Abstract
Lung cancer is a leading cause of cancer-related mortality worldwide and in the People’s Republic of China. Recently, the pathological proportions of the various forms of lung cancer have changed. A shift to a preponderance of adenocarcinoma at the expense of squamous cell carcinoma is observable. Treatment decisions have historically been based on tumor histology, and evolution of our molecular understanding of cancer has led to development of targeted therapeutic agents. It is essential to further understand mutations that drive cancer development (driver mutations) in relevant genes and their effects on cancer cell proliferation and survival. The epidemiology of lung cancer in the People’s Republic of China has been extensively reviewed elsewhere. However, molecular epidemiological data from mainland China are scarce. Consequently, we herein review the prevalence of driver mutations in Chinese patients.
Keywords: lung cancer, driver mutation, prevalence, EGFR, EML4-ALK, KRAS, ROS1, PIK3CA, BRAF, RET, HER2
Introduction
As reported in the Chinese Cancer Registry Annual Report in 2012,1 lung cancer is the leading cause of cancer-related death in the People’s Republic of China. Mortality from this cancer was 45.57 per 100,000 of the national population, being 61.00 for 100,000 males and 29.77 for 100,000 females. Male mortality was 2.05-fold that of female mortality. A shift in the relative frequency of lung cancer type from squamous cell cancer to adenocarcinoma is evident both domestically and overseas in countries where numbers of male smokers remain high.2 Most Chinese women do not smoke. However, in most areas, the frequency of smoking is increasing among women. Wang et al3 studied 32,845 patients with newly diagnosed lung cancer between 1998 and 2007 (Table 1). In terms of cancer subtype, the proportion of squamous cell cancers decreased annually from 30.41% (333/1,095) in 1998 to 24.16% (638/2,641) in 2007, while the proportion of adenocarcinomas increased from 42.83% (469/1,095) to 46.80% (1,236/2,641) over the same period. In women, the decline was more marked; the proportions of squamous cell cancer and adenocarcinoma were changed to 14.77% (925/6,262) and 60.83% (3,809/6,262), respectively, between 1998 and 2007. Alamoudi et al4 and Chang et al5 also found that the principal subtype of lung cancer that was histologically categorized apparently changed from squamous cell cancer to adenocarcinoma. However, the reason for this change requires further assessment.
Table 1.
Distribution of pathological types of lung cancer according to gender in Beijing 1998–2007
| Pathological type | Male
|
Female
|
||||||
|---|---|---|---|---|---|---|---|---|
| Patients (n) | Ratio (%) | Crude incidence rate (per 100,000) | Adjusted rate (per 100,000) | Patient (n) | Ratio (%) | Crude incidence rate (per 100,000) | Adjusted rate (per 100,000) | |
| AD | 4,276 | 36.68 | 12.25 | 8.13 | 3,809 | 60.83 | 11.27 | 7.12 |
| SCC | 4,096 | 35.13 | 11.73 | 7.69 | 925 | 14.77 | 2.74 | 1.62 |
| SCLC | 1,630 | 13.98 | 4.67 | 3.08 | 580 | 9.26 | 1.72 | 1.04 |
Abbreviations: SCC, squamous cell cancer; AD, adenocarcinoma; SCLC, small cell lung cancer.
Over the past few decades, translational research has clarified many of the molecular mechanisms for development, growth, and metastasis of lung cancer. Driver mutations occur in genes encoding signaling proteins critical in terms of cellular proliferation and survival. In lung adenocarcinomas, such mutations occur in the EGFR, EML4-ALK, KRAS, ROS1, PIK3CA, BRAF, RET, and HER2 genes. Tyrosine kinase inhibitors (TKIs) targeting these genes have been developed. Such drugs include gefitinib, erlotinib, and crizotinib, which were used in the IPASS, BR.21, and Profile 1007 clinical trials, respectively.6–9 Many other new compounds, including the MET inhibitor tivantinib10 and the phosphatidylinositol 3-kinase inhibitor XL147,11 which directly or indirectly target these driver mutations, have been developed and are undergoing clinical trials. Non-smoking-related adenocarcinoma of the lung has its peculiar epidemiologic, clinical, and biological characteristics. Many experts, including Sun et al12 and Yano et al,13 have named it as a distinct entity. In recent years, many studies have shown that EGFR, EML4-ALK, HER2, KRAS, and BRAF mutations are mainly found in non-smoking-related adenocarcinoma.
The People’s Republic of China has the highest population of any country. However, molecular epidemiological data from mainland China remain scarce. A comprehensive review of the prevalence of driver mutations is essential for development of personalized therapy targeting non-small-cell lung cancer (NSCLC). Consequently, we herein review the prevalence of driver mutations and the epidemiology of resistance mechanisms in Chinese patients with NSCLC.
Prevalence of driver mutations in lung cancers
EGFR mutations
Currently, a promising treatment strategy for advanced NSCLC features small molecules targeting epidermal growth factor receptor (EGFR) mutations.14 The prevalence of EGFR mutations in pulmonary adenocarcinoma among female non-smoking Asian patients was first revealed by a subgroup analysis of the Iressa Survival Evaluation in Lung Cancer trial.15 Further, several Phase III studies7,16,17 have compared first-line EGFR TKIs such as gefitinib and erlotinib with double platinum-based chemotherapy, used to treat NSCLC patients harboring activating EGFR mutations. It was found that EGFR TKIs were associated with significantly higher response rates and afforded longer progression-free survival than did traditional chemotherapy in selected populations.
Currently, adenocarcinoma of the lung in non-smokers is recognized as a distinct disease entity because of its peculiar epidemiological, clinical, and biological characteristics; many studies have comprehensively analyzed the major known driver mutations in female non-smoking Asian patients with pulmonary adenocarcinoma. EGFR mutations are associated with smoking status and particular histological subtypes of disease. Wu et al18 performed a meta-analysis of up-to-date individual patient data from six medical centers of mainland China. Of 506 patients with NSCLC, the EGFR mutation rate in smokers was 15.1%, and lower than in non-smokers (45.5%; P<0.00001). Male patients showed a lower mutation rate than female patients (23.1% versus 42.9%; P<0.0001). An et al19 found that EGFR mutation rates varied by smoking status and the histological lung cancer subtype; these authors analyzed differences in EGFR mutations among patients varying in these parameters. In non-smokers with adenocarcinoma, EGFR was the most frequently altered gene (in 49.8%, 114/229). The mutation rates of EGFR in patients with adenocarcinoma and squamous cell cancer were 40.3% (140/347) and 4.4% (6/144), respectively.
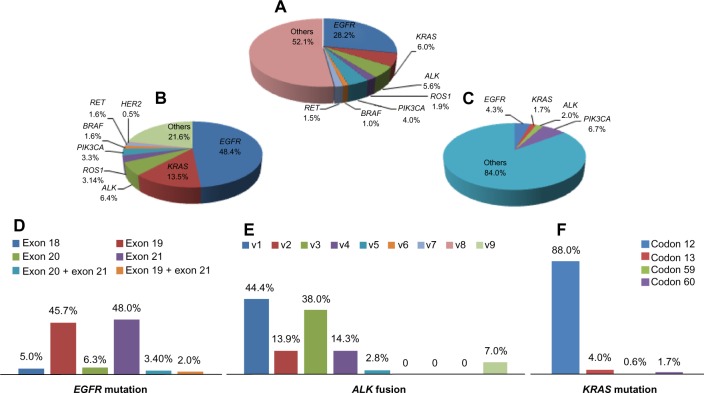
In summary, EGFR mutations occur at high frequencies in female patients, non-smokers, those of Asian ethnicity, and patients with adenocarcinoma. Earlier Chinese studies19–27 recorded the frequencies of EGFR mutations in patients with NSCLC, adenocarcinoma, and squamous cell cancer (Table 2). Such mutations include exon 18 G719A, G719V, and G719D; exon 19 E746–A750 del, E746–S752 del, and L747–A750 (751, 753) del; exon 20 T790M and R776H; and exon 21 L858R, L858M, and L861R. As reported earlier, most EGFR mutations occur in exons 19 and 21. We have summarized all previous Chinese data in Figure 1D.
Table 2.
Frequency of EGFR and KRAS mutation and ALK rearrangements in different subgroups
| Study | Techniques |
EGFR mutation
|
KRAS mutation
|
ALK rearrangements
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AD | SCC | NSCLC | AD | SCC | NSCLC | Study | Techniques | AD | SCC | NSCLC | ||
| Ren et al21 | Sequencing | 70.2% (73/104) | 1.9% (2/104) | RT-PCR | 9.6% (10/104) | |||||||
| Sun et al44 | Sequencing | 78.8% (41/52) | 1.9% (1/52) | RT-PCR | 5.8% (3/52) | |||||||
| An et al19 | Sequencing | 40.3% (140/347) | 4.4% (6/144) | 28.4% (147/517) | 7.1% (24/340) | 1.5% (2/132) | 5.4% (27/498) | RT-PCR | 7.7% (10/130) | 4.1% (4/93) | 6.3% (15/239) | |
| Li et al20 | Sequencing | 43.5% (100/230) | 16.5% (38/230) | RT-PCR | 3.0% (7/230) | |||||||
| Xu et al67 | ARMS | 48.1% (149/310) | 5.2% (16/310) | RT-PCR | 7.4% (23/310) | |||||||
| Li et al61 | Sequencing | 44.2% (42/95) | 5.2% (5/96) | 24.5% (51/208) | 4.2% (4/95) | 1.0% (1/96) | 3.37% (7/208) | RT-PCR | 6.32% (6/95) | 0 (0/96) | 3.37% (7/208) | |
| Shaozhang et al62 | Sequencing | 45.8% (33/72) | 43.0% (44/102) | 11.1% (8/72) | 16,7% (17/102) | RT-PCR | 6.9% (5/72) | 7.1% (1/14) | 8.0% (8/102) | |||
| Sun et al63 | Sequencing | 32.5% (26/80) | 8.75% (7/80) | Zhang et al23 | RACE-coupled sequencing | 16.13% (10/62) | 6.9% (2/29) | 11.6% (12/103) | ||||
| Wu et al24 | Sequencing | 57.3% (90/157) | 4.69% (3/64) | 40.8% (96/235) | 5.03% (8/159) | 1.56% (1/64) | 3.80% (9/237) | Wong et al42 | RT-PCR | 5.3% (11/209) | 0/34 | 4.9% (13/266) |
| Xu et al26 | Sequencing | 47.5% (317/667) | 41.0% (353/861) | 9.9% (151/667) | 1.5% (3/194) | 8.0% (69/861) | ||||||
| Gao et al64 | ARMS | 40.9% (38/93) | 36.7% (44/120) | 10.4% (8/77) | 8.7% (9/104) | Wang et al43 | FISH, IHC | 10.5% (10/95) | 5.6% (1/18) | 9.7% (11/113) | ||
| Li et al34 | HRM | 4.3% (3/69) | 3.1% (1/32) | 5.8% (6/103) | An et al19 | RT-PCR | 5.4% (22/409) | |||||
| Guan et al35 | HRM | 5.06% (98/1935) | Zhang et al41 | FISH, IHC, qPCR | 5.6% (19/341) | 0/112 | 4.2% (20/473) | |||||
| Wang et al22 | ARMS | 5.8% (16/103) | ||||||||||
| Sun et al27 | Sequencing | 4.7% (14/301) | ||||||||||
| Total | 48.4% (675/1,395) | 4.3% (9/208) | 28.2% (570/2,021) | 13.5% (264/1,955) | 1.7% (12/422) | 6.0% (274/4,585) | 6.7% (114/1,700) | 2.0% (8/396) | 5.6% (108/1,913) | |||
Abbreviations: NSCLC, non-small-cell lung cancer; AD, adenocarcinoma; SCC, squamous cell carcinoma; ARMS, amplification refractory mutation system; HRM, high resolution melting analysis; RT-PCR, reverse transcriptase polymerase chain reaction; qPCR, quantitative polymerase chain reaction; IHC, immunohistochemistry; FISH, fluorescence in situ hybridization; RACE, rapid cloning of cDNA ends.
Figure 1.
(A) Frequency of driver mutation in Chinese non-small-cell lung cancer, (B) frequency of driver mutations in Chinese lung adenocarcinoma, (C) frequency of driver mutations in Chinese squamous cell carcinoma, (D) frequency of EGFR mutation in different subtypes, (E) frequency of ALK fusion in different subtypes, and (F) frequency of KRAS mutation in different subtypes.
Abbreviation: EGFR, epidermal growth factor receptor.
KRAS mutations
The KRAS oncogene is often mutated during carcinogenesis. Many reports have linked KRAS mutations to lung cancer.28,29 However, the frequency of KRAS mutations is lower in Eastern Asia than in the West. The KRAS mutation frequency is 30% or over in NSCLC patients from the USA and some other Western countries, but less than 10% in patients from Japan, Korea, Taiwan, and Hong Kong (Table 2).20,24,25,30–34 Other research19,26,35 showed that KRAS mutations occurred more often in males than females, in patients with adenocarcinoma compared with other forms of lung cancer, and in smokers compared with non-smokers.
Several KRAS mutations are known. Codon 12 mutations include G12A, G12C, G12D, G12R, G12S, G12V, G12V/C, and G12V/D. Codon 13 mutations include G13C and G13D. Guan et al35 found KRAS mutations in codons 12 (81.32%, 74/1,935), 13 (5.49%, 5/1,935), 59 (1.10%, 1/1,935), 60 (3.30%, 3/1,935), and both 12 and 13 (5.49%, 5/1,935). The only codon 59 mutation was G59G. The codon 60 mutations were Q60H and Q60L. Mutations in both codons 12 and 13 consisted of G12D/G13D (n=3), G12C/G13C (n=1), and G12V/G13C (n=1). An et al19 enrolled 524 patients and found that 27 had KRAS mutations, all in codon 12, including G12C (44.4%, 12/27), G12V (18.5%, 5/27), G12D (18.5%, 5/27) and others (18.5%, 5/27).
In summary, most studies support the idea that KRAS mutations occur in Chinese smokers with adenocarcinoma. We analyzed studies19,22–27,34,35 performed in Chinese patients with lung cancer to identify the subtypes and frequencies of KRAS mutations, and found that most occurred in codons 12, 13, 59, and 60, especially codon 12 (88%; Figure 1F). Further, EGFR mutations were never found in tumors with KRAS mutations, suggesting that these mutations are mutually exclusive.
ALK fusion genes
ALK is a receptor tyrosine kinase that is not normally expressed in the lung.36 Fusions of ALK with an upstream gene, EML4, were found in NSCLC patients in 2007.37 The frequency of EML4-ALK translocations ranges from 3% to 7% in unselected patients with NSCLC.38–40 Detection methods include reverse-transcriptase polymerase chain reaction (RT-PCR), fluorescence in situ hybridization, and immunohistochemistry. As with EGFR mutations, the frequency of EML4-ALK fusions is elevated in patients with adenocarcinoma, in younger adult patients, and in those who have never smoked (<100 cigarettes in a lifetime) or who are light smokers (≤15 pack-years). EML4-ALK translocations are generally found in tumors wild-type for EGFR and KRAS.38 The EML4-ALK translocation occurs in a low proportion of lung cancers, ie, in 4%–11.6% of unselected Asian patients with early stage NSCLC.23,41–43 However, only small numbers of EML4-ALK-positive patients have been studied, because patient enrolment in most cited studies was non-selective. Thus, the relationship between gene fusion and clinical and pathological profiles requires further study.
A careful review of the literature in the Chinese language revealed that ALK fusion proteins are frequently present in patients with lung cancer (Table 2). This table lists the frequencies of ALK fusions in patients with NSCLC, adenocarcinoma, and squamous cell cancer, respectively. In summary, the results show that risk factors for development of the EML4-ALK fusion are female gender, adenocarcinoma, non-smoking status, and relative youth. Patients with EML4-ALK fusions are always wild-type for EGFR and KRAS.
EML4-ALK fusions are caused by various small inversions within the short arm of chromosome 2.38–40 At least nine variants have been identified.38 Exons 20–29 of ALK may be fused to EML4 exon 13 (variant 1, V1), exon 20 (V2), exon 6 (V3a), exon 6 with an 11 amino acid insertion (V3b), exon 14 with an additional 11 nucleotide insertion of unknown origin at nucleotide 50 of exon 20 of ALK (V4), exon 2 (V5), exon 13 (V6), exon 14 with the fusion beginning at nucleotide 13 of exon 20 of ALK (V7), exon 15 (also termed “V4” [V8]), and exon 18 (also termed “V5” [V9]). The details can be found in a review by Horn and Pao.38 For example, Wong et al42 used RT-PCR to show that the frequencies of the fusion variants 1, 2, 3, and 9 were 2/13, 2/13, 8/13, and 1/13, respectively, in Chinese patients with lung cancer. Zhang et al23 found that the frequencies of fusion variants 1, 2, 3, 5, and 9 were 4/12, 1/12, 3/12, 1/12, and 3/12, respectively. Sun et al44 found that variants 1, 2, and 3 were each present alone in individual Chinese patients with NSCLC using RT-PCR. Li et al20 found that the frequencies of EML4-ALK variants 1, 2, 3, and 4 were 2/7, 1/7, 3/7, and 1/7, in Chinese patients with lung adenocarcinoma using RT-PCR. Similarly, Li et al61 found that the frequencies of fusion variants 1, 2, 3, 5, and 9 were 14/22, 3/22, 3/22, 1/22, and 1/22, respectively. No Chinese patient has been found to be positive for variant 6, 7, or 8 (Figure 1E).
ROS1 mutation
ROS1 rearrangements were initially identified in a human glioblastoma cell line.46 Such rearrangements have been identified in approximately 1%–2% of NSCLC patients using different genotyping techniques (Table 3).47,48 Several recent studies used whole-transcriptome and whole-genome sequencing to detect ROS1 rearrangements, and several novel fusion partners were identified.47,48 Such methods are not yet readily applicable in the clinic. Therefore, ROS1 screening strategies have been largely informed by experience with ALK testing, for which the three most commonly used methods of detection are fluorescence in situ hybridization, RT-PCR, and immunohistochemistry. ROS1 fusions appear to be more common in patients with adenocarcinoma. Cai et al48 found that the ratios of females to males and never-smokers to smokers in a ROS1 fusion-positive group were 5:3. No significant differences in age (P=0.866), gender (P=0.479), smoking history (P=1.0), histological tumor type (P=0.148), or pathological stage (P=0.475) were evident between ROS1 fusion-positive and fusion-negative patients.
Table 3.
Frequency of new driver mutations in non-small-cell lung cancers
| Study | Techniques | Prevalence
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
ROS1
|
PIK3CA
|
BRAF
|
RET
|
HER2
|
|||||||||
| AD | SCC | NSCLC | AD | SCC | NSCLC | AD | SCC | NSCLC | AD | NSCLC | AD | ||
| Xu et al26 | Sequencing | 2.7% (18/667) | 7.2% (14/194) | 3.7% (32/861) | 0.7% (6/861) | ||||||||
| An et al19 | HRM | 4.2% (13/307) | 5.8% (7/121) | 4.4% (20/452) | 2.3% (7/307) | 0% (0/121) | 1.5% (7/452) | ||||||
| Ren et al21 | Sequencing | 1.9% (2/104) | 1.9% (2/104) | 0% (0/104) | |||||||||
| Sun et al44 | Sequencing | 7.6% (4/52) | 0 (0/52) | 3.8% (2/52) | |||||||||
| Li et al20 | Sequencing | 3.5% (8/230) | 3.0% (7/230) | 0% (0/230) | |||||||||
| Zheng et al66 | Sequencing | 2.2% (9/409) | 1.5% (6/409) | ||||||||||
| Xu et al67 | Sequencing | 0% (0/310) | 1.6% (5/310) | ||||||||||
| Cai et al48 | RT-PCR | 3.0% (7/231) | 0 (0/19) | 2.0% (8/392) | |||||||||
| Rimkunas et al47 | FISH, IHC | 3.3% (8/246) | 0 (0/226) | 1.6% (9/556) | |||||||||
| Total | 3.14% (15/477) | 0 | 1.9% (26/1,357) | 3.3% (45/1,360) | 6.7% (21/315) | 4.0% (52/1,313) | 1.6% (16/1,003) | 0 | 1.0% (13/1,313) | 1.6% (5/310) | 1.5% (6/409) | 0.5% (2/386) | |
Abbreviations: NSCLC, non-small-cell lung cancer; AD, adenocarcinoma; SCC, squamous cell carcinoma; HRM, high resolution melting analysis; RT-PCR, reverse transcriptase polymerase chain reaction; IHC, immunohistochemistry; FISH, fluorescence in situ hybridization.
General spectra of mutations in PIK3CA, BRAF, RET, and HER2
Recently, many studies have analyzed the spectra of well known driver mutations in non-smokers with adenocarcinoma of the lung. Table 3 shows the oncogenic driver mutations in the ROS1, BRAF, PIK3CA, RET, and HER2 genes of such highly selected Chinese patients. Targeted therapies are developing rapidly, and molecular data in conjunction with clinical and pathological features suggest that prospective genotyping of lung adenocarcinomas in smokers, in terms of changes in the genes listed above, may facilitate targeted therapy in almost all cases.
As with EGFR, the HER2 protein is a member of the HER family of receptor tyrosine kinases. The protein forms homodimers or heterodimers with other members of the same family. HER2 is overexpressed in about 20% of patients with NSCLC, but gene amplification occurs in only 2%.49, 50 Early clinical trials with trastuzumab, a humanized monoclonal antibody against HER2 (which is effective to treat breast cancers in which HER2 is amplified), exerted only slight effects in unselected patients with NSCLC.51
Xu et al67 and others20,21,26 have shown that the frequency of the PIK3CA mutation in NSCLC patients is 2%–4%, and the mutation occurs less often in those with adenocarcinomas compared with other forms of lung cancer.
We have summarized the data from the Chinese studies, and describe the general spectrum of mutations in EGFR, ALK, KRAS, PIK3CA, BRAF, RET, and HER2 in patients with NSCLC, adenocarcinoma, and squamous cell cancer, respectively (Figure 1A–C).
Mechanism of development of drug resistance in lung cancers
Most advanced NSCLCs with activating EGFR mutations initially respond to the TKIs gefitinib or erlotinib. However, after 6–12 months, most tumors acquire resistance to these agents. Elucidation of the mechanism underlying this process is important. T790M mutation, MET amplification, overexpression of hepatocyte growth factor, and activation of the insulin-like growth factor 1 receptor (IGF1R) and other factors have been reported to be associated with acquired resistance to EGFR TKIs.52 Most previous clinical reports indicated that acquisition of the T790M mutation explained approximately 50% of acquired TKI resistance in both Caucasian and Asian populations.53–55 Many of these studies were retrospective and enrolled small numbers of heterogeneous patients. In addition, whether the T790M mutation is present before treatment, the mechanism of mutation, and the optimal detection method remain controversial. Other secondary resistance mutations seem to be rare. Generally, MET amplification accounts for about 20% of TKI resistance; the molecular mechanism differs from that of T790M. In some patients, both mutations are present.
Studies of acquired resistance to EGFR TKIs in Chinese patients with lung cancer seem to be rare. We have extensively reviewed the literature on acquired resistance to TKIs in such patients and have summarized the data (Table 4).21,56–59 This table contains information on T790M mutations developing in 44 of 1,579 (2.8%) TKI-naïve patients studied in six clinical reports. However, 45.4% (54/119) were positive after failure of TKI therapy.
Table 4.
Prevalence of T790M mutation and c-MET amplification in Chinese patients with TKI-resistant NSCLC
| Study | Year | Technique |
T790M mutation
|
c-MET amplification
|
c-MET coexistent T790M
|
||
|---|---|---|---|---|---|---|---|
| TKI-naïve | TKI acquired-resistant | TKI-naïve | TKI acquired-resistant | TKI acquired-resistant | |||
| Chen et al65 | 2009 | HRM | 0% (0/53) | 48.3% (14/29) | 3.8% (2/53) | 10.3% (3/29) | 6.9% (2/29) |
| Chen et al56 | 2011 | FISH | 10.58% (22/208) | ||||
| Wu et al57 | 2011 | RT-PCR | 0.5% (6/1,261) | ||||
| He et al58 | 2013 | RT-PCR | 36.4% (24/33) | ||||
| Sequencing | 6.1% (2/33) | ||||||
| Su et al59 | 2012 | MALDI-TOF MS | 25.2% (27/107) | 83.3% (10/12) | |||
| Sequencing | 2.8% (3/107) | 33.3% (4/12) | |||||
| Ren et al21 | 2012 | Sequencing | 7.7% (8/104) | ||||
| Total | 2.8% (44/1,579) | 45.4% (54/119) | 9.2% (24/261) | 10.3% (3/29) | 6.9% (2/29) | ||
Abbreviations: MALDI-TOF MS, matrix-assisted laser desorption ionization-time of flight mass spectrometry; NSCLC, non-small-cell lung cancer; HRM, high resolution melting analysis; RT-PCR, reverse transcriptase polymerase chain reaction; IHC, immunohistochemistry; FISH, fluorescence in situ hybridization; TKI, tyrosine kinase inhibitor.
The increasing amount of preclinical data on EGFR-mutated NSCLCs that have acquired resistance to TKIs has enhanced interest in the development of novel drugs inhibiting the effects of T790M, MET, or IGF-1R mutations, to be used in combination with EGFR TKIs. A pan-HER TKI, PF00299804, to which resistance does not develop, has been tested in xenograft models and shows promise for use in humans to overcome T790M-mediated TKI resistance.60 Ongoing clinical research on EGFR-mutated NSCLCs will improve the survival of patients with somatic mutations such as T790M, MET, or IGF-1R.
Conclusion
Currently, non-smoker lung adenocarcinoma is the principal subtype of lung cancer, and is recognized as a distinct entity because of its peculiar biological characteristics. The frequency of EGFR mutations ranges from 28.0% in unselected Chinese patients with NSCLC to 48.5% in those with lung adenocarcinoma. The KRAS mutation frequency is lower in the People’s Republic of China than in Western countries. The frequency of the EML4-ALK fusion is 6.4% and this mutation is often found in females, patients with adenocarcinoma, non-smokers, and younger patients. ROS1 fusions appear to be more common in patients with adenocarcinoma. These observations have changed the treatment strategies for lung cancer. Genetic testing prior to treatment is now considered essential, to allow the best treatment option to be selected. First, EGFR mutation status should be tested; almost 70% of adenocarcinoma tumors have such a mutation. If the result is negative, further molecular testing is required. Treatment options include EGFR TKIs such as gefitinib or erlotinib or irreversible TKIs such as afatinib (if the T790M mutation is detected). If the tumor is wild-type in terms of EGFR, ALK translocations, found in about 30% of EGFR-negative tumors, should be sought. If a translocation is present, treatment would include an ALK inhibitor. If the tumor is negative for an ALK translocation, more rare mutations (in KRAS, ROS1, PIK3CA, BRAF, RET, or HER2) should be sought. Also, new driver mutations remain to be discovered.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.He J, Chen WQ. The Chinese Cancer Registry Annual Report 2012[R] 2012. pp. 65–72. [Google Scholar]
- 2.Harkness EF, Brewster DH, Kerr KM, Fergusson RJ, MacFarlane GJ. Changing trends in incidence of lung cancer by histologic type in Scotland. Int J Cancer. 2002;102(2):179–183. doi: 10.1002/ijc.10661. [DOI] [PubMed] [Google Scholar]
- 3.Wang N, Chen WQ, Zhu WX, Xing XM, Lu AP, Yang L. Incidence trends and pathological characteristics of lung cancer in urban Beijing during period of 1998–2007. Zhonghua Yu Fang Yi Xue Za Zhi. 2011;45(3):249–254. [PubMed] [Google Scholar]
- 4.Alamoudi OS. Lung cancer at a University Hospital in Saudi Arabia: A four-year prospective study of clinical, pathological, radiological, bronchoscopic, and biochemical parameters. Ann Thorac Med. 2010;5(1):30–36. doi: 10.4103/1817-1737.58957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang S, Dai M, Ren JS, Chen YH, Guo LW. Estimates and prediction on incidence, mortality and prevalence of lung cancer in China in 2008. Zhonghua Liu Xing Bing Xue Za Zhi. 2012;33(4):391–394. [PubMed] [Google Scholar]
- 6.Clark GM, Zborowski DM, Santabarbara P, et al. Smoking history and epidermal growth factor receptor expression as predictors of survival benefit from erlotinib for patients with non-small-cell lung cancer in the National Cancer Institute of Canada Clinical Trials Group study BR.21. Clin Lung Cancer. 2006;7(6):389–394. doi: 10.3816/clc.2006.n.022. [DOI] [PubMed] [Google Scholar]
- 7.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 8.Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368(25):2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 9.Camidge DR, Bang YJ, Kwak EL, et al. Activity and safety of crizotinib in patients with ALK-positive non-small-cell lung cancer: updated results from a phase 1 study. Lancet Oncol. 2012;13(10):1011–1019. doi: 10.1016/S1470-2045(12)70344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sequist LV, von Pawel J, Garmey EG, et al. Randomized phase II study of erlotinib plus tivantinib versus erlotinib plus placebo in previously treated non-small-cell lung cancer. J Clin Oncol. 2011;29(24):3307–3315. doi: 10.1200/JCO.2010.34.0570. [DOI] [PubMed] [Google Scholar]
- 11.Shapiro GI, Rodon J, Bedell C, et al. Phase I safety, pharmacokinetic, and pharmacodynamic study of SAR245408 (XL147), an oral pan-class I PI3K inhibitor, in patients with advanced solid tumors. Clin Cancer Res. 2014;20(1):233–245. doi: 10.1158/1078-0432.CCR-13-1777. [DOI] [PubMed] [Google Scholar]
- 12.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers – a different disease. Nat Rev Cancer. 2007;7(10):778–790. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 13.Yano T, Haro A, Shikada Y, Maruyama R, Maehara Y. Non-small cell lung cancer in never smokers as a representative ‘non-smoking-associated lung cancer’: epidemiology and clinical features. Int J Clin Oncol. 2011;16(4):287–293. doi: 10.1007/s10147-010-0160-8. [DOI] [PubMed] [Google Scholar]
- 14.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12(2):175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- 15.Thatcher N, Chang A, Parikh P, et al. Gefitinib plus best supportive care in previously treated patients with refractory advanced non-small-cell lung cancer: results from a randomised, placebo-controlled, multicentre study (Iressa Survival Evaluation in Lung Cancer) Lancet. 2005;366(9496):1527–1537. doi: 10.1016/S0140-6736(05)67625-8. [DOI] [PubMed] [Google Scholar]
- 16.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362(25):2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 17.Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12(8):735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 18.Wu YL, Zhong WZ, Li LY, et al. Epidermal growth factor receptor mutations and their correlation with gefitinib therapy in patients with non-small cell lung cancer: a meta-analysis based on updated individual patient data from six medical centers in mainland China. J Thorac Oncol. 2007;2(5):430–439. doi: 10.1097/01.JTO.0000268677.87496.4c. [DOI] [PubMed] [Google Scholar]
- 19.An SJ, Chen ZH, Su J, et al. Identification of enriched driver gene alterations in subgroups of non-small cell lung cancer patients based on histology and smoking status. PLoS One. 2012;7(6):e40109. doi: 10.1371/journal.pone.0040109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Pan Y, Li Y, et al. Frequency of well-identified oncogenic driver mutations in lung adenocarcinoma of smokers varies with histological subtypes and graduated smoking dose. Lung Cancer. 2013;79(1):8–13. doi: 10.1016/j.lungcan.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 21.Ren S, Kuang P, Zheng L, et al. Analysis of driver mutations in female non-smoker Asian patients with pulmonary adenocarcinoma. Cell Biochem Biophys. 2012;64(2):155–160. doi: 10.1007/s12013-012-9384-8. [DOI] [PubMed] [Google Scholar]
- 22.Wang Z, Wu YL, Zhang GC, Zhou Q, Xu CR, Guo AL. EGFR/KRAS mutations and gefitinib therapy in Chinese NSCLC patients. Onkologie. 2008;31(4):174–178. doi: 10.1159/000116736. [DOI] [PubMed] [Google Scholar]
- 23.Zhang X, Zhang S, Yang X, et al. Fusion of EML4 and ALK is associated with development of lung adenocarcinomas lacking EGFR and KRAS mutations and is correlated with ALK expression. Mol Cancer. 2010;9:188. doi: 10.1186/1476-4598-9-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu CC, Hsu HY, Liu HP, et al. Reversed mutation rates of KRAS and EGFR genes in adenocarcinoma of the lung in Taiwan and their implications. Cancer. 2008;113(11):3199–3208. doi: 10.1002/cncr.23925. [DOI] [PubMed] [Google Scholar]
- 25.Tam IY, Chung LP, Suen WS, et al. Distinct epidermal growth factor receptor and KRAS mutation patterns in non-small cell lung cancer patients with different tobacco exposure and clinicopathologic features. Clin Cancer Res. 2006;12(5):1647–1653. doi: 10.1158/1078-0432.CCR-05-1981. [DOI] [PubMed] [Google Scholar]
- 26.Xu J, He J, Yang H, et al. Somatic mutation analysis of EGFR, KRAS, BRAF and PIK3CA in 861 patients with non-small cell lung cancer. Cancer Biomark. 2011;10(2):63–69. doi: 10.3233/CBM-2012-0233. [DOI] [PubMed] [Google Scholar]
- 27.Sun LN, Luan HL, Zang FL, et al. Relationship between EGFR and K-ras mutations and clinicopathological characteristics and response to erlotinib treatment in 301 Chinese patients with non-small cell lung cancer. Zhonghua Zhong Liu Za Zhi. 2010;32(9):667–670. [PubMed] [Google Scholar]
- 28.Der CJ, Krontiris TG, Cooper GM. Transforming genes of human bladder and lung carcinoma cell lines are homologous to the ras genes of Harvey and Kirsten sarcoma viruses. Proc Natl Acad Sci U S A. 1982;79(11):3637–3640. doi: 10.1073/pnas.79.11.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.To MD, Wong CE, Karnezis AN, Del Rosario R, Di Lauro R, Balmain A. Kras regulatory elements and exon 4A determine mutation specificity in lung cancer. Nat Genet. 2008;40(10):1240–1244. doi: 10.1038/ng.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keohavong P, DeMichele MA, Melacrinos AC, Landreneau RJ, Weyant RJ, Siegfried JM. Detection of K-ras mutations in lung carcinomas: relationship to prognosis. Clin Cancer Res. 1996;2(2):411–418. [PubMed] [Google Scholar]
- 31.Rosell R, Monzo M, Pifarre A, et al. Molecular staging of non-small cell lung cancer according to K-ras genotypes. Clin Cancer Res. 1996;2(6):1083–1086. [PubMed] [Google Scholar]
- 32.Sakuma Y, Matsukuma S, Yoshihara M, et al. Distinctive evaluation of nonmucinous and mucinous subtypes of bronchioloalveolar carcinomas in EGFR and K-ras gene-mutation analyses for Japanese lung adenocarcinomas: confirmation of the correlations with histologic subtypes and gene mutations. Am J Clin Pathol. 2007;128(1):100–108. doi: 10.1309/WVXFGAFLAUX48DU6. [DOI] [PubMed] [Google Scholar]
- 33.Bae NC, Chae MH, Lee MH, et al. EGFR, ERBB2, and KRAS mutations in Korean non-small cell lung cancer patients. Cancer Genet Cytogenet. 2007;173(2):107–113. doi: 10.1016/j.cancergencyto.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 34.Li M, Liu L, Liu Z, et al. The status of KRAS mutations in patients with non-small cell lung cancers from mainland China. Oncol Rep. 2009;22(5):1013–1020. doi: 10.3892/or_00000529. [DOI] [PubMed] [Google Scholar]
- 35.Guan JL, Zhong WZ, An SJ, et al. KRAS mutation in patients with lung cancer: a predictor for poor prognosis but not for EGFR-TKIs or chemotherapy. Ann Surg Oncol. 2013;20(4):1381–1388. doi: 10.1245/s10434-012-2754-z. [DOI] [PubMed] [Google Scholar]
- 36.Morris SW, Naeve C, Mathew P, et al. ALK, the chromosome 2 gene locus altered by the t(2;5) in non-Hodgkin’s lymphoma, encodes a novel neural receptor tyrosine kinase that is highly related to leukocyte tyrosine kinase (LTK) Oncogene. 1997;14(18):2175–2188. doi: 10.1038/sj.onc.1201062. [DOI] [PubMed] [Google Scholar]
- 37.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 38.Horn L, Pao W. EML4-ALK: honing in on a new target in non-small-cell lung cancer. J Clin Oncol. 2009;27(26):4232–4235. doi: 10.1200/JCO.2009.23.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koivunen JP, Mermel C, Zejnullahu K, et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res. 2008;14(13):4275–4283. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi YL, Takeuchi K, Soda M, et al. Identification of novel isoforms of the EML4-ALK transforming gene in non-small cell lung cancer. Cancer Res. 2008;68(13):4971–4976. doi: 10.1158/0008-5472.CAN-07-6158. [DOI] [PubMed] [Google Scholar]
- 41.Zhang YG, Jin ML, Li L, et al. Evaluation of ALK rearrangement in Chinese non-small cell lung cancer using FISH, immunohistochemistry, and real-time quantitative RT- PCR on paraffin-embedded tissues. PLoS One. 2013;8(5):e64821. doi: 10.1371/journal.pone.0064821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong DW, Leung EL, So KK, et al. The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer. 2009;115(8):1723–1733. doi: 10.1002/cncr.24181. [DOI] [PubMed] [Google Scholar]
- 43.Wang Z, Zhang X, Bai H, et al. EML4-ALK rearrangement and its clinical significance in Chinese patients with advanced non-small cell lung cancer. Oncology. 2012;83(5):248–256. doi: 10.1159/000341381. [DOI] [PubMed] [Google Scholar]
- 44.Sun Y, Ren Y, Fang Z, et al. Lung adenocarcinoma from east asian never-smokers is a disease largely defined by targetable oncogenic mutant kinases. Journal of Clinical Oncology. 2010;28(30):4616–4620. doi: 10.1200/JCO.2010.29.6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bertolini G, Roz L, Perego P, et al. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci U S A. 2009;106(38):16281–16286. doi: 10.1073/pnas.0905653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Birchmeier C, Sharma S, Wigler M. Expression and rearrangement of the ROS1 gene in human glioblastoma cells. Proc Natl Acad Sci U S A. 1987;84(24):9270–9274. doi: 10.1073/pnas.84.24.9270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rimkunas VM, Crosby KE, Li D, et al. Analysis of receptor tyrosine kinase ROS1-positive tumors in non-small cell lung cancer: identification of a FIG-ROS1 fusion. Clin Cancer Res. 2012;18(16):4449–4457. doi: 10.1158/1078-0432.CCR-11-3351. [DOI] [PubMed] [Google Scholar]
- 48.Cai W, Li X, Su C, et al. ROS1 fusions in Chinese patients with non-small-cell lung cancer. Ann Oncol. 2013;24(7):1822–1827. doi: 10.1093/annonc/mdt071. [DOI] [PubMed] [Google Scholar]
- 49.Heinmoller P, Gross C, Beyser K, et al. HER2 status in non-small cell lung cancer: results from patient screening for enrollment to a phase II study of herceptin. Clin Cancer Res. 2003;9(14):5238–5243. [PubMed] [Google Scholar]
- 50.Stephens P, Hunter C, Bignell G, et al. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature. 2004;431(7008):525–526. doi: 10.1038/431525b. [DOI] [PubMed] [Google Scholar]
- 51.Hirsch FR, Langer CJ. The role of HER2/neu expression and trastuzumab in non-small cell lung cancer. Semin Oncol. 2004;31(1 Suppl 1):75–82. doi: 10.1053/j.seminoncol.2003.12.018. [DOI] [PubMed] [Google Scholar]
- 52.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2(3):e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Balak MN, Gong Y, Riely GJ, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res. 2006;12(21):6494–6501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- 54.Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104(52):20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang SX, Yamashita K, Yamamoto M, et al. EGFR genetic heterogeneity of nonsmall cell lung cancers contributing to acquired gefitinib resistance. Int J Cancer. 2008;123(11):2480–2486. doi: 10.1002/ijc.23868. [DOI] [PubMed] [Google Scholar]
- 56.Chen YT, Chang JW, Liu HP, et al. Clinical implications of high MET gene dosage in non-small cell lung cancer patients without previous tyrosine kinase inhibitor treatment. J Thorac Oncol. 2011;6(12):2027–2035. doi: 10.1097/JTO.0b013e3182307e92. [DOI] [PubMed] [Google Scholar]
- 57.Wu JY, Yu CJ, Chang YC, Yang CH, Shih JY, Yang PC. Effectiveness of tyrosine kinase inhibitors on “uncommon” epidermal growth factor receptor mutations of unknown clinical significance in non-small cell lung cancer. Clin Cancer Res. 2011;17(11):3812–3821. doi: 10.1158/1078-0432.CCR-10-3408. [DOI] [PubMed] [Google Scholar]
- 58.He C, Zheng L, Xu Y, Liu M, Li Y, Xu J. Highly sensitive and noninvasive detection of epidermal growth factor receptor T790M mutation in non-small cell lung cancer. Clin Chim Acta. 2013;425:119–124. doi: 10.1016/j.cca.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 59.Su KY, Chen HY, Li KC, et al. Pretreatment epidermal growth factor receptor (EGFR) T790M mutation predicts shorter EGFR tyrosine kinase inhibitor response duration in patients with non-small-cell lung cancer. J Clin Oncol. 2012;30(4):433–440. doi: 10.1200/JCO.2011.38.3224. [DOI] [PubMed] [Google Scholar]
- 60.Doebele RC, Oton AB, Peled N, Camidge DR, Bunn PA., Jr New strategies to overcome limitations of reversible EGFR tyrosine kinase inhibitor therapy in non-small cell lung cancer. Lung Cancer. 2010;69(1):1–12. doi: 10.1016/j.lungcan.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 61.Li Y, Yang T, Wei S, et al. Clinical significance of EML4-ALK fusion gene and association with EGFR and KRAS gene mutations in 208 Chinese patients with non-small cell lung cancer. PLoS One. 2013;8(1):e52093. doi: 10.1371/journal.pone.0052093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang SZ, Li XM, Zhang AP, Sun XQ, Yang QT. Detection of EML4-ALK fusion genes in non-small cell lung cancer patients with clinical features associated with EGFR mutations. Genes Chromosomes Cancer. 2012;51(10):925–932. doi: 10.1002/gcc.21976. [DOI] [PubMed] [Google Scholar]
- 63.Sun L, Zhang Q, Luan H, Zhan Z, Wang C, Sun B. Comparison of KRAS and EGFR gene status between primary non-small cell lung cancer and local lymph node metastases: implications for clinical practice. J Exp Clin Cancer Res. 2011;30:30. doi: 10.1186/1756-9966-30-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gao J, Chen JQ, Zhang L, Liang ZY. Relationship between EGFR and KRAS mutations and prognosis in Chinese patients with non-small cell lung cancer: a mutation analysis with real-time polymerase chain reaction using scorpion amplification refractory mutation system. Zhonghua Bing Li Xue Za Zhi. 2012;41(10):652–656. doi: 10.3760/cma.j.issn.0529-5807.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 65.Chen HJ, Mok TS, Chen ZH, et al. Clinicopathologic and molecular features of epidermal growth factor receptor T790M mutation and c-MET amplification in tyrosine kinase inhibitor-resistant Chinese non-small cell lung cancer. Pathol Oncol Res. 2009;15(4):651–658. doi: 10.1007/s12253-009-9167-8. [DOI] [PubMed] [Google Scholar]
- 66.Zheng LM, David BW, Li R, et al. Screening of NSCLC samples from Chinese lung cancer patients for activating rearrangements of the ALK, RET, and ROS1 genes[abstract] J Clin Oncol. 2013;(Suppl 31):e22066. [Google Scholar]
- 67.Xu R, Ma J, Xie LQ, et al. Frequency of oncogenic alterations in five driver genes in Chinese female lung adenocarcinoma[abstract] J Clin Oncol. 2013;(Suppl 31):1588. [Google Scholar]