Abstract
Accurate yet efficient computational models of solvent environment are central for most calculations that rely on atomistic modeling, such as prediction of protein-ligand binding affinities. In this study, we evaluate the accuracy of a recently developed generalized Born implicit solvent model, GBNSR6 (Aguilar et al. J. Chem. Theory Comput. 2010, 6, 3613–3639), in estimating the electrostatic solvation free energies (ΔGpol) and binding free energies (ΔΔGpol) for small protein-ligand complexes. We also compare estimates based on three different explicit solvent models (TIP3P, TIP4PEw and OPC). The two main findings are as follows.
First, the deviation (RMSD=7.04 kcal/mol) of GBNSR6 binding affinities from commonly used TIP3P reference values is comparable to the deviations between explicit models themselves, e.g. TIP4PEw vs. TIP3P (RMSD=5.30 kcal/mol). A simple uniform adjustment of the atomic radii by a single scaling factor reduces the RMS deviation of GBNSR6 from TIP3P to within the above “error margin” – differences between ΔΔGpol estimated by different common explicit solvent models. The simple radii scaling virtually eliminates the systematic deviation (ΔΔGpol) between GBNSR6 and two out of the three explicit water models, and significantly reduces the deviation from the third explicit model.
Second, the differences between electrostatic binding energy estimates from different explicit models is disturbingly large; for example, the deviation between TIP4PEw and TIP3P estimates of ΔΔGpol values can be up to ~50% in relative error, or ~9 kcal/mol in absolute error, which is significantly larger than “chemical accuracy” goal of ~1 kcal/mol. The absolute ΔGpol calculated with different explicit models could differ by tens of kcal/mol. These discrepancies point to unacceptably high sensitivity of binding affinity estimates to the choice of common explicit water models. The absence of a clear “gold standard” among these models strengthens the case for the use of accurate implicit solvation models for binding energetics, which may be orders of magnitude faster.
Graphical Abstract
1 Introduction
An accurate yet efficient determination of the electrostatics in protein-ligand interactions is of profound importance in molecular design and drug discovery.1–6 The computational prediction of binding free energies is however complex and challeng-ing,7,8 and its outcomes can depend strongly on the molecular modeling technique used.9 In particular, accuracy of solvation and binding free energies calculations depends critically on the quality of the underlying solvent model.10–12
Extensive studies have been performed to evaluate the accuracy of solvent models in predicting solvation free energies for small molecular systems;10–13 in many cases the desirable “chemical accuracy” of 1 kcal/mol was reported ,10,11,14–16 at least on average. Yet, the high accuracy in predicting individual solvation free energies does not necessarily translate into high accuracy in atomistic binding free energy calculations.17–19 High accuracy and robustness of the force fields and solvent models in these calculations has proven difficult to achieve.20–25 Errors in even some of the highly accurate calculated binding energies can represent a significant percentage of the target binding free energies, sometimes as large as ~50% in relative error or ~7 kcal/mol in absolute error,26 in particular when the number of interactions in a protein-ligand complex increases.19 Even for small and relatively rigid host-guest systems, predicting binding affinities within typical chemical accuracy of ~1 kcal/mol remains elusive.22,23,27 Perhaps not surprisingly, binding free energies can be sensitive to the method parameters: the relatively small binding affinities are the difference between large free energy terms corresponding to the bound and unbound states.17 The interactions between proteins and ligands are short ranged and strong, which leads to the strong dependence of energy functions on the details of molecular conformation.22 Effects of systematic error cancellation can also be consequential in binding free energy calculations.17,18 Since long-range electrostatic interactions play a dominant role in biomolecular simulations, a careful treatment of electrostatic interactions is essential,28–34 and merits especial attention: this is the focus of the current work.
Implicit solvent models are currently routinely employed for evaluation of electrostatic interactions in many scenarios of biomolecular modeling.35,36,36–52 By replacing discrete water models with a continuum medium using the average dielectric properties of water, implicit solvents provide significant decrease in the computational cost of simulations. Within the implicit solvent framework, the Generalized Born (GB) model43,53–75 provides a relatively simple, efficient and robust estimate to calculate the long-range electrostatic interactions in molecular simulations.43,76 Methods based on the implicit solvent framework, such as MMPB(GB)/SA,77,78 are extensively used in estimates of solvation free energies and protein-ligand binding interactions.31,37,41,62,79
Recently, a new flavor of GB model, the so-called GBNSR6,80 was reported; unlike most of its predecessors, the model relies on the “R6”81–84 effective Born radius, which is calculated as a single r/|r|6 integral over the Lee-Richard molecular surface.80 A good agreement of the electrostatic component of the solvation free energies (ΔGpol) by the R6 flavor compared to the more fundamental Poisson-Boltzman (PB) model for small proteins and DNA was previously reported.84 It was also shown that GBNSR6 and the TIP3P explicit model are in close agreement for different conformations of alanine polypeptide.80 In a recent study, it was shown that GBNSR6 (with an appropriate non-polar contribution added) reproduces experimentally measured solvation free energies of small molecules with near “chemical” accuracy,13 on average.
Given the high promise of GBNSR6 method in predicting solvation free energies of some molecular systems, here we evaluate the accuracy of the model in predicting protein-ligand binding energies, crucial for rational drug design. To address this question, in this study we evaluate the binding free energies from GBNSR6 for a set of 15 small protein-ligand complexes, using explicit solvent free energies as reference.
While using explicit solvent as accuracy reference for an implicit solvent model is natural, the question arises which of the great many85 available explicit water models should be used. The question is non-trivial, as none of the current models can be considered as uncontested “gold standard”.86–88 Explicit water models are built to reproduce bulk properties, but being imperfect,86–88 improved performance in pure water properties does not necessarily translate into better performance in solvated systems. For example, TIP4PEw is more accurate than TIP3P in predicting water bulk properties,88 but less accurate in predicting hydration energies of small molecules;11 TIP5P is superior to both in reproducing details of water structure,89 but trails behind TIP4PEw in accuracy of predicted small molecule hydration energies.11 Besides, the transferability of the observed accuracy of these explicit models in predicting solvation free energies of small molecules (RMS errors slightly over 1 kcal/mol) to macromolecular systems is not guaranteed,19 nor is it certain whether the same level of accuracy is achievable in binding free energy calculations, which is of main interest to us here. The question would be moot, however, if commonly used water models showed consistent performance in these calculations, say within 1 kcal/mol of each other. Whether or not common (and some new) explicit water models are equivalent in this respect is the second main question we address in this work. For this purpose we compare ΔGpol and ΔΔGpol computed with two highly popular, fixed-charge, rigid explicit water models for which free energy calculation protocols are well-established and their computational expense is reasonable: TIP3P90 and TIP4PEw.91 We also make a comparison with a recently developed 4-point rigid explicit water model, OPC,92 which is arguably the first model of this class that predicts hydration free energies of small molecules with RMSD accuracy of less than 1 kcal/mol.92
The remainder of the paper is organized as follows. In Section 2 we present the specifics of the protein-ligand complexes used for the comparative studies. The details of implicit solvent calculations and explicit solvent calculations using Thermodynamics Integration are provided in Section 2. The comparative studies are presented in Section 3. A summary of our findings is discussed in Section 4.
2 METHODS
2.1 Preparation of Complexes
A set of 15 protein-ligand complexes was selected (Table 1). One feature of the selected complexes is their small size (~1635–1995 atoms), essential to ensure convergence of the (lengthy) free energy perturbation (FEP) estimates. As a result of the limitation on the structure size, the diversity of the set is limited in terms of biological function of the complexes. However, the set is diverse with respect to values of electrostatic binding free energies: it covers a wide range of those (see Section 3), which is sufficient for our purpose. Another feature of the collected set is that ligands are neutral and proteins are either neutral or are forced to be neutral. The neutralization is performed to avoid various uncertainties and complications93 due to the use of Ewald summation and periodic boundary conditions in explicit solvent simulations. For each component that needed to be neutralized, the neutralization was performed as follows. First, its isoelectric point pI was computed. Then, protonation state and charge states of each titratable group was set according to its computed pK value at pH = pI, which forced over-all neutrality of the structure. The calculations of pK, pI, the titration curves, and the protonation state adjustments were performed using H++ server94 with the default settings; the server employs a continuum electrostatic approach to pK prediction. In principle, the explicit solvent box could alternatively be neutralized by adding counterions, however we did not follow this approach due to notably slow convergence of counterion distributions in MD simulations95 (tens of nanoseconds for monovalent ions), which would make our TI-based estimates of ΔGpol prohibitively expensive here.
Table 1.
Specifics of protein and ligand components for the set of 15 small complexes studied here
| PDB ID | Protein name | Number of atoms in protein |
Ligand name | Number of atoms in ligand |
|---|---|---|---|---|
| 1b11 | FELINE IMMUNODEFICIENCY VIRUS PROTEASE |
1824 | TL- 3–093 | 66 |
| 1bkf | FK506 BINDING PROTEIN FKBP MUTANT R42K/H87V |
1659 | FK506 | 128 |
| 1f40 | FKBP12 | 1662 | GPI-1046 | 54 |
| 1fb7 | HIV-1 PROTEASE MUTANT | 1566 | SAQUINAVIR | 99 |
| 1fkb | HUMAN IMMUNOPHILIN FKBP-12 | 1662 | RAPAMYCIN | 144 |
| 1fkf | IMMUNOPHILIN FKBP | 1662 | FK506 | 126 |
| 1fkg | FKBP | 1662 | SB3 | 68 |
| 1fkh | FKBP | 1662 | SBX | 74 |
| 1fkj | FKBP12 | 1662 | FK506 | 128 |
| 1fkl | FKBP12 | 1661 | RAPAYMYCIN | 146 |
| 1pbk | FKBP25 | 1851 | RAP | 144 |
| 1zp8 | HIV PROTEASE | 1566 | INHIBITOR AB-2 | 88 |
| 2fke | FK-506-BINDING PROTEIN | 1662 | 8-DEETHYL-8-[BUT-3- ENYL]-ASCOMYCIN |
126 |
| 3kfp | HIV PROTEASE | 1569 | INHIBITOR TL-3 | 66 |
| 2hah | FIV/HIV chimeric protease | 1800 | broad-based inhibitor, TL 3 | 66 |
For setting up the structures we used H++94 server that creates topology and coordinates files in Amber96 format. The ff99bsc0 parameters and the GAFF force field, both part of Amber12,97 were used for preparing the topology and coordinate files which includes partial charges. We performed 500 steps of minimization on the neutral complexes (without restraint) in vacuum to relax the structure. The minimization was performed in SANDER molecular dynamics module of Amber with a 12 cutoff distance. After minimization, the complex structures were broken down to the protein and ligand components to be used for the binding free energy calculations. The Amber format topology and coordinate files as well as the corresponding ΔGpol values are available in the Supporting Information.
2.2 Implicit Solvent Details
GBNSR6
GBNSR6 is an implementation of the Generalized Born (GB) model in which the effective Born radii are computed numerically, via the so-called “R6” integration,13,80 over the Lee-Richards molecular surface.98 The polar component of the solvation energy, ΔGpol, is calculated by the ALPB model,99 which introduces physically correct dependence on dielectric constants into the original GB model of Still et al.,54 while maintaining the efficiency of the original:
| (1) |
where εin and εout are the dielectric constants of the solute and the solvent respectively, β = εin/εout, α = 0.571412, and A is the electrostatic size of the molecule, which is essentially the overall size of the structure, that can be computed analytically. Here, qi is the partial charge of atom i. The most widely used functional form76 of is employed, where Ri is the effective Born radius of atom i, and rij is the distance between atoms i and j. We set εin = 1 and εout = 80 in Eq. (1). Note that to mitigate uncertainties related to conformational sampling,23,100 and to facilitate direct comparison between implicit with explicit solvent model predictions, we eliminated structural fluctuations by keeping all of the structures fixed by strong coordinate restraints in all of the explicit solvent simulations performed in this study (see Section 2.3). As a result, there was no dielectric response from the protein. This scenario is consistent with a value of unity for the solute dielectric constant (εin = 1) in the corresponding implicit solvent modeling62,101 (Eq. (1)). The assignment of solute dielectric constant can, however, be different for a direct comparison to experiment.
The effective Born radii Ri are calculated via:
| (2) |
where ∂V represents the molecular surface of the molecule, dS is the infinitesimal surface element vector, ri is the position of atom i, and r represents the position of the infinitesimal surface element. In contrast to most GB practical models, GBNSR6 model is essentially parameter-free in the same sense as the numerical PB framework is. Thus, accuracy of GBNSR6 relative to the PB standard is unaffected by the choice of input atomic radii. Here we use the simple, standard Bondi102 radii set to determine the surface of the molecule. The solvent probe radius is equal to 1.4 Å. We use the same constant offset B = 0.028 Å−1 to the inverse radii as in Mongan et al.84
The GBNSR6 model exploits the Cartesian grid developed previously for PBSA module of Amber,103 to build a numerical discretization of the Lee–Richards molecular surface.98 The spacing between two neighboring grid points is uniformly set to h=0.3 Å, for the molecular surface resolution. The arc resolution (arcres), defined as the arc length between two neighboring solvent probe sites as the probe rolls over the atoms,103 is set to 0.1 Å. This implementation of GBNSR6 is currently available as a part of Amber Tools suit of programs in Amber15.104
PB model
The Adaptive Poisson-Boltzmann Solver (APBS) software package105 was used for evaluating the polar part of solvation energies. The solute dielectric constant was set to 1 and the solvent dielectric constant was 80, which are consistent with the values chosen for GBNSR6. The grid spacing was set to 0.3 Å. To set the dimensions of the grids, we keep a distance equal to the size of the structure between the protein boundary and the grid boundary for the largest structure. Accordingly, the grid dimension size was set to 449 in x, y and z directions for all of the structures. The solvent probe radius is 1.4 Å. We use APBS default values for the remaining parameters, and assume no monovalent salt present, as in both the GB and the explicit solvent calculations.
2.3 Explicit Solvent Calculations
Electrostatic components ΔGpol of explicit solvation free energies are computed by using the Thermodynamic Integration (TI) method of the SANDER module in Amber12.97 Here we only compute the free energy transformations where the charges on protein-ligand complexes are removed: state 0 represents all solute atomic charges “on”, and state 1 represents all solute atomic charges “off”. We have used 5 values of lambda for TI calculations, λ = 0.04691, 0.23076, 0.50000, 0.76923, 0.95308. The TI values were obtained from Gaussian integration over the λ values. TI calculations were performed in water (TIP3P, TIP4PEw, or OPC explicit model) and in vacuum. Then the corresponding free energy values were subtracted to cancel out the intrasolute charge interactions as well as the restraint energies.106 In all the simulations, the bonds to hydrogen atoms were constrained with the SHAKE algorithm using a geometrical tolerance of 0.000001 Å. The nonbonded interaction cutoff was 9 Å for simulations in water and 99 Å (effectively infinite) for simulations in vacuum. A time step of 2 fs is used (reduced to 1.8 fs if numerical instability was encountered). The following process is performed for each value of lambda: first, we run 1000 steps of minimization using steepest decent method. Then, we run 30 ps of NVT ensemble by gradually increasing the temperature from 0 K to 300 K. Then we run 1 ns of NPT ensemble at 300 K, for density equilibration. For the production we run 2 ns of NVT ensemble at 300 K. We run 200 ps MD simulation in vacuum. Protein-ligand complexes have many degrees of freedom that makes exploring all potentially relevant conformations computationally intractable.23,100 Thus, 200 kcal/mol/Å2 harmonic Cartesian coordinate restraints were imposed to all atoms, except during the minimization step for which 500 kcal/mol/Å2 harmonic Cartesian coordinate restraints were applied to all atoms.
Robustness of the Protocol and Error Estimate
The standard deviation of computed ΔGpol values is smaller than ± 0.7 kcal/mol for complexes and protein components, and smaller than 0.14 kcal/mol for ligands components. The standard deviations are calculated by assuming a correlation time of 1 ps, which is a conservative assumption considering that it is usually smaller than 0.8 ps.107 The analysis is performed on the last 1.5 ns of 2 ns long simulation to ensure convergence. To test sensitivity to initial conditions we repeated the calculations for two complexes using different random seeds for the random number generator, and obtained differences less than the average standard deviation of computed ΔGpol values given above. To further test that the TI results have reached convergence, we extended the simulation time from 2 ns to 5 ns for two randomly selected complexes and noticed the resulting TI values differ by less than the standard deviation above.
2.4 The Electrostatic Component of Binding Free Energies, ΔΔGpol
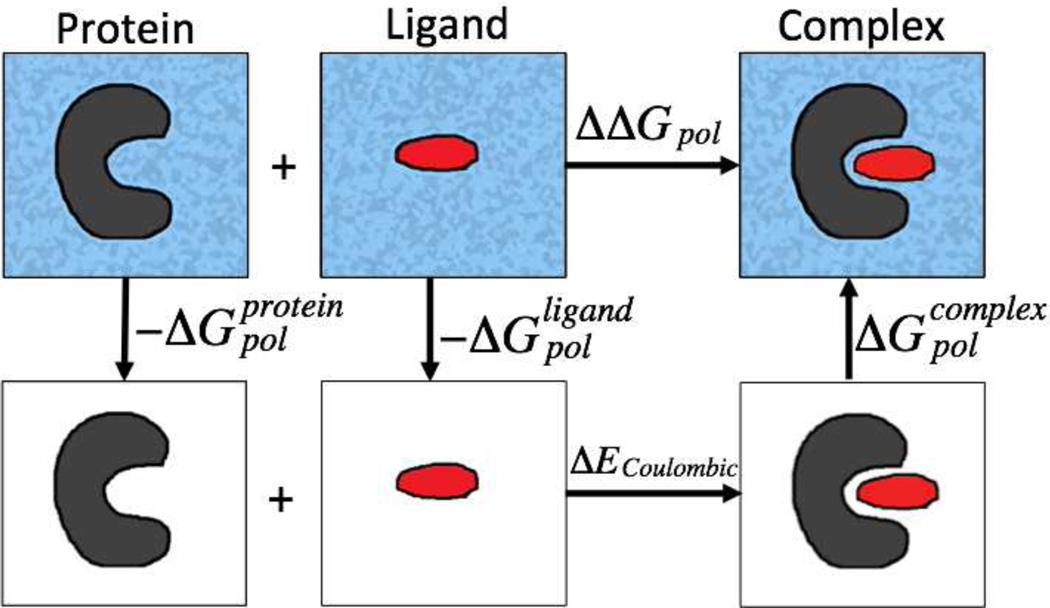
The electrostatic component of the binding free energies can be calculated using the thermodynamics cycle shown in Figure 1. The first step is to transfer the individual protein and ligand from the solvent into vaccum, with the energy costs of and , respectively. The second step is to combine the protein and the ligand into a complex in vaccum. The energy cost would be the difference in Coulombic energies in vaccum (). Calculation of ΔECoulombic is the same for all models. The final step is to solvate the complex into the solvent, where the corresponding energy cost would be . Using this cycle, the electrostatic component of binding free energies (ΔΔGpol) can be computed as
| (3) |
Figure 1.
Illustration of the thermodynamic cycle for the decomposition of the electrostatic component of the binding free energies. The surrounding dielectric medium is shaded for water and is white for vaccum.
The standard deviation of computed ΔΔGpol values from TI is smaller than ± 0.58 kcal/mol.
2.5 Computational Expense
A general performance comparison of the different methods used here to compute the electrostatic solvation free energies of the protein-ligand complexes is given in Table 2. The computations of GBNSR6 and PB are performed on a commodity PC with Intel(R) Core(TM) i7–3770 CPU 3.40GHz processor and 16 GB of RAM memory. All of the explicit solvent free energy TI calculations were performed on Virginia Tech’s HokieSpeed supercomputing cluster (http://www.arc.vt.edu) on a single node that has 12 processors. Expectedly, GB-NSR6 is significantly faster than the PB and explicit models studied here. Our studies show that the computational time required for grid-based calculation of the molecular surface needed by GB-NSR6 is similar to that of MSMS-based calculations; a detailed and exhaustive performance analysis of GBNSR6 based on MSMS molecular surface108 is presented in Ref.13
Table 2.
Average computational time for calculating ΔGpol per complex
| Method | Computational time |
|---|---|
| explicit solvent TI | ≈ 12 hours |
| PB | ≈ 15 minutes |
| GBNSR6 | ≈ 6 seconds |
3 Results and Discussion
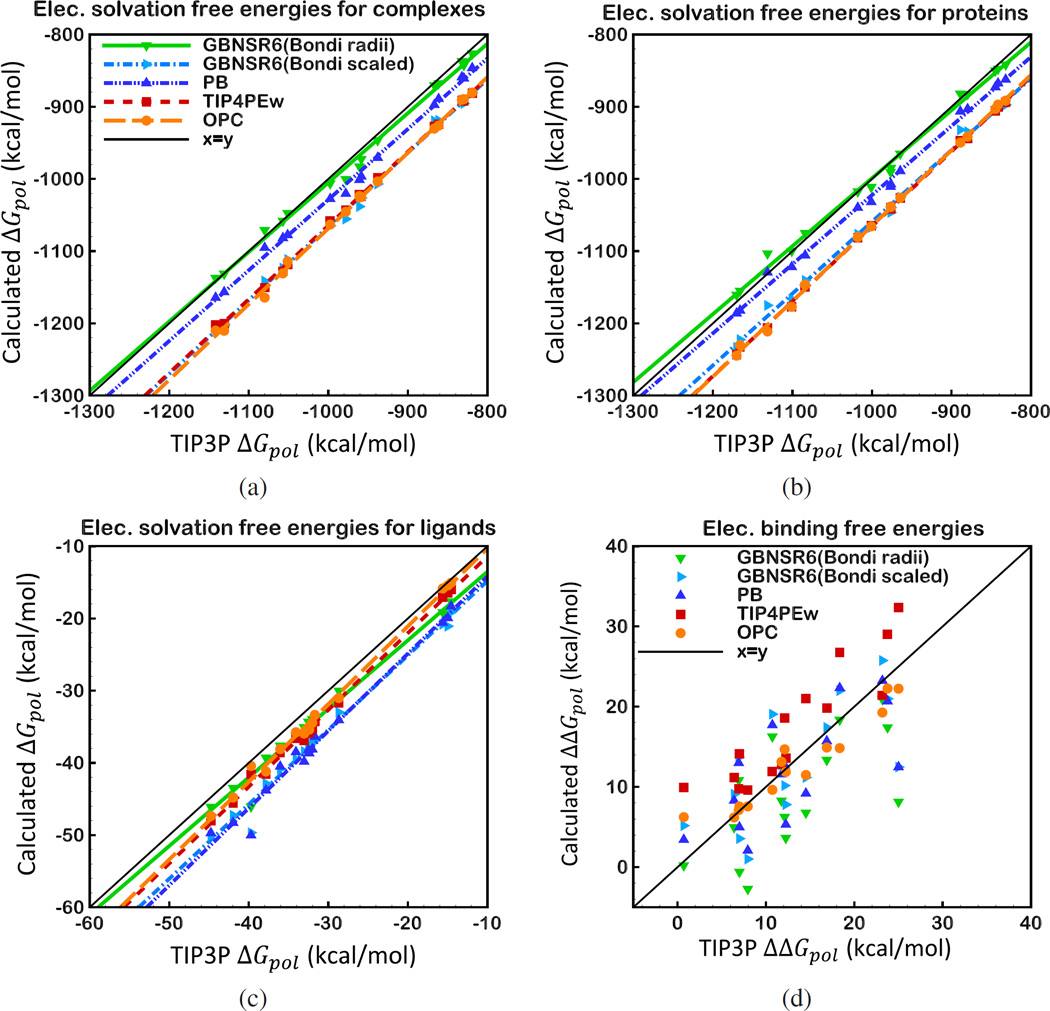
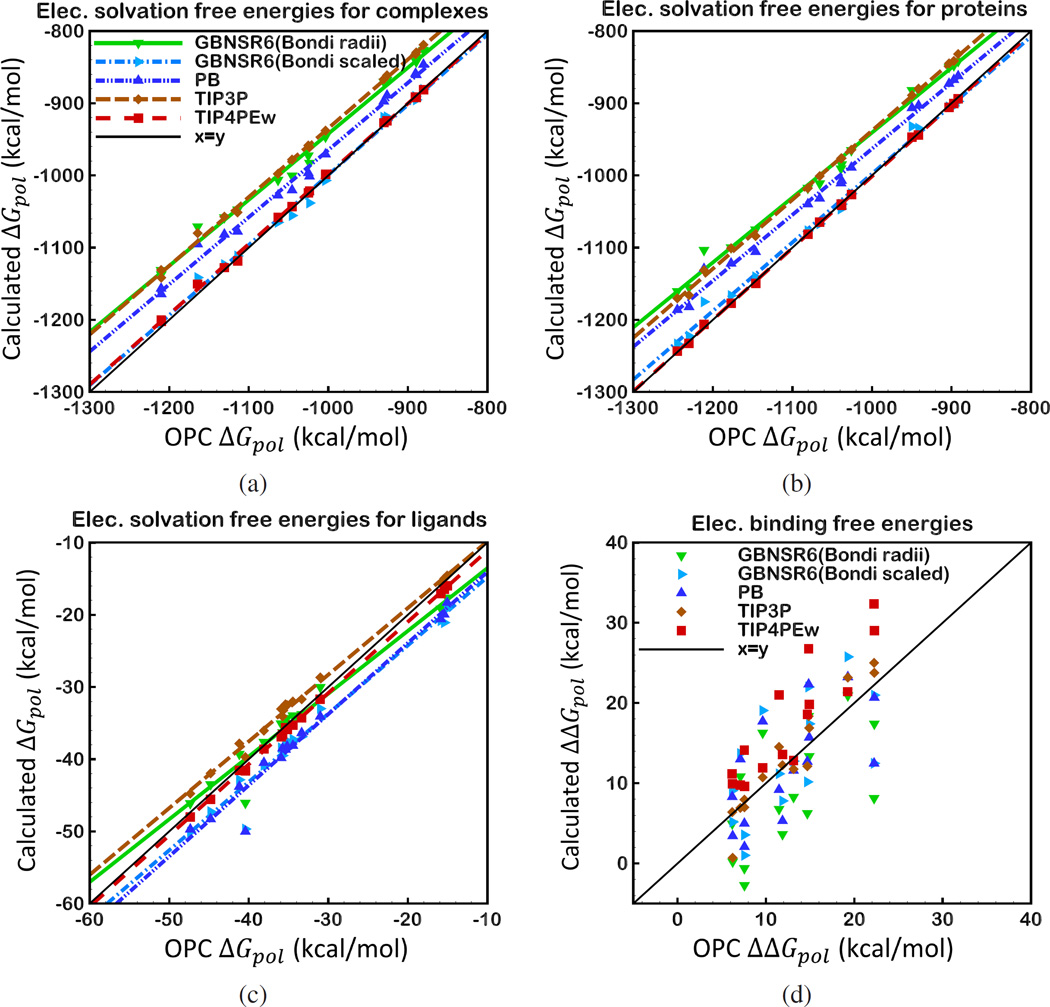
At the moment, comparison with explicit solvent predictions is a natural way to evaluate accuracy of implicit solvent models such as the GB. Among the most commonly used explicit water models, TIP3P is known to give better accuracy in hydration free energy calculations than many other water models tested previously;11 TIP3P has been commonly used for benchmarking implicit solvent models. However, recent developments in building explicit water models yielded a model (OPC water model92) that shows better agreement with experiment in hydration free energy calculations of small molecules. To be consistent with earlier works, we first compare deviation of the implicit models relative to TIP3P as reference, although we do not imply that TIP3P is any better or worse than the other explicit models studied here. We also benchmark computed electrostatic solvation free energy ΔGpol and electrostatic binding free energy ΔΔGpol values against free energy estimates performed in OPC water. Below, we give a brief summary of the agreement of GBNSR6 compared with the explicit solvent and also the numerical PB model. The correlation of ΔGpol and ΔΔGpol values relative to TIP3P and OPC are presented in Figure 2 and Figure 4, respectively. The statistics of ΔGpol and ΔΔGpol values relative to TIP3P are given in Table 3 and Table 4, and relative to OPC are given in Table 6 and Table 7, respectively.
Figure 2.
Correlation between ΔGpol and ΔΔGpol computed by GBNSR6, PB, TIP4PEw and OPC solvent models relative to TIP3P for 15 small protein-ligand complexes specified in Table 1, a) ΔGpol of protein-ligand complexes b) ΔGpol of protein components, c) ΔGpol of ligand components, d) Electrostatic binding free energies, ΔΔGpol
Figure 4.
Correlation between ΔGpol and ΔΔGpol computed by GBNSR6, PB, TIP4PEw and TIP3P solvent models relative to OPC for 15 small protein-ligand complexes specified in Table 1, a) ΔGpol of protein-ligand complexes b) ΔGpol of protein components, c) ΔGpol of ligand components, d) Electrostatic binding free energies, ΔΔGpol
Table 3.
Deviation of (ΔGpol) values (kcal/mol) from those computed with TIP3P explicit solvent model. Atomic radii sets used in implicit solvent estimates are given in parenthesis.
| RMSD | TIP4PEw | OPC | PB(Bondi) | GBNSR6(Bondi) | GBNSR6(Bondi scaled) | |
|---|---|---|---|---|---|---|
| 63.9 | 67.3 | 30.2 | 10.8 | 65.8 | ||
| 65.6 | 64.8 | 24.3 | 10.5 | 59.8 | ||
| 2.9 | 2.2 | 5.9 | 2.8 | 5.6 | ||
Table 4.
Deviation of (ΔΔGpol) values (kcal/mol) from those computed with TIP3P explicit solvent model. Atomic radii sets used in implicit solvent estimates are given in parenthesis.
| TIP4PEw | OPC | PB(Bondi) | GBNSR6(Bondi) | GBNSR6(Bondi scaled) | |
|---|---|---|---|---|---|
| RMSD | 5.30 | 2.47 | 5.14 | 7.04 | 5.31 |
| avg | 4.29 | −0.57 | −0.99 | −4.37 | −0.36 |
| corr. coef. (r2) | 0.81 | 0.91 | 0.52 | 0.47 | 0.50 |
| RMS of worst 20% | 8.36 | 4.41 | 9.2 | 12.56 | 9.57 |
Table 6.
Deviation of (ΔGpol) values (kcal/mol) from those computed with OPC explicit solvent model. Atomic radii sets used in implicit solvent estimates are given in parenthesis.
| RMSD | TIP3P | TIP4PEw | PB(Bondi) | GBNSR6(Bondi) | GBNSR6(Bondi scaled) | |
|---|---|---|---|---|---|---|
| 67.3 | 5.62 | 39.38 | 62.14 | 8.92 | ||
|
|
64.8 | 2.49 | 44.00 | 66.82 | 11.99 | |
| 2.2 | 0.83 | 4.08 | 2.26 | 3.98 | ||
Table 7.
Deviation of (ΔΔGpol) values (kcal/mol) from those computed with OPC explicit solvent model. Atomic radii sets used in implicit solvent estimates are given in parenthesis.
| TIP3P | TIP4PEw | PB(Bondi) | GBNSR6(Bondi) | GBNSR6(Bondi scaled) | |
|---|---|---|---|---|---|
| RMSD | 2.47 | 5.92 | 5.00 | 6.80 | 5.38 |
| avg | −0.57 | 4.86 | −0.42 | −3.80 | 0.20 |
| corr. coef. (r2) | 0.91 | 0.80 | 0.44 | 0.37 | 0.41 |
| RMS of worst 20% | 4.41 | 10.56 | 8.5 | 11.20 | 8.86 |
3.1 Implicit models
ΔGpol Deviations from TIP3P
The computed values of ΔGpol for the protein-ligand complexes and their components obtained from GBNSR6 are compared with the corresponding TIP3P (TI) values. Figure 2(a) and (b) show that among different implicit and explicit models, and values from GBNSR6 agree best with TIP3P ΔGpol values, with RMSD value of 10.76 and 10.53 kcal/mol, respectively (Table 3). Deviation of values computed using GBNSR6 from TIP3P is comparable to that of OPC from TIP3P (Figure 2(c)). The next model to best reproduce TIP3P’s and values is the PB model, although it shows the lowest agreement with TIP3P for , among all the solvent models studied here. At the same time, the better agreement of GBNSR6 with TIP3P values holds for the ligands as well.
ΔΔGpol Deviations from TIP3P
The RMSD error of PB’s ΔΔGpol relative to TIP3P is 5.14 kcal/mol, which is slightly lower than the deviation of TIP4PEw from TIP3P (RMSD = 5.30 kcal/mol) (Table 4). The RMSD error of GBNSR6 based on Bondi radii relative to TIP3P (7.04 kcal/mol) is higher than that of the PB (5.14 kcal/mol). However, the difference between the RMSD errors of GBNSR6 and PB is within the differences in ΔΔGpol between the explicit water model calculations; e. g. the deviation of TIP4PEw or OPC from TIP3P. As can be seen in Table 4, the average error in ΔΔGpol values of GBNSR6 is relatively large (−4.37 kcal/mol) which indicates a systematic error19 relative to TIP3P. The systematic deviation reflects the uncertainties associated with the “best” definition of the dielectric boundary needed by the GB and PB models.109 To confirm the boundary definition origin of the systematic component of the deviation between GBNSR6 and TIP3P electrostatic binding energies, we show that the average error can be virtually eliminated (−0.36 kcal/mol) by a uniform scaling (multiplication) of the Bondi radii101,109 with a single coefficient of 0.968 (Table 4 and Figure 2). The resulting RMSD error of GBNSR6 based on scaled Bondi radii relative to TIP3P is reduced to 5.31 kcal/mol, and becomes comparable to the RMS deviations of TIP4PEw and PB models relative to TIP3P (5.3 kcal/mol). We stress that uniform radii scaling by a single multiplicative factor101,109 is not tantamount to full reoptimization110,111 of the radii intended for best fit against a specific explicit solvent reference. The same scaling of Bondi radii by 0.968 also virtually eliminates the systematic deviation between ΔΔGpol from GBNSR6 and a very different explicit solvent model (OPC) (as we shall see later), and nearly halves the deviation between GBNSR6 and TIP4PEw (from −8.66 kcal/mol to −4.65 kcal/mol).
3.2 Explicit models
ΔGpol Deviations from TIP3P
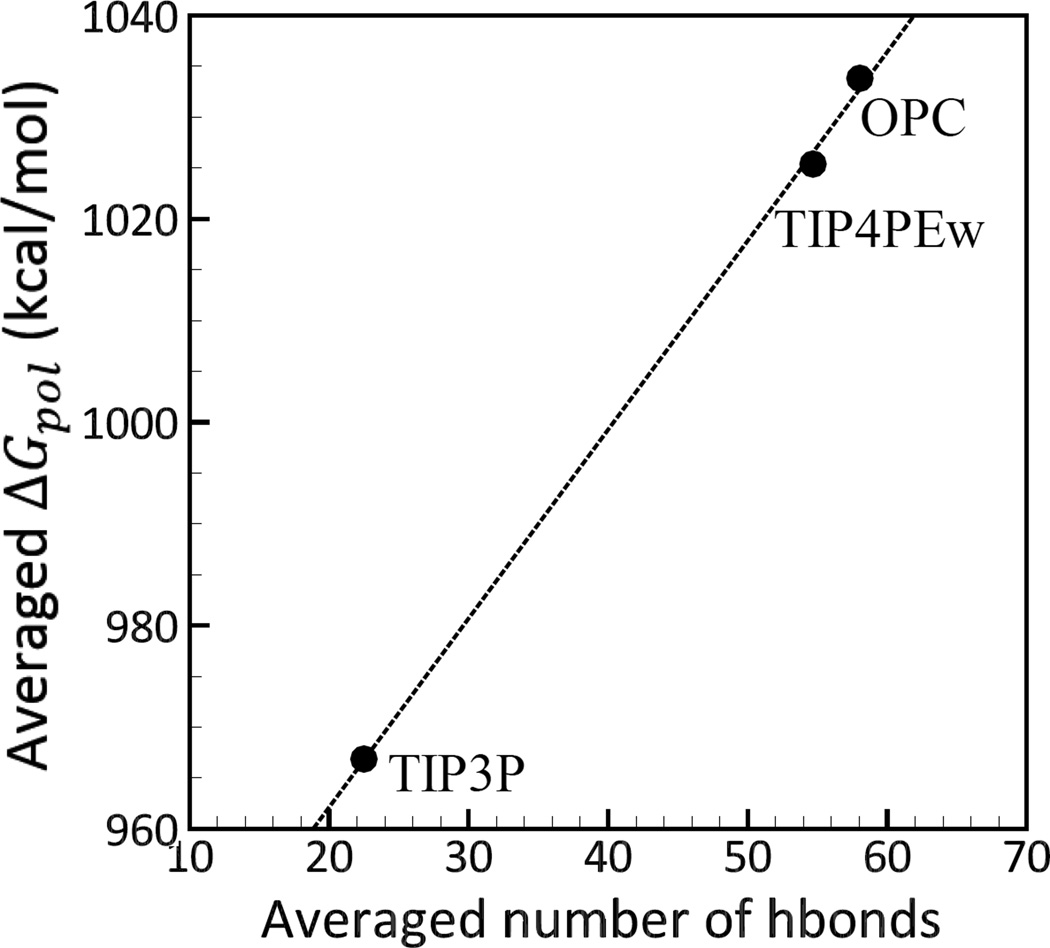
The absolute ΔGpol values calculated using explicit water models differ significantly between themselves, Figure 2 and Table 3. Surprisingly, TIP4PEw and OPC show the lowest agreement with TIP3P in and values compared to implicit models. As seen from Figure 2, ΔGpol values from TIP4PEw and OPC deviate systematically from the TIP3P values, with over-all average errors of −43.96 and −44.45 kcal/mol, and RMSD errors of 52.89 and 53.96 kcal/mol, respectively. Significant discrepancies between the values from OPC and TIP4PEw relative to TIP3P is likely due to stronger electrostatic interactions in OPC and TIP4PEw compared to TIP3P. For instance, while the dipole moment for TIP3P and TIP4PEw are close to each other (2.35D vs 2.32D, respectively) (Table 5), TIP4PEw’s square quadrupole is significantly larger than that of TIP3P (2.16DÅ vs 1.72DÅ) (Table 5). OPC’s dipole and square quadrupole moments are both larger (and closer to Quantum Mechanical predictions, Table 5) than those of TIP3P and TIP4PEw (Table 5). As a result of the differences in the strength of electrostatic interactions,30 the numbers of hydrogen bonds formed by the three explicit water models differ (Figure 3). Specifically, the average number of hydrogen bonds formed between the solute and the solvent for our molecular systems in TIP4PEw is higher than that in TIP3P, and it is the highest for OPC (Figure 3). Solvation free energies increase almost linearly with the average number of hydrogen bonds, Figure 3. As a result, OPC yields the largest ΔGpol values, followed by TIP4PEw and TIP3P. We did not find a correlation between the value of the static dielectric constant and electrostatic solvation free energies for the explicit solvent models studied here.
Table 5.
Three lowest order multipole moments of the water molecule; the values found in explicit water models are compared to experiment (EXP) where available, and liquid phase quantum calculations (QM). Moments are computed relative to oxygen center: dipole (µ), linear (Q0) and square (QT) quadrupole, linear (Ω0) and square (ΩT) octupole.
Figure 3.
Correlation between the electrostatic sol-vation free energies ΔGpol and the number of hydrogen bonds formed between the complexes and the explicit solvent models (TIP3P, TIP4PEw and OPC). The ΔGpol values shown for each model are averages over complexes, and the number of hydrogen bonds represents averages over MD trajectory and over complexes. The hydrogen bond is considered to be formed if the distance between the acceptor (A) and the donor (D) atoms is smaller than 3Å, and angle D-H-A is greater than 135°.114 Connecting lines are shown to guide the eye.
ΔΔGpol Deviations from TIP3P
The RMSD error in ΔΔGpol calculated with OPC and TIP4PEw water models relative to the TIP3P reference are 2.47 and 5.3 kcal/mol, respectively. The RMSD of “worst” (largest deviation) 20% of TIP4PEw’s ΔΔGpol values relative to TIP3P is as large as 8.36 kcal/mol. The deviations of ΔΔGpol from explicit models appear much smaller than the deviations in ΔGpol; however, ΔΔGpol values are relatively much smaller (tens of kcal/mol) than ΔGpol (thousands of kcal/mol), see Figure 2 and Figure 4, which results in large relative errors in ΔΔGpol. For instance, deviation of TIP4PEw from TIP3P can be up to 50% in relative error of ΔΔGpol values. Note that, expectedly, the correlation between ΔΔGpol estimated by solvent models of the same class (e.g. TIP4PEw vs TIP3P) is considerably better than that between very different solvent models such as the GB and TIP3P (Table 4). At the same time, the average deviations between ΔΔGpol computed by explicit models (e.g., TIP4PEw vs TIP3P) is still large, essentially comparable to the deviations between implicit and explicit models. The high correlation between the explicit solvent estimates suggests that the deviations between them may be systematic. This observation is further strengthened by the fact that the systematic error between the implicit and explicit solvent ΔΔGpol can be virtually eliminated by a one-parameter adjustment of the dielectric boundary used in the implicit estimates, as discussed earlier.
Another interesting observation is that the ability of one explicit model to emulate estimates of ΔGpol by another model can be independent of its ability to emulate ΔΔGpol; for instance, among all implicit and explicit models studied here, OPC shows closest agreement with TIP3P in ΔΔGpol while its ΔGpol is furthest from TIP3P.
3.3 ΔGpol and ΔΔGpol Deviations from OPC
Here we investigate deviation of computed electrostatic solvation free energy ΔGpol and electrostatic binding free energy ΔΔGpol values from the values estimated with a recently developed explicit water model, OPC. The GBNSR6’s ΔGpol values based on Bondi radii are systematically shifted from the OPC reference values (Figure 4 (a), (b) and (c)). Yet, deviation of GBNSR6’s ΔΔGpol values from OPC is comparable to that of TIP4PEw from OPC. The same simple uniform scaling (multiplication) of all the radii in the Bondi set by 0.968 introduced earlier also virtually eliminates the systematic deviation between GBNSR6 and OPC in ΔGpol and ΔΔGpol values simultaneously (Figure 4 and Table 6). Interestingly, the radii rescaling, which amounts to the dielectric boundary adjustment, makes the deviation of GBNSR6’s ΔΔGpol from OPC even smaller than that of TIP4PEw from OPC (average error 0.2 kcal/mol vs 4.86 kcal/mol).
Optimizing atomic radii, including uniform scaling, was used earlier101,109–111,115–117 to better reproduce solvation free energies from explicit solvent models. Here the scaling is used mainly to show that the apparent systematic deviation between the GB and explicit solvent is a consequence of a (radii-specific) definition of the dielectric boundary, which can be removed by a uniform “shift” of the latter. Still, achieving a good agreement with 3 different explicit water models simultaneously by a single-parameter uniform scaling of the Bondi radii, one of the smallest and simplest radii sets available in literature, seems noteworthy. Obviously, transferability of the scaled Bondi radii set optimized for the limited set protein-ligand complexes is not guaranteed, which motivates future studies.
To further illustrate the sensitivity of electrostatic binding free energies to the choice of explicit water model, we have also compared the TIP4PEw to the OPC-based estimates, (Figure 4, Table 6 and Table 7). In this comparison, TIP4PEw and OPC water models are more similar to each other than to TIP3P: both are 4-point models parametrized for use in long-range electrostatics interactions, and the polarization correction is included in calculations of heat of vaporization in the parametrization pro-cedure.88,91,92 It is evident from Figure 4 (a), (b) and (c) that ΔGpol values estimated with TIP4PEw and OPC are highly correlated, and the RMSD error of ΔGpol calculations using TIP4PEw relative to OPC is relatively small. Yet, ΔΔGpol values from TIP4PEw substantially deviate from that of OPC (RMSD= 5.92 kcal/mol). Despite the much smaller absolute ΔΔGpol values compared to ΔGpol values, the RMS deviation of TIP4PEw from OPC in ΔΔGpol values is even larger than that in ΔGpol (5.92 kcal/mol for ΔΔGpol vs 5.62 kcal/mol for ). The deviation of TIP4PEw from OPC is even larger than the one between TIP4PEw and TIP3P – water models that are parametrized quite differently (Table 3). Surprisingly, the RMSD error of TIP4PEw relative to OPC is similar to the RMSD error of implicit models (GBNSR6 based on scaled Bondi radii and PB) relative to TIP3P.
4 Conclusion
An accurate representation of the solvent is crucial for realistic and physically rigorous calculations of solvation and protein-ligand binding free energies. In this work, we have evaluated the accuracy of a recently developed generalized Born model, GB-NSR6, in predicting the electrostatic binding free energies ΔΔGpol and electrostatic solvation free energies ΔGpol of small protein-ligand complexes and their components. The estimates from GBNSR6 (and also the standard numerical PB) were compared to the estimates based on three explicit solvent models: RMS deviations of GBNSR6 and the PB from the explicit models were found to be comparable. It was shown that RMS deviation from TIP3P of GBNSR6 (Bondi radii) is comparable to the “error margin” of the explicit models themselves – the differences between the ΔΔGpol values obtained from the explicit models (e.g. TIP4PEw vs TIP3P). Expectedly, the r2 correlation between either of the implicit models and the explicit solvent is lower than between different explicit solvent models. GBNSR6’s ΔΔGpol is closer to estimates based on OPC – a new 4-point rigid water model shown to give higher accuracy in estimation of solvation free energies of small molecules compared to TIP3P.92 A simple uniform scaling of Bondi radii set was shown to bring GBNSR6 RMS deviation essentially within the “error margin” of the three explicit models. The same simple scaling of Bondi radii was shown to virtually eliminate the systematic deviation of GBNSR6 from two out of the three explicit models and reduce the deviation from the third one by about fifty percent. Although the scaled Bondi radii set presented here is not guaranteed to be transferable to protein-ligand systems other than the ones studied here, the fact that a single-parameter uniform scaling of radii significantly improves the agreement of implicit solvent GBNSR6 with all three explicit models simultaneously is noteworthy.
A perhaps unexpected finding is that computed binding and solvation free energies using explicit water models can deviate significantly from each other. Also counterintuitively, lowest RMS deviation from TIP3P’s ΔGpol is achieved by GBNSR6, rather than by the solvent models of the same class such as TIP4PEw and OPC, with RMS errors being up to tens of kcal/mol smaller. The results show that RMS deviations of ΔΔGpol values obtained from different explicit models can be larger than that of ΔGpol, although ΔΔGpol is often orders of magnitude smaller than ΔGpol in absolute values. The discrepancies between results from explicit models indicate the high sensitivity of electrostatic solvation and protein-ligand binding free energy calculations to the choice of explicit water models. Other studies have previously reported that Poisson-Boltzamn based approaches that are accurate in calculating ΔGpol may not be equally successful at predicting ΔΔGpol.17 The high sensitivity of ΔΔGpol values to the choice of explicit water models observed in this work suggests that the sensitivity is not necessarily inherent to implicit solvent models.
In the absence of a “gold standard” explicit solvent model, the large discrepancy in free energy estimates from explicit water models is of paramount concern as it is unclear which of these models is most accurate in these calculations. Some of the water models most commonly used for solvation free energy calculations (e. g. TIP3P) can misrepresent key bulk properties by as much as 250 percent off the experimental values, suggesting the presence of serious physical flaws in these models. At the same time, these explicit models are often treated as accuracy “gold standard” for implicit solvents such as the GB, justified by the idea that implicit solvent models are designed to mimic the effects of explicit models as their higher level predecessors in the hierarchy of approximations leading to these models. In fact, the approach of fitting GB models to explicit solvent models seems reasonable because going directly from GB models to experimental observations with multiple levels of approximation can lead to over-fitting of the GB model parameters. Yet, given the significantly improved accuracy of most recently developed GB models, the strategy is called into question by the large discrepancies between free energy estimates obtained from commonly used explicit models. Efforts to develop more accurate explicit models,92,118,119 or identifying the best among the existing ones, will ultimately help improve accuracy of implicit solvent models as well. In the meantime, adjusting implicit solvent theory to provide best match with several explicit models simultaneously might be the best practical strategy.
A direct comparison with experiment is needed for a decisive accuracy evaluation of explicit models in protein-ligand interactions. However, these comparisons are not straightforward. Protein-ligand complexes are very flexible and have many degrees of freedom introducing large uncertainties in calculations of entropy that make a direct comparison with experiment often difficult. An appealing alternative is to compare the computed binding enthalpies with experimental enthalpies for small host-guest systems.100,120 Many fewer degrees of freedom and relative rigidity of host-guest systems compared to protein-ligand systems make these calculations computationally more straightforward and robust, albeit being still time consuming.23,100
Supplementary Material
Acknowledgments
We would like to thank Ekaterina Katkova for the help with the preparation of the small protein-ligand complexes used in this work. We are also grateful to Marcia O. Fenley and Robert C. Harris for valuable comments and suggestions. This work was supported by the NIH GM076121, and in part by NSF grant CNS-0960081 and the HokieSpeed supercomputer at Virginia Tech.
Footnotes
Supporting Information Available: Amber format topology and coordinate files as well as the corresponding ΔGpol values for 15 small protein-ligand complexes and their components computed using TIP3P, TIP4PEw, OPC, APBS and GBNSR6. This material is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Jorgensen WL. Science. 2004;303:1813–1818. doi: 10.1126/science.1096361. [DOI] [PubMed] [Google Scholar]
- 2.Mobley DL, Dill KA. Structure. 2009;17:489–498. doi: 10.1016/j.str.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shirts MRLMD, Brown SP. In: Free energy calculations in structure-based drug design. 1st. Merz KM, Ringe D, Reynolds CH, editors. Cambridge, New York USA: Lecture Notes in Computer Science; Cambridge University Press; 2010. pp. 61–85. [Google Scholar]
- 4.Skandani AA, Zeineldin R, Al-Haik M. Langmuir. 2012;28:7872–7879. doi: 10.1021/la3011162. PMID: 22545729. [DOI] [PubMed] [Google Scholar]
- 5.Lau AY, Roux BA. Nat Struct Mol Biol. 2011;18:283–287. doi: 10.1038/nsmb.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin Y-L, Meng Y, Jiang W, Roux B. Proc. Natl. Acad. Sci. U. S.A. 2013;110:201214330–201211669. doi: 10.1073/pnas.1214330110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shirts M, Mobley D. Biomolecular Simulations. Vol. 924. Humana Press; 2013. pp. 1271–1311. [Google Scholar]
- 8.Gallicchio E, Levy RM. Recent theoretical and computational advances for modeling protein-ligand binding affinities. In: Christov C, editor. Computational chemistry methods in structural biology. Vol. 85. Academic Press; 2011. pp. 27–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rocklin GJ, Mobley DL, Dill KA. J. Chem. Theory Comput. 2013;9:3072–3083. doi: 10.1021/ct400315q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mobley DL, Dill KA, Chodera JD. J. Phys. Chem. B. 2008;112:938–946. doi: 10.1021/jp0764384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mobley DL, Bayly CI, Cooper MD, Shirts MR, Dill KA. J. Chem. Theory Comput. 2009;5:350–358. doi: 10.1021/ct800409d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shivakumar D, Deng Y, Roux B. J. Chem. Theory Comput. 2009;5:919–930. doi: 10.1021/ct800445x. [DOI] [PubMed] [Google Scholar]
- 13.Aguilar B, Onufriev AV. J. Chem. Theory Comput. 2012;8:2404–2411. doi: 10.1021/ct200786m. [DOI] [PubMed] [Google Scholar]
- 14.Bartlett RJ, Musiał M. Rev. Mod. Phys. 2007;79:291–352. [Google Scholar]
- 15.Helgaker T, Klopper W, Tew DP. Molecular Physics. 2008;106:2107–2143. [Google Scholar]
- 16.Fennell CJ, Kehoe CW, Dill KA. Proc. Natl. Acad. Sci. U. S.A. 2011;108:3234–3239. doi: 10.1073/pnas.1017130108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris RC, Mackoy T, Fenley MO. J. Chem. Theory Comput. 2015;11:705–712. doi: 10.1021/ct5005017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris RC, Mackoy T, Fenley MO. Molecular Based Mathematical Biology. 2013;1:63–74. [Google Scholar]
- 19.Merz KM. J. Chem. Theory Comput. 2010;6:1769–1776. doi: 10.1021/ct100102q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friesner RA, Repasky M, Farid RI. In: Small Molecule Docking. Computational Structural Biology. Schwede T, Peitsch MC, editors. World Scientific: Switzerland; 2008. pp. 469–500. [Google Scholar]
- 21.Kolb P, Irwin J. Current Topics in Medicinal Chemistry. 2009;9:755–770. doi: 10.2174/156802609789207091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilson MK, Zhou HX. Annu. Rev. Biophys. Biomol. Struct. 2007;36:21–42. doi: 10.1146/annurev.biophys.36.040306.132550. [DOI] [PubMed] [Google Scholar]
- 23.Chen W, Chang C-E, Gilson MK. Biophysical Journal. 2004;87:3035–3049. doi: 10.1529/biophysj.104.049494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mobley DL, Graves AP, Chodera JD, McReynolds AC, Shoichet BK, Dill KA. Journal of Molecular Biology. 2007;371:1118–1134. doi: 10.1016/j.jmb.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Onufriev AV, Alexov E. Quarterly Reviews of Biophysics. 2013;46:181–209. doi: 10.1017/S0033583513000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng Y, Roux B. J. Phys. Chem. B. 2009;113:2234–2246. doi: 10.1021/jp807701h. PMID: 19146384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moghaddam S, Inoue Y, Gilson MK. J. Am. Chem. Soc. 2009;131:4012–4021. doi: 10.1021/ja808175m. PMID: 19133781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis ME, McCammon JA. Chemical Reviews. 1990;90:509–521. [Google Scholar]
- 29.Jayaram B, Sharp KA, Honig B. Biopolymers. 1989;28:975–993. doi: 10.1002/bip.360280506. [DOI] [PubMed] [Google Scholar]
- 30.Morokuma K. Accounts Chem Res. 1977;10:294–300. [Google Scholar]
- 31.Qin S, Zhou H-X. Biopolymers. 2007;86:112–118. doi: 10.1002/bip.20708. [DOI] [PubMed] [Google Scholar]
- 32.Anandakrishnan R, Baker C, Izadi S, Onufriev AV. PLoS ONE. 2013;8:e67715. doi: 10.1371/journal.pone.0067715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z, Witham S, Alexov E. Physical Biology. 2011;8:035001. doi: 10.1088/1478-3975/8/3/035001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kundrotas PJ, Alexov E. Biophys. J. 2006;91:1724–1736. doi: 10.1529/biophysj.106.086025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cramer CJ, Truhlar DG. Chem. Rev. 1999;99:2161–2200. doi: 10.1021/cr960149m. [DOI] [PubMed] [Google Scholar]
- 36.Honig B, Nicholls A. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- 37.Beroza P, Case DA. Methods Enzymol. 1998;295:170–189. doi: 10.1016/s0076-6879(98)95040-6. [DOI] [PubMed] [Google Scholar]
- 38.Madura JD, Davis ME, Gilson MK, Wade RC, Luty BA, McCammon JA. Rev. Comp. Chem. 1994;5:229–267. [Google Scholar]
- 39.Gilson MK. Curr. Opin. Struct. Biol. 1995;5:216–223. doi: 10.1016/0959-440x(95)80079-4. [DOI] [PubMed] [Google Scholar]
- 40.Scarsi M, Apostolakis J, Caflisch A. J. Phys. Chem. A. 1997;101:8098–8106. [Google Scholar]
- 41.Luo R, David L, Gilson MK. J. Comput. Chem. 2002;23:1244–1253. doi: 10.1002/jcc.10120. [DOI] [PubMed] [Google Scholar]
- 42.Simonson T. Rep. Prog. Phys. 2003;66:737–787. [Google Scholar]
- 43.Onufriev A, Bashford D, Case DA. Proteins. 2004;55:383–394. doi: 10.1002/prot.20033. [DOI] [PubMed] [Google Scholar]
- 44.Labute P. J. Comput. Chem. 2008;29:1693–1698. doi: 10.1002/jcc.20933. [DOI] [PubMed] [Google Scholar]
- 45.Nicholls A, Honig B. J. Comput. Chem. 1991;12:435–445. [Google Scholar]
- 46.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Proc. Natl. Acad. Sci. U. S.A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bashford D, Karplus M. Biochemistry. 1990;29:10219–10225. doi: 10.1021/bi00496a010. [DOI] [PubMed] [Google Scholar]
- 48.Im W, Beglov D, Roux B. Computer Physics Communications. 1998;111:59–75. [Google Scholar]
- 49.Madura JD, Davist ME, Gilson MK, Wades RC, Luty BA, McCammon JA. Biological Applications of Electrostatic Calculations and Brownian Dynamics Simulations. John Wiley and Sons, Inc; 2007. pp. 229–267. [Google Scholar]
- 50.Altman MD, Bardhan JP, White JK, Tidor B. J. Comput. Chem. 2009;30:132–153. doi: 10.1002/jcc.21027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li B, Cheng X, Zhang Z. SIAM journal on applied mathematics. 2011;71:2093–2111. doi: 10.1137/110826436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simonov NA, Mascagni M, Fen-ley MO. J. Chem. Phys. 2007;127:185105. doi: 10.1063/1.2803189. [DOI] [PubMed] [Google Scholar]
- 53.Feig M, Brooks CL. Curr. Opin. Struct. Biol. 2004;14:217–224. doi: 10.1016/j.sbi.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 54.Still WC, Tempczyk A, Hawley RC, Hendrickson T. J. Am. Chem. Soc. 1990;112:6127–6129. [Google Scholar]
- 55.Hawkins GD, Cramer CJ, Truhlar DG. Chem. Phys. Lett. 1995;246:122–129. [Google Scholar]
- 56.Hawkins GD, Cramer CJ, Truhlar DG. J. Phys. Chem. 1996;100:19824–19836. [Google Scholar]
- 57.Schaefer M, Karplus M. J. Phys. Chem. 1996;100:1578–1599. [Google Scholar]
- 58.Qiu D, Shenkin P, Hollinger F, Still WC. J. Phys. Chem. A. 1997;101:3005–3014. [Google Scholar]
- 59.Edinger S, Cortis C, Shenkin P, Fries-ner R. J. Phys. Chem. B. 1997;101:1190–1197. [Google Scholar]
- 60.Jayaram B, Liu Y, Beveridge DL. J. Chem. Phys. 1998;109:1465–1471. [Google Scholar]
- 61.Ghosh A, Rapp CS, Friesner RA. J. Phys. Chem. B. 1998;102:10983–10990. [Google Scholar]
- 62.Bashford D, Case DA. Annu. Rev. Phys. Chem. 2000;51:129–152. doi: 10.1146/annurev.physchem.51.1.129. [DOI] [PubMed] [Google Scholar]
- 63.Lee MS, Salsbury FR, Brooks CL. J. Chem. Phys. 2002;116:10606–10614. [Google Scholar]
- 64.Felts AK, Harano Y, Gallicchio E, Levy RM. Proteins. 2004;56:310–321. doi: 10.1002/prot.20104. [DOI] [PubMed] [Google Scholar]
- 65.Romanov AN, Jabin SN, Mar-tynov YB, Sulimov AV, Grigoriev FV, Sulimov VB. J. Phys. Chem. A. 2004;108:9323–9327. [Google Scholar]
- 66.Dominy BN, Brooks CL. J. Phys. Chem. B. 1999;103:3765–3773. [Google Scholar]
- 67.David L, Luo R, Gilson MK. J. Comput. Chem. 2000;21:295–309. doi: 10.1002/jcc.10120. [DOI] [PubMed] [Google Scholar]
- 68.Tsui V, Case D. J. Am. Chem. Soc. 2000;122:2489–2498. [Google Scholar]
- 69.Calimet N, Schaefer M, Simonson T. Proteins: Structure, Function, and Genetics. 2001;45:144–158. doi: 10.1002/prot.1134. [DOI] [PubMed] [Google Scholar]
- 70.Spassov VZ, Yan L, Szalma S. J. Phys. Chem. B. 2002;106:8726–8738. [Google Scholar]
- 71.Simmerling C, Strockbine B, Roit-berg AE. J. Am. Chem. Soc. 2002;124:11258–11259. doi: 10.1021/ja0273851. [DOI] [PubMed] [Google Scholar]
- 72.Wang T, Wade R. Proteins. 2003;50:158–169. doi: 10.1002/prot.10248. [DOI] [PubMed] [Google Scholar]
- 73.Nymeyer H, Garcia AE. Proc. Natl. Acad. Sci. U. S.A. 2003;100:13934–13949. doi: 10.1073/pnas.2232868100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gallicchio E, Levy RM. J. Comput. Chem. 2004;25:479–499. doi: 10.1002/jcc.10400. [DOI] [PubMed] [Google Scholar]
- 75.Lee MC, Duan Y. Proteins. 2004;55:620–634. doi: 10.1002/prot.10470. [DOI] [PubMed] [Google Scholar]
- 76.Onufriev A. In: Continuum Electrostatics Solvent Modeling with the Generalized Born Model. 1st. Feig M, editor. Wiley: USA; 2010. pp. 127–165. [Google Scholar]
- 77.Srinivasan J, Cheatham TE, Cieplak P, Kollman PA, Case DA. J. Am. Chem. Soc. 1998;120:9401–9409. [Google Scholar]
- 78.Gohlke H, Kiel C, Case DA. J Mol Biol. 2003;330:891–913. doi: 10.1016/s0022-2836(03)00610-7. [DOI] [PubMed] [Google Scholar]
- 79.Dong F, Zhou H-X. Proteins. 2006;65:87–102. doi: 10.1002/prot.21070. [DOI] [PubMed] [Google Scholar]
- 80.Aguilar B, Anandakrishnan R, Rus-cio JZ, Onufriev AV. Biophys. J. 2010;98:872–880. doi: 10.1016/j.bpj.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Svrcek-Seiler A. Personal communication. 2001 [Google Scholar]
- 82.Grycuk T. J. Chem. Phys. 2003;119:4817–4826. [Google Scholar]
- 83.Tjong H, Zhou HX. J. Phys. Chem. B. 2007;111:3055–3061. doi: 10.1021/jp066284c. [DOI] [PubMed] [Google Scholar]
- 84.Mongan J, Svrcek-Seiler A, Onufriev A. J. Chem. Phys. 2007;127:185101–185101. doi: 10.1063/1.2783847. [DOI] [PubMed] [Google Scholar]
- 85.Guillot B. J Mol Liq. 2002;101:219–260. [Google Scholar]
- 86.Wu Y, Tepper HL, Voth GA. J Chem Phys. 2006;124:024503. doi: 10.1063/1.2136877. [DOI] [PubMed] [Google Scholar]
- 87.Vega C, Abascal JLF, Conde MM, Aragones JL. Faraday Discuss. 2009;141:251–276. doi: 10.1039/b805531a. [DOI] [PubMed] [Google Scholar]
- 88.Vega C, Abascal JLF. Phys Chem Chem Phys. 2011;13:19663–19688. doi: 10.1039/c1cp22168j. [DOI] [PubMed] [Google Scholar]
- 89.Mahoney MW, Jorgensen WL. J Chem Phys. 2000;112:8910–8922. [Google Scholar]
- 90.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 91.Horn HW, Swope WC, Pitera JW, Madura JD, Dick TJ, Hura GL, Head-Gordon T. J. Chem. Phys. 2004;120:9665–9678. doi: 10.1063/1.1683075. [DOI] [PubMed] [Google Scholar]
- 92.Izadi S, Anandakrishnan R, Onufriev AV. J Phys Chem Lett. 2014;5:3863–3871. doi: 10.1021/jz501780a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lin Y-L, Aleksandrov A, Simonson T, Roux B. J. Chem. Theory Comput. 2014;10:2690–2709. doi: 10.1021/ct500195p. [DOI] [PubMed] [Google Scholar]
- 94.Anandakrishnan R, Aguilar B, Onufriev AV. Nucleic acids research. 2012;40:W537–W541. doi: 10.1093/nar/gks375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kirmizialtin S, Elber R. J. Phys. Chem. B. 2010;114:8207–8220. doi: 10.1021/jp911992t. PMID: 20518549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Case DA, Cheatham TE, Dar-den T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Case D, et al. San Francisco: University of California; 2012. [Google Scholar]
- 98.Lee B, Richards FM. J. Mol. Biol. 1971;55:379. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 99.Sigalov G, Fenley A, Onufriev A. J. Chem. Phys. 2006;124:124902. doi: 10.1063/1.2177251. [DOI] [PubMed] [Google Scholar]
- 100.Fenley AT, Henriksen NM, Mud-dana HS, Gilson MK. J. Chem. Theory Comput. 2014;10:4069–4078. doi: 10.1021/ct5004109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang LY, Gallicchio E, Friesner RA, Levy RM. J. Comput. Chem. 2001;22:591–607. [Google Scholar]
- 102.Bondi A. J. Phys. Chem. 1964;68:441–451. [Google Scholar]
- 103.Cai Q, Ye X, Wang J, Luo R. J. Chem. Theory Comput. 2011;7:3608–3619. doi: 10.1021/ct200389p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Case D, et al. AMBER 2015. San Francisco: University of California; 2015. [Google Scholar]
- 105.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Proc. Natl. Acad. Sci. U. S.A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Roe DR, Okur A, Wickstrom L, Hor-nak V, Simmerling C. J. Phys. Chem. B. 2007;111:1846–1857. doi: 10.1021/jp066831u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tan C, Yang L, Luo R. J. Phys. Chem. B. 2006;110:18680–18687. doi: 10.1021/jp063479b. PMID: 16970499. [DOI] [PubMed] [Google Scholar]
- 108.Sanner MF, Olson AJ, Spehner JC. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 109.Onufriev AV, Aguilar B. Journal of Theoretical and Computational Chemistry. 2014;13:1440006. doi: 10.1142/S0219633614400069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nina M, Beglov D, Roux B. J. Phys. Chem. B. 1997;101:5239–5248. [Google Scholar]
- 111.Swanson JMJ, Mongan J, McCam-mon JA. J. Phys. Chem. B. 2005;109:14769–14772. doi: 10.1021/jp052883s. [DOI] [PubMed] [Google Scholar]
- 112.Gregory JK, Clary DC, Liu K, Brown MG, Saykally RJ. Science. 1997;275:814–817. doi: 10.1126/science.275.5301.814. [DOI] [PubMed] [Google Scholar]
- 113.Coutinho K, Guedes R, Cabral BC, Canuto S. Chem. Phys. Lett. 2003;369:345–353. [Google Scholar]
- 114.Roe DR, Cheatham T., III J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 115.Banavali NK, Roux B. J. Phys. Chem. B. 2002;106:11026–11035. [Google Scholar]
- 116.Rashin AA, Honig B. J. Phys. Chem. 1985;89:5588–5593. [Google Scholar]
- 117.Chocholousova J, Feig M. J. Phys. Chem. B. 2006;110:17240–17251. doi: 10.1021/jp0627675. PMID: 16928023. [DOI] [PubMed] [Google Scholar]
- 118.Wang LP, Martinez TJ, Pande VS. J Phys Chem Lett. 2014;5:1885–1891. doi: 10.1021/jz500737m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fennell CJ, Li L, Dill KA. J. Phys. Chem. B. 2012;116:6936–6944. doi: 10.1021/jp3002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wickstrom L, He P, Gallicchio E, Levy RM. J. Chem. Theory Comput. 2013;9:3136–3150. doi: 10.1021/ct400003r. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.