Abstract
Background
Little is known about the association of the single nucleotide polymorphism (SNP) of rs364585 near serine palmitoyl-transferase long-chain base subunit 3 gene (SPTLC3) and serum lipid profiles. The present study was detected the association of the SPTLC3 rs364585 SNP and several environmental factors with serum lipid profiles in the Han and Jing populations.
Methods
Genotyping of the SPTLC3 rs364585 SNP was performed in 824 unrelated individuals of Han and 783 participants of Jing by polymerase chain reaction and restriction fragment length polymorphism combined with gel electrophoresis, and then confirmed by direct sequencing.
Results
There was no significant difference in either genotypic or allelic frequencies between Han and Jing, or between males and females of the both ethnic groups. The levels of serum low-density lipoprotein cholesterol (LDL-C) and the ratio of apolipoprotein (Apo) A1 to ApoB in Han; and total cholesterol (TC), high-density lipoprotein cholesterol (HDL-C) and LDL-C in Jing were different between the A allele carriers and the A allele non-carriers (P < 0.05-0.001). Subgroup analysis according to sex showed that the levels of LDL-C in Han males; TC and LDL-C in Jing males; and HDL-C and LDL-C in Jing females were different between the A allele carriers and the A allele non-carriers (P < 0.05-0.001), the A allele carriers had higher LDL-C and TC levels, and lower HDL-C levels than the A allele non-carriers. Serum lipid traits were also associated with several environmental factors in the Han and Jing populations, or in males and females of the both ethnic groups.
Conclusions
The present study demonstrates the association between the SPTLC3 rs364585 SNP and serum TC, HDL-C and LDL-C levels in our study populations. These associations might have ethnic- and/or sex-specificity.
Trial registration
Retrospectively registered.
Keywords: Lipids, Serine palmitoyl-transferase long-chain base subunit 3, Single nucleotide polymorphism, Environmental factors
Background
Cardiovascular disease (CVD) is the leading cause of morbidity and mortality worldwide [1, 2]. The increased incidence of CVD in the world has been linked to dyslipidemia [3–6]. Unfavorable lipid profiles including high levels of serum total cholesterol (TC) [7], triglyceride (TG) [8], low-density lipoprotein cholesterol (LDL-C) [9] and apolipoprotein (Apo) B [10], and low levels of high-density lipoprotein cholesterol (HDL-C) [11] and ApoA1 [10] play a significant role for CVD, and are the main target for therapeutic intervention. Epidemiological studies have consistently showed that dyslipidemia is a complex trait resulted from the joint effects of multiple genetic and environmental factors [12–14]. The heritability estimates of the interindividual variations in serum lipid levels from both twin and family studies are in the range of 40–70%, suggesting a considerable genetic contribution [15, 16]. Therefore, the understanding of the association of single nucleotide polymorphisms (SNPs) and serum lipid levels has become crucial in the pursuit of reducing CVD [17].
Recently, several genome wide association studies (GWAS) have reported the association of many SNPs near the serine palmitoyl-transferase long chain base subunit 3 gene (SPTLC3; also known as: LCB3, SPT3; Gene ID: 55304; OMIM:611120; chromosomal location: 20p12.1) with one or more lipid traits [18–20] though the biological function of SPTLC3 in lipid metabolism is unknown. So far, the well-known function of SPTLC3 is that it encodes a functional subunit of the SPT enzyme-complex that catalyzes the first and rate-limiting step of de novo sphingolipid synthesis, which is involved in lipid metabolism [21, 22]. A recent GWAS about SNPs affecting serum metabolomic traits showed that the SPTLC3 rs168622 SNP was significantly correlated with hepatic lipid content [19]. Another study conducted in European populations mentioned that a locus (rs3848751) in SPTLC3 has association with HDL-C and LDL-C in males [23]. However, the association of the SPTLC3 rs364585 SNP and serum lipid levels and the mechanism were yet unclear. Furthermore, the reproducibility of this association has not been detected in the Chinese population so far.
China is a multi-ethnic country of 56 ethnic groups. Han is the largest ethnic group and Jing is one of the 55 ethnic minorities in south China with a small population of 28199 according to the sixth national census statistics of China in 2010. In the early 16th century, the Jing ancestors emigrated from Vietnam to China, and firstly settled on the three islands of Wanwei, Wutou and Shanxin in Dongxing City, where almost all of the Jing population now live [24]. The Jing population is the only oceanic ethnic group in China and preserves their custom of intra-ethnic marriage, which suggests that there are lots of differences between Jing and Han (as well as the other landlocked nationalities) nationalities in diet custom and culture characteristics. Several previous studies have showed that the associations of variants in several lipid-related genes and serum lipid profiles were significantly different between the Jing and Han populations and their gender subgroups [25–27]. However, to the best of our knowledge, the association between the SPTLC3 rs364585 SNP and serum lipid levels has not been previously reported in this population. Therefore, the present study was evaluated the association between the rs364585 SNP and several environmental factors with serum lipid levels in the Guangxi Han and Jing populations.
Methods
Subjects
A total of 824 unrelated subjects (405 males, 49.15% and 419 females, 50.85%) of Han nationality and 783 unrelated participants (389 males, 49.68% and 394 females, 50.32%) of Jing nationality were randomly selected from our previous stratified randomized samples. All participants were rural agricultural (Han) and/or fishery (Jing) workers living in Jiangping Down, Dongxing City, Guangxi Zhuang Autonomous Region, People’s Republic of China. The participants’ age ranged from 15–80 years with a mean age of 57.27 ± 12.40 years in Han and 57.19 ± 12.50 years in Jing, respectively. All participants were essentially healthy and had no evidence of diseases related to atherosclerosis, CVD and diabetes. Any participant had a history of taking medications known to affect serum lipid levels (lipid-lowering drugs such as statins or fibrates, beta blockers, diuretics, or hormones) was excluded before the blood sample was taken. The study design was approved by the Ethics Committee of the First Affiliated Hospital, Guangxi Medical University. Informed consent was obtained from all participants.
Epidemiological survey
The survey was carried out using internationally standardized methods, following a common protocol [28]. Information on demographics, socioeconomic status, and lifestyle factors was collected with standardized questionnaires. The intake of alcohol was quantified as the number of liang (about 50 g) of rice wine, corn wine, rum, beer, or liquor consumed during the preceding 12 months. Alcohol consumption was categorized into groups of grams of alcohol per day: 0, < 25 and ≥ 25. Smoking status was categorized into groups of cigarettes per day: 0, < 20 and ≥ 20. In the physical examination, several parameters such as height, weight, and waist circumference were measured. Blood pressure of the subjects in a sitting position was measured taking the mean of 3 separated intevals after the subjects had a 5-min rest using a mercury sphygmomanometer. Body mass index (BMI) was calculated as weight/height2 (kg/m2).
Biochemical measurements
A fasting venous blood sample of 5 ml was drawn from the participants. The levels of TC, TG, HDL-C and LDL-C in the samples were determined by enzymatic methods with commercially available kits. Serum ApoA1 and ApoB levels were assessed by the immune-turbidimetric immunoassay [29, 30]. Fasting blood glucose was determined by glucose meter.
DNA amplification and genotyping
Genomic DNA was isolated from peripheral blood leukocytes using the phenol-chloroform method [31, 32]. The extracted DNA was stored at 4 °C until analysis. The SPTLC3 rs364585 SNP was genotyped by polymerase chain reaction and restriction fragment length polymorphism (PCR-RFLP). PCR amplification was performed using 5’-TGCCACCTGACCATTTCTCC-3’ as the forward and 5’-AACAAACTTCTGCCTGCCTG-3’ as reversed primer pair. Each amplification reaction was performed in a total volume of 25 μL, 12.5 μL of 2 × Taq PCR MasterMix (constituent: 0.1 U Taq polymerase/μL, 500 μM dNTP each and PCR buffer), nuclease-free water 8.5 μL, 1 μL each primer (10 pmol/L) and 2 μL genomic DNA, processing started with 5 min of pre-denaturing at 95 °C and followed by 30 s of denaturing at 95 °C, 30 s of annealing at 59 °C and 35 s of elongation at 72 °C for 33 cycles. The amplification was completed by a final extension at 72 °C for 7 min. Following electrophoresis on a 2.0% agarose gel with 0.5 μg/mL ethidium bromide, the amplification products were visualized under ultraviolet light. For the restriction digestion, 5μL of amplified DNA and 5 U of MlyI restriction enzyme were added to each reaction mix, and samples were digested at 37 °C for 30 min. After restriction enzyme digestion of the amplified DNA, genotypes were identified by electrophoresis on 2% ethidium bromide stained agarose gels and visualized under ultraviolet light. Six samples (each genotype in two; respectively) detected by the PCR-RFLP were also confirmed by direct sequencing. The PCR products were purified by low melting point gel electrophoresis and phenol extraction, and then the DNA sequences were analyzed using an ABI Prism 3100 (Applied Biosystems) in Shanghai Sangon Biological Engineering Technology & Services Co., Ltd., People’s Republic of China.
Diagnostic criteria
The normal values of serum TC, TG, HDL-C, LDL-C, ApoA1 and ApoB levels, and the ratio of ApoA1 to ApoB in our Clinical Science Experiment Center were 3.10-5.17, 0.56-1.70, 1.16-1.42, 2.70-3.10 mmol/L, 1.20-1.60, 0.80-1.05g/L, and 1.00-2.50; respectively. The individuals with TC > 5.17 mmol/L and/or TG > 1.70 mmol/L were defined as hyperlipidemic [33, 34]. Hypertension was diagnosed according to the criteria of 1999 World Health Organization-International Society of Hypertension Guide lines for the management of hypertension [35, 36]. The diagnostic criteria of overweight and obesity were according to the Cooperative Meta-analysis Group of China Obesity Task Force. Normal weight, overweight and obesity were defined as a BMI < 24, 24-28, and > 28 kg/m2; respectively [37].
Statistical analysis
The statistical analyses were performed with the statistical software package SPSS 17.0 (SPSS Inc., Chicago, Illinois). The quantitative variables were presented as mean ± standard deviation (serum TG levels were presented as medians and interquartile ranges). Allele frequency was determined via direct counting, and the Hardy-Weinberg equilibrium was verified with the standard goodness-of-fit test. The genotype distribution between the groups was analyzed by the chi-square test. General characteristics between two ethnic groups were compared by the Student’s unpaired t-test. The association between genotypes and serum lipid parameters was tested by analysis of covariance (ANCOVA) with age, sex, BMI, cigarette smoking, and alcohol consumption as covariates. Multivariable linear regression analyses with stepwise modeling were used to determine the correlation between genotypes (GG = 1, GA/AA = 2) and several environmental factors with serum lipid levels in males and females of Han and Jing populations. Two sided P value < 0.05 was considered statistically significant.
Results
General characteristics and serum lipid profiles
The general characteristics and serum lipid profiles between the two ethnic groups and between males and females of the Han and Jing populations are shown in Tables 1 and 2. The values of height, weight, BMI, waist circumference, blood glucose and the levels of TC were higher in Jing than in Han (P < 0.05-0.001), whereas the percentage of alcohol consumption, the levels of diastolic blood pressure, LDL-C, and ApoA1 were lower in Jing than in Han (P < 0.05-0.001). Subgroup analyses according to sex showed that males had higher height, weight, waist circumference, the percentage of cigarette smoking and alcohol consumption than females in both ethnic groups (P < 0.05-0.001). Han males had lower systolic blood pressure, pulse pressure, TC, TG and HDL-C levels than females (P < 0.01 for all). Jing men had lower HDL-C and ApoA1 levels and the ApoA1/ApoB ratio than women (P < 0.05-0.001). Han males had higher percentage of cigarette smoking and alcohol consumption and serum ApoA1 level and lower values of weight, BMI, waist circumference, pulse pressure, blood glucose and serum TC level than Jing males (P < 0.05-0.001). Han females had higher pulse pressure and LDL-C levels and lower values of height, weight, BMI and waist circumference than Jing females (P < 0.05-0.001).
Table 1.
Comparison of demographic, lifestyle characteristics, serum lipid profiles and the genotype and allele frequencies of the SPTLC3 rs364585 SNP between the Han and Jing populations
| Parameter | Han | Jing | t (x 2) | P |
|---|---|---|---|---|
| No. of patients | 824 | 783 | ||
| Male/female | 405 / 419 | 389 / 394 | 0.045 | 0.832 |
| Age (years) | 57.27 ± 12.40 | 57.19 ± 12.50 | 0.122 | 0.903 |
| Height (cm) | 157.28 ± 8.27 | 158.08 ± 7.87 | −1.984 | 0.047 |
| Weight (kg) | 56.35 ± 9.41 | 58.80 ± 10.09 | −5.018 | 0.000 |
| Body mass index (kg/m 2) | 22.74 ± 3.19 | 23.46 ± 3.20 | −4.517 | 0.000 |
| Waist circumference (cm) | 77.46 ± 8.90 | 80.38 ± 9.20 | −6.647 | 0.000 |
| Smoking status (n%) | ||||
| 0 g/day (non-smoker) | 666 (80.8) | 656 (83.8) | ||
| ≤ 20 cigarettes/day | 31 (3.8) | 34 (4.3) | ||
| > 20 cigarettes/day | 127 (15.4) | 93 (11.9) | 4.425 | 0.109 |
| Alcohol consumption (n%) | ||||
| 0 g/day (non-drinker) | 648 (78.6) | 681 (87.0) | ||
| ≤ 25 g/day | 40 (4.9) | 83 (10.6) | ||
| > 25 g/day | 136 (16.5) | 19 (2.4) | 103.189 | 0.000 |
| Systolic blood pressure (mmHg) | 132.40 ± 18.90 | 132.18 ± 21.91 | 0.215 | 0.830 |
| Diastolic blood pressure (mmHg) | 81.78 ± 10.45 | 80.34 ± 10.30 | 2.778 | 0.006 |
| Pulse pressure (mmHg) | 50.62 ± 15.13 | 51.84 ± 17.81 | −1.481 | 0.139 |
| Glucose (mmol/l) | 6.63 ± 1.06 | 6.86 ± 1.62 | −3.390 | 0.001 |
| Total cholesterol (mmol/l) | 4.98 ± 0.85 | 5.08 ± 0.90 | −2.286 | 0.022 |
| Triglyceride (mmol/l) | 1.39(0.67) | 1.43(0.75) | −0.349 | 0.727 |
| HDL-C (mmol/l) | 1.80 ± 0.52 | 1.79 ± 0.46 | 0.419 | 0.676 |
| LDL-C (mmol/l) | 2.87 ± 0.44 | 2.82 ± 0.43 | 2.593 | 0.010 |
| Apo A1 (g/l) | 1.33 ± 0.20 | 1.28 ± 0.22 | 4.116 | 0.000 |
| ApoB (g/l) | 1.05 ± 0.24 | 1.04 ± 0.23 | 1.023 | 0.307 |
| ApoA1/ApoB | 1.32 ± 0.36 | 1.29 ± 0.38 | 1.481 | 0.139 |
| Genotype [n(%)] | ||||
| GG | 223(27.1) | 235(30.0) | ||
| GA | 392(47.6) | 371(47.4) | ||
| AA | 209(25.4) | 177(22.6) | 2.501 | 0.286 |
| Allele [n(%)] | ||||
| G | 838(50.9) | 841(53.7) | ||
| A | 810(49.1) | 725(46.3) | 2.622 | 0.105 |
HDL-C high-density lipoprotein cholesterol, LDL-C low-density lipoprotein cholesterol, Apo Apolipoprotein. The value of triglyceride was presented as median (interquartile range), the difference between the two ethnic groups was determined by the Wilcoxon-Mann-Whitney test
Table 2.
Comparison of demographic, lifestyle characteristics, serum lipid profiles and the genotype and allele frequencies of the SPTLC3 rs364585 SNP between males and females in the Han and Jing populations
| Parameter | Han | t (x 2) | P | Jing | t (x 2) | P | ||
|---|---|---|---|---|---|---|---|---|
| Male | Female | Male | Female | |||||
| No. of patients | 405 | 419 | 389 | 394 | ||||
| Age (years) | 56.88 ± 11.64 | 57.65 ± 13.10 | −0.888 | 0.375 | 57.77 ± 14.20 | 56.64 ± 12.27 | 1.129 | 0.234 |
| Height (cm) | 162.99 ± 5.56 | 151.76 ± 6.51 | 26.538 | 0.000 | 162.94 ± 6.60 | 153.29 ± 5.82c | 21.728 | 0.000 |
| Weight (kg) | 60.29 ± 8.53 | 52.54 ± 8.83 | 12.820 | 0.000 | 62.35 ± 10.02b | 55.30 ± 8.87c | 10.437 | 0.000 |
| Body mass index (kg/m2) | 22.68 ± 2.81 | 22.80 ± 3.52 | −0.560 | 0.567 | 23.42 ± 3.10c | 23.50 ± 3.30b | −0.333 | 0.740 |
| Waist circumference (cm) | 78.31 ± 8.19 | 76.63 ± 9.49 | 2.730 | 0.006 | 81.59 ± 9.83c | 79.19 ± 8.40c | 3.677 | 0.000 |
| Smoking status (n%) | ||||||||
| Non-smoker | 248 (61.2) | 418 (99.8) | 264 (67.9) | 392 (99.5) | ||||
| Smoker | 157 (38.8) | 1(0.2) | 196.00 | 0.000 | 115 (32.1)a | 2 (0.5) | 133.70 | 0.000 |
| Alcohol consumption (n%) | ||||||||
| Non- drinker | 232 (57.3) | 416 (99.3) | 292(75.1) | 389 (98.7) | ||||
| Drinker | 173 (42.7) | 3 (0.7) | 216.01 | 0.000 | 97(24.9)c | 5 (1.3) | 96.65 | 0.000 |
| Systolic blood pressure (mmHg) | 130.14 ± 16.58 | 134.58 ± 20.79 | −3.392 | 0.001 | 132.26 ± 20.77 | 132.11 ± 23.00 | 0.096 | 0.924 |
| Diastolic blood pressure (mmHg) | 81.99 ± 10.38 | 81.58 ± 10.50 | 0.554 | 0.580 | 80.68 ± 10.61 | 80.02 ± 10.00a | 0.901 | 0.368 |
| Pulse pressure (mmHg) | 48.16 ± 13.21 | 53.00 ± 16.44 | −4.651 | 0.000 | 51.59 ± 16.87b | 52.09 ± 18.71 | −0.403 | 0.687 |
| Glucose (mmol/l) | 6.58 ± 1.08 | 6.68 ± 1.04 | −1.348 | 0.178 | 6.94 ± 1.75c | 6.79 ± 1.49 | 1.307 | 0.191 |
| Total cholesterol (mmol/l) | 4.90 ± 0.83 | 5.06 ± 0.87 | −2.734 | 0.006 | 5.04 ± 0.82a | 5.11 ± 0.97 | −1.165 | 0.244 |
| Triglyceride (mmol/l) | 1.39(1.71) | 1.40(1.63) | −2.720 | 0.007 | 1.48(1.83) | 1.40(1.63) | −0.963 | 0.336 |
| HDL-C (mmol/l) | 1.74 ± 0.55 | 1.87 ± 0.48 | −2.417 | 0.000 | 1.75 ± 0.46 | 1.83 ± 0.45 | −2.417 | 0.016 |
| LDL-C (mmol/l) | 2.86 ± 0.43 | 2.89 ± 0.45 | −3.615 | 0.382 | 2.82 ± 0.37 | 2.81 ± 0.48a | 0.183 | 0.855 |
| Apo A1 (g/l) | 1.33 ± 0.21 | 1.32 ± 0.20 | 0.263 | 0.793 | 1.26 ± 0.21c | 1.30 ± 0.23 | −2.764 | 0.006 |
| ApoB (g/l) | 1.06 ± 0.23 | 1.05 ± 0.26 | 0.465 | 0.642 | 1.04 ± 0.22 | 1.03 ± 0.24 | 1.011 | 0.312 |
| ApoA1/ApoB | 1.31 ± 0.36 | 1.33 ± 0.37 | −0.697 | 0.486 | 1.26 ± 0.40 | 1.32 ± 0.37 | −2.102 | 0.036 |
| Genotype [n(%)] | ||||||||
| GG | 105(25.9) | 118(28.2) | 108(27.8) | 127(32.2) | ||||
| GA | 188(46.4) | 204(48.7) | 188(48.3) | 183(46.4) | ||||
| AA | 112(27.7) | 97(23.2) | 2.250 | 0.325 | 93(23.9) | 84(21.3) | 2.029 | 0.363 |
| Allele [n(%)] | ||||||||
| G | 398(49.1) | 440(52.6) | 404(51.9) | 437(55.5) | ||||
| A | 412(50.9) | 398(47.4) | 1.972 | 0.171 | 374(48.1) | 351(44.5) | 1.961 | 0.161 |
HDL-C high-density lipoprotein cholesterol, LDL-C low-density lipoprotein cholesterol, Apo Apolipoprotein. The value of triglyceride was presented as median (interquartile range), the difference between the two groups was determined by the Wilcoxon-Mann-Whitney test. a P < 0.001, b P < 0.01 and c P < 0.05 in comparison with the same sex subgroup of the Han ethnic group
Results of genotyping
After the genomic DNA of the samples was amplified by PCR, the purpose gene of 519-bp nucleotide sequences could be seen in all samples (Fig. 1). The genotypes identified were labeled according to the presence or absence of the enzyme restriction sites. Thus, AA genotype is homozygote for the absence of the site (band at 519-bp), GA genotype is heterozygote for the presence and absence of the site (bands at 519-, 306- and 213-bp) and GG genotype is homozygote for the presence of the site (bands at 306- and 213-bp, Fig. 2). The GG, GA and AA genotypes detected by PCR-RFLP were also confirmed by direct sequencing (Fig. 3).
Fig. 1.
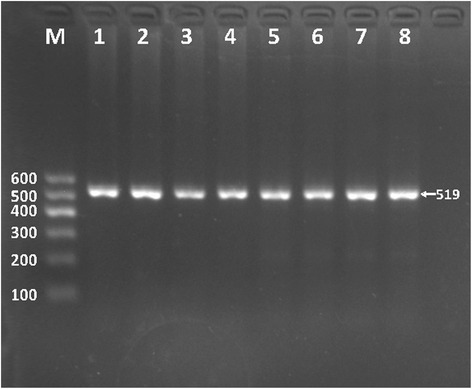
Electrophoresis of polymerase chain reaction products of the samples. Lane M is the 100-bp marker ladder; Lanes 1-8 are samples, the 519-bp bands are the target genes
Fig. 2.
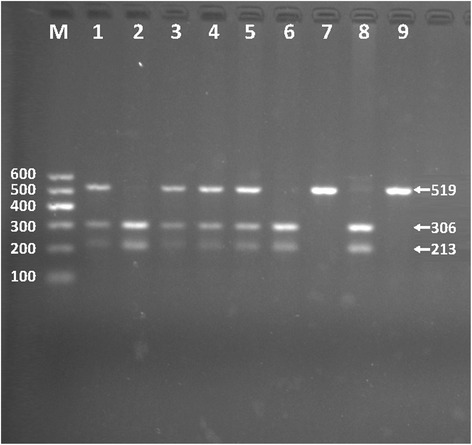
Genotyping of the SPTLC3 rs364585 SNP. Lane M is the 100-bp Marker Ladder; lanes 2, 6 and 8, GG genotype (306- and 213-bp); lanes 1, 3, 4 and 5, GA genotype (519-, 306- and 213-bp); and lanes 7 and 9, AA genotype (519-bp)
Fig. 3.

A part of the nucleotide sequence of the SPTLC3 rs364585 SNP. a AA genotype; b GA genotype; and c GG genotype
Genotypic and allelic frequencies
The genotypic and allelic distribution of the rs364585 SNP is also revealed in Tables 1 and 2. The genotypic distribution was followed Hardy-Weinberg equilibrium (HWE). The frequency of SPTLC3 rs364585-A allele was 49.1% in Han and 46.3% in Jing. There was no significant difference in either genotypic or allelic frequencies between Han and Jing (Table 1), or between males and females (Table 2) of the both ethnic groups.
Genotypes and serum lipid levels
Tables 3 and 4 describe the association between genotypes and serum lipid levels in the two ethnic groups. The levels of LDL-C and the ratio of ApoA1 to ApoB in Han were different between the genotypes (P < 0.05-0.001), the A allele carriers had higher LDL-C levels and lower the ApoA1/ApoB ratio than the A allele non-carriers. For the Jing population, serum TC, HDL-C and LDL-C levels were different between the genotypes (P < 0.05-0.001), the A allele carriers had higher TC and LDL-C levels and lower HDL-C levels than the A allele non-carriers. In the sex subgroup analyses, the A allele carriers in Han males had higher serum LDL-C levels than the A allele non-carriers (P < 0.05); the A allele carriers in Jing had higher TC and LDL-C levels in males; and higher LDL-C levels and lower HDL-C levels in females than the A allele non-carriers (P < 0.05 for all).
Table 3.
Comparison of the genotypes and serum lipid levels in the Han and Jing populations
| Genotype | n | TC (mmol/L) |
TG (mmol/L) |
HDL-C (mmol/L) |
LDL-C (mmol/L) |
ApoA1 (g/L) |
ApoB (g/L) |
ApoA1 /ApoB |
|---|---|---|---|---|---|---|---|---|
| Han | ||||||||
| GG | 223 | 4.96 ± 0.90 | 1.38(0.69) | 1.84 ± 0.46 | 2.80 ± 0.45 | 1.33 ± 0.20 | 1.04 ± 0.25 | 1.35 ± 0.39 |
| GA/AA | 601 | 4.98 ± 0.83 | 1.40(0.67) | 1.79 ± 0.54 | 2.89 ± 0.43 | 1.32 ± 0.20 | 1.06 ± 0.24 | 1.31 ± 0.36 |
| F | 0.796 | 1.429 | 2.395 | 14.391 | 0.698 | 1.572 | 4.243 | |
| P | 0.373 | 0.153 | 0.122 | 0.000 | 0.404 | 0.210 | 0.039 | |
| Jing | ||||||||
| GG | 235 | 5.00 ± 0.88 | 1.40(0.72) | 1.85 ± 0.43 | 2.73 ± 0.46 | 1.29 ± 0.22 | 1.04 ± 0.25 | 1.34 ± 0.46 |
| GA/ AA | 548 | 5.12 ± 0.91 | 1.44(0.75) | 1.77 ± 0.47 | 2.84 ± 0.43 | 1.27 ± 0.22 | 1.04 ± 0.22 | 1.28 ± 0.35 |
| F | 6.391 | 0.062 | 6.612 | 16.788 | 0.734 | 0.002 | 2.705 | |
| P | 0.012 | 0.979 | 0.013 | 0.000 | 0.392 | 0.961 | 0.100 | |
TC total cholesterol, TG triglyceride, HDL-C high-density lipoprotein cholesterol, LDL-C low-density lipoprotein cholesterol, ApoA1 apolipoprotein A1, ApoB apolipoprotein B, ApoA1/ApoB the ratio of apolipoprotein A1 to apolipoprotein B. The value of TG was presented as median (interquartile range), the difference between the genotypes was determined by the Wilcoxon-Mann-Whitney test
Table 4.
Comparison between the SPTLC3 rs364585 SNP genotypes and serum lipid levels between males and females in the Han and Jing populations
| Ethnic/ Genotype | n | TC (mmol/L) |
TG (mmol/L) |
HDL-C (mmol/L) |
LDL-C (mmol/L) |
ApoA1 (g/L) |
ApoB (g/L) |
ApoA1 /ApoB |
|---|---|---|---|---|---|---|---|---|
| Han/male | ||||||||
| GG | 105 | 4.85 ± 0.88 | 1.32(0.62) | 1.80 ± 0.45 | 2.75 ± 0.41 | 1.34 ± 0.22 | 1.03 ± 0.21 | 1.35 ± 0.38 |
| GA / AA | 300 | 4.92 ± 0.80 | 1.41(0.72) | 1.72 ± 0.57 | 2.90 ± 0.42 | 1.32 ± 0.20 | 1.07 ± 0.23 | 1.30 ± 0.35 |
| F | 1.560 | 1.674 | 2.028 | 11.855 | 1.117 | 2.088 | 2.563 | |
| P | 0.212 | 0.094 | 0.155 | 0.001 | 0.291 | 0.149 | 0.110 | |
| Han/female | ||||||||
| GG | 118 | 5.07 ± 0.90 | 1.41(0.68) | 1.87 ± 0.46 | 2.83 ± 0.47 | 1.32 ± 0.18 | 1.05 ± 0.29 | 1.34 ± 0.40 |
| GA / AA | 301 | 5.05 ± 0.86 | 1.40(0.60) | 1.87 ± 0.48 | 2.93 ± 0.44 | 1.32 ± 0.20 | 1.05 ± 0.25 | 1.33 ± 0.36 |
| F | 0.004 | 0.358 | 0.509 | 2.434 | 0.128 | 0.004 | 1.096 | |
| P | 0.951 | 0.720 | 0.476 | 0.120 | 0.721 | 0.948 | 0.296 | |
| Jing/male | ||||||||
| GG | 108 | 4.86 ± 0.72 | 1.40(0.95) | 1.81 ± 0.42 | 2.75 ± 0.36 | 1.29 ± 0.23 | 1.06 ± 0.23 | 1.31 ± 0.48 |
| GA/ AA | 281 | 5.12 ± 0.85 | 1.50(0.81) | 1.73 ± 0.48 | 2.85 ± 0.38 | 1.25 ± 0.20 | 1.05 ± 0.21 | 1.25 ± 0.37 |
| F | 14.266 | 0.448 | 0.278 | 6.305 | 1.200 | 0.616 | 0.465 | |
| P | 0.000 | 0.654 | 0.598 | 0.012 | 0.274 | 0.433 | 0.496 | |
| Jing/female | ||||||||
| GG | 127 | 5.11 ± 0.98 | 1.40(0.59) | 1.92 ± 0.42 | 2.72 ± 0.53 | 1.31 ± 0.21 | 1.02 ± 0.27 | 1.36 ± 0.44 |
| GA/ AA | 267 | 5.12 ± 0.97 | 1.39(0.66) | 1.79 ± 0.45 | 2.86 ± 0.48 | 1.30 ± 0.25 | 1.04 ± 0.23 | 1.31 ± 0.33 |
| F | 0.035 | 0.669 | 7.450 | 6.235 | 0.018 | 0.005 | 1.308 | |
| P | 0.852 | 0.503 | 0.007 | 0.013 | 0.892 | 0.945 | 0.253 | |
TC total cholesterol, TG triglyceride, HDL-C high-density lipoprotein cholesterol, LDL-C low-density lipoprotein cholesterol, ApoA1 apolipoprotein A1, ApoB apolipoprotein B, ApoA1/ApoB the ratio of apolipoprotein A1 to apolipoprotein B. The values of triglyceride were presented as median (interquartile range), and the difference between the GG and GA/AA genotypes was determined by the Wilcoxon-Mann-Whitney test
Relative factors for serum lipid parameters
Several environmental factors such as age, gender, height, weight, waist circumference, alcohol consumption and cigarette smoking, and traditional cardiovascular risk factors such as BMI, fasting blood glucose and blood pressure levels were also correlated with serum lipid parameters in the Han and Jing populations and in males and females of both ethnic groups (P < 0.05-0.001, Tables 5 and 6).
Table 5.
The risk factors for serum lipid parameters in the Han and Jing populations
| Lipid | Risk factor | B | Std.error | Beta | t | P |
|---|---|---|---|---|---|---|
| Han and Jing | ||||||
| TC | Glucose | 0.140 | 0.016 | 0.220 | 8.922 | 0.000 |
| Age | 0.009 | 0.002 | 0.130 | 4.838 | 0.000 | |
| Gender | 0.198 | 0.047 | 0.113 | 4.206 | 0.000 | |
| Diastolic blood pressure | 0.005 | 0.002 | 0.056 | 2.312 | 0.021 | |
| Genotype | 0.097 | 0.046 | 0.050 | 2.091 | 0.037 | |
| Pulse pressure | −0.003 | 0.001 | −0.060 | −2.233 | 0.026 | |
| Ethnic group | 0.102 | 0.043 | 0.058 | 2.369 | 0.018 | |
| Alcohol consumption | 0.077 | 0.038 | 0.055 | 2.034 | 0.042 | |
| TG | Waist circumference | 0.038 | 0.004 | 0.406 | 8.415 | 0.000 |
| Cigarette smoking | 0.257 | 0.031 | 0.211 | 8.403 | 0.000 | |
| Glucose | 0.071 | 0.015 | 0.113 | 4.784 | 0.000 | |
| Height | −0.015 | 0.003 | −0.143 | −4.952 | 0.000 | |
| Diastolic blood pressure | 0.007 | 0.002 | 0.085 | 3.575 | 0.000 | |
| Body mass index | −0.029 | 0.012 | −0.109 | −2.338 | 0.020 | |
| Age | −0.003 | 0.002 | −0.053 | −2.139 | 0.033 | |
| HDL-C | Waist circumference | −0.017 | 0.001 | −0.321 | −12.942 | 0.000 |
| Alcohol consumption | 0.143 | 0.021 | 0.183 | 6.662 | 0.000 | |
| Gender | 0.193 | 0.034 | 0.198 | 5.602 | 0.000 | |
| Cigarette smoking | −0.053 | 0.019 | −0.076 | −2.717 | 0.007 | |
| Height | 0.008 | 0.002 | 0.124 | 3.641 | 0.000 | |
| Genotype | −0.070 | 0.025 | −0.065 | −2.778 | 0.006 | |
| Age | 0.003 | 0.001 | 0.070 | 2.691 | 0.007 | |
| Ethnic group | 0.061 | 0.024 | 0.063 | 2.590 | 0.010 | |
| LDL-C | Glucose | 0.037 | 0.008 | 0.116 | 4.650 | 0.000 |
| Genotype | 0.123 | 0.024 | 0.127 | 5.223 | 0.000 | |
| Age | 0.003 | 0.001 | 0.091 | 3.658 | 0.000 | |
| Diastolic blood pressure | 0.003 | 0.001 | 0.076 | 3.081 | 0.002 | |
| Ethnic group | −0.056 | 0.021 | −0.064 | −2.611 | 0.009 | |
| ApoA1 | Waist circumference | −0.003 | 0.001 | −0.137 | −3.059 | 0.002 |
| Alcohol consumption | 0.077 | 0.009 | 0.224 | 8.472 | 0.000 | |
| Gender | 0.052 | 0.011 | 0.121 | 4.553 | 0.000 | |
| Glucose | −0.011 | 0.004 | −0.068 | −2.818 | 0.005 | |
| Systolic blood pressure | 0.001 | 0.000 | 0.067 | 2.721 | 0.007 | |
| Body mass index | −0.006 | 0.003 | −0.089 | −2.006 | 0.045 | |
| ApoB | Waist circumference | 0.005 | 0.001 | 0.200 | 7.974 | 0.000 |
| Age | 0.002 | 0.000 | 0.089 | 3.364 | 0.001 | |
| Ethnic group | −0.027 | 0.012 | −0.056 | −2.299 | 0.022 | |
| Systolic blood pressure | 0.001 | 0.000 | 0.061 | 2.294 | 0.022 | |
| ApoA1/ApoB | Waist circumference | −0.007 | 0.002 | −0.172 | −3.447 | 0.001 |
| Glucose | −0.023 | 0.007 | −0.084 | −3.485 | 0.001 | |
| Alcohol consumption | 0.065 | 0.016 | 0.108 | 4.112 | 0.000 | |
| Age | −0.002 | 0.001 | −0.059 | −2.241 | 0.025 | |
| Genotype | −0.049 | 0.020 | −0.058 | −2.470 | 0.014 | |
| Weight | −0.039 | 0.011 | −1.020 | −3.672 | 0.000 | |
| Height | 0.029 | 0.008 | 0.634 | 3.801 | 0.000 | |
| Body mass index | 0.082 | 0.026 | 0.702 | 3.145 | 0.002 | |
| Gender | 0.063 | 0.026 | 0.084 | 2.414 | 0.016 | |
| Han | ||||||
| TC | Glucose | 0.210 | 0.027 | 0.262 | 7.710 | 0.000 |
| Cigarette smoking | −0.117 | 0.039 | −0.101 | −3.042 | 0.002 | |
| Pulse pressure | 0.001 | 0.000 | 0.072 | 2.141 | 0.033 | |
| Systolic blood pressure | 0.003 | 0.002 | 0.073 | 2.123 | 0.034 | |
| TG | Waist circumference | 0.038 | 0.006 | 0.386 | 6.678 | 0.000 |
| Cigarette smoking | 0.226 | 0.041 | 0.190 | 5.535 | 0.000 | |
| Glucose | 0.107 | 0.027 | 0.131 | 3.978 | 0.000 | |
| Diastolic blood pressure | 0.007 | 0.003 | 0.089 | 2.639 | 0.008 | |
| Weight | −0.014 | 0.005 | −0.150 | −2.558 | 0.011 | |
| HDL-C | Waist circumference | −0.015 | 0.002 | −0.265 | −7.742 | 0.000 |
| Cigarette smoking | −0.125 | 0.028 | −0.177 | −4.404 | 0.000 | |
| Alcohol consumption | 0.135 | 0.028 | 0.196 | 4.803 | 0.000 | |
| Gender | 0.212 | 0.051 | 0.205 | 4.162 | 0.000 | |
| Height | 0.009 | 0.003 | 0.138 | 3.008 | 0.003 | |
| Genotype | −0.089 | 0.038 | −0.076 | −2.306 | 0.021 | |
| LDL-C | Glucose | 0.073 | 0.014 | 0.175 | 5.042 | 0.000 |
| Systolic blood pressure | 0.003 | 0.001 | 0.133 | 3.853 | 0.000 | |
| Genotype | 0.122 | 0.033 | 0.123 | 3.658 | 0.000 | |
| ApoA1 | Alcohol consumption | 0.071 | 0.009 | 0.264 | 7.538 | 0.000 |
| Weight | −0.004 | 0.001 | −0.186 | −5.318 | 0.000 | |
| ApoB | Waist circumference | 0.005 | 0.001 | 0.185 | 5.415 | 0.000 |
| Systolic blood pressure | 0.002 | 0.000 | 0.132 | 3.763 | 0.000 | |
| Glucose | 0.019 | 0.008 | 0.083 | 2.393 | 0.017 | |
| ApoA1/ApoB | Body mass index | −0.028 | 0.004 | −0.245 | −7.259 | 0.000 |
| Glucose | −0.042 | 0.012 | −0.122 | −3.570 | 0.000 | |
| Alcohol consumption | 0.063 | 0.019 | 0.130 | 3.376 | 0.001 | |
| Gender | 0.078 | 0.028 | 0.107 | 2.775 | 0.006 | |
| Systolic blood pressure | −0.002 | 0.001 | −0.080 | −2.290 | 0.022 | |
| Genotype | −0.056 | 0.027 | −0.069 | −2.065 | 0.039 | |
| Jing | ||||||
| TC | Glucose | 0.103 | 0.019 | 0.186 | 5.414 | 0.000 |
| Age | 0.017 | 0.003 | 0.262 | 6.767 | 0.000 | |
| Pulse pressure | −0.008 | 0.002 | −0.151 | −4.064 | 0.000 | |
| Gender | 0.348 | 0.073 | 0.193 | 4.754 | 0.000 | |
| Cigarette smoking | 0.198 | 0.053 | 0.146 | 3.767 | 0.000 | |
| Alcohol consumption | 0.296 | 0.077 | 0.139 | 3.825 | 0.000 | |
| Body mass index | 0.056 | 0.017 | 0.198 | 3.257 | 0.001 | |
| Genotype | 0.166 | 0.066 | 0.085 | 2.524 | 0.012 | |
| Waist circumference | −0.015 | 0.006 | −0.158 | −2.539 | 0.011 | |
| Diastolic blood pressure | 0.006 | 0.003 | 0.069 | 1.964 | 0.050 | |
| TG | Waist circumference | 0.034 | 0.003 | 0.373 | 10.642 | 0.000 |
| Cigarette smoking | 0.286 | 0.045 | 0.226 | 6.436 | 0.000 | |
| Height | −0.016 | 0.004 | −0.149 | −4.041 | 0.000 | |
| Glucose | 0.049 | 0.017 | 0.096 | 2.896 | 0.004 | |
| HDL-C | Waist circumference | −0.018 | 0.002 | −0.364 | −11.101 | 0.000 |
| Alcohol consumption | 0.250 | 0.037 | 0.232 | 6.739 | 0.000 | |
| Gender | 0.103 | 0.032 | 0.113 | 3.259 | 0.001 | |
| Genotype | −0.089 | 0.032 | −0.090 | −2.758 | 0.006 | |
| LDL-C | Genotype | 0.136 | 0.033 | 0.144 | 4.123 | 0.000 |
| Age | 0.005 | 0.001 | 0.149 | 4.028 | 0.000 | |
| Cigarette smoking | 0.090 | 0.026 | 0.139 | 3.503 | 0.000 | |
| Gender | 0.080 | 0.034 | 0.092 | 2.308 | 0.021 | |
| Glucose | 0.019 | 0.009 | 0.073 | 2.054 | 0.040 | |
| Diastolic blood pressure | 0.003 | 0.001 | 0.071 | 2.031 | 0.043 | |
| ApoA1 | Waist circumference | −0.006 | 0.001 | −0.261 | −7.425 | 0.000 |
| Alcohol consumption | 0.067 | 0.019 | 0.127 | 3.469 | 0.001 | |
| Gender | 0.048 | 0.016 | 0.107 | 2.933 | 0.003 | |
| Systolic blood pressure | 0.001 | 0.000 | 0.079 | 2.278 | 0.023 | |
| ApoB | Waist circumference | 0.005 | 0.001 | 0.202 | 5.802 | 0.000 |
| Age | 0.002 | 0.001 | 0.117 | 3.348 | 0.001 | |
| ApoA1/ApoB | Waist circumference | −0.008 | 0.003 | −0.196 | −3.198 | 0.001 |
| Age | −0.003 | 0.001 | −0.122 | −3.482 | 0.001 | |
| Alcohol consumption | 0.087 | 0.031 | 0.095 | 2.776 | 0.006 | |
| Weight | −0.005 | 0.002 | −0.136 | −2.187 | 0.029 | |
TC total cholesterol, TG triglyceride, HDL-C high-density lipoprotein cholesterol, LDL-C low-density lipoprotein cholesterol, ApoA1 apolipoprotein A1, ApoB apolipoprotein B, ApoA1/ApoB the ratio of apolipoprotein A1 to apolipoprotein B
Table 6.
The risk factors for serum lipid parameters in the males and females of the Han and Jing populations
| Lipid | Risk factor | B | Std.error | Beta | t | P |
|---|---|---|---|---|---|---|
| Han/male | ||||||
| TC | Glucose | 0.224 | 0.036 | 0.295 | 6.291 | 0.000 |
| Pulse pressure | 0.001 | 0.000 | 0.114 | 2.444 | 0.015 | |
| Diastolic blood pressure | 0.009 | 0.004 | 0.110 | 2.357 | 0.019 | |
| Cigarette smoking | −0.089 | 0.042 | −0.100 | −2.123 | 0.034 | |
| TG | Waist circumference | 0.049 | 0.010 | 0.405 | 4.765 | 0.000 |
| Cigarette smoking | 0.219 | 0.052 | 0.201 | 4.182 | 0.000 | |
| Glucose | 0.168 | 0.044 | 0.181 | 3.770 | 0.000 | |
| Diastolic blood pressure | 0.014 | 0.005 | 0.142 | 2.998 | 0.003 | |
| Age | −0.013 | 0.004 | −0.146 | −2.823 | 0.005 | |
| Weight | −0.023 | 0.010 | −0.198 | −2.256 | 0.025 | |
| HDL-C | Waist circumference | −0.021 | 0.003 | −0.307 | −6.068 | 0.000 |
| Alcohol consumption | 0.129 | 0.031 | 0.217 | 4.171 | 0.000 | |
| Cigarette smoking | −0.128 | 0.031 | −0.213 | −4.164 | 0.000 | |
| Genotype | −0.138 | 0.059 | −0.111 | −2.347 | 0.019 | |
| Height | 0.011 | 0.005 | 0.109 | 2.244 | 0.025 | |
| Diastolic blood pressure | 0.005 | 0.003 | 0.101 | 2.061 | 0.040 | |
| LDL-C | Glucose | 0.095 | 0.019 | 0.241 | 5.060 | 0.000 |
| Genotype | 0.165 | 0.046 | 0.170 | 3.570 | 0.000 | |
| Diastolic blood pressure | 0.005 | 0.002 | 0.111 | 2.324 | 0.021 | |
| ApoA1 | Alcohol consumption | 0.082 | 0.011 | 0.366 | 7.783 | 0.000 |
| Waist circumference | −0.005 | 0.001 | −0.196 | −4.153 | 0.000 | |
| ApoB | Waist circumference | 0.005 | 0.001 | 0.184 | 3.767 | 0.000 |
| Systolic blood pressure | 0.002 | 0.001 | 0.179 | 3.513 | 0.000 | |
| Glucose | 0.031 | 0.011 | 0.146 | 2.913 | 0.004 | |
| Age | −0.002 | 0.001 | −0.118 | −2.296 | 0.022 | |
| ApoA1/ApoB | Waist circumference | −0.011 | 0.002 | −0.246 | −5.086 | 0.000 |
| Alcohol consumption | 0.087 | 0.019 | 0.221 | 4.512 | 0.000 | |
| Systolic blood pressure | −0.003 | 0.001 | −0.153 | −2.986 | 0.003 | |
| Age | 0.005 | 0.002 | 0.166 | 3.246 | 0.001 | |
| Glucose | −0.045 | 0.017 | −0.133 | −2.692 | 0.007 | |
| Han/female | ||||||
| TC | Glucose | 0.223 | 0.039 | 0.266 | 5.645 | 0.000 |
| TG | Waist circumference | 0.024 | 0.004 | 0.320 | 6.964 | 0.000 |
| Glucose | 0.094 | 0.032 | 0.135 | 2.936 | 0.004 | |
| HDL-C | Body mass index | −0.034 | 0.006 | −0.253 | −5.330 | 0.000 |
| LDL-C | Glucose | 0.057 | 0.021 | 0.131 | 2.684 | 0.008 |
| Age | 0.005 | 0.002 | 0.156 | 3.209 | 0.001 | |
| ApoA1 | Body mass index | −0.009 | 0.003 | −0.153 | −3.170 | 0.002 |
| ApoB | Height | 0.006 | 0.001 | 0.215 | 4.562 | 0.000 |
| Age | 0.003 | 0.001 | 0.166 | 3.349 | 0.001 | |
| Height | −0.005 | 0.002 | −0.129 | −2.580 | 0.010 | |
| ApoA1/ApoB | Age | −0.003 | 0.001 | −0.113 | −2.330 | 0.020 |
| Glucose | −0.052 | 0.016 | −0.147 | −3.208 | 0.001 | |
| Body mass index | 0.193 | 0.049 | 1.846 | 3.928 | 0.000 | |
| Height | 0.074 | 0.015 | 1.311 | 4.827 | 0.000 | |
| Weight | −0.095 | 0.021 | −2.293 | −4.464 | 0.000 | |
| Jing/male | ||||||
| TC | Alcohol consumption | 0.313 | 0.072 | 0.212 | 4.356 | 0.000 |
| Glucose | 0.073 | 0.023 | 0.156 | 3.185 | 0.002 | |
| Cigarette smoking | 0.154 | 0.050 | 0.159 | 3.097 | 0.002 | |
| Age | 0.017 | 0.003 | 0.292 | 5.273 | 0.000 | |
| Genotype | 0.315 | 0.087 | 0.172 | 3.634 | 0.000 | |
| Pulse pressure | −0.011 | 0.003 | −0.232 | −4.453 | 0.000 | |
| Body mass index | 0.097 | 0.026 | 0.368 | 3.800 | 0.000 | |
| Waist circumference | −0.024 | 0.008 | −0.284 | −2.857 | 0.005 | |
| TG | Waist circumference | 0.040 | 0.005 | 0.426 | 8.633 | 0.000 |
| Cigarette smoking | 0.214 | 0.052 | 0.198 | 4.119 | 0.000 | |
| Height | −0.025 | 0.007 | −0.179 | −3.484 | 0.001 | |
| Age | −0.011 | 0.003 | −0.168 | −3.249 | 0.001 | |
| Glucose | 0.066 | 0.025 | 0.125 | 2.666 | 0.008 | |
| HDL-C | Waist circumference | −0.019 | 0.002 | −0.402 | −8.995 | 0.000 |
| Alcohol consumption | 0.259 | 0.037 | 0.310 | 6.939 | 0.000 | |
| LDL-C | Cigarette smoking | 0.061 | 0.022 | 0.139 | 2.778 | 0.006 |
| Genotype | 0.091 | 0.042 | 0.109 | 2.161 | 0.031 | |
| Body mass index | 0.012 | 0.006 | 0.101 | 2.001 | 0.046 | |
| ApoA1 | Waist circumference | −0.008 | 0.001 | −0.357 | −7.594 | 0.000 |
| Alcohol consumption | 0.072 | 0.018 | 0.191 | 4.056 | 0.000 | |
| ApoB | Weight | 0.007 | 0.001 | 0.314 | 6.223 | 0.000 |
| Age | 0.002 | 0.001 | 0.112 | 2.228 | 0.026 | |
| ApoA1/ ApoB | Waist circumference | −0.015 | 0.002 | −0.375 | −7.981 | 0.000 |
| Alcohol consumption | 0.109 | 0.034 | 0.150 | 3.184 | 0.002 | |
| Jing/female | ||||||
| TC | Glucose | 0.163 | 0.031 | 0.251 | 5.250 | 0.000 |
| Age | 0.021 | 0.004 | 0.266 | 5.145 | 0.000 | |
| Pulse pressure | −0.006 | 0.003 | −0.106 | −2.046 | 0.041 | |
| TG | Waist circumference | 0.026 | 0.004 | 0.291 | 6.007 | 0.000 |
| Cigarette smoking | 1.164 | 0.318 | 0.174 | 3.661 | 0.000 | |
| Height | −0.027 | 0.006 | −0.207 | −4.240 | 0.000 | |
| HDL-C | Waist circumference | −0.018 | 0.003 | −0.347 | −7.170 | 0.000 |
| Genotype | −0.131 | 0.045 | −0.138 | −2.920 | 0.004 | |
| Diastolic blood pressure | 0.004 | 0.002 | 0.099 | 2.041 | 0.042 | |
| LDL-C | Genotype | 0.123 | 0.050 | 0.119 | 2.463 | 0.014 |
| Age | 0.008 | 0.002 | 0.210 | 4.293 | 0.000 | |
| Glucose | 0.045 | 0.016 | 0.139 | 2.855 | 0.005 | |
| ApoA1 | Body mass index | −0.011 | 0.004 | −0.159 | −3.194 | 0.002 |
| ApoB | Age | 0.004 | 0.001 | 0.191 | 3.893 | 0.000 |
| Body mass index | 0.010 | 0.004 | 0.138 | 2.805 | 0.005 | |
| ApoA1/ApoB | Body mass index | −0.023 | 0.005 | −0.205 | −4.176 | 0.000 |
| Age | −0.004 | 0.001 | −0.144 | −2.943 | 0.003 | |
TC total cholesterol, TG triglyceride, HDL-C high-density lipoprotein cholesterol, LDL-C low-density lipoprotein cholesterol, ApoA1 apolipoprotein A1, ApoB apolipoprotein B, ApoA1/ApoB the ratio of apolipoprotein A1 to apolipoprotein B
Discussion
In the present study, we found that the levels of TC were higher but the levels of LDL-C and ApoA1 were lower in Jing than in Han. In the sex subgroup analyses, Han males had lower serum TC, TG and HDL-C levels than Han females, whereas Jing men had lower HDL-C and ApoA1 levels and the ApoA1/ApoB ratio than Jing women. Han males had higher ApoA1 levels and lower TC levels than Jing males, whereas Han females had higher LDL-C levels than Jing females. It was widely realized that dyslipidemia as a serious risk factor for CVD is a multifactorial and complicated disease caused by genetic factors, including lipid-associated gene variants and environmental factors, including age, sex, diet, alcohol consumption, cigarette smoking, obesity, exercise, hypertension [38, 39], and their interactions [12, 13].
Han nationality is the largest ethnic group among the 56 ethnic groups in China and is widely distributed in 2/3 regions of China. Its economy was dominated by agriculture and it abounds in rice, corn and wheat. Jing, one of the 55 official ethnic minorities in China is the only Chinese minority for coastal fisheries and is the only sea people in China. They live in a relatively isolated environment and share local similar recipes. In this case, it has a very special lifestyle and dietary habits compared with the other landlocked nationalities. Their marriages were family-arranged in the old days when they sing antiphonal songs to look for the other half. After antiphonal singing, if the boy’s into the girl he would kick sand toward her while approaching her. If the girl feel the same she would kick back, which means engagement. While the formal engagement ceremony and wedding they need pork, cake, tea, wine, glutinous rice as gifts. Jing stays endogamy, intermarriage with Han or Zhuang people is seldom happened. Owing to its own strict intra-ethnic marriage customs and unique traditions, we speculate that some hereditary characteristics and genotypes of lipid metabolism-related genes in this population might be different from those in Han Chinese.
The genotypic and allelic frequencies of the SPTLC3 rs364585 SNP in diverse racial/ethnic groups are significantly different. According to the International HapMap Project’s data-base, the frequencies of A allele and AA, AG genotypes were 34.4%, 15.6% and 37.8% in Han Chinese in Beijing; 40.8%, 13.3% and 55.0% in European; 48.9%, 27.3% and 43.2% in Japanese; and 10.8%, 1.7% and 18.3% in Yoruba; respectively. In the present study, we showed that the frequencies of A allele and AA, AG genotypes were 49.1%, 25.4% and 47.6% in Han; and 46.3%, 22.6% and 47.4% in Jing; respectively. There were no conspicuous differences in the genotypic and allelic frequencies of the rs364585 SNP between the Jing and Han populations, or between males and females in the both ethnic groups. As compared with the data in the International HapMap Project’s data-base, we found that the frequencies of the A allele and AA, AG genotypes in our study populations were higher than those in Han Chinese from Beijing, which may be caused by different sample sizes and regions (Beijing vs. Guangxi).
There were hardly any previous studies presented the direct relationship between the SPTLC3 rs364585 SNP and blood lipid levels in humans except a newly GWAS which showed that the SPTLC3 rs364585 SNP was significant associated with LDL-C concentrations (P < 5 × 10−8) in the population of European descent [40]. In the present study, we found that the SPTLC3 rs364585 SNP was significant associated with multiple serum lipid parameters in our study populations. The A allele carriers had higher LDL-C levels and lower the ApoA1/ApoB ratio in Han; and higher TC and LDL-C levels and lower HDL-C levels in Jing than the A allele non-carriers. When serum lipid parameters were analyzed according to gender, we found that the A allele carriers had higher LDL-C levels in Han males; higher TC and LDL-C levels in Jing males; and higher LDL-C levels and lower HDL-C levels in Jing females than the A allele non-Carriers. These results indicated that the association of the SPTLC3 rs364585 SNP and serum lipid levels may have racial/ethnic and/or sex specificity.
SPTLC3 is a 552 amino acid single-pass membrane protein and is a member of the class-II pyridoxal-phosphate-dependent amino transferase family. SPTLC3 encodes a functional subunit of the SPT enzyme-complex that catalyzes the first and rate-limiting step of de novo sphingolipid synthesis [21], which is involved in lipid metabolism. Direct experimental evidence indicates a role for sphingolipids in several common complex chronic disease processes including atherosclerotic plaque formation, myocardial infarction, cardiomyopathy, pancreatic beta cell failure, insulin resistance and type 2 diabetes mellitus [41]. A genome-wide analysis identified that the SPTLC3 played a functional role in the hepatic lipid accumulation, which is a common example of imbalance in lipid and energy homeostasis [19]. Another study demonstrated that the SNP of rs3848751 in SPTLC3 was associated with HDL-C and LDL-C in males and variants (rs3848751 and rs6078888) within SPTLC3 had influence on the risk of myocardial infarction [23]. The results may be due to mutations causing changes in circulating sphingolipid concentrations. Taken together, we speculate that the SPTLC3 rs364585 mutation may act in the subunit of the SPT enzyme-complex to influence the de novo sphingolipid synthesis and bring about the cascade of events in lipid metabolism. However, the biological function and detailed role of the SPTLC3 rs364585 SNP in lipid metabolism need to be further explored.
The gonadal hormone is also a contribution factor on lipid metabolism although the reasons for sex differences in serum lipid levels are still unclear. It is commonly accepted that androgens induce changes in serum lipid concentrations that would predispose towards CVD, while oestrogens have the opposite effects [42]. Oestrogens share structural similarities with vitamin E and other lipophilic antioxidants and are thus able to function as scavengers for lipid peroxyl radicals and interrupt the chain reaction of lipid peroxidation. Oestrogens also protect HDL from oxidation, an effect that should preserve the beneficial functions of HDL. In the present study, we found the levels of HDL-C in Han was lower in males than in females and the levels of HDL-C, ApoA1 and the ApoA1/ApoB ratio in Jing were lower in males than females. It is well known that both HDL-C and ApoA1 are protective factors for CVD and the ratio of ApoA1 to ApoB reflects the cholesterol balance between antiatherogenic and atherogenic lipoprotein particles. These results are consistent with previous findings that females have more favorate serum lipid profiles to reduce the risk of CVD than males. In addition, we also found sex difference in the association of the SPTLC3 rs364585 SNP and serum lipid levels. The reasons for the discrepancy are still not well understood. Other unknown genetic factors may also be involved in determining this complex status. Besides, the sample size may be not large enough to detect the association of the SPTLC3 rs364585 SNP and serum lipid levels in the subgroup analyses. Further studies are needed to clarify.
Furthermore, it is well recognized that environmental factors such as dietary patterns, lifestyle and physical inactivity are all strongly related with serum lipid levels [39, 43]. In the present study, multiple linear regression analysis showed that serum lipid parameters were also affected by several environmental factors such as age, gender, BMI, waist circumference, alcohol consumption, cigarette smoking, blood pressure and blood glucose. These data suggest that the environmental factors also play an important role in determining serum lipid profiles in our study populations. Fishery is the major source of income for Jing population and fish is appeared most frequently dish on their tables. A kind of fish sauce called nuoc-mam is also popular on Jing people’s dinner table, which contains 17 amino acids (8 essential amino acids included of course). Fish rich in omrga-3 polyunsaturated fatty acids (N-3PUFA) have been suggested to have a favorable effect on serum concentrations of pleiotropic lipid traits. However, previous researches also demonstrated the effects of N-3PUFA on key metabolic functions, including significant rise in TC, TG and LDL-C levels and decrease in HDL-C levels [44, 45].
In addition, we also found that the levels of weight and BMI were higher in Jing than in Han and the percentages of alcohol consumption were lower in Jing males than in Han males (P < 0.05 for each). A previous study reported that for every 1-kg decrease in body weight, TG decreased by 0.011 mmol/L and HDL-C increased by 0.011 mmol/L [46]. In another study, Rimm et al. documented that consuming of 30 g of ethanol per day increased the concentrations of HDL-C by 3.99 mg/ dL, ApoA1 by 8.82 mg/dL, and TG by 5.69 mg/ dL [47]. What’s more, Yin et al. also showed that BMI and alcohol consumption could interact with certain lipid-related gene variants to modify the serum lipid levels in Bai Ku Yao and Han Chinese ethnic groups. Therefore, the results of exposure to different environmental factors may further modify the effect of genetic variation on serum lipid levels in our study populations.
There are several major strengths in our study. First, the study is an investigation of a representative random sample of the Jing population, which retains its regional and special customs in China and may be a useful subgroup for population genetic studies. Second, the sample size of the study is moderate with 783 subjects of Jing and 824 subjects of Han Chinese. Third, a recent GWAS has reported the association between the SPTLC3 rs364585 SNP and serum LDL-C levels [40] and our present study is the first replication of GWAS signals in the Chinese population to provide significant evidence for the association of the SPTLC3 rs364585 SNP with serum lipid traits. To interpret the findings, however, several potential limitations in our study should be acknowledged. Firstly, we were not able to alleviate the effect of diet and several environmental factors during the statistical analysis. Secondly, although we have detected the effects of the SPTLC3 rs364585 SNP on serum lipid levels in this study, there are still many lipid-related SNPs and the interactions of SNP-SNP and/or SNP-environmental factors. Finally, we recognize the limited power to provide a more significant advance in understanding the full impact of rs364585 SNP on lipoprotein metabolism. To confirm our findings, further in-depth studies on the biological actions of SPTLC3 rs364585 variation and the interactions of gene-environment are necessary.
Conclusions
This study showed that the association of the SPTLC3 rs364585 SNP and serum lipid profiles is different between the Jing and Han populations and between males and females in the both ethnic groups. These results suggest that there may be a racial/ethnic- and/ or sex-specific association of the SPTLC3 rs364585 SNP and serum lipid parameters.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural Science Foundation of China (No: 81160111).
Availability of data and materials
The datasets generated during the present study are not publicly available, because detailed genetic information of each participant were included in these materials.
Authors’ contributions
QHZ conceived the study, participated in the design, undertook genotyping, performed the statistical analyses, and drafted the manuscript. RXY conceived the study, participated in the design, carried out the epidemiological survey, collected the samples, and helped to draft the manuscript. HG collaborated to the genotyping. FH, JZW, SLP, WXL and DZY carried out the epidemiological survey, and collected the samples. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Informed consent including consent to publish was obtained from all participants by signature or by fingerprint (to express consent), as approved by the ethical review committee.
Ethics approval and consent to participate
All procedures of the investigation were carried out following the rules of the Declaration of Helsinki of 1975 (http://www.wma.net/en/30publications/10policies/b3/), revised in 2008. The study design was approved by the Ethics Committee of the First Affiliated Hospital, Guangxi Medical University (No: Lunshen-2011-KY-Guoji-001; Mar. 7, 2011). Informed consent was obtained from all participants.
Abbreviations
- ANCOVA
Analysis of covariance
- Apo
Apolipoprotein
- BMI
Body mass index
- CVD
Cardiovascular disease
- GWAS
Genome-wide association study
- HDL-C
High-density lipoprotein cholesterol
- HWE
Hardy-Weinberg equilibrium
- LDL-C
Low-density lipoprotein cholesterol
- N-3PUFA
Omrga-3 polyunsaturated fatty acids
- PCR
Polymerase chain reaction
- RFLP
Restriction fragment length polymorphism
- SNP
Single nucleotide polymorphism
- SPTLC3
The serine palmitoyl-transferase long chain base subunit 3
- TC
Total cholesterol
- TG
Triglyceride
Contributor Information
Qing-Hui Zhang, Email: zhangqinghuixx@163.com.
Rui-Xing Yin, Email: yinruixing@163.com.
Hui Gao, Email: gaohuiyouxiang@126.com.
Feng Huang, Email: huangfeng3000@126.com.
Jin-Zhen Wu, Email: wujinzhengx@sohu.com.
Shang-Ling Pan, Email: sl.pan.berkeley@gmail.com.
Wei-Xiong Lin, Email: lin78018@163.com.
De-Zhai Yang, Email: ydz969@126.com.
References
- 1.Moran A, Forouzanfar M, Sampson U, Chugh S, Feigin V, Mensah G. The epidemiology of cardiovascular diseases in sub-Saharan Africa: the Global Burden of Diseases, Injuries and Risk Factors 2010 Study. Prog Cardiovasc Dis. 2013;56:234–9. doi: 10.1016/j.pcad.2013.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pjanic M, Miller CL, Wirka R, Kim JB, DiRenzo DM, Quertermous T. Genetics and genomics of coronary artery disease. Curr Cardiol Rep. 2016;18:102. doi: 10.1007/s11886-016-0777-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ezzati M, Lopez AD, Rodgers A, Vander Hoorn S, Murray CJ. Selected major risk factors and global and regional burden of disease. Lancet. 2002;360:1347–60. doi: 10.1016/S0140-6736(02)11403-6. [DOI] [PubMed] [Google Scholar]
- 4.Law MR, Wald NJ, Rudnicka AR. Quantifying effect of statins on low density lipoprotein cholesterol, ischaemic heart disease, and stroke: systematic review and meta-analysis. BMJ. 2003;326:1423. doi: 10.1136/bmj.326.7404.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke R, Emberson JR, Parish S, Palmer A, Shipley M, Linksted P, et al. Cholesterol fractions and apolipoproteins as risk factors for heart disease mortality in older men. Arch Intern Med. 2007;167:1373–8. doi: 10.1001/archinte.167.13.1373. [DOI] [PubMed] [Google Scholar]
- 6.Alberti KG, Eckel RH, Grundy SM, Zimmet PZ, Cleeman JI, Donato KA, et al. Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation. 2009;120:1640–5. doi: 10.1161/CIRCULATIONAHA.109.192644. [DOI] [PubMed] [Google Scholar]
- 7.Matthan NR, Zhu L, Pencina M, D'Agostino RB, Schaefer EJ, Lichtenstein AH. Sex-specific differences in the predictive value of cholesterol homeostasis markers and 10-year cardiovascular disease event rate in Framingham Offspring Study participants. J Am Heart Assoc. 2013;2 doi: 10.1161/JAHA.112.005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindman AS, Veierod MB, Tverdal A, Pedersen JI, Selmer R. Nonfasting triglycerides and risk of cardiovascular death in men and women from the Norwegian Counties Study. Eur J Epidemiol. 2010;25:789–98. doi: 10.1007/s10654-010-9501-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orekhov AN, Bobryshev YV, Sobenin IA, Melnichenko AA, Chistiakov DA. Modified low density lipoprotein and lipoprotein-containing circulating immune complexes as diagnostic and prognostic biomarkers of atherosclerosis and type 1 diabetes macrovascular disease. Int J Mol Sci. 2014;15:12807–41. doi: 10.3390/ijms150712807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwiterovich PO, Jr, Coresh J, Smith HH, Bachorik PS, Derby CA, Pearson TA. Comparison of the plasma levels of apolipoproteins B and A-1, and other risk factors in men and women with premature coronary artery disease. Am J Cardiol. 1992;69:1015–21. doi: 10.1016/0002-9149(92)90856-T. [DOI] [PubMed] [Google Scholar]
- 11.Silbernagel G, Schottker B, Appelbaum S, Scharnagl H, Kleber ME, Grammer TB, et al. High-density lipoprotein cholesterol, coronary artery disease, and cardiovascular mortality. Eur Heart J. 2013;34:3563–71. doi: 10.1093/eurheartj/eht343. [DOI] [PubMed] [Google Scholar]
- 12.Ordovas JM, Shen AH. Genetics, the environment, and lipid abnormalities. Curr Cardiol Rep. 2002;4:508–13. doi: 10.1007/s11886-002-0115-4. [DOI] [PubMed] [Google Scholar]
- 13.Ordovas JM, Robertson R, Cleirigh EN. Gene-gene and gene-environment interactions defining lipid-related traits. Curr Opin Lipidol. 2011;22:129–36. doi: 10.1097/MOL.0b013e32834477a9. [DOI] [PubMed] [Google Scholar]
- 14.Chaput JP, Perusse L, Despres JP, Tremblay A, Bouchard C. Findings from the Quebec Family Study on the Etiology of Obesity: Genetics and Environmental Highlights. Curr Obes Rep. 2014;3:54–66. doi: 10.1007/s13679-013-0086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lusis AJ, Mar R, Pajukanta P. Genetics of atherosclerosis. Annu Rev Genomics Hum Genet. 2004;5:189–218. doi: 10.1146/annurev.genom.5.061903.175930. [DOI] [PubMed] [Google Scholar]
- 16.Perusse L, Rice T, Despres JP, Bergeron J, Province MA, Gagnon J, et al. Familial resemblance of plasma lipids, lipoproteins and postheparin lipoprotein and hepatic lipases in the HERITAGE Family Study. Arterioscler Thromb Vasc Biol. 1997;17:3263–9. doi: 10.1161/01.ATV.17.11.3263. [DOI] [PubMed] [Google Scholar]
- 17.Roberts R. Personalized medicine: a reality within this decade. J Cardiovasc Transl Res. 2008;1:11–6. doi: 10.1007/s12265-007-9001-1. [DOI] [PubMed] [Google Scholar]
- 18.Illig T, Gieger C, Zhai G, Romisch-Margl W, Wang-Sattler R, Prehn C, et al. A genome-wide perspective of genetic variation in human metabolism. Nat Genet. 2010;42:137–41. doi: 10.1038/ng.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mirkov S, Myers JL, Ramirez J, Liu W. SNPs affecting serum metabolomic traits may regulate gene transcription and lipid accumulation in the liver. Metabolism. 2012;61:1523–7. doi: 10.1016/j.metabol.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemaitre RN, King IB, Kabagambe EK, Wu JH, McKnight B, Manichaikul A, et al. Genetic loci associated with circulating levels of very long-chain saturated fatty acids. J Lipid Res. 2015;56:176–84. doi: 10.1194/jlr.M052456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hornemann T, Penno A, Rutti MF, Ernst D, Kivrak-Pfiffner F, Rohrer L, et al. The SPTLC3 subunit of serine palmitoyltransferase generates short chain sphingoid bases. J Biol Chem. 2009;284:26322–30. doi: 10.1074/jbc.M109.023192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hornemann T, Richard S, Rutti MF, Wei Y, von Eckardstein A. Cloning and initial characterization of a new subunit for mammalian serine-palmitoyltransferase. J Biol Chem. 2006;281:37275–81. doi: 10.1074/jbc.M608066200. [DOI] [PubMed] [Google Scholar]
- 23.Hicks AA, Pramstaller PP, Johansson A, Vitart V, Rudan I, Ugocsai P, et al. Genetic determinants of circulating sphingolipid concentrations in European populations. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin JH, Liu ZH, Lv FJ, Fu YG, Fan XL, Li SY, et al. Molecular analyses of HLA-DRB1, -DPB1, and -DQB1 in Jing ethnic minority of Southwest China. Hum Immunol. 2003;64:830–4. doi: 10.1016/S0198-8859(03)00128-9. [DOI] [PubMed] [Google Scholar]
- 25.Sun JQ, Yin RX, Shi GY, Shen SW, Chen X, Bin Y, et al. Association of the ARL15 rs6450176 SNP and serum lipid levels in the Jing and Han populations. Int J Clin Exp Pathol. 2015;8:12977–94. [PMC free article] [PubMed] [Google Scholar]
- 26.Guo T, Yin RX, Huang F, Yao LM, Lin WX, Pan SL. Association between the DOCK7, PCSK9 and GALNT2 Gene Polymorphisms and Serum Lipid levels. Sci Rep. 2016;6:19079. doi: 10.1038/srep19079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo T, Yin RX, Lin WX, Wang W, Huang F, Pan SL. Association of the variants and haplotypes in the DOCK7, PCSK9 and GALNT2 genes and the risk of hyperlipidaemia. J Cell Mol Med. 2016;20:243–65. doi: 10.1111/jcmm.12713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.People’s Republic of China--United States Cardiovascular and Cardiopulmonary Epidemiology Research Group. An epidemiological study of cardiovascular and cardiopulmonary disease risk factors in four populations in the People's Republic of China. Baseline report from the P.R.C.-U.S.A. Collaborative Study. Circulation. 1992;85:1083-96. [DOI] [PubMed]
- 29.Ruixing Y, Qiming F, Dezhai Y, Shuquan L, Weixiong L, Shangling P, et al. Comparison of demography, diet, lifestyle, and serum lipid levels between the Guangxi Bai Ku Yao and Han populations. J Lipid Res. 2007;48:2673–81. doi: 10.1194/jlr.M700335-JLR200. [DOI] [PubMed] [Google Scholar]
- 30.Ruixing Y, Dezhai Y, Shuquan L, Yuming C, Hanjun Y, Qiming F, et al. Hyperlipidaemia and its risk factors in the Guangxi Bai Ku Yao and Han populations. Public Health Nutr. 2009;12:816–24. doi: 10.1017/S1368980008003273. [DOI] [PubMed] [Google Scholar]
- 31.Wu DF, Yin RX, Aung LH, Li Q, Yan TT, Zeng XN, et al. Sex-specific association of ACAT-1 rs1044925 SNP and serum lipid levels in the hypercholesterolemic subjects. Lipids Health Dis. 2012;11:9. doi: 10.1186/1476-511X-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Q, Yin RX, Yan TT, Miao L, Cao XL, Hu XJ, et al. Association of the GALNT2 gene polymorphisms and several environmental factors with serum lipid levels in the Mulao and Han populations. Lipids Health Dis. 2011;10:160. doi: 10.1186/1476-511X-10-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramazauskiene V, Petkeviciene J, Klumbiene J, Kriaucioniene V, Sakyte E. Diet and serum lipids: changes over socio-economic transition period in Lithuanian rural population. BMC Public Health. 2011;11:447. doi: 10.1186/1471-2458-11-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gronroos P, Raitakari OT, Kahonen M, Hutri-Kahonen N, Juonala M, Marniemi J, et al. Relation of apolipoprotein E polymorphism to markers of early atherosclerotic changes in young adults--the Cardiovascular Risk in Young Finns Study. Circ J. 2008;72:29–34. doi: 10.1253/circj.72.29. [DOI] [PubMed] [Google Scholar]
- 35.Ishii M. The sixth report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure, and 1999 World Health Organization-International Society of Hypertension Guidelines for the Management of Hypertension. Nihon Rinsho. 2000;58(Suppl 1):267–75. [PubMed] [Google Scholar]
- 36.Whitworth JA. 2003 World Health Organization (WHO)/International Society of Hypertension (ISH) statement on management of hypertension. J Hypertens. 2003;21:1983–92. doi: 10.1097/00004872-200311000-00002. [DOI] [PubMed] [Google Scholar]
- 37.Muller MJ, Lagerpusch M, Enderle J, Schautz B, Heller M, Bosy-Westphal A. Beyond the body mass index: tracking body composition in the pathogenesis of obesity and the metabolic syndrome. Obes Rev. 2012;13(Suppl 2):6–13. doi: 10.1111/j.1467-789X.2012.01033.x. [DOI] [PubMed] [Google Scholar]
- 38.Hata Y, Nakajima K. Life-style and serum lipids and lipoproteins. J Atheroscler Thromb. 2000;7:177–97. doi: 10.5551/jat1994.7.177. [DOI] [PubMed] [Google Scholar]
- 39.Barnard RJ. Effects of life-style modification on serum lipids. Arch Intern Med. 1991;151:1389–94. doi: 10.1001/archinte.1991.00400070141019. [DOI] [PubMed] [Google Scholar]
- 40.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–83. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holland WL, Summers SA. Sphingolipids, insulin resistance, and metabolic disease: new insights from in vivo manipulation of sphingolipid metabolism. Endocr Rev. 2008;29:381–402. doi: 10.1210/er.2007-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mudali S, Dobs AS, Ding J, Cauley JA, Szklo M, Golden SH. Endogenous postmenopausal hormones and serum lipids: the atherosclerosis risk in communities study. J Clin Endocrinol Metab. 2005;90:1202–9. doi: 10.1210/jc.2004-0744. [DOI] [PubMed] [Google Scholar]
- 43.Erkkila AT, Sarkkinen ES, Lehto S, Pyorala K, Uusitupa MI. Dietary associates of serum total, LDL, and HDL cholesterol and triglycerides in patients with coronary heart disease. Prev Med. 1999;28:558–65. doi: 10.1006/pmed.1998.0478. [DOI] [PubMed] [Google Scholar]
- 44.Sala-Vila A, Guasch-Ferre M, Hu FB, Sanchez-Tainta A, Bullo M, Serra-Mir M, et al. Dietary alpha-Linolenic Acid, Marine omega-3 Fatty Acids, and Mortality in a Population With High Fish Consumption: Findings From the PREvencion con DIeta MEDiterranea (PREDIMED) Study. J Am Heart Assoc. 2016; 5: doi:10.1161/jaha.115.002543. [DOI] [PMC free article] [PubMed]
- 45.Dias CB, Wood LG, Garg ML. Effects of dietary saturated and n-6 polyunsaturated fatty acids on the incorporation of long-chain n-3 polyunsaturated fatty acids into blood lipids. Eur J Clin Nutr. 2016;70:812–8. doi: 10.1038/ejcn.2015.213. [DOI] [PubMed] [Google Scholar]
- 46.Yu-Poth S, Zhao G, Etherton T, Naglak M, Jonnalagadda S, Kris-Etherton PM. Effects of the National Cholesterol Education Program's Step I and Step II dietary intervention programs on cardiovascular disease risk factors: a meta-analysis. Am J Clin Nutr. 1999;69:632–646. doi: 10.1093/ajcn/69.4.632. [DOI] [PubMed] [Google Scholar]
- 47.Rimm EB, Williams P, Fosher K, Criqui M, Stampfer MJ. Moderate alcohol intake and lower risk of coronary heart disease: meta-analysis of effects on lipids and haemostatic factors. BMJ. 1999;319:1523–8. doi: 10.1136/bmj.319.7224.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during the present study are not publicly available, because detailed genetic information of each participant were included in these materials.


