Abstract
Background
Bloodstream infections due to bacterial pathogens are a major cause of morbidity and mortality in Bangladesh and other developing countries. In these countries, most patients are treated empirically based on their clinical symptoms. Therefore, up to date etiological data for major pathogens causing bloodstream infections may play a positive role in better healthcare management. The aim of this study was to identify the bacterial pathogens causing major bloodstream infections in Dhaka, Bangladesh and determine their antibiotic susceptibility pattern.
Methods
From January 2005 to December 2014, a total of 103,679 single bottle blood samples were collected from both hospitalized and domiciliary patients attending Dhaka hospital, icddrb, Bangladesh All the blood samples were processed for culture using a BACT/Alert blood culture machine. Further identification of bacterial pathogens and their antimicrobial susceptibility test were performed using standard microbiological procedures.
Results
Overall, 13.6% of the cultured blood samples were positive and Gram-negative (72.1%) bacteria were predominant throughout the study period. Salmonella Typhi was the most frequently isolated organism (36.9% of samples) in this study and a high percentage of those strains were multidrug-resistant (MDR). However, a decreasing trend in the S. Typhi isolation rate was observed and, noticeably, the percentage of MDR S. Typhi isolated declined sharply over the study period. An overall increase in the presence of Gram-positive bacteria was observed, but most significantly we observed the percentage of MDR Gram-positive bacteria to double over the study period. Overall, Gram positive bacteria were more resistant to most of the commonly used antibiotics than Gram-negative bacteria, but the MDR level was high in both groups.
Conclusions
This study identified the major bacterial pathogens involved with BSI in Dhaka, Bangladesh and also revealed their antibiotic susceptibility patterns. We expect our findings to help healthcare professionals to make informed decisions and provide better care for their patients. Also, we hope this study will assist researchers and policy makers to prioritize their research options to face the future challenges of infectious diseases.
Electronic supplementary material
The online version of this article (doi:10.1186/s13756-016-0162-z) contains supplementary material, which is available to authorized users.
Keywords: Bloodstream infection, BSI, Antimicrobial resistance, Multidrug-resistance (MDR), Epidemiology, Gram-positive bacteria, Gram-negative bacteria, Dhaka, Bangladesh
Background
Bloodstream infection (BSI) due to bacterial pathogens is a global concern. It is often associated with increased length of hospital stay, a significant amount of healthcare related costs and most significantly, a high rate of morbidity and mortality [1]. Depending on the age, severity of infection and other risk factors, the mortality rate for BSI varies between 4.0 and 41.5% [2–7]. Recent studies have reported a rapid increase in the number of bloodstream infections from both community and nosocomial sources [8, 9]. Staphylococcus aureus, Streptococcus pneumoniae and Escherichia coli have been found to be the most commonly isolated pathogens associated with BSI worldwide [3, 6].
The epidemiology of BSI varies depending on the geographic location, age and co-morbid illnesses. As an example, Salmonella enterica is a frequently isolated pathogen from blood samples in both African and Asian regions, however their serotypes differ substantially [10]. S. Paratyphi is the predominant organism in the Salmonella group in Africa whereas S. Typhi is the most frequently isolated organism in Asia. Besides their isolation rate, their antibiotic susceptibility pattern varies substantially [10]. So, understanding of local epidemiology may play an important role in making proper empirical treatment choices before laboratory test results are available. This is especially true for Bangladesh and other developing countries where healthcare systems operate on poor hygiene system and lack proper facilities to contain infections. In these countries, early treatment is usually based on the patient’s clinical symptoms rather than diagnostic results. Therefore, patient’s early prognosis to final outcome might be much improved by available epidemiologic data for the most frequently isolated pathogenic organisms. However, complete data on BSI causing organisms from Dhaka, Bangladesh is scarce.
In this study, we aimed to identify the most prevalent bacterial pathogens involved in BSI in hospital and domiciliary patients from Dhaka, Bangladesh. We also determined pathogen antibiotic susceptibility patterns to evaluate the changing trend of antimicrobial susceptibility in this region.
Methods
In this retrospective study, blood samples were obtained from patients attending out-patient and in-patient services at Dhaka hospital, icddrb, Bangladesh which is a primary care hospital with 200 in-patient beds. A total of 103679 blood samples were processed from January 2005 to December 2014. In the consecutive ten years of the study 9600, 9364, 9189, 9523, 9523, 11578, 11179, 10822, 10161 and 12740 samples were received and processed from 2005 to 2014 respectively. All the blood samples were processed for culture using a BACT/Alert blood culture machine to identify the presence of bacterial pathogens. Antimicrobial susceptibility tests were performed on the isolated pathogens using standard microbiological procedures [11].
Bacterial isolation
Collected blood samples were directly inoculated into adult (more than 12 years of age) and pediatric (up to 12 years of age) FAN blood culture bottle. Bottles were incubated in the BACT/Alert machine for up to 5 days. Positive culture samples were directly inoculated onto MacConkey (MC) agar, chocolate agar and blood agar (5% sheep blood) plates. MC plates were then incubated at 35 °C in aerobic condition. Chocolate and blood agar plates were incubated at 35 °C in microaerophilic condition (containing 5% CO2). Bacterial pathogens were identified using standard bacteriological procedures [11]. API identification strips (bioMérieux, France) were used as supportive tests for further identification.
Antimicrobial susceptibility testing
Antimicrobial susceptibility tests were performed by using the disk diffusion method and susceptibility patterns were determined following CLSI guidelines [11–14]. The study was begun following CLSI, 2004 guidelines [14] and later the relevant changes made in 2010, 2012 and 2013 by CLSI were incorporated. Breakpoint changes made in 2010 and 2013 for carbapenems in the case of Enterobacteriaceae [12, 13] were adapted, and also the revised ciprofloxacin breakpoint for Salmonella in 2012 [11]. Antibiotic susceptibility was tested for CN (10 μg), SXT (25 μg), Cip (5 μg), CRO (30 μg), AMP (10 μg), Caz (30 μg), Imp (10 μg), Net (30 μg), Ak (30 μg), CFM (5 μg), Azi (15 μg), Pen G (10 μg), E (15 μg), C (30 μg), Van (30 μg) and Tet (30 μg). All the antibiotic disks were obtained from Oxoid, UK. E. coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as quality control strains.
We were not able to find any standard definition of multidrug-resistance (MDR) and observed that many previous studies have used the definition of MDR as resistance against three or more classes of antibiotics both for Gram-positive [15–17] and Gram-negative [18–21] bacteria. Consequently, the definition of MDR as acquired non-susceptibility against at least three classes of antibiotics was adopted. Salmonella species were tested against six classes of antibiotics; penicillin and cephalosporin (AMP, CRO, CFM), aminoglycosides (CN), fluor/quinolones (Cip, NA), sulfonamides (SXT), macrolides (Azi) and chloramphenicol. Other Gram-negative bacteria were tested against seven classes of antibiotics; penicillin and cephalosporin (AMP, CRO, CFM, Caz), carbapenems (Imp, Mem), aminoglycosides (CN, Net, Ak), fluoro/quinolones (Cip, NA), sulfonamides (SXT), macrolides (Azi) and chloramphenicol. Gram-positive bacteria were tested against seven classes of antibiotics; penicillin and cephalosporin (AMP, Pen G, Oxa, CRO, CFM), carbapenems (Imp), aminoglycosides (CN), fluor/quinolones (Cip), sulfonamides (SXT), macrolides (Azi, E) and others (C, Van, Rif).
Statistical analysis
Blood-borne pathogen trends for isolation and antimicrobial susceptibility over the last ten years was determined using χ2 (chi-square) test for trend in SPSS version 16.0. P value ≤ 0.05 was considered significant. Standard error of the mean (SEM) was calculated for mean isolation rates. Odd ratios for age and sex were calculated to show their association with infection by particular organisms, and SPSS version 16.0 was used for this purpose. For age, an odds ratio greater than 1.0 indicated the association of a pathogen with age group of less than five years old, and an odds ratio of less than 1.0 indicated the association of a pathogen with age group of more than five years old. For sex, an odds ratio of less than 1.0 indicated the association of a pathogen with female patient group and an odds ratio of more than 1.0 indicated an association of that pathogen with male patient group.
Results
From January 2005 to December 2014, a total of 103,679 blood samples were received from both hospitalized and domiciliary patients and among them 14015 samples were found to be culture positive. Over these past ten years 11.9, 12.2, 13.9, 16.5, 16.0, 17.3, 12.7, 12.9, 10.7 and 11.9% of the samples were found to be culture positive (P < 0.001) respectively. The mean culture positive rate was 13.6 ± 0.7%. Table 1 shows the distribution of organisms found throughout this study period. Blood samples were received from patients with age range of 1 day to 115 years and the mean age was 18 years. S. Typhi was the most frequently isolated blood-borne bacterial pathogen in this study, accounting for 36.9% of the total isolates. Other frequently isolated pathogens included coagulase-negative Staphylococcus species (21.5%), Pseudomonas species (12.5%), S. Paratyphi A, B (8.9%) and Acinetobacter species (5.1%).
Table 1.
Bacterial pathogens isolated from blood cultures in Dhaka, Bangladesh from 2005 to 2014
| Organisms | Year | Total (14015) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2005 (1126) | 2006 (1132) | 2007 (1266) | 2008 (1566) | 2009 (1522) | 2010 (2003) | 2011 (1418) | 2012 (1391) | 2013 (1077) | 2014 (1514) | ||
| Salmonella | 566 a (50.3) b | 747 (66.0) | 707 (55.8) | 755 (48.2) | 593 (39.0) | 828 (41.3) | 592 (41.7) | 663 (47.7) | 474 (44.0) | 623 (41.1) | 6548 (46.7) |
| Salmonella Typhi | 457 (40.6) | 595 (52.6) | 571 (45.1) | 600 (38.3) | 483 (31.7) | 643 (32.1) | 451 (31.8) | 527 (37.9) | 355 (33.0) | 509 (33.6) | 5191 (37.0) |
| Salmonella Paratyphi A, B | 85 (7.5) | 133 (11.7) | 127 (10.0) | 146 (9.3) | 102 (6.7) | 179 (8.9) | 135 (9.5) | 129 (9.3) | 107 (9.9) | 110 (7.3) | 1253 (8.9) |
| Non-typhoidal Salmonella species | 24 (2.1) | 19 (1.7) | 9 (0.7) | 9 (0.6) | 8 (0.5) | 6 (0.3) | 6 (0.4) | 7 (0.5) | 12 (1.1) | 4 (0.3) | 104 (0.7) |
| Non-fermenter | 178 (15.8) | 196 (17.3) | 232 (18.3) | 290 (18.5) | 276 (18.1) | 338 (16.9) | 248 (17.5) | 232 (16.7) | 187 (17.4) | 307 (20.3) | 2484 (17.7) |
| Acinetobacter species | 98 (8.7) | 45 (4.0) | 101 (8.0) | 86 (5.5) | 82 (5.4) | 68 (3.4) | 55 (3.9) | 55 (4.0) | 56 (5.2) | 76 (5.0) | 722 (5.2) |
| Pseudomonas species | 80 (7.1) | 151 (13.3) | 131 (10.3) | 204 (13.0) | 194 (12.7) | 270 (13.5) | 193 (13.6) | 177 (12.7) | 131 (12.2) | 231 (15.3) | 1762 (12.6) |
| Gram Negative | 146 (13.0) | 66 (5.8) | 78 (6.2) | 85 (5.4) | 130 (8.5) | 138 (6.9) | 114 (8.0) | 128 (9.2) | 74 (6.9) | 116 (7.7) | 1075 (7.7) |
| Escherichia coli | 35 (3.1) | 29 (2.6) | 34 (2.7) | 46 (2.9) | 51 (3.4) | 64 (3.2) | 48 (3.4) | 45 (3.2) | 39 (3.6) | 36 (2.4) | 427 (3.0) |
| Klebsiella species | 33 (2.9) | 27 (2.4) | 31 (2.4) | 27 (1.7) | 37 (2.4) | 51 (2.5) | 35 (2.5) | 46 (3.3) | 29 (2.7) | 59 (3.9) | 375 (2.7) |
| Enterobacter species | 30 (2.7) | 8 (0.7) | 11 (0.9) | 8 (0.5) | 42 (2.8) | 17 (0.8) | 24 (1.7) | 27 (1.9) | 4 (0.4) | 17 (1.1) | 188 (1.3) |
| Serratia species | 48 (4.3) | 2 (0.2) | 2 (0.2) | 4 (0.3) | 0 (0.0) | 6 (0.3) | 7 (0.5) | 10 (0.7) | 2 (0.2) | 4 (0.3) | 85 (0.6) |
| Gram Positive | 236 (21.0) | 123 (10.9) | 249 (19.7) | 436 (27.8) | 523 (34.4) | 699 (34.9) | 464 (32.7) | 368 (26.5) | 342 (31.8) | 468 (30.9) | 3908 (27.9) |
| Coagulase-negative Staphylococci | 147 (13.1) | 54 (4.8) | 179 (14.1) | 350 (22.2) | 416 (27.3) | 591 (29.5) | 364 (25.7) | 271 (19.5) | 248 (23.0) | 372 (24.6) | 2992 (21.3) |
| Staphylococcus aureus | 14 (1.2) | 10 (0.9) | 15 (1.2) | 23 (1.5) | 26 (1.7) | 30 (1.5) | 25 (1.8) | 23 (1.7) | 23 (2.1) | 30 (2.0) | 219 (1.6) |
| Streptococcus pneumoniae | 49 (4.4) | 32 (2.8) | 23 (1.8) | 23 (1.5) | 25 (1.6) | 25 (1.2) | 29 (2.0) | 23 (1.7) | 21 (1.9) | 26 (1.7) | 276 (2.0) |
| Streptococcus species | 21 (1.9) | 22 (1.9) | 17 (1.3) | 23 (1.5) | 30 (2.0) | 27 (1.3) | 29 (2.0) | 29 (2.1) | 35 (3.2) | 23 (1.5) | 256 (1.8) |
| Enterococcus faecalis | 5 (0.4) | 5 (0.4) | 15 (1.2) | 17 (1.1) | 26 (1.7) | 26 (1.3) | 17 (1.2) | 22 (1.6) | 15 (1.4) | 17 (1.1) | 165 (1.2) |
aValues in parentheses indicate the number of isolates found in each year; bValues in parentheses indicate the percentage of isolates found in each year
The bolded data indicates the total number and percentage of that group
Pseudomonas species and Acinetobacter species were the two major non-fermenter bacteria isolated between 2005 and 2014. Pseudomonas species showed a sharp increase in isolation rate, from 40.6 to 79.2% (χ2 = 105.1, P < 0.001) of the total non-fermenter bacteria, between 2005 and 2010. However, for the next four years their isolation rate decreased slightly and reached 74.3% in 2014. Acinetobacter species showed a decreasing trend in their isolation rate over this study period, from 49.8 to 24.4% (χ2 = 95.5, P < 0.001). Salmonella species accounted for 46.7% of the total blood-borne pathogens. We observed an overall decreasing trend in their isolation rate from 2005 to 2014 (χ2 = 306.5, P < 0.001). Besides non-fermenter and Salmonella species, other Gram-negative bacteria constituted only 7.7% (χ2 = 72.0, P < 0.001) of the total blood-borne pathogens. E. coli, Enterobacter species and Serratia species had a steady isolation rate over the ten year period, while Klebsiella species showed an increasing trend in their isolation rate, from 22.6 to 50.9% (χ2 = 29.1, P < 0.01) of the total Gram-negative bacterial group (excluding Salmonella species). Gram-positive bacteria accounted for 27.7% of the total blood-borne pathogens. Their isolation rate increased from 20.6 to 30.8% (χ2 = 351.7, P < 0.001) over study period. From 2005 to 2014, Streptococcus pneumoniae showed a decreasing trend in their isolation rate, from 20.8 to 5.6% (χ2 = 158.7, P < 0.001) of the total Gram-positive bacteria.
We observed significant associations between different age groups and infection from specific pathogenic organisms (Additional file 1: Table S7). Pseudomonas species (OR: 0.383, 0.341- 0.430, P < 0.001), S. Paratyphi A, B (0.510, 0.449-0.579, P < 0.001) and Serratia species (0.592, 0.373-0.941, P < 0.05) had significant associations with age group of more than five years old. On the other hand, non-typhoidal Salmonella species (5.082, 3.171-8.146, P < 0.001) and S. pneumoniae (3.827, 2.922-5.012, P < 0.001) had significant association with age group of less than five years old. We also observed the association of sex with infection from different pathogenic bacteria (Additional file 2: Table S8). S. aureus (1.348, 1.019-1.783, P < 0.05) infection was significantly associated with the male patient group, while E. coli (0.705, 0.581-0.855, P < 0.001) was more associated with the female patient group. Seasonal fluctuation was also observed in the incidence rate of a few pathogens (Fig. 1). Acinetobacter species (April-May), Enterobacter species (July), S. aureus (March and November) and S. pneumoniae (March) showed distinct seasonal peaks.
Fig. 1.
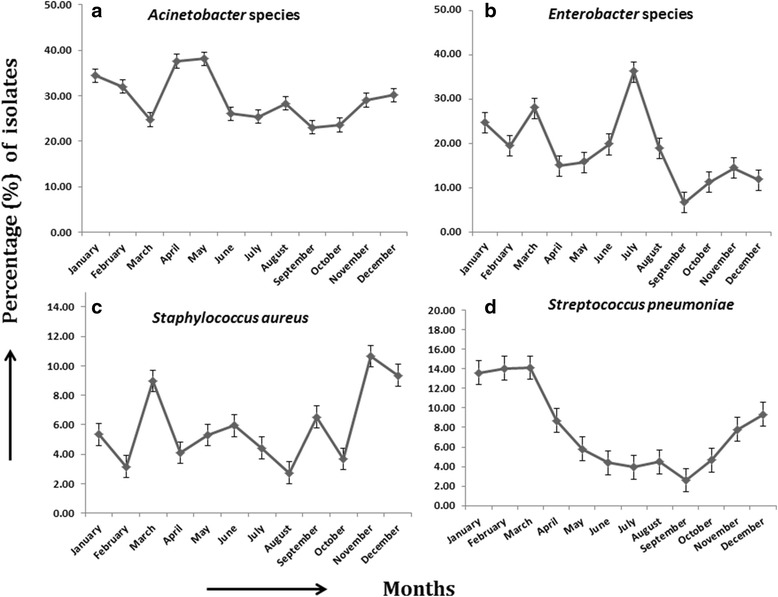
Seasonal variability of BSI causing pathogens- a Acinetobacter species, b Enterobacter species, c Staphylococcus aureus, d Streptococcus pneumoniae
Acinetobacter species showed an increasing trend of resistance against gentamicin (χ2 = 101.6, P < 0.001), ceftriaxone (χ2 = 51.8, P < 0.001) and ciprofloxacin (χ2 = 135.2, P < 0.001) (Additional file 3: Table S1). Pseudomonas species also showed an increasing trend of resistance against gentamicin (χ2 = 127.5, P < 0.001), and ciprofloxacin (χ2 = 141.7, P < 0.001) (Additional file 4: Table S2). Additionally, Acinetobacter species and Pseudomonas species showed an overall 56.2, 47.6, 32.8, 51.4 and 23.0, 17.4, 56.2, 50.8% resistance against ceftazidime, imipenem, netilmicin and amikacin respectively. Table 2 shows the distribution of their MDR strains over this study period.
Table 2.
Percentage of MDR strains isolated from blood cultures in Dhaka, Bangladesh from 2005 to 2014
| Organisms | Year | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2005 | 2006 | 2007 | 2008 | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | |
| Salmonella | 286 a (50.5) b | 383 (51.3) | 308 (43.6) | 269 (35.7) | 189 (31.9) | 219 (26.4) | 131 (22.1) | 109 (16.4) | 87 (18.4) | 120 (19.3) |
| Salmonella Typhi | 282 (61.7) | 379 (63.7) | 307 (53.8) | 268 (44.7) | 188 (38.9) | 219 (34.1) | 130 (28.8) | 109 (20.7) | 87 (24.5) | 120 (23.6) |
| Salmonella Paratyphi A, B | 1 (1.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Non-typhoidal Salmonella species | 3 (12.5) | 4 (21.1) | 1 (11.1) | 1 (12.5) | 1 (12.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Non-Fermenter | 67 (37.6) | 81 (41.3) | 85 (36.6) | 138 (47.6) | 112 (40.6) | 101 (29.9) | 52 (21.0) | 51 (22.0) | 57 (30.5) | 99 (32.2) |
| Acinetobacter species | 24 (24.5) | 17 (37.8) | 25 (24.8) | 37 (43.0) | 42 (51.2) | 35 (51.5) | 23 (41.8) | 30 (54.5) | 34 (60.7) | 50 (65.8) |
| Pseudomonas species | 43 (53.8) | 64 (42.4) | 60 (45.8) | 101 (49.5) | 70 (36.1) | 66 (24.4) | 29 (15.0) | 21 (11.9) | 23 (17.6) | 49 (21.2) |
| Gram-Negative | 53 (36.3) | 40 (60.6) | 45 (57.7) | 47 (55.3) | 79 (60.8) | 98 (71.0) | 57 (50.0) | 76 (59.4) | 53 (71.6) | 86 (74.1) |
| Escherichia coli | 22 (62.9) | 19 (65.5) | 19 (55.9) | 23 (50.0) | 37 (72.5) | 38 (59.4) | 25 (52.1) | 31 (68.9) | 27 (69.2) | 26 (72.2) |
| Klebsiella species | 24 (72.7) | 18 (66.7) | 16 (51.6) | 20 (74.1) | 21 (56.8) | 43 (84.3) | 19 (54.3) | 24 (52.2) | 22 (75.9) | 48 (81.4) |
| Enterobacter species | 6 (20.0) | 3 (37.5) | 8 (72.7) | 1 (12.5) | 21 (50.0) | 12 (70.6) | 9 (37.5) | 12 (44.4) | 3 (75.0) | 12 (70.6) |
| Serratia species | 1 (2.1) | 0 (0.0) | 2 (100) | 3 (75.0) | - | 5 (84.3) | 4 (57.1) | 9 (90.0) | 1 (50.0) | 0 (0.0) |
| Gram-Positive | 16 (18.0) | 16 (23.2) | 26 (37.1) | 37 (43.0) | 51 (47.7) | 36 (33.3) | 30 (30.0) | 27 (27.8) | 27 (28.7) | 34 (35.4) |
| Staphylococcus aureus | 4 (28.6) | 2 (20.0) | 5 (33.3) | 16 (69.6) | 15 (57.7) | 16 (53.3) | 14 (56.0) | 12 (52.2) | 10 (43.5) | 19 (63.3) |
| Streptococcus pneumoniae | 3 (6.1) | 3 (9.4) | 3 (13.0) | 1 (4.3) | 4 (16.0) | 4 (16.0) | 2 (6.9) | 3 (13.0) | 6 (28.6) | 4 (15.4) |
| Streptococcus species | 6 (28.6) | 10 (45.5) | 6 (35.3) | 8 (34.8) | 13 (43.3) | 10 (37.0) | 8 (27.6) | 9 (31.0) | 9 (25.7) | 7 (30.4) |
| Enterococcus faecalis | 3 (60) | 1 (20.0) | 12 (80.0) | 12 (70.6) | 19 (73.1) | 6 (23.1) | 6 (35.3) | 3 (13.6) | 2 (13.3) | 4 (23.5) |
aIndicates the number of MDR strain isolated at that particular year; bindicates the percentage of MDR strains of that particular strain
The bolded data indicates the total number and percentage of that group
S. Typhi was found to be consistently sensitive to ceftriaxone and cefixime over this study period; however, a strain of ESBL S. Typhi was reported [22]. At the same time, decreasing trends of resistance against ampicillin (χ2 = 540.4, P < 0.001) and co-trimoxazole (χ2 = 740.3, P < 0.001) were observed. S. Paratyphi A, B was found to be consistently susceptible to ampicillin, co-trimoxazole, ceftriaxone and cefixime over the ten year period. Noticeably, both S. Typhi and S. Paratyphi A, B showed a trend of increasingly reduced susceptibility against ciprofloxacin (χ2 = 24.3 and 66.1 respectively, P < 0.001) throughout this study period (Table 3).
Table 3.
Percentage of antimicrobial resistance in Salmonella Typhi and Salmonella Paratyphi A, B strains isolated from blood cultures
| Salmonella Typhi | Salmonella Paratyphi A, B | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2005 | 2006 | 2007 | 2008 | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | 2005 | 2006 | 2007 | 2008 | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | ||
| (457)a | (595) | (571) | (600) | (483) | (643) | (451) | (527) | (354) | (509) | (85)a | (133) | (127) | (146) | (102) | (179) | (135) | (129) | (107) | (110) | ||
| AMP | 61 | 62 | 54 | 50 | 41 | 41 | 42 | 24 | 28 | 26 | AMP | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 |
| SXT | 61 | 63 | 54 | 44 | 39 | 34 | 29 | 21 | 24 | 24 | SXT | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 |
| CipR | 3 | 4 | 2 | 1 | 0 | 2 | 2 | 1 | 1 | 4 | CipR | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| CipI | 87 | 89 | 88 | 93 | 94 | 94 | 92 | 96 | 92 | 94 | CipI | 91 | 93 | 97 | 99 | 100 | 99 | 98 | 100 | 98 | 99 |
| CRO | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | CRO | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CFM | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | CFM | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
AMP ampicillin, SXT cotrimoxazole, Cip ciprofloxacin, CRO ceftriaxone, CFM cefixime, aValues in parentheses indicate the number of isolates tested each year
E. coli and Enterobacter species showed an increasing trend of resistance against gentamicin (χ2 = 52.44 and statistically insignificant respectively, P < 0.001) and ceftriaxone (χ2 = 52.4 and 16.5 respectively, P < 0.001). A reducing trend of susceptibility against ciprofloxacin (χ2 = 89.9 and 86.8 respectively, P < 0.001) was also observed (Table 4 and Additional file 5: Table S4). Additionally, E. coli showed an overall increase in resistance, from 69.0 to 90.0%, against co-trimoxazole and ampicillin between 2005 and 2014. Klebsiella species showed an increasing trend of resistance against imipenem (χ2 = 79.6, P < 0.001), and a reducing trend of susceptibility against ciprofloxacin (χ2 = 36.1, P < 0.001) (Additional file 6: Table S3). Over the same time period an overall increase in resistance, from 62.0 to 76.0%, was observed against gentamicin and ceftriaxone respectively for Klebsiella species. Additionally, Klebsiella species showed an overall 86.0, 33.0 and 70.0% resistance against cefixime, meropenem and azithromycin respectively from 2010 to 2014.
Table 4.
Percentage of antimicrobial resistance in E. coli strains isolated from blood cultures
| E. coli | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2005 | 2006 | 2007 | 2008 | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | 2010 | 2011 | 2012 | 2013 | 2014 | ||
| (35)a | (29) | (34) | (46) | (51) | (64) | (48) | (45) | (39) | (36) | (64) | (48) | (45) | (39) | (36) | ||
| Amp | 88 | 96 | 71 | 79 | 97 | 96 | 93 | 94 | 92 | 92 | Net | 8 | 7 | 7 | 8 | 14 |
| SXT | 74 | 75 | 76 | 61 | 73 | 70 | 68 | 73 | 56 | 67 | Ak | 7 | 4 | 4 | 8 | 14 |
| CN | 15 | 28 | 24 | 33 | 37 | 36 | 38 | 30 | 44 | 36 | Imp | 3 | 2 | 4 | 8 | 11 |
| CipR | 51 | 48 | 41 | 54 | 63 | 73 | 49 | 71 | 72 | 72 | Caz | 35 | 30 | 38 | 36 | 56 |
| CipI | 0 | 0 | 6 | 2 | 6 | 2 | 2 | 4 | 0 | 0 | CFM | 69 | 70 | 79 | 68 | 78 |
| CRO | 34 | 55 | 32 | 46 | 55 | 63 | 66 | 76 | 69 | 75 | ||||||
AMP ampicillin, SXT cotrimoxazole, CN gentamicin, Cip ciprofloxacin, CRO ceftriaxone, Net netilmicin, Ak amikacin, Imp imipenem, Caz ceftazidime, CFM cefixime, aValues in parentheses indicate the number of isolates tested each year
S. pneumoniae was found to be consistently susceptible to ampicillin and ceftriaxone between 2005 and 2014 (Table 5). Over this period, their resistance to co-trimoxazole reduced (χ2 = 92.5, P < 0.001). However, it increased against penicillin G (χ2 = 195.3, P < 0.001) and erythromycin (χ2 = 293.2, P < 0.001). Also, a trend of increased susceptibility was observed against ciprofloxacin (χ2 = 79.2, P < 0.001). Other Streptococcus species showed an increasing trend of resistance against gentamicin (χ2 = 44.9, P < 0.001) and erythromycin (χ2 = 70.0, P < 0.001), while there was a reducing trend in susceptibility against ciprofloxacin (χ2 = 67.3, P < 0.001) (Additional file 7: Table S5). S. aureus was found to be highly resistant to ampicillin (up to 100%) throughout this study (Table 6). For this species, resistance against erythromycin increased from 36.0% in 2005 to 75.0% in 2014 (P < 0.09) while susceptibility to ciprofloxacin reduced (χ2 = 37.2, P < 0.001). S. aureus remained consistently susceptible to vancomycin for the last five years of the study and during the same time period they showed an overall 42.0% resistance against ceftriaxone. E. faecalis was found consistently susceptible to vancomycin between 2010 and 2014, while this species showed an overall 31.0, 45.0, 60.0, 39.0 and 81.0% resistance against ampicillin, gentamicin (120), ciprofloxacin, penicillin G and co-trimoxazole respectively (Additional file 8: Table S6).
Table 5.
Percentage of antimicrobial resistance in Streptococcus pneumoniae strains isolated from blood cultures
| Streptococcus pneumoniae | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2005 | 2006 | 2007 | 2008 | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | |
| (49)a | (32) | (23) | (23) | (25) | (25) | (29) | (23) | (21) | (26) | |
| Amp | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| SXT | 91 | 90 | 87 | 73 | 76 | 91 | 78 | 70 | 71 | 73 |
| CipR | 6 | 0 | 5 | 0 | 4 | 0 | 8 | 0 | 0 | 0 |
| CipI | 22 | 38 | 24 | 5 | 16 | 0 | 8 | 0 | 9 | 9 |
| CRO | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pen G | 2 | 0 | 0 | 0 | 9 | 8 | 3 | 4 | 5 | 8 |
| E | 4 | 0 | 0 | 0 | 8 | 5 | 24 | 23 | 48 | 31 |
| Azi | - | - | - | - | - | 7 | 39 | 14 | 55 | 31 |
| CFM | - | - | - | - | - | 17 | 0 | 7 | 0 | 8 |
AMP ampicillin, SXT cotrimoxazole, Cip ciprofloxacin, CRO ceftriaxone, Pen G penicillin G, E erythromycin, Azi azithromycin, CFM cefixime. aValues in parentheses indicate the number of isolates tested each year; − indicates absence of data in respective years
Table 6.
Percentage of antimicrobial resistance in Staphylococcus aureus strains isolated from blood cultures
| Staphylococcus aureus | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2005 | 2006 | 2007 | 2008 | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | |
| (14)a | (10) | (15) | (23) | (26) | (30) | (25) | (23) | (23) | (30) | |
| Amp | 100 | 100 | 88 | 90 | 88 | 67 | 100 | 100 | 100 | 71 |
| SXT | 29 | 20 | 20 | 52 | 35 | 33 | 36 | 23 | 17 | 38 |
| C | 7 | 0 | 7 | 4 | 4 | 4 | 0 | 13 | 4 | 17 |
| E | 36 | 20 | 33 | 78 | 81 | 67 | 72 | 73 | 64 | 75 |
| CipR | 25 | 20 | 40 | 65 | 54 | 57 | 64 | 48 | 39 | 67 |
| CipI | 8 | 0 | 0 | 0 | 4 | 3 | 4 | 9 | 13 | 0 |
| CN | 25 | 11 | 20 | 26 | 27 | 33 | 24 | 13 | 22 | 27 |
| CRO | - | - | - | - | - | 43 | 29 | 50 | 41 | 45 |
| Van | - | - | - | - | - | 4 | 0 | 0 | 0 | 0 |
AMP ampicillin, SXT cotrimoxazole, C chloramphenicol, E erythromycin, Cip ciprofloxacin, CN gentamicin, CRO ceftriaxone, Van vancomycin, aValues in parentheses indicate the number of isolates tested each year; − indicates absence of data in respective years
Discussion
In this retrospective study, we aimed to identify the most prevalent pathogenic organisms involved in bloodstream infections (BSI) over a ten year period (2005–14) in patients in Dhaka, Bangladesh. We also aimed to determine the antimicrobial susceptibility of the isolated pathogens against multiple antibiotics to achieve a clear outlook on the changing trend of their antibiotic susceptibility.
S. Typhi, the causative agent of typhoid fever, is a major public health concern in Bangladesh and other developing Asian countries. Several studies from Bangladesh have already identified S. Typhi as a common cause of bloodstream infection in this region [23–25]. We found Salmonella species to be responsible for almost half of the disease burden associated with BSI in Dhaka, Bangladesh and about 80% of these infections were due to S. Typhi. However, we have observed an overall decrease in Salmonella species isolation rate over this study period. This decrease may be attributed to the improved urban water management system and sanitation practices in Dhaka city over the past years.
A significant change in the epidemiology of S. Typhi was observed in early 1990’s as those years experienced a dramatic rise of MDR strains in Dhaka, Bangladesh [26]. By mid-1990s, about half of the S. Typhi strains were MDR; these were resistant against three first line antibiotics - ampicillin, cotrimoxazole and chloramphenicol [26]. Noticeably, after a few years of an initial epidemic period, a decreasing trend in the isolation rate of MDR S. Typhi strains was observed [23]. Our study confirms this trend to continue; from 2005 to 2010, the percentage of MDR S. Typhi strains isolated declined from 61.7 to 23.7% (Fig. 2). As indicated by recent studies from Bangladesh, S. Typhi still shows a high level of resistance against first line antibiotics [23]. However, we have observed a steady decrease in resistance against ampicillin and cotrimoxazole over the ten year study period. Hopefully, if this trend continues, then using the cheaper, first line antibiotics again to treat S. Typhi infections might be a possibility in near future. Ciprofloxacin or ceftriaxone is usually the choice of treatment against MDR Salmonella strains [27]. We found all Salmonella isolates were consistently susceptible to ceftriaxone, but a very high level of reduced susceptibility exists against ciprofloxacin. Studies suggest that the presence of a mutation in the quinolone resistance-determining region (QRDR) of the gyrA gene is responsible for the emergence of this reduced susceptibility [28].
Fig. 2.
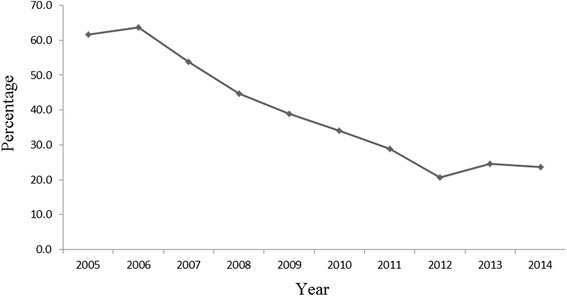
Decrease of multidrug-resistant (MDR) Salmonella Typhi isolation rate from blood cultures
Non-fermenter Gram-negative bacilli are well known for their ability to cause nosocomial infections [29, 30]. We identified Pseudomonas species and Acinetobacter species to be the third (12.6%) and fifth (5.2%) most prevalent organisms associated with BSI in Dhaka, Bangladesh. A gradual decrease in the presence of Acinetobacter species and a significant increase in the presence of Pseudomonas species were observed over the study period. This increase in BSI from Pseudomonas species was possibly from the patients increased access to the health care facilities over these years. Also noticeably, Pseudomonas species was found to be more associated with patients above five years of age; almost 70% of the infections occurring among adults (≥18 years of age). A recent study from Bangladesh also suggested that there is a very low amount of bacteremia cases due to Pseudomonas species among diarrheal children less than five years of age [31].
Acinetobacter species and Pseudomonas species are well known for their high degree of resistance against all classes of antibiotics, and the emergence of MDR strains makes it very difficult to treat them. An aminoglycosides combined with a β-lactamase-stable β-lactam is the typical choice of treatment for Acinetobacter species infection [32], and carbapenems are known to be most effective against their MDR strains [33]. We observed Acinetobacter species to develop increasingly high levels of antibiotic resistance against all aminoglycosides, and noticeably, very high levels of resistance against imipenem (64%). Usually, in the case of resistance against carbapenems, the expert’s choice of treatment for Acinetobacter species infections entails colistin and tigecycline [34]. We found only 3.73% of the MDR Acinetobacter species strains to be resistant against polymyxin B (300 u), therefore it might be considered as a good treatment choice against MDR Acinetobacter species. Colistin and polymyxin B (300 u) are usually considered as the last resort for treatment of infections due to MDR Pseudomonas species [35], but we observed high levels of resistance against them. In contrast, we found imipenem to be consistently effective against all Pseudomonas species so it might be considered as a good choice of treatment.
Besides non-fermenter and Salmonella species, other clinically important Gram-negative bacteria were E. coli, Enterobacter species, Klebsiella species and Serratia species. E. coli has been reported as the leading cause of BSI in developed countries [1]; however, data is relatively unavailable for Bangladesh. We found E. coli to be responsible for only 3.05% of the total BSI cases. It is likely that the presence of E. coli is overshadowed by the overwhelming presence of Salmonella species in Dhaka, Bangladesh, so it would be unwise to ignore them as clinically insignificant.
A recent study from Bangladesh has shown a very high level of antibiotic resistance and MDR level among E. coli isolates from environmental samples [36]. In this study we identified a much higher level of antibiotic resistance and MDR level (36% MDR in environment samples vs. 62.38% MDR in our clinical samples) among BSI causing E. coli strains. We found carbapenems to be the most active antibiotics against E. coli. Overall, 3.2% (including MDR strains) of the E. coli strains showed resistance against them. So, carbapenems may be considered as a good choice of treatment for BSI caused by E. coli. Klebsiella species showed the highest level of resistance against β-lactams, especially penicillins and third generation cephalosporins. Previous studies suggested that carbapenems were highly effective against Klebsiella species until the early years of 2000s. According to Centers for Disease Control and Prevention (CDC) in the USA, Klebsiella species acquired the highest amount of carbapenem resistance (from 1.6 to 10.4%) in the decade from 2001–2011. In accordance with this finding, we observed a very dramatic rise in Klebsiella species resistance against imipenem (0 to 46%) and meropenem (0 to 46.5%) from 2005 to 2014. In fact, we found Klebsiella species to show the highest level of MDR (68%) among all the isolated pathogens. Overall, E. coli, Klebsiella species and Enterobacter species had relatively low prevalence throughout this study period. However, they should be considered important as they had the highest amount of MDR strains and thus the potential to cause serious health threats.
Gram-positive bacteria also pose serious threats as their incidence in BSI is increasing steadily worldwide. The problem is particularly acute in nosocomial settings where methicillin-resistant S. aureus (MRSA) and vancomycin resistant Enterococcus (VRE) are considered serious health threats [37, 38]. In this study, we observed an increasing trend in the isolation rate of Gram-positive bacteria. This trend may be attributed to the patient’s increased access to the health care facilities where BSI from Gram-positive bacteria is common. We identified S. aureus and S. pneumoniae as the two major Gram-positive pathogenic bacteria associated with BSI in Dhaka, Bangladesh. S. pneumoniae reportedly kills about 0.7 to 1 million children under five years of age every year worldwide and 90% of these deaths occur in developing countries [39]. Pneumonia is the primary cause of childhood death in Bangladesh [40] and studies suggest that the incidence of pneumonia among preschool children in Bangladesh is surprisingly higher than in developed countries [41, 42]. We found almost 75% of the BSI cases from S. pneumoniae to occur among children less than five years of age. However, a decreasing trend in their isolation rate was observed throughout this study period.
Increasing levels of antibiotic resistance are a big concern for S. pneumoniae, especially their resistance against macrolides and β-lactams, which are the first choice of treatment for most pneumonia cases. One recent review indicates a very high level of penicillin resistance among S. pneumoniae strains in different Asian countries [43]. However, we observed only a small increase in penicillin resistance in Dhaka, Bangladesh. Another study from Dhaka indicates the presence of high level of resistance against ciprofloxacin. However our study found a much lower level of resistance against ciprofloxacin and also we observed a decreasing trend in their susceptibility pattern [44]. Ampicillin and ceftriaxone were found to be consistently active against S. pneumoniae, so they might be considered as good treatment choices for them. Emergence of MRSA strain marks another very important epidemiological change for the past few decades [45]. Naturally, S. aureus are susceptible to commonly used antibiotics. However methicillin resistance is associated with resistance against a broad range of antibiotics [46]. Over the study period, we found overall 50.2% of S. aureus strains to be MDR. In the first five years of our study, we identified 34.4% of the S. aureus strains to be oxacillin (a synthetic form of methicillin) resistant; however, the percentage was much higher (58.7%) amongst the MDR strains. Vancomycin has been widely used as the preferred choice of treatment for MRSA strains [47], and we also found vancomycin to be highly effective against all S. aureus strains; only 2.1% strains were resistant. Linezolid was also found to be very effective against S. aureus; only one resistant strain was detected over the study period. Therefore, both vancomycin and linezolid might be considered as a good treatment choices against S. aureus which are resistant to commonly used antibiotics.
Limitations of the study
Due to the lack of resources, we were not able to differentiate the samples received from our hospital patients and from domiciliary patients. As a result we could not show the difference in epidemiology between nosocomial and community acquired BSI. Also due to lack of resources, we were not able to collect patient data on the clinical manifestations or any other patient characteristics, other than age and sex, which could be considered as risk factors for BSI. However, we confirm that each patient was diagnosed with signs of bacteremia by their respective physicians. Also, we were not able to perform any molecular tests on received samples due to lack of required resources.
Conclusion
This study clearly identifies the bacterial pathogens involved with bloodstream infections (BSI) occurring in Dhaka, Bangladesh between January 2005 to December 2014. It also reveals their antibiotic susceptibility patterns for commonly used antibiotics. We expect our findings to help healthcare professionals to make informed decisions and provide better care for their patients. Also, we hope this study to help researchers and policy makers to prioritize their research options to face future challenges of infectious diseases both at home and abroad.
Acknowledgement
This research study was funded by core donors which provided unrestricted support to icddr,b for its operations and research. Current donors providing unrestricted support include: Government of the People’s Republic of Bangladesh; Global Affairs Canada (GAC); Swedish International Development Cooperation Agency (Sida) and the Department for International Development (UK Aid). We gratefully acknowledge these donors for their support and commitment to icddr,b’s research efforts. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We also thank Dr. Daniel Leung, from University of Utah, for his valuable comments on draft versions of this manuscript.
Funding
This research study was funded by core donors which provide unrestricted support to icddr,b for its operations and research. But the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The raw data can be made available to the interested researchers by the authors of this article if requested.
Authors’ contributions
DA designed the study and reviewed the manuscript for important intellectual content. MAN analyzed the data and wrote the manuscript. ABS took part in data analysis and reviewed the manuscript. FH, NA, TS, MSR performed experimental works. MSBE was responsible for collecting the data. MMR critically revised the manuscript. All the authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The study protocol was approved by the Ethical Review Committee of the icddr,b (PR-14042). Patient consent for sample handling was not required as samples were subject of routine management and samples were further anonymised for research use.
Abbreviations
- FAN
Fastidious antibiotic neutralization
- API
Analytical profile index
Additional files
Association of two age groups with distinct bacterial pathogens causing BSI in Dhaka, Bangladesh. (DOC 38 kb)
Association of sex with distinct bacterial pathogens causing BSI in Dhaka, Bangladesh. (DOC 40 kb)
Percentage of antimicrobial resistance in Acinetobacter species strains isolated from blood cultures. (DOC 35 kb)
Percentage of antimicrobial resistance in Pseudomonas species strains isolated from blood cultures. (DOC 35 kb)
Percentage of antimicrobial resistance in Enterobacter species strains isolated from blood samples. (DOC 33 kb)
Percentage of antimicrobial resistance in Klebsiella species strains isolated from blood cultures. (DOC 38 kb)
Percentage of antimicrobial resistance in Streptococcus species strains isolated from blood cultures. (DOC 36 kb)
Percentage of antimicrobial resistance in Enterococcus faecalis strains isolated from blood cultures. (DOC 36 kb)
Contributor Information
Dilruba Ahmed, Phone: +880-2-9827001-10, Email: dahmed@icddrb.org.
Md Ausrafuggaman Nahid, Email: a.nahid@icddrb.org.
Abdullah Bashar Sami, Email: sami_khangb@yahoo.com.
Farhana Halim, Email: farhana_mb@gmail.com.
Nasrin Akter, Email: nakter@icddrb.org.
Tuhin Sadique, Email: tuhin.sadique@icddrb.org.
Md Sohel Rana, Email: sohelrana.icddrb@gmail.com.
Md Shahriar Bin Elahi, Email: sbelahi@icddrb.org.
Md Mahbubur Rahman, Email: mahbubur@icddrb.org.
References
- 1.Tumbarello M, et al. Costs of bloodstream infections caused by Escherichia coli and influence of extended-spectrum-beta-lactamase production and inadequate initial antibiotic therapy. Antimicrob Agents Chemother. 2010;54(10):4085–4091. doi: 10.1128/AAC.00143-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanoksil M, et al. Epidemiology, microbiology and mortality associated with community-acquired bacteremia in northeast Thailand: a multicenter surveillance study. PLoS One. 2013;8(1):e54714. doi: 10.1371/journal.pone.0054714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kollef MH, et al. Epidemiology, microbiology and outcomes of healthcare-associated and community-acquired bacteremia: a multicenter cohort study. J Infect. 2011;62(2):130–135. doi: 10.1016/j.jinf.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 4.Reddy EA, Shaw AV, Crump JA. Community-acquired bloodstream infections in Africa: a systematic review and meta-analysis. Lancet Infect Dis. 2010;10(6):417–432. doi: 10.1016/S1473-3099(10)70072-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sogaard M, et al. Temporal changes in the incidence and 30-day mortality associated with bacteremia in hospitalized patients from 1992 through 2006: a population-based cohort study. Clin Infect Dis. 2011;52(1):61–69. doi: 10.1093/cid/ciq069. [DOI] [PubMed] [Google Scholar]
- 6.Valles J, et al. Evolution over a 15-year period of clinical characteristics and outcomes of critically ill patients with community-acquired bacteremia. Crit Care Med. 2013;41(1):76–83. doi: 10.1097/CCM.0b013e3182676698. [DOI] [PubMed] [Google Scholar]
- 7.Valles J, et al. Community-acquired bloodstream infection in critically ill adult patients: impact of shock and inappropriate antibiotic therapy on survival. Chest. 2003;123(5):1615–1624. doi: 10.1378/chest.123.5.1615. [DOI] [PubMed] [Google Scholar]
- 8.Friedman ND, et al. Health care--associated bloodstream infections in adults: a reason to change the accepted definition of community-acquired infections. Ann Intern Med. 2002;137(10):791–797. doi: 10.7326/0003-4819-137-10-200211190-00007. [DOI] [PubMed] [Google Scholar]
- 9.Siegman-Igra Y, et al. Reappraisal of community-acquired bacteremia: a proposal of a new classification for the spectrum of acquisition of bacteremia. Clin Infect Dis. 2002;34(11):1431–1439. doi: 10.1086/339809. [DOI] [PubMed] [Google Scholar]
- 10.Deen J, et al. Community-acquired bacterial bloodstream infections in developing countries in south and southeast Asia: a systematic review. Lancet Infect Dis. 2012;12(6):480–487. doi: 10.1016/S1473-3099(12)70028-2. [DOI] [PubMed] [Google Scholar]
- 11.(CLSI), C.L.S.I . Performance standards for antimicrobial susceptibility testing: twenty second informational supplement ed. Wayne: CLSI; 2012. [Google Scholar]
- 12.(CLSI), C.L.S.I . Performance standards for antimicrobial susceptibility testing: 23rd informational supplement. Wayne: CLSI; 2013. [Google Scholar]
- 13.(CLSI), C.L.S.I . Performance standards for antimicrobial susceptibility testing: 20th informational supplement. Wayne: CLSI; 2010. [Google Scholar]
- 14.(CLSI), C.L.S.I . Performance standards for antimicrobial susceptibility testing: fourteenth informational supplement ed. Wayne: CLSI; 2004. [Google Scholar]
- 15.Pillar CM, et al. Prevalence of multidrug-resistant, methicillin-resistant staphylococcus aureus in the United States: findings of the stratified analysis of the 2004 to 2005 LEADER surveillance programs. Diagn Microbiol Infect Dis. 2008;60(2):221–224. doi: 10.1016/j.diagmicrobio.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 16.Cohen AL, et al. Recommendations for metrics for multidrug-resistant organisms in healthcare settings: SHEA/HICPAC Position paper. Infect Control Hosp Epidemiol. 2008;29(10):901–913. doi: 10.1086/591741. [DOI] [PubMed] [Google Scholar]
- 17.Critchley IA, et al. Activity of daptomycin against susceptible and multidrug-resistant gram-positive pathogens collected in the SECURE study (Europe) during 2000–2001. J Antimicrob Chemother. 2003;51(3):639–649. doi: 10.1093/jac/dkg130. [DOI] [PubMed] [Google Scholar]
- 18.Kallen AJ, et al. Multidrug resistance among gram-negative pathogens that caused healthcare-associated infections reported to the National Healthcare Safety Network, 2006–2008. Infect Control Hosp Epidemiol. 2010;31(5):528–531. doi: 10.1086/652152. [DOI] [PubMed] [Google Scholar]
- 19.O’Fallon E, Gautam S, D’Agata EM. Colonization with multidrug-resistant gram-negative bacteria: prolonged duration and frequent cocolonization. Clin Infect Dis. 2009;48(10):1375–1381. doi: 10.1086/598194. [DOI] [PubMed] [Google Scholar]
- 20.Paterson DL, Doi Y. A step closer to extreme drug resistance (XDR) in gram-negative bacilli. Clin Infect Dis. 2007;45(9):1179–1181. doi: 10.1086/522287. [DOI] [PubMed] [Google Scholar]
- 21.Falagas ME, Koletsi PK, Bliziotis IA. The diversity of definitions of multidrug-resistant (MDR) and pandrug-resistant (PDR) Acinetobacter baumannii and Pseudomonas aeruginosa. J Med Microbiol. 2006;55(Pt 12):1619–1629. doi: 10.1099/jmm.0.46747-0. [DOI] [PubMed] [Google Scholar]
- 22.Ahmed D, et al. Salmonella enterica serovar typhi strain producing extended-spectrum beta-lactamases in Dhaka, Bangladesh. J Med Microbiol. 2012;61(Pt 7):1032–1033. doi: 10.1099/jmm.0.044065-0. [DOI] [PubMed] [Google Scholar]
- 23.Brooks WA, et al. Bacteremic typhoid fever in children in an urban slum, Bangladesh. Emerg Infect Dis. 2005;11(2):326–329. doi: 10.3201/eid1102.040422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naheed A, et al. Burden of typhoid and paratyphoid fever in a densely populated urban community, Dhaka, Bangladesh. Int J Infect Dis. 2010;14(Suppl 3):e93–e99. doi: 10.1016/j.ijid.2009.11.023. [DOI] [PubMed] [Google Scholar]
- 25.Saha SK, et al. Typhoid fever in Bangladesh: implications for vaccination policy. Pediatr Infect Dis J. 2001;20(5):521–524. doi: 10.1097/00006454-200105000-00010. [DOI] [PubMed] [Google Scholar]
- 26.Rahman M, Ahmad A, Shoma S. Decline in epidemic of multidrug resistant Salmonella typhi is not associated with increased incidence of antibiotic-susceptible strain in Bangladesh. Epidemiol Infect. 2002;129(1):29–34. doi: 10.1017/S0950268802007203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Threlfall EJ, Ward LR. Decreased susceptibility to ciprofloxacin in Salmonella enterica serotype typhi, United Kingdom. Emerg Infect Dis. 2001;7(3):448–450. doi: 10.3201/eid0703.017315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hassing RJ, et al. Analysis of mechanisms involved in reduced susceptibility to ciprofloxacin in Salmonella enterica serotypes typhi and paratyphi a isolates from travellers to southeast Asia. Int J Antimicrob Agents. 2011;37(3):240–243. doi: 10.1016/j.ijantimicag.2010.10.026. [DOI] [PubMed] [Google Scholar]
- 29.Cisneros JM, Rodriguez-Bano J. Nosocomial bacteremia due to Acinetobacter baumannii: epidemiology, clinical features and treatment. Clin Microbiol Infect. 2002;8(11):687–693. doi: 10.1046/j.1469-0691.2002.00487.x. [DOI] [PubMed] [Google Scholar]
- 30.Cross A, et al. Nosocomial infections due to Pseudomonas aeruginosa: review of recent trends. Rev Infect Dis. 1983;5(Suppl 5):S837–S845. doi: 10.1093/clinids/5.Supplement_5.S837. [DOI] [PubMed] [Google Scholar]
- 31.Akram F, et al. Prevalence, clinical features, and outcome of pseudomonas bacteremia in under-five diarrheal children in bangladesh. ISRN Microbiol. 2014;2014:469758. doi: 10.1155/2014/469758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brauers J, et al. Activities of various beta-lactams and beta-lactam/beta-lactamase inhibitor combinations against Acinetobacter baumannii and Acinetobacter DNA group 3 strains. Clin Microbiol Infect. 2005;11(1):24–30. doi: 10.1111/j.1469-0691.2004.01015.x. [DOI] [PubMed] [Google Scholar]
- 33.Fournier PE, Richet H. The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin Infect Dis. 2006;42(5):692–699. doi: 10.1086/500202. [DOI] [PubMed] [Google Scholar]
- 34.Manchanda V, Sanchaita S, Singh N. Multidrug resistant acinetobacter. J Glob Infect Dis. 2010;2(3):291–304. doi: 10.4103/0974-777X.68538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zavascki AP, et al. Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J Antimicrob Chemother. 2007;60(6):1206–1215. doi: 10.1093/jac/dkm357. [DOI] [PubMed] [Google Scholar]
- 36.Talukdar PK, et al. Antimicrobial resistance, virulence factors and genetic diversity of Escherichia coli isolates from household water supply in Dhaka, Bangladesh. PLoS One. 2013;8(4):e61090. doi: 10.1371/journal.pone.0061090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arias CA, Murray BE. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol. 2012;10(4):266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boucher HW, Corey GR. Epidemiology of methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 2008;46(Suppl 5):S344–S349. doi: 10.1086/533590. [DOI] [PubMed] [Google Scholar]
- 39.Williams BG, et al. Estimates of world-wide distribution of child deaths from acute respiratory infections. Lancet Infect Dis. 2002;2(1):25–32. doi: 10.1016/S1473-3099(01)00170-0. [DOI] [PubMed] [Google Scholar]
- 40.Baqui AH, et al. Causes of childhood deaths in Bangladesh: an update. Acta Paediatr. 2001;90(6):682–690. doi: 10.1111/j.1651-2227.2001.tb02434.x. [DOI] [PubMed] [Google Scholar]
- 41.Brooks WA, et al. Effect of weekly zinc supplements on incidence of pneumonia and diarrhoea in children younger than 2 years in an urban, low-income population in Bangladesh: randomised controlled trial. Lancet. 2005;366(9490):999–1004. doi: 10.1016/S0140-6736(05)67109-7. [DOI] [PubMed] [Google Scholar]
- 42.Farha T, Thomson AH. The burden of pneumonia in children in the developed world. Paediatr Respir Rev. 2005;6(2):76–82. doi: 10.1016/j.prrv.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 43.Mamishi S, et al. Penicillin-resistant trend of Streptococcus pneumoniae in Asia: a systematic review. Iran J Microbiol. 2014;6(4):198–210. [PMC free article] [PubMed] [Google Scholar]
- 44.Brooks WA, et al. Invasive pneumococcal disease burden and implications for vaccine policy in urban Bangladesh. Am J Trop Med Hyg. 2007;77(5):795–801. [PubMed] [Google Scholar]
- 45.Klevens RM, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298(15):1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 46.Chambers HF, Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 2009;7(9):629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rasmussen RV, et al. Future challenges and treatment of Staphylococcus aureus bacteremia with emphasis on MRSA. Future Microbiol. 2011;6(1):43–56. doi: 10.2217/fmb.10.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data can be made available to the interested researchers by the authors of this article if requested.


