Abstract
Clavibacter michiganensis, subsp. michiganensis is a Gram-positive plant pathogen infecting tomato (Solanum lycopersicum). Despite a considerable economic importance due to significant losses of infected plants and fruits, knowledge about virulence factors of C. michiganensis subsp. michiganensis and host-pathogen interactions on a molecular level are rather limited. In the study presented here, the proteome of culture supernatants from C. michiganensis subsp. michiganensis NCPPB382 was analyzed. In total, 1872 proteins were identified in M9 and 1766 proteins in xylem mimicking medium. Filtration of supernatants before protein precipitation reduced these to 1276 proteins in M9 and 976 proteins in the xylem mimicking medium culture filtrate. The results obtained indicate that C. michiganensis subsp. michiganensis reacts to a sucrose- and glucose-depleted medium similar to the xylem sap by utilizing amino acids and host cell polymers as well as their degradation products, mainly peptides, amino acids and various C5 and C6 sugars. Interestingly, the bacterium expresses the previously described virulence factors Pat-1 and CelA not exclusively after host cell contact in planta but already in M9 minimal and xylem mimicking medium.
Keywords: canker, Clavibacter, plant pathogen, secretome, wilt, xylem mimicking medium, xylem sap
1. Introduction
The genus Clavibacter belongs to the group of Actinobacteria, and comprises a single species, Clavibacter michiganensis, with five established subspecies. Recently, the existence of a sixth subspecies was proposed [1]. All subspecies are plant pathogenic but differ significantly in their respective host spectrum. Despite the economic importance due to significant losses of the different crops infected, knowledge about virulence factors of C. michiganensis subspecies and host-pathogen interactions on a molecular level are rather limited. The best investigated subspecies in this respect is C. michiganensis subsp. michiganensis, which causes wilt and canker of tomato plants (Solanum lycopersicum) and bird’s eye spots on their fruits [2]. The plant is colonized either by wounds, foliar infection through stomata and trichomes or through hydatodes [3,4,5,6]. Resistant cultivars or effective chemical or antibiotic controls in field and greenhouse cultivation are unknown [7]. The infection is spread by contaminated seeds or transplants [8,9] and, consequently, testing of seeds and plant tissue as well as quarantine measurements are the most effective disease control measures.
Even in the case of C. michiganensis subsp. michiganensis, only three genes and their respective products have been studied in detail: pat-1, encoding a serine protease [5], celA, encoding a β-1,4-endoglucanase [10] and tomA, coding for a tomatinase which degrades the antimicrobial saponin tomatin [11]. The genes pat-1 and celA are located on two natural plasmids, which are crucial for virulence and symptom development. In addition to pat-1 and celA, three more putative serine proteases (ppaJ, phpA and phpB), which may also play an important role in pathogenicity, are encoded on these plasmids designated pCM1 and pCM2. The tomA gene is chromosomally encoded and located within the chp/tomA region, a putative pathogenicity island [12], which comprises also the serine protease-encoding genes chpC, chpG, ppaA and ppaC, further putative virulence factors of C. michiganensis subsp. michiganensis [13,14]. The loss of this pathogenicity island, as well as the loss of pCM1 and pCM2, results in reduced pathogenicity and symptom development in tomato [14]. Furthermore, Savidor and co-workers showed that the transcriptional regulators Vatr1 and Vatr2 are crucial for virulence in tomato [15].
When the genomes of C. michiganensis subsp. michiganensis [12] and C. michiganensis subsp. sepedonicus [16] were compared, the gene products of the pat-1 and celA homologs were recognized as common virulence factors. Furthermore, a pat-1 homolog was also observed in the genome of the closely related sugarcane pathogen Leifsonia xyli subsp. xyli [17].
The publication of the C. michiganensis subsp. michiganensis genome sequence has not only promoted bioinformatic analyses, but allowed global analysis approaches. Unfortunately, from 3080 detected genes in core genome and plasmids pCM1 and pCM2, 1030 were annotated as hypothetical, making an elucidation of their physiological function difficult [12]. Global studies might help to give at least basic information about expression of these proteins and, in fact, several studies using proteome and transcriptome analyses gave valuable information in this respect [18,19,20].
In the study presented here, the proteome of C. michiganensis subsp. michiganensis wild type strain NCPPB382382 was studied in order to identify proteins secreted to the culture supernatant. This strain carries the two natural Clavibacter plasmids pCM1 and pCM2, which are important for virulence [12].
2. Experimental Section
2.1. Strains and Growth Conditions
C. michiganensis subsp. michiganensis was grown at 26 to 28 °C in M9 minimal medium (5 g/L glucose, 15 g/L Na2HPO4 × 12 H2O, 3 g/L KH2PO4, 0,5 g/L NaCl, 1 g/L NH4Cl, 0.3 g/L methionine, 11 mg/L CaCl2, 240 mg/L MgSO4, 500 µg/L thiamine, 500 µg/L nicotinic acid, 0.2 mg/L FeCl3 × 6 H2O, 0.01 mg/L CuCl2 × 2 H2O, 0.01 mg/L Na2B4O7 × 10 H2O, 0.01 mg/L MnCl2 × 2 H2O, 0.01 mg/L (NH4)6Mo7 × 4 H2O, 0.04 mg/L ZnCl2) [21] and xylem mimicking medium XMM (1.168 g/L NaCl, 2.02 g/L KNO3, 1.233 g/L MgSO4, 0.147 g/L CaCl2, 22 mg/L KH2PO4, 56 mg/L K2HPO4, 1.5 µg/L FeSO4, 2 g/L casamino acids) [20]. For solid media, 15 g/L Bacto-agar (Oxoid, Basinstoke, UK) was added.
Bacteria were inoculated from an agar plate for overnight culture, which was used for inoculation of a new culture to an OD600 of 0.1–0.2. For this purpose, bacteria were harvested by centrifugation to avoid carry-over from the overnight culture. Subsequently, the cultures were incubated at 28 °C until the exponential growth phase was reached (OD600 approx. 0.5) [20] and further processed as described below.
2.2. Isolation and Filtration of Extracellular Proteins
The extraction of extracellular proteins was carried out as described for Corynebacterium glutamicum [22]. In short, mid-exponential phase cultures were centrifuged (25 min, 5500× g, 4 °C), the supernatant was collected and centrifuged again (1 h, 7000× g, 4 °C). To avoid proteolytic degradation, COMPLETE EDTA-free protease inhibitor cocktail was added as recommended by the supplier (Roche, Mannheim, Germany). Proteins were precipitated from the second supernatant by addition of trichloroacetic acid (10% w/v final concentration) and incubation at 4 °C (up to 3 days). Precipitated proteins were harvested by centrifugation (1 h, 8000× g, 4 °C), the pellet were washed first with 80% and subsequently with 100% acetone, dried on ice and resuspended in dehydration buffer (8 M urea, 50 mM Tris, 20 mM DTT, 1% sodium desoxycholate).
To improve the separation of cytoplasmic and extracellular proteins and to prevent contamination by cells and cell debris, the supernatant was filtered before precipitation using non-pyrogenic, 0.22 µm filters mounted on syringes (Sterile R, Sarstedt, Nümbrecht, Germany).
2.3. In-Solution Tryptic Digest
About 1.5 µg soluble proteins secreted in XMM and M9 medium were transferred to 10 KDa Hydrosart® membrane filters (Sartorius Stedim biotech, Göttingen, Germany) for modified filter-aided sample preparation (FASP) [23]. Flow-through was discarded by spinning the tubes at 15,000× g for 20 min and proteins were reduced with 250 µL of reduction buffer containing 8 M urea, 100 mM triethylammonium bicarbonate buffer (TEAB, Sigma-Aldrich, Taufkirchen, Germany) and 20 mM dithiothreitol (DTT) for 30 min at room temperature. Alkylation of sulfhydryl groups was carried out in the same buffer containing 40 mM chloroacetamide instead of DTT for an additional 30 min in the dark. Proteins were then washed with 250 µL of 1 M urea in 50 mM TEAB and trypsinized with 1 µg of sequencing grade trypsin (Promega, Mannheim, Germany) in 1 M urea in 50 mM TEAB for 18 h at 37 °C. Peptides were extracted through centrifugation and desalted on C18 stage tips (5 min, 500× g). Prior to LC-MS/MS analysis, tryptic peptides were vacuum dried and resuspended in 0.1% trifluoroacetic acid (TFA).
2.4. NanoLC-MS/MS Analysis
Tryptic peptides of a 5 µL aliquot from each sample were analyzed by Orbitrap Fusion™ Tribrid™ mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Samples were loaded (300 µm i.d. × 5 mm, 5 μm, 100 Å) and separated (75µm i.d. × 50 cm, 3 µm, 100 Å) on PepMap100 columns using an UltiMate 3000 RSLC nano UHPLC system (Dionex, Sunnyvale, CA) with a gradient of 3%–35% system B (acetonitrile in 0.1% formic acid, system A: 1% acetonitrile in 0.05% formic acid/water) in 160 min at a flow rate of 300 nL/min at 35 °C. Eluted peptides were ionized with nanospray Flex™ ion source (Thermo Fisher Scientific) using nano-bore emitters and measured with a survey scan range of 300–2000 m/z. Most intense peptides of each scan cycle were selected and fragmented using collision-induced dissociation (normalized collision energy 35%, activation time 250 ms, isolation width 1.6 m/z). Resulting MS/MS scans were analyzed in ion trap. Dynamic exclusion was enabled for 60 sec and excluded after 1 time. Raw data files were processed against C. michiganensis subsp. michiganensis NCPPB382 database (Proteome Id: UP000001564) comprised in UniProt [24] using Peaks v7.5 (released on 16 June, 2015, Bioinformatics Solutions Inc., Waterloo, ON, Canada). Theoretical masses for peptides were generated by trypsin with a maximum of two missed cleavages and their product ions were compared to the measured spectra with following parameters: carbamidomethyl modification was set as fixed and oxidation of methionine residues and carbamidomethylated lysines were set as an optional modification. Mass tolerance was set to 10 ppm for survey scans and 0.5 Da for fragment mass measurements. Peptide charges of 2–7 were allowed. Only resulting peptides with false discovery rate (FDR) below 0.01 were regarded as identified [25,26].
Label free quantification was performed with Peaks Q (Bioinformatics Solutions Inc.) out of six samples in two groups with a FDR threshold of 1%. Samples were not normalized and proteins considered for calculation had at least two unique peptides. Proteins were only regarded as identified, when these were found in two out of three samples.
2.5. Analysis of Functional Categories and Localization of Proteins
Classification of proteins in functional categories was based on the lists provided by Flügel and co-workers [18]. Analysis of protein localization was carried out by two approaches, (i) manually based on the protein annotation; and (ii) automatically using the proteome discoverer version 1.14 software package (Thermo Fisher Scientific, Bremen, Germany), SignalP 4.1 and the SecretomeP 2.0 server [27,28].
3. Results and Discussion
The functions of the genes of C. michiganensis subsp. michiganensis were predicted before and the genes were dedicated to specific COG groups by Flügel and coworkers, who introduced a fifth COG-related group designated as “potentially relevant in the phytopathogenic interaction” [18]. Most of the predicted gene products fulfil their functions in processes like metabolism, cellular processes and information and storage. Some proteins, like proteases and hydrolytic enzymes, seem to be important for pathogenicity. First global experiments to identify genes and proteins, which are important for pathogenicity, were carried out before. In these studies, C. michiganensis subsp. michiganensis was grown in planta or in vitro in a minimal medium with 5% tomato homogenate, 10% xylem sap or in a xylem surrogate medium [18,19,20]. In the study presented here, culture supernatants of cells grown in M9 and xylem mimicking medium [20] were studied in a proteomic approach.
3.1. Analysis of C. michiganensis Subsp. michiganensis M9 and XMM Culture Supernatants
In this study, the extracellular proteome of C. michiganensis subsp. michiganensis was analyzed after growth in minimal medium and xylem surrogate medium. In total, 1872 proteins were identified in M9 and 1766 proteins in XMM (for a complete list, see supplementary material Table S1). Since the C. michiganensis subsp. michiganensis genome encodes only 3107 annotated genes [29], this high number can only be explained when cells lysis during growth and release proteins into the medium is considered.
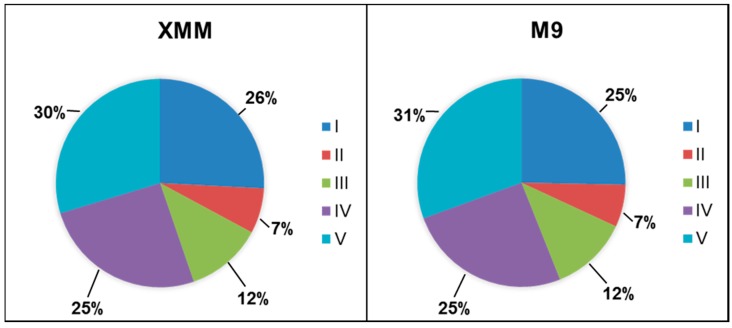
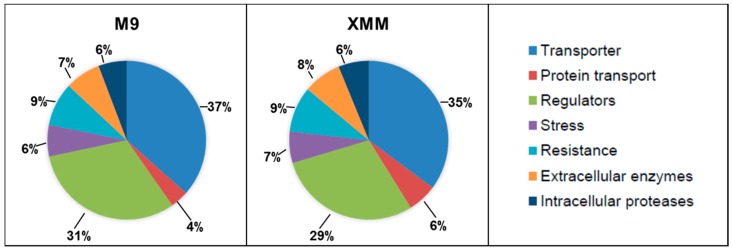
While 203 proteins were exclusively detected in M9 and 97 proteins in XMM, 1669 proteins were detected under both growth conditions, indicating a rather small difference in protein expression and the use of rather similar protein sets in the two media. When these proteins were classified in functional categories according to Flügel and co-workers [18], a very similar distribution was found for M9- and XMM-isolated proteins (Figure 1). Thirty percent of all detected proteins were members of COG V (poorly characterized), followed by about 25% attributed to COG I (metabolism) and COG IV (potentially relevant in the phytopathogenic interaction), respectively, and lower shares of COG III (information storage and processing) and COG II (cellular processes). When COG IV was analyzed in more detail (Figure 2), again the distribution of M9 and XMM-isolated proteins was highly similar with the majority classified as transporters, followed by regulators and smaller shares of proteins involved in protein transport, regulation, stress response, resistance and the groups of extracellular enzymes and intracellular proteases.
Figure 1.
Distribution of functional categories of proteins isolated from M9 and XMM medium. COG I, metabolism; COG II, cellular processes; COG III, information storage and processing; COG IV, potentially relevant in phytopathogenic interaction; COG V, poorly characterized.
Figure 2.
Distribution of COG IV subgroups of proteins isolated from M9 and XMM medium. For explanation of COG IV groups, see the right panel.
This result is consistent with recently carried out transcriptome analyses [20], which also emphasized the role of transport processes for C. michiganensis subsp. michiganensis.
3.2. Analysis of the Putative C. michiganensis Subsp. michiganensis Secretome
The detected proteins were analyzed in respect to their predicted localization according to their annotation (Table 1). Based on their annotation only thirteen proteins had a clear cell wall or surface localization and might be either actively secreted into the medium or have been sheared from the cell surface during incubation in the shaking flasks. These were putative penicillin binding proteins, putative sortases, putative cell surface proteins, a capsular polysaccharide synthesis protein, and a putative NplC/P60 protein.
Table 1.
Predicted functions of C. michiganensis subsp. michiganensis proteins detected in culture supernatants. Expression in the respective media is indicated by + absence by -, proteins identified only after filtration are indicated in bold.
| Identifier | Annotated Function | M9/XMM |
|---|---|---|
| CMM_0013 | putative sortase | +/+ |
| CMM_0017 | putative penicillin binding protein | +/+ |
| CMM_0039 | putative extracellular serine protease | +/+ |
| CMM_0041 | putative extracellular serine protease | +/+ |
| CMM_0053 | putative extracellular serine protease | +/+ |
| CMM_0059 | putative extracellular serine protease | +/+ |
| CMM_0075 | putative extracellular serine protease | +/+ |
| CMM_0090 | β-xylanase | +/+ |
| CMM_0109 | putative sugar ABC transporter substrate-binding protein | +/+ |
| CMM_0129 | putative sortase | +/− |
| CMM_0141 | putative secreted protein | +/− |
| CMM_0166 | putative iron-siderophore ABC transporter substrate-binding protein | +/+ |
| CMM_0289 | conserved secreted/exported protein | +/+ |
| CMM_0296 | putative sugar ABC transporter substrate-binding protein | +/+ |
| CMM_0345 | conserved secreted/exported protein | +/+ |
| CMM_0359 | putative sugar ABC transporter substrate-binding protein | +/+ |
| CMM_0363 | putative iron-siderophore ABC transporter substrate-binding protein | +/+ |
| CMM_0423 | putative sugar ABC transporter substrate-binding protein | +/+ |
| CMM_0430 | putative cell surface protein | +/+ |
| CMM_0431 | putative hemagglutinin/hemolysin-related protein | +/+ |
| CMM_0435 | putative iron-siderophore ABC transporter substrate-binding protein | +/+ |
| CMM_0613 | levanase | +/+ |
| CMM_0667 | secreted phosphoesterase | +/+ |
| CMM_0706 | putative penicillin binding protein | +/− |
| CMM_0792 | putative sugar ABC transporter substrate-binding protein | +/+ |
| CMM_0795 | putative extracellular nuclease/phosphatase | +/+ |
| CMM_0799 | putative peptide ABC transporter substrate-binding protein | +/+ |
| CMM_0819 | putative cell surface protein | +/+ |
| CMM_0825 | putative cell surface protein | +/+ |
| CMM_0827 | capsular polysaccharide synthesis protein | +/+ |
| CMM_0840 | putative NplC/P60 protein | +/+ |
| CMM_0866 | putative α-glucoside ABC transporter substrate-binding protein | +/+ |
| CMM_0879 | putative sugar ABC transporter substrate-binding protein | +/+ |
| CMM_0915 | putative penicillin binding protein | +/+ |
| CMM_0919 | putative penicillin binding protein | +/+ |
| CMM_0944 | putative sugar ABC transporter substrate-binding protein | +/+ |
| CMM_0975 | putative ABC transporter substrate-binding protein | +/+ |
| CMM_0976 | putative ABC transporter substrate-binding protein | +/+ |
| CMM_1022 | putative secreted protein | +/+ |
| CMM_1031 | putative secreted protein | +/+ |
| CMM_1032 | putative secreted protein | +/+ |
| CMM_1129 | putative siderophore interacting protein | +/− |
| CMM_1243 | putative sugar ABC transporter substrate-binding protein | +/+ |
| CMM_1250 | putative secreted protein | +/+ |
| CMM_1262 | putative sugar ABC transporter substrate-binding protein | +/+ |
| CMM_1304 | conserved secreted protein | −/+ |
| CMM_1314 | putative iron ABC transporter substrate binding protein | +/+ |
| CMM_1389 | conserved secreted/exported proteins | +/+ |
| CMM_1405 | conserved secreted protein | +/+ |
| CMM_1406 | putative secreted protein | +/+ |
| CMM_1411 | putative cell surface protein | +/+ |
| CMM_1450 | putative secreted protein | +/+ |
| CMM_1478 | putative peptide ABC transporter substrate-binding protein | +/+ |
| CMM_1532 | putative proline/glycine/betaine/choline ABC transporter substrate-binding protein | +/+ |
| CMM_1557 | putative secreted protein | +/+ |
| CMM_1673 | β-xylanase | +/+ |
| CMM_1674 | β-xylanase | +/+ |
| CMM_1790 | putative anion ABC transporter substrate-binding protein | +/+ |
| CMM_1865 | putative penicillin binding protein | +/+ |
| CMM_1947 | putative extracellular serine protease | +/+ |
| CMM_1948 | putative extracellular serine protease | +/+ |
| CMM_1960 | putative peptide ABC transporter substrate-binding protein | +/+ |
| CMM_2106 | putative sugar ABC transporter substrate-binding protein | +/+ |
| CMM_2169 | putative RTX toxin | +/+ |
| CMM_2176 | conserved secreted lipoprotein | +/+ |
| CMM_2178 | putative secreted protein | +/+ |
| CMM_2180 | putative peptide ABC transporter substrate-binding protein | +/+ |
| CMM_2185 | putative peptide ABC transporter substrate-binding protein | +/+ |
| CMM_2238 | putative sugar ABC transporter substrate-binding protein | −/+ |
| CMM_2283 | putative metal ABC transporter substrate-binding protein | +/+ |
| CMM_2349 | putative iron-siderophore ABC transporter substrate-binding protein | +/+ |
| CMM_2410 | putative sugar ABC transporter substrate-binding protein | +/+ |
| CMM_2434 | putative extracellular serine protease | +/+ |
| CMM_2438 | putative L-arabinose ABC transporter substrate-binding protein | +/+ |
| CMM_2467 | putative secreted hydrolase | +/+ |
| CMM_2505 | phosphate binding protein PstS | +/+ |
| CMM_2566 | putative branched-chain amino acid ABC transporter substrate-binding protein | +/+ |
| CMM_2628 | putative polar amino acid ABC transporter substrate-binding protein | +/+ |
| CMM_2676 | putative β-lactamase | +/+ |
| CMM_2699 | putative sugar ABC transporter substrate-binding protein | +/+ |
| CMM_2733 | putative sugar ABC transporter substrate-binding protein | +/+ |
| CMM_2842 | putative sugar ABC transporter substrate-binding protein | +/+ |
| CMM_2941 | putative metal ABC transporter substrate-binding protein | +/+ |
| pCM1_0018 | putative secreted protein | +/+ |
| pCM1_0020 | CelA | +/+ |
| pCM1_0023 | putative extracellular serine protease | |
| pCM2_0025 | putative secreted protein | +/+ |
| pCM2_0028 | conserved secreted protein | +/+ |
| pCM2_0053 | putative extracellular serine protease | +/+ |
| pCM2_0054 | Pat-1 | +/+ |
In addition to these, a high number of proteins related to sugar, amino acid and peptide uptake were identified, mainly substrate-binding subunits of ABC transporters, which are located at the outside of the cytoplasmic membrane. Among these, sugar transporter subunits were the most prominent group. Fifteen not further characterized putative sugar ABC transporter substrate-binding proteins were detected in addition to a putative l-arabinose ABC transporter substrate-binding protein and a putative α-glucoside ABC transporter substrate-binding protein.
Slightly less abundant were amino acid and peptide uptake components with a putative polar amino acid ABC transporter substrate-binding protein, a putative branched-chain amino acid ABC transporter substrate-binding protein, a putative proline/glycine/betaine/choline ABC transporter substrate-binding protein, and five putative peptide ABC transporter substrate-binding proteins.
The third major group of proteins were related to the uptake of iron and other ions; namely, a putative siderophore interacting protein, four putative iron-siderophore ABC transporter substrate-binding proteins, a putative iron ABC transporter substrate binding protein, a putative anion ABC transporter substrate-binding protein, two putative metal ABC transporter substrate-binding proteins, and the phosphate binding protein PstS.
Additionally, two putative ABC transporter substrate-binding protein without further annotation were found.
Twenty-three proteins were annotated to be secreted. Nine putative secreted proteins, three conserved secreted proteins, nine putative extracellular serine proteases, a conserved secreted lipoprotein, a putative secreted hydrolase. Furthermore, five polymer degrading enzymes were found; namely a levanase, three β-xylanases, and a putative extracellular nuclease/phosphatase, in addition to two proteins, which might be involved in host interaction, a putative RTX toxin and a putative hemagglutinin/hemolysin-related protein.
From these protein, the putative siderophore interacting protein CMM_1129, the putative penicillin binding protein CMM_0706, the putative sortase CMM_0129, and the putative secreted protein CMM_0141 were exclusively found in M9 supernatants, while the putative sugar ABC transporter substrate binding protein CMM_2238 and the conserved secreted protein CMM_1304 were only detected after XMM incubation.
In summary, C. michiganensis subsp. michiganensis grown in either standard M9 minimal medium or xylem-mimicking medium XMM secrete a cocktail of proteases and other polymer degrading enzymes, while a wide variety of transporters is expressed in parallel to take up the degradation products.
3.3. Effect of Filtration on the Detectable Secretome
As described before, application of 0.22 µm filters might improve enrichment of secreted protein. In case of xylem sap filtration, the number of proteins was reduced from 1013 to 165 after introduction of a filtration step [19]. Unexpectedly, the effect was less drastic in this study. In total 1276 proteins were detected in the M9 culture filtrate and 976 proteins in the XMM culture filtrate, which results in a reduction of detectable proteins by only one third, while Savidor and co-workers found a massive reduction of 84% [19]. Nevertheless, also in this study an enrichment of secreted proteins was observed even by simple inspection of protein lists. In the filtrate—but not in the unfiltered samples—the following secreted proteins were found: three putative secreted proteins, three conserved secreted/exported proteins, a secreted phosphoesterase, a putative β-lactamase, and a putative extracellular serine protease. Moreover, the previously described virulence factors Pat-1 and CelA were identified now both, in M9 and XMM supernatants (Table 1). The expression of these virulence factors in minimal media was already observed in previous studies [18,20,21], indicating that plant contact is not crucial for an induction of virulence genes.
In parallel to the data analysis described above, bioinformatics analyses were carried out. More than 76% of the proteins identified in M9 and XMM filtrates had no annotation, and only about 0.4% were identified as extracellular or cell surface proteins by the proteome discoverer. Further analyses of proteins differentially expressed proteins in M9 and XMM medium using Peaks 7.5 (Bioinformatics Solution Inc., Canada), SignalP and Secretome 2.0 revealed 4 proteins with classical signal sequence and 10 non-classically secreted proteins (Table 2). Half of these proteins lacked an annotated function. However, Pat-1 and a putative extracellular serine protease, which might also contribute to virulence were more abundant in XMM.
Table 2.
Differentially expressed proteins in filtered culture supernatants.
| Identifier | Annotated Function | Upregulated in |
|---|---|---|
| Proteins with classical signal peptide | ||
| CMM_0166 | putative Fe3+ siderophore | M9 |
| pCM2_0042 | uncharacterized protein | XMM |
| pCM2_0054 | Pat-1 | XMM |
| CMM_0052 | putative extracellular serine protease | XMM |
| Non-classically secreted proteins | ||
| CMM_1654 | uncharacterized protein | M9 |
| CMM_1841 | putative cytochrome c oxidase | M9 |
| CMM_2064 | uncharacterized protein | M9 |
| CMM_2634 | uncharacterized protein | XMM |
| CMM_1793 | elongation factor P | M9 |
| pCM2_0042 | uncharacterized protein | XMM |
| CMM_0166 | putative Fe3+ siderophore | M9 |
| CMM_0486 | uncharacterized protein | M9 |
| CMM_0517 | putative transcriptional regulator | M9 |
| CMM_0581 | uncharacterized protein | M9 |
4. Conclusions
C. michiganensis subsp. michiganensis is an important tomato pathogen, which is, however, hardly studied. Only 116 entries for the query “Clavibacter michiganensis subsp. michiganensis” were found in a PubMed search end of October 2015. Proteome studies might help to improve the understanding of this pathogen. As shown in this study, bacteria grown in either standard M9 minimal medium or xylem-mimicking medium XMM secrete a cocktail of proteases and other polymer degrading enzymes into the surrounding, while a wide variety of transporters is expressed in parallel to take up the degradation products, especially sugars and amino acids. In planta, this will lead to efficient utilization of xylem sap components, mainly amino acids [20], and host cell polymer degradation products, i.e., amino acids, peptides, pentoses and hexoses. In fact, Savidor and co-workers found different proteases (ChpE, Pat-1, PpaC, PpaH, SbtC), the cellulose CelA, a pectate lyase and a polygalacturonidase in filtered xylem sap of C. michiganensis subsp. michiganensis-infected tomato plants [19].
Supernatants of minimal medium cultures were significantly contaminated with cytoplasmic proteins in this study, despite the fact that carry-over from the inoculum was avoided and exponential growth phase cells were used. In contrast to previous work [19], filtration of supernatants had only a relatively small effect, indicating a considerable amount of cell lysis during growth of C. michiganensis subsp. michiganensis in shaking flasks, which was unexpected due to the rigid Gram-positive cell wall structure of this species. The reason for this observation has to be investigated in future studies.
Nevertheless, filtration of culture supernatants led to an enrichment of secreted proteins and under these conditions, the known major virulence factors Pat-1 and CelA were identified as well. The identification of these and other virulence factors indicate that C. michiganensis subsp. michiganensis expresses virulence determinants not exclusively after host cell contact in planta but already in minimal medium, i.e., in an in vitro situation.
In summary, proteome studies are a valuable tool to understand physiology and adaptations of the C. michiganensis subsp. Michiganensis, however, these studies are hampered by the high number of proteins lacking an annotated function and the overall very limited knowledge about this bacterium. Especially studies on protein export in C. michiganensis subsp. michiganensis would be interesting not only in respect to this study but also due to the fact that effector proteins might be secreted into the host to manipulate plant metabolism and pathogen defense.
Acknowledgments
C. michiganensis subsp. michiganensis was kindly provided by R. Eichenlaub (Bielefeld). The project was supported by the Deutsche Forschungsgemeinschaft in frame of SFB796.
Supplementary Materials
Author Contributions
Isolation of proteins was carried out by EH and TM, AP, BA and JH were involved in sample preparation and mass spectrometry, AB was responsible for supervision of EH and TM and wrote the draft of the manuscript, which was finalized with the help of all co-authors.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Gonzalez A.J., Trapiello E. Clavibacter michiganensis subsp. phaseoli subsp. nov., pathogenic in bean. Int. J. Syst. Evol. Microbiol. 2014;64:1752–1755. doi: 10.1099/ijs.0.058099-0. [DOI] [PubMed] [Google Scholar]
- 2.Strider D.L. Bacterial Canker of Tomato Caused by Corynebacterium Michiganense: A Literature Review and Bibliography. North Carolina Agricultural Experiment Station; Releigh, NC, USA: 1969. [Google Scholar]
- 3.Smith E.F. An Introduction of Bacterial Diseases of Plants. Saunders; Philadelphia, PA, USA: 1920. [Google Scholar]
- 4.Carlton W.M., Braun E.J., Gleason M.L. Ingress of Clavibacter michiganensis subsp. michiganensis into tomato leaves through hydathodes. Phytopathology. 1998;88:525–529. doi: 10.1094/PHYTO.1998.88.6.525. [DOI] [PubMed] [Google Scholar]
- 5.Dreier J., Meletzus D., Eichenlaub R. Characterization of the plasmid encoded virulence region pat-1 of phytopathogenic clavibacter michiganensis subsp. michiganensis. Mol. Plant Microbe Interact. 1997;10:195–206. doi: 10.1094/MPMI.1997.10.2.195. [DOI] [PubMed] [Google Scholar]
- 6.Tancos M.A., Chalupowicz L., Barash I., Manulis-Sasson S., Smart C.D. Tomato fruit and seed colonization by Clavibacter michiganensis subsp. michiganensis through external and internal routes. Appl. Environ. Microbiol. 2013;79:6948–6957. doi: 10.1128/AEM.02495-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hausbeck M.K., Bell J., Medina-Mora C., Podolsky R., Fulbright D.W. Effect of bactericides on population sizes and spread of Clavibacter michiganensis subsp. michiganensis on tomatoes in the greenhouse and on disease development and crop yield in the field. Phytopathology. 2000;90:38–44. doi: 10.1094/PHYTO.2000.90.1.38. [DOI] [PubMed] [Google Scholar]
- 8.Fatmi M.S.N.W. Semiselective agar medium for isolation of Clavibacter michiganense subsp. michiganense from tomato seed. Phytopathology. 1988;78:121–126. doi: 10.1094/Phyto-78-121. [DOI] [Google Scholar]
- 9.Gitaitis R.D., Beaver R.W., Voloudakis A.E. Detection of Clavibacter michiganensis subsp. michiganensis in symptomless tomato transplants. Plant Dis. 1991;74:834–838. doi: 10.1094/PD-75-0834. [DOI] [Google Scholar]
- 10.Jahr H., Dreier J., Meletzus D., Bahro R., Eichenlaub R. The endo-β-1,4-glucanase CelA of Clavibacter michiganensis subsp. michiganensis is a pathogenicity determinant required for induction of bacterial wilt of tomato. Mol. Plant Microbe Interact. 2000;13:703–714. doi: 10.1094/MPMI.2000.13.7.703. [DOI] [PubMed] [Google Scholar]
- 11.Kaup O., Gräfen I., Zellermann E.M., Eichenlaub R., Gartemann K.H. Identification of a tomatinase in the tomato-pathogenic actinomycete Clavibacter michiganensis subsp. michiganensis NCPPB382. Mol. Plant Microbe Interact. 2005;18:1090–1098. doi: 10.1094/MPMI-18-1090. [DOI] [PubMed] [Google Scholar]
- 12.Gartemann K.H., Abt B., Bekel T., Burger A., Engemann J., Flügel M., Gaigalat L., Goesmann A., Gräfen I., Kalinowski J., et al. The genome sequence of the tomato-pathogenic actinomycete Clavibacter michiganensis subsp. michiganensis NCPPB382 reveals a large island involved in pathogenicity. J. Bacteriol. 2008;190:2138–2149. doi: 10.1128/JB.01595-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stork I., Gartemann K.H., Burger A., Eichenlaub R. A family of serine proteases of Clavibacter michiganensis subsp. michiganensis: ChpC plays a role in colonization of the host plant tomato. Mol. Plant Pathol. 2008;9:599–608. doi: 10.1111/j.1364-3703.2008.00484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eichenlaub R., Gartemann K.H. The Clavibacter michiganensis subspecies: Molecular investigation of gram-positive bacterial plant pathogens. Annu. Rev. Phytopathol. 2011;49:445–464. doi: 10.1146/annurev-phyto-072910-095258. [DOI] [PubMed] [Google Scholar]
- 15.Savidor A., Chalupowicz L., Teper D., Gartemann K.H., Eichenlaub R., Manulis-Sasson S., Barash I., Sessa G. Clavibacter michiganensis subsp. michiganensis Vatr1 and Vatr2 transcriptional regulators are required for virulence in tomato. Mol. Plant Microbe Interact. 2014;27:1035–1047. doi: 10.1094/MPMI-02-14-0061-R. [DOI] [PubMed] [Google Scholar]
- 16.Bentley S.D., Corton C., Brown S.E., Barron A., Clark L., Doggett J., Harris B., Ormond D., Quail M.A., May G., et al. Genome of the actinomycete plant pathogen Clavibacter michiganensis subsp. sepedonicus suggests recent niche adaptation. J. Bacteriol. 2008;190:2150–2160. doi: 10.1128/JB.01598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monteiro-Vitorello C.B., Camargo L.E., van Sluys M.A., Kitajima J.P., Truffi D., do Amaral A.M., Harakava R., de Oliveira J.C., Wood D., de Oliveira M.C., et al. The genome sequence of the gram-positive sugarcane pathogen Leifsonia xyli subsp. xyli. Mol. Plant Microbe Interact. 2004;17:827–836. doi: 10.1094/MPMI.2004.17.8.827. [DOI] [PubMed] [Google Scholar]
- 18.Flügel M., Becker A., Gartemann K.H., Eichenlaub R. Analysis of the interaction of Clavibacter michiganensis subsp. michiganensis with its host plant tomato by genome-wide expression profiling. J. Biotechnol. 2012;160:42–54. doi: 10.1016/j.jbiotec.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 19.Savidor A., Teper D., Gartemann K.H., Eichenlaub R., Chalupowicz L., Manulis-Sasson S., Barash I., Tews H., Mayer K., Giannone R.J., et al. The Clavibacter michiganensis subsp. michiganensis-tomato interactome reveals the perception of pathogen by the host and suggests mechanisms of infection. J. Proteome Res. 2012;11:736–750. doi: 10.1021/pr200646a. [DOI] [PubMed] [Google Scholar]
- 20.Hiery E., Adam S., Reid S., Hofmann J., Sonnewald S., Burkovski A. Genome-wide transcriptome analysis of clavibacter michiganensis subsp. michiganensis grown in xylem mimicking medium. J. Biotechnol. 2013;168:348–354. doi: 10.1016/j.jbiotec.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 21.Chalupowicz L., Cohen-Kandli M., Dror O., Eichenlaub R., Gartemann K.H., Sessa G., Barash I., Manulis-Sasson S. Sequential expression of bacterial virulence and plant defense genes during infection of tomato with Clavibacter michiganensis subsp. michiganensis. Phytopathology. 2010;100:252–261. doi: 10.1094/PHYTO-100-3-0252. [DOI] [PubMed] [Google Scholar]
- 22.Hansmeier N., Chao T.C., Pühler A., Tauch A., Kalinowski J. The cytosolic, cell surface and extracellular proteomes of the biotechnologically important soil bacterium Corynebacterium efficiens YS-314 in comparison to those of Corynebacterium glutamicum ATCC 13032. Proteomics. 2006;6:233–250. doi: 10.1002/pmic.200500144. [DOI] [PubMed] [Google Scholar]
- 23.Wiśniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 24.Proteomes–Clavibacter michiganensis subsp. michiganensis (strain NCPPB 382) [(accessed on 15 April 2015)]. Available online: http://www.uniprot.org/proteomes/UP000001564.
- 25.Amin B., Maurer A., Voelter W., Melms A., Kalbacher H. New poteintial serum biomarkers in multiple sclerosis identified by proteomic strategies. Curr. Med. Chem. 2014;21:1544–1556. doi: 10.2174/09298673113206660311. [DOI] [PubMed] [Google Scholar]
- 26.Maurer A., Zeyher C., Amin B., Kalbacher H. A periodate-cleavable linker for functional proteomics under slightly acidic conditions: Application for the analysis of intracellular aspartic proteases. J. Proteome Res. 2012;12:199–207. doi: 10.1021/pr300758c. [DOI] [PubMed] [Google Scholar]
- 27.SignalP 4.1 Server. [(accessed on 27 October 2015)]. Available online: http://www.cbs.dtu.dk/services/SignalP/
- 28.SecretomeP 2.0 Server. [(accessed on 27 October 2015)]. Available online: http://www.cbs.dtu.dk/services/SecretomeP/
- 29.Comprehensive Microbial Resource. [(accessed on 17 July 2015)]. Available online: http://cmr.jcvi.org/tigr-scripts/CMR/CmrHomePage.cgi.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.