Abstract
Plants are a sustainable resource for valuable natural chemicals best illustrated by large-scale farming centered on specific products. Here, we review recent discoveries of plant metabolic pathways producing natural products with unconventional biomolecular structures. Prenylation of polyketides by aromatic prenyltransferases (aPTases) ties together two of the major groups of plant specialized chemicals, terpenoids and polyketides, providing a core modification leading to new bioactivities and downstream metabolic processing. Moreover, PTases that biosynthesize Z-terpenoid precursors for small molecules such as lycosantalene have recently been found in the tomato family. Gaps in our understanding of how economically important compounds such as cannabinoids are produced are being identified using next-generation ‘omics’ to rapidly advance biochemical breakthroughs at an unprecedented rate. For instance, olivetolic acid cyclase, a polyketide synthase (PKS) co-factor from Cannabis sativa, directs the proper cyclization of a polyketide intermediate. Elucidations of spatial and temporal arrangements of biosynthetic enzymes into metabolons, such as those used to control the efficient production of natural polymers such as rubber and defensive small molecules such as linamarin and lotaustralin, provide blueprints for engineering streamlined production of plant products.
Introduction
Plants are a rich source of commercially relevant natural products commonly used as agricultural chemicals, nutraceuticals, therapeutics, flavors, and fragrances [1,2]. In particular, terpenes and terpenoids, polyketides and associated phenylpropanoids, and alkaloids (reviewed by O’Connor et al. in this issue) are commonly associated with plant metabolism [3,4]. As analytical methods improve with increasing sensitivities and lower costs, examinations of well-studied plant models, as well as new plant specimens, continue to reveal unexpected chemical structures. Often these molecules contain building blocks derived from two or more distinct classes of plant-specialized metabolites. The isolation and structure elucidation of such compounds portend yet to be discovered enzymes, mechanisms, and assemblies of biosynthetic pathways [5].
As biotic and abiotic factors fluctuate appreciably across a myriad of plant ecological niches, these specialized metabolic pathways provide host populations with enhanced fitness while exhibiting both spatial and temporal control across specific tissues and cells (Figure 1). An increasing appreciation of plant natural product biosynthesis, driven by high-content and cost effective metabolomic, transcriptomic, and genomic surveys integrated with ecology, biochemistry and heterologous expression, has expanded our understanding of mechanistic and organizational strategies that have evolved across the green plant lineage to produce structurally complex and specialized bioactive metabolites. This review highlights recent advances in plant biochemistry that utilize multidimensional approaches and disciplines to uncover and manipulate these newly discovered biosynthetic pathways.
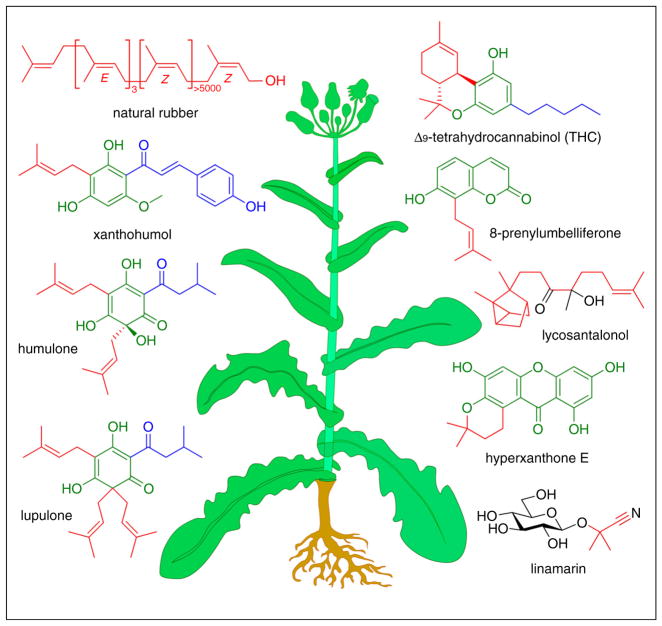
Figure 1.
Specialized metabolites from plants. Segments of the molecules depicted are color-coded based on biosynthetic origin. Molecules in green are derived from T3PKSs, red derived from PTases, blue derived from fatty acid or amino acid anabolic and catabolic pathways, and black derived from an assortment of other biosynthetic pathways. E or Z configurations are shown for natural rubber to highlight the absolute stereochemistry of its double bonds.
Natural products arising from aromatic prenyltransferases (aPTases)
Aromatic polyketides formed by type III polyketide synthases (T3PKSs) and terpenes/terpenoids formed by terpene synthases (TPSs) are well-known and abundant classes of plant natural products with a wide variety of uses by humans and functions in planta [6]. These two classes of compounds have been extensively studied with respect to their structural diversity, economic value, pharmacological properties, and biosynthetic origins [7••,8]. Recently, plant molecules that contain multiple molecular motifs originating from disparate biosynthetic pathways are garnering widespread interest for their novelty and commercial value [9–11]. Prenylated aromatic polyketides are prevalent throughout the plant lineage, suggesting an important biosynthetic and evolutionary role for the aPTases responsible for their production (Figures 1 and 2a). While aPTases from other organisms, such as bacteria and fungi, are often cytosolic and soluble, plant aPTases tend to be integral membrane proteins [12,13]. The installed prenyl moieties are essential for tuning and expanding the bioactivity and downstream metabolic processing of aromatic products, as demonstrated for the plant terpene-flavonoid polyketide 8-prenylnaringenin (Figure 2a) [14–16].
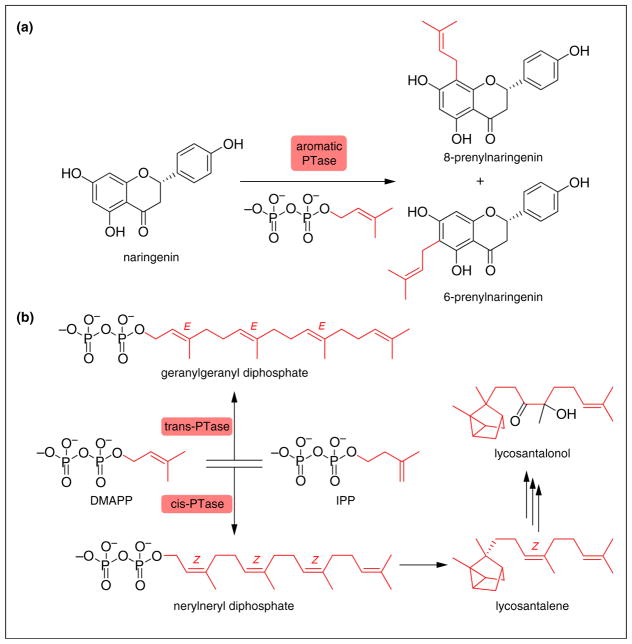
Figure 2.
PTases employed in plant natural product biosynthesis. (a) Aromatic PTases (aPTases) catalyze the addition of a terpenoid-derived prenyl group onto an electron-rich aromatic phenolic core, as exemplified by the prenylation of naringenin to form 6-prenylnaringenin and 8-prenylnaringenin. Notably, aPTases often exhibit relaxed substrate specificity and/or relaxed regiospecificity as shown here. As reported, when isolated from hops, the amounts of 6-prenylnaringenin and 8-prenylnaringenin are approximately equal. However, this ratio may not hold true in other plants, which may vary from plant to plant. Additionally, the ratios of each prenylated compound account for 1–2% of total prenylflavonoids in hops and vary dependent on growth conditions making accurate quantification difficult. (b) cis-PTases catalyze the formation of Z-terpenoids, thus increasing the structural diversity and potential beneficial activity of the broad family of plant terpenoids.
Investigation of Hypericum calycinum (Great St. John’s Wort), a source of pharmacologically active specialized metabolites, led to the identification of a membrane-bound aPTase that elaborates xanthone polyketides [17]. This aPTase transfers a 5-carbon dimethylallyl moiety onto 1,2,6,7-tetrahydroxyxanthone to redirect polyketide biosynthesis to tailored natural products such as hyperxanthone E (Figure 1), a compound exhibiting anti-inflammatory properties [18].
In Petroselinum crispum (parsley), an aPTase named PcPT was discovered that is specific for the modification of coumarin derivatives [19•]. PcPT adds a dimethylallyl moiety to umbelliferone, the product of which serves as the substrate for downstream enzymes that together generate bioactive furanocoumarin molecules such as angelicin [20]. Like many specialized metabolic enzymes of plants that exhibit relaxed regiospecificity (Figure 2a), the dimethylallyl group can be added at either of two positions to produce a 6-prenyl or 8-prenyl product. While many aPTases will prenylate a variety of substrates [21,22], PcPT, while displaying relaxed regiospecificity, maintains high selectivity for umbelliferone.
A phylogenetically similar aPTase was discovered in Citrus limon (lemon), dubbed ClPT [23••]. In contrast to PcPT, ClPT transfers a 10-carbon geranyl unit onto the C-8 of umbelliferone. While many aPTases will accept a variety of substrates, ClPT is very specific towards coumarin molecules. CIPT does not exhibit transferase activity with furanocoumarins, coumaric acid derivatives, flavonols or isoflavones.
Psychoactive cannabinoids of Cannabis sativa are derived from an aromatic polyketide, olivetolic acid, biosynthesized by an unexpected heteromeric T3PKS system discussed later. Olivetolic acid is poised for geranylation catalyzed by the C. sativa encoded aPTase CsPT1 to form cannabigerolic acid [24]. The geranyl moiety then serves as a reactive handle that undergoes carbocation-mediated cyclization catalyzed by Δ9-tetrahydrocannabinolic acid synthase (Figure 3) [25].
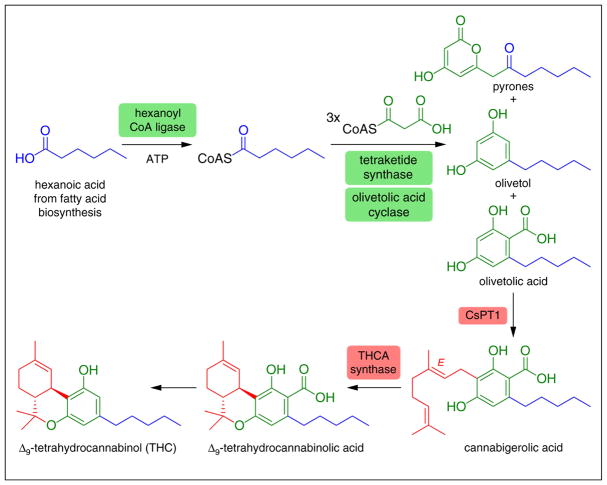
Figure 3.
Biosynthesis of Δ9-tetrahydrocannabinolic acid. This biosynthetic pathway features enzymes that likely possess species-specific enzymatic activity, namely the hexanoyl-CoA ligase, tetraketide synthase and olivetolic acid cyclase that are all required for the formation of the precursor molecule olivetolic acid. In the absence of olivetolic acid cyclase, 2-pyrones (α-pyrones) form as the major products. 2-Pyrones derived from both triketide and tetraketide intermediates occur upon lactonization of extended polyketide intermediates. For clarity, the 2-pyrone derived from a fully extended tetraketide intermediate is shown. Olivetolic acid is then geranylated by CsPT1 to form cannabigerolic acid. Cannabigerolic acid then undergoes cyclization catalyzed by THCA synthase to form Δ9-tetrahydrocannabinolic acid. The psychotropic activity of these compounds comes about upon heating of Δ9-tetrahydrocannabinolic acid, which undergoes decarboxylation to Δ9-tetrahydrocannabinol.
Acyclic terpenoids and cis-PTases
Terpenoids that arise from Z-olefinic, rather than E-olefinic, acyclic terpenoid precursors are being discovered now with regularity, and the corresponding cis-PTases identified, most recently exemplified by discoveries made in Solanum lycopersicum (tomato) [26,27]. These cis-PTases biosynthesize linear terpenoid precursors that contain Z-olefins, as opposed to the canonical E-olefins found in many linear and cyclic terpenes and terpenoids (Figure 2b). The genes encoding the biosynthetic route to the tomato natural product lycosantalonol (Figure 1) are co-localized in a small gene cluster containing the cis-PTase CPT2 gene. CPT2 catalyzes the biosynthesis of nerylneryl pyrophosphate (NNPP) (Figure 2b) [28]. NNPP then undergoes ionization and cyclization catalyzed by TPS21, followed by oxidative modification catalyzed by the cytochrome P450 enzyme CYP71D51 to form lycosantalonol [29••].
Three cis-PTases, TkCPT1, TkCPT2 and TkCPT3, have been implicated in the biosynthesis of natural rubber in Taraxacum koksaghyz (dandelion). TkCPT1, TkCPT2 and TkCPT3 produce natural rubber-like molecules in vitro (Figure 1) [30]. Each TkCPT was transfected into Nicotiana tabacum mesophyll protoplasts individually and exhibited pronounced isopentenyl pyrophosphate (IPP) transferase activity, suggesting that each TkCPT is catalytically active [31]. Recently, in planta genetic experiments in the closely related species Taraxacum brevicorniculatum provided support for three homologous TbCPTases required for natural rubber biosynthesis [32•]. Notably, RNAi knockdown of the CPTs elicited a decrease in natural rubber production most noticeable by a decrease in rubber particles formed. Concurrently, pathways that utilize the same 5-carbon terpenoid precursors were found to be more productive in RNAi treated plants. Specifically, triterpene abundance rose substantially, most likely resulting from an increased pool of the 5-carbon terpenoid building block IPP.
Non-canonical enzymes involved in specialized metabolite biosynthesis
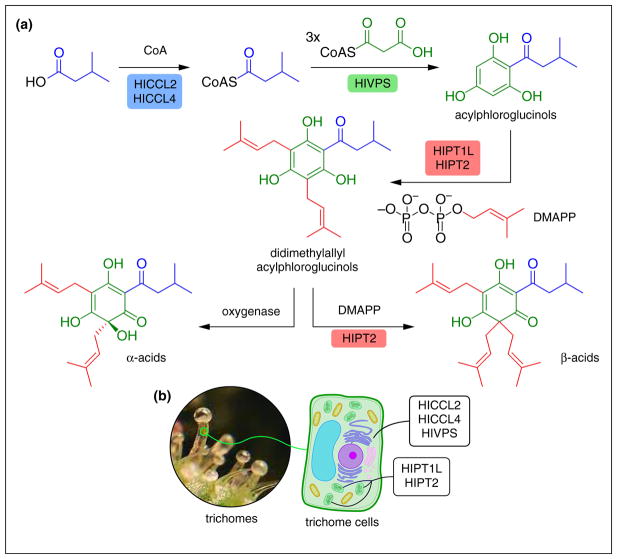
CoA ligases are common components of natural product biosynthesis in plants, especially in relation to T3PKS-derived polyketides. These PKSs generally require CoA-linked substrates, such as coumaroyl-CoA, to initiate and malonyl-CoA to carry forward the iterative biosyntheses of plant polyketides such as stilbenes, chalcones and pyrones through aldol-based, Claisen-based and lactonization-based cyclizations, respectively [33]. In Humulus lupus (hops), a variety of prenylated polyketide-derived products are produced, including xanthohumol and the bitter acids humulone and lupulone (Figure 1) [34,35]. However, bitter acid biosynthesis requires CoA thioester substrates derived from branched alkyl amino acids as opposed to the more ubiquitous cinnamic acid-based starter molecules. Recently, a screen of all putative CoA ligases from H. lupus uncovered two CoA ligases, HlCCL2 and HlCCL4, that are highly specific for branched, short-chain fatty acids [36••]. HlCCL2 and HlCCL4 produce isovaleryl-CoA and isobutyryl-CoA, respectively, each of which is then shuttled into bitter acid biosynthesis (Figure 4a).
Figure 4.
Production of bitter acids in hops trichomes. (a) Biosynthetic pathway to α-acids and β-acids, including the enzymatic pathways discussed in the text. The oxygenase required for α-acid formation has not yet been functionally characterized to date. While both isovaleryl-CoA and isobutyryl-CoA are utilized by HlVPS, for clarity, only the isovaleryl-CoA transformation is depicted. (b) Depiction and cellular location of the metabolon that produces β-acids in hops. Many plant natural products are sequestered in specialized cells and tissues. Glandular trichomes are notable developmental innovations that have arisen during land plant evolution. Practically, they also provide an economical route to downstream processing to enrich for particular plant chemicals.
A salient example of a plant natural product biosynthetic pathway of high commercial relevance that utilizes several non-canonical enzymes is the metabolic route to Δ9-tetrahydrocannabinol (THC) and related cannabinoids in Cannabis sativa (hemp) (Figure 3). The first step of THC biosynthesis is the production of olivetolic acid, which requires hexanoyl-CoA as a starter unit [37]. A gene encoding a hexanoyl-CoA ligase was putatively identified in the genome of C. sativa. The encoded enzyme was then shown biochemically to produce hexanoyl-CoA from hexanoic acid, CoA and ATP in a Mg2+-dependent manner [38].
The next enzymatic step in THC biosynthesis, the formation of olivetolic acid, requires a T3PKS, termed tetraketide synthase (TKS), that utilizes hexanoyl-CoA and 3 molecules of malonyl-CoA to form olivetolic acid, hypothetically through an aldol-based cyclization of the acyclic tetraketide intermediate. Initial in vitro efforts to reconstitute olivetolic acid biosynthesis using putative T3PKSs from C. sativa were unsuccessful. Notably, incubation of TKS with hexanoyl-CoA and malonyl-CoA gave rise to substantial amounts of 2-pyrones (α-pyrones) derived from incorporation of 2 and 3 malonyl-CoAs, triketide and tetraketide intermediates, respectively, and detectable quantities of olivetol [39]. It was hypothesized that an additional ‘accessory’ enzyme was possibly required for the efficient formation of olivetolic acid by TKS. Using next-generation omics to narrow down the candidate gene pool, heterologous expression and biochemical reconstitution, this posited activity was shown to be encoded by a single gene, and the resultant protein product dubbed olivetolic acid cyclase (Figure 3) [40••].
In addition to discoveries made in dandelion previously discussed [30,31,32•], a recent study uncovered several genes encoding protein products correlated with natural rubber biosynthesis in Lactuca sativa (lettuce) [41•]. Specifically, CPT3 was found to be a cis-PTase, and CPTL2 was described as a PTase-like protein. Notably, CPTL2 does not contain the conserved catalytic residues that are commonly associated with cis-PTase activity [42]. It was shown using a yeast two-hybrid assay that CPTL2 and CPT3 form a heteromeric complex. Additionally, knockdown of CPTL2 in lettuce resulted in a substantial drop in natural rubber production. Collectively, these results lead to the hypothesis that CPTL2, while not possessing PTase activity, is essential for rubber biosynthesis. The authors speculate that this inactive PTase-like protein recruits CPT3 to the endoplasmic reticulum (ER) through membrane association and protein–protein interactions. It is at the ER where the hydrophobic substrates, intermediates and products of natural rubber are produced and packaged. A homologous pair of proteins, CPTL1 and CPT1, was also found in L. sativa, and while they have not been characterized to date, comparison to CPTL2/CPT3 suggests a similar potential for protein–protein interactions [41•].
Spatial and temporal organization of biosynthetic pathways
An important aspect of natural product biosynthesis that is attracting widespread attention is the spatial and temporal organization of biosynthetic enzymes that participate in particular metabolic pathways. These often loosely or transiently associated complexes termed ‘metabolons,’ provide a plant with the ability to tune biosynthetic specificity and efficiency at the right place and the right time in response to external and internal biotic and abiotic stimuli. While the nature of these interactions appears to be weakly associating, this phenomenon affords complex organisms such as plants with an interchangeable and dynamic set of catalytic units that can be assembled and disassembled into distinct biosynthetic factories. The timing and constituents of a particular metabolon, together with the availability of the appropriate starting materials, then leads to the efficient production of both primary and specialized natural products at high levels [5]. For instance, primary metabolic pathways, such as those involved in lignin and sporopollenin biosynthesis, require specific metabolons that localize to the ER [43,44].
The biosynthetic pathway leading to the defensive cyanogenic glucosides linamarin and lotaustralin was recently elucidated in Manihot esculenta (cassava) [45•]. Two uridine diphosphate glucosyltransferases (UGTs) associated with linamarin (Figure 1) and lotaustralin, UGT85K4 and UGT85K5, respectively, possess broad substrate tolerances in vitro. In vivo, the two enzymes localize in mesophyll and xylem parenchyma cells in the first unfolded leaves of cassava. Moreover, these two UGTs are co-expressed in planta with the cytochrome P450s CYP79D1 and two CYP71E7 paralogs, which together catalyze the initial steps of linamarin and lotaustralin biosynthesis. More importantly, all of these enzymes are co-localized to the same tissue and cell types, including the cortex, xylem, and phloem parenchyma, and in the endodermis of the petiole of the first unfolded leaf [45•]. This provides strong evidence for the existence and highly specialized organization of a metabolon required for cyanogenic glucoside biosynthesis.
In the related biosynthetic pathway of dhurrin (structure not shown), a cyanogenic glucoside from Sorghum bicolor (sorghum), computational docking studies of homologs of CYP79D1 and CYP71E7 revealed putative interacting surfaces that may facilitate formation of a membrane bound complex [46]. A series of modeling, docking, and membrane association experiments all provided evidence for a three-enzyme complex that, together with downstream enzymes, forms a cyanogenic glucoside metabolon in sorghum.
H. lupus serves as a source for a variety of important flavoring molecules including α-acids and β-acids such as humulone and lupulone, respectively (Figure 1). The biosynthesis of these molecules requires a complex interplay between several different enzymes, including aPTases [47] (Figure 4a). From recent studies, the aPTase HlPT2 was shown to interact with the aPTase-like protein HlPT1L, and this interaction is essential for bitter acid production [48,49,50••]. This complex performs sequential prenylation reactions (typically 2–3) that decorate precursors produced by HlCCL2, HlCCL4, and the T3PKS HlPVS. Three prenylations leads to β-acids, while oxidation by a yet to be discovered oxygenase leads to α-acids (Figure 4a). The translational importance of achieving a spatial and temporal understanding of a metabolon is most notably demonstrated by the success at assembling these five enzymes in a heterologous host to produce β-acids [50••].
Conclusion
Genomic, metabolomic, and proteomic analyses, together with the knowledge bases and toolsets developed for ecology, structural biology, biochemistry and synthetic biology, are providing an unprecedented opportunity to rapidly harness and extract substantial value out of plant specialized metabolic pathways. These integrative approaches are also leading to an atomic-level understanding of the underlying principles governing the spatial, temporal and mechanistic complexity of plant natural product biosynthesis. Moreover, as evidenced by several studies discussed here, homologous biosynthetic enzymes exist across the green plant lineage, especially between phylogenetically related species. This, coupled with the continually falling costs of next-generation technologies and the power of natural selection, rapidly advance the discovery and expansion of a treasure trove of natural molecules, tuned by 500 million years of land plant evolution [51].
These few examples underscore the importance of cutting-edge technologies that enable the discovery and exploitation of important natural compounds with speed and economic efficiency in genomically complex organisms such as plants. Achieving a multifaceted understanding of these pathways is critical to the future expansion of plant resources for agriculture, health-promotion, therapeutic intervention, nutrition, flavors, and fragrances. Here, uncovering the parts list and organization principles of biosynthetic pathways leading to specific compounds enables heterologous expression and bio-production of plant metabolites that may accumulate only at low levels in the native plant host, or may otherwise be economically inaccessible [52]. More broadly, understanding plant metabolic pathways enables generalized platforms for the study and harnessing of plant and animal resilience related to plant-based diets in a globally sustainable manner.
Acknowledgments
Research in our laboratories is supported by NSF EEC-0813570 and MCB-0645794 to JPN and NIH GM095970 to MDB. JPN is an investigator with the Howard Hughes Medical Institute.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Atanasov AG, Waltenberger B, Pferschy-Wenzig E-M, Linder T, Wawrosch C, Uhrin P, Temml V, Wang L, Schwaiger S, Heiss EH, et al. Discovery and resupply of pharmacologically active plant-derived natural products: a review. Biotechnol Adv. 2015;33:1582–1614. doi: 10.1016/j.biotechadv.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayseli MT, Ayseli Yİ. Flavors of the future: health benefits of flavor precursors and volatile compounds in plant foods. Trends Food Sci Technol. 2015 http://dx.doi.org/10.1016/j.tifs.2015.11.005.
- 3.Lange BM. The evolution of plant secretory structures and emergence of terpenoid chemical diversity. Annu Rev Plant Biol. 2015;66:139–159. doi: 10.1146/annurev-arplant-043014-114639. [DOI] [PubMed] [Google Scholar]
- 4.Stewart C, Vickery CR, Burkart MD, Noel JP. Confluence of structural and chemical biology: plant polyketide synthases as biocatalysts for a bio-based future. Curr Opin Plant Biol. 2013;16:365–372. doi: 10.1016/j.pbi.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 5.Laursen T, Møller BL, Bassard J-E. Plasticity of specialized metabolism as mediated by dynamic metabolons. Trends Plant Sci. 2015;20:20–32. doi: 10.1016/j.tplants.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Dewick PM. Medicinal Natural Products: A Biosynthetic Approach. Wiley, A John Wiley and Sons, Ltd., Publication; 2009. [Google Scholar]
- 7••.Anarat-Cappillino G, Sattely ES. The chemical logic of plant natural product biosynthesis. Curr Opin Plant Biol. 2014;19:51–58. doi: 10.1016/j.pbi.2014.03.007. This paper provides a cogent overview of plant natural product pathways, concentrating on structural diversity, thus serving as a great resource for the types of chemical complexity found in plant natural products. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalez-Burgos E, Gomez-Serranillos MP. Terpene compounds in nature: a review of their potential antioxidant activity. Curr Med Chem. 2012;19:5319–5341. doi: 10.2174/092986712803833335. [DOI] [PubMed] [Google Scholar]
- 9.Yang X, Jiang Y, Yang J, He J, Sun J, Chen F, Zhang M, Yang B. Prenylated flavonoids, promising nutraceuticals with impressive biological activities. Trends Food Sci Technol. 2015;44:93–104. [Google Scholar]
- 10.Hanáková Z, Hošek J, Babula P, Dall’Acqua S, Václavík J, Šmejkal K. C-geranylated flavanones from Paulownia tomentosa fruits as potential anti-inflammatory compounds acting via inhibition of TNF-α production. J Nat Prod. 2015;78:850–863. doi: 10.1021/acs.jnatprod.5b00005. [DOI] [PubMed] [Google Scholar]
- 11.Šmejkal K. Cytotoxic potential of C-prenylated flavonoids. Phytochem Rev. 2014;13:245–275. [Google Scholar]
- 12.Tello M, Kuzuyama T, Heide L, Noel JP, Richard SB. The ABBA family of aromatic prenyltransferases: broadening natural product diversity. Cell Mol Life Sci. 2008;65:1459–1463. doi: 10.1007/s00018-008-7579-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winkelblech J, Fan A, Li S-M. Prenyltransferases as key enzymes in primary and secondary metabolism. Appl Microbiol Biotechnol. 2015;99:7379–7397. doi: 10.1007/s00253-015-6811-y. [DOI] [PubMed] [Google Scholar]
- 14.Ming L-G, Lv X, Ma X-N, Ge B-F, Zhen P, Song P, Zhou J, Ma H-P, Xian CJ, Chen K-M. The prenyl group contributes to activities of phytoestrogen 8-prenynaringenin in enhancing bone formation and inhibiting bone resorption in vitro. Endocrinology. 2013;154:1202–1214. doi: 10.1210/en.2012-2086. [DOI] [PubMed] [Google Scholar]
- 15.Terao J, Mukai R. Prenylation modulates the bioavailability and bioaccumulation of dietary flavonoids. Arch Biochem Biophys. 2014;559:12–16. doi: 10.1016/j.abb.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 16.Marín M, Máñez S. Recent trends in the pharmacological activity of isoprenyl phenolics. Curr Med Chem. 2013;20:272–279. doi: 10.2174/092986713804806676. [DOI] [PubMed] [Google Scholar]
- 17.Fiesel T, Gaid M, Müller A, Bartels J, El-Awaad I, Beuerle T, Ernst L, Behrends S, Beerhues L. Molecular cloning and characterization of a xanthone prenyltransferase from Hypericum calycinum cell cultures. Molecules. 2015;20:15616–15630. doi: 10.3390/molecules200915616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H, Zhang D-D, Lao Y-Z, Fu W-W, Liang S, Yuan Q-H, Yang L, Xu H-X. Cytotoxic and anti-inflammatory prenylated benzoylphloroglucinols and xanthones from the twigs of Garcinia esculenta. J Nat Prod. 2014;77:1700–1707. doi: 10.1021/np5003498. [DOI] [PubMed] [Google Scholar]
- 19•.Karamat F, Olry A, Munakata R, Koeduka T, Sugiyama A, Paris C, Hehn A, Bourgaud F, Yazaki K. A coumarin-specific prenyltransferase catalyzes the crucial biosynthetic reaction for furanocoumarin formation in parsley. Plant J. 2014;77:627–638. doi: 10.1111/tpj.12409. This paper describes the biochemistry underlying furanocoumarin formation in parsley. This pathway provides insight into the biosynthesis of other furanocoumarin and furanocoumarin derived compounds that utilize aPTases in plants. [DOI] [PubMed] [Google Scholar]
- 20.Cho H-J, Jeong S-G, Park J-E, Han J-A, Kang H-R, Lee D, Song MJ. Antiviral activity of angelicin against gammaherpesviruses. Antivir Res. 2013;100:75–83. doi: 10.1016/j.antiviral.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 21.Kuzuyama T, Noel JP, Richard SB. Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products. Nature. 2005;435:983–987. doi: 10.1038/nature03668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumano T, Richard SB, Noel JP, Nishiyama M, Kuzuyama T. Chemoenzymatic syntheses of prenylated aromatic small molecules using Streptomyces prenyltransferases with relaxed substrate specificities. Bioorgan Med Chem. 2008;16:8117–8126. doi: 10.1016/j.bmc.2008.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23••.Munakata R, Inoue T, Koeduka T, Karamat F, Olry A, Sugiyama A, Takanashi K, Dugrand A, Froelicher Y, Tanaka R, et al. Molecular cloning and characterization of a geranyl diphosphate-specific aromatic prenyltransferase from lemon. Plant Physiol. 2014;166:80–90. doi: 10.1104/pp.114.246892. The authors describe an unusually specific aPTase, in contrast to the relaxed specificity of other plant aPTases. Additionally, the aPTase is specific for geranyl diphosphate, as opposed to dimethyallyl diphosphate, the typical prenyl donor for plant aPTases. This paper highlights the diversity of plant aPTase activity that continues to be uncovered as more plants are examined metabolomically. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Page JE, Boubakir Z, inventors. National Research Council of Canada, University of Saskatchewan, assignee. Aromatic prenyltransferase from cannabis. US20150128301 A1. US patent. 2014 Oct 9;
- 25.Taura F, Morimoto S, Shoyama Y, Mechoulam R. First direct evidence for the mechanism of Delta1-tetrahydrocannabinolic acid biosynthesis. J Am Chem Soc. 1995;117:9766–9767. [Google Scholar]
- 26.Akhtar TA, Matsuba Y, Schauvinhold I, Yu G, Lees HA, Klein SE, Pichersky E. The tomato cis-prenyltransferase gene family. Plant J. 2013;73:640–652. doi: 10.1111/tpj.12063. [DOI] [PubMed] [Google Scholar]
- 27.Schilmiller AL, Schauvinhold I, Larson M, Xu R, Charbonneau AL, Schmidt A, Wilkerson C, Last RL, Pichersky E. Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. Proc Natl Acad Sci U S A. 2009;106:10865–10870. doi: 10.1073/pnas.0904113106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuba Y, Zi J, Jones AD, Peters RJ, Pichersky E. Biosynthesis of the diterpenoid lycosantalonol via nerylneryl diphosphate in Solanum lycopersicum. PLoS One. 2015;10:e0119302. doi: 10.1371/journal.pone.0119302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29••.Zi J, Matsuba Y, Hong YJ, Jackson AJ, Tantillo DJ, Pichersky E, Peters RJ. Biosynthesis of lycosantalonol, a cis-prenyl derived diterpenoid. J Am Chem Soc. 2014;136:16951–16953. doi: 10.1021/ja508477e. The authors describe the formation of lycosantalonol from the linear cis-terpene nerylneryl diphosphate, and the required tailoring enzymes that complete the biosynthesis. The complete characterization of this biosynthetic gene cluster in tomato is an important contribution to the general understanding of plant cis-PTases and their associated pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt T, Hillebrand A, Wurbs D, Wahler D, Lenders M, Schulze Gronover C, Prüfer D. Molecular cloning and characterization of rubber biosynthetic genes from Taraxacum koksaghyz. Plant Mol Biol Rep. 2010;28:277–284. [Google Scholar]
- 31.Schmidt T, Lenders M, Hillebrand A, van Deenen N, Munt O, Reichelt R, Eisenreich W, Fischer R, Prufer D, Schulze Gronover C. Characterization of rubber particles and rubber chain elongation in Taraxacum koksaghyz. BMC Biochem. 2010;11:11. doi: 10.1186/1471-2091-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.Post J, van Deenen N, Fricke J, Kowalski N, Wurbs D, Schaller H, Eisenreich W, Huber C, Twyman RM, Prufer D, et al. Laticifer-specific cis-prenyltransferase silencing affects the rubber, triterpene, and inulin content of Taraxacum brevicorniculatum. Plant Physiol. 2012;158:1406–1417. doi: 10.1104/pp.111.187880. The authors describe a set of three cis-prenyltransferases directly involved in natural rubber biosynthesis. In planta knockdown of these PTases results in a decrease in rubber content, but also an increase in other DMAPP-derived natural products, providing convincing evidence that these three enzymes are responsible for natural rubber biosynthesis using terpenoid precursors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Austin MB, Noel JP. The chalcone synthase superfamily of type III polyketide synthases. Nat Prod Rep. 2003;20:79–110. doi: 10.1039/b100917f. [DOI] [PubMed] [Google Scholar]
- 34.Stevens JF, Page JE. Xanthohumol and related prenylflavonoids from hops and beer: to your good health! Phytochemistry. 2004;65:1317–1330. doi: 10.1016/j.phytochem.2004.04.025. [DOI] [PubMed] [Google Scholar]
- 35.Van Cleemput M, Cattoor K, De Bosscher K, Haegeman G, De Keukeleire D, Heyerick A. Hop (Humulus lupulus)-derived bitter acids as multipotent bioactive compounds. J Nat Prod. 2009;72:1220–1230. doi: 10.1021/np800740m. [DOI] [PubMed] [Google Scholar]
- 36••.Xu H, Zhang F, Liu B, Huhman DV, Sumner LW, Dixon RA, Wang G. Characterization of the formation of branched short-chain fatty acid:CoAs for bitter acid biosynthesis in hop glandular trichomes. Mol Plant. 2013;6:1301–1317. doi: 10.1093/mp/sst004. The authors performed a thorough, systematic search for CoA ligases though to play a role in bitter acid biosynthesis. The two identified CoA ligases were found to be highly expressed in the cytoplasm of glandular trichome cells, along with the bitter acid enzyme component HlVPS. These three enzymes, when expressed in yeast together, produced acylphluoroglucinols, which are direct precursors of hops bitter acids. [DOI] [PubMed] [Google Scholar]
- 37.Marks MD, Tian L, Wenger JP, Omburo SN, Soto-Fuentes W, He J, Gang DR, Weiblen GD, Dixon RA. Identification of candidate genes affecting Delta9-tetrahydrocannabinol biosynthesis in Cannabis sativa. J Exp Bot. 2009;60:3715–3726. doi: 10.1093/jxb/erp210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stout JM, Boubakir Z, Ambrose SJ, Purves RW, Page JE. The hexanoyl-CoA precursor for cannabinoid biosynthesis is formed by an acyl-activating enzyme in Cannabis sativa trichomes. Plant J. 2012;71:353–365. doi: 10.1111/j.1365-313X.2012.04949.x. [DOI] [PubMed] [Google Scholar]
- 39.Taura F, Tanaka S, Taguchi C, Fukamizu T, Tanaka H, Shoyama Y, Morimoto S. Characterization of olivetol synthase, a polyketide synthase putatively involved in cannabinoid biosynthetic pathway. FEBS Lett. 2009;583:2061–2066. doi: 10.1016/j.febslet.2009.05.024. [DOI] [PubMed] [Google Scholar]
- 40••.Gagne SJ, Stout JM, Liu E, Boubakir Z, Clark SM, Page JE. Identification of olivetolic acid cyclase from Cannabis sativa reveals a unique catalytic route to plant polyketides. Proc Natl Acad Sci U S A. 2012;109:12811–12816. doi: 10.1073/pnas.1200330109. This paper reports the first instance of a chaperone-like function for an enzyme required for proper formation of a T3PKS-derived polyketide. Biochemical characterization of TKS with and without olivetolic acid cyclase confirms the latter protein’s role in catalyzing olivetolic acid formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41•.Qu Y, Chakrabarty R, Tran HT, Kwon E-JG, Kwon M, Nguyen T-D, Ro D-K. A lettuce (Lactuca sativa) homolog of human nogo-B receptor interacts with cis-prenyltransferase and is necessary for natural rubber biosynthesis. J Biol Chem. 2015;290:1898–1914. doi: 10.1074/jbc.M114.616920. This paper provides a prime example of an auxiliary function in organizing a multiprotein complex that putatively originated from a catalytically active enzyme fold. It highlights the essential nature of enzymes-substrates co-localization for metabolic pathway efficiency and specificity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miao RQ, Gao Y, Harrison KD, Prendergast J, Acevedo LM, Yu J, Hu F, Strittmatter SM, Sessa WC. Identification of a receptor necessary for Nogo-B stimulated chemotaxis and morphogenesis of endothelial cells. Proc Natl Acad Sci U S A. 2006;103:10997–11002. doi: 10.1073/pnas.0602427103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bassard J-E, Richert L, Geerinck J, Renault H, Duval F, Ullmann P, Schmitt M, Meyer E, Mutterer J, Boerjan W, et al. Protein–protein and protein–membrane associations in the lignin pathway. Plant Cell. 2012;24:4465–4482. doi: 10.1105/tpc.112.102566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lallemand B, Erhardt M, Heitz T, Legrand M. Sporopollenin biosynthetic enzymes interact and constitute a metabolon localized to the endoplasmic reticulum of tapetum cells. Plant Physiol. 2013;162:616–625. doi: 10.1104/pp.112.213124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45•.Kannangara R, Motawia MS, Hansen NKK, Paquette SM, Olsen CE, Møller BL, Jørgensen K. Characterization and expression profile of two UDP-glucosyltransferases, UGT85K4 and UGT85K5, catalyzing the last step in cyanogenic glucoside biosynthesis in cassava. Plant J. 2011;68:287–301. doi: 10.1111/j.1365-313X.2011.04695.x. This paper describes a small metabolon of two glucosyltransferases that complete the biosynthesis of a natural product. Most importantly, the authors confirm the tissue-specific co-localization of the two glucosyltransferases with other catalytic members of the metabolic pathway producing cyanogenic glycosides. [DOI] [PubMed] [Google Scholar]
- 46.Jensen K, Osmani SA, Hamann T, Naur P, Møller BL. Homology modeling of the three membrane proteins of the dhurrin metabolon: catalytic sites, membrane surface association and protein–protein interactions. Phytochemistry. 2011;72:2113–2123. doi: 10.1016/j.phytochem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 47.Clark SM, Vaitheeswaran V, Ambrose SJ, Purves RW, Page JE. Transcriptome analysis of bitter acid biosynthesis and precursor pathways in hop (Humulus lupulus) BMC Plant Biol. 2013;13:12. doi: 10.1186/1471-2229-13-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsurumaru Y, Sasaki K, Miyawaki T, Momma T, Umemoto N, Yazaki K. An aromatic prenyltransferase-like gene HlPT-1 preferentially expressed in lupulin glands of hop. Plant Biotechnol. 2010;27:199–204. [Google Scholar]
- 49.Tsurumaru Y, Sasaki K, Miyawaki T, Uto Y, Momma T, Umemoto N, Momose M, Yazaki K. HlPT-1, a membrane-bound prenyltransferase responsible for the biosynthesis of bitter acids in hops. Biochem Biophys Res Commun. 2012;417:393–398. doi: 10.1016/j.bbrc.2011.11.125. [DOI] [PubMed] [Google Scholar]
- 50••.Li H, Ban Z, Qin H, Ma L, King AJ, Wang G. A heteromeric membrane-bound prenyltransferase complex from hop catalyzes three sequential aromatic prenylations in the bitter acid pathway. Plant Physiol. 2015;167:650–659. doi: 10.1104/pp.114.253682. Biochemical characterization of the two aPTases required for bitter acid molecules in hops is the penultimate step in fully understanding bitter hops biosynthesis. Only one oxygenase remains to be discovered. Furthermore, design of a heterologous host capable of producing bitter acids provides not only a platform for reconstitution and full characterization of bitter acid biosynthetic pathways, but paves the way for production of economically valuable natural and unnatural analogs of bitter acids from hops using synthetic biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weng J-K, Philippe RN, Noel JP. The rise of chemodiversity in plants. Science. 2012;336:1667–1670. doi: 10.1126/science.1217411. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki S, Koeduka T, Sugiyama A, Yazaki K, Umezawa T. Microbial production of plant specialized metabolites. Plant Biotechnol. 2014;31:465–482. [Google Scholar]