Abstract
Background
Carbapenem-resistant Acinetobacter baumannii poses a significant threat to hospitalized patients, as few therapeutic options remain. Thus, we investigated the molecular epidemiology and mechanism of resistance of carbapenem-resistant A.baumannii isolates in Beijing, China.
Methods
Carbapenem-resistant A.baumannii isolates (n = 101) obtained between June 2009 and November 2014 were used. Multilocus sequence typing (MLST) and PCR assays for class C and D β-lactamase were performed on all isolates. S1 nuclease pulsed-field gel electrophoresis (PFGE) and Southern blot hybridization were performed to identify the resistance gene location.
Results
All 101 A.baumannii isolates were highly resistant to frequently used antimicrobials, and were considered multidrug resistant. A total of 12 sequence types (STs) were identified, including 10 reported STs and 2 novel STs. Eighty-seven isolates were classified to clonal complex 92 (CC92), among which ST191 and ST195 were the most common STs. The bla OXA-23 gene was positive in most (n = 95) of the A.baumannii isolates. Using S1-nuclease digestion PFGE and Southern blot hybridization, 3 patterns of plasmids carrying bla OXA-23 were confirmed. ST191 and ST195 (both harboring bla OXA-23) caused outbreaks during the study period, and this is the first report of outbreaks caused by ST191 and ST195 in north China.
Conclusion
bla OXA-23-producing A.baumannii ST191 and ST 195 isolates can disseminate in a hospital and are potential nosocomial outbreak strains. Surveillance of imipenem-resistant A.baumannii and antimicrobial stewardship should be strengthened.
Keywords: Acinetobacter baumannii, Carbapenem resistant, blaOXA-23, CC92, Outbreak
Background
Acinetobacter baumannii is an opportunistic pathogen involved in outbreaks occurring in burn units, surgical wards or intensive care units (ICUs), as well as important cause of nosocomial septicemia, pneumonia and urinary tract infections [1]. A.baumannii is of interest due to increasing the increase in antimicrobial resistance [2]. This organism is generally intrinsically resistant to many frequently-used antibiotics, including aminopenicillin, first- and second-generation cephalosporins and chloramphenicol [3, 4]. Carbapenems are important antibiotics to treat A.baumannii because they are highly efficacious and have low toxicity [5]. However, the emergent and rapid spread of carbapenem-resistant A.baumannii isolates pose a severe threat to public health and are a global concern [6]. Carbapenem resistance, such as to imipenem, increased in China from 31.0% in 2005 to 62.4% in 2014 [7]. Recent studies also suggest high resistant of A.baumannii against carbapenems across the world [8–11].
Carbapenem resistance in A.baumannii is mainly mediated by the production of carbapenem-hydrolyzing enzymes [6]. Class D OXA-type enzymes are the most prevalent carbapenemases in A.baumannii [12]. In addition to the intrinsic OXA-51-like enzymes, 3 unrelated groups of these carbapenem-hydrolysing enzymes have been identified OXA-23-like, -40-like and -58-like [13]. Outbreaks of bla OXA-23-producing A.baumannii have been reported across the world [14–16] and a previously study has pointed out that bla OXA-23 was the predominant group of carbapenem-hydrolysing enzymes in China [17].
Multilocus sequence typing (MLST) is used for global and long-term epidemiological studies [18], and data from MLST show that CC92 was the most widely distributed A.baumannii clone globally [19–21]. Studies from China indicate that bla OXA-23-producing CC92clones are prevalent in most provinces of China [17, 22]. Although the molecular epidemiology of carbapenem-resistant A.baumannii has been investigated, the epidemiology of carbapenem-resistant A.baumannii over long time periods in single institution may allow new insights into the behavior of this pathogen.
Thus, we sought to investigate carbapenem-resistance mechanisms and the molecular epidemiology of carbapenem-resistant A. baumannii in a single hospital over a 65-month period.
Methods
Bacterial isolates
Between June 2009 and November 2014, a total of 101 nonduplicate carbapenem-resistant (Zone Diameter of imipenem ≤18 mm; Clinical Laboratory Standards Institute [CLSI] breakpoint) A. baumannii (CRAB) isolates were collected from a single hospital in Beijing, China. A single isolate per patient was included. All isolates were identified by conventional biochemical techniques using VITEK 2 system (BioMérieux France). PCR confirmation of the bla OXA-51-like carbapenemase gene was performed to help identify A. baumannii simultaneously, because this gene is intrinsic to A. baumannii [23, 24].
Antimicrobial susceptibility testing
The disk diffusion method was used to evaluate susceptibility to the following antimicrobial agents: imipenem (IPM: 10 μg), ceftazidime (CAZ: 30 μg), amikacin (AMK: 30 μg), piperacillin/tazobactam (TZP: 100/10 μg), levofloxacin (LVX: 5 μg), ticarcillin/Clavulanic acid (TCC: 75/10 μg), minocycline (MNO: 30 μg) (Oxoid, UK). Results were interpreted in accordance with CLSI guidelines from 2011. Isolates with intermediate susceptibility were classified as non-susceptible.
Molecular typing methods
Multilocus sequence typing (MLST) was performed on all A.baumannii isolates as described previously [18]. Analysis of allele sequences and sequence type (ST) assignment made use of the Oxford Acinetobacter baumannii MLST website (http://pubmlst.org/abaumannii/). The eBURST algorithm (version 3; http://eburst.mlst.net/) was used to assign clonal complexes (CCs).
Screening of ambler classes C and D β-lactamase genes
PCR experiments were carried out using primers specific for the genes encoding Ambler C and D β-lactamase (AmpC, MOX-1, MOX-2, CMY-1 to CMY-11, BIL-1, DHA-1, DHA-2, ACC, ACT-1, MIR-1 T, FOX-1 to FOX-5b, blaOXA-23-like, blaOXA-40-like, blaOXA-51-like, and blaOXA-58-like and blaOXA-143)as described previously [25–28]. Primers are depicted in Table 1. For each gene detected, some PCR products were randomly selected, and then sequenced to confirm genes.
Table 1.
Primers used in this study
| Primer | Sequence(5′ to 3′) | Target | Reference |
|---|---|---|---|
| MOXMF | GCTGCTCAAGGAGCACAGGAT | MOX-1, MOX-2, CMY-1, CMY-8 to CMY-11; |
[25] |
| MOXMR | CACATTGACATAGGTGTGGTGC | ||
| CITMF | TGGCCAGAACTGACAGGCAAA | LAT-1 to LAT-4, CMY-2 to CMY-7, BIL-1; |
[25] |
| CITMR | TTTCTCCTGAACGTGGCTGGC | ||
| DHAMF | AACTTTCACAGGTGTGCTGGGT | DHA-1, DHA-2 | [25] |
| DHAMR | CCGTACGCATACTGGCTTTGC | ||
| ACCMF | AACAGCCTCAGCAGCCGGTTA | ACC | [25] |
| ACCMR | TTCGCCGCAATCATCCCTAGC | ||
| EBCMF | TCGGTAAAGCCGATGTTGCGG | MIR-1 T ACT-1 | [25] |
| EBCMR | CTTCCACTGCGGCTGCCAGTT | ||
| FOXMF | AACATGGGGTATCAGGGAGATG | FOX-1 to FOX-5b | [25] |
| FOXMR | CAAAGCGCGTAACCGGATTGG | ||
| AmpCF | ACAGAGGAGCTAATCATGCG | AmpC | [26] |
| AmpCR | GTTCTTTTAAACCATATACC | ||
| OXA-23-likeF | GATCGGATTGGAGAACCAGA' | blaOXA-23-like | [27] |
| OXA-23-likeR | ATTTCTGACCGCATTTCCAT | ||
| OXA-40-likeF | GGTTAGTTGGCCCCCTTAAA | blaOXA-40-like | [27] |
| OXA-40-likeR | AGTTGAGCGAAAAGGGGATT | ||
| OXA-51-likeF | TAATGCTTTGATCGGCCTTG | blaOXA-51-like | [27] |
| OXA-51-likeR | TGGATTGCACTTCATCTTGG | ||
| OXA-58-likeF | AAGTATTGGGGCTTGTGCTG | blaOXA-58-like | [27] |
| OXA-58-likeR | CCCCTCTGCGCTCTACATAC | ||
| OXA-143 | TGGCACTTTCAGCAGTTCCT | blaOXA-143 | [28] |
| OXA-143 | TAATCTTGAGGGGGCCAACC |
PFGE and Southern blot hybridization
To detect plasmids of A.baumannii isolates, an agarose gel plug containing total cellular DNA was prepared and digested with S1 nuclease (Takara, Japan) as described previously [29]. Digested plugs were subjected to PFGE using a CHEF-Mapper system (pulse times, 5 to 30 s; running time, 15 h; 6 V/cm). Gels were blotted onto nylon membranes (Millipore, USA) using standard techniques. The membrane was hybridized with a digoxigenin-labeled probe consisting of a bla OXA-23 fragment which was amplified by primers.
Results
A total of 101 A.baumannii isolates were resistant to imipenem and considered carbapenem-resistant and enrolled in our study. CRAB isolates were obtained from various sources, including sputum (n = 72 isolates), blood (n = 14 isolates), abdominal fluid (n = 9 isolates), secretion (n = 2 isolates), catheter (n = 1 isolates), eyes (n = 1 isolates), pus (n = 1 isolates) and throat swabs (n = 1 isolates). Of 101 CRAB isolates, 87% (n = 88) were collected from the ICU. The temporal distribution of CRAB isolates is showed as follows. 2, 3, 45, 4, 18 and 29 isolates were obtained in 2009, 2010, 2011, 2012, 2013 and 2014, respectively. CRAB isolates resistance data appear in Table 2. All CRAB isolates were resistant to at least 3 classes of antibiotic and were considered multidrug resistant. CRAB isolate data for AmpC and bla OXA-51-like genes appear in Table 2.
Table 2.
Details of A. baumannii isolates, by sequence type
| Non-susceptible to (%): | Resistant determinants | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ST | No. | Allelic profile | Time course | IPM | TZP | TCC | CAZ | AMK | LVX | MNO | |
| 191 | 32 | 1-3-3-2-2-94-3 | Aug. 2009 Feb. 2011–May. 2012 |
100 | 100 | 100 | 100 | 38 | 100 | 23 | OXA-51, OXA-23, AMPC |
| 195 | 31 | 1-3-3-2-2-96-3 | Mar. 2013–Nov. 2014 | 100 | 100 | 100 | 100 | 97 | 100 | 100 | OXA-51, OXA-23, AMPC |
| 208 | 15 | 1-3-3-2-2-97-3 | Jul. 2010–Oct. 2014 | 100 | 100 | 100 | 93 | 100 | 100 | 100 | OXA-51, OXA-23, AMPC |
| 218 | 2 | 1-3-3-2-2-102-3 | Jul. 2011–Aug. 2011 | 100 | 100 | 100 | 100 | 100 | 100 | 0 | OXA-51, OXA-23, AmpC |
| 368 | 6 | 1-3-3-2-2-140-3 | Jun. 2009–Sep. 2014 | 100 | 100 | 100 | 100 | 67 | 67 | 33 | OXA-51, OXA-23, AmpC |
| 369 | 1 | 1-3-3-2-2-106-3 | Jun. 2013 | 100 | 100 | 100 | 100 | 0 | 100 | - | OXA-51, OXA-23, AmpC |
| 373 | 2 | 1-12-12-11-4-103-3 | Mar. 2011, Apr. 2011 | 100 | 100 | 100 | 100 | 0 | 100 | 0 | OXA-51, AmpC |
| 383 | 2 | 1-12-56-1-4-149-45 | Aug. 2011 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | OXA-51, AmpC |
| 429 | 2 | 1-34-56-1-4-144-45 | Jul. 2011, Sep. 2011 | 100 | 100 | 100 | 100 | 50 | 50 | 100 | OXA-51, OXA-23, AmpC |
| 469 | 6 | 1-12-3-2-2-103-3 | Dec. 2012-Nov. 2013 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | OXA-51, OXA-23, AmpC |
| 1302n | 1 | 2-52-80-6-23-140-4 | Jul. 2014 | 100 | 100 | 100 | 0 | 0 | 0 | 0 | OXA-51, OXA-40, AmpC |
| 1309n | 1 | 27-155n-99-55-25 -270n-60 | Aug. 2011 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | OXA-51, AmpC |
| Total | 101 | - | Jun. 2009–Nov. 2014 | 100 | 100 | 100 | 98 | 80 | 96 | 68 | - |
nNovel; IPM imipenem, TZP piperacillin/tazobactam, TCC ticarcillin/clavulanic acid, CAZ ceftazidime, AMK amikacin, LVX levofloxacin, MNO minocycline
To investigate the molecular epidemiology of isolates, MLST was performed to characterize CRABs and data are summarized in Table 2. The eBURST analysis data appear in Fig. 1.
Fig. 1.
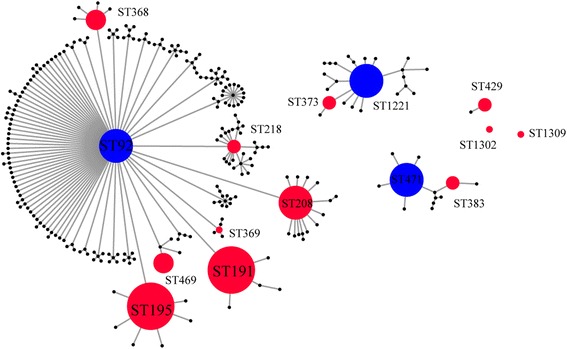
eBURST results of 12 STs presented in this study. The 1309 STs in the entire A.baumannii MLST database were analyzed using the most stringent definition (6/7 shared alleles). Four groups and 2 singletons that included STs found in our study are displayed as an eBURST diagram together. Each circle represents an ST. STs in a group are considered to belong to same clonal complex. Blue STs are founders of corresponding group. Red STs were found in this study. Circle size reflects the number of strains. Other ST labels have been removed for clarity
ST191 was the largest sequence type (32 of 101 isolates) and was found in our institution from August 2009 to May 2012. Only 1 isolate was obtained in August 2009 from the Liver Failure ward, but most (96.9%) of ST191 isolates were collected between February 2011 and May 2012. Importantly, 28 of the 32 ST191 strains were isolated from ICU ward. All ST191 isolates were resistant to piperacillin/tazobactam, ticarcillin/clavulanic acid, ceftazidime and levofloxacin but had variable susceptibilities to amikacin and minocycline (Table 2). All of ST191 isolates were bla OXA-23-positive. One strain was selected randomly to be subjected to PFGE digested with S1 nuclease, and results show that this ST strain contains a plasmid of approximately 78 Kb. Southern blot hybrid hybridization assays confirmed that bla OXA-23 gene was located on this plasmid (Fig. 2).
Fig. 2.
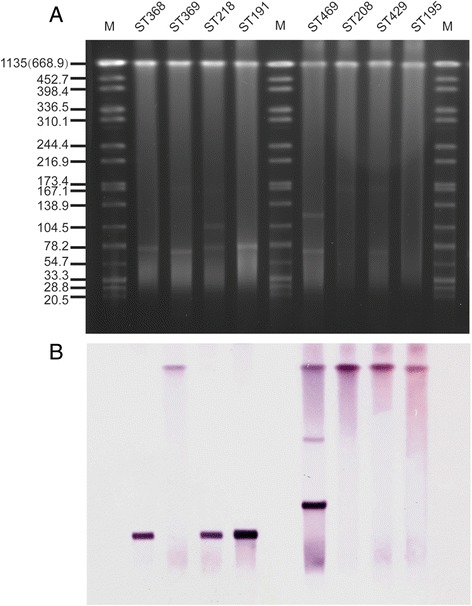
Results of plasmid measurement and hybridization. a S1 nulcease digestion of total DNA of A.baumannii isolates was followed by PFGE. Plasmid bands are shown as linearized fragments on the gel. For each A.baumannii ST, only one strain was selected randomly for this assay. b Southern blot hybridization for the bla oxa-23 gene. Lane M, reference standard strain H9812 restricted with XbaI. MW are in KB
Thirty-one (30.7%) isolates were defined as ST195, and this ST was first detected in the infectious liver diseases ward in March 2013. Twenty-eight ST195 isolates were collected from ICU ward from then on. It is notable that all ST195 isolates were resistant to almost all antibiotics tested in this study except 1 isolate was susceptible to amikacin. All of ST195 isolates were also bla OXA-23 positive. This ST strain does not contain any plasmid, and the bla OXA-23 gene is located on the chromosome (Fig. 2).
Another 4 sequence types, ST208, ST368, ST218 and ST369, were found in 15 (14.8%), 6 (5.9%), 2 (2.0%) and 1 (1.0%) isolates, respectively, and carried the bla OXA−23 gene but had different resistance profiles. The bla OXA−23 gene is located on the plasmid in the A.baumannii ST218 and ST368 strain, but is found on the chromosome in ST208 and ST369.
Six isolates were ST469. This is a double-locus variant (DLV) of multiple STs within CC92, implying a close relationship. However, it does not agree with the conservative definition of sharing alleles at 6/7 of the loci, and thus ST469 cannot be considered a CC92 member. All ST469 isolates were collected during December 2012 and November 2013. ST469 isolates were bla OXA-23 positive and were resistant to allantibiotics. PFGE and hybridization results show that this ST isolate harbored 2 plasmids. The bla OXA-23 gene was located on a ca. 120 kb plasmid and on a plasmid of approximately 245 kb, with another copy of bla OXA-23 on a chromosome (Fig. 2).
Two unreported singleton STs were identified. ST1309 presented in this hospital in August 2011 only and was resistant to all the 7 classes of antimicrobials. The carbapenem-resistant determinant of this isolate remains unclear. The ST1302 isolate was unique among imipenem-resistant strains; it was ceftazidime, amikacin, levofloxacin and minocycline susceptible and was the only isolate that carried the bla OXA-40 gene.
Discussion
This study offers insight into the longitudinal evaluation of the molecular epidemiology of carbapenem-resistant A.baumannii in a single institution over a 65-month period.
The bla OXA-23 gene was positive in most (n = 95) of the A.baumannii isolates in this institution. The first report of this enzyme in A.baumannii was ARI-1, which was identified in an isolate from Scotland collected in 1985 [30]. In 2000, enzyme sequence analysis (re-named OXA-23) indicated that it was a member of the ambler class D group of β-lactamases [31]. Since then, outbreaks of OXA-23 carbapenemase-producing A.baumannii have been reported all over the world [32–36]. Our finding was consistent with other reports from China. Zhou’s group investigated resistance determinants of 342 imipenem-resistant A.baumannii isolates which were collected from 16 Chinese cities in 2005, and found that most CRAB isolates contained the bla OXA-23 gene [37]. Recent studies confirm a high prevalence of the bla OXA-23 gene in carbapenem-resistant A.baumannii in different Chinese cities (80.6-100%) [38–40]. Southern blotting revealed that the bla oxa-23 gene is plasmid-mediated in some STs (ST191, ST218 and ST368), but chromosome borne in others (ST195, ST208, ST369 and ST429). Chromosomal locations of bla oxa-23 make it less likely for A.baumannii to lose caarbapenem resistance. Investigation of OXA-23 producing A.baumannii isolates collected from 28 hospitals in 18 provinces of China showed that OXA-23 was mainly located on a ca.78-kb plasmid or on a chromosome [41].
We were concerned that A.baumanii isolates harboring the bla OXA-23 gene were multidrug resistant in our study and had few antibiotic therapeutic options for treating CRAB infection. Thus, controlling the spread of bla OXA-23 producing A.baumannii is important.
We identified ST191, ST195, ST208, ST218, ST368 and ST369 as classified into CC92 which was the largest and most widely distributed A.baumannii clone in China [17]. CC92 represented the most epidemic CRAB STs in this hospital, accounting for 86.1% of isolates in this study. For bla OXA-23-producing CRAB of CC92, the ability to disseminate in a single institution for a long time suggests that adaptation to the hospital environment may be important for the success of A.baumannii.
It has been suggested that any clinical A.baumannii isolates with resistance to multiple antibiotics can cause a nosocomial outbreak [42]. We found that imipenem-resistant A.baumannii of CC92, compared with other clonal complexes, may be more prone to cause severe outbreaks during long-term dissemination. Two outbreaks of CRAB CC92 were observed in our institution. Most ST191 isolates (31/32) were identified between February 2011 and May 2012 in the ICU ward, suggesting an outbreak of bla OXA-23-producing ST191. Deng’s group has reported the prevalence of an A.baumannii ST191 clone in a southern Chinese hospital [43]. To our knowledge, this is the first identification of an outbreak of bla OXA-23 harboring A.baumannii ST191 isolate in north China. A second outbreak of bla OXA-23-producing CRAB occurred in this ICU later. Thirty-one bla OXA-23-producing ST195 isolates were also found in the ICU between March 2013 and November 2014and this sequence type was more resistant to frequently-used antimicrobial agents compared with ST191. ST195 has frequently been identified in Asian countries, including Japan, Vietnam, and Malaysia [44–46]. To our knowledge, Li’s group was first to identify ST195 in a teaching hospital in Guangzhou, in southern China [47]. Since then, ST195 clones has been identified in western and eastern China [38, 48]. Here, we offer the first report of an outbreak of bla OXA-23-producing ST195 in north China, suggesting that ST195 has been successfully disseminated in this country.
We collected no environmental strains from the work place, so we lack surveillance for source identification, which is a significant limitation of our study.
Conclusions
In summary, bla OXA-23-producing CC92 isolates were prevalent in this hospital over a 65-month period. Successive outbreaks of ST191 and ST195 demonstrated that persisting clinical carbapenem-resistant A.baumannii isolate can cause a nosocomial outbreak. Periodic investigation of molecular epidemiology and resistance determinant of A.baumannii is necessary.
Acknowledgements
This publication made use of the Acinetobacter baumannii MLST website (http://pubmlst.org/abaumannii/) sited at the University of Oxford (Jolley & Maiden 2010, BMC Bioinformatics, 11:595). The development of this site has been funded by the Wellcome Trust.
Funding
This work was supported by National Key Basic Research Program (973) of China (2015CB554202), the State Key Research Development Program Of China (2016YFC1200301), the National Science and Technology Major Project for Creation of Major New Drugs of China (2013ZX09304101),National Natural Science Foundation of China (81401643), the State Key Laboratory of Pathogen and BioSecurity Program (SKLPBS1530 and SKLPBS1424).
Availability of data and materials
The datasets generated during and/or analysed during the current study are available in the PubMLST repository, [http://pubmlst.org/abaumannii/].
Authors’ contributions
HW, EBC, FQ and TL designed and supervised the experiments. NZN, JH, CMB, SMC, EBC, JLZ and FHC performed experiments. NZN, XL and FQ interpreted and analyzed the data. NZN and XL wrote the paper. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Microbiology isolation and identification were routine work in our hospital. Only strains that have been routinely collected for diagnosis were studied. No extra sampling from the patients was performed. No personal information about patients was requested. Therefore, a written personal informed consent and ethics committee approval were not required and Chinese law was strictly complied.
Abbreviations
- CC
Clonal complex 92
- CRAB
Carbapenem-resistant A. baumannii
- DLVs
Double-locus variants
- MLST
Multilocus sequence typing
- PFGE
Pulsed-field gel electrophoresis
- SLVs
Single-locus variants
- ST
Sequence type
Contributor Information
Nian-zhi Ning, Email: ningnianzhi@163.com.
Xiong Liu, Email: liuxiong714@163.com.
Chun-mei Bao, Email: bcm2003@sina.com.
Su-ming Chen, Email: chensuming987@163.com.
En-bo Cui, Email: cebo_bj@sina.com.
Ju-ling zhang, Email: zhangjuling0913@163.com.
Jie Huang, Email: s1006317@163.com.
Fang-hong Chen, Email: cfh880314@163.com.
Tao Li, Email: litaobmi@126.com.
Fen Qu, Email: qf302@163.com.
Hui Wang, Phone: +00 860 106 694 8587, Email: geno0109@vip.sina.com.
References
- 1.Bergogne-Berezin E, Towner KJ. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev. 1996;9(2):148–165. doi: 10.1128/cmr.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51(10):3471–3484. doi: 10.1128/AAC.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vila J, Marcos A, Marco F, Abdalla S, Vergara Y, Reig R, Gomez-Lus R, Jimenez de Anta T. In vitro antimicrobial production of beta-lactamases, aminoglycoside-modifying enzymes, and chloramphenicol acetyltransferase by and susceptibility of clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother. 1993;37(1):138–141. doi: 10.1128/AAC.37.1.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seifert H, Stefanik D, Wisplinghoff H. Comparative in vitro activities of tigecycline and 11 other antimicrobial agents against 215 epidemiologically defined multidrug-resistant Acinetobacter baumannii isolates. J Antimicrob Chemother. 2006;58(5):1099–1100. doi: 10.1093/jac/dkl383. [DOI] [PubMed] [Google Scholar]
- 5.Evans BA, Hamouda A, Amyes SG. The rise of carbapenem-resistant Acinetobacter baumannii. Curr Pharm Des. 2013;19(2):223–238. doi: 10.2174/138161213804070285. [DOI] [PubMed] [Google Scholar]
- 6.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21(3):538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu FP, Guo Y, Zhu DM, Wang F, Jiang XF, Xu YC, Zhang XJ, Zhang CX, Ji P, Xie Y, et al. Resistance trends among clinical isolates in China reported from CHINET surveillance of bacterial resistance, 2005-2014. Clin Microbiol Infect. 2016;22(Suppl 1):S9–S14. doi: 10.1016/j.cmi.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Kempf M, Rolain JM. Emergence of resistance to carbapenems in Acinetobacter baumannii in Europe: clinical impact and therapeutic options. Int J Antimicrob Agents. 2012;39(2):105–114. doi: 10.1016/j.ijantimicag.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Shivaprasad A, Antony B, Shenoy P. Comparative Evaluation of Four Phenotypic Tests for Detection of Metallo-beta-Lactamase and Carbapenemase Production in Acinetobacter baumannii. J Clin Diagn Res. 2014;8(5):DC05–08. doi: 10.7860/JCDR/2014/6447.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hasan B, Perveen K, Olsen B, Zahra R. Emergence of carbapenem-resistant Acinetobacter baumannii in hospitals in Pakistan. J Med Microbiol. 2014;63(Pt 1):50–55. doi: 10.1099/jmm.0.063925-0. [DOI] [PubMed] [Google Scholar]
- 11.Bertrand X, Dowzicky MJ. Antimicrobial susceptibility among gram-negative isolates collected from intensive care units in North America, Europe, the Asia-Pacific Rim, Latin America, the Middle East, and Africa between 2004 and 2009 as part of the Tigecycline Evaluation and Surveillance Trial. Clin Ther. 2012;34(1):124–137. doi: 10.1016/j.clinthera.2011.11.023. [DOI] [PubMed] [Google Scholar]
- 12.Poirel L, Nordmann P. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin Microbiol Infect. 2006;12(9):826–836. doi: 10.1111/j.1469-0691.2006.01456.x. [DOI] [PubMed] [Google Scholar]
- 13.Dijkshoorn L, Nemec A, Seifert H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol. 2007;5(12):939–951. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 14.Principe L, Piazza A, Giani T, Bracco S, Caltagirone MS, Arena F, Nucleo E, Tammaro F, Rossolini GM, Pagani L, et al. Epidemic diffusion of OXA-23-producing Acinetobacter baumannii isolates in Italy: results of the first cross-sectional countrywide survey. J Clin Microbiol. 2014;52(8):3004–3010. doi: 10.1128/JCM.00291-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merino M, Poza M, Roca I, Barba MJ, Sousa MD, Vila J, Bou G. Nosocomial outbreak of a multiresistant Acinetobacter baumannii expressing OXA-23 carbapenemase in Spain. Microb. Drug Resist. (Larchmont, NY) 2014;20(4):259–263. doi: 10.1089/mdr.2013.0127. [DOI] [PubMed] [Google Scholar]
- 16.Lee Y, Kim YR, Kim J, Park YJ, Song W, Shin JH, Uh Y, Lee K, Lee SH, Cho JH, et al. Increasing prevalence of blaOXA-23-carrying Acinetobacter baumannii and the emergence of blaOXA-182-carrying Acinetobacter nosocomialis in Korea. Diagn Microbiol Infect Dis. 2013;77(2):160–163. doi: 10.1016/j.diagmicrobio.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 17.Ruan Z, Chen Y, Jiang Y, Zhou H, Zhou Z, Fu Y, Wang H, Wang Y, Yu Y. Wide distribution of CC92 carbapenem-resistant and OXA-23-producing Acinetobacter baumannii in multiple provinces of China. Int J Antimicrob Agents. 2013;42(4):322–328. doi: 10.1016/j.ijantimicag.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 18.Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodriguez-Valera F. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol. 2005;43(9):4382–4390. doi: 10.1128/JCM.43.9.4382-4390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karah N, Sundsfjord A, Towner K, Samuelsen O. Insights into the global molecular epidemiology of carbapenem non-susceptible clones of Acinetobacter baumannii. Drug Resist Updat. 2012;15(4):237–247. doi: 10.1016/j.drup.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 20.Kim DH, Choi JY, Kim HW, Kim SH, Chung DR, Peck KR, Thamlikitkul V, So TM, Yasin RM, Hsueh PR, et al. Spread of carbapenem-resistant Acinetobacter baumannii global clone 2 in Asia and AbaR-type resistance islands. Antimicrob Agents Chemother. 2013;57(11):5239–5246. doi: 10.1128/AAC.00633-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams-Haduch JM, Onuoha EO, Bogdanovich T, Tian GB, Marschall J, Urban CM, Spellberg BJ, Rhee D, Halstead DC, Pasculle AW, et al. Molecular epidemiology of carbapenem-nonsusceptible Acinetobacter baumannii in the United States. J Clin Microbiol. 2011;49(11):3849–3854. doi: 10.1128/JCM.00619-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Qiao F, Yu R, Gao Y, Zong Z. Clonal diversity of Acinetobacter baumannii clinical isolates revealed by a snapshot study. BMC Microbiol. 2013;13:234. doi: 10.1186/1471-2180-13-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turton JF, Woodford N, Glover J, Yarde S, Kaufmann ME, Pitt TL. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J Clin Microbiol. 2006;44(8):2974–2976. doi: 10.1128/JCM.01021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hojabri Z, Pajand O, Bonura C, Aleo A, Giammanco A, Mammina C. Molecular epidemiology of Acinetobacter baumannii in Iran: endemic and epidemic spread of multiresistant isolates. J Antimicrob Chemother. 2014;69(9):2383–2387. doi: 10.1093/jac/dku045. [DOI] [PubMed] [Google Scholar]
- 25.Perez-Perez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40(6):2153–2162. doi: 10.1128/JCM.40.6.2153-2162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heritier C, Poirel L, Nordmann P. Cephalosporinase over-expression resulting from insertion of ISAba1 in Acinetobacter baumannii. Clin Microbiol Infect. 2006;12(2):123–130. doi: 10.1111/j.1469-0691.2005.01320.x. [DOI] [PubMed] [Google Scholar]
- 27.Turton JF, Ward ME, Woodford N, Kaufmann ME, Pike R, Livermore DM, Pitt TL. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol Lett. 2006;258(1):72–77. doi: 10.1111/j.1574-6968.2006.00195.x. [DOI] [PubMed] [Google Scholar]
- 28.Higgins PG, Poirel L, Lehmann M, Nordmann P, Seifert H. OXA-143, a novel carbapenem-hydrolyzing class D beta-lactamase in Acinetobacter baumannii. Antimicrob Agents Chemother. 2009;53(12):5035–5038. doi: 10.1128/AAC.00856-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barton BM, Harding GP, Zuccarelli AJ. A general method for detecting and sizing large plasmids. Anal Biochem. 1995;226(2):235–240. doi: 10.1006/abio.1995.1220. [DOI] [PubMed] [Google Scholar]
- 30.Paton R, Miles RS, Hood J, Amyes SG, Miles RS, Amyes SG. ARI 1: beta-lactamase-mediated imipenem resistance in Acinetobacter baumannii. Int J Antimicrob Agents. 1993;2(2):81–87. doi: 10.1016/0924-8579(93)90045-7. [DOI] [PubMed] [Google Scholar]
- 31.Donald HM, Scaife W, Amyes SG, Young HK. Sequence analysis of ARI-1, a novel OXA beta-lactamase, responsible for imipenem resistance in Acinetobacter baumannii 6B92. Antimicrob Agents Chemother. 2000;44(1):196–199. doi: 10.1128/AAC.44.1.196-199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dalla-Costa LM, Coelho JM, Souza HA, Castro ME, Stier CJ, Bragagnolo KL, Rea-Neto A, Penteado-Filho SR, Livermore DM, Woodford N. Outbreak of carbapenem-resistant Acinetobacter baumannii producing the OXA-23 enzyme in Curitiba, Brazil. J Clin Microbiol. 2003;41(7):3403–3406. doi: 10.1128/JCM.41.7.3403-3406.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeon BC, Jeong SH, Bae IK, Kwon SB, Lee K, Young D, Lee JH, Song JS, Lee SH. Investigation of a nosocomial outbreak of imipenem-resistant Acinetobacter baumannii producing the OXA-23 beta-lactamase in korea. J Clin Microbiol. 2005;43(5):2241–2245. doi: 10.1128/JCM.43.5.2241-2245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naas T, Levy M, Hirschauer C, Marchandin H, Nordmann P. Outbreak of carbapenem-resistant Acinetobacter baumannii producing the carbapenemase OXA-23 in a tertiary care hospital of Papeete, French Polynesia. J Clin Microbiol. 2005;43(9):4826–4829. doi: 10.1128/JCM.43.9.4826-4829.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kohlenberg A, Brummer S, Higgins PG, Sohr D, Piening BC, de Grahl C, Halle E, Ruden H, Seifert H. Outbreak of carbapenem-resistant Acinetobacter baumannii carrying the carbapenemase OXA-23 in a German university medical centre. J Med Microbiol. 2009;58(Pt 11):1499–1507. doi: 10.1099/jmm.0.012302-0. [DOI] [PubMed] [Google Scholar]
- 36.Mosqueda N, Espinal P, Cosgaya C, Viota S, Plasensia V, Alvarez-Lerma F, Montero M, Gomez J, Horcajada JP, Vila J, et al. Globally expanding carbapenemase finally appears in Spain: nosocomial outbreak of acinetobacter baumannii producing plasmid-encoded OXA-23 in Barcelona, Spain. Antimicrob Agents Chemother. 2013;57(10):5155–5157. doi: 10.1128/AAC.01486-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou H, Yang Q, Yu YS, Wei ZQ, Li LJ. Clonal spread of imipenem-resistant Acinetobacter baumannii among different cities of China. J Clin Microbiol. 2007;45(12):4054–4057. doi: 10.1128/JCM.00343-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang Y, Luan G, Xu Y, Wang Y, Shen M, Zhang C, Zheng W, Huang J, Yang J, Jia X, et al. Characterization of carbapenem-resistant Acinetobacter baumannii isolates in a Chinese teaching hospital. Front Microbiol. 2015;6:910. doi: 10.3389/fmicb.2015.00910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang D, Yan D, Hou W, Zeng X, Qi Y, Chen J. Characterization of bla(OxA-23) gene regions in isolates of Acinetobacter baumannii. J Microbiol Immunol Infect. 2015;48(3):284–290. doi: 10.1016/j.jmii.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 40.Zhou Y, Wu X, Zhang X, Hu Y, Yang X, Yang Z, Wang M. Genetic Characterization of ST195 and ST365 Carbapenem-Resistant Acinetobacter baumannii Harboring blaOXA-23 in Guangzhou, China. Microb. Drug Resist. (Larchmont, NY) 2015;21(4):386–390. doi: 10.1089/mdr.2014.0183. [DOI] [PubMed] [Google Scholar]
- 41.Liu LL, Ji SJ, Ruan Z, Fu Y, Fu YQ, Wang YF, Yu YS. Dissemination of blaOXA-23 in Acinetobacter spp. in China: main roles of conjugative plasmid pAZJ221 and transposon Tn2009. Antimicrob Agents Chemother. 2015;59(4):1998–2005. doi: 10.1128/AAC.04574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koeleman JG, van der Bijl MW, Stoof J, Vandenbroucke-Grauls CM, Savelkoul PH. Antibiotic resistance is a major risk factor for epidemic behavior of Acinetobacter baumannii. Infect Control Hosp Epidemiol. 2001;22(5):284–288. doi: 10.1086/501901. [DOI] [PubMed] [Google Scholar]
- 43.Deng M, Zhu MH, Li JJ, Bi S, Sheng ZK, Hu FS, Zhang JJ, Chen W, Xue XW, Sheng JF, et al. Molecular epidemiology and mechanisms of tigecycline resistance in clinical isolates of Acinetobacter baumannii from a Chinese university hospital. Antimicrob Agents Chemother. 2014;58(1):297–303. doi: 10.1128/AAC.01727-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Endo S, Yano H, Hirakata Y, Arai K, Kanamori H, Ogawa M, Shimojima M, Ishibashi N, Aoyagi T, Hatta M, et al. Molecular epidemiology of carbapenem-non-susceptible Acinetobacter baumannii in Japan. J Antimicrob Chemother. 2012;67(7):1623–1626. doi: 10.1093/jac/dks094. [DOI] [PubMed] [Google Scholar]
- 45.Tada T, Miyoshi-Akiyama T, Kato Y, Ohmagari N, Takeshita N, Hung NV, Phuong DM, Thu TA, Binh NG, Anh NQ, et al. Emergence of 16S rRNA methylase-producing Acinetobacter baumannii and Pseudomonas aeruginosa isolates in hospitals in Vietnam. BMC Infect Dis. 2013;13:251. doi: 10.1186/1471-2334-13-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lean SS, Yeo CC, Suhaili Z, Thong KL. Whole-genome analysis of an extensively drug-resistant clinical isolate of Acinetobacter baumannii AC12: insights into the mechanisms of resistance of an ST195 clone from Malaysia. Int J Antimicrob Agents. 2015;45(2):178–182. doi: 10.1016/j.ijantimicag.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 47.Li Y, Pan C, Zhao Z, Zhao Z, Chen H, Lu W. Effects of a combination of amlodipine and imipenem on 42 clinical isolates of Acinetobacter baumannii obtained from a teaching hospital in Guangzhou, China. BMC Infect Dis. 2013;13:548. doi: 10.1186/1471-2334-13-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ying C, Li Y, Wang Y, Zheng B, Yang C. Investigation of the molecular epidemiology of Acinetobacter baumannii isolated from patients and environmental contamination. J Antibiot. 2015;68(9):562–567. doi: 10.1038/ja.2015.30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analysed during the current study are available in the PubMLST repository, [http://pubmlst.org/abaumannii/].


