arietinum | 2 | 95 | 15.2 | GSSNSYAIK, AQCPIVER, VNQAIYLLTTGAR | 3 | V |
| 5,6 | PDC2_ORYSI | Pyruvate decarboxylase isozyme 2 OS = Oryza sativa subsp. Indica | 2 | 94 | 4.5 | AVKPVLVGGPK, ILHHTIGLPDFSQELR | 2 | II |
| 8 | HSP14_SOYBN | 17.5 kDa class I heat shock protein OS = Glycine max | 4 | 92 | 24.7 | AIEISG, ADIPGLK, VLQISGER, FRLPENAK | 4 | IX |
| 6 | AMPL1_ARATH | Leucine aminopeptidase 1 OS = Arabidopsis thaliana | 2 | 92 | 4.6 | GLTFDSGGYNIK, TIEVNNTDAEGR | 2 | I/IX |
| 6 | ACT5_ARATH | Putative actin-5 OS = Arabidopsis thaliana | 1 | 92 | 15.9 | AGFAGDDAPR, IWHHTFYNELR | 2 | VII |
| 8 | RS14_CHLRE | 40S ribosomal protein S14 OS = Chlamydomonas reinhardtii | 1 | 87 | 15.7 | TPGPGAQSALR, IEDVTPIPTDSTR | 2 | V |
| 8 | RS3A1_VITVI | 40S ribosomal protein S3a-1 OS = Vitis vinifera | 2 | 86 | 6.5 | TTDNYTLR, LRAEDVQGR | 2 | V |
| 7 | AAT3_ARATH | Aspartate aminotransferase, chloroplastic OS = Arabidopsis thaliana | 1 | 83 | 4.9 | LNLGVGAYR, TEEGKPLVLNVVR | 2 | I |
| 1 | COB21_ORYSJ | Coatomer subunit beta-1 OS = Oryza sativa subsp. Japonica | 1 | 83 | 4.5 | HNEIQTVNIK, DTNTFASASLDR | 2 | VI |
| 8 | GRDH1_ARATH | Glucose and ribitol dehydrogenase homolog 1 OS = Arabidopsis thaliana | 3 | 83 | 8 | GAIVAFTR, EGSSIINTTSVNAYK | 2 | II |
| 8 | ANXD1_ARATH | Annexin D1 OS = Arabidopsis thaliana | 1 | 80 | 5 | AQINATFNR, SKAQINATFNR | 2 | IX |
| 7 | PDI21_ORYSJ | Protein disulfide isomerase-like 2-1 OS = Oryza sativa subsp. Japonica | 1 | 80 | 10.9 | KLAPEYEK, YGVSGFPTLK, YGVSGYPTIQWFPK | 3 | V |
| 8,(6) | ATPAM_NICPL | ATP synthase subunit alpha, mitochondrial OS = Nicotiana plumbaginifolia | 4 | 79 | 9.4 | VVSVGDGIAR, TAIAIDTILNQK | 2 | I |
| 8,(1) | CB2_PHYPA | Chlorophyll a-b binding protein, chloroplastic OS = Physcomitrella patens subsp. Patens | 1 | 75 | 3.7 | ELEVIHAR, NRELEVIHAR | 2 | II |
| 8 | HSP12_SOYBN | Class I heat shock protein (Fragment) OS = Glycine max | 1 | 75 | 18.9 | AIEISG, ILQISGER | 2 | IX |
| 8 | BAS1_ORYSJ | 2-Cys peroxiredoxin BAS1, chloroplastic OS = Oryza sativa subsp. Japonica | 1 | 69 | 9.6 | LSDYIGKK, SGGLGDLKYPLISDVTK | 2 | IX |
| 8 | RLA0_LUPLU | 60S acidic ribosomal protein P0 OS = Lupinus luteus | 1 | 69 | 7.5 | VGSSEAALLAK, GTVEIITPVELIK | 2 | V |
| 7 | EF1G2_ORYSJ | Elongation factor 1-gamma 2 OS = Oryza sativa subsp. Japonica | 1 | 68 | 5 | NPLDLLPPSK, SFTSEFPHVER | 2 | V |
| 1 | MDAR_SOLLC | Monodehydroascorbate reductase OS = Solanum lycopersicum | 1 | 68 | 9.7 | AYLFPEGAAR, IVGAFLESGSPEENKAIAK | 2 | IX |
| 7 | RSSA_BRANA | 40S ribosomal protein SA OS = Brassica napus | 1 | 65 | 10.3 | LLILTDPR, VIVAIENPQDIIVQSARPYGQR | 2 | V |
| 6,8 | IF4A1_ARATH | Eukaryotic initiation factor 4A-1 OS = Arabidopsis thaliana | 1 | 65 | 8.3 | ELAQQIEK, VLITTDLLAR | 2 | V |
| 7 | HSP11_PEA | 18.1 kDa class I heat shock protein OS = Pisum sativum | 1 | 64 | 14.6 | SIEISG, VLQISGER | 2 | IX |
| 8 | RS16_FRIAG | 40S ribosomal protein S16 OS = Fritillaria agrestis | 1 | 64 | 12.4 | ALVAYYQK, AFEPILLLGR | 2 | V |
| 8 | RL51_ARATH | 60S ribosomal protein L5-1 OS = Arabidopsis thaliana | 1 | 64 | 7.3 | KLTYEER, GALDGGLDIPHSDKR | 2 | V |
| 4 | HSP7M_PHAVU | Heat shock 70 kDa protein, mitochondrial OS = Phaseolus vulgaris | 1 | 64 | 6.1 | HLNITLTR, SSGGLSEDEIEK | 2 | IX |
| 8,7 | HSP12_MEDSA | 18.2 kDa class I heat shock protein OS = Medicago sativa | 1 | 63 | 22.8 | TIDISG, VLQISGER, FRLPENAK | 3 | IX |
| 8 | RS102_ARATH | 40S ribosomal protein S10-2 OS = Arabidopsis thaliana | 1 | 61 | 8.9 | TYLNLPSEIVPATLK, TYLNLPSEIVPATLKK | 2 | V |
| 8 | RS193_ARATH | 40S ribosomal protein S19-3 OS = Arabidopsis thaliana | 1 | 60 | 15.4 | DVSPHEFVK, ELAPYDPDWYYIR | 2 | V |
| 1 | CYF_AETCO | Apocytochrome f OS = Aethionema cordifolium | 2 | 60 | 8.7 | NILVIGPVPGQK, SNNTVYNATAGGIISK | 2 | II |
| 8 | UBIQP_ACECL | Polyubiquitin (Fragment) OS = Acetabularia cliftonii | 1 | 60 | 8.7 | IIFAGK, TLADYNIQK, ESTLHLVLR | 3 | V |
| 8 | RL40A_ARATH | Ubiquitin-60S ribosomal protein L40-1 OS = Arabidopsis thaliana | 1 | 60 | 37.5 | LIFAGK, TLADYNIQK, ESTLHLVLR | 3 | V |
| 3 | UREA_CANEN | Urease OS = Canavalia ensiformis | 1 | 59 | 3 | NYFLF, TIHTYHSEGAGGGHAPDIIK | 2 | I |
| 8 | RS13_PEA | 40S ribosomal protein S13 OS = Pisum sativum | 1 | 58 | 17.2 | DSHGIAQVK, AHGLAPEIPEDLYHLIK | 2 | V |
| 1,8 | RAN_VICFA | GTP-binding nuclear protein Ran/TC4 OS = Vicia faba | 2 | 58 | 13.1 | HLTGEFEK, NLQYYEISAK | 2 | III/VI |
| 7 | PYRB_ARATH | Aspartate carbamoyltransferase, chloroplastic OS = Arabidopsis thaliana | 1 | 57 | 5.4 | GETLEDTIR, LGGEVLTTENAR | 2 | I |
| 8 | HSP11_CHERU | 18.3 kDa class I heat shock protein OS = Chenopodium rubrum | 1 | 52 | 18.6 | FRLPENAK, IDWKETPEAHVFK | 2 | IX |
| 5 | CLAH1_ARATH | Clathrin heavy chain 1 OS = Arabidopsis thaliana | 1 | 51 | 0.9 | ILALK, SPEQVSAAVK | 2 | VI |
| 6 | PDI_RICCO | Protein disulfide-isomerase OS = Ricinus communis | 1 | 49 | 4.2 | FFNSPDAK, SEPIPEVNNEPVK | 2 | V |
| 7 | PDIA6_MEDSA | Probable protein disulfide-isomerase A6 OS = Medicago sativa | 1 | 48 | 7.4 | KLAPEYEK, YGVSGYPTIQWFPK | 2 | V |
| 4 | SUSY_MEDSA | Sucrose synthase OS = Medicago sativa | 1 | 46 | 2.9 | NITGLVEWYGK, SGFHIDPYHGDR | 2 | II |
| 7 | FKB62_ARATH | Peptidyl-prolyl cis-trans isomerase FKBP62 OS = Arabidopsis thaliana | 1 | 42 | 4.4 | SDGVEFTVK, FTLGQGQVIK | 2 | V |
| 5 | DLDH2_ARATH | Dihydrolipoyl dehydrogenase 2, mitochondrial OS = Arabidopsis thaliana | 1 | 40 | 3.4 | AAQLGLK, SLPGITIDEK | 2 | II |
| 7 | WIT2_ARATH | WPP domain-interacting tail-anchored protein 2 OS = Arabidopsis thaliana | 1 | 39 | 3.3 | ELELEK, AESGEAKIK | 2 | III |
| 8 | TBA_PRUDU | Tubulin alpha chain OS = Prunus dulcis | 2 | 37 | 7.8 | DVNAAVATIK, LVSQVISSLTASLR | 2 | VII |
| 7 | PER1B_ARMRU | Peroxidase C1B OS = Armoracia rusticana | 1 | 35 | 5.7 | VPLGR, MGNITPLTGTQGEIR | 2 | IX |
| 7,(8,6) | YCF1_IPOPU | Putative membrane protein ycf1 OS = Ipomoea purpurea | 3 | 35 | 1.3 | ALILK, IVIEK, VIQEKER | 3 | X |
| 6 | RFS_ORYSJ | Galactinol--sucrose galactosyltransferase OS = Oryza sativa subsp. Japonica | 1 | 27 | 2.8 | VELAK, LMEEK | 2 | II |
| 7 | Y1497_ARATH | Probable receptor-like protein kinase At1g49730 OS = Arabidopsis thaliana | 1 | 20 | 1.7 | FLLAK, NLVALK | 2 | V |
1 Fraction corresponding to slice of the 1-D gel in which matches for the protein were found. Numbers in parenthesis indicate fractions where additional similar matches (see 2.) were found. 2 Number of protein matches of high taxonomical and sequence similarity grouped together with this match. (Match displayed was the top-scored one.) 3 MASCOT score. 4 I: metabolism, II: energy, III: cell growth/division, IV: transcription, V: protein synthesis/destination, VI: transporters, VII: cell structure, VIII: signal transduction, IX: disease/stress defense, and X: unclassified.
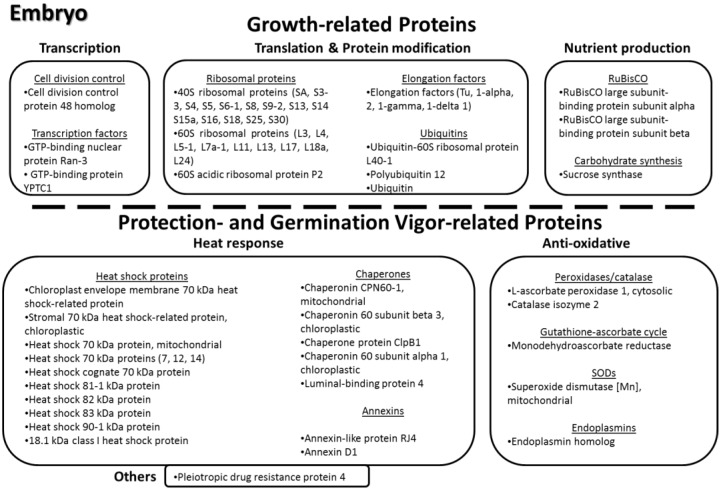
3.3. Lotus Embryo Proteins Identified by 1-DGE and MS/MS Analyses
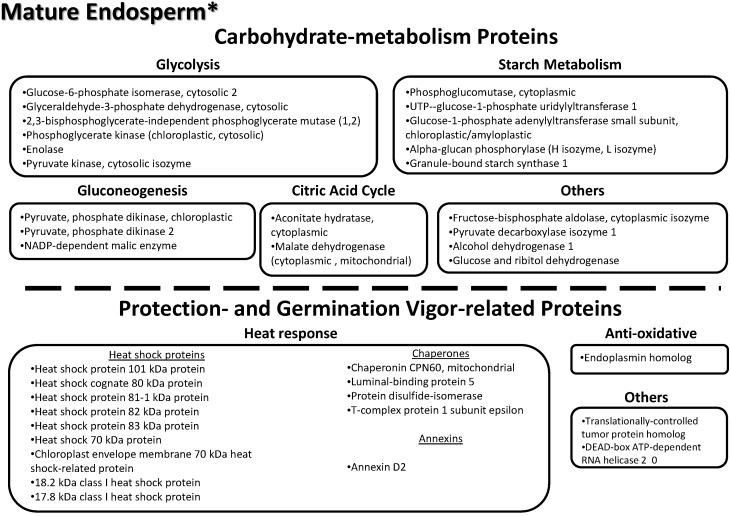
The 1-DGE-MS analysis of the lotus embryo protein extract was performed following the same methodology, green plant database, and same parameters as for the immature endosperm extract. For the sample consisting of a purified extract of lotus embryo proteins, after separation by SDS-PAGE, the sample was divided into eight fractions, analyzed by LC-MS/MS, and matched against a green plant database, as above. From the initial results, 500+ protein matches with at least two confirmed peptide fragment matches were identified. After removing duplicate results from different gel fractions, there were 373 unique protein matches remaining. After grouping results likely to be the same protein in the sample, based on protein taxonomy and similarity of identified peptide sequences, 141 nr protein matches were listed (Table 2).
Table 2.
List of top-scored non-redundant (nr) protein matches of the lotus embryo 1-D shotgun mass spectroscopy results, as matched to Green Plant proteome database (SwissProt 57.0).
| Fractions 1 | Protein Accession | Protein Description | Similar 2 | Score 3 | Cover (%) | Peptide sequences | Sig. Peptide Number | Func. Cat. 4 |
|---|---|---|---|---|---|---|---|---|
| 6,(5,2,1,7,3) | ENO1_HEVBR | Enolase 1 OS = Hevea brasiliensis | 19 | 1125 | 42 | TAIAK, YNQLLR, LTSEIGEK, DGGSDYLGK, AGWGVMASHR, MGAEVYHHLK, DGGSDYLGKGVSK, VQIVGDDLLVTNPK, VNQIGSVTESIEAVK, EAMKMGAEVYHHLK, AAVPSGASTGIYEALELR, LAMQEFMILPVGASSFK, SGETEDTFIADLSVGLATGQIK, YGQDATNVGDEGGFAPNIQENK, KYGQDATNVGDEGGFAPNIQENK, YGQDATNVGDEGGFAPNIQENKEGLELLK | 16 | II |
| 4,(1,2,3,5) | HSP7C_PETHY | Heat shock cognate 70 kDa protein OS = Petunia hybrida | 33 | 922 | 33.6 | IEEVD, DISGNPR, NTTIPTKK, ITITNDKGR, DAGVIAGLNVMR, MVNHFVQEFK, NALENYAYNMR, MVNHFVQEFKR, TTPSYVGFTDTER, ARFEELNMDLFR, IINEPTAAAIAYGLDK, NQVAMNPINTVFDAK ATAGDTHLGGEDFDNR, NQVAMNPINTVFDAK, NAVVTVPAYFNDSQR, EQVFSTYSDNQPGVLIQVYEGER | 16 | IX |
| 1,2,3,4,5,6,7 | ACT_GOSHI | Actin OS = Gossypium hirsutum | 14 | 903 | 50.7 | DLTDALMK, AGFAGDDAPR, IKVVAPPER, GYSFTTTAER, HTGVMVGMGQK, EITALAPSSMK, DAYVGDEAQSK, AVFPSIVGRPR, IWHHTFYNELR, LDLAGRDLTDALMK, GYSFTTTAEREIVR, SYELPDGQVITIGAER, VAPEEHPVLLTEAPLNPK, VAPEEHPVLLTEAPLNPK, TTGIVLDSGDGVSHTVPIYEGYALPHAILR | 15 | VII |
| 6,(1,2,3,7) | ACT12_SOLTU | Actin-100 (Fragment) OS = Solanum tuberosum | 5 | 872 | 53.5 | AGFAGDDAPR, IKVVAPPER, HTGVMVGMGQK, EITALAPSSMK, DAYVGDEAQSK, AVFPSIVGRPR, DAYVGDEAQSKR, GEYDESGPSIVHR, IWHHTFYNELR, SYELPDGQVITIGAER, LAYVALDYEQELETAK, YPIEHGIVSNWDDMEK, TTGIVLDSGDGVSHTVPIYEGYALPHAILR | 13 | VII |
| 7,(8,2) | G3PC_ANTMA | Glyceraldehyde-3-phosphate dehydrogenase, cytosolic OS = Antirrhinum majus | 20 | 749 | 43.6 | VALQR, SSIFDAK, KATYEQIK, AAIKEESEGK, AGIALNDNFVK, DAPMFVVGVNEK, AASFNIIPSSTGAAK, VPTVDVSVVDLTVR, VPTVDVSVVDLTVRLEK, FGIVEGLMTTVHSITATQK, GILGYTEDDVVSTDFVGDSR, LTGMSFRVPTVDVSVVDLTVR, LKGILGYTEDDVVSTDFVGDSR, VINDRFGIVEGLMTTVHSITATQK | 14 | II |
| 4,(1,5,2) | HSP83_IPONI | Heat shock protein 83 OS = Ipomoea nil | 4 | 717 | 31 | VIVTTK, VVVSDR, KLVSATK, AILFVPK, EMLQQNK, DVDGEQLGR, FESLTDKSK, APFDLFDTR, AVENSPFLER, LGIHEDSQNR, DIYYITGESK, LDAQPELFIR, RAPFDLFDTR, ADLVNNLGTIAR, ELISNASDALDK, KAVENSPFLER, HFSVEGQLEFK, GVVDSDDLPLNISR, ELISNASDALDKIR, SGDELTSLKDYVTR, KPEEITKEEYASFYK, HSEFISYPIYLWTEK, ITLFLKEDQLEYLEER | 23 | IX |
| 6 | ATPBM_MAIZE | ATP synthase subunit beta, mitochondrial OS = Zea mays | 3 | 712 | 31.8 | IGLFGGAGVGK, VVDLLAPYQR, TIAMDGTEGLVR, AHGGFSVFAGVGER, VGLTGLTVAEHFR, VLNTGSPITVPVGR, TVLIMELINNVAK, FTQANSEVSALLGR, QISELGIYPAVDPLDSTSR, EAPAFVEQATEQQILVTGIK, IPSAVGYQPTLATDLGGLQER | 11 | I |
| 4,(5,2,1,3) | HSP7E_SPIOL | Chloroplast envelope membrane 70 kDa heat shock-related protein OS = Spinacia oleracea | 5 | 631 | 31.2 | NTTIPTKK, LSKEEIEK, TRDNNLLGK, DAGVISGLNVMR, EIAEAYLGSTVK, NALENYAYNMR, TTPSYVAFTDSER, IINEPTAAAIAYGLDK, ATAGDTHLGGEDFDNR, NQVAMNPINTVFDAK, NAVVTVPAYFNDSQR, EQVFSTYSDNQPGVLIQVYEGER | 12 | IX |
| 4,(2,1,5) | HSP81_ORYSI | Heat shock protein 81-1 OS = Oryza sativa subsp. Indica | 9 | 610 | 35.1 | NLVKK, VVVTTK, IAELLR, KLVSATK, EMLQQNK, FESLTDKSK, APFDLFDTR, DSSMAGYMSSK, RAPFDLFDTR, KAVENSPFLEK, SDLVNNLGTIAR, HFSVEGQLEFK, EVSHEWSLVNK, GIVDSEDLPLNISR, SLTNDWEEHLAVK, SGDELTSLKDYVTR, LDAQPELFIHIVPDK, HSEFISYPISLWTEK, KPEEITKEEYAAFYK, MKEGQNDIYYITGESK, KHSEFISYPISLWTEK | 21 | IX |
| 6,(3,1,2) | TBB_HORVU | Tubulin beta chain OS = Hordeum vulgare | 21 | 583 | 39.1 | YLTASAMFR, IREEYPDR, LAVNLIPFPR, VSEQFTAMFR, YTGTSDLQLER, MMLTFSVFPSPK, EVDEQMINVQNK, LHFFMVGFAPLTSR, AVLMDLEPGTMDSVR, LHFFMVGFAPLTSR, NSSYFVEWIPNNVK, ALTVPELTQQMWDAK, GHYTEGAELIDSVLDVVRK, TGPYGQIFRPDNFVFGQSGAGNNWAK | 14 | VII |
| 6,(1,7,3,4,2) | EF1A_TOBAC | Elongation factor 1-alpha OS = Nicotiana tabacum | 17 | 574 | 38.5 | YDEIVK, GFVASNSK, EVSSYLK, QTVAVGVIK, EVSSYLKK, RGFVASNSK, LPLQDVYK, ARYDEIVK, IGGIGTVPVGR, STNLDWYK, STTTGHLIYK, EHALLAFTLGVK, GFVASNSKDDPAK, YYCTVIDAPGHR, YDEIVKEVSSYLK, YYCTVIDAPGHRDFIK, MIPTKPMVVETFSEYPPLGR, NMITGTSQADCAVLIIDSTTGGFEAGISK | 18 | V |
| 4,(1,2,5,4) | HS901_ARATH | Heat shock protein 90-1 OS = Arabidopsis thaliana | 6 | 539 | 26.4 | VVVTTKVVVTTK, VVVSDR, KLVSATK, AILFVPK, FESLTDKSK, APFDLFDTR, AVENSPFLER, LGIHEDSQNR, DSSMSGYMSSK, RAPFDLFDTR, ADLVNNLGTIAR, KAVENSPFLER, HFSVEGQLEFK, TLSIIDSGIGMTK, GVVDSDDLPLNISR, KPEEITKEEYAAFYK, HSEFISYPIYLWTEK | 17 | IX |
| 4,(2,1,3) | METE_ARATH | 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase OS = Arabidopsis thaliana | 12 | 536 | 20.9 | AAAALK, VVEVNALAK, SWLAFAAQK, AVNEYKEAK, YLFAGVVDGR, SDEKLLSVFR, FALESFWDGK, GNASVPAMEMTK, YGAGIGPGVYDIHSPR, GMLTGPVTILNWSFVR | 10 | I |
| 4 | ENPL_CATRO | Endoplasmin homolog OS = Catharanthus roseus | 3 | 472 | 12.6 | NLGTIAK, FWNEFGK, YGWSSNMER, ELISNASDALDK, IMQSQTLSDASK, GLVDSDTLPLNVSR, ELISNASDALDKIR, VFISDEFDELLPK, RVFISDEFDELLPK, LMDIIINSLYSNKDIFLR | 10 | IX |
| 4,(5,2) | BIP4_TOBAC | Luminal-binding protein 4 OS = Nicotiana tabacum | 6 | 454 | 21 | LIGEAAK, NTVIPTKK, IMEYFIK, LSQEEIER, ITITNDKGR, ALSSQHQVR, EAEEFAEEDKK, IVNKDGKPYIQVK, ARFEELNNDLFR, IINEPTAAAIAYGLDK, IKDAVVTVPAYFNDAQR | 11 | IX |
| 4,(1,3,2,7) | EF2_BETVU | Elongation factor 2 OS = Beta vulgaris | 6 | 454 | 16.6 | DLYVK, VASDLPK, GGGQIIPTAR, MIPASDKGR, IRPVLTVNK, EGALAEENMR, NMSVIAHVDHGK, FGVDESKMMER, VFYASQLTAKPR, LWGENFFDPATK, IRPVLTVNKMDR, RVFYASQLTAKPR, GHVFEEMQRPGTPLYNIK, RGHVFEEMQRPGTPLYNIK | 15 | V |
| 4,(1,2,3) | HSP82_MAIZE | Heat shock protein 82 OS = Zea mays | 5 | 429 | 16.1 | VVVSDR, KLVSATK, APFDLFDTR, AVENSPFLER, LGIHEDSQNR, RAPFDLFDTR, SDLVNNLGTIAR, ELISNASDALDK, KAVENSPFLER, HFSVEGQLEFK, GVVDSDDLPLNISR, ELISNASDALDKIR, HSEFISYPIYLWTEK | 13 | IX |
| 6,(2,3) | IF4A1_ORYSJ | Eukaryotic initiation factor 4A-1 OS = Oryza sativa subsp. Japonica | 4 | 418 | 31.4 | ALGDYLGVK, ELAQQIEK, KGVAINFVTR, VLITTDLLAR, QSLRPDYIK, RDELTLEGIK, GLDVIQQAQSGTGK, GIYAYGFEKPSAIQQR, GFKDQIYDIFQLLPSK | 9 | V |
| 3,(2,1) | CAPPC_FLATR | Phosphoenolpyruvate carboxylase 2 OS = Flaveria trinervia | 32 | 417 | 18 | GIAAGMQNTG, MNIGSRPSK, VILGDVRDK, KPSGGIESLR, LSAAWQLYK, SPEEVFDALK, RPLFGPDLPK, TPPTPQDEMR, QVSTFGLSLVR, VTIDLVEMVFAK, AGMSYFHETIWK, AIPWIFAWTQTR, VPYNAPLIQFSSWMGGDRDGNPR | 13 | I |
| 6,(2) | ENO2_ARATH | Bifunctional enolase 2/transcriptional activator OS = Arabidopsis thaliana | 2 | 391 | 33.3 | YNQLLR, DGGSDYLGK, ISGDALKDLYK, DGGSDYLGKGVSK, VNQIGSVTESIEAVK, IVLPVPAFNVINGGSHAGNK, SGETEDTFIADLAVGLSTGQIK, YGQDATNVGDEGGFAPNIQENK, KYGQDATNVGDEGGFAPNIQENK, YGQDATNVGDEGGFAPNIQENKEGLELLK | 10 | IV |
| 5,(2) | CH61_CUCMA | Chaperonin CPN60-1, mitochondrial OS = Cucurbita maxima | 6 | 387 | 33.33 | ISSINAVVK, VTDALNATK, VTKDGVTVAK, KISSINAVVK, IGGASEAEVGEK, IGVQIIQNALK, IGGASEAEVGEKK, GYISPYFITNQK, AAVEEGIVPGGGVALLYASK, TPVHTIASNAGVEGAVVVGK | 10 | IX |
| 4 | HSP7L_ARATH | Heat shock 70 kDa protein 12 OS = Arabidopsis thaliana | 1 | 382 | 16.8 | NTVIPTKK, IMEYFIK, ALSSQHQVR, EAEEFAEEDKK, ARFEELNNDLFR, ARFEELNNDLFR, IINEPTAAAIAYGLDK, IKDAVVTVPAYFNDAQR | 8 | IX |
| 2,(1) | CLAH1_ARATH | Clathrin heavy chain 1 OS = Arabidopsis thaliana | 4 | 374 | 10.3 | TVDNDLALK, SPEQVSAAVK, VANVELYYK, DPTLAVVAYR, FQELFAQTK, VEEDAVWSQVAK, GNLPGAENLVVQR, EGLVSDAIESFIR, GNMQLFSVDQQR, KNLLENWLAEDK, RGNLPGAENLVVQR, QLIDQVVSTALPESK, YKEAAELAAESPQGILR | 13 | VI |
| 6,(2,5,1) | ATPAM_PEA | ATP synthase subunit alpha, mitochondrial OS = Pisum sativum | 1 | 364 | 29.4 | VVSVGDGIAR, TGSIVDVPAGK, AAELTTLLESR, VVDALGVPIDGR, TAIAIDTILNQK, KSVHEPMQTGLK, GIRPAINVGLSVSR, EAFPGDVFYLHSR, ITNFYTNFQVDEIGR, LTEVLKQPQYAPLPIEK, EVAAFAQFGSDLDAATQALLNR | 11 | I |
| 5 | RUBB_PEA | RuBisCO large subunit-binding protein subunit beta, chloroplastic OS = Pisum sativum | 2 | 357 | 23.98 | IAALK, VVLTK, NVVLESK, VEDALNATK, IVNDGVTVAK, KGVVTLEEGK, LADLVGVTLGPK, GYISPYFVTDSEK, EVELEDPVENIGAK, TNDLAGDGTTTSVVLAQGLIAEGVK, IVNDGVTVAKEVELEDPVENIGAK | 11 | I |
| 5,(2) | PGMC_PEA | Phosphoglucomutase, cytoplasmic OS = Pisum sativum | 5 | 333 | 41.16 | YLFEDGSR, FFEVPTGWK, LSGTGSEGATIR, SMPTSAALDVVAK, YDYENVDAGAAK | 5 | II |
| 6,(2) | SAHH_MESCR | Adenosylhomocysteinase OS = Mesembryanthemum crystallinum | 6 | 330 | 11.1 | ATDVMIAGK, HSLPDGLMR, ITIKPQTDR, TEFGPSQPFK, LVGVSEETTTGVK, TEFGPSQPFKGAK, LVGVSEETTTGVKR | 7 | I |
| 7 | PGKY_TOBAC | Phosphoglycerate kinase, cytosolic OS = Nicotiana tabacum | 7 | 302 | 21.7 | LAELSGK, YSLKPLVPR, YLKPAVAGFLMQK, GVSLLLPTDVVIADK, GVTTIIGGGDSVAAVEK, LASLADLYVNDAFGTAHR, KLASLADLYVNDAFGTAHR | 7 | II |
| 4 | SUSY_SOYBN | Sucrose synthase OS = Glycine max | 12 | 295 | 13.4 | YLEMFYALK, VVHGIDVFDPK, NITGLVEWYGK, ELVNLVVVAGDR, LLPDAVGTTCGQR, SGFHIDPYHGDR, LGVTQCTIAHALEK | 7 | II |
| 5 | CPNB3_ARATH | Chaperonin 60 subunit beta 3, chloroplastic OS = Arabidopsis thaliana | 1 | 292 | 25.35 | VVLTK, NVVLESK, VEDALNATK, KGVVTLEEGK, LADLVGVTLGPK, GYISPYFVTDSEK, EVELEDPVENIGAK, TNDLAGDGTTTSVVLAQGLIAEGVK | 8 | IX |
| 7,(8) | MDHC2_ARATH | Malate dehydrogenase, cytoplasmic 2 OS = Arabidopsis thaliana | 2 | 288 | 23.2 | GAAIIK, NVSIYK, SQASALEK, EFAPSIPEK, MELVDAAFPLLK, VLVVANPANTNALILK, VLVTGAAGQIGYALVPMIAR | 7 | II |
| 8,(1) | 1433E_TOBAC | 14-3-3-like protein E OS = Nicotiana tabacum | 19 | 284 | 29.8 | NVIGAR, VFYLK, YLAEFK, MKGDYHR, NLLSVAYK, IISSIEQK, TVDVEELTVEER, IISSIEQKEESR, SAQDIALAELAPTHPIR | 9 | VIII |
| 5 | VATA_GOSHI | V-type proton ATPase catalytic subunit A OS = Gossypium hirsutum | 2 | 272 | 49.1 | SGDVYIPR, TVISQALSK, LAADTPLLTGQR, LAEMPADSGYPAYLAAR, LTTFEDSEKESEYGYVR, LVSQKFEDPAEGEEALVAK | 6 | VI |
| 4,(3,2,1) | CD48A_ARATH | Cell division control protein 48 homolog A OS = Arabidopsis thaliana | 3 | 265 | 18.5 | TLLAK, KGDLFLVR, RSVSDADIR, DFSTAILER, LAEDVDLER, GILLYGPPGSGK, LAGESESNLRK, IVSQLLTLMDGLK, ELVELPLRHPQLFK, NAPSIIFIDEIDSIAPK | 10 | III/IV |
| 7 | ALF_CICAR | Fructose-bisphosphate aldolase, cytoplasmic isozyme OS = Cicer arietinum | 1 | 245 | 11.1 | GILAADESTGTIGK, GILAADESTGTIGKR, YHDELIANAAYIGTPGK | 3 | II |
| 7 | MDHM_CITLA | Malate dehydrogenase, mitochondrial OS = Citrullus lanatus | 2 | 235 | 17.6 | LFGVTTLDVVR, TQDGGTEVVEAK, DDLFNINAGIVK, RTQDGGTEVVEAK, VAVLGAAGGIGQPLALLMK | 5 | II |
| 6,7 | ACT5_ARATH | Putative actin-5 OS = Arabidopsis thaliana | 1 | 213 | 20.4 | AGFAGDDAPR, IKVVAPPER, IWHHTFYNELR, TTGIVLDSGDGVSHTVPIYEGYALPHAILR | 4 | VII |
| 6 | UGPA_MUSAC | UTP--glucose-1-phosphate uridylyltransferase OS = Musa acuminata | 3 | 199 | 18 | VANFLSR, GGTLISYEGR, VLQLETAAGAAIR, FFDHAIGINVPR, LQSAVAELNQISENEK | 5 | II |
| 8 | ADT1_GOSHI | ADP, ATP carrier protein 1, mitochondrial OS = Gossypium hirsutum | 2 | 194 | 8.8 | SSLDAFSQILK, LLIQNQDEMIK, YFPTQALNFAFK | 3 | VI |
| 8 | RAN_VICFA | GTP-binding nuclear protein Ran/TC4 OS = Vicia faba | 1 | 193 | 26.2 | NVPTWHR, HLTGEFEK, AKQVTFHR, LVIVGDGGTGK, NLQYYEISAK, SNYNFEKPFLYLAR | 6 | VIII |
| 3 | CLPB1_ARATH | Chaperone protein ClpB1 OS = Arabidopsis thaliana | 2 | 192 | 11.1 | TAVVEGLAQR, YRGEFEER, TKNNPVLIGEPGVGK, KVESASGDTNFQALK, VQLDSQPEEIDNLER, LIGAPPGYVGHEEGGQLTEAVR | 6 | IX |
| 5 | CPNA1_ARATH | Chaperonin 60 subunit alpha 1, chloroplastic OS = Arabidopsis thaliana | 1 | 189 | 82.82 | VVNDGVTIAR, NVVLDEFGSPK, VGAATETELEDR | 3 | IX |
| 3 | ACOC_CUCMA | Aconitate hydratase, cytoplasmic OS = Cucurbita maxima | 6 | 187 | 9.2 | NFEGR, ILLESAIR, STYESITK, DFNSYGSR, RGNDEVMAR, TSLAPGSGVVTK, ATIANMSPEYGATMGFFPVDHVTLQYLK | 7 | II |
| 3 | SYA_ARATH | Alanine--tRNA ligase OS = Arabidopsis thaliana | 1 | 181 | 5.4 | LTSVLQNK, HVDTGMGFER, ESDGSLKPLPAK, AFALLSEEGIAK, AVFGEVYPDPVR | 5 | IV |
| 6,(7) | PRS6A_SOLLC | 26S protease regulatory subunit 6A homolog OS = Solanum lycopersicum | 4 | 170 | 8.5 | IIKEELQR, GVLLYGPPGTGK, LAGPQLVQMFIGDGAK | 3 | I |
| 7 | AATM_LUPAN | Aspartate aminotransferase P2, mitochondrial (Fragment) OS = Lupinus angustifolius | 1 | 170 | 10.8 | IADVIQEK, NLGLYAER, LNLGVGAYR, ISLAGLSLAK, VATVQGLSGTGSLR | 5 | I |
| 6 | UGDH_SOYBN | UDP-glucose 6-dehydrogenase OS = Glycine max | 1 | 168 | 6.5 | IAILGFAFK, LAANAFLAQR, AADLTYWESAAR | 3 | II |
| 2,8 | ANX4_FRAAN | Annexin-like protein RJ4 OS = Fragaria ananassa | 1 | 163 | 9.6 | VGTDEDALTR, LLVALVTAYR | 2 | IX |
| 8 | RS4_GOSHI | 40S ribosomal protein S4 OS = Gossypium hirsutum | 4 | 163 | 22.9 | LSIIEEAR, LGNVFTIGK, GIPYLNTYDGR, LGGAFAPKPSSGPHK, TDKTYPAGFMDVVSIPK | 5 | V |
| 7,(8) | RSSA_SOYBN | 40S ribosomal protein SA OS = Glycine max | 2 | 161 | 19 | LLILTDPR, YVDIGIPANNK, HTPGTFTNQLQTSFSEPR, VIVAIENPQDIIVQSARPYGQR | 4 | V |
| 8 | RAA1D_ARATH | Ras-related protein RABA1d OS = Arabidopsis thaliana | 9 | 161 | 26.6 | AITSAYYR, VVLIGDSGVGK, STIGVEFATR, HSTFENVER, AQIWDTAGQER | 5 | VIII |
| 8 | RS18_ARATH | 40S ribosomal protein S18 OS = Arabidopsis thaliana | 1 | 155 | 23.7 | LRDDLER, VLNTNVDGK, IPDWFLNR, YSQVVSNALDMK | 4 | V |
| 1 | AVP_VIGRR | Pyrophosphate-energized vacuolar membrane proton pump OS = Vigna radiata var. Radiata | 1 | 151 | 9.5 | TDALDAAGNTTAAIGK, AAVIGDTIGDPLKDTSGPSLNILIK | 2 | VI |
| 5 | ILV5_ARATH | Ketol-acid reductoisomerase, chloroplastic OS = Arabidopsis thaliana | 1 | 150 | 27.45 | SDIVVK, SVVLAGR, QIGVIGWGSQGPAQAQNLR | 3 | I |
| 7 | AATC_DAUCA | Aspartate aminotransferase, cytoplasmic OS = Daucus carota | 2 | 149 | 10.4 | ISMAGLSSR, LNLGVGAYR, LIFGADSPAIQENR | 3 | I |
| 8 | RL13_TOBAC | 60S ribosomal protein L13 OS = Nicotiana tabacum | 1 | 149 | 23.3 | GFSLEELK, TWFNQPAR, SLEGLQTNVQR, KLAPTIGIAVDHR | 4 | V |
| 5 | ACLB1_ORYSJ | ATP-citrate synthase beta chain protein 1 OS = Oryza sativa subsp. Japonica | 1 | 148 | 35.6 | FNNIPQVK, FGGAIDDAAR, SEVQFGHAGAK, SIGLIGHTFDQKR, VVAIIAEGVPESDTK | 5 | I |
| 2,(3) | COPA1_ARATH | Coatomer subunit alpha-1 OS = Arabidopsis thaliana | 3 | 147 | 4.4 | VWDIGALR, YVLEGHDR, AWEVDTLR, VVIFDLQQR, TLDVPIYITK, QDIIVSNSEDK | 6 | VI |
| 6 | CATA2_RICCO | Catalase isozyme 2 OS = Ricinus communis | 2 | 146 | 9.3 | FSTVIHER, APGVQTPVIVR, EGNFDIVGNNFPVFFIR | 3 | IX |
| 8 | RAN3_ORYSI | GTP-binding nuclear protein Ran-3 OS = Oryza sativa subsp. Indica | 1 | 145 | 14.2 | HITGEFEK, NLQYYEISAK, SNYNFEKPFLYLAR | 3 | III/VI |
| 6 | MDAR_SOLLC | Monodehydroascorbate reductase OS = Solanum lycopersicum | 1 | 139 | 12.5 | AYLFPEGAAR, LSDFGVQGADSK, IVGAFLESGSPEENKAIAK | 3 | IX |
| 6,(1) | RL3_ORYSJ | 60S ribosomal protein L3 OS = Oryza sativa subsp. Japonica | 4 | 138 | 9.3 | VIAHTQIR, HGSLGFLPR, LALEEIKLK, GKGYEGVVTR | 4 | V |
| 5 | PMGI_RICCO | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase OS = Ricinus communis | 4 | 136 | 43.5 | ARDAILSGK, LVDLALASGK, LDQLQLLLK, AHGTAVGLPTEDDMGNSEVGHNALGAGR | 4 | II |
| 5 | RUBA_RICCO | RuBisCO large subunit-binding protein subunit alpha (Fragment) OS = Ricinus communis | 2 | 134 | 71.19 | NVVLDEFGSPK, VGAATETELEDR, LGLLSVTSGANPVSIK | 3 | I |
| 7 | RL4_PRUAR | 60S ribosomal protein L4 OS = Prunus armeniaca | 1 | 134 | 14.5 | AGHQTSAESWGTGR, YAVVSAIAASAVPSLVLAR | 2 | V |
| 5 | G6PI_SPIOL | Glucose-6-phosphate isomerase, cytosolic OS = Spinacia oleracea | 2 | 131 | 27 | SQQPVYLK, FLANVDPIDVAK, TFTTAETMLNAR | 3 | II |
| 8 | RS8_MAIZE | 40S ribosomal protein S8 OS = Zea mays | 1 | 130 | 21.7 | LDTGNYSWGSEAVTR, ILDVVYNASNNELVR | 2 | V |
| 8 | RL11_MEDSA | 60S ribosomal protein L11 OS = Medicago sativa | 1 | 129 | 17.7 | YEGVILNK, AMQLLESGLK, VLEQLSGQTPVFSK | 3 | V |
| 8,(7) | H4_ARATH | Histone H4 OS = Arabidopsis thaliana | 2 | 128 | 46.6 | TLYGFGG, IFLENVIR, DAVTYTEHAR, ISGLIYEETR | 4 | VII |
| 5 | TCPE_ARATH | T-complex protein 1 subunit epsilon OS = Arabidopsis thaliana | 1 | 123 | 73.41 | IAEGYEMASR, QQQILLATQVVK | 2 | V |
| 5 | TCPA_ARATH | T-complex protein 1 subunit alpha OS = Arabidopsis thaliana | 1 | 119 | 71.23 | YFVEAGAIAVR, NKIHPTSIISGYR | 2 | V |
| 4 | TKTC_SPIOL | Transketolase, chloroplastic OS = Spinacia oleracea | 4 | 118 | 6.1 | FLAIDAVEK, ALPTYTPETPGDATR, VIPGLLGGSADLASSNMTLLK | 3 | II |
| 8 | TPIS_MAIZE | Triosephosphate isomerase, cytosolic OS = Zea mays | 1 | 117 | 11.9 | FFVGGNWK, VAYALSQGLK, VIACVGETLEQR | 3 | VII |
| 8 | PROF3_ARATH | Profilin-3 OS = Arabidopsis thaliana | 2 | 117 | 17.2 | LGDYLLEQGL, YMVIQGEPGAVIR | 2 | VII |
| 8 | RS92_ARATH | 40S ribosomal protein S9-2 OS = Arabidopsis thaliana | 1 | 115 | 19.3 | LVGEYGLR, ERLDAELK, RPYEKER, RLQTIVFK, IFEGEALLR | 5 | V |
| 6 | VATB1_ARATH | V-type proton ATPase subunit B1 OS = Arabidopsis thaliana | 1 | 115 | 10.5 | YQEIVNIR, TVSGVAGPLVILDK, QIYPPINVLPSLSR | 3 | VI |
| 6 | ERF1X_ARATH | Eukaryotic peptide chain release factor subunit 1-1 OS = Arabidopsis thaliana | 1 | 113 | 9.9 | GFGGIGGILR, QSVLGAITSAQQR | 2 | V |
| 5 | HSP7M_PHAVU | Heat shock 70 kDa protein, mitochondrial OS = Phaseolus vulgaris | 3 | 111 | 24.87 | HLNITLTR, VIENSEGAR, TTPSVVAFNQK, SSGGLSEDEIEK | 4 | IX |
| 8 | ANXD1_ARATH | Annexin D1 OS = Arabidopsis thaliana | 1 | 110 | 5 | AQINATFNR, SKAQINATFNR | 2 | IX |
| 8 | RS16_FRIAG | 40S ribosomal protein S16 OS = Fritillaria agrestis | 3 | 109 | 13.8 | ALVAYYQK, AFEPILLLGR, YKAFEPILLLGR | 3 | V |
| 8 | RS5_CICAR | 40S ribosomal protein S5 (Fragment) OS = Cicer arietinum | 1 | 108 | 15.7 | IGSAGVVRR, GSSNSYAIK, VNQAIYLLTTGAR | 3 | V |
| 8 | ARF_VIGUN | ADP-ribosylation factor OS = Vigna unguiculata | 1 | 104 | 28.7 | ILMVGLDAAGK, NISFTVWDVGGQDK | 2 | VIII |
| 4 | SYGM1_ARATH | Glycine--tRNA ligase 1, mitochondrial OS = Arabidopsis thaliana | 1 | 103 | 5.1 | LFYIPSFK, VFTPSVIEPSFGIGR | 2 | IV/VI |
| 7 | RGP1_ORYSJ | UDP-arabinopyranose mutase 1 OS = Oryza sativa subsp. Japonica | 1 | 101 | 6.9 | ILGPK, ASNPFVNLK, ASNPFVNLKK, YVDAVMTVPK | 4 | I |
| 3 | HSP7O_ARATH | Heat shock 70 kDa protein 14 OS = Arabidopsis thaliana | 1 | 101 | 5.1 | ILSHAFDR, NAVESYVYDMR, AVLDAATIAGLHPLR | 3 | IX |
| 8 | RS15A_DAUCA | 40S ribosomal protein S15a OS = Daucus carota | 1 | 100 | 29.2 | VSVLNDALK, HGYIGEFEYVDDHR | 2 | V |
| 8 | RS61_ARATH | 40S ribosomal protein S6-1 OS = Arabidopsis thaliana | 2 | 100 | 18 | LVTPLTLQR, KGENDLPGLTDTEKPR, ISQEVSGDALGEEFKGYVFK | 3 | V |
| 6 | ACCC2_POPTR | Biotin carboxylase 2, chloroplastic OS = Populus trichocarpa | 1 | 97 | 7.8 | LLEEAPSPALTPELR, ALDDTVITGVPTTIDYHK | 2 | I |
| 8 | RLA2_PARAR | 60S acidic ribosomal protein P2 OS = Parthenium argentatum | 1 | 97 | 10.5 | DITELIASGR, GKDITELIASGR | 2 | V |
| 8 | RS33_ARATH | 40S ribosomal protein S3-3 OS = Arabidopsis thaliana | 3 | 96 | 10.9 | ELAEDGYSGVEVR, FKFPQDSVELYAEK | 2 | V |
| 5 | KPYC_SOYBN | Pyruvate kinase, cytosolic isozyme OS = Glycine max | 1 | 94 | 55.66 | KGSDLVNVR, STPLPMSPLESLASSAVR | 2 | II |
| 7 | GMD1_ARATH | GDP-mannose 4,6 dehydratase 1 OS = Arabidopsis thaliana | 2 | 93 | 5 | RGENFVTR, LFLGNIQASR | 2 | II |
| 4 | HSP7S_SPIOL | Stromal 70 kDa heat shock-related protein, chloroplastic (Fragment) OS = Spinacia oleracea | 2 | 93 | 5.3 | HIETTLTR, IINEPTAASLAYGFEK | 2 | IX |
| 6 | EF1G2_ORYSJ | Elongation factor 1-gamma 2 OS = Oryza sativa subsp. Japonica | 3 | 93 | 16.7 | EVAIK, LYSNTK, NPLDLLPPSK, MILDEWKR, SFTSEFPHVER | 5 | V |
| 8 | RLA0_LUPLU | 60S acidic ribosomal protein P0 OS = Lupinus luteus | 1 | 90 | 9.9 | EYLKDPSK, VGSSEAALLAK | 2 | V |
| 7 | GCST_PEA | Aminomethyltransferase, mitochondrial OS = Pisum sativum | 1 | 89 | 8.3 | GGAIDDSVITK, TGYTGEDGFEISVPSEHGVELAK | 2 | I |
| 8 | APX1_ORYSJ | L-ascorbate peroxidase 1, cytosolic OS = Oryza sativa subsp. Japonica | 1 | 89 | 15.2 | TGGPFGTMK, LSELGFADA, ALLSDPAFRPLVEK | 3 | IX |
| 8 | RS193_ARATH | 40S ribosomal protein S19-3 OS = Arabidopsis thaliana | 1 | 88 | 23.1 | AYAAHLKR, TVKDVSPHEFVK, ELAPYDPDWYYIR | 3 | V |
| 1,(2) | RPN1A_ARATH | 26S proteasome non-ATPase regulatory subunit 2 1A OS = Arabidopsis thaliana | 3 | 87 | 5.5 | VGQAVDVVGQAGRPK, NLAGEIAQEYTKR | 2 | I |
| 4,(3) | PPDK_FLABR | Pyruvate, phosphate dikinase, chloroplastic OS = Flaveria brownii | 8 | 87 | 2.9 | SDFEGIFR, AALIADEIAK, AMDGLPVTIR | 3 | II |
| 8 | RS13_PEA | 40S ribosomal protein S13 OS = Pisum sativum | 1 | 87 | 25.8 | DSHGIAQVK, GLTPSQIGVILR, KGLTPSQIGVILR, AHGLAPEIPEDLYHLIK | 4 | V |
| 8 | RS14_CHLRE | 40S ribosomal protein S14 OS = Chlamydomonas reinhardtii | 1 | 85 | 18.3 | TPGPGAQSALR, IEDVTPIPTDSTRR | 2 | V |
| 6 | VATB2_GOSHI | V-type proton ATPase subunit B 2 (Fragment) OS = Gossypium hirsutum | 1 | 84 | 10.1 | FVTQGAYDTR, QIYPPINVLPSLSR | 2 | VI |
| 8,(5) | CYPH_MAIZE | Peptidyl-prolyl cis-trans isomerase OS = Zea mays | 3 | 83 | 16.3 | SGKPLHYK, VFFDMTVGGAPAGR | 2 | V |
| 8 | NDK1_ARATH | Nucleoside diphosphate kinase 1 OS = Arabidopsis thaliana | 1 | 82 | 9.4 | NVIHGSDSVESAR, NVIHGSDSVESARK | 2 | I |
| 7 | GLN11_ORYSJ | Glutamine synthetase cytosolic isozyme 1-1 OS = Oryza sativa subsp. Japonica | 1 | 79 | 7.6 | DIVDSHYK, HKEHISAYGEGNER | 2 | I |
| 7 | SERC_SPIOL | Phosphoserine aminotransferase, chloroplastic OS = Spinacia oleracea | 1 | 79 | 5.3 | FGLIYAGAQK, NVGPSGVTIVIVR | 2 | I |
| 7 | PDI21_ARATH | Protein disulfide-isomerase like 2-1 OS = Arabidopsis thaliana | 1 | 78 | 8.9 | KLAPEYEK, YGVSGFPTLK, YGVSGYPTIQWFPK | 3 | V |
| 8 | RS254_ARATH | 40S ribosomal protein S25-4 OS = Arabidopsis thaliana | 2 | 76 | 29.6 | LITPSILSDR, MVAAHSSQQIYTR | 2 | V |
| 6 | IDHC_TOBAC | Isocitrate dehydrogenase [NADP] OS = Nicotiana tabacum | 2 | 75 | 7.5 | HAFGDQYR, DLALIIHGSK, TIEAEAAHGTVTR | 3 | II |
| 6 | OPD22_ARATH | Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial OS = Arabidopsis thaliana | 1 | 73 | 3.9 | ISVNDLVIK, VIDGAIGAEWLK | 2 | II |
| 8 | RL40A_ARATH | Ubiquitin-60S ribosomal protein L40-1 OS = Arabidopsis thaliana | 1 | 70 | 45.3 | ESTLHLVLR, TITLEVESSDTIDNVK | 2 | V |
| 8 | RL24_PRUAV | 60S ribosomal protein L24 OS = Prunus avium | 1 | 69 | 7 | SIVGATLEVIQK, SIVGATLEVIQKR | 2 | V |
| 7,(5) | SAPK6_ORYSJ | Serine/threonine-protein kinase SAPK6 OS = Oryza sativa subsp. Japonica | 2 | 68 | 7.4 | DIGSGNFGVAR, STVGTPAYIAPEVLSR | 2 | III |
| 6 | GME2_ORYSJ | GDP-mannose 3,5-epimerase 2 OS = Oryza sativa subsp. Japonica | 1 | 67 | 7 | NSDNTLIKEK, ISITGAGGFIASHIAR | 2 | II |
| 8 | EF1D1_ORYSJ | Elongation factor 1-delta 1 OS = Oryza sativa subsp. Japonica | 2 | 66 | 7.9 | LVPVGYGIK, KLDEYLLTR | 2 | V |
| 8 | IF5A1_ARATH | Eukaryotic translation initiation factor 5A-1 OS = Arabidopsis thaliana | 1 | 64 | 12 | VVEVSTSK, TYPQQAGTIR, TYPQQAGTIRK | 3 | V |
| 8 | H2B11_ARATH | Histone H2B.11 OS = Arabidopsis thaliana | 1 | 62 | 30 | LVLPGELAK, QVHPDIGISSK, YNKKPTITSR | 3 | VII |
| 8 | PSA3_ARATH | Proteasome subunit alpha type-3 OS = Arabidopsis thaliana | 1 | 60 | 7.6 | VFQIEYAAK, VPDDLLEEAK | 2 | I |
| 7 | AAT3_ARATH | Aspartate aminotransferase, chloroplastic OS = Arabidopsis thaliana | 1 | 60 | 4.9 | LNLGVGAYR, TEEGKPLVLNVVR | 2 | I |
| 1 | PDR4_ORYSJ | Pleiotropic drug resistance protein 4 OS = Oryza sativa subsp. Japonica | 1 | 60 | 1.5 | TTLLLALAGK, VTTGEMLVGPAR | 2 | IX |
| 2,(8) | UBQ12_ARATH | Polyubiquitin 12 OS = Arabidopsis thaliana | 3 | 60 | 23.9 | MQIFLKTLTGK, IQDKEGIPPDQQR, TITLEVESSDTIDNVK | 3 | V |
| 2,(1) | UBIQ_AVESA | Ubiquitin OS = Avena sativa | 1 | 60 | 57.9 | TLADYNIQK, IQDKEGIPPDQQR, TITLEVESSDTIDNVK | 3 | V |
| 5,(1) | DIM_PEA | Delta(24)-sterol reductase OS = Pisum sativum | 2 | 59 | 45.72 | NILDIDKER, SDLEAPLRPK | 2 | I |
| 8 | HSP11_PEA | 18.1 kDa class I heat shock protein OS = Pisum sativum | 2 | 56 | 8.9 | SIEISG, VLQISGER | 2 | IX |
| 6 | MPPA_SOLTU | Mitochondrial-processing peptidase subunit alpha OS = Solanum tuberosum | 1 | 52 | 3.4 | QLLTYGER, MVASEDIGR | 2 | I |
| 8 | RL17_MAIZE | 60S ribosomal protein L17 OS = Zea mays | 1 | 52 | 10.5 | NAESNADVK, YLEDVIAHK | 2 | V |
| 8 | RL51_ARATH | 60S ribosomal protein L5-1 OS = Arabidopsis thaliana | 2 | 52 | 9.3 | VFGALK, KLTYEER, GALDGGLDIPHSDKR | 3 | V |
| 3 | PHSL1_SOLTU | Alpha-1,4 glucan phosphorylase L-1 isozyme, chloroplastic/amyloplastic OS = Solanum tuberosum | 1 | 50 | 1.8 | NDVSYPIK, AFATYVQAK | 2 | II |
| 6 | PRS4A_ARATH | 26S proteasome regulatory subunit 4 homolog A OS = Arabidopsis thaliana | 1 | 49 | 9.5 | VVGSELIQK, GVILYGEPGTGK | 2 | II |
| 8 | YPTC1_CHLRE | GTP-binding protein YPTC1 OS = Chlamydomonas reinhardtii | 2 | 49 | 17.2 | TITSSYYR, LLLIGDSGVGK | 2 | III/VI |
| 4 | HSP7G_ARATH | Heat shock 70 kDa protein 7, chloroplastic OS = Arabidopsis thaliana | 1 | 47 | 9.5 | HIETTLTR, TTPSVVAYTK, QAVVNPENTFFSVKR | 3 | IX |
| 8 | SODM_HEVBR | Superoxide dismutase [Mn], mitochondrial OS = Hevea brasiliensis | 1 | 46 | 11.2 | HHQTYITNYNK, LVVETTANQDPLVTK | 2 | IX |
| 5 | CALX2_ARATH | Calnexin homolog 2 OS = Arabidopsis thaliana | 2 | 45 | 30.3 | NPAYK, SEGHDDYGLLVSEK | 2 | V |
| 8 | RS30_ARATH | 40S ribosomal protein S30 OS = Arabidopsis thaliana | 1 | 44 | 30.6 | GKVHGSLAR, FVTAVVGFGK | 2 | V |
| 8 | RL7A1_ARATH | 60S ribosomal protein L7a-1 OS = Arabidopsis thaliana | 1 | 44 | 9.3 | TLDKNLATSLFK, LKVPPALNQFTK | 2 | V |
| 7 | METK4_POPTR | S-adenosylmethionine synthase 4 OS = Populus trichocarpa | 1 | 42 | 12.3 | FVIGGPHGDAGLTGR, VLVNIEQQSPDIAQGVHGHLTK | 2 | I |
| 8 | RL18A_CASSA | 60S ribosomal protein L18a OS = Castanea sativa | 1 | 41 | 9.6 | ASRPNLFM, FHQYQVVGR | 2 | V |
| 7 | EFTM_ARATH | Elongation factor Tu, mitochondrial OS = Arabidopsis thaliana | 1 | 41 | 9.7 | QAILK, VLAEEGKAK, GITIATAHVEYETAKR | 3 | V |
| 1 | CALSB_ARATH | Callose synthase 11 OS = Arabidopsis thaliana | 1 | 38 | 1.5 | ILFNEAFSR, LGEGKPENQNHALIFTR | 2 | IX |
| 3 | APBLB_ARATH | Beta-adaptin-like protein B OS = Arabidopsis thaliana | 1 | 27 | 2 | EAENIVER, DSQDPNPLIR | 2 | VII |
1 Fraction corresponding to slice of the 1-D gel in which matches for the protein were found. Numbers in parenthesis indicate fractions where additional similar matches (see 2) were found. 2 Number of protein matches of high taxonomical and sequence similarity grouped together with this match. (Match displayed was the top-scored one.) 3 MASCOT score. 4 I: metabolism, II: energy, III: cell growth/division, IV: transcription, V: protein synthesis/destination, VI: transporters, VII: cell structure, VIII: signal transduction, IX: disease/stress defense, and X: unclassified
3.4. Comparative Analysis of Lotus Seed (Immature Endosperm, Mature Endosperm, and Embryo) Proteins
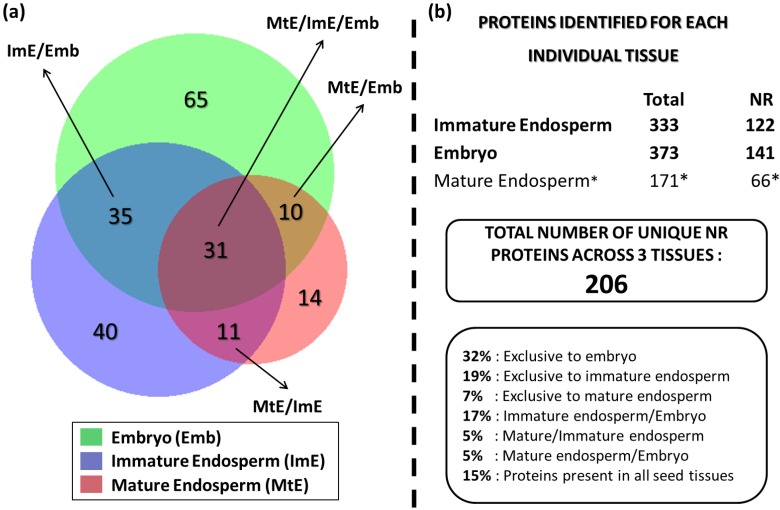
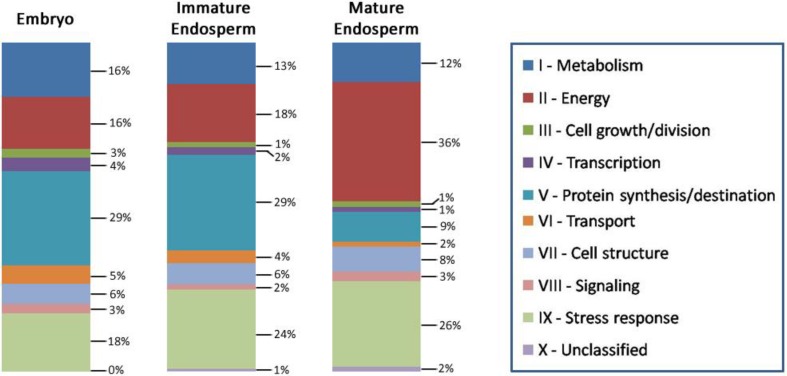
As is to be expected, there were many proteins in common found among the immature endosperm and embryo tissues, as well as with the mature endosperm previously analyzed [17]. Amongst all three seed tissues, a total of 206 nr proteins were identified against the plant database (Figure 3). Of these, 31 (15%) were common to all three tissues, 40 (19%) were unique to the immature endosperm, and 65 (32%) were unique to the embryo; only 14 (7%) were exclusively found in the mature endosperm. To note, the larger share of embryo-only proteins is a consequence of the embryo tissue being much more involved in plant metabolism, and therefore is expected to express a larger number of functional proteins than the endosperm, which, especially in its mature phase, has nutrient storage as its primary function. The immature endosperm, as a developing tissue, also expresses a larger number of proteins than its mature form, and also shares a significant number of proteins with the embryo—35 (17%) of the identified ones. Common proteins between mature and immature endosperm only amounted to 5% of the identified ones (same as for between the mature endosperm and embryo). Although, considering that both immature endosperm and immature embryo are much softer and with a higher water content than their mature stages, there is a possibility that some of the proteins in common with the embryo identified in the immature endosperm might have originated from the embryo and diffused through the endosperm, despite the care taken to remove embryo fragments and the endosperm immediately around them in the sample preparation.
Figure 3.
Venn diagram displaying distribution of non-redundant (nr) proteins amongst lotus seed immature endosperm (ImE), mature endosperm (MtE), and embryo (Emb) (a); Listing of the total and nr protein matches found for each lotus seed tissue analyzed (b); * see reference [17].
3.5. Functional Significance of the Identified Seed Proteins
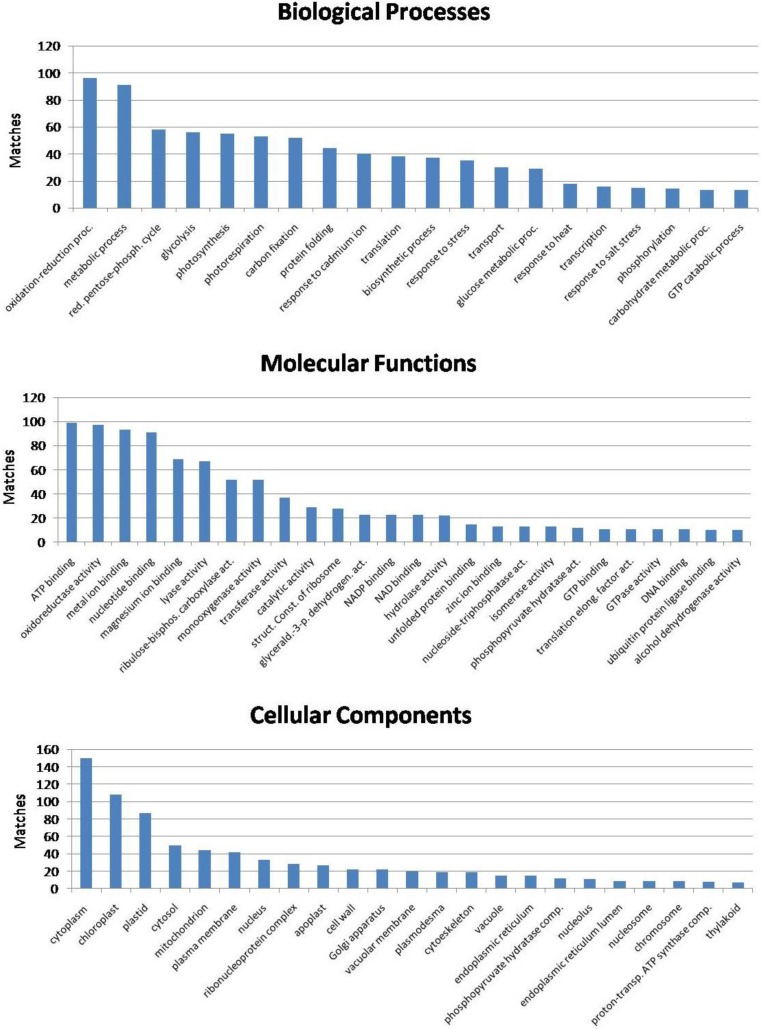
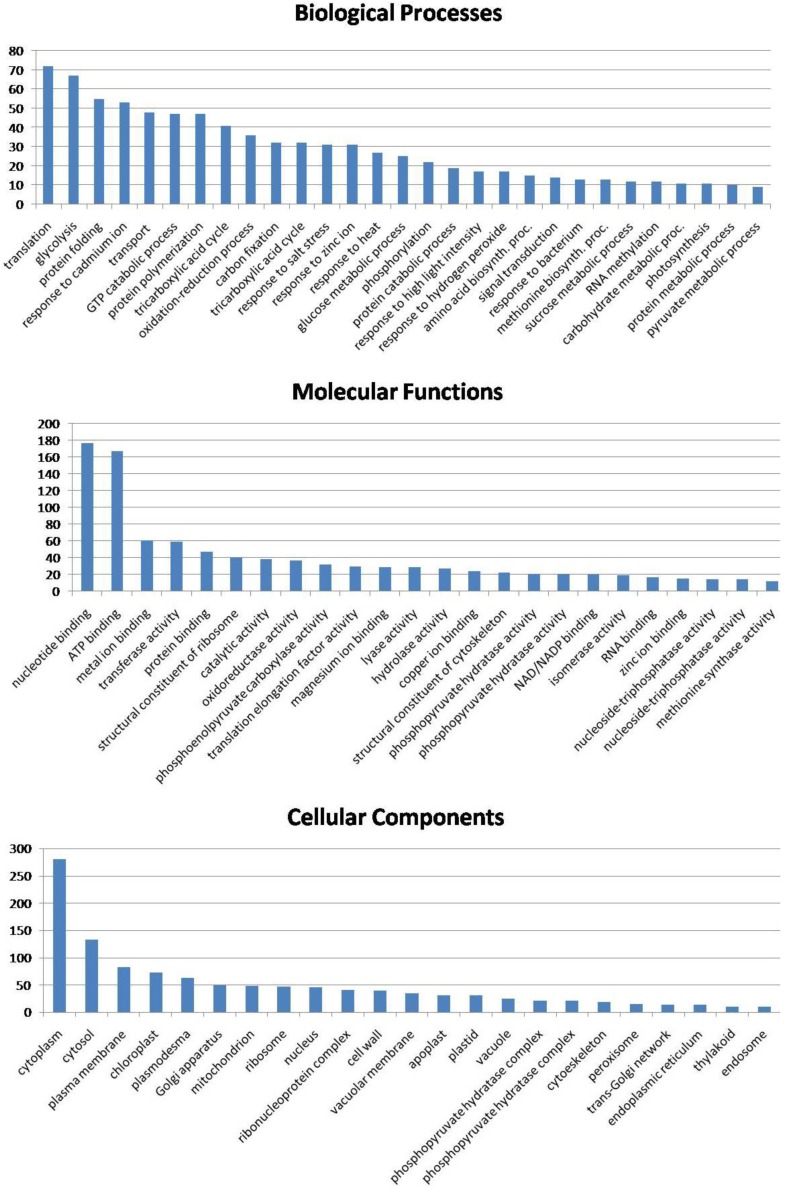
Gene ontology data (biological processes, molecular functions and cellular localization) for all identified proteins were obtained from the UniProtKB database, using the EMBL-EBI (www.ebi.ac.uk) search tool (Table 3).
Table 3.
List of all 206 non-redundant (nr) proteins found across the three tissues of the lotus seed (embryo, immature endosperm and mature endosperm).
| Protein Accession | Protein Description | Tissues 1 |
|---|---|---|
| 1433E_TOBAC | 14-3-3-like protein E OS = Nicotiana tabacum | M/I/E |
| HSP14_SOYBN | 17.5 kDa class I heat shock protein OS = Glycine max | I |
| HSP11_SOLLC | 17.8 kDa class I heat shock protein OS = Solanum lycopersicum | M |
| HSP11_PEA | 18.1 kDa class I heat shock protein OS = Pisum sativum | M/I/E |
| HSP12_MEDSA | 18.2 kDa class I heat shock protein OS = Medicago sativa | M/I |
| HSP11_CHERU | 18.3 kDa class I heat shock protein OS = Chenopodium rubrum | I |
| PMG1_ARATH | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase 1 OS = Arabidopsis thaliana | M/I/E |
| PRS6A_SOLLC | 26S protease regulatory subunit 6A homolog OS = Solanum lycopersicum | E |
| RPN1A_ARATH | 26S proteasome non-ATPase regulatory subunit 2 1A OS = Arabidopsis thaliana | E |
| PRS4A_ARATH | 26S proteasome regulatory subunit 4 homolog A OS = Arabidopsis thaliana | E |
| BAS1_ORYSJ | 2-Cys peroxiredoxin BAS1, chloroplastic OS = Oryza sativa subsp. Japonica | I |
| RS102_ARATH | 40S ribosomal protein S10-2 OS = Arabidopsis thaliana | I |
| RS13_PEA | 40S ribosomal protein S13 OS = Pisum sativum | I/E |
| RS14_CHLRE | 40S ribosomal protein S14 OS = Chlamydomonas reinhardtii | I/E |
| RS15A_DAUCA | 40S ribosomal protein S15a OS = Daucus carota | E |
| RS16_FRIAG | 40S ribosomal protein S16 OS = Fritillaria agrestis | I/E |
| RS18_ARATH | 40S ribosomal protein S18 OS = Arabidopsis thaliana | I/E |
| RS193_ARATH | 40S ribosomal protein S19-3 OS = Arabidopsis thaliana | I/E |
| RS254_ARATH | 40S ribosomal protein S25-4 OS = Arabidopsis thaliana | E |
| RS30_ARATH | 40S ribosomal protein S30 OS = Arabidopsis thaliana | E |
| RS33_ARATH | 40S ribosomal protein S3-3 OS = Arabidopsis thaliana | E |
| RS3A1_VITVI | 40S ribosomal protein S3a-1 OS = Vitis vinifera | I |
| RS4_GOSHI | 40S ribosomal protein S4 OS = Gossypium hirsutum | I/E |
| RS5_CICAR | 40S ribosomal protein S5 (fragment) OS = Cicer arietinum | I/E |
| RS6_ASPOF | 40S ribosomal protein S6 OS = Asparagus officinalis | I |
| RS61_ARATH | 40S ribosomal protein S6-1 OS = Arabidopsis thaliana | E |
| RS8_MAIZE | 40S ribosomal protein S8 OS = Zea mays | E |
| RS91_ARATH | 40S ribosomal protein S9-1 OS = Arabidopsis thaliana | M |
| RS92_ARATH | 40S ribosomal protein S9-2 OS = Arabidopsis thaliana | E |
| RSSA_SOYBN | 40S ribosomal protein SA OS = Glycine max | I/E |
| METE_ARATH | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase OS = Arabidopsis thaliana | M/I/E |
| RLA0_LUPLU | 60S acidic ribosomal protein P0 OS = Lupinus luteus | I/E |
| RLA2_PARAR | 60S acidic ribosomal protein P2 OS = Parthenium argentatum | E |
| RL10_VITRI | 60S ribosomal protein L10 OS = Vitis riparia | I |
| RL11_MEDSA | 60S ribosomal protein L11 OS = Medicago sativa | E |
| RL12_PRUAR | 60S ribosomal protein L12 OS = Prunus armeniaca | I |
| RL13_TOBAC | 60S ribosomal protein L13 OS = Nicotiana tabacum | I/E |
| RL17_MAIZE | 60S ribosomal protein L17 OS = Zea mays | E |
| RL182_ARATH | 60S ribosomal protein L18-2 OS = Arabidopsis thaliana | I |
| RL18A_CASSA | 60S ribosomal protein L18a OS = Castanea sativa | E |
| RL24_PRUAV | 60S ribosomal protein L24 OS = Prunus avium | E |
| RL3_ORYSJ | 60S ribosomal protein L3 OS = Oryza sativa subsp. Japonica | I/E |
| RL4_PRUAR | 60S ribosomal protein L4 OS = Prunus armeniaca | I/E |
| RL51_ARATH | 60S ribosomal protein L5-1 OS = Arabidopsis thaliana | I/E |
| RL6_MESCR | 60S ribosomal protein L6 OS = Mesembryanthemum crystallinum | I |
| RL7A1_ARATH | 60S ribosomal protein L7a-1 OS = Arabidopsis thaliana | E |
| ACOC_CUCMA | Aconitate hydratase, cytoplasmic OS = Cucurbita maxima | M/E |
| ACT_GOSHI | Actin OS = Gossypium hirsutum | I/E |
| ACT1_ORYSI | Actin-1 OS = Oryza sativa subsp. Indica | M |
| ACT12_SOLTU | Actin-100 (fragment) OS = Solanum tuberosum | M/E |
| ACT1_SOLLC | Actin-41 (fragment) OS = Solanum lycopersicum | M |
| ACT7_ARATH | Actin-7 OS = Arabidopsis thaliana | M |
| SAHH_MEDSA | Adenosylhomocysteinase OS = Medicago sativa | M/I/E |
| ADT1_GOSHI | ADP, ATP carrier protein 1, mitochondrial OS = Gossypium hirsutum | M/I/E |
| ARF_VIGUN | ADP-ribosylation factor OS = Vigna unguiculata | E |
| SYA_ARATH | Alanine--tRNA ligase OS = Arabidopsis thaliana | E |
| ADH1_SOLTU | Alcohol dehydrogenase 1 OS = Solanum tuberosum | M/I |
| PHSL_IPOBA | Alpha-1,4 glucan phosphorylase L isozyme, chloroplastic/amyloplastic OS = Ipomoea batatas | M/E |
| PHSH_ARATH | Alpha-glucan phosphorylase, H isozyme OS = Arabidopsis thaliana | M/I |
| GCST_PEA | Aminomethyltransferase, mitochondrial OS = Pisum sativum | I/E |
| ANXD1_ARATH | Annexin D1 OS = Arabidopsis thaliana | M/I/E |
| ANX4_FRAAN | Annexin-like protein RJ4 OS = Fragaria ananassa | E |
| CYF_AETCO | Apocytochrome f OS = Aethionema cordifolium | I |
| AATM_LUPAN | Aspartate aminotransferase P2, mitochondrial (fragment) OS = Lupinus angustifolius | E |
| AATM_LUPAN | Aspartate aminotransferase P2, mitochondrial (fragment) OS = Lupinus angustifolius | I |
| AAT3_ARATH | Aspartate aminotransferase, chloroplastic OS = Arabidopsis thaliana | I/E |
| AATC_DAUCA | Aspartate aminotransferase, cytoplasmic OS = Daucus carota | E |
| PYRB_ARATH | Aspartate carbamoyltransferase, chloroplastic OS = Arabidopsis thaliana | I |
| ATPAM_HELAN | ATP synthase subunit alpha, mitochondrial OS = Helianthus annuus | M/I/E |
| ATPBM_NICPL | ATP synthase subunit beta, mitochondrial OS = Nicotiana plumbaginifolia | M/I/E |
| ACLB1_ORYSJ | ATP-citrate synthase beta chain protein 1 OS = Oryza sativa subsp. Japonica | E |
| CLPA_BRANA | ATP-dependent Clp protease ATP-binding subunit clpA homolog, chloroplastic (fragment) OS = Brassica napus | I |
| APBLB_ARATH | Beta-adaptin-like protein B OS = Arabidopsis thaliana | E |
| ENO2_ARATH | Bifunctional enolase 2/transcriptional activator OS = Arabidopsis thaliana | I/E |
| ACCC2_POPTR | Biotin carboxylase 2, chloroplastic OS = Populus trichocarpa | E |
| CALSB_ARATH | Callose synthase 11 OS = Arabidopsis thaliana | E |
| CALX2_ARATH | Calnexin homolog 2 OS = Arabidopsis thaliana | E |
| CALR_BERST | Calreticulin OS = Berberis stolonifera | I |
| CATA2_RICCO | Catalase isozyme 2 OS = Ricinus communis | E |
| CD48A_ARATH | Cell division control protein 48 homolog A OS = Arabidopsis thaliana | M/I/E |
| CLPB1_ARATH | Chaperone protein ClpB1 OS = Arabidopsis thaliana | E |
| CLPC1_ARATH | Chaperone protein ClpC1, chloroplastic OS = Arabidopsis thaliana | I |
| CPNA1_ARATH | Chaperonin 60 subunit alpha 1, chloroplastic OS = Arabidopsis thaliana | I/E |
| CPNB3_ARATH | Chaperonin 60 subunit beta 3, chloroplastic OS = Arabidopsis thaliana | E |
| CH60A_ARATH | Chaperonin CPN60, mitochondrial OS = Arabidopsis thaliana | M/I/E |
| CB2_PHYPA | Chlorophyll a-b binding protein, chloroplastic OS = Physcomitrella patens subsp. patens | I |
| HSP7E_SPIOL | Chloroplast envelope membrane 70 kDa heat shock-related protein OS = Spinacia oleracea | M/I/E |
| HSP12_SOYBN | Class I heat shock protein (fragment) OS = Glycine max | I |
| CLAH1_ARATH | Clathrin heavy chain 1 OS = Arabidopsis thaliana | I/E |
| COPA1_ARATH | Coatomer subunit alpha-1 OS = Arabidopsis thaliana | E |
| COB21_ORYSJ | Coatomer subunit beta-1 OS = Oryza sativa subsp. Japonica | I |
| RH2_ORYSJ | DEAD-box ATP-dependent RNA helicase 2 OS = Oryza sativa subsp. Japonica | M |
| DIM_PEA | Delta(24)-sterol reductase OS = Pisum sativum | E |
| DLDH2_ARATH | Dihydrolipoyl dehydrogenase 2, mitochondrial OS = Arabidopsis thaliana | I |
| OPD22_ARATH | Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial OS = Arabidopsis thaliana | E |
| EF1A_TOBAC | Elongation factor 1-alpha OS = Nicotiana tabacum | M/I/E |
| EF1D1_ORYSJ | Elongation factor 1-delta 1 OS = Oryza sativa subsp. Japonica | E |
| EF1G2_ORYSJ | Elongation factor 1-gamma 2 OS = Oryza sativa subsp. Japonica | M/I/E |
| EF2_BETVU | Elongation factor 2 OS = Beta vulgaris | M/I/E |
| EFTM_ARATH | Elongation factor Tu, mitochondrial OS = Arabidopsis thaliana | E |
| ENPL_CATRO | Endoplasmin homolog OS = Catharanthus roseus | M/I/E |
| ENO1_HEVBR | Enolase 1 OS = Hevea brasiliensis | M/I/E |
| IF4A1_ARATH | Eukaryotic initiation factor 4A-1 OS = Arabidopsis thaliana | M/I/E |
| ERF1X_ARATH | Eukaryotic peptide chain release factor subunit 1-1 OS = Arabidopsis thaliana | E |
| IF5A1_ARATH | Eukaryotic translation initiation factor 5A-1 OS = Arabidopsis thaliana | E |
| ALF_CICAR | Fructose-bisphosphate aldolase, cytoplasmic isozyme OS = Cicer arietinum | M/I/E |
| RFS_ORYSJ | Galactinol--sucrose galactosyltransferase OS = Oryza sativa subsp. Japonica | I |
| GME2_ORYSJ | GDP-mannose 3,5-epimerase 2 OS = Oryza sativa subsp. Japonica | E |
| GMD1_ARATH | GDP-mannose 4,6 dehydratase 1 OS = Arabidopsis thaliana | E |
| GRDH1_ARATH | Glucose and ribitol dehydrogenase homolog 1 OS = Arabidopsis thaliana | M/I |
| GLGS_BETVU | Glucose-1-phosphate adenylyltransferase small subunit, chloroplastic/amyloplastic (fragment) OS = Beta vulgaris | M |
| G6PI2_CLACO | Glucose-6-phosphate isomerase, cytosolic 2 OS = Clarkia concinna | M/E |
| GPT2_ARATH | Glucose-6-phosphate/phosphate translocator 2, chloroplastic OS = Arabidopsis thaliana | M |
| GLN11_ORYSJ | Glutamine synthetase cytosolic isozyme 1-1 OS = Oryza sativa subsp. Japonica | E |
| G3PC_ANTMA | Glyceraldehyde-3-phosphate dehydrogenase, cytosolic OS = Antirrhinum majus | M/I/E |
| SYGM1_ARATH | Glycine--tRNA ligase 1, mitochondrial OS = Arabidopsis thaliana | E |
| SSG1_HORVU | Granule-bound starch synthase 1, chloroplastic/amyloplastic OS = Hordeum vulgare | M/I |
| RAN_VICFA | GTP-binding nuclear protein Ran/TC4 OS = Vicia faba | M/I/E |
| RAN3_ORYSI | GTP-binding nuclear protein Ran-3 OS = Oryza sativa subsp. Indica | E |
| YPTC1_CHLRE | GTP-binding protein YPTC1 OS = Chlamydomonas reinhardtii | E |
| GBLPA_ORYSJ | Guanine nucleotide-binding protein subunit beta-like protein A OS = Oryza sativa subsp. Japonica | I |
| HSP7L_ARATH | Heat shock 70 kDa protein 12 OS = Arabidopsis thaliana | I/E |
| HSP7O_ARATH | Heat shock 70 kDa protein 14 OS = Arabidopsis thaliana | I/E |
| HSP7N_ARATH | Heat shock 70 kDa protein 18 OS = Arabidopsis thaliana | I |
| HSP7D_ARATH | Heat shock 70 kDa protein 4 OS = Arabidopsis thaliana | I |
| HSP7F_ARATH | Heat shock 70 kDa protein 6, chloroplastic OS = Arabidopsis thaliana | I |
| HSP7G_ARATH | Heat shock 70 kDa protein 7, chloroplastic OS = Arabidopsis thaliana | E |
| HSP70_DAUCA | Heat shock 70 kDa protein OS = Daucus carota | M/I/E |
| HSP7M_PHAVU | Heat shock 70 kDa protein, mitochondrial OS = Phaseolus vulgaris | I/E |
| HSP80_SOLLC | Heat shock cognate protein 80 OS = Solanum lycopersicum | M/I |
| HS101_ARATH | Heat shock protein 101 OS = Arabidopsis thaliana | M |
| HS101_ORYSJ | Heat shock protein 101 OS = Oryza sativa subsp. Japonica | M |
| HSP81_ORYSI | Heat shock protein 81-1 OS = Oryza sativa subsp. Indica | M/I/E |
| HSP82_TOBAC | Heat shock protein 82 (fragment) OS = Nicotiana tabacum | M |
| HSP82_MAIZE | Heat shock protein 82 OS = Zea mays | M/I/E |
| HSP83_IPONI | Heat shock protein 83 OS = Ipomoea nil | M/I/E |
| HS901_ARATH | Heat shock protein 90-1 OS = Arabidopsis thaliana | E |
| HS903_ARATH | Heat shock protein 90-3 OS = Arabidopsis thaliana | I |
| H2AX_CICAR | Histone H2AX OS = Cicer arietinum | I |
| H2B_GOSHI | Histone H2B OS = Gossypium hirsutum | I/E |
| H4_ARATH | Histone H4 OS = Arabidopsis thaliana | I/E |
| IDHC_TOBAC | Isocitrate dehydrogenase [NADP] OS = Nicotiana tabacum | E |
| ILV5_ARATH | Ketol-acid reductoisomerase, chloroplastic OS = Arabidopsis thaliana | E |
| APX1_ORYSJ | L-ascorbate peroxidase 1, cytosolic OS = Oryza sativa subsp. Japonica | E |
| LE194_HORVU | Late embryogenesis abundant protein B19.4 OS = Hordeum vulgare | I |
| AMPL1_ARATH | Leucine aminopeptidase 1 OS = Arabidopsis thaliana | M/I |
| BIP4_TOBAC | Luminal-binding protein OS = Nicotiana tabacum | M/I/E |
| MDHC2_ARATH | Malate dehydrogenase, cytoplasmic 2 OS = Arabidopsis thaliana | M/E |
| MDHM_CITLA | Malate dehydrogenase, mitochondrial OS = Citrullus lanatus | M/I/E |
| MPPA_SOLTU | Mitochondrial-processing peptidase subunit alpha OS = Solanum tuberosum | E |
| MDAR_SOLLC | Monodehydroascorbate reductase OS = Solanum lycopersicum | I/E |
| MAOX_POPTR | NADP-dependent malic enzyme OS = Populus trichocarpa | M |
| NDK1_ARATH | Nucleoside diphosphate kinase 1 OS = Arabidopsis thaliana | M/I/E |
| FKB62_ARATH | Peptidyl-prolyl cis-trans isomerase FKBP62 OS = Arabidopsis thaliana | I/E |
| PER1B_ARMRU | Peroxidase C1B OS = Armoracia rusticana | I |
| CAPPC_FLATR | Phosphoenolpyruvate carboxylase 2 OS = Flaveria trinervia | E |
| PGMC_PEA | Phosphoglucomutase, cytoplasmic OS = Pisum sativum | M/I/E |
| PGKH_TOBAC | Phosphoglycerate kinase, chloroplastic OS = Nicotiana tabacum | M/I |
| PGKY_TOBAC | Phosphoglycerate kinase, cytosolic OS = Nicotiana tabacum | M/E |
| SERC_SPIOL | Phosphoserine aminotransferase, chloroplastic OS = Spinacia oleracea | E |
| PDR4_ORYSJ | Pleiotropic drug resistance protein 4 OS = Oryza sativa subsp. Japonica | E |
| PARP3_SOYBN | Poly [ADP-ribose] polymerase 3 OS = Glycine max | I |
| UBIQP_ACECL | Polyubiquitin (fragment) OS = Acetabularia cliftonii | I |
| UBQ12_ARATH | Polyubiquitin 12 OS = Arabidopsis thaliana | E |
| PMG2_ARATH | Probable 2,3-bisphosphoglycerate-independent phosphoglycerate mutase 2 OS = Arabidopsis thaliana | M/I |
| SSG1_ARATH | Probable granule-bound starch synthase 1, chloroplastic/amyloplastic OS = Arabidopsis thaliana | I |
| H2B1_MEDTR | Probable histone H2B.1 OS = Medicago truncatula | I |
| PDIA6_MEDSA | Probable protein disulfide-isomerase A6 OS = Medicago sativa | I |
| Y1497_ARATH | Probable receptor-like protein kinase At1g49730 OS = Arabidopsis thaliana | I |
| PROF3_ARATH | Profilin-3 OS = Arabidopsis thaliana | E |
| PSA3_ARATH | Proteasome subunit alpha type-3 OS = Arabidopsis thaliana | E |
| PDI21_ORYSJ | Protein disulfide isomerase-like 2-1 OS = Oryza sativa subsp. Japonica | I |
| PDI21_ARATH | Protein disulfide-isomerase like 2-1 OS = Arabidopsis thaliana | M/I/E |
| ACT5_ARATH | Putative actin-5 OS = Arabidopsis thaliana | I/E |
| YCF1_IPOPU | Putative membrane protein ycf1 OS = Ipomoea purpurea | M/I |
| AVP_VIGRR | Pyrophosphate-energized vacuolar membrane proton pump OS = Vigna radiata var. radiata | I/E |
| PDC1_TOBAC | Pyruvate decarboxylase isozyme 1 (fragment) OS = Nicotiana tabacum | M/I |
| KPYC_SOYBN | Pyruvate kinase, cytosolic isozyme OS = Glycine max | M/I/E |
| PPDK2_ORYSJ | Pyruvate, phosphate dikinase 2 OS = Oryza sativa subsp. Japonica | M |
| PPDK_FLABR | Pyruvate, phosphate dikinase, chloroplastic OS = Flaveria brownii | M/E |
| RAA1D_ARATH | Ras-related protein RABA1d OS = Arabidopsis thaliana | E |
| RBL_MAIZE | Ribulose bisphosphate carboxylase large chain OS = Zea mays | I |
| RUBA_RICCO | RuBisCO large subunit-binding protein subunit alpha (fragment) OS = Ricinus communis | I/E |
| RUBB_PEA | RuBisCO large subunit-binding protein subunit beta, chloroplastic OS = Pisum sativum | E |
| METK4_POPTR | S-adenosylmethionine synthase 4 OS = Populus trichocarpa | E |
| SAPK6_ORYSJ | Serine/threonine-protein kinase SAPK6 OS = Oryza sativa subsp. Japonica | E |
| HSP7S_SPIOL | Stromal 70 kDa heat shock-related protein, chloroplastic (fragment) OS = Spinacia oleracea | I/E |
| SUSY_SOYBN | Sucrose synthase OS = Glycine max | I/E |
| SODM_HEVBR | Superoxide dismutase [Mn], mitochondrial OS = Hevea brasiliensis | E |
| TCPA_ARATH | T-complex protein 1 subunit alpha OS = Arabidopsis thaliana | I/E |
| TCPE_ARATH | T-complex protein 1 subunit epsilon OS = Arabidopsis thaliana | M/E |
| TKTC_SPIOL | Transketolase, chloroplastic OS = Spinacia oleracea | E |
| TCTP_TOBAC | Translationally-controlled tumor protein homolog OS = Nicotiana tabacum | M |
| TPIS_MAIZE | Triosephosphate isomerase, cytosolic OS = Zea mays | I/E |
| TBA_PRUDU | Tubulin alpha chain OS = Prunus dulcis | I |
| TBB_HORVU | Tubulin beta chain OS = Hordeum vulgare | E |
| UBIQ_ARATH | Ubiquitin OS = Arabidopsis thaliana | M/E |
| RL40A_ARATH | Ubiquitin-60S ribosomal protein L40-1 OS = Arabidopsis thaliana | I/E |
| RGP1_ORYSJ | UDP-arabinopyranose mutase 1 OS = Oryza sativa subsp. Japonica | E |
| UGDH_SOYBN | UDP-glucose 6-dehydrogenase OS = Glycine max | E |
| UREA_CANEN | Urease OS = Canavalia ensiformis | I |
| UGPA1_ARATH | UTP--glucose-1-phosphate uridylyltransferase 1 OS = Arabidopsis thaliana | M/E |
| VATA_GOSHI | V-type proton ATPase catalytic subunit A OS = Gossypium hirsutum | I/E |
| VATB2_GOSHI | V-type proton ATPase subunit B 2 (fragment) OS = Gossypium hirsutum | E |
| VATB1_ARATH | V-type proton ATPase subunit B1 OS = Arabidopsis thaliana | E |
| WIT2_ARATH | WPP domain-interacting tail-anchored protein 2 OS = Arabidopsis thaliana | I |
1 M: Mature endosperm; I: Immature endosperm; E: Embryo
Analysis of the annotations referent to the immature endosperm revealed that functions related to protein synthesis (translation, protein folding and polymerization, etc.), general metabolism (amino acid, carbon fixation) and carbohydrate metabolism (glycolysis, etc.) are all considerably represented, with the proteins in the first category being relatively more numerous (Figure 4).
Figure 4.
Distribution of the top gene ontology (GO) data for lotus immature endosperm proteome based on 1-DGE-MS analysis.
On the other hand, the embryo proteome shows considerable prevalence of proteins involved in protein synthesis, followed then by carbohydrate and general metabolism processes (Figure 5).
Figure 5.
Distribution of the top gene ontology (GO) data for lotus embryo proteome based on 1-DGE-MS analysis.
3.6. Biological Function of the Identified Seed Proteins
Furthermore, the nr protein matches were also classified according to their broader biological function [22,23], divided into 10 categories: metabolism, energy, cell growth/division, transcription, protein synthesis/destination, transporters, cell structure, signal transduction, stress response, and unclassified (Figure 6).
Figure 6.
Bar charts displaying the division according to functional categories, of the non-redundant (nr) protein matches found in the lotus seed embryo, immature endosperm, and mature endosperm, as determined by 1-DGE-MS.
A comparison of the distribution of protein functionality between the seed immature endosperm and embryo, and the previous results obtained from the mature endosperm shows that immature endosperm and embryo have a quite similar functionality profile of the mature endosperm. However, in the embryo the identified proteins related to general cell housekeeping functions (non-energy metabolism, cell growth, transcription, transport, and signaling) were slightly more apparent than in the immature endosperm. In contrast with the mature endosperm, both immature endosperm and embryo show a larger percentage of the identified proteins related to protein synthesis. This correlated well with the fact that the tissues are either in a growing phase, i.e., immature endosperm or have growth as their main function, i.e., embryo. The mature endosperm, on the other hand, having its primary function as energy and nutrient storage, has the larger share of its proteins related to energy metabolism. A common element for all the lotus seed tissues is the large presence of stress-/defense-related proteins across all samples.
3.7. Lotus Seed Proteome Compared with Other Seed Proteomes
Unlike some seeds, such as tomato, where non-germinating embryo and endosperm were shown to have very similar proteomes [24], the analysis of lotus seed proteomes showed some remarkable difference in proteins identified/function between the non-germinating embryo and mature endosperm. Contrary to other seed proteomes like Jatropha curcas [23] and sugarbeet [25], the lotus embryo in its pre-germination stage did not seem to have a considerably higher expression of metabolism- and energy-related proteins compared to the mature endosperm. Structural proteins, however, did seem to be at least slightly more represented in the endosperm, as in the case of J. curcas. Compared with other embryo proteomes, such as Brassica campestri [26], and sugarbeet, the lotus embryo appears to have a larger percentage of proteins related to protein synthesis in comparison to primary and energy metabolism, as well as a much greater presence of defense related proteins. We further discuss below the key proteins identified in this study.
3.8. Key Proteins of the Lotus Immature Seed Endosperm
Contrary to the mature endosperm, the key functional proteins identified in the lotus immature endosperm mostly consisted of proteins related to plant growth and development (Figure 7).
Figure 7.
Key functional proteins identified in the lotus seed immature endosperm, and subdivided according to their role in plant metabolism.
Amongst the identified proteins were several transcription proteins (cell division control and transcription factors), translation (ribosomal) proteins, post-translational modification proteins (elongation factors and ubiquitins) and nutrient production proteins (RuBisCO subunits and sucrose synthase). Many stress response- and plant defense-related proteins were also present in the immature endosperm. Of these, the largest subgroup is the heat shock response proteins (high- and low-molecular weight heat shock proteins (HSPs), as well as chaperone and annexin proteins). Anti-oxidative stress (peroxidases, endoplasmin, and monodehydroascorbate reductase) are also present, more so than in the mature endosperm (see below section). Proteins related to carbohydrate metabolism are also present in the immature endosperm, but in a smaller number.
3.9. Key Proteins Previously Identified in the Lotus Mature Seed Endosperm
In the case of mature endosperm proteome [17], the two most significant groups of proteins identified were related to energy/carbohydrate metabolism, and stress response and plant protection (Figure 8). In the first group, several proteins that are part of glycolysis, gluconeogenesis, citric acid cycle and starch metabolism including other carbohydrate metabolism proteins, were identified. Of the stress response proteins, HSPs, along with other heat response proteins (chaperones, annexin), constituted the most numerous category. Anti-oxidative stress proteins were not greatly represented. Of note is the identification of storage proteins (such as globulins, castanins) for the mature endosperm by 2-D MS and N-terminal sequencing, but not by 1-D MS [17,27], which might indicate a possible detection gap of this technique.
Figure 8.
Key functional proteins identified in the lotus seed mature endosperm, and subdivided according to their role in plant metabolism. * for original protein lists, see reference [17].
3.10. Proteome Changes between Mature and Immature Stages of the Endosperm
Despite constituting the endosperm tissue samples, protein extracts from the mature and immature seed presented a notably different proteome composition (Figure 9).
Figure 9.
Depiction of the changes in biological function, nutrient content and functional proteome composition from the immature (left) to the mature (right) endosperm in the lotus seed.
This reflects the changes the endosperm undergoes during the maturation process, where it develops from a soft wet tissue to a dry one with a large amount per weight of both carbohydrates and proteins [6]. The endosperm’s main role in the seed is as a nutrient storage tissue, so it is expected that during the maturation phase, these nutrients are going to be produced for later storage, hence the larger number of functional proteins related to the protein and carbohydrate synthesis categories. In the mature endosperm, a large percentage of the total protein content is expected to be seed storage proteins (SSPs). Although not many SSPs were identified by MS analysis of the mature endosperm, several possible matches were found by N-terminal sequencing analysis [27]. The prevalence of carbohydrate metabolism proteins amongst the identified functional proteins in the mature endosperm could be a result of production in the late maturation stage, with such proteins playing a quasi-dormant role in managing the nutrient content of the seed before and during germination.
3.11. Key Proteins of the Lotus Seed Embryo
In the case of the embryo proteins identified by database matching, the distribution of key proteins was similar to that of the immature endosperm, in that they can be divided in the same main groups: proteins related to plant growth, and proteins responsible for plant protection and germination vigor (Figure 10). Of the first group, those also include the same subgroups of transcription, translation, and post-translation proteins as well as nutrient production proteins. In the case of stress/defense-related proteins, the embryo was also found to possess the largest number heat shock response proteins (12 HSPs, mostly of high-molecular weight, five chaperone proteins and two annexins). However, the embryo also contained a larger number of anti-oxidative stress proteins, including l-ascorbate peroxidase, catalase, monodehydroascorbate reductase, superoxide dismutase [Mn], and endoplasmin. S-adenosylmethionine synthase and adenosylhomocysteinase (also found in the endosperm tissues), and two proteins from the active methyl cycle, which is of great importance to plant metabolism as well as their nutritional value [28], were also identified in the lotus embryo.
Figure 10.
Key functional proteins identified in the lotus seed embryo, and subdivided according to their role in plant metabolism.
4. Conclusions
Analysis of protein extracts from the lotus seed embryo and immature seed endosperm was performed following 1-DGE separation in conjunction with LC-MS/MS analysis. This “bottom-up” proteomics analysis, represented by the SDS-PAGE technique, has been shown to be a good approach for identifying the lotus seed proteins [17]. For both tissues, a great number of proteins were identified by database matching. A total of 141 nr protein matches were identified in the embryo, and 122 in the immature endosperm. Together with the 66 proteins previously identified for the mature endosperm, a total of 206 nr proteins have been identified to date.
Combined datasets are a resource in itself towards complete proteomics analysis of lotus seeds and plants. By producing more extensive datasets, these results help toward forming a complete proteomic picture of the lotus seeds. The analysis of protein makeup and functionality across different tissues within the seed also permits a comparison of metabolic functions across different tissues and developmental stages of the lotus seed, as well as allowing for the comparison with similar tissues from other plants. Furthermore, the identification of proteins of interest—such as key proteins in the metabolism, proteins that confer resistance against stress or germination vigor—opens up several possibilities for more specific studies on these proteins and their possible use in producing transgenic varieties of interest.
Future work will both strive to expand the lotus proteome to other developmentally important tissues, such as seedling and rhizome, as well as to isolate and characterize functional proteins of interest in the seed proteome. Moreover, 2-DGE-MS analysis of individual proteins, especially by de novo proteome analysis techniques, coupled with genome comparison, can help obtain more detailed sequences of lotus-specific proteins, since the high taxonomical distance of the lotus in relation to other modern plants hinders the achievement of higher homology values when database-matching proteins.
Acknowledgments
CFM greatly appreciates and acknowledges the financial support of the Japanese Ministry of Education, Culture, Sports, Science and Technology (MEXT) in the conducting this research. Authors appreciate the International Plant Proteomics Organization (INPPO) initiative (www.inppo.com) for connecting plant proteomic researchers between Brazil and Japan, and continuing collaborations between INPPO-India-Nepal chapter and INPPO-Japan.
Author Contributions
C.F.M., R.R., G.K.A., S.S., Y.K. and M.Y. were responsible for the conception of the study; C.F.M. and J.S. performed sample preparation and gel analysis; C.F.M. and Y.F. were responsible for mass spectrometry analyses; C.F.M. and R.R. performed the data analysis; C.F.M., R.R., G.K.A. and M.Y. wrote or contributed to the manuscript; figures and tables were prepared by C.F.M. and R.R. All authors read and approved the final version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Sridhar K.R., Rajeev B. Lotus—A potential nutraceutical source. J. Agric. Technol. 2007;3:143–155. [Google Scholar]
- 2.Pandey B.P. Economic Botany. 5th ed. Chand (S.) & Co. Ltd.; New Delhi, India: 1999. p. 61. [Google Scholar]
- 3.Loewer H.P. Seeds: The Definitive Guide to Growing, History and Lore. 1st ed. Timber Press; Cambridge, UK: 2005. p. 56. [Google Scholar]
- 4.Shen-Miller J., Schopf J.W., Harbottle G., Cao R.J., Ouyang S., Zhou K.S., Southon J.R., Liu G.H. Long-living lotus: germination and soil γ-irradiation of centuries-old fruits, and cultivation, growth, and phenotypic abnormalities of offspring. Am. J. Bot. 2002;89:236–247. doi: 10.3732/ajb.89.2.236. [DOI] [PubMed] [Google Scholar]
- 5.Shen-Miller J., Aung L.H., Turek J., Schopf J.W., Tholandi M., Yang M., Czaja M. Centuries-old viable fruit of sacred lotus Nelumbo nucifera Gaertn var. China antique. Trop. Plant Biol. 2013;6:53–68. doi: 10.1007/s12042-013-9125-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MEXT—Ministry of Education, Culture, Sports, Science and Technology . Standard Tables of Food Composition in Japan. 5th ed. MEXT; Tokyo, Japan: 2000. [Google Scholar]
- 7.Guo H.B. Cultivation of lotus (Nelumbo nucifera Gaertn. ssp. nucifera) and its utilization in China. Gen. Res. Crop Evol. 2009;56:323–330. doi: 10.1007/s10722-008-9366-2. [DOI] [Google Scholar]
- 8.Komatsu E., Tsukahara A., Amagaya H., Okazawa N., Noguchi T., Okuyama T. Lotus. In: Izaki M., editor. The Cultivation and Management in Aquatic Vegetables. Volume 1. Ie-No-Hikari Kyokai Press; Tokyo, Japan: 1975. pp. 9–94. [Google Scholar]
- 9.Ling Z.Q., Xie B.J., Yang E.L. Isolation, characterization, and determination of antioxidative activity of oligomeric procyanidins from the seedpod of Nelumbo nucifera Gaertn. J. Agric. Food Chem. 2005;53:2441–2445. doi: 10.1021/jf040325p. [DOI] [PubMed] [Google Scholar]
- 10.Ou M. Chinese-English Manual of Commonly-Used in Traditional Chinese Medicine. Joint Publishing Co. Ltd.; Hong Kong, China: 1989. [Google Scholar]
- 11.Moro C.F., Yonekura M., Kouzuma Y., Agrawal G.K., Rakwal R. Lotus—A source of food and medicine: Current status and future perspectives in context of the seed proteomics. Int. J. Life Sci. 2013;7:1–5. doi: 10.3126/ijls.v7i1.6394. [DOI] [Google Scholar]
- 12.Ming R., Vanburen R., Liu Y., Yang M., Han Y., Li L.T., Zhang Q., Kim M.J., Schatz M.C., Campbell M., et al. Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.) Genome Bio. 2013;14 doi: 10.1186/gb-2013-14-5-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong C., Zheng X., Li G., Zhu H., Zhou M., Hu Z. Molecular cloning and expression of two cytosolic copper–zinc superoxide dismutases genes from Nelumbo nucifera. Appl. Biochem. Biotechnol. 2010;163:679–691. doi: 10.1007/s12010-010-9074-1. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Y., Chen H., Chu P., Li Y., Tan B., Ding Y., Tsang E.W.T., Jiang L., Wu K., Huang S. NnHSP17.5, a cytosolic class II small heat shock protein gene from Nelumbo nucifera, contributes to seed germination vigor and seedling thermotolerance in transgenic Arabidopsis. Plant Cell Rep. 2012;31:379–389. doi: 10.1007/s00299-011-1173-0. [DOI] [PubMed] [Google Scholar]
- 15.Liu Z., Gu G., Chen F., Yang D., Wu K., Chen S., Jiang J., Zhang Z. Heterologous expression of a Nelumbo nucifera phytochelatin synthase gene enhances cadmium tolerance in Arabidopsis thaliana. Appl. Biochem. Biotechnol. 2012;166:722–734. doi: 10.1007/s12010-011-9461-2. [DOI] [PubMed] [Google Scholar]
- 16.Chu P., Chen H., Zhou Y., Li Y., Ding Y., Jiang L., Tsang E.W., Wu K., Huang S. 2012. Proteomic and functional analyses of Nelumbo nucifera annexins involved in seed thermotolerance and germination vigor. Planta. 2012;235:1271–1288. doi: 10.1007/s00425-011-1573-y. [DOI] [PubMed] [Google Scholar]
- 17.Moro C.F., Fukao Y., Shibato J., Rakwal R., Timperio A.M., Zolla L., Agrawal G.K., Shioda S., Kouzuma Y., Yonekura M. Unraveling the seed endosperm proteome of the lotus (Nelumbo nucifera Gaertn.) utilizing 1DE and 2DE separation in conjunction with tandem mass spectrometry. Proteomics. 2015;15:1717–1735. doi: 10.1002/pmic.201400406. [DOI] [PubMed] [Google Scholar]
- 18.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 19.Wang X., Li X., Li Y. A modified Coomassie Brilliant Blue staining method at nanogram sensitivity compatible with proteomic analysis. Biotech. Lett. 2007;29:1599–1603. doi: 10.1007/s10529-007-9425-3. [DOI] [PubMed] [Google Scholar]
- 20.Horie K., Rakwal R., Hirano M., Shibato J., Nam H.W., Kim Y.S., Kouzuma Y., Agrawal G.K., Masuo Y., Yonekura M. Proteomics of two cultivated mushrooms Sparassis crispa and Hericium erinaceum provides insight into their numerous functional protein components and diversity. J. Proteome Res. 2008;7:1819–1835. doi: 10.1021/pr070369o. [DOI] [PubMed] [Google Scholar]
- 21.Yates J.R., Ruse C.I., Nakorchevsky A. Proteomics by mass spectrometry: Approaches, advances, and applications. Ann. Rev. Biomed. Eng. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- 22.Bevan M., Bancroft I., Bent E., Love K., Goodman H., Dean C., Bergkamp R., Dirkse W., van Staveren M., Stiekema W., et al. Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature. 1998;391:485–488. doi: 10.1038/35140. [DOI] [PubMed] [Google Scholar]
- 23.Liu H., Yang Z., Yang M, Shen S. The differential proteome of endosperm and embryo from mature seed of Jatropha curcas. Plant Sci. 2011;181:660–666. doi: 10.1016/j.plantsci.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 24.Sheoran I.S., Olson D.J., Ross A.R., Sawhney V.K. Proteome analysis of embryo and endosperm from germinating tomato seeds. Proteomics. 2005;5:3752–3764. doi: 10.1002/pmic.200401209. [DOI] [PubMed] [Google Scholar]
- 25.Catusse J., Strub J.M., Job C., Dorsselaer A., Job D. Proteome-wide characterization of sugarbeet seed vigor and its tissue specific expression. Proc. Natl. Acad. Sci. U.S.A. 2008;105:10262–10267. doi: 10.1073/pnas.0800585105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li W., Gao Y., Xu H., Zhang Y., Wang J. A proteomic analysis of seed development in Brassica campestri L. PLoS ONE. 2012;7:e50290. doi: 10.1371/journal.pone.0050290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moro C.F. Ph.D. Thesis. Tokyo University of Agriculture and Technology; Tokyo, Japan: Mar, 2015. Study of the Lotus (Nelumbo nucifera Gaertn.) Seed Proteome. [Google Scholar]
- 28.Ravanel S., Gakière B., Job D., Douce R. The specific features of methionine biosynthesis and metabolism in plants. Proc. Natl. Acad. Sci. U.S.A. 1998;95:7805–7812. doi: 10.1073/pnas.95.13.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]