Abstract
In most eukaryotes, telomere maintenance relies on telomeric repeat synthesis by a reverse transcriptase named telomerase. To synthesize telomeric repeats, the catalytic subunit telomerase reverse transcriptase (TERT) uses the RNA subunit (TER) as a template. In the ciliate Tetrahymena thermophila, the telomerase holoenzyme consists of TER, TERT, and eight additional proteins, including the telomeric repeat single-stranded DNA-binding protein Teb1 and its heterotrimer partners Teb2 and Teb3. Teb1 is paralogous to the large subunit of the general single-stranded DNA binding heterotrimer replication protein A (RPA). Little is known about the function of Teb2 and Teb3, which are structurally homologous to the RPA middle and small subunits, respectively. Here, epitope-tagging Teb2 and Teb3 expressed at their endogenous gene loci enabled affinity purifications that revealed that, unlike other Tetrahymena telomerase holoenzyme subunits, Teb2 and Teb3 are not telomerase-specific. Teb2 and Teb3 assembled into other heterotrimer complexes, which when recombinantly expressed had the general single-stranded DNA binding activity of RPA complexes, unlike the telomere-specific DNA binding of Teb1 or the TEB heterotrimer of Teb1, Teb2, and Teb3. TEB had no more DNA binding affinity than Teb1 alone. In contrast, heterotrimers reconstituted with Teb2 and Teb3 and two other Tetrahymena RPA large subunit paralogs had higher DNA binding affinity than their large subunit alone. Teb1 and TEB, but not RPA, increased telomerase processivity. We conclude that in the telomerase holoenzyme, instead of binding DNA, Teb2 and Teb3 are Teb1 assembly factors. These findings demonstrate that Tetrahymena telomerase holoenzyme and RPA complexes share subunits and that RPA subunits have distinct functions in different heterotrimer assemblies.
Keywords: chromosomes, DNA, protozoan, telomerase, telomere
Introduction
Telomeres, which are the DNA-protein complexes at the ends of eukaryotic chromosomes, are essential for genome stability and long term cellular proliferation (1, 2). Generally, telomeric DNA is composed of simple sequence repeats arranged as a tract of duplex repeats followed by a single-stranded 3′ overhang (3). These telomeric repeats recruit sequence-specific double-stranded and single-stranded DNA-binding proteins to nucleate the assembly of telomere-specific protein complexes, which sequester chromosome termini from DNA damage sensors (3, 4). The accessibility of strand termini is strictly regulated, and as a consequence, the 3′ overhang has a fixed length range in any given species. This 3′ overhang is critical for telomere end protection, but it must be created anew after genome replication in a manner that obliges a loss of telomeric repeats with each round of cell division (5). Single-celled organisms have a relatively short telomeric 3′ overhang and consequently lose a few or tens of base pairs per cell division, whereas human cells have relatively long overhangs on the order of ∼100 nucleotides (nt)3 and correspondingly lose more base pairs of telomeric repeats per cell division (6, 7).
To compensate for incomplete telomere replication by conventional DNA polymerases, most eukaryotes rely on the ribonucleoprotein (RNP) telomerase (8). Each telomeric repeat array is maintained in a dynamic equilibrium of attrition from genome replication and telomerase-mediated de novo synthesis. Telomerase acts by reverse transcribing the integral RNA component, TER, with the catalytic telomerase reverse transcriptase protein, TERT (9, 10). By copying a short template sequence within its RNA moiety, telomerase synthesizes the guanosine-rich telomeric DNA strand (G-strand) running 5′ to 3′ toward a chromosome terminus (e.g. repeats of TTGGGG in the ciliate Tetrahymena or TTAGGG in vertebrates). TERT and TER assembled in a heterologous cell extract can reconstitute repeat synthesis activity; therefore, an RNP with these two subunits is considered the minimal recombinant RNP (11, 12). For biologically functional telomerase holoenzyme, TER and TERT require a number of other subunits to properly fold TER, assemble TER with TERT, and allow active RNP to elongate telomeres (13, 14). Although telomerase holoenzyme subunits are evolutionarily divergent in sequence, studies across model organisms have illuminated recurrent functionalities for holoenzyme proteins in RNA stabilization, intracellular RNP trafficking, and RNP recruitment to telomeres (15, 16).
Telomerase binds a chromosome 3′ overhang in competition, and also coordination, with other single-stranded DNA (ssDNA)-binding proteins (17, 18). Throughout most of the cell cycle, the telomere 3′ overhang is sequestered by DNA binding and telomere remodeling activities of the ssDNA-binding protein Pot1 (18, 19). The Pot1 N-terminal pair of oligonucleotide/oligosaccharide-binding fold (OB-fold) domains interacts sequence-specifically with the telomeric repeat G-strand, whereas the Pot1 C-terminal region interacts with vertebrate TPP1/fission yeast Tpz1/Tetrahymena Tpt1 (19, 20). TPP1 and Tpz1 bridge Pot1 with proteins assembled on the double-stranded telomeric DNA repeats (21).
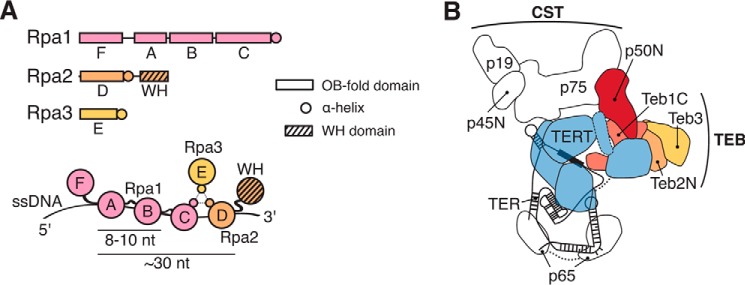
Telomeric repeat ssDNA is also bound, at least transiently, by the general ssDNA-binding RPA heterotrimer of ∼70-kDa Rpa1, ∼30-kDa Rpa2, and ∼15-kDa Rpa3. RPA serves essential roles in DNA replication and repair, recruiting myriad cellular factors to bound ssDNA with specificities that are incompletely understood (22, 23). The RPA heterotrimer has six OB-fold domains, four of which contact DNA: domains A, B, and C in the large subunit Rpa1 and domain D in the middle subunit Rpa2 (Fig. 1A). DNA-binding domains A and B initially engage 8–10 nt of ssDNA, and then subsequent DNA binding by domains C and D extends the footprint to ∼30 nt (23, 24). Domains A–D are oriented from 5′ to 3′ on ssDNA, with interdomain contacts and linker structuring induced by DNA binding (25). Despite high DNA binding affinity from this interdomain cooperation, RPA can diffuse along a bound DNA by a series of individual domain dissociations (23, 24, 26).
FIGURE 1.
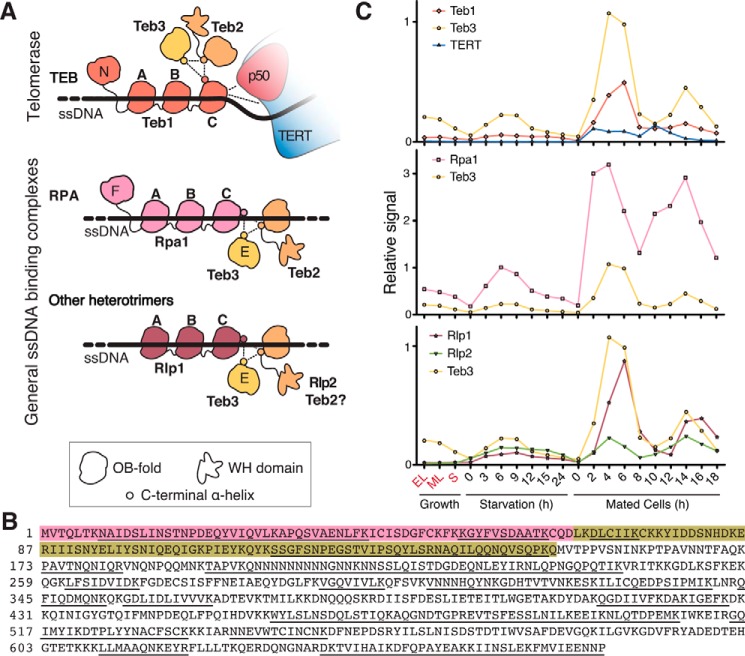
RPA domains and TEB subunit organization in telomerase holoenzyme. A, schematic diagram of RPA subunit domains (top) and their DNA contact (bottom). B, simplified diagram of Tetrahymena telomerase holoenzyme architecture (34). Subunits are colored as follows: TERT (blue), p50 N-terminal domain (red), Teb1C (pink), Teb2 N-terminal domain (gold), and Teb3 (yellow). The remaining holoenzyme proteins and TER are labeled without color; two p65 RNA-binding domains are connected by a dashed line, and only the N-terminal OB-fold domain of p45 (p45N) is included in the model.
Telomeric repeat ssDNA is also bound by the RPA-like CST complex, which is composed of large subunit Cdc13 (in budding yeast), CTC1 (in vertebrate cells), or p75 (in Tetrahymena; see below) as well as the middle and small subunits Stn1 and Ten1, respectively. CST has evolutionarily variable ssDNA binding properties and variable biological roles linked to a high degree of large subunit divergence (18, 27). Vertebrate CST contributes to DNA replication at sites throughout the genome and, with distinct structural requirements, to telomere-specific processes, such as the post-replication cytidine-rich strand (C-strand) fill-in by polymerase α-primase (28–30). Vertebrate CST also has been proposed to inhibit telomerase access to chromosome ends, although this role is not uniformly evident across different studies (30, 31). Budding yeast CST function is telomere-specific; it stimulates C-strand fill-in and contributes to chromosome end-capping, and, when disassembled in S-phase, its Cdc13 subunit recruits telomerase holoenzyme (18). A single OB-fold domain within budding yeast Cdc13 is necessary and sufficient for sequence-specific recognition of G-strand ssDNA (32), whereas detectable binding of vertebrate or Tetrahymena CST to DNA requires all three subunits (28, 31, 33). Tetrahymena CST assembles as a stable subcomplex of the telomerase holoenzyme, where it is proposed to couple G-strand synthesis to C-strand fill-in (33, 34).
Remarkably, the Tetrahymena telomerase holoenzyme contains another RPA-like heterotrimer in addition to CST: the TEB heterotrimer composed of Teb1, Teb2, and Teb3 (34)(Fig. 1B). The holoenzyme RNP catalytic core (TERT, TER, and the RNA-binding protein p65) interacts with the central hub protein p50 (with an OB-fold domain structurally and functionally related to TPP1), which in turn binds independently to the CST and TEB heterotrimers (34–36). The RPA-like TEB complex includes large subunit Teb1, middle subunit Teb2, and small subunit Teb3 (34, 37). Teb1 has an N-terminal OB-fold domain that does not contribute to DNA binding, two central DNA-binding OB-fold domains (Teb1A and Teb1B), and a C-terminal OB-fold domain (Teb1C) that improves DNA binding by Teb1AB (37, 38). Teb1A and Teb1B each bind sequence-specifically to the G-strand of Tetrahymena telomeric repeats (38, 39). Teb1C interacts with p50, probably threading ssDNA between the telomerase active site and Teb1AB (34, 35). In cells, high affinity ssDNA binding by Teb1 is a major determinant of telomerase association to telomeres. Teb1C mutations that disrupt p50 interaction do not reduce Teb1 binding to telomeric DNA, but Teb1 interaction with p50 is required for robust telomere association of the rest of the telomerase holoenzyme (40).
In comparison with Teb1, little is known about the function(s) of Teb2 or Teb3. A structural model from cryo-electron microscopy (34) indicates that the Teb2 OB-fold domain (the N-terminal half of the protein) and the Teb3 OB-fold domain (the full-length protein) interact with Teb1C to form the heterotrimer core, which is stabilized by RPA-like bundling of the α-helices immediately following the OB-fold domains (Fig. 1A). Teb1 alone or TEB heterotrimer dramatically increases telomerase repeat addition processivity (RAP), but for proteins expressed in rabbit reticulocyte lysate (RRL), TEB mediates this stimulation more effectively than Teb1 (34, 36, 37). We suggested previously that Teb2 and Teb3 could be Tetrahymena RPA subunits as well as subunits of telomerase holoenzyme (34). Phylogenetic analysis grouped Teb2 with Rpa2 and Teb3 with Rpa3 proteins of other species (34). Furthermore, Teb2 and Teb3 have mRNA expression levels higher than Teb1 and the other telomerase-specific holoenzyme subunits, approaching the mRNA level of the previously characterized RPA large subunit Rpa1 (34, 38). Together, these observations raise the hypothesis that Teb2 and Teb3 could be shared subunits of telomerase holoenzyme and RPA.
To test this hypothesis and to better understand the function(s) of Teb2 and Teb3 in cells, we investigated their endogenous interaction partners by purification of cellular complexes. These studies and complementary ssDNA-binding assays performed with purified recombinant heterotrimers establish that Teb2 and Teb3 are subunits of telomerase holoenzyme and Tetrahymena RPA complexes. Curiously, the same proteins make different functional contributions to different heterotrimers. Understanding the complexity of RPA and RPA-like complexes in Tetrahymena provides a precedent for studies of alternative RPA subunits in other organisms, including humans (41–43).
Results
Teb2 and Teb3 Are Not Telomerase-specific Proteins
To further our functional understanding of Teb2 and Teb3, we sought to characterize endogenously assembled complexes containing these proteins. In addition to the telomerase holoenzyme (Fig. 1B), Teb2 and Teb3 could be subunits of RPA or RPA-like complexes likely to be more abundant than telomerase. We performed unbiased affinity purification of Teb2 and Teb3 using N-terminally tagged versions of the proteins expressed from their endogenous gene loci. We targeted each endogenous gene locus to insert an N-terminal tag of tandem Protein A domains (ZZ) and a triple FLAG peptide (F), optimized for affinity purifications from Tetrahymena cell extracts (37). The integrated construct also contained a blasticidin resistance cassette (Fig. 2A), allowing for a standard protocol of assortment to maximal recombinant chromosome copy number in replacement of the endogenous locus chromosome (44). Cells released from selection were used for genomic DNA analysis to discriminate whether the recombinant chromosome had entirely substituted for the wild-type chromosome. Southern blotting hybridization confirmed full macronuclear replacement of endogenous chromosomes with ZZF-Teb2 or ZZF-Teb3 chromosomes (Fig. 2B). The silent micronuclear gene locus gives a very faint wild-type locus signal that is discriminated as micronuclear because it does not rebound in copy number with release of cells from selection in blasticidin (44).
FIGURE 2.
Teb2 and Teb3 are not telomerase-specific. A, genomic locus targeting strategy to generate ZZF-Teb2 and ZZF-Teb3 cell lines. B, genomic DNA Southern blots showing macronuclear replacement of an endogenous locus with the recombinant locus. C, affinity purifications from extracts of cells in synchronized vegetative growth (left) or mated cells (right) profiled by colloidal Coomassie staining after SDS-PAGE. D, telomerase activity assays for the purifications shown in C. RC, recovery control for telomerase product precipitation. Numbers of nt added to the primer to complete the first three telomeric repeats are indicated at the right.
We performed tagged protein affinity purifications from extracts of cells in synchronized, asexual (vegetative) growth or mated cells in the process of sexual reproduction, two conditions that demand high rates of new DNA synthesis (45). Tandem affinity purification of ZZF-Teb2 or ZZF-Teb3 from whole-cell lysates co-purified several polypeptides not recovered in parallel mock purifications from cell extracts without tagged protein (Fig. 2C). Proteins with the SDS-polyacrylamide gel mobilities of F-Teb2 and Teb3 or F-Teb3 and Teb2 were readily detectable (note that the ZZ portion of the tag was removed during purification). Aside from Teb2 and Teb3, the proteins in the purifications did not co-migrate with telomerase holoenzyme proteins (37), such as the ∼130-kDa TERT. Although it contributes a minority of the total associated proteins, telomerase holoenzyme was co-purified with F-Teb2 and F-Teb3, as judged by specific enrichment of telomerase activity assayed using direct telomeric primer extension with radiolabeled dGTP and dTTP (Fig. 2D). Telomerase enriched by ZZF-Teb2 or ZZF-Teb3 had more low RAP activity with the purifications from cells in vegetative growth versus the purifications from mated cells, which, based on our experience, is likely to reflect more proteolysis in extracts of growing cells.
To identify unknown proteins that interact with Teb2 and Teb3, we submitted the entire pool of proteins associated with F-Teb2 or F-Teb3 for mass spectrometry (MS), using the purifications from mated cells. Proteins detected by MS in the mock purification from the parental strain were subtracted from the list of proteins specifically associated with Teb2 or Teb3, which we then rank-ordered by number of unique sequence peptides (Table 1). Both purifications co-enriched Tetrahymena Rpa1 as the top-ranked associated protein. We had previously characterized Rpa1 as a general ssDNA-binding subunit genetically essential for Tetrahymena growth (38). In addition to Rpa1, the Teb2 purification co-enriched Teb3, and the Teb3 purification co-enriched Teb2. The representation of Rpa1, Teb2, and Teb3 clustered them together as a top-ranking group (Table 1). Also well represented were likely RPA-interacting factors involved in DNA repair (22, 23), including DNA mismatch repair proteins (Tetrahymena gene names MSH2 and MSH6) and other DNA repair factors (Ku80 and Ku70). The telomerase holoenzyme proteins p75, p65, p50, and p45 were detected at low representation in the Teb2 purification (Table 1), which had a higher yield of total protein than the Teb3 purification in the samples used for MS. Due to the scarce amount of telomerase compared with DNA replication and repair factors, it is not surprising that only a subset of telomerase holoenzyme subunits was detected in only the higher yield affinity purification.
TABLE 1.
Proteins co-purified with Teb2 or Teb3 as detected by MS
The column labeled “ZZF” indicates the tagged protein used for affinity purification. Listing order of MS-identified proteins associated with ZZF-Teb2 or ZZF-Teb3 is by decreasing number of unique-sequence peptides (sequence count) detected for the top 15 associated proteins, followed by any telomerase holoenzyme subunits. Teb2 recovered 70 total proteins and 35 hits with more than one peptide read. Two additional proteins fall between p75 and the top 15 associated proteins, whereas 28 additional proteins fall between p45 and the top 15 associated proteins. Teb3 recovered 40 total proteins and 24 with more than one peptide read. Boldface type indicates an RPA subunit paralog or telomerase holoenzyme protein.
| ZZF | Locus | Gene name | Description | Sequence count | Protein coverage |
|---|---|---|---|---|---|
| % | |||||
| Teb2 | TTHERM_00106890 | RFA1 | Replication protein A large subunit | 73 | 53.8 |
| TTHERM_001113129 | TEB2 | Teb2 | 32 | 39.8 | |
| TTHERM_00439320 | TEB3 | Teb3 | 19 | 47.6 | |
| TTHERM_00295920 | MSH2 | MutS domain III family protein | 17 | 20.2 | |
| TTHERM_00194810 | MSH6 | MutS domain III family protein | 17 | 14.4 | |
| TTHERM_00502600 | PARP6 | WGR domain-containing protein | 11 | 6.4 | |
| TTHERM_00046920 | RVB2 | DNA helicase RBV2 homolog | 8 | 21.3 | |
| TTHERM_00865240 | None | DNA ligase I | 8 | 15.6 | |
| TTHERM_00125640 | SSA3 | HSP70a paralog | 8 | 14.1 | |
| TTHERM_00558440 | SSA5 | HSP70a paralog | 8 | 11.7 | |
| TTHERM_00216140 | None | DNA topoisomerase family protein | 8 | 9.6 | |
| TTHERM_00633360 | HTB1 | Histone H2B | 7 | 29.5 | |
| TTHERM_00492460 | TKU80 | Ku80 ortholog | 7 | 12.2 | |
| TTHERM_00105110 | HSP70 | Putative 70-kDa heat shock protein | 7 | 11.8 | |
| TTHERM_00043780 | POLN1 | DNA polymerase I family protein | 7 | 8.0 | |
| TTHERM_00059040 | TAP75 | p75 | 3 | 8.2 | |
| TTHERM_00083360 | TAP45 | p45 | 1 | 4.3 | |
| TTHERM_000318539 | TAP65 | p65 | 1 | 2.6 | |
| TTHERM_01049190 | TAP50 | p50 | 1 | 2.4 | |
| Teb3 | TTHERM_00106890 | RFA1 | Replication Protein A large subunit | 70 | 44.1 |
| TTHERM_001113129 | TEB2 | Teb2 | 42 | 57.6 | |
| TTHERM_00439320 | TEB3 | Teb3 | 28 | 92.4 | |
| TTHERM_00194810 | MSH6 | MutS domain III family protein | 21 | 18.8 | |
| TTHERM_00216140 | None | DNA topoisomerase family protein | 19 | 21.7 | |
| TTHERM_00672190 | THD13 | Histone deacetylase 13 | 17 | 11.8 | |
| TTHERM_00316500 | HTA2 | Histone H2A | 7 | 24.8 | |
| TTHERM_00295920 | MSH2 | MutS domain III family protein | 7 | 13.7 | |
| TTHERM_00726370 | RLP1 | RPA-like protein 1 | 5 | 9.7 | |
| TTHERM_00502600 | PARP6 | WGR domain-containing protein | 4 | 2.1 | |
| TTHERM_00691710 | None | Toprim domain-containing protein | 4 | 3.3 | |
| TTHERM_00561799 | TKU70 | Ku70 ortholog | 4 | 5.1 | |
| TTHERM_00070820 | None | U-box domain containing protein | 4 | 8.9 | |
| TTHERM_00459400 | RLP2 | RPA-like protein 2 | 4 | 15.2 | |
| TTHERM_00328620 | None | Hypothetical protein | 4 | 17.1 |
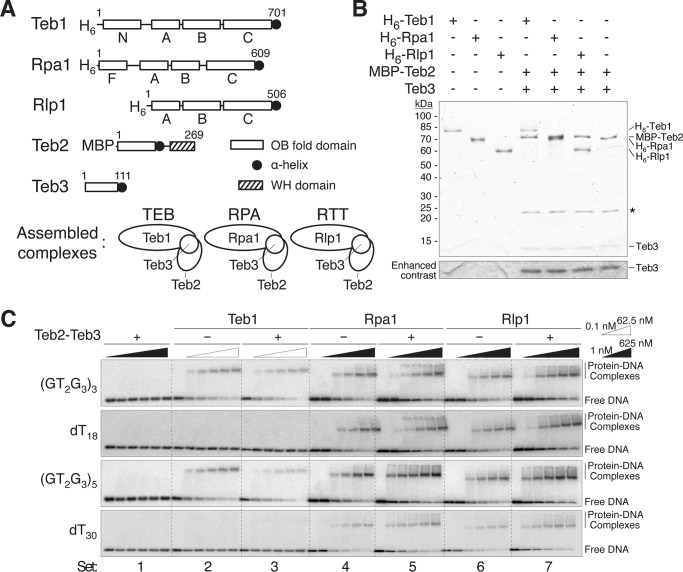
Interestingly, Teb3 co-purified two RPA-like proteins not associated with Teb2: the RPA large subunit paralog Rlp1 (RPA-like protein 1), which we previously characterized in parallel with Rpa1 in cells and as recombinant protein (38), and a putative RPA middle subunit paralog TTHERM_00459400, here designated Rlp2 (RPA-like protein 2). Rlp1 has a sequence and predicted domain structure similar to Rpa1 and Teb1, except that Rlp1 lacks a regulatory N-terminal OB-fold domain (Fig. 3A). Unlike Rpa1 and Teb1, Rlp1 was not genetically essential for cell growth (37, 38). Rlp2 is a hypothetical protein predicted from genome sequence (46). BLAST of Rlp2 against all sequences in GenBankTM identified Tetrahymena Teb2 and the protist Phytophthora infestans Rpa2 in the top 10 scores, the remainder of which were hypothetical proteins, including a marine ciliate protein annotated only as “nucleic acid-binding OB-fold.” Several domain prediction methods confirm that Rlp2 could harbor an OB-fold, which will require future direct structural analysis to confirm. Because both Teb2 and Teb3 co-purified Rpa1 but only Teb3 co-purified Rlp1 and Rlp2, we suggest that Tetrahymena cells assemble at least two RPA or RPA-like heterotrimers beyond the TEB and CST complexes of telomerase holoenzyme: an abundant RPA complex containing Rpa1, Teb2, and Teb3 and complex(es) containing Rlp1, Rlp2, and Teb3 (see “Discussion”). Cells could also assemble a complex of Rlp1, Teb2, and Teb3 that we did not detect by MS. An Rlp1, Teb2, and Teb3 heterotrimer could be low in abundance or not assembled at the specific state of sexual reproduction that cells were harvested in for large scale purifications. Because the goal of this work is to characterize Teb2 and Teb3, we compared heterotrimers containing these two subunits and each alternative large subunit. For convenience, we will refer to a recombinant complex containing Rlp1, Teb2, and Teb3 as RTT.
FIGURE 3.
Comparison of DNA binding by Teb1, TEB, Rpa1, RPA, Rlp1, and RTT. A, domain structures for Teb1, Rpa1, Rlp1, Teb2, and Teb3 and complexes formed by co-assembly that were assayed for DNA binding affinity. B, colloidal Coomassie staining after SDS-PAGE of proteins and complexes used for DNA binding assays. *, a background protein in the Teb2-Teb3 purification. Enhanced contrast for the bottom of the gel is shown below the main gel panel. C, DNA binding by Teb2-Teb3, Teb1, TEB, Rpa1, RPA, Rlp1, and RTT. A fixed concentration of the indicated end-labeled ssDNA oligonucleotide (∼10 pm) was incubated with protein or protein complex added at steps of final concentration over the indicated range of 0.1–62.5 or 1–625 nm.
Teb2 and Teb3 Contribute to the DNA Binding Affinity of RPA but Not TEB
We next investigated the DNA binding properties of the RPA large subunit paralogs Teb1, Rpa1, and Rlp1 with or without co-assembled Teb2-Teb3. We expressed and purified N-terminally six-histidine (His6)-tagged Teb1, Rpa1, and Rlp1 from Escherichia coli either alone, as done previously for Teb1 and Rpa1 (38), or co-expressed with maltose-binding protein (MBP)-tagged Teb2 and untagged Teb3 (Fig. 3A). MBP-Teb2 and Teb3 were also co-expressed in the absence of an RPA large subunit paralog. His6-tagged large subunit proteins were purified using Ni-NTA resin. Teb2-Teb3 complex was purified using amylose resin. Heterotrimer complexes were purified using NI-NTA resin and then amylose resin in series. Overall, these recombinant protein purifications (Fig. 3B) indicate that each large subunit protein can assemble with Teb2-Teb3, at least in the absence of competing cellular factors. Furthermore, Teb1, Rpa1, and Rlp1 each formed complexes with roughly stoichiometric amounts of MBP-Teb2 and Teb3 (Fig. 3B).
To investigate the functional contribution of Teb2-Teb3 to each heterotrimer, we tested the recombinant protein preparations above for binding to a panel of four ssDNA oligonucleotides differing in length (18 or 30 nt) and in telomeric versus non-telomeric sequence (Fig. 3C). In electrophoretic mobility shift assays, the Teb2-Teb3 complex had undetectable DNA binding affinity for any ssDNA (Fig. 3C, set 1). This finding parallels results observed for human Rpa2 and Rpa3 (47). Teb1 alone bound 18- and 30-nt telomeric ssDNAs with similar affinity and did not bind either length of polythymidine (Fig. 3C (set 2) and Table 2), as expected from previous observations (38). Also consistent with previous assays (38), Rpa1 alone bound all four ssDNAs with approximately equal affinity (Fig. 3C (set 4) and Table 2), as did Rlp1 (Fig. 3C (set 6) and Table 2). The TEB complex had DNA binding properties indistinguishable from Teb1 alone, even on the longer ssDNAs (Fig. 3C (set 3) and Table 2), indicating very little or no contribution of Teb2-Teb3 to DNA contact by TEB. In contrast, RPA and RTT each bound 30-nt ssDNA with higher affinity than the corresponding large subunit alone (5–7-fold for RPA, 3–4-fold for RTT; Fig. 3C (sets 4–7) and Table 2). Binding of RPA and RTT to the shorter 18-nt ssDNAs was not improved relative to the corresponding large subunit alone (Fig. 3C (sets 4–7) and Table 2). These results strongly support the conclusion that Teb2 and Teb3 are canonical RPA subunits when in complex with Rpa1; they increase DNA binding affinity and in parallel extend the length of ssDNA that contributes to protein interaction (Fig. 1A) (23, 24).
TABLE 2.
Dissociation constants for oligonucleotide binding to recombinant proteins and complexes
The column labeled “Protein” indicates the recombinant protein or protein complex used for assays of binding to the column header DNA oligonucleotides. Numbers for Teb1, TEB, Rpa1, RPA, Rlp1, and RTT are given in nm as calculated from three experimental replicates of the gel mobility shift assays. S.E. was calculated for each mean to give an estimate of the variation among the replicates. All values had p < 0.06 for goodness of fit for the one-site binding model used to calculate the dissociation constant. −, binding affinity too low to quantify using the gel mobility shift assay conditions. Rows labeled “-Fold change” indicate the relative increase in binding comparing the preceding large subunit protein alone to the heterotrimer.
| Protein | (GT2G3)3 | dT18 | (GT2G3)5 | dT30 |
|---|---|---|---|---|
| Teb2-Teb3 | − | − | − | − |
| Teb1 (nm) | 3.4 ± 1.3 | − | 2.5 ± 1.0 | − |
| TEB (nm) | 2.9 ± 0.8 | − | 2.2 ± 1.7 | − |
| -Fold change | 1.2 | − | 1.1 | − |
| Rpa1 (nm) | 36 ± 11 | 41 ± 20 | 36 ± 15 | 39 ± 16 |
| RPA (nm) | 24 ± 9 | 30 ± 12 | 7.4 ± 3.0 | 5.3 ± 0.6 |
| -Fold change | 1.5 | 1.4 | 4.8 | 7.3 |
| Rlp1 (nm) | 68 ± 22 | 75 ± 9 | 73 ± 28 | 81 ± 14 |
| RTT (nm) | 45 ± 16 | 56 ± 25 | 27 ± 13 | 21 ± 7 |
| -Fold change | 1.5 | 1.3 | 2.7 | 3.8 |
Teb2-Teb3 Function in Telomerase Holoenzyme Is Dependent on Teb1
When expressed in RRL, Teb2 and Teb3 without Teb1 did not stimulate telomerase activity or RAP (34). The very low amount of protein produced in RRL and its uncertain folding presented caveats to the interpretation of this finding. Bacterially expressed proteins generated in this study offered new opportunity to investigate whether Teb2 and Teb3 directly contribute to RAP stimulation. We also sought to investigate whether Tetrahymena RPA, like TEB, stimulates telomerase RAP in vitro. RRL-assembled telomerase RNP catalytic core was combined with p50 and CST (p75, p45, p19) containing C-terminally F-tagged p45, which has biological function (37). The telomerase subunit complex was purified from RRL using anti-FLAG antibody resin and mixed with purified Teb2-Teb3, Teb1, TEB, Rpa1, RPA, Rlp1, or RTT added at final concentrations of 40 or 200 nm. These reconstitutions were then assayed for telomerase product synthesis by direct primer extension. Human RPA (48) or Tetrahymena RPA (data not shown) added at high concentration to an activity assay sequestered primer from telomerase. Here we sought to test the functional interplay of elongating telomerase and the ssDNA binding factors, so we used a 200 nm concentration of an 18-nt telomeric repeat primer with 40 or 200 nm added protein or protein complex.
The addition of Teb1 or TEB converted the RNP catalytic core with p50 to high RAP activity (Fig. 4, lanes 1 and 3–6). The addition of the Teb2-Teb3 complex alone had no influence (Fig. 4, lane 2). The addition of Rpa1 did not affect activity or RAP, consistent with previous studies (38), and neither did the addition of RPA (Fig. 4, lanes 7–10). For Rlp1 or RTT, protein(s) added at a concentration equimolar with the 200 nm DNA primer resulted in modest inhibition of overall activity but no change in RAP (Fig. 4, lanes 11–14). Thus, only Teb1 or TEB stimulates RAP, not other ssDNA binding proteins or protein complexes containing Teb2 and Teb3. These activity assays, along with the gel mobility shift assays, suggest that the role of Teb2-Teb3 in telomerase holoenzyme is indirect through Teb1.
FIGURE 4.
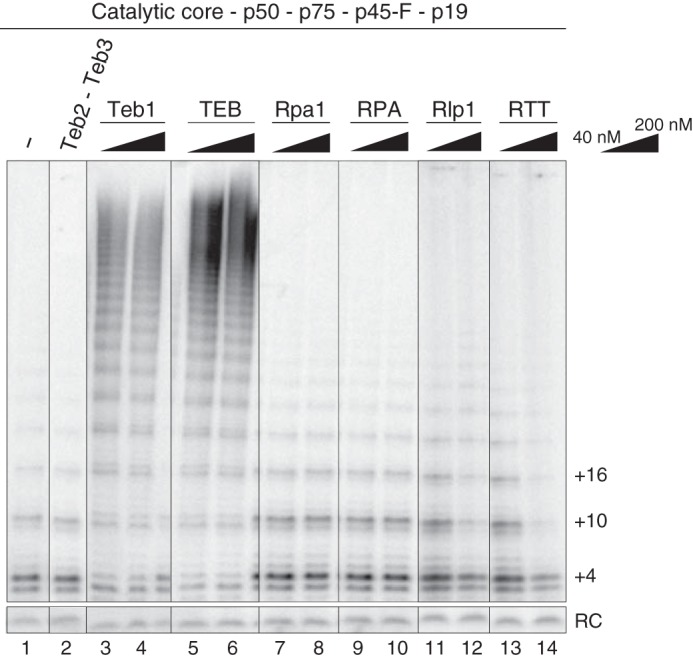
Comparison of telomerase activity stimulation by Teb2-Teb3, Teb1, TEB, Rpa1, RPA, Rlp1, or RTT. The same amount of RRL-reconstituted telomerase complex (composed of the RNP catalytic core, p50, p75, p45-F, and p19) was assayed in the presence or absence of the indicated recombinant proteins at a concentration of 40 or 200 nm. Teb2-Teb3 was added at 200 nm.
TEB Heterotrimer Formation Enhances Teb1 Assembly in Telomerase Holoenzyme
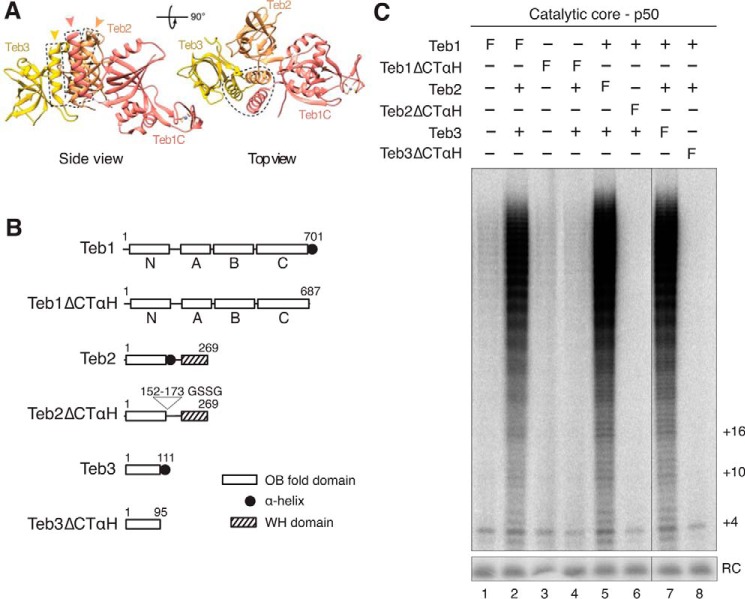
We turned to the hypothesis that Teb2 and Teb3 favor a conformation of Teb1 optimal for its holoenzyme assembly and high RAP stimulation. If Teb2-Teb3 function is indirect through Teb1, it would be dependent on TEB heterotrimer assembly. In RPA, the C-terminal α-helix (CTαH) of Rpa1C, Rpa2 OB-fold domain, and Rpa3 together form the trimerization interface (49). Similarly, the OB-fold domains of Teb1C, Teb2, and Teb3 are each followed by an α-helix that forms their trimerization interface (Fig. 5, A and B), which is on the far side of Teb1C from the contact surface with TERT (34). Reconstitution of high RAP telomerase activity in assays with bacterially expressed Teb1 was robust without Teb2 and Teb3 and showed no requirement for the Teb1C CTαH (38). However, deletion of this α-helix precluded assembly of Teb1 with other telomerase holoenzyme subunits in vivo (40). Because RRL expression and assembly of Teb1 sensitized high RAP activity reconstitution for stimulation by Teb2 and Teb3 (34), we used this system to test the significance of the TEB heterotrimer interface.
FIGURE 5.
Roles of the TEB subunit OB-fold domain C-terminal α-helices. A, model of Teb1C-Teb2N-Teb3 based on fitting of the human RPA heterotrimer (RPA70C-RPA32N-RPA14) (Protein Data Bank entry 1L1O) into a cryo-electron microscopy map, followed by replacement of RPA70C with Teb1C, except for the RPA70C C-terminal α-helix (34). Human Rpa2 OB-fold domain and Rpa3 fit the density of Teb2 OB-fold domain and Teb3, respectively. The three-helix bundle is highlighted with a dashed line. B, diagram of proteins without or with OB-fold CTαH removal. Numbers, amino acids in the full-length proteins; GSSG, a glycine/serine linker added to replace the CTαH of the Teb2 OB-fold domain. C, telomerase activity reconstituted using the proteins diagrammed in B after purification using anti-FLAG antibody resin.
We generated expression constructs that removed the CTαH of Teb1C, Teb2 OB-fold domain, or Teb3 (Fig. 5B). Recombinant telomerase complexes were assembled containing the RNP catalytic core, p50, and N-terminally F-tagged Teb1, Teb2, or Teb3 in the presence or absence of the other TEB subunits. Complexes containing an F-tagged protein were enriched by binding to anti-FLAG-agarose and then assayed for co-purified telomerase catalytic activity (Fig. 5C). Each F-tagged TEB subunit co-purified a similar amount of high RAP activity in the presence of all three TEB subunits (Fig. 5C, lanes 2, 5, and 7), indicating that the tagged proteins assembled into telomerase holoenzyme. F-Teb1 recovered a low amount of high RAP activity in the absence of Teb2-Teb3 and more high RAP activity in the presence of Teb2 and Teb3 (Fig. 5C, lanes 1 and 2). F-Teb1ΔCTαH co-purified the same level of activity co-purified by Teb1 without Teb2 and Teb3 even if Teb2 and Teb3 were present (Fig. 5C, lanes 3 and 4). In comparison, purification through F-Teb2ΔCTαH or F-Teb3ΔCTαH failed to recover any telomerase activity, even in the presence of other TEB subunits (Fig. 5C, lanes 6 and 8). These findings establish that assembly of Teb2-Teb3 into telomerase holoenzyme requires the formation of the heterotrimer α-helix bundle. Deletion of any one of the three α-helices precluded Teb2-Teb3 association with or stimulation of high RAP activity.
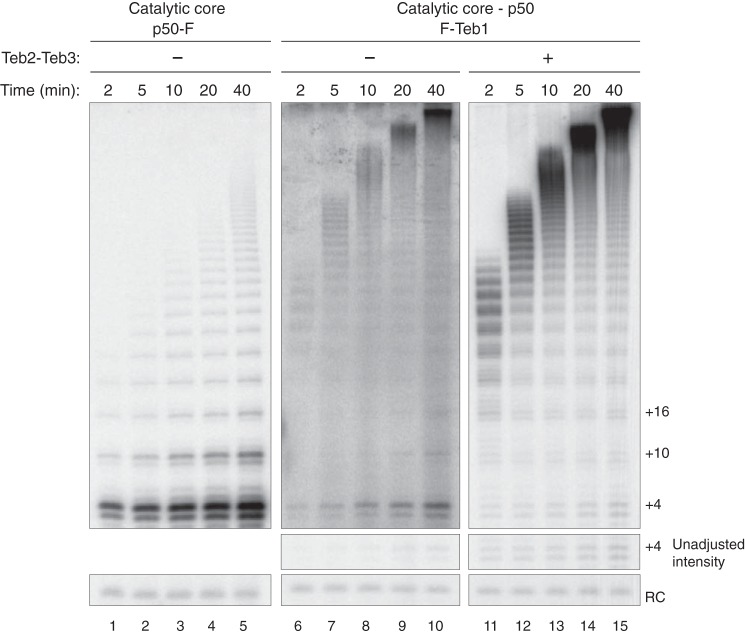
Previous assays were not designed to detect a subtle influence of Teb2-Teb3 on RAP or the rate of repeat synthesis. Therefore, we profiled repeat synthesis over a time course of 40 min for telomerase enzymes reconstituted with Teb1 or TEB. We purified Teb1- or TEB-containing telomerase complexes from reconstitutions with the p50-bound RNP catalytic core using F-Teb1. In comparison, we purified the p50-bound RNP catalytic core reconstituted without TEB subunits using p50-F. Activity assays confirmed that both Teb1 and TEB dramatically increase the rate of tandem repeat synthesis and the amount of high RAP product relative to the p50-bound RNP catalytic core alone (Fig. 6). There was no major difference in the profile of high RAP repeat synthesis with Teb1 alone versus TEB. However, F-Teb1 co-purified much less activity in the absence than in the presence of Teb2-Teb3 (Fig. 6, lanes 6–10 versus lanes 11–15 from the same gel, but the top panel of lanes 6–10 is shown with amplified signal intensity relative to lanes 1–5 and 11–15). With equal inputs of F-Teb1 and p50-bound RNP catalytic core, the simplest explanation for the results is that Teb2-Teb3 increased the amount of Teb1 associated with active telomerase. Background binding of RRL-expressed telomerase subunits to purification resin makes direct quantification of this influence unreliable (data not shown). In addition, some activity of p50-bound RNP catalytic core alone was detectable in assays of enzyme purified by F-Teb1 without Teb2-Teb3, but not enzyme with TEB (Fig. 6, compare low RAP with high RAP activity in lanes 6–10 and 11–15). We suggest that some p50-bound RNP catalytic core dissociated from Teb1, but not TEB, during the activity assay reaction.
FIGURE 6.
Comparison of telomerase assembly and activity with Teb1 or TEB. A time course of product synthesis was monitored for enzyme without Teb1 (lanes 1–5), with Teb1 alone (lanes 6–10), or with TEB (lanes 11–15). All lanes are from the same gel, but the top panel of the middle set of lanes is shown at amplified signal intensity relative to the flanking panels.
Altogether, results of this work implicate Teb2 and Teb3 as assembly factors for Teb1 incorporation into telomerase holoenzyme. The importance of this Teb2-Teb3 activity in vivo is supported by the inability of Teb1ΔCTαH to interact with active telomerase in cells (40).
Discussion
In this work, we characterized the proteins Teb2 and Teb3 previously isolated as subunits of the Tetrahymena telomerase holoenzyme (34). Although the other Tetrahymena telomerase holoenzyme subunits are telomerase-specific (37), we show here that Teb2 and Teb3 are not (Fig. 7A). Teb2 and Teb3 have all of the properties expected for RPA middle and small subunits, respectively. Phylogenetic alignments clustered Teb2 with Rpa2 and Teb3 with Rpa3, and their mRNA abundance indicated an expression level higher than other telomerase proteins (34). More definitively, in this study, we demonstrate that in cells, Teb2 and Teb3 are bound to Tetrahymena Rpa1 as the majority fraction of their cellular pool. In addition to this physical interaction evidence, DNA binding assays indicate that Teb2 and Teb3 increase the DNA binding affinity of heterotrimeric Rpa1-Teb2-Teb3 relative to Rpa1 alone. Specifically, Teb2 and Teb3 improved Rpa1 interaction with 30-nt ssDNAs but not 18-nt ssDNAs, regardless of ssDNA sequence. In summary, our purification and reconstitution approaches strongly support a complex of Rpa1, Teb2, and Teb3 as the general ssDNA-binding Tetrahymena RPA (Fig. 7A, middle).
FIGURE 7.
RPA and RPA-like complexes in Tetrahymena. A, illustration of the inferred DNA contacts within TEB, RPA, and Rlp1-containing heterotrimer(s). Teb2-Teb3 interaction with Teb1 stabilizes the interaction of Teb1C with p50 and the RNP catalytic core. Teb2-Teb3 interaction with Rpa1 or Rlp1 instead extends the DNA-binding surface of the heterotrimer compared with the large subunit alone. B, Tetrahymena Rpa1 coding region. Pink, extra sequence in ∼80-kDa versus ∼70-kDa predicted proteins; yellow, extra sequence in ∼70-kDa versus ∼60-kDa predicted proteins. The original ∼70-kDa predicted protein had three additional N-terminal amino acids not shown. Peptide sequences identified by MS are underlined. We note that the protein open reading frame is validated by peptide sequence to the C terminus that we annotated, instead of the C terminus from an alternative reading frame predicted in the most recent genome database update. C, microarray mRNA expression profiles for Teb3 (yellow) compared with other telomerase subunits (top), other RPA subunits (middle), and RPA-like protein subunits (bottom). There are no microarray expression data for Teb2. Graphs were generated using information from the Tetrahymena Gene Expression Database (52, 53). Vertical axes, relative microarray signal intensity. Horizontal axis, a progression of life cycle states: in vegetative growth at low density (EL), log phase (ML), or stationary phase (S); in starvation medium for the indicated time in hours following transfer from growth media; mating time points in hours following mixing of two mating types.
Telomerase “appropriation” of Teb2 and Teb3 from RPA raises the question of what the subunits of a general ssDNA-binding factor contribute to telomeric repeat synthesis. The Teb2 OB-fold domain is displaced from the RPA-like configuration on DNA by Teb1C interaction with the telomerase RNP catalytic core (Fig. 7A, top). Also, DNA binding by TEB-assembled Teb2 would be unnecessary given the high affinity of Teb1 alone for telomeric ssDNA. We suggest that Teb2 and Teb3 influence Teb1 function indirectly by favoring a Teb1 conformation productive for holoenzyme assembly. Teb2-Teb3 function depends on the Teb1C CTαH, because Teb1ΔCTαH stimulation of high RAP activity lost its enhancement by Teb2-Teb3. Teb1ΔCTαH expressed in Tetrahymena does not co-purify telomerase activity from cell extracts (40), yet bacterially expressed Teb1ΔCTαH can reconstitute high RAP telomerase activity in vitro (38). Combined, these findings support the conclusions that the telomerase holoenzyme role of Teb2-Teb3 is indirect through Teb1, is critical under biological conditions, and is better recapitulated by TEB expressed and assembled with active RNP in RRL than by bacterially expressed and purified TEB. Our findings also reveal that the same proteins make different functional contributions in an RPA versus TEB heterotrimer.
The original annotation of the open reading frame for Tetrahymena Rpa1 suggested an ∼70-kDa protein, which did not match the mobility of a major ZZF-Teb2- or ZZF-Teb3-associated protein. We initially suspected that a post-translation modification of endogenous Rpa1 altered its SDS-PAGE migration. However, upon further investigation, GenBankTM-deposited Tetrahymena mRNA expressed sequence tags support the possibility of an extended N terminus. The longest Rpa1 mRNA would encode an ∼80-kDa protein and from an alternative start site an ∼60-kDa protein (Fig. 7B), which are the sizes of the predominant polypeptides co-purified with Teb2 and Teb3 (Fig. 2C). The ∼60-kDa Rpa1 protein could also result from proteolysis in the linker between the regulatory N-terminal OB-fold domain and the DNA-binding OB-fold domains. MS analysis here validated expression of the longest open reading frame, because sequenced peptides mapped within its unique N-terminal region (Fig. 7B). Because C-terminal tagging of Tetrahymena Rpa1 inactivated its biological function (38), a different approach will be required to test whether a start codon other than that for the ∼80-kDa protein is also used for Rpa1 translation in cells. Our biochemical characterizations used Rpa1 expressed from a synthetic gene encoding the originally annotated sequence (38), which is truncated for the N-terminal region of the N-terminal OB-fold domain. For in vitro assays the ∼70-kDa protein should be representative, because the Rpa1 N-terminal OB-fold does not influence DNA binding affinity or assembly of Rpa1 into heterotrimer (23, 24).
Beyond the general ssDNA-binding Tetrahymena RPA composed of Rpa1, Teb2, and Teb3, our findings suggest that cells undergoing sexual reproduction assemble an additional RPA-related complex composed of Rlp1, Rlp2, and Teb3. Also, it remains possible that Rlp1 assembles with Teb2 and Teb3 in cells under conditions not studied in this work. Based on the sequence-nonspecific DNA binding specificity of Rlp1, any heterotrimer with Rlp1 is likely to have general ssDNA binding activity. Rlp1 lacks an N-terminal regulatory OB-fold domain, as do some predicted Rpa1 paralogs in protozoan parasites (50). Rpa1, Teb1, Teb2, and Teb3 mRNAs are detectable in cells in vegetative growth, starvation, and sexual reproduction (Fig. 7C, top and middle; Teb2 abundance could be analyzed only by expressed sequence tags, not shown). In contrast, Rlp1 and Rlp2 mRNAs are expressed at extremely low, if any, level in vegetative growth (Fig. 7C, bottom). Consistent with this expression pattern, RLP1 knock-out was not deleterious for vegetative growth (38). The expression specificity of Rlp1 and Rlp2 suggests that they and Teb3 could function together specifically in mated cells. The complexity of RPA and RPA-like complexes in Tetrahymena provides new insights and opportunities to understand the function of alternative RPA subunits, which, based on genome sequencing, appear widespread across organisms ranging from apicomplexan parasites to plants to mammals (41–43).
Experimental Procedures
Tetrahymena Strain Construction and Growth
Tetrahymena strains expressing tagged Teb2 or tagged Teb3 instead of the endogenous untagged protein were generated by cassette integration at the respective genomic loci using the BSR2 cassette (44). N-terminal tag fusion was chosen due to predicted protein domain structures and the loss of function imposed by fusion of the same tag to the C terminus of Tetrahymena Rpa1 (38). Cells were grown in modified Neff medium (0.25% proteose peptone, 0.25% yeast extract, 0.2% dextrose, 30 μm FeCl3) to mid-log phase (3 × 105 cells/ml). For mating, cells were starved in 10 mm Tris (pH 8.0) for 16 h and mixed in a 1:1 ratio with complementary mating type SB1969 at 2 × 105 cells/ml. To maximally synchronize mating, cells were shaken at 180 rpm for 30 min followed by a 30-min rest period three times. Conjugating cells were harvested 5 h after the final shake period. For cells synchronized in vegetative growth, starved cell cultures described above were re-fed with modified Neff medium at 3 × 105 cells/ml for 4 h.
Affinity Purification and Mass Spectrometry
Cell lysis (completed at 4 °C) and subsequent steps (completed at room temperature) used T2EG50 (20 mm Tris, pH 8.0, 2 mm EGTA, 10% glycerol, 50 mm NaCl) supplemented with 0.2% Igepal CA-630 and 2 mm dithiothreitol (DTT). After binding to IgG-agarose (Sigma) and washing in T2EG50 with 0.03% Igepal CA-630 and 2 mm DTT, bound complexes were incubated with 30 μg/ml tobacco etch virus protease for 0.5–1 h. Eluted samples were bound in batch to 10 μl of EZView Red anti-FLAG M2 resin (Sigma) per 50-ml initial extract volume in low retention tubes for 1 h. Washed resin was eluted into T2MG (20 mm Tris, pH 8.0, 2 mm MgCl2, 10% glycerol) with 1 mm DTT and 150 ng/μl of 3× FLAG peptide for 60 min. For mass spectrometry, samples were washed with T2EG50 supplemented with 2 mm DTT but no Igepal CA-630 before elution and dialyzed into the same buffer to remove the FLAG peptide.
MS peptide digests used sequencing grade trypsin (Promega). The IP2 program suite (Integrated Proteomics) was used for peptide and protein identification. The proLuCid search engine was used with the Tetrahymena thermophila predicted proteome database (ciliate.org) downloaded on June 27, 2014. The set of three samples was purified and processed for MS in parallel, with MS data shown from one biological replicate (of multiple replicates that had a similar SDS-PAGE protein profile).
Recombinant Protein Expression and Purification
Recombinant Teb1, Rpa1, Rlp1, Teb2, and Teb3 (34, 37, 38) were expressed in E. coli BL21 (DE3) using synthetic open reading frames, which is necessary due to expanded Tetrahymena codon usage. Each RPA large subunit paralog was expressed from pET28a with an N-terminal His6 tag, as described previously (38), or in combination with pCDFDuet vector expressing Teb2 with an N-terminal MBP tag and untagged Teb3. Cells were lysed by sonication for 3 min at 4 °C in T2MG50 (20 mm Tris, pH 8.0, 2 mm MgCl2, 10% glycerol, 50 mm NaCl) supplemented with 0.1% Igepal CA-630, 1 mm DTT, and 20 mm imidazole. His6-tagged proteins were isolated by purification on Ni-NTA-agarose (Qiagen) and eluted into T2MG with 2 mm DTT and 500 mm imidazole. Complexes that also contained an MBP-tagged subunit were isolated by subsequent purification on amylose resin (New England Biolabs) and eluted into T2MG with 1 mm DTT and 10 mm maltose. The complex of MBP-Teb2 and Teb3 was purified using amylose resin only.
Telomerase Activity and DNA Binding Assays
Activity assays used a standard Tetrahymena telomerase reaction buffer containing 50 mm Tris acetate (pH 8.0), 2 mm MgCl2, 10 mm spermidine, and 5 mm β-mercaptoethanol. Reactions additionally contained 24 nm [α-32P]dGTP, 300 nm dGTP, 200 μm dTTP, and 200 nm DNA primer (GT2G3)3. Reactions were allowed to proceed at room temperature for 5 min for cell-assembled telomerase or 15 min for telomerase assembled in RRL, unless indicated otherwise. A 5′-labeled oligonucleotide DNA (the recovery control) was added to telomerase products before precipitation and denaturing PAGE. Electrophoretic mobility shift assays were performed as described previously (38). Binding affinities were calculated based on free probe signal using ImageQuant software.
Telomerase Reconstitutions
Recombinant telomerase was reconstituted by RRL expression of synthetic open reading frames for TERT, p50, p75, p45, p19, Teb1, Teb2, and/or Teb3 (34). Bacterially expressed, purified p65 (51) and in vitro transcribed TER were added at 25 nm each to the TERT RRL reaction before protein synthesis to assemble the RNP catalytic core. A separate RRL reaction was performed to produce the p75-p45-p19 CST complex or p50, which here was the p50N30 domain sufficient for p50 biological function (36). RRL-expressed telomerase subunits were combined, bound to anti-FLAG M2 affinity resin (Sigma), and washed into T2MG with 2 mm DTT. Afterward, purified proteins other than p65 were added to a final concentration of 200 nm (unless indicated otherwise) and allowed to bind for 20 min at room temperature, followed by an activity assay.
Sequence Depositions
GenBankTM accession numbers for the Tetrahymena thermophila protein sequences expressed recombinantly in this work are as follows: Rpa1, ADB03555.1; Rlp1, GU384877; Teb1 (encoded by gene TAP82), EU873081; Teb2, BK009378; Teb3, BK009379. Revised Rpa1 sequence has GenBankTM accession number ADB03555.2. Rlp2 sequence has GenBankTM accession number KX987301.
Author Contributions
H. E. U. and K. C. conceived and interpreted the experiments and wrote the paper. H. E. U. performed all experiments. H. C. and J. F. shared reagents and information from parallel studies. All authors revised and approved the final version of the manuscript.
Acknowledgment
This work used the Vincent J. Coates Proteomics/Mass Spectrometry Laboratory at UC Berkeley, supported in part by National Institutes of Health Grant S10RR025622.
The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) ADB03555.1, GU384877, EU873081, BK009378, BK009379, and ADB03555.2.
- nt
- nucleotide(s)
- RPA
- replication protein A
- RNP
- ribonucleoprotein
- G-strand
- guanosine-rich telomeric DNA strand
- OB-fold
- oligonucleotide/oligosaccharide-binding fold
- C-strand
- cytidine-rich telomeric DNA strand
- RAP
- repeat addition processivity
- RRL
- rabbit reticulocyte lysate
- ZZ
- tandem protein A domains
- F
- triple FLAG peptide
- MBP
- maltose-binding protein
- CTαH
- C-terminal α-helix
- Ni-NTA
- nickel-nitrilotriacetic acid.
References
- 1. Doksani Y., and de Lange T. (2014) The role of double-strand break repair pathways at functional and dysfunctional telomeres. Cold Spring Harb. Perspect. Biol. 6, a016576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shay J. W. (2016) Role of telomeres and telomerase in aging and cancer. Cancer Discov. 6, 584–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stewart J. A., Chaiken M. F., Wang F., and Price C. M. (2012) Maintaining the end: roles of telomere proteins in end-protection, telomere replication and length regulation. Mutat. Res. 730, 12–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arnoult N., and Karlseder J. (2015) Complex interactions between the DNA-damage response and mammalian telomeres. Nat. Struct. Mol. Biol. 22, 859–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Martínez P., and Blasco M. A. (2015) Replicating through telomeres: a means to an end. Trends Biochem. Sci. 40, 504–515 [DOI] [PubMed] [Google Scholar]
- 6. Jacob N. K., Skopp R., and Price C. M. (2001) G-overhang dynamics at Tetrahymena telomeres. EMBO J. 20, 4299–4308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhao Y., Hoshiyama H., Shay J. W., and Wright W. E. (2008) Quantitative telomeric overhang determination using a double-strand specific nuclease. Nucleic Acids Res. 36, e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blackburn E. H., Greider C. W., and Szostak J. W. (2006) Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat. Med. 12, 1133–1138 [DOI] [PubMed] [Google Scholar]
- 9. Greider C. W., and Blackburn E. H. (1989) A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337, 331–337 [DOI] [PubMed] [Google Scholar]
- 10. Lingner J., Hughes T. R., Shevchenko A., Mann M., Lundblad V., and Cech T. R. (1997) Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276, 561–567 [DOI] [PubMed] [Google Scholar]
- 11. Weinrich S. L., Pruzan R., Ma L., Ouellette M., Tesmer V. M., Holt S. E., Bodnar A. G., Lichtsteiner S., Kim N. W., Trager J. B., Taylor R. D., Carlos R., Andrews W. H., Wright W. E., Shay J. W., Harley C. B., and Morin G. B. (1997) Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 17, 498–502 [DOI] [PubMed] [Google Scholar]
- 12. Podlevsky J. D., and Chen J. J. (2016) Evolutionary perspectives of telomerase RNA structure and function. RNA Biol. 13, 720–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Londoño-Vallejo J. A., and Wellinger R. J. (2012) Telomeres and telomerase dance to the rhythm of the cell cycle. Trends Biochem. Sci. 37, 391–399 [DOI] [PubMed] [Google Scholar]
- 14. MacNeil D. E., Bensoussan H. J., and Autexier C. (2016) Telomerase regulation from beginning to the end. Genes 7, E64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Egan E. D., and Collins K. (2012) Biogenesis of telomerase ribonucleoproteins. RNA 18, 1747–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nandakumar J., and Cech T. R. (2013) Finding the end: recruitment of telomerase to telomeres. Nat. Rev. Mol. Cell Biol. 14, 69–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hockemeyer D., and Collins K. (2015) Control of human telomerase action at telomeres. Nat. Struct. Mol. Biol. 22, 848–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lloyd N. R., Dickey T. H., Hom R. A., and Wuttke D. S. (2016) Tying up the ends: plasticity in the recognition of single-stranded DNA at telomeres. Biochemistry 55, 5326–5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baumann P., and Price C. (2010) Pot1 and telomere maintenance. FEBS Lett. 584, 3779–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Linger B. R., Morin G. B., and Price C. M. (2011) The Pot1a-associated proteins Tpt1 and Pat1 coordinate telomere protection and length regulation in Tetrahymena. Mol. Biol. Cell 22, 4161–4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Linger B. R., and Price C. M. (2009) Conservation of telomere protein complexes: shuffling through evolution. Crit. Rev. Biochem. Mol. Biol. 44, 434–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fanning E., Klimovich V., and Nager A. R. (2006) A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 34, 4126–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen R., and Wold M. S. (2014) Replication protein A: single-stranded DNA's first responder. Bioessays 36, 1156–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sugitani N., and Chazin W. J. (2015) Characteristics and concepts of dynamic hub proteins in DNA processing machinery from studies of RPA. Prog. Biophys. Mol. Biol. 117, 206–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fan J., and Pavletich N. P. (2012) Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev. 26, 2337–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gibb B., Ye L. F., Gergoudis S. C., Kwon Y., Niu H., Sung P., and Greene E. C. (2014) Concentration-dependent exchange of replication protein A on single-stranded DNA revealed by single-molecule imaging. PLoS One 9, e87922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rice C., and Skordalakes E. (2016) Structure and function of the telomeric CST complex. Comput. Struct. Biotechnol. J. 14, 161–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miyake Y., Nakamura M., Nabetani A., Shimamura S., Tamura M., Yonehara S., Saito M., and Ishikawa F. (2009) RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol. Cell 36, 193–206 [DOI] [PubMed] [Google Scholar]
- 29. Surovtseva Y. V., Churikov D., Boltz K. A., Song X., Lamb J. C., Warrington R., Leehy K., Heacock M., Price C. M., and Shippen D. E. (2009) Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol. Cell 36, 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bhattacharjee A., Stewart J., Chaiken M., and Price C. M. (2016) STN1 OB fold mutation alters DNA binding and affects selective aspects of CST function. PLoS Genet. 12, e1006342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen L. Y., Redon S., and Lingner J. (2012) The human CST complex is a terminator of telomerase activity. Nature 488, 540–544 [DOI] [PubMed] [Google Scholar]
- 32. Mitton-Fry R. M., Anderson E. M., Hughes T. R., Lundblad V., and Wuttke D. S. (2002) Conserved structure for single-stranded telomeric DNA recognition. Science 296, 145–147 [DOI] [PubMed] [Google Scholar]
- 33. Wan B., Tang T., Upton H., Shuai J., Zhou Y., Li S., Chen J., Brunzelle J. S., Zeng Z., Collins K., Wu J., and Lei M. (2015) The Tetrahymena telomerase p75-p45-p19 subcomplex is a unique CST complex. Nat. Struct. Mol. Biol. 22, 1023–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jiang J., Chan H., Cash D. D., Miracco E. J., Ogorzalek Loo R. R., Upton H. E., Cascio D., O'Brien Johnson R., Collins K., Loo J. A., Zhou Z. H., and Feigon J. (2015) Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions. Science 350, aab4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jiang J., Miracco E. J., Hong K., Eckert B., Chan H., Cash D. D., Min B., Zhou Z. H., Collins K., and Feigon J. (2013) The architecture of Tetrahymena telomerase holoenzyme. Nature 496, 187–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hong K., Upton H., Miracco E. J., Jiang J., Zhou Z. H., Feigon J., and Collins K. (2013) Tetrahymena telomerase holoenzyme assembly, activation, and inhibition by domains of the p50 central hub. Mol. Cell Biol. 33, 3962–3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Min B., and Collins K. (2009) An RPA-related sequence-specific DNA-binding subunit of telomerase holoenzyme is required for elongation processivity and telomere maintenance. Mol. Cell 36, 609–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Min B., and Collins K. (2010) Multiple mechanisms for elongation processivity within the reconstituted Tetrahymena telomerase holoenzyme. J. Biol. Chem. 285, 16434–16443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zeng Z., Min B., Huang J., Hong K., Yang Y., Collins K., and Lei M. (2011) Structural basis for Tetrahymena telomerase processivity factor Teb1 binding to single-stranded telomeric-repeat DNA. Proc. Natl. Acad. Sci. U.S.A. 108, 20357–20361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Upton H. E., Hong K., and Collins K. (2014) Direct single-stranded DNA binding by Teb1 mediates the recruitment of Tetrahymena thermophila telomerase to telomeres. Mol. Cell Biol. 34, 4200–4212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rider S. D. Jr., and Zhu G. (2008) Differential expression of the two distinct replication protein A subunits from Cryptosporidium parvum. J. Cell Biochem. 104, 2207–2216 [DOI] [PubMed] [Google Scholar]
- 42. Sakaguchi K., Ishibashi T., Uchiyama Y., and Iwabata K. (2009) The multi-replication protein A (RPA) system: a new perspective. FEBS J. 276, 943–963 [DOI] [PubMed] [Google Scholar]
- 43. Mason A. C., Roy R., Simmons D. T., and Wold M. S. (2010) Functions of alternative replication protein A in initiation and elongation. Biochemistry 49, 5919–5928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Couvillion M. T., and Collins K. (2012) Biochemical approaches including the design and use of strains expressing epitope-tagged proteins. Methods Cell Biol. 109, 347–355 [DOI] [PubMed] [Google Scholar]
- 45. Ruehle M. D., Orias E., and Pearson C. G. (2016) Tetrahymena as a unicellular model eukaryote: genetic and genomic tools. Genetics 203, 649–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Coyne R. S., Thiagarajan M., Jones K. M., Wortman J. R., Tallon L. J., Haas B. J., Cassidy-Hanley D. M., Wiley E. A., Smith J. J., Collins K., Lee S. R., Couvillion M. T., Liu Y., Garg J., Pearlman R. E., et al. (2008) Refined annotation and assembly of the Tetrahymena thermophila genome sequence through EST analysis, comparative genomic hybridization, and targeted gap closure. BMC Genomics 9, 562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sibenaller Z. A., Sorensen B. R., and Wold M. S. (1998) The 32- and 14-kilodalton subunits of replication protein A are responsible for species-specific interactions with single-stranded DNA. Biochemistry 37, 12496–12506 [DOI] [PubMed] [Google Scholar]
- 48. Rubtsova M. P., Skvortsov D. A., Petruseva I. O., Lavrik O. I., Spirin P. V., Prasolov V. S., Kisseljov F. L., and Dontsova O. A. (2009) Replication protein A modulates the activity of human telomerase in vitro. Biochemistry 74, 92–96 [DOI] [PubMed] [Google Scholar]
- 49. Bochkareva E., Korolev S., Lees-Miller S. P., and Bochkarev A. (2002) Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J. 21, 1855–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rider S. D. Jr, Cai X., Sullivan W. J. Jr, Smith A. T., Radke J., White M., and Zhu G. (2005) The protozoan parasite Cryptosporidium parvum possesses two functionally and evolutionarily divergent replication protein A large subunits. J. Biol. Chem. 280, 31460–31469 [DOI] [PubMed] [Google Scholar]
- 51. O'Connor C. M., and Collins K. (2006) A novel RNA binding domain in Tetrahymena telomerase p65 initiates hierarchical assembly of telomerase holoenzyme. Mol. Cell Biol. 26, 2029–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Miao W., Xiong J., Bowen J., Wang W., Liu Y., Braguinets O., Grigull J., Pearlman R. E., Orias E., and Gorovsky M. A. (2009) Microarray analyses of gene expression during the Tetrahymena thermophila life cycle. PLoS One 4, e4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xiong J., Lu X., Lu Y., Zeng H., Yuan D., Feng L., Chang Y., Bowen J., Gorovsky M., Fu C., and Miao W. (2011) Tetrahymena Gene Expression Database (TGED): a resource of microarray data and co-expression analyses for Tetrahymena. Sci. China Life Sci. 54, 65–67 [DOI] [PubMed] [Google Scholar]