Abstract
Mammalian target of rapamycin complex 1 (mTORC1) is involved in anabolic metabolism in both osteoblasts and chondrocytes, but the role of mTORC1 in osteoclast biology in vivo remains to be elucidated. In this study, we showed that deletion of regulatory-associated protein of mTOR (Raptor) in osteoclasts led to an increase in bone mass with decreased bone resorption. Raptor-deficient bone marrow-derived macrophages exhibited lower mTORC1-S6K1 signaling and retarded osteoclast differentiation, as determined by the number of osteoclasts, tartrate-resistant acid phosphatase activity, and expression of osteoclast-specific genes. Enforced expression of constitutively active S6K1 rescued the impaired osteoclast differentiation in Raptor-deficient bone marrow-derived macrophages. Furthermore, pharmacological inhibition of mTORC1 signaling by rapamycin could also inhibit osteoclast differentiation and osteoclast-specific gene expression. Taken together, our findings demonstrate that mTORC1 plays a key role in the network of catabolic bone resorption in osteoclasts and may serve as a potential pharmacological target for the regulation of osteoclast activity in bone metabolic disorders.
Keywords: bone, cell differentiation, mTOR complex (mTORC), osteoclast, osteoporosis, S6 kinase
Introduction
Bone is a rigid yet metabolically active organ that is molded, shaped, and repaired continuously (1). After bone is formed, bone undergoes a process known as remodeling by which bone is turned over throughout life (2). Bone remodeling acts as the predominant metabolic regulator of both the physical structure and physiological function of bone. Remodeling is a complex process involving osteoclasts, which are responsible for removing old mineralized matrix, and osteoblasts, which synthesize and secrete new bone matrix (1, 3). An imbalance in bone remodeling can induce perturbation of bone structure and function and potentially result in disease (1). In adults, most bone diseases, such as osteoporosis, rheumatoid arthritis, and periodontal disease, are the result of bone loss secondary to excess osteoclast activity (4). Prevention and treatment of these pathological disorders highlight the study of the underlying mechanisms by which osteoclasts differentiate from their precursors.
Osteoclasts are tissue-specific giant multinucleated cells that differentiate from monocyte/macrophage precursor cells at or near the bone surface (1). It is known that the differentiation of osteoclasts is under the control of two important cytokines, receptor activator of nuclear factor κB ligand (RANKL)5 and M-CSF (3). RANKL and macrophage colony-stimulating factor (M-CSF) may activate a set of signaling pathways, including AKT and NF-κB, that promote the differentiation, multinucleation, activation, and survival of osteoclasts (5). However, the precise mechanism regulating osteoclast differentiation is not fully understood.
Mammalian/mechanistic target of rapamycin (mTOR) is an evolutionarily conserved protein kinase (6, 7). mTOR functions in two structurally and functionally distinct multiprotein complexes, mTORC1 and mTORC2, that are distinct in their unique components and downstream targets (7). mTORC1 contains regulatory-associated protein of mTOR (Raptor) and is sensitive to rapamycin, whereas mTORC2 contains rapamycin-insensitive companion of mTOR (Rictor) and is resistant to rapamycin. mTORC1 constitutes a molecular node that regulates cell differentiation, growth, and survival though a series of downstream effectors, including S6 kinase1 (S6K1). The mTORC1/S6K1 axis transmits and integrates important signals that have been found to be critical for bone biology, including nutrients, growth factors, and energy metabolism.
Recently, mTORC1 has been found to play roles in bone biology and pathology (3, 8–10). Everolimus, a derivative of rapamycin, has been reported to have beneficial effects on bone when used as an anticancer ancillary in postmenopausal women with breast cancer (11, 12). Although the underlying mechanism of the protective effects of everolimus on bone is still unclear, osteoclasts may be its target, considering that blocking of mTORC1 in osteoblasts results in decreased bone mass in mice (4). It is reported that mTOR signaling is critical in osteoclast survival and bone resorption in vitro (13, 14). However, the role of mTORC1 in osteoclasts in vivo has not been completely elucidated. Here we report that osteoclast-specific deletion of Raptor (an indispensable component of mTORC1) results in increased bone mass with decreased bone resorption. We found that the Raptor/mTORC1-S6K1 axis played a determinative role in osteoclast differentiation and may be a potent drug target for regulation of osteoclasts.
Results
Raptor Deficiency in Osteoclasts Results in Increased Bone Mass with Impaired Osteoclast Differentiation
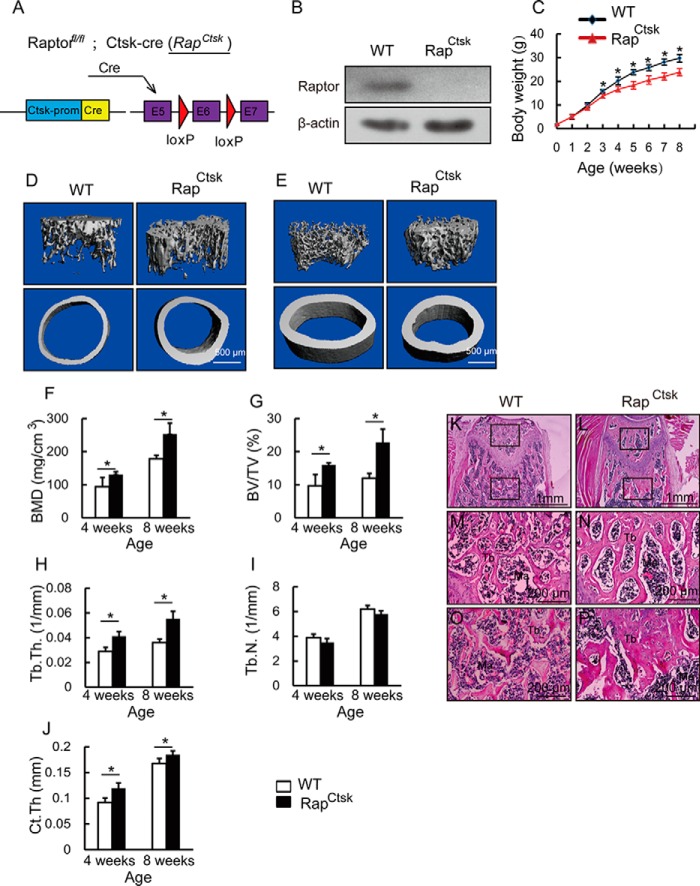
To determine the role of mTORC1 signaling in osteoclasts in vivo, we generated conditional Raptor knockout mice (Rapfl/fl; Ctsk-cre, hereafter called RapCtsk) (Fig. 1A) by crossing Rapfl/fl mice with Ctsk-cre mice, a transgenic line in which Cre expression is driven by the promoter of cathepsin K to achieve osteoclast-specific expression of Cre. To confirm the gene depletion of Raptor, we isolated BMMs from 4-week-old WT and RapCtsk mice and treated BMMs with 20 ng/ml M-CSF and 250 ng/ml RANKL. The cell lysates were collected and subjected to immunoblotting with Raptor antibody, which confirmed the loss of Raptor protein in RapCtsk BMMs (Fig. 1B). Compared with WT mice, RapCtsk mice showed a slower growth rate (Fig. 1C).
FIGURE 1.
Deletion of Raptor in osteoclasts led to an increase of bone mass. A, illustration of Raptor deletion in Ctsk-expressing osteoclasts. B, Western blotting assay of Raptor of WT and RapCtsk BMMs cultured with osteoclast differentiation medium for 6 days. C, body weight of male WT and RapCtsk littermates measured at different age points. Data represent mean ± S.D. *, p < 0.05; n = 5. D, three-dimensional reconstruction of micro-CT images of trabecular bone close to the distal growth plate and cortical bone at the middle of femora from 4-week-old WT and RapCtsk littermates. E, representative view of micro-CT of femora from 8-week-old WT and RapCtsk mice. F–J, quantitative parameters of micro-CT. The trabecular bone close to the distal growth plate and cortical bone at the middle of the femur was analyzed. Data represent mean ± S.D. *, p < 0.05; n = 5. K–P, H&E staining of femora from 4-week-old male WT and RapCtsk littermates. M and N, high-power images of the secondary spongiosa marked in K and L, respectively. O and P, high-power images of the primary spongiosa marked in K and L, respectively. Ma, bone marrow.
To determine the effect of Raptor depletion in osteoclasts on bone, femora from 4- and 8-week-old male WT and RapCtsk mice were subjected to micro computed tomography (micro-CT) analysis. Trabecular bone in the distal femur of RapCtsk mice was more compact than that in WT mice at 4 weeks (Fig. 1D) and 8 weeks (Fig. 1E). Microstructure parameter analysis showed that RapCtsk mice displayed augmentation of bone mineral density (BMD), bone volume fraction (BV/TV), and trabecular thickness (Tb.Th.) in comparison with WT mice at both 4 and 8 weeks of age (Fig. 1, F and H). Trabecular number (Tb.N.) was comparable between WT and RapCtsk mice (Fig. 1I). We also examined midshaft cortical bone and found an increase in cortical thickness in RapCtsk mice at 4 and 8 weeks (Ct.Th) (Fig. 1, D, E, and J). H&E staining showed an increase in bone mass and trabecular thickness in both primary and secondary spongiosa of the femur from 4-week-old male RapCtsk mice in comparison with WT mice (Fig. 1, K–P). Furthermore, we analyzed bone structure changes of femora from 4-week-old female and 20-week-old male RapCtsk mice by histology. As shown in Fig. 2, A–D, bone mass and trabecular thickness increased in the primary spongiosa of femora from 4-week-old female RapCtsk mice compared with WT mice. Twenty-week-old male RapCtsk mice exhibited increased bone volume and trabecular thickness too (Fig. 2, E–H). These results indicated that inactivation of mTORC1 in osteoclasts led to increased bone mass.
FIGURE 2.
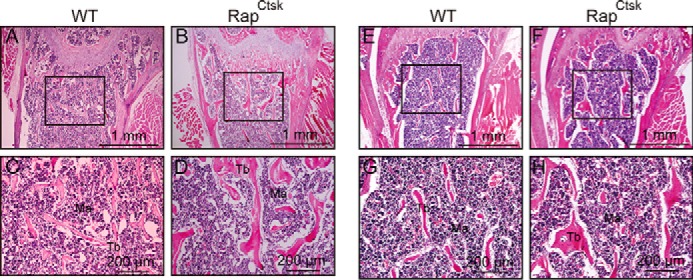
Raptor deficiency in osteoclasts induced increased bone volume. A–D, H&E staining of femora from 4-week-old female WT and RapCtsk littermates. C and D, high-power images of the primary spongiosa marked in A and B, respectively. E–H, H&E staining of femora from 20-week-old male WT and RapCtsk littermates. G and H, high-power images of the primary spongiosa marked in E and F, respectively. Ma, bone marrow.
This increase in bone mass in RapCtsk mice might arise through decreased bone resorption, elevated bone formation, or a combination of both. To distinguish these possibilities, we first performed TRAP staining and immunohistochemical analysis to examine osteoclast function in femora from 4-week-old WT and RapCtsk mice. As shown in Fig. 3, A–D, osteoclasts were TRAP-positive multinuclear cells present along the trabecular bone surface. Histomorphometric analysis revealed that the number of TRAP-positive osteoclasts of trabecular bone in femora of RapCtsk mice decreased in comparison with WT mice (Fig. 3, A–D and G). Immunohistochemistry staining with an antibody specific to Ctsk demonstrated a robust Ctsk signal in osteoclasts of WT mice, whereas Ctsk staining was also reduced in RapCtsk mice (Fig. 3, E, F, and H). These results indicated reduced osteoclast activity and bone resorption in RapCtsk mice, which may be the main contributor to the increase in bone mass. We next analyzed the bone-forming activity by alizarin red and calcein double-labeling. As shown in Fig. 2, I–K, there was no significant difference in mineral apposition rate (MAR) and bone formation rate (BFR) between of the distal femur trabecular bone in WT and RapCtsk mice, indicating that the increased bone mass was not a result of a change in bone formation. It is similar to the results of trabecular bone that the cortical bone of the femur from RapCtsk mice exhibited decreased osteoclast numbers (Fig. 3, L–P) with comparable MAR and BFR (Fig. 3, Q–S) in comparison with WT mice. These results confirmed that the bone mass increase in RapCtsk mice resulted from decreased bone resorption.
FIGURE 3.
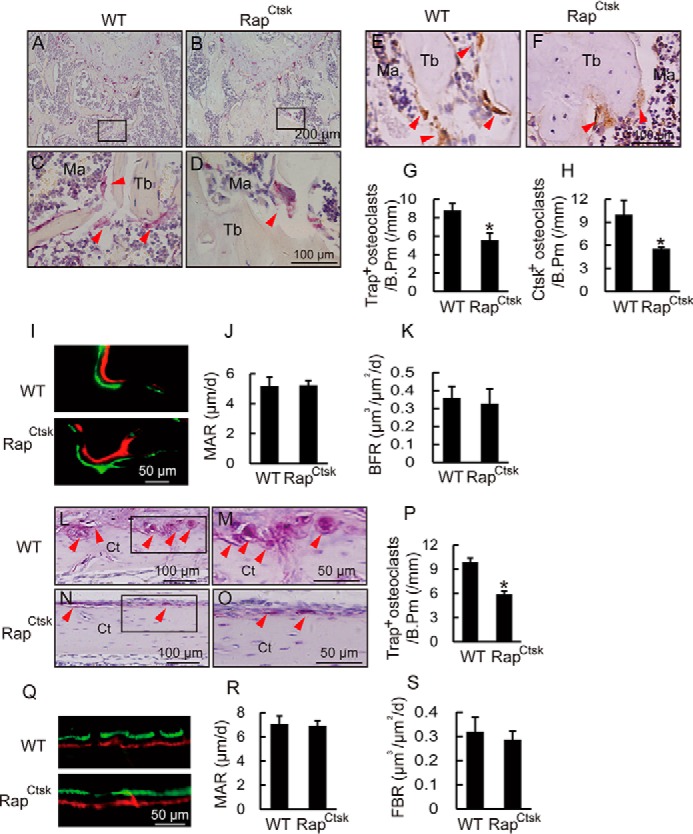
Ablation of Raptor in osteoclasts resulted in decreased bone resorption. A–D, representative view of TRAP staining of distal femur trabecular bone of 4-week-old male WT and RapCtsk mice. C and D, high-power images of the marked region in A and B, respectively. Arrowheads indicate TRAP-positive osteoclasts. Ma, bone marrow. E and F, immunohistochemical staining of Ctsk in distal femur trabecular bone of 4-week-old male WT and RapCtsk mice. Arrowheads indicate Ctsk-positive osteoclasts. G and H, numbers of TRAP- and Ctsk-positive osteoclasts on the trabecular bone surface, measured as cells per millimeter of perimeter (/B.Pm). Data are mean ± S.D. *, p < 0.05; n = 3. I, alizarin red and calcein double-labeling of femora showed the bone turnover rate of distal femur trabecular bone in 8-week-old WT and RapCtsk littermates. J and K, quantitative parameters of MAR and BFR of trabecular bone in WT and RapCtsk mice. Data represent mean ± S.D.; n = 3. L–O, representative view of TRAP staining of distal femur cortical bone from of 4-week-old male WT and RapCtsk mice. M and O, high-power images of the marked regions in L and N, respectively. P, numbers of TRAP-positive osteoclasts on the cortical bone surface, measured as cells per millimeter of perimeter (/B.Pm). Data are mean ± S.D. *, p < 0.05; n = 3. Q, alizarin red and calcein double-labeling of distal femur cortical bone from 8-week-old WT and RapCtsk littermates. R and S, quantitative parameters of MAR and BFR in WT and RapCtsk mice. Data represent mean ± S.D.
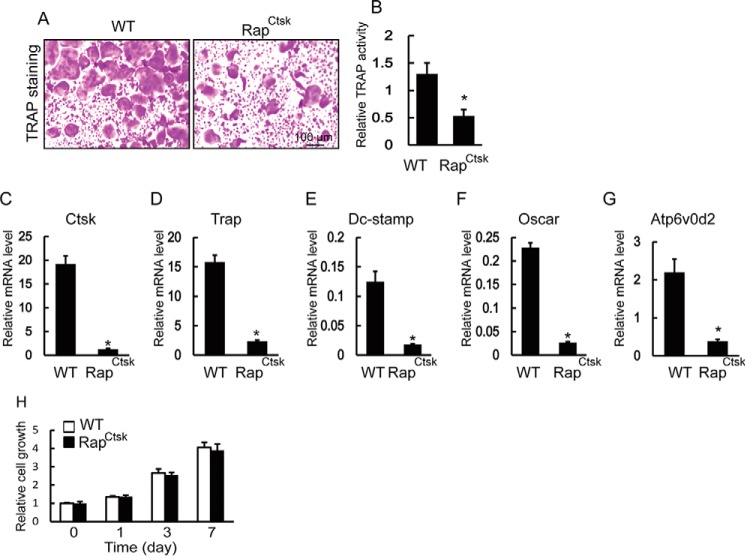
We next examined whether the reduced bone resorption in RapCtsk mice was a result of inadequate osteoclast differentiation. As shown in Fig. 4A, BMMs from WT mice fused to one another to form giant TRAP-positive multinucleated osteoclasts. In contrast, BMMs from RapCtsk mice showed impaired osteoclast differentiation and formation, as indicated by the decreased number of TRAP-positive osteoclasts and reduction in TRAP activity of the culture supernatant (Fig. 4, A and B). Because a specific set of genes is up-regulated during osteoclast differentiation (1), we used quantitative PCR (qPCR) to analyze and quantify the expression of the RANKL-induced osteoclast-specific genes Ctsk, Trap, Dc-stamp, Oscar, and Atp6v0d2. Consistent with the TRAP staining results, the expression of these osteoclast marker genes was suppressed in RapCtsk BMMs compared with WT cells (Fig. 4, C–J), confirming the inhibitory effects of Raptor deficiency on osteoclast differentiation.
FIGURE 4.
Raptor deficiency impaired osteoclast differentiation. A, BMMs were seeded in 96-well plates (5 × 104 cells/cm2) and treated with 20 ng/ml M-CSF and 250 ng/ml RANKL for 6 days. Cells were fixed with 4% paraformaldehyde and subjected to TRAP staining. B, relative TRAP activity of culture supernatant of WT and RapCtsk BMMs. Data represent means ± S.D. *, p < 0.05; n = 3. C–G, osteoclast-specific gene expression of WT and RapCtsk BMMs cultured with M-CSF and RANKL. Data represent mean ± S.D. *, p < 0.05; n = 3. H, relative growth of WT and RapCtsk BMMs determined by crystal violet staining. Data represent mean ± S.D., n = 3.
Further, to determine whether Raptor deficiency affects osteoclast progenitor cell proliferation, the growth of BMMs from WT and RapCtsk mice was determined by crystal violet staining. As is showed in Fig. 4H, there was no significant difference in cell proliferation between WT and RapCtsk BMMs. All of these results indicated that the increased bone mass in RapCtsk mice was due to reduced bone resorption secondary to impaired osteoclast differentiation.
S6K1 Is a Downstream Factor of Raptor/mTORC1 in Osteoclasts
S6K1 is the most important downstream regulator in mTORC1 signaling and plays crucial roles in development and aging (6). To determine whether S6K1 also functions downstream of Raptor/mTORC1 in osteoclasts, we examined the activity of S6K1 in BMMs isolated from WT and RapCtsk mice by immunoblotting analysis. As shown in Fig. 5A, although the expression level of S6K1 in BMMS was comparable between WT and RapCtsk mice, the phosphorylation level of S6K1, which reflects its activation, was dramatically reduced in the BMMs of RapCtsk mice in comparison with that in BMMs of WT mice. To prove that this attenuated S6K1 phosphorylation is responsible for the impaired osteoclast differentiation caused by the inactivation of mTORC1 signaling, we analyzed the effects of enforced expression of constitutively active S6K1 (CAS6K1) on the osteoclast differentiation of Raptor-deficient BMMs. A Western blotting assay confirmed CAS6K1 overexpression in RapCtsk BMMs by both antibodies of P-S6K1 (Thr389) and FLAG (Fig. 5B). The impaired osteoclast differentiation of RapCtsk BMMs was significantly improved by CAS6K1 overexpression, as demonstrated by the increased number of TRAP-positive cells (Fig. 5C) together with elevated TRAP activity in the culture supernatant (Fig. 5D). Consistent with this, the suppressed expression of the osteoclast-specific genes Ctsk, Trap, Dc-stamp, Oscar, and Atp6v0d2 in Raptor-deficient BMMs was rescued by CAS6K1 overexpression, as shown in Fig. 5, E–I. The rescue of osteoclast differentiation of Raptor-deficient BMMs by CAS6K1 overexpression demonstrates that reduced S6K1 activity is responsible for the impaired osteoclast differentiation in Raptor-deficient BMMs and confirms that Raptor/mTORC1-S6K1 signaling is essential for osteoclast differentiation.
FIGURE 5.
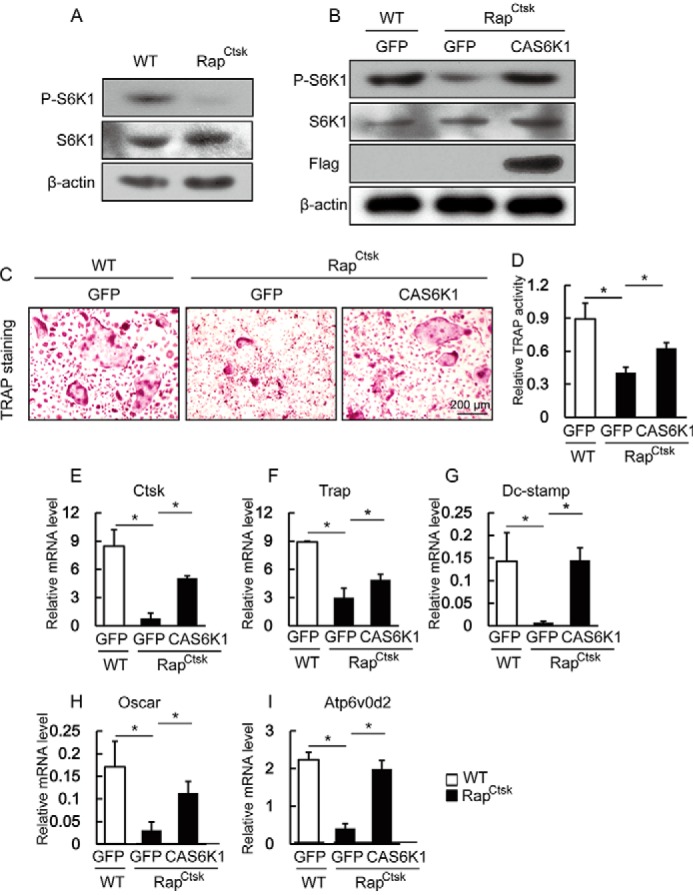
CAS6K1 rescue impaired osteoclast differentiation of RapCtsk BMMs. A, WT and RapCtsk BMMs were cultured with M-CSF and RANKL for 6 days and subjected to a Western blotting assay. B, WT and RapCtsk BMMs were infected with lentiviruses expressing GFP and CAS6K1, which was followed by osteoclast differentiation and Western blotting assay. C, WT and RapCtsk BMMs infected with Lenti-GFP or Lenti-S6K1. Cells were treated with RANKL for 6 days and subjected to TRAP staining. D, relative TRAP activity of culture supernatant. Data represent mean ± S.D. *, p < 0.05; n = 3. E–I, osteoclast-specific gene expression of WT and RapCtsk BMMs infected with Lenti-GFP or Lenti-S6K1. Data represent mean ± S.D. *, p < 0.05; n = 3.
Inhibition of mTORC1 by Rapamycin Suppresses Osteoclast Differentiation of BMMs
Our above data demonstrate that inactivation of mTORC1 signaling by Raptor depletion in osteoclasts leads to decreased osteoclast activity. This finding is significant as it provides the rationale for clinical intervention in mTORC1 signaling in the treatment of osteoclast-related diseases. To explore this possibility, we asked whether pharmacological inhibition of mTORC1 impairs osteoclast differentiation in a similar way as gene deletion of Raptor. BMMs from 4-week-old WT mice were treated with M-CSF and RANKL in the absence or presence of different concentrations of rapamycin, a selective inhibitor of mTORC1. As shown in Fig. 6B, although the expression level of S6K1 is comparable between non-rapamycin treated and rapamycin-treated groups, the level of phosphorylated S6K1 was decreased in the rapamycin-treated groups. TRAP staining revealed that rapamycin treatment inhibited giant osteoclast formation, as the giant osteoclasts were barely detectable in the rapamycin-treated mice, whereas giant osteoclasts were obvious in untreated mice (Fig. 6A). Notably, the decrease in phosphor-S6K1 level was rapamycin dose-dependent and correlated with the decrease in both the number of TRAP-positive osteoclasts and in TRAP activity (Fig. 6, A and C). In addition, the expression level of Ctsk, Trap, Dc-stamp, Oscar, and Atp6v0d2 was suppressed by rapamycin in a dose-dependent manner (Fig. 6, D–H). Thus, our data show that pharmacological inhibition of mTORC1 inhibits osteoclast formation and suppresses the expression of osteoclast-specific genes.
FIGURE 6.
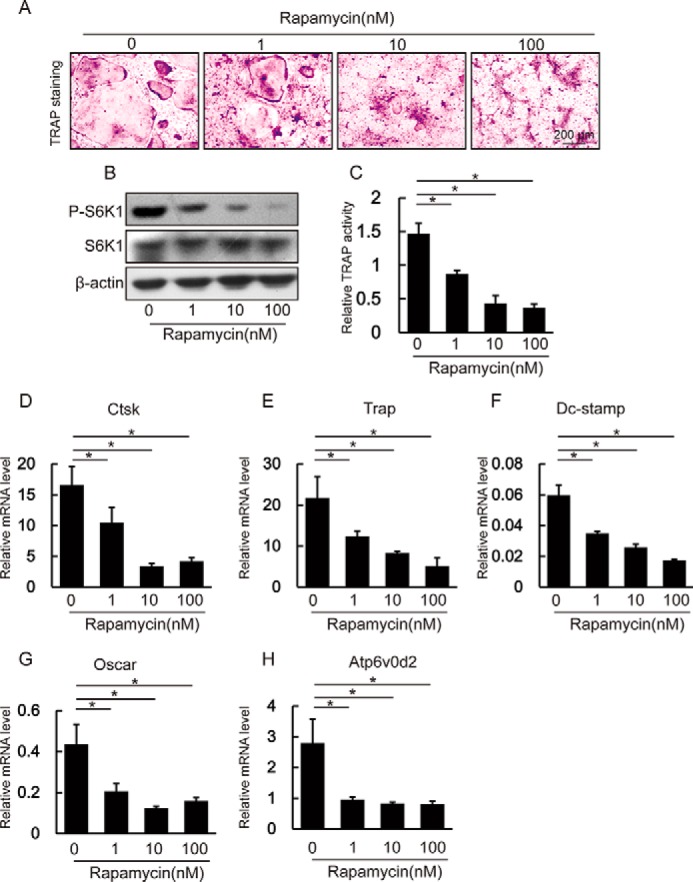
Rapamycin inhibited osteoclast differentiation of BMMs. A, WT BMMs were cultured with 20 ng/ml M-CSF or 250 ng/ml RANKL in the absence or presence of different concentrations of rapamycin for 6 days. Cells were subjected to TRAP staining. B, Western blotting assay of BMMs cultured in the absence or presence of different concentrations of rapamycin. C, relative TRAP activity of culture supernatant. Data represent mean ± S.D. *, p < 0.05; n = 3. D–H, rapamycin decreased the expression of osteoclast-specific genes in BMMs. Data represent mean ± S.D. *, p < 0.05; n = 3.
Discussion
This study provides several lines of evidence demonstrating that mTORC1 is required for osteoclast differentiation and bone metabolism. Conditional deletion of Raptor in osteoclasts led to increased bone mass accompanied by decreased osteoclast activity in mice. Osteoclast differentiation of Raptor-deficient BMMs was retarded with lower mTORC1-S6K1 signaling. Enforced expression of constitutively active S6K1 rescued the impaired osteoclast differentiation of Raptor-deficient BMMs. Further, pharmacological inhibition of mTORC1 signaling could also inhibit osteoclast differentiation.
The skeleton is a dynamic organ that undergoes continuous remodeling by osteoclasts (bone resorption) and osteoblasts (bone formation) (4). The balance of bone resorption and bone formation enables bone to fulfill the physiological needs of the organism with the minimum of mass. To maintain bone homeostasis, the process of bone remodeling is regulated in an orderly manner at both the systemic and local level, including nutrients, growth factors, and energy metabolism (4). On the other hand, an imbalance in bone remodeling can result in various metabolic diseases such as osteoporosis, rheumatoid arthritis, and periodontal disease, in which bone resorption disproportionately exceeds bone formation. Therefore, it is important for prevention and treatment of metabolic bone disease that the molecular mechanism that drives the differentiation of osteoclasts during both biological and pathological bone remodeling is better understood (5). In this study, we found that mTORC1 is a molecular node that is critical for osteoclast differentiation and bone metabolism.
mTORC1 signaling has been reported to regulate bone anabolism in both osteoblasts and chondrocytes (12, 15–18). However, evidence for the role of mTORC1 in osteoclast catabolism is limited. A recent study suggested that mTOR signaling in B cells could indirectly regulate osteoclast formation through regulation of β-catenin and RANKL/osteoprotegerin (OPG) (9) and another study showed everolimus can restrain the paracrine pro-osteoclast activity of breast cancer cells (19). mTOR signaling may also play an important role in osteoclast survival and differentiation of osteoclast progenitors (8, 20, 21). In this study, we found that mTORC1 regulates osteoclast differentiation and formation in a cell-autonomous manner with a conditional knockout mice model.
In the current mouse model, we found that blocking of mTORC1 in osteoclasts by deletion of Raptor led to increased bone mass, as determined by increases in BMD and BV/TV in the femur. Histological analysis showed that deletion of Raptor in osteoclasts did not influence osteoblast activity, as indicated by consistent MAR and BFR in RapCtsk and WT mice. These results excluded the possibility that deletion of Raptor in osteoclasts promoted bone formation and increased bone mass indirectly through actions on osteoblasts, although mTORC1 may play an important but as yet undetermined role in osteoblasts (15, 16, 18). Further, reduced osteoclast activity, determined by decreased TRAP activity and Ctsk expression in RapCtsk mice, was the chief cause of the increased bone mass. This is consistent with the observation that an mTOR inhibitor could rescue bone loss partly through suppression of bone resorption in ovariectomized rats (11) and protect bone health in postmenopausal women with breast cancer (12, 22). Meanwhile, Xian et al. (23) reported that rapamycin decreased bone mass in WT mice, which may be caused by the fact that the effects of rapamycin on osteoblasts masked those on osteoclasts. Taken together, these data show that deletion of Raptor in osteoclasts impaired bone resorption but did not affect bone formation and, ultimately, led to increased bone mass in mice.
In our mouse model, we found that the RapCtsk mice exhibited decreased body weight with increased bone density of distal femur. Raptor deletion in osteoclasts impaired bone resorption, which is important for bone modeling during bone development (3, 24), which may disturb the normal developmental process and, ultimately, induce decreased body weight. This phenomenon was found in other mouse models too (25, 26).
The differentiation and formation of osteoclasts from monocyte/macrophage precursor cells is regulated precisely to maintain the balance of bone remodeling (1–3). Our study found that primary Raptor-deficient BMMs displayed reduced osteoclast differentiation, as determined by decreased numbers of TRAP-positive osteoclasts, reduced TRAP activity, and lower expression of osteoclast-specific genes. This finding is supported by a previous in vitro study showing that mTORC1 plays a dominant role in osteoclast development as a nutrient sensor (21). There is limited information available concerning the molecular mechanisms by which mTORC1 controls osteoclast differentiation. It has been demonstrated that S6K1, a downstream regulator of mTORC1, can positively regulate the differentiation of osteoblasts (15) and chondrocytes (17). In this study, we provide evidence that the mTORC1-S6K1 axis can promote osteoclast differentiation. S6K1 activity was reduced in Raptor-deficient BMMs, accompanied by impaired osteoclast differentiation in vitro. Moreover, impaired osteoclast differentiation in RapCtsk BMMs was rescued by enforced overexpression of CAS6K1, indicating that S6K1 acts as a major downstream regulator of mTORC1 in osteoclasts.
mTOR signaling has been used as a target for drug treatment for a series of diseases, including cancer and aging (6). In this study, we found that rapamycin could inhibit osteoclast differentiation of primary BMMs in a dose-dependent manner, as determined by a decrease in the number of TRAP-positive osteoclasts, TRAP activity, and expression of osteoclast-specific genes. These findings suggest that mTORC1-S6K1 may be a drug target for the treatment of bone diseases related to osteoclasts. On the other hand, mTORC1 may play an important role in osteoblasts, although the results are still controversial. The mTORC1 inhibitor rapamycin can promote (27) or inhibit (28, 29) osteogenesis, which may be depended on cell type or cell differentiation stages. Further, blocking mTORC1 in preosteoblasts induced a decrease in bone mass in mice (15). Hence, further pharmacological studies about the effects of mTORC1 signaling on bone metabolism are needed in the future.
In summary, we reveal for the first time, to our knowledge, that specific inhibition of mTORC1 in osteoclasts results in increased bone mass in mice with decreased bone resorption. The Raptor/mTORC1-S6K1 axis constitutes a molecular node in the network of catabolic bone resorption of osteoclasts. These findings not only offer a mechanistic insight into the way in which mTORC1 drives osteoclast differentiation and bone resorption but also suggest an alternative pharmacological target for the regulation of osteoclasts in bone metabolic disorders.
Experimental Procedures
Mice
Rapfl/fl mice bearing loxP sites flanking exon 6 of the Raptor gene (stock no. 013188) were purchased from The Jackson Laboratory. Rapfl/fl mice were crossed with Cathepsin K-Cre mice (Ctsk-cre; a gift from S. Kato, University of Tokyo, Tokyo, Japan) to generate Raptorfl/fl; Ctsk-Cre (RapCtsk). All mice were bred and maintained under specific pathogen-free conditions in the institutional animal facility of the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences. All experiments were performed with the protocol approved by the Animal Care and Use Committee of the Shanghai Institute of Biochemistry and Cell Biology.
Micro-CT Analysis
Femora from 4- and 8-week-old male mice were used for micro-CT analysis at a resolution of 10 μm (Skyscan μCT80, Bruker MicroCT, Kontich, Belgium). One hundred slices (total 1 mm) close to the distal growth plate of the femur were used to analyze trabecular microarchitectural parameters, including BMD, bone volume fraction (BV/TV), Tb.Th., and Tb.N. Fifty slices from the middle of the femur were used to analyze Ct.Th.
Histological Analysis
Femora from 4- and 200-week-old mice were fixed in 4% paraformaldehyde for 48 h, followed by decalcification in 10% EDTA for 4 weeks. Specimens were embedded in paraffin and cut into sections of 4-μm thickness. H&E staining and tartrate-resistant acid phosphatase (TRAP, Sigma) staining were performed according to methods described previously (30, 31).
Immunohistochemical staining was performed following a protocol described previously (31). Sections were dewaxed and rehydrated. A solution of 3% H2O2 was used to block the activity of endogenous peroxidase. Antigen retrieval was performed with protease K at 37 °C for 15 min. Antibodies against Ctsk (1:200, Santa Cruz Biotechnology, Santa Cruz, CA) were added and incubated overnight at 4 °C. Corresponding biotinylated secondary antibodies were then added and incubated for 1 h at room temperature, followed by color development with an ABC kit (Vector Labs, Peterborough, UK).
To label the mineralization fronts, 8-week-old WT and RapCtsk mice received an intraperitoneal injection of 20 mg/kg body weight calcein (Sigma) and 25 mg/kg BW alizarin red (Sigma) 6 and 3 days before sacrifice (32, 33). The femora were embedded in polymethylmethacrylate and cut into 2-μm-thick sections (Leica, Wetzlar, Germany). Both MAR and BFR were measured following a method described previously (34).
Cell Culture and Osteoclast Differentiation
For osteoclast differentiation analysis, 4-week-old mice were used to obtain BMMs from whole bone marrow following a method described previously (26, 35). Briefly, bone marrow was washed out from the femora and tibiae and centrifuged at 500 × g for 10 min. Cells were resuspended and cultured in α-MEM supplemented with 10% FBS, 1% penicillin/streptomycin in a 37 °C, 5% CO2 incubator overnight. Then the non-adherent cells were seeded into α-MEM supplemented with 10 ng/ml M-CSF. After 48 h, the non-adherent cells were discarded and adherent cells were used as BMMs. For osteoclast differentiation, BMMs were reseeded into 24-well plate (for protein and RNA assay) and 96-well plate (for TRAP staining) at a density of 5 × 104 cells/cm2. The BMMs were cultured in osteoclast differentiation medium consisting of α-MEM supplemented with 20 ng/ml M-CSF and 250 ng/ml RANKL in the absence or presence of different concentrations of rapamycin. The culture medium was replaced every 2 days until formation of mature multinuclear osteoclasts was observed (5–7 days). Then the culture supernatant was harvested for examination of TRAP activity. Cells were washed twice with PBS, fixed with 4% paraformaldehyde for 5 min, and stained for TRAP using a diagnostic acid phosphatase kit (Sigma). TRAP-positive osteoclasts with more than three nuclei were counted in each well of a 96-well plate.
To determine the proliferation of BMMs, BMMs from WT and RapCtsk mice were seeded in 96-well plates (2.5 × 104 cells/cm2) with 10 ng/ml M-CSF. Crystal violet staining was used to detect cell proliferation on days 0, 1, 3, and 7.
Plasmid and Lentivirus
cDNA of S6K1 was cloned into a phage-based plasmid. Constitutively active S6K1 was constructed following methods described previously by converting Thr390 to glutamic acid (36). The CAS6K1 vector and package vectors were transfected into HEK 293T/17 cells to generate a lentivirus expressing FLAG-CAS6K1(Lenti-CAS6K1), and a lentivirus expressing green fluorescent protein (Lenti-GFP) was used as control. To force expression of CAS6K1, RapCtsk BMMs were infected with Lenti-CAS6K1 24 h after seeding and then cultured in osteoclast differentiation medium until formation of osteoclasts.
TRAP Activity Quantification
For the quantification of TRAP activity, cultured supernatant collected from the differentiated BMMs was incubated with a 0.33 m tartrate solution containing phosphatase substrate (Sigma) at 37 °C for 2 h before the reaction was terminated with 3 n NaOH. TRAP activity was measured by colorimetric analysis at a maximum wavelength of 405 nm.
Western Blotting
BMMs seeded into 24-well plates and cultured with osteoclast differentiation medium were used for Western blotting. Total proteins were extracted from cultured cells using 1× SDS lysis buffer (Takara Bio Inc., Shiga, Japan) containing protease inhibitor mixture. Lysates were centrifuged at 12,000 × g at 4 °C for 10 min, and the supernatants containing proteins were collected. Lysates containing 30 μg of protein were separated by 10% SDS-PAGE followed by Western blotting according to a standard protocol. The antibodies used were Raptor (Cell Signaling Technology, Danvers, MA, USA), P-S6K1 (Thr389, Merck Millipore, Darmstadt, Germany), S6K1 (Cell Signaling Technology), FLAG (Sigma), and β-actin (Santa Cruz Biotechnology).
qPCR
Total RNA was extracted from BMMs after osteoclastic differentiation using TRIzol following a standard protocol (Invitrogen). An aliquot of 500 ng of RNA was reverse-transcribed to cDNA using Takara PrimeScript reverse transcriptase (Takara Bio Inc., Shiga, Japan). qPCR was performed using a SYBR Green mixture (Takara) to detect expression of osteoclastic-specific genes. The primers used were displayed as follows: Ctsk, 5′-GAAGAAGACTCACCAGAAGCAG-3′ and 5′-TCCAGGTTATGGGCAGAGATT-3′; Trap, 5′-GCGACCATTGTTAGCCACATACG-3′ and 5′-CGTTGATGTCGCACAGAGGGAT-3′; dendritic cell-specific transmembrane protein (Dc-stamp), 5′-GGGGACTTATGTGTTTCCACG-3′ and 5′-ACAAAGCAACAGACTCCCAAAT-3′; osteoclast-associated receptor (Oscar), 5′-CCTAGCCTCATACCCCCAG-3′ and 5′-CGTTGATCCCAGGAGTCACAA-3′; Atp6v0d2, 5′-ACGGTGATGTCACAGCAGACGT-3′ and 5′-CTCTGGATAGAGCCTGCCGCA-3′; and Hprt, 5′-CTGGTGAAAAGGACCTCTCGAAG-3′ and 5′-CCAGTTTCACTAATGACACAAACG-3′.
Statistical Analysis
Data are expressed as the mean ± S.D. Two groups were compared using independent sample t tests. One-way analysis of variance was performed for multiple comparisons. p < 0.05 indicated a significant difference. Data analysis was performed with SPSS 13.0 analysis software (SPSS Inc., Chicago, IL).
Author Contributions
Study design: J. W., W. Z., and L. J. Study conduct: Q. D., F. X., Y. H., X. M., and S. Z. Data collection: Q. D. and Y. H. Data analysis and interpretation: Q. D. and X. M. Drafting of the manuscript: Q. D. and Y. H. Revision of the manuscript: W. Z. and J. W.
Acknowledgments
We thank Dr. Minghan Tong and S. Kato for reagents and mice and the members of the Zou laboratory for useful discussions.
This work was supported in part by grants from the 973 Program from the Chinese Ministry of Science and Technology (2014CB964704 and 2015CB964503), the Science and Technology Commission of Shanghai (124119b0101), the National Natural Science Foundation of China (31371463, 81371121, and 81570950), Shanghai Summit and Plateau Disciplines, and the “Chen Xing” project from Shanghai Jiaotong University. The authors declare that they have no conflicts of interest with the contents of this article.
- RANKL
- receptor activator of nuclear factor κB ligand
- mTOR
- mammalian/mechanistic target of rapamycin
- Raptor
- regulatory-associated protein of mTOR
- mTORC
- mammalian/mechanistic target of rapamycin complex
- BMM
- bone marrow-derived macrophage
- BMD
- bone mineral density
- BV
- bone volume
- TV
- total volume
- Tb.Th.
- trabecular thickness
- Tb.N.
- trabecular number
- Ct.Th.
- cortical thickness
- MAR
- mineral apposition rate
- BFR
- bone formation rate
- qPCR
- quantitative PCR
- CA
- constitutively active
- TRAP
- tartrate-resistant acid phosphatase
- α-MEM
- α-minimum Eagle's medium
- M-CSF
- macrophage colony-stimulating factor
- micro-CT
- micro computed tomography.
References
- 1. Boyle W. J., Simonet W. S., and Lacey D. L. (2003) Osteoclast differentiation and activation. Nature 423, 337–342 [DOI] [PubMed] [Google Scholar]
- 2. Raggatt L. J., and Partridge N. C. (2010) Cellular and molecular mechanisms of bone remodeling. J. Biol. Chem. 285, 25103–25108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hadjidakis D. J., and Androulakis I. I. (2006) Bone remodeling. Ann. N.Y. Acad. Sci. 1092, 385–396 [DOI] [PubMed] [Google Scholar]
- 4. Rodan G. A., and Martin T. J. (2000) Therapeutic approaches to bone diseases. Science 289, 1508–1514 [DOI] [PubMed] [Google Scholar]
- 5. Boyce B. F. (2013) Advances in osteoclast biology reveal potential new drug targets and new roles for osteoclasts. J. Bone Miner. Res. 28, 711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zoncu R., Efeyan A., and Sabatini D. M. (2011) mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12, 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bhaskar P. T., and Hay N. (2007) The two TORCs and Akt. Dev. Cell 12, 487–502 [DOI] [PubMed] [Google Scholar]
- 8. Yan B., Zhang Z., Jin D., Cai C., Jia C., Liu W., Wang T., Li S., Zhang H., Huang B., Lai P., Wang H., Liu A., Zeng C., Cai D., et al. (2016) mTORC1 regulates PTHrP to coordinate chondrocyte growth, proliferation and differentiation. Nat. Commun. 7, 11151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu S., Zhang Y., Liu B., Li K., Huang B., Yan B., Zhang Z., Liang K., Jia C., Lin J., Zeng C., Cai D., Jin D., Jiang Y., and Bai X. (2016) Activation of mTORC1 in B lymphocytes promotes osteoclast formation via regulation of β-catenin and RANKL/OPG. J. Bone Miner. Res. 31, 1320–1333 [DOI] [PubMed] [Google Scholar]
- 10. Chen J., and Long F. (2014) mTORC1 signaling controls mammalian skeletal growth through stimulation of protein synthesis. Development 141, 2848–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kneissel M., Luong-Nguyen N. H., Baptist M., Cortesi R., Zumstein-Mecker S., Kossida S., O'Reilly T., Lane H., and Susa M. (2004) Everolimus suppresses cancellous bone loss, bone resorption, and cathepsin K expression by osteoclasts. Bone 35, 1144–1156 [DOI] [PubMed] [Google Scholar]
- 12. Hadji P., Coleman R., and Gnant M. (2013) Bone effects of mammalian target of rapamycin (mTOR) inhibition with everolimus. Crit. Rev. Oncol. Hematol. 87, 101–111 [DOI] [PubMed] [Google Scholar]
- 13. Glantschnig H., Fisher J. E., Wesolowski G., Rodan G. A., and Reszka A. A. (2003) M-CSF, TNFα and RANK ligand promote osteoclast survival by signaling through mTOR/S6 kinase. Cell Death Differ. 10, 1165–1177 [DOI] [PubMed] [Google Scholar]
- 14. Sugatani T., and Hruska K. A. (2005) Akt1/Akt2 and mammalian target of rapamycin/Bim play critical roles in osteoclast differentiation and survival, respectively, whereas Akt is dispensable for cell survival in isolated osteoclast precursors. J. Biol. Chem. 280, 3583–3589 [DOI] [PubMed] [Google Scholar]
- 15. Chen J., and Long F. (2015) mTORC1 signaling promotes osteoblast differentiation from preosteoblasts. PLoS ONE 10, e0130627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen C., Akiyama K., Wang D., Xu X., Li B., Moshaverinia A., Brombacher F., Sun L., and Shi S. (2015) mTOR inhibition rescues osteopenia in mice with systemic sclerosis. J. Exp. Med. 212, 73–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martin S. K., Fitter S., Dutta A. K., Matthews M. P., Walkley C. R., Hall M. N., Ruegg M. A., Gronthos S., and Zannettino A. C. (2015) Brief report: the differential roles of mTORC1 and mTORC2 in mesenchymal stem cell differentiation. Stem Cells 33, 1359–1365 [DOI] [PubMed] [Google Scholar]
- 18. Huang B., Wang Y., Wang W., Chen J., Lai P., Liu Z., Yan B., Xu S., Zhang Z., Zeng C., Rong L., Liu B., Cai D., Jin D., and Bai X. (2015) mTORC1 prevents preosteoblast differentiation through the Notch signaling pathway. PLoS Genet. 11, e1005426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Simone V., Ciavarella S., Brunetti O., Savonarola A., Cives M., Tucci M., Opinto G., Maiorano E., and Silvestris F. (2015) Everolimus restrains the paracrine pro-osteoclast activity of breast cancer cells. BMC Cancer 15, 692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hu Y., Carraro-Lacroix L. R., Wang A., Owen C., Bajenova E., Corey P. N., Brumell J. H., and Voronov I. (2016) Lysosomal pH plays a key role in regulation of mTOR activity in osteoclasts. J. Cell. Biochem. 117, 413–425 [DOI] [PubMed] [Google Scholar]
- 21. Indo Y., Takeshita S., Ishii K. A., Hoshii T., Aburatani H., Hirao A., and Ikeda K. (2013) Metabolic regulation of osteoclast differentiation and function. J. Bone Miner. Res. 28, 2392–2399 [DOI] [PubMed] [Google Scholar]
- 22. Gnant M., Baselga J., Rugo H. S., Noguchi S., Burris H. A., Piccart M., Hortobagyi G. N., Eakle J., Mukai H., Iwata H., Geberth M., Hart L. L., Hadji P., El-Hashimy M., Rao S., et al. (2013) Effect of everolimus on bone marker levels and progressive disease in bone in BOLERO-2. J. Natl. Cancer Inst. 105, 654–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xian L., Wu X., Pang L., Lou M., Rosen C. J., Qiu T., Crane J., Frassica F., Zhang L., Rodriguez J. P., Jia X., Yakar S., Xuan S., Efstratiadis A., Wan M., and Cao X. (2012) Matrix IGF-1 maintains bone mass by activation of mTOR in mesenchymal stem cells. Nat. Med. 18, 1095–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Feng X., and McDonald J. M. (2011) Disorders of bone remodeling. Annu. Rev. Pathol. 6, 121–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Y. P., Chen W., Liang Y., Li E., and Stashenko P. (1999) Atp6i-deficient mice exhibit severe osteopetrosis due to loss of osteoclast-mediated extracellular acidification. Nat. Genet. 23, 447–451 [DOI] [PubMed] [Google Scholar]
- 26. Greenblatt M. B., Park K. H., Oh H., Kim J. M., Shin D. Y., Lee J. M., Lee J. W., Singh A., Lee K. Y., Hu D., Xiao C., Charles J. F., Penninger J. M., Lotinun S., Baron R., et al. (2015) CHMP5 controls bone turnover rates by dampening NF-κB activity in osteoclasts. J. Exp. Med. 212, 1283–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee K. W., Yook J. Y., Son M. Y., Kim M. J., Koo D. B., Han Y. M., and Cho Y. S. (2010) Rapamycin promotes the osteoblastic differentiation of human embryonic stem cells by blocking the mTOR pathway and stimulating the BMP/Smad pathway. Stem Cells Dev. 19, 557–568 [DOI] [PubMed] [Google Scholar]
- 28. Singha U. K., Jiang Y., Yu S., Luo M., Lu Y., Zhang J., and Xiao G. (2008) Rapamycin inhibits osteoblast proliferation and differentiation in MC3T3-E1 cells and primary mouse bone marrow stromal cells. J. Cell. Biochem. 103, 434–446 [DOI] [PubMed] [Google Scholar]
- 29. Yeh L. C., Ma X., Ford J. J., Adamo M. L., and Lee J. C. (2013) Rapamycin inhibits BMP-7-induced osteogenic and lipogenic marker expressions in fetal rat calvarial cells. J. Cell. Biochem. 114, 1760–1771 [DOI] [PubMed] [Google Scholar]
- 30. Andersson G. N., and Marks S. C. Jr. (1989) Tartrate-resistant acid ATPase as a cytochemical marker for osteoclasts. J. Histochem. Cytochem. 37, 115–117 [DOI] [PubMed] [Google Scholar]
- 31. Zou W., Greenblatt M. B., Brady N., Lotinun S., Zhai B., de Rivera H., Singh A., Sun J., Gygi S. P., Baron R., Glimcher L. H., and Jones D. C. (2013) The microtubule-associated protein DCAMKL1 regulates osteoblast function via repression of Runx2. J. Exp. Med. 210, 1793–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zou D., Zhang Z., He J., Zhu S., Wang S., Zhang W., Zhou J., Xu Y., Huang Y., Wang Y., Han W., Zhou Y., Wang S., You S., Jiang X., and Huang Y. (2011) Repairing critical-sized calvarial defects with BMSCs modified by a constitutively active form of hypoxia-inducible factor-1α and a phosphate cement scaffold. Biomaterials 32, 9707–9718 [DOI] [PubMed] [Google Scholar]
- 33. Wang S., Zhang W., Zhao J., Ye D., Zhu C., Yang Y., Zhang X., Sun X., Yang C., Jiang X., and Zhang Z. (2011) Long-term outcome of cryopreserved bone-derived osteoblasts for bone regeneration in vivo. Biomaterials 32, 4546–4555 [DOI] [PubMed] [Google Scholar]
- 34. Parfitt A. M., Drezner M. K., Glorieux F. H., Kanis J. A., Malluche H., Meunier P. J., Ott S. M., and Recker R. R. (1987) Bone histomorphometry: standardization of nomenclature, symbols, and units: report of the ASBMR Histomorphometry Nomenclature Committee. J. Bone Miner. Res. 2, 595–610 [DOI] [PubMed] [Google Scholar]
- 35. Liu X., Qu X., Wu C., Zhai Z., Tian B., Li H., Ouyang Z., Xu X., Wang W., Fan Q., Tang T., Qin A., and Dai K. (2014) The effect of enoxacin on osteoclastogenesis and reduction of titanium particle-induced osteolysis via suppression of JNK signaling pathway. Biomaterials 35, 5721–5730 [DOI] [PubMed] [Google Scholar]
- 36. Xia T., Cheng Y., Zhang Q., Xiao F., Liu B., Chen S., and Guo F. (2012) S6K1 in the central nervous system regulates energy expenditure via MC4R/CRH pathways in response to deprivation of an essential amino acid. Diabetes 61, 2461–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]