Abstract
SAMHD1 is a phosphohydrolase maintaining cellular dNTP homeostasis but also acts as a critical regulator in innate immune responses due to its antiviral activity and association with autoimmune disease, leading to aberrant activation of interferon. SAMHD1 expression is differentially regulated by interferon in certain primary cells, but the underlying mechanism is not understood. Here, we report a detailed characterization of the promotor region, the 5′- and 3′-untranslated region (UTR) of SAMHD1, and the mechanism responsible for the cell type-dependent up-regulation of SAMHD1 protein by interferon. We demonstrate that induction of SAMHD1 by type I and II interferons depends on 3′-UTR post-transcriptional regulation, whereas the promoter drives basal expression levels. We reveal novel functional target sites for the microRNAs miR-181a, miR-30a, and miR-155 in the SAMHD1 3′-UTR. Furthermore, we demonstrate that down-regulation of endogenous miR-181a and miR-30a levels inversely correlates with SAMHD1 protein up-regulation upon type I and II interferon stimulation in primary human monocytes. These miRNAs are not modulated by interferon in macrophages or dendritic cells, and consequently protein levels of SAMHD1 remain unchanged. These results suggest that SAMHD1 is a non-classical interferon-stimulated gene regulated through cell type-dependent down-regulation of miR-181a and miR-30a in innate sentinel cells.
Keywords: human immunodeficiency virus (HIV), innate immunity, interferon, microRNA (miRNA), promoter, SAM domain and HD domain-containing protein 1 (SAMHD1), dNTPase, innate sentinel cells, interferon stimulated gene, regulation of expression
Introduction
Sterile α motif (SAM) and HD domain-containing protein 1 (SAMHD1)2 is a deoxynucleoside triphosphate triphosphohydrolase that catalyzes the hydrolytic reaction of dNTPs into NTPs and free triphosphate in a deoxyguanosine triphosphate (dGTP)-dependent manner (1, 2), leading to a reduction of cellular dNTP pools (3–6). SAMHD1 is involved in regulation of dNTP homeostasis (7) and affects cell cycle distribution (7, 8), cell proliferation, apoptosis (8), and genome integrity (9). More importantly, SAMHD1 plays a critical role in innate immunity and infection as a negative regulator of innate sensing (10, 11). Mutations at the SAMHD1 locus cause Aicardi-Goutières syndrome, a severe autoimmune disease leading to aberrant activation of the innate immune system (12). It was suggested that SAMHD1 prevents the accumulation and sensing of endogenous nucleic acid species that would otherwise lead to induction of interferon (IFN) (11, 13). Moreover, SAMHD1 has been identified as a potent anti-HIV-1 restriction factor in myeloid cells (14–16). During HIV-1 infection, SAMHD1 prevents sensing and activation of innate sentinel cells and consequently influences the magnitude of the adaptive responses (11, 17).
SAMHD1 is a broadly expressed protein found in almost all human tissues (18, 19). Several reports find human SAMHD1 expression to be induced upon infection with viral species (20–25) or innate stimuli (26, 27). Current knowledge suggests that SAMHD1 is induced by IFN stimulation in a cell type-dependent manner in innate sentinel cells, the critical mediators for innate sensing and co-stimulation for adaptive responses. SAMHD1 protein is induced by type I IFNα in human monocytes (16) and in various cell lines (6, 28) and by IFNα and IFNγ in microglia (29). On the contrary, SAMHD1 expression does not change upon type I IFN stimulation in immunocompetent cells, such as THP-1 cells (30, 31), and by type I or II IFN treatment of monocyte-derived macrophages (MDMs) (19, 30, 31) or monocyte-derived dendritic cells (MDDCs) (6, 31).
Recently, it was proposed that the basal level of SAMHD1 mRNA expression is negatively regulated by microRNAs (miRNAs) (32–34). miRNAs are small non-coding RNAs of 22 nucleotides (nt) in length that are part of the post-transcriptional regulatory network. They act mostly as negative regulators that recognize complementary sequences in mRNAs and induce translational repression or deadenylation of the respective mRNA (35, 36). Alternatively, extensive complementarity to the target sequence can induce an RNAi-like mechanism by which miRNAs induce cleavage of the target mRNA (35, 36). Furthermore, miRNAs have also been described to mediate repressive chromatin modifications and transcriptional gene silencing (37). Jin et al. (33) demonstrated that the miR-181 family acts on the SAMHD1 3′-UTR in reporter assays, most potently miR-181a. Furthermore, it has been shown in astrocytes that miRNA mimics or inhibitors of miR-181a and miR-155 influence SAMHD1 basal expression levels (34). An earlier report on a screen for targets of the Kaposi's sarcoma-associated herpesvirus-encoded miRNA miR-K12-11 revealed that both miR-K12-11 and its cellular homologue miR-155 negatively regulate SAMHD1 basal expression in reporter assays (32). In these reports, however, only one functional target site of miR-181 has been mapped. miRNAs target the 5′-UTR, coding DNA sequences (38, 39), or the 3′-UTR (36), with the latter being the most frequent and effective target site. miRNAs are thought to control expression of up to 60% of all mammalian genes (40). It is known that expression patterns of miRNAs are cell type/tissue-dependent (41, 42). Moreover, IFNs have been shown to modulate the expression levels of miRNAs, suggesting a significant potential for post-transcriptional regulation by miRNAs in the innate immune response (43–46). A recent study suggests that miR-181a levels and SAMHD1 mRNA levels are inversely correlated in microglia by IFNα and -γ (29). Nonetheless, a comprehensive investigation of the mechanisms of SAMHD1 regulation upon interferon stimulation, specifically the regulatory elements, specific miRNA species involved, and their exact target sites, has not yet been undertaken. Moreover, the regulatory mechanisms affecting differential SAMHD1 expression in sentinel cells of the innate immune response are still unknown.
Here, we show, for the first time, a detailed analysis of the promoter, 5′-UTR, and 3′-UTR of SAMHD1 and provide new insights into the underlying mechanism of IFN-mediated stimulation of SAMHD1 expression in innate sentinel cells. We demonstrate that the SAMHD1 3′-UTR is responsible for the IFN induction of SAMHD1, whereas the promoter drives basal expression levels. The SAMHD1 locus reveals a dispersed pattern of transcription initiation, common for broadly expressed proteins, as reported for SAMHD1. We define novel active target sites for miR-181a, miR-30a, and miR-155 in the SAMHD1 3′-UTR. The cell type-dependent IFN stimulation of SAMHD1 is mediated by modulation of levels of miR-181a and miR-30a, but not miR-155. In conclusion, we demonstrate an inverse correlation of both miRNA levels and protein levels of SAMHD1 in primary human monocytes upon IFN stimulation, whereas both miRNA and protein levels stay unchanged in monocyte-derived macrophages or dendritic cells.
Results
Type I and II Interferon Stimulation Increases SAMHD1 Protein Expression in Sentinel Cells
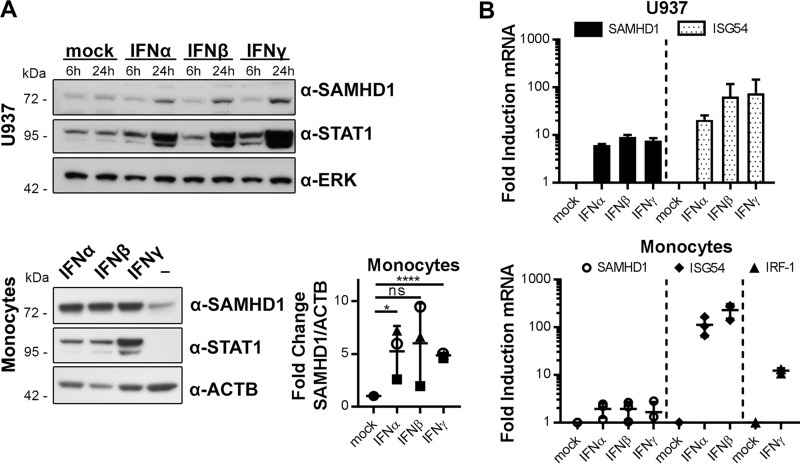
Current data suggest that SAMHD1 is an interferon-stimulated gene (ISG) in certain cell types. We and others described up-regulation of SAMHD1 protein level by IFNα in monocytes (16) and monocytic cell lines (30), by IFNα and IFNγ in microglia (29), and by IFNα and IFNβ in various cell lines (6). To corroborate these findings, we comprehensively analyzed the up-regulation of SAMHD1 protein and mRNA levels upon stimulation with type I and II IFNs in sentinel cells. In the monocytic cell line U937 as well as in primary human monocytes, both type I (IFNα and IFNβ) and type II IFNγ led to an increase in SAMHD1 protein (Fig. 1A) and mRNA levels (Fig. 1B) after 24 h. Of note, induction of SAMHD1 mRNA was less pronounced than mRNA levels of the known ISGs ISG54 or IRF1. Specifically, in primary human monocytes, we observed SAMHD1 mRNA induction of about 2-fold by any of the IFNs (Fig. 1B) accompanied by protein up-regulation of 2–9-fold (Fig. 1A), depending on the donor and the type of IFN.
FIGURE 1.
Interferons stimulate SAMHD1 expression. A, monocytic U937 cells and primary monocytes were treated with 1000 units/ml IFNα, IFNβ, IFNγ for 6 and 24 h (U937, top) and for 24 h (monocytes, bottom left). Protein samples were subjected to immunoblotting analysis and probed for SAMHD1, STAT1 to monitor successful stimulation, and ERK or ACTB as a loading control. Blots of three independent donors were quantified and corrected for ACTB signal and normalized to unstimulated controls (means ± S.D., bottom right). Immunoblots are representative of three independent experiments (n = 3). B, SAMHD1, ISG54, and IRF1 mRNA levels were quantified by real-time quantitative RT-PCR in U937 cells (top) and primary monocytes (bottom) after treatment with 1000 units/ml IFN for 24 h. Top, -fold changes of mRNA levels to the untreated samples were calculated for each individual experiment based on the mean of three technical replicates. The means ± S.D. of the -fold changes of three independent experiments are depicted (n = 3). Bottom, -fold changes of mRNA levels to the untreated samples were calculated for each individual donor. For two donors, the mean -fold change was based on one biological replicate each, measured in technical triplicates. For one donor, the mean -fold change was based on two biological replicates, each measured in two technical replicates. The means ± S.D. (error bars) of the -fold changes of three donors are depicted (n = 3). ns, p > 0.05; *, p ≤ 0.05; ****, p ≤ 0.0001.
Characterization of the SAMHD1 Promoter and the 5′- and 3′-Untranslated Regions
To evaluate the possible regulatory mechanisms of SAMHD1 expression, we performed a detailed characterization of the promotor region and the 5′- and 3′-UTR of SAMHD1. IFN stimulation could lead to activation of transcription factors (TFs) specifically targeting the SAMHD1 promoter, leading to an enhanced transcriptional activity. Additionally, there is the possibility of post-transcriptional regulation of the SAMHD1 mRNA upon IFN stimulation. Because one major post-transcriptional mechanism is the transcript regulation by IFN-controlled miRNAs (43–46), we were interested in analyzing the SAMHD1 sequence for potential miRNA target sites.
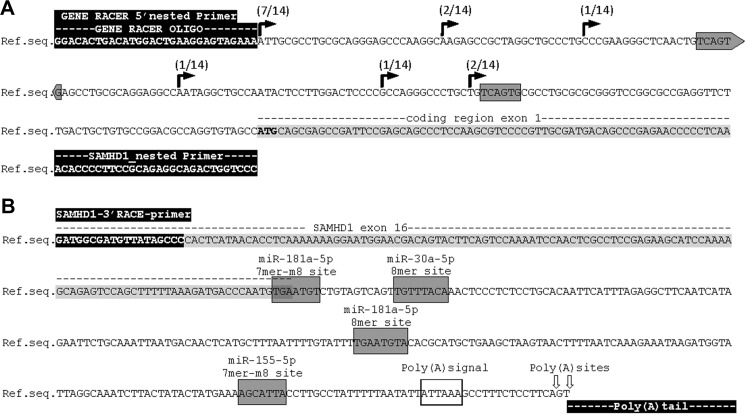
First, we characterized in detail the 5′-UTR of SAMHD1 using rapid amplification of cDNA ends (RACE) on mRNA from the monocytic cell lines THP-1 and U937 (Fig. 2A). Six transcriptional start sites (TSSs) were detected at positions 69, 82, 112, 152, 173, and 200 nt upstream of the coding region. One of these TSSs has been reported previously (18). The TSS at position 200 was present in 7 of 14 clones and matches the putative TSS indicated by the NCBI sequence deposit NM_015474.3. We conclude that the SAMHD1 locus displays a dispersed pattern of transcription initiation, which is common among TATA-less promoters containing CpG islands and typical for type II promoters of genes that are broadly expressed (47), in concordance with the expression pattern of SAMHD1 in the majority of human tissues (19).
FIGURE 2.
Determination of SAMHD1 transcription start site by 5′-RACE and SAMHD1 3′-UTR by 3′-RACE. A, determination of SAMHD1 transcription start site by 5′-RACE was performed on mRNA from THP-1 and U937 cells. TSS-derived sequences of 5′-RACE of 10 THP-1s and 4 U937s are indicated on the genomic SAMHD1 database sequence NC_000020.11, reference GRCh38.p2 primary assembly. Occurrence of the TSSs in the analyzed data sets is indicated. The SAMHD1 5′-UTR has a length of 69, 82, 112, 152, 173, and 200 nt. GENE RACER 5′ nested primer and SAMHD1 exon 1 primer (SAMHD1_nested primer) are highlighted in black. The coding region of SAMHD1 exon 1 is highlighted in light gray, and the ATG start codon is displayed in boldface letters. Gray boxes mark putative non-canonical offset 6-mer seed regions of miR-181a-3p, as found by seed sequence comparison. B, SAMHD1 3′-UTR was determined by 3′-RACE on mRNA from THP-1 and U937 cells. The SAMHD1 3′-UTR has a total length of 239 nt; in half of the captured sequences, the poly(A)-tail is initiated 2 nt farther downstream, resulting in a 241-nt-long 3′-UTR sequence, as indicated by arrows on the genomic SAMHD1 reference sequence (Ref.seq.) as given in NC_000020.11 reference GRCh38.p2 primary assembly. 3′-RACE sequencing results of nine clones from THP-1 cells and one from U937 cells were analyzed. The SAMHD1 3′-RACE primer is indicated and highlighted in black. The nucleotide sequence belonging to SAMHD1 exon 16 is highlighted in gray. Gray boxes mark canonical seed regions of miR-181a-5p, miR-155–5p, and miR-30a-5p, as found by miRNA target predictions of Miranda, miRcode, TargetScan version 7.1, and seed sequence comparison, and the type of target site is indicated. Poly(A) signal is marked by an open box, poly(A)sites are indicated by arrows, and the poly(A) tail is highlighted in black at the end of the sequence.
This finding is substantiated by in silico analysis of the SAMHD1 promoter sequence ranging from 2000 nt upstream to 500 nt downstream relative to the TSS at position 200. The in silico profile revealed the lack of a TATA-box and canonical core promoter regulatory elements but shows high GC content in the presumable SAMHD1 promoter sequence, including a CpG island and GC-boxes (Table 1). In silico transcription factor binding site prediction using the SAMHD1 promoter and 5′-UTR sequence yielded several putative sites specific for interferon signaling, such as STAT1, IRF1, and GAS consensus sequences as well as binding sites for general transcription factor SP1 (Table 1).
TABLE 1.
Promoter element prediction −2000 to +500 nt
Shown are prediction and analysis of promoter elements in the −2000 to +500 nt region relative to the TSS 200 nt upstream of the SAMHD1 start codon in NCBI deposit NM_015474.3. Only hits with high stringency (GPMiner: core score ≥ 0.999 and matrix score ≥ 0.95) are listed.
| Software | Predicted elements with nucleotide positions |
|---|---|
| EPD | GC-boxes at −89 to −77, −67 to −53 |
| Alibaba2.1 | SP1 binding sites at +155 to +164, +145 to +158, +124 to +133, +113 to +124, +79 to +88, +43 to +52, +10 to +19 |
| CpGPlot | CpG island at −232 to +17 |
| GPMiner | GC-boxes at −194 to −186, −31 to −26, +158 to +163, +431 to +436, +439 to +444, +451 to −456 |
| CpG island at −351 to +500 | |
| STAT1 binding sites at −1854 to −1847, −1586 to −1579, −1522 to −1515, −1240 to −1233, −1116 to −1109, −1079 to −1072, −987 to −980, −789 to −782, −607 to −600, −296 to −289, −233 to −226 | |
| IRF1 binding sites at −1495 to −1489, −568 to −560, −478 to −470, −280 to −272 | |
| VectorNTI | GAS consensus sequences at −976 to −984, −233 to −225 |
We next sought to define the exact length of the SAMHD1 3′-UTR by 3′-RACE on mRNA from THP-1 and U937 cells. The 3′-RACE revealed a polyadenylation signal at 219 nt downstream of the coding sequence that initiated poly(A) sites at position 239 or 241 nt (Fig. 2B) in concordance with the reference sequence (NC_000020.11) and data from dendritic cells (18), including a close variant of consensus poly(A) signal (Fig. 2B) that commonly initiate polyadenylation in ∼17% of human genes (48, 49). Predictions of miRNA target sites in the 3′-UTR were performed using the miRanda algorithm (50) and mirSVR regression model (51), TargetScan (52), and miRcode (53). We predicted two putative target sites of the miR-181 family: one 8-mer site, reported previously (33), and one novel putative 7-mer-m8 site (Fig. 2B and Table 2). Interestingly, the 7-mer-m8 miR-181 target site is located at the transition of the coding region to the 3′-UTR and has not been described before. Furthermore, we detected one novel putative 7-mer-m8 target site for miR-155 and one novel putative 8-mer target site for the miR-30 family (Fig. 2B and Table 2), concluding that the 3′-UTR harbors multiple miRNA seed sequences that may mediate translational repression. Moreover, we analyzed the SAMHD1 5′-UTR using miRcode or direct seed sequence comparison. By sequence comparison, we found that the 5′-UTR contains two putative non-canonical offset 6-mer seed regions for the 3p strand of miR-181a (Fig. 2A). The miR-181-3p hits were not reported by miRcode because this algorithm neglects 6-mer hits. We did not detect additional target sites for either the 3p or 5p strand of miR-155 or miR-30a within the SAMHD1 5′-UTR.
TABLE 2.
Prediction and analysis of putative miRNA binding sites Miranda (via microrna.org) and miRcode target predictions were used to identify putative miRNA target sites on the SAMHD1 mRNA (NM_015474)
Search criteria were stringent, listing only hits of conserved miRNAs. Only putative hits by Miranda with a mirSVR score of ≤−0.5, a PhastCons conservation score of ≥0.5, and at least 6-mer seed sequence complementarity are reported. MiRcode was used to search for sites with scores ≥200. Given target sites are according to Ref. 31.
| MicroRNA | Target site | Position in 3′-UTR | Miranda (mirSVR score) | miRcode (score) |
|---|---|---|---|---|
| miR-181a/b/c/d | 7-mer-m8 | −3 to 4 | 356 | |
| miR-181a/b/c/d | 8-mer | 110 to 117 | −1, 2448 | 444 |
| miR-30a/b/c/d/w | 8-mer | 16 to 23 | Ø −1, 3042 | 267 |
| miR-155 | 7-mer-m8 | 193 to 199 | −1, 0451 | 255 |
The Promoter Region and the 5′-UTR Are Not Responsive to Interferon Stimulation
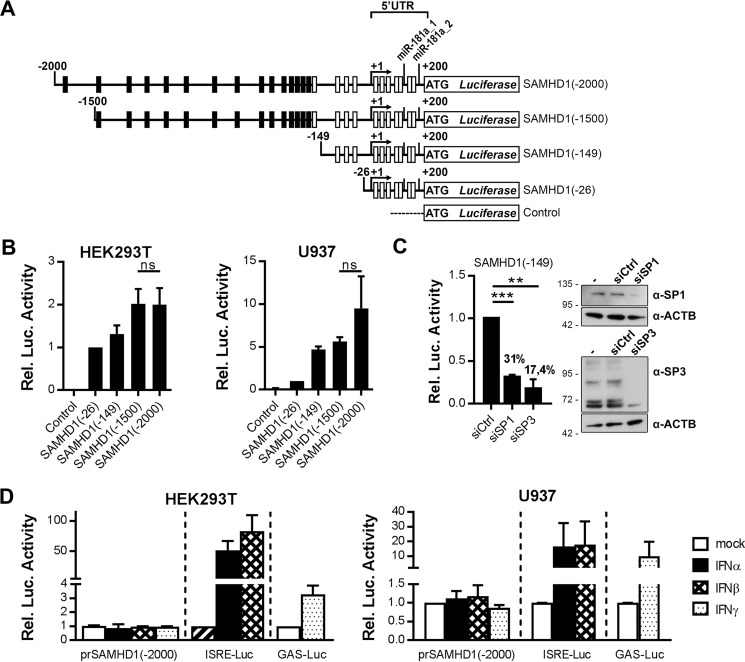
To assess the transcriptional regulation of the promoter and the 5′-UTR of SAMHD1 in more detail, we generated luciferase reporter constructs encompassing the 200 nt 5′-UTR harboring the putative miR-181-3p target sites and additionally up to 2000 nt upstream (Fig. 3A). Constructs encompassing shorter upstream sequences of 26 or 149 nt were generated to include either all TSSs or most putative GC-boxes without putative IFN-responsive elements, respectively (Fig. 3A). We chose a CpG-free plasmid system to avoid inadvertent activation of the innate immune response upon transfection of the plasmids. The reporter constructs exhibited basal transcriptional activity, which reached saturated levels at 1500 nt upstream sequence length (Fig. 3B). The core promoter encompassed by the SAMHD1(−149) reporter construct was regulated by general transcription factor SP1 or SP3 (Fig. 3C). In contrast to successful stimulation of control constructs harboring the type I IFN-responsive ISRE promoter element or the IFNγ-responsive element GAS, there was no substantial up-regulation of SAMHD1 promoter transcriptional activity detected either in HEK293T cells or in U937 cells (Fig. 3D). A range of shorter reporter constructs were also tested to exclude activity of negative regulatory elements in the 2000-nt promoter sequence (data not shown). None of the tested shorter constructs demonstrated changes in transcriptional activity upon IFN stimulation, indicating that the SAMHD1 promoter is not affected by IFN-induced transcription factors; nor do potentially IFN-modulated miRNAs influence the 5′-UTR of SAMHD1.
FIGURE 3.
The promoter and the 5′-UTR of SAMHD1 drive basal expression but are not responsive to IFNs. A, schematic representation of SAMHD1 promoter luciferase reporter constructs, including the SAMHD1 5′-UTR and 26-, 149-, 1500-, or 2000-nt upstream sequence. Numbers indicate the nucleotide position relative to the TSS 200 nt upstream of the start codon. White boxes indicate positions of putative GC-box elements and SP1/SP3 binding sites, and black boxes indicate positions of putative interferon signaling-related transcription factors (Table 1), as predicted by EPD, GPMiner, and AliBaba2.1. Putative non-canonical 6-mer offset seed regions of miR-181a-3p, as found by seed sequence comparison, are indicated. B, SAMHD1 promoter reporter constructs were co-transfected with a Renilla luciferase as internal control into HEK293T or in U937, and the control reporter vector containing no promoter sequence served as negative control. The transcriptional activity of the SAMHD1 promoter elements was assessed by measuring the luciferase activities 48 h post-transfection. Luciferase activity was corrected for Renilla luciferase activity, and -fold changes to SAMHD1(−26) were calculated based on the mean of at least two technical replicates for each individual experiment. Shown are means ± S.D. (error bars) of the -fold changes of three independent experiments (n = 3). C, HeLa cells were transfected with well established siRNAs targeting SP1 (siSP1) or SP3 (siSP3) or two non-targeting control siRNAs (siCtrl_1 and siCtrl_2; values are combined to siCtrl). 48 h after transfection of siRNAs, protein samples were taken (right), and the SAMHD1(−149) reporter was transfected (left). The transcriptional activity of the SAMHD1 promoter element was assessed by measuring the luciferase activity 24 h post-transfection. Luciferase activity was corrected for protein content, and -fold changes to the mean of siCtrl samples (siCtrl_1 and siCtrl_2) were calculated based on the mean of three technical replicates for each individual experiment. Shown are means ± S.D. of the -fold changes from two independent experiments (n = 2). Percent values indicate the residual luciferase activity compared with the siCtrl samples (left). Protein samples with no transfected siRNA (−), the non-targeting siRNA siCtrl_1 (siCtrl) or siRNAs targeting SP1 (siSP1) or SP3 (siSP3) were subjected to immunoblotting analysis and probed for SP1 or SP3 and ACTB to visualize knockdown efficiency (n = 1) (right). D, SAMHD1-promoter reporter construct SAMHD1(−2000) containing 200-nt 5′-UTR and 2000-nt upstream sequence or ISRE/GAS elements containing control plasmids were co-transfected with an internal Renilla control in HEK293T cells or in U937 cells. IFNs were applied 24 h post-transfection at concentrations of 1000 units/ml. Luciferase activities were assessed 24 h post-stimulation. Luciferase activity was corrected for Renilla luciferase activity, and -fold changes to the untreated samples were calculated based on the mean of three technical replicates for each individual experiment. The means ± S.D. of the -fold change of three independent experiments are depicted (n = 3). ns, p > 0.05; **, p ≤ 0.01; ***, p ≤ 0.001.
Interferon-induced SAMHD1 Expression Depends on the 3′-UTR
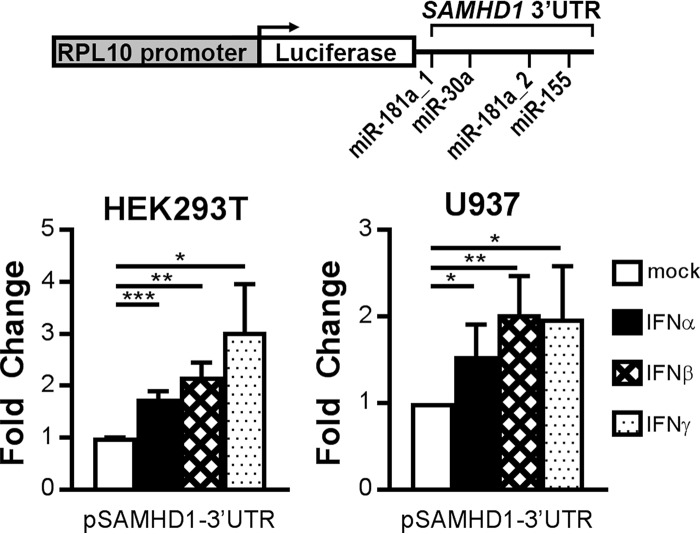
Following promoter and 5′-UTR analysis, we also investigated the influence of IFN stimulation on the 3′-UTR of SAMHD1. To this end, we used a reporter construct harboring the SAMHD1 3′-UTR, including the putative indicated miRNA seed sequences, fused to a constitutively expressed luciferase gene (Fig. 4, top). Upon transfection into HEK293T or U937 cells and stimulation with type I or II IFNs, luciferase activity was evaluated (Fig. 4). Interestingly, IFNα, -β, and -γ stimulation led to significant up-regulation of the luciferase activity of the reporter construct in both cell types (Fig. 4, bottom). These results demonstrate that the 3′-UTR contains regulatory elements that are targets of type I and II IFN stimulation.
FIGURE 4.
Interferon-responsive elements are located in the SAMHD1 3′-UTR. Shown is a schematic representation of the SAMHD1-3′-UTR reporter plasmid containing luciferase; SAMHD1-3′-UTR lies under control of the constitutive RPL10 promoter (SAMHD1-3′-UTR). The indicated putative miRNA target sites are seed regions of miR-181a-5p, miR-155–5p, and miR-30a-5p (top). The reporter was transfected in HEK293T (bottom left) or U937 cells (bottom right), and interferons were applied 24 h post-transfection at 1000 or 2000 units/ml in U937 and HEK293T cells, respectively. 24 h post-stimulation, luciferase activities were measured. -Fold changes to the untreated samples were calculated based on the mean of three technical replicates for each individual experiment. Depicted are means ± S.D. (error bars) of the -fold change of three independent experiments for type I IFN stimulation (n = 3) and of two independent experiments for type II IFN stimulation (n = 2) in HEK293T cells and of three independent experiments in U937 cells (n = 3). *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001.
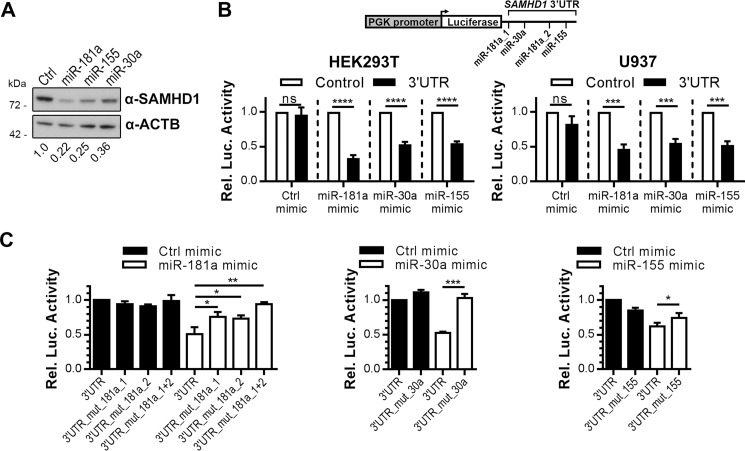
miRNAs Negatively Regulate SAMHD1 Expression by Targeting the 3′-UTR
We next asked whether overexpression of specific miRNAs (mimics of the miRNAs) with predicted seed sequences in the 3′-UTR affected SAMHD1 expression. First, we analyzed the influence of miRNA mimics on endogenous levels of SAMHD1 in HEK293T cells. miRNA mimics of miR-181a, miR-155, and miR-30a led to down-regulation of SAMHD1 protein expression in HEK293T cells in comparison with transfection of a control miRNA mimic (Fig. 5A). Interestingly, miR-181a mimic was the most potent mimic compared with miR-155 and miR-30a mimics (Fig. 5A). To prove that the down-regulation of SAMHD1 protein expression by miRNA mimics is mediated by the 3′-UTR of SAMHD1, the miRNA mimics were co-transfected together with the 3′-UTR luciferase reporter (black) or a control reporter construct (white) into HEK293T or U937 cells (Fig. 5B). The SAMHD1 3′-UTR luciferase reporter plasmid activity was considerably inhibited by miR-181a, miR-30a, and miR-155 mimics compared with the controls in both cell lines (Fig. 5B). Inhibition by miR-181a mimic was more pronounced compared with the other mimics (Fig. 5B), in concordance with Fig. 5A. We sought to verify the direct interaction and map the miRNA target sites inside the 3′-UTR. Therefore, we mutated the putative binding sites of the miRNAs in the 3′-UTR of the reporter construct one-by-one and tested the mutated constructs in comparison with the wild type side by side with the respective miRNA mimics and negative controls (Fig. 5C). The impact of miR-181a mimics on the reporter activity was successfully rescued by mutation of either miR-181 site to a similar extent. Mutation of both miR-181 sites together abolished the negative effect of miR-181a mimic and restored luciferase activity to that of control mimics. Intriguingly, mutation of the miR-30a or the miR-155 seed region completely abrogated the negative regulatory impact of the respective miRNA mimics and rescued the luciferase activity (Fig. 5C). miR-155 mimic, however, displayed only a small effect on the wild-type 3′-UTR but was still significantly rescued when the putative target site was mutated. In summary, these data indicate that the tested miRNAs target novel functional sites in the 3′-UTR directly and regulate SAMHD1 expression, whereby miR-181a and miR-30a display higher efficacy than miR-155.
FIGURE 5.
miR-181a, miR-30a, and miR-155 specifically target the SAMHD1 3′-UTR. A, HEK293T cells were transfected with mimics of miR-181a, miR-155, or miR-30a or negative control mimics (Ctrl). Cells were harvested 48 h post-transfection and analyzed by immunoblotting for protein levels of SAMHD1 and ACTB as loading control. The immunoblot signals were densitometrically quantified, and the ratios of SAMHD1/ACTB normalized to the ratio of transfected Ctrl mimics are displayed. The presented immunoblots are representative of two independent experiments (n = 2). B, the SAMHD1-3′-UTR reporter plasmid (black bars) or a control reporter plasmid (white bars) expressed under control of the human phosphoglycerate kinase (PGK) promoter (scheme depicted in top panel) were co-transfected with mimics of miR-181a, miR-155, or miR-30a or negative control mimics in HEK293T (left) and U937 cells (right). Luciferase activities were quantified 48 h post-transfection. Luciferase activities were corrected for Renilla luciferase activity, and -fold changes to the control reporter treated with the corresponding miRNA mimics were calculated based on the mean of three technical replicates for each individual experiment. Shown are means ± S.D. (error bars) of the -fold changes of three independent experiments (n = 3). C, 3′-UTR reporter plasmid carrying the complete SAMHD1 3′-UTR (3′UTR) or the SAMHD1 3′-UTR with mutations in either the first miR-181a seed regions (3′UTR mut181a_1) or second (3′UTR mut181a_2) or both (3′UTR mut181a_1+2) or with the miR-30-a seed region mutated (3′UTR_mut_30a) or with the miR-155-a seed region mutated (3′UTR_mut_155) were transfected in HEK293T cells together with control mimic (Ctrl mimic) or mimics of miR-181a (left), miR-30a (middle), or miR-155 (right). Luciferase activities were quantified 48 h post-transfection. Luciferase activities were corrected for Renilla luciferase activity, and -fold changes to the 3′-UTR reporter treated with control mimic were calculated based on the mean of at least three technical replicates for each individual experiment. Depicted are the means ± S.D. of the -fold changes of three independent experiments for the left and middle panel (n = 3) and of four independent experiments for the right panel (n = 4). ns, p > 0.05; *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001.
IFN Induces Down-regulation of miR-181a and miR-30a in Monocytes
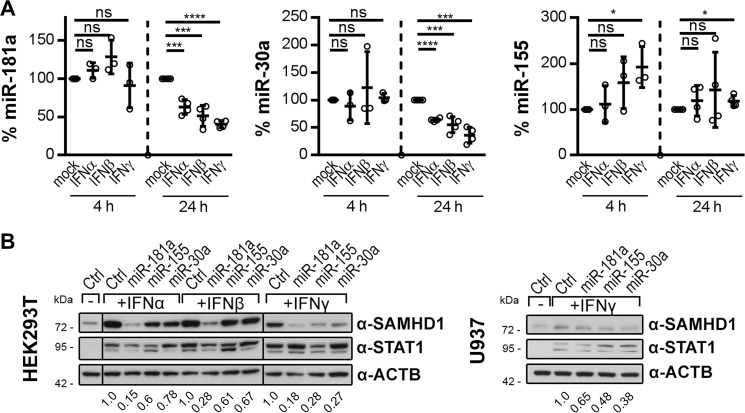
Because the 3′-UTR of SAMHD1 is sensitive to IFN stimulation (Fig. 4) and a target of miR-181a, miR-155, and miR-30a (Fig. 5), we asked whether the miRNAs themselves are subject to IFN regulation. Therefore, we assessed whether IFN stimulation would lead to down-regulation of these miRNAs. We analyzed the endogenous levels of miR-181a, miR-30a, and miR-155 in primary monocytes after treatment with both type I and II IFNs (Fig. 6A). Interestingly, miR-181a and miR-30a levels were significantly and consistently reduced at 24 h after stimulation with any of the IFNs (Fig. 6A, two left panels). The strongest down-regulation of miR-181a and miR-30a by up to 60% was observed upon IFNγ stimulation. At an earlier time point, at 4 h after stimulation, we observed some donor and stimulus variability but did not observe any significant changes in miRNA-181a or miR-30a levels (Fig. 6A, two left panels). By contrast, miR-155 was not down-regulated upon IFN stimulation, which is in concordance with the previous finding that miR-155 displayed the smallest effect on the 3′-UTR of SAMHD1 (Fig. 5C). In fact, for miR-155, we observed no significant change upon type I IFN stimulation, only upon IFNγ stimulation. Importantly, at time points of marked miR-181a and miR-30a down-modulation, miR-155 was only slightly up-regulated (Fig. 6A, right). These data suggest that IFNs induce down-regulation of specific endogenous miRNAs, leading to up-regulation of SAMHD1.
FIGURE 6.
miR-181a and miR-30a are down-regulated by IFNs in monocytes and prevent IFN-induced up-regulation of SAMHD1 protein. A, primary human monocytes were stimulated with 1000 units/ml IFNα, IFNβ, or IFNγ for 4 and 24 h, and levels of endogenous miR-181a (left), miR-30a (middle), miR-155 (right) and small nuclear RNA controls RNU6B or snRNAU6 were quantified. Data were corrected for RNU6B. miR-181a quantification of one donor was corrected for snRNAU6. For every individual donor, the -fold changes to the untreated samples of the respective time point were calculated based on the mean of two biological replicates per donor, each measured in three technical replicates. The means ± S.D. (error bars) of the -fold changes of three donors for the 4 h (n = 3) and four donors for the 24 h (n = 4) time points are depicted. B, mimics of miR-181a, miR-155, or miR-30a or negative control mimics (Ctrl) were delivered into HEK293T (left) and U937 cells (right). The cells were stimulated with 1000 units/ml IFNα, IFNβ, or IFNγ 3 h post-transfection or were left mock-treated. Cells were harvested 48 h post-transfection and analyzed by immunoblotting for protein levels of SAMHD1, STAT1 to monitor successful stimulation, and ACTB as loading control. The immunoblot signals were densitometrically quantified, and the ratios of SAMHD1/ACTB normalized to the respective ratio of transfected Ctrl mimics are displayed. Dividing lines indicate splicing of the original image. The presented immunoblots are representative of two independent experiments (n = 2). ns, p > 0.05; *, p ≤ 0.05; ***, p ≤ 0.001; ****, p ≤ 0.0001.
Next, we were interested to see whether supply of these miRNAs might abolish the IFN-induced up-regulation of SAMHD1 expression. Mimics of miR-181a, miR-155, and miR-30a were delivered into HEK293T or U937 cells, followed by stimulation with IFNα, -β, or -γ. Any of the transfected miRNA mimics were able to at least partially inhibit the type I or II IFN-induced SAMHD1 protein up-regulation in HEK293T cells (Fig. 6B, left). Interestingly, miR-181a mimics displayed the highest capacity to overcome the IFN-induced increase in SAMHD1 protein level in HEK293T cells (Fig. 6B, left). In the presence of IFNγ, all three miRNA mimics were able to abrogate the IFN effect in HEK293T cells (Fig. 6B, left). In U937 cells, the effect of miRNA mimics on overcoming IFN-induced increase in SAMHD1 protein level is less pronounced (Fig. 6B, right), very likely attributable to lower efficiency of miRNA delivery in this cell type. In summary, mimics of miR-181a, miR-155, and miR-30a abrogated either partly or completely the IFN-induced up-regulation of SAMHD1 (Fig. 6B).
miR-181a and miR-30a Levels Are Not Modulated upon IFN Stimulation in MDMs and MDDCs, Resulting in Unchanged Expression of SAMHD1
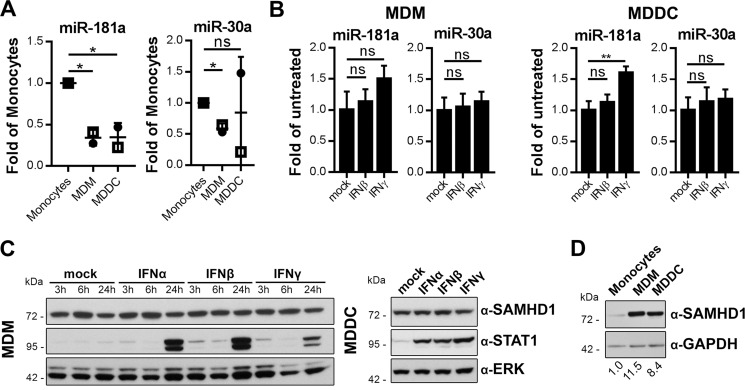
Recent data suggest that SAMHD1 is not IFN-inducible in all primary innate cells. SAMHD1 protein levels are unaffected by IFNα in MDMs and MDDCs (6, 30, 31) and additionally by IFNβ or -γ in MDDCs (6). We asked whether primary cells, such as MDMs and MDDCs, that do not display a regulation of SAMHD1 expression by IFNs might differ from monocytes in their ability to modulate miRNA expression levels. We focused on miR-181a and miR-30a because these were found to be more active on the SAMHD1 3′-UTR than miR-155 (Fig. 5C). Furthermore, miR-181a and miR-30a levels were down-regulated after IFN stimulation in monocytes in contrast to miR-155 (Fig. 6A). We found that the basal levels of miR-181a in MDMs and MDDCs were 41 and 34% of the levels in monocytes, respectively, whereas the levels of miR-30a in MDMs were 65% of those in monocytes and display donor variability in MDDCs (Fig. 7A). Stimulation of MDMs and MDDCs with IFNβ or -γ did not further reduce these levels (Fig. 7B), which is consistent with the finding that SAMHD1 protein levels are not up-regulated by type I or II IFNs in MDM (30, 31) or in MDDCs (6, 31) (Fig. 7C). We were interested in whether the observed differential expression of miR-181a and miR-30a in monocytes compared with MDMs and MDDCs is reflected in the SAMHD1 protein expression levels in these cell types. Indeed, monocytes express less SAMHD1 protein compared with MDMs or MDDCs (Fig. 7D) (54). In conclusion, these data indicate that cell type-dependent induction of SAMHD1 by IFNs correlated with expression levels of miRNAs.
FIGURE 7.
IFNs do not down-regulate miR-181a or miR-30a in MDMs or MDDCs and do not up-regulate SAMHD1 protein levels. A, endogenous levels of miR-181a (left), miR-30a (right), and RNU6B were quantified in monocytes, MDMs, and MDDCs. Data were corrected for RNU6B. -Fold changes in MDMs and MDDCs to monocytes were calculated for each individual donor based on the mean of three technical replicates. The means ± S.D. (error bars) of the -fold changes of two donors are depicted (n = 2). B, MDMs (left) and MDDCs (right) were stimulated with 1000 units/ml IFNβ or IFNγ for 24 h, and relative levels of endogenous miR-181a, miR-30a, and RNU6B as internal control were quantified. Data were corrected for RNU6B. -Fold changes to unstimulated samples were calculated based on the mean of three technical replicates. Shown are the mean -fold changes ± S.D. of one representative result of three donors (n = 3). C, MDMs (left) and MDDCs (right) were stimulated with 1000 units/ml IFNα, IFNβ, or IFNγ or left mock-treated. Cells were harvested at the indicated time points and analyzed by immunoblotting for protein levels of SAMHD1, STAT1 to monitor successful stimulation, and ERK as loading control. The presented immunoblots are representative of three independent experiments (n = 3). D, monocytes, MDMs, and MDDCs generated from the same donor were cultured for 24 h and analyzed by immunoblotting for protein levels of SAMHD1 and GAPDH as loading control. The immunoblot signals were densitometrically quantified, and the ratios of SAMHD1/GAPDH were normalized to that of monocytes. The presented immunoblot is representative of two independent experiments (n = 2). ns, p > 0.05; *, p ≤ 0.05; **, p ≤ 0.01.
Discussion
This study provides comprehensive data on the miRNAs involved in the IFN-induced up-regulation of SAMHD1. Our results support a model where SAMHD1 is a non-classical ISG regulated on a post-transcriptional level in a cell type-dependent manner. Similarly to SAMHD1, other well known ISGs have been reported to be regulated by non-classical mechanisms and not on the transcriptional level. For instance, up-regulation of BST2/tetherin by an IFNα-induced lncRNA and down-regulation of CDK6 by the IFNγ-induced miR-29 have been demonstrated (55–57).
In accordance with the described ubiquitous expression pattern of SAMHD1 in most human tissues (18, 19), the human SAMHD1 promoter is disperse (Fig. 2A) and lacks canonical core promoter elements (Table 1), reflective of ubiquitously expressed genes (58–60), and its transcriptional activity is mediated by basal TFs SP1/SP3 (Fig. 3C).
Although in silico analysis of the SAMHD1 5′ sequence suggested IFN responsiveness of this promoter (Table 1) (24) we could not detect any substantial IFN-induced up-regulation using CpG-free luciferase reporter constructs covering the 5′-UTR and up to 2000-nt (Fig. 3A) upstream sequence in HEK293T or U937 cells (Fig. 3D), suggesting that neither 5′-UTR nor the upstream genomic sequences of SAMHD1 are mediating the IFN stimulation of SAMHD1. Recently, Yang et al. (25) provided data indicating that IRF3 might be involved in mediating IFNα-induced SAMHD1 protein up-regulation. We cannot formally exclude the possibility that the promoter does react to IFN stimulation in an endogenous context, as suggested by Yang et al. (25), or, for example, through distant enhancers/silencers that may be located up to millions of nt (61) remote from the SAMHD1 promoter but may still have an influence in vivo. This is in concordance with our finding that endogenous SAMHD1 protein up-regulation was accompanied by mRNA induction, although untypically low for a promoter-driven ISG (Fig. 1, A and B). Nevertheless, we are confident that core and proximal SAMHD1 promoter are not IFN-responsive. Although in silico analysis yields potential IFN-related TF binding sites (Table 1), their influence could not be functionally validated in our luciferase reporter assays, indicating a high rate of false positive predictions. Furthermore, we would like to point out that no IFN-related TFs have been associated in experimental data sets curated in the Open Regulatory Annotation (ORegAnno) project in the 10-kb sequence upstream of SAMHD1 TSS beyond the sequence analyzed in our promoter reporters (62–64).
We could clearly demonstrate a post-transcriptional regulation of the SAMHD1 transcript mediated by its 3′-UTR upon type I and II IFN stimulation (Fig. 4). miRNAs target 3′-UTRs of mRNAs in particular and primarily induce halt of translation, inhibiting successful protein expression, but may also affect mRNA levels. miRNA-dependent regulation results in highly physiologically relevant fine tuning of gene expression in a rheostat-like manner (36, 65). Importantly, the vast majority of miRNA-mediated gene regulation commonly results in modulation of about 1.5–2-fold changes of the target protein (66, 65), further indicating that regulation of the SAMHD1 3′-UTR by IFNs (Fig. 4) might be related to miRNAs. Indeed, several predicted miRNAs (Fig. 2B and Table 2) are acting on the SAMHD1 3′-UTR (Fig. 5).
In light of these findings, we hypothesize that IFN stimulation may affect miRNAs levels, which subsequently would mediate SAMHD1 regulation in a post-transcriptional manner. In that sense, SAMHD1 protein translation is inhibited by miRNAs. Their negative pressure on SAMHD1 levels is relieved upon IFN stimulation. In support of this hypothesis, miR-181a and miR-30a are down-regulated by type I and II IFNs in primary human monocytes (Fig. 6A), correlating with induced expression levels of SAMHD1 protein in these cells (Fig. 1A). Furthermore, IFN-induced SAMHD1 protein up-regulation was inhibited by application of miR-181a, miR-155, and miR-30a (Fig. 6B).
We propose a minor role for miR-155 in SAMHD1 regulation because it was not substantially altered upon IFN treatment at the relevant time point (Fig. 6A), and it displayed only a weak impact on SAMHD1 expression (Fig. 5C), possibly due to its 7-mer-m8 target site that is associated with about 10-fold lower efficiency compared with the 8-mer type (36). This is further substantiated by observations on miRNA abundance demonstrating that miR-155 is among the lesser expressed miRNAs with ∼100 times lower expression when compared with miR-181a in peripheral blood mononuclear cells (41). In concordance with our study, a recent report found miR-181a down-regulated upon IFNα and -γ stimulation in astrocytes and microglia, influencing SAMHD1 expression (29).
Importantly, in this study, we demonstrate that the regulation of SAMHD1 by IFNs through miRNAs is a cell type-specific mechanism. Type I and type II IFN treatment of MDMs and MDDCs modulated neither SAMHD1 protein (Fig. 7C) (31) nor miR-181a and miR-30a levels (Fig. 7B). Interestingly, MDMs and MDDCs already contain lower levels of miR-181a and miR-30a (in one donor in MDDC) (Fig. 7A) but severalfold higher basal SAMHD1 protein expression compared with monocytes (Fig. 7D). This further supports our model in which miR-181a and miR-30a levels are inversely correlated with SAMHD1 protein expression. We suggest that a low endogenous threshold of miR-181a and miR-30a has been reached in MDMs and MDDCs that cannot be further down-regulated upon IFN stimulation (Fig. 7B). Similarly, high SAMHD1 expression and low miR-181a levels are inversely correlated in monocytic THP1 cells (33) and primary effector CD4+ T cells (3, 67). IFN stimulation of THP1 (30, 31) and CD4+ T cells (6, 31) does not induce SAMHD1 protein levels in support of our model.
In summary, we propose that IFN stimulation induces SAMHD1 in a non-classical way through modulation of miRNA levels. Thus, we provide for the first time a model to explain the cell type-dependent stimulation of SAMHD1 by IFNs. IFNs induce down-regulation of miRNAs miR-181a and -30a and possibly also the levels of their family members or other not yet defined miRNAs but not miR-155 in a cell type-dependent manner. Thereby, their negative regulatory pressure on the SAMHD1 mRNA is relieved, and SAMHD1 translation is increased.
Monocytes may benefit from lower SAMHD1 expression compared to MDMs and MDDCs (Fig. 7D) because SAMHD1 has been recently proposed to be a proapoptotic factor (8). Nevertheless, SAMHD1 expression may be up-regulated in monocytes upon IFN stimulation or innate immune sensing, contributing to an antiviral state, if required.
The presence of SAMHD1 protein is critical for regulation of the innate immune response in myeloid sentinel cells, and consequently, the expression levels may influence the adaptive immune responses (11). Furthermore, changes in expression levels of SAMHD1 can have beneficial effects for HIV patients. For instance, studies in HIV-1 elite controllers, a specific patient group that can maintain undetectable levels of HIV-1 viral replication in the absence of antiretroviral therapy, have revealed that the expression level of SAMHD1 is maintained at a lower level in dendritic cells in comparison with other patient and healthy donor cohorts where SAMHD1 is up-regulated upon HIV-1 infection. In consequence, the dendritic cells of elite controllers may allow for sufficient HIV replication to trigger sensing (11) and are therefore more prone to prime the innate and adaptive immune system and enhance CD8 T cell responses to infection (68).
Therefore, the knowledge of the regulatory mechanisms impacting SAMHD1 expression during innate immunity or upon therapeutic IFN treatments could be used to open up novel strategies for therapeutic approaches or developing effective HIV-1 vaccines. Moreover, detailed knowledge on the regulation of SAMHD1 will help to provide insight into the critical cellular functions of SAMHD1.
Experimental Procedures
Ethics Statement
Buffy coats obtained from anonymous blood donors were purchased from the German Red Cross Blood Donor Service Baden-Württemberg Hessen. The research was performed according to the principles expressed in the Declaration of Helsinki.
Cell Lines
The adherent cell lines human embryonic kidney cells (HEK293T, RRID: CVCL_0063), cervix carcinoma cell line (HeLa Kyoto, RRID: CVCL_1922), and hepatocellular carcinoma cell line (Huh7, RRID: CVCL_0336) were cultured in DMEM (Lonza). Suspension cell lines of monocytic U937 (RRID: CVCL_0007) and THP-1 (RRID: CVCL_0006) were cultured in Roswell Park Memorial Institute medium (RPMI 1640, Biowest SAS). Both media were supplemented with 2 mm l-glutamine (Biochrom), 10% FCS (Sigma-Aldrich), 1% (v/v) penicillin-streptomycin (Life Technologies, Inc.).
Identification of Transcription Start and Termination Sites (5′- and 3′-RACE)
Total RNA was isolated from cell lines THP1 and U937 using TriFast (Peqlab). 75 μg of total RNA were subjected to Poly(A)+ RNA isolation with the Dynabeads mRNA purification kit (Life Technologies) according to the manufacturer's instructions. SAMHD1 transcription start and termination sites were identified using the GeneRacerTM SuperScript-III kit (Invitrogen) according to the manufacturer's protocol.
5′-RACE cDNA synthesis with gene-specific 5′_SAMHD1-primer (5′-GTCGGGATGGAGTTCCAG-3′) was performed at 55 °C for 1 h. The first round of PCR for amplification of transcription start sites was accomplished with primers 5′_SAMHD1-primer (5′-GTCGGGATGGAGTTCCAG-3′) and GeneRacerTM_5′-primer. To identify the polyadenylation site of SAMHD1 transcripts, first PCR was performed with primer 3′_SAMHD1-primer (5′-GATGGCGATGTTATAGCCC-3′) in combination with GeneRacerTM_3′-primer.
For both experimental approaches, nested PCR was performed by using either GeneRacer 5′_nested primer or GeneRacer 3′_nested primer together with the gene-specific primers already mentioned. All PCR protocols were designed as touchdown PCRs: 2′ initial denaturation steps at 94 °C followed by five cycles of 30 s at 94 °C and 72 °C for 3 min; five cycles of 30 s at 94 °C and 2 min at 70 °C; and 25 cycles of 30 s at 94 °C, 30 s at 65 °C, and 2 min at 72 °C, followed by a final extension step at 72 °C for 10 min. PCR fragments were excised and extracted from agarose gel and cloned into pGEM-T Easy vector (Promega) for sequencing.
Preparations of Monocytes, MDMs, MDDCs, and Interferon Treatment
Human buffy coats were obtained from German Red Cross Blood Donor Service Baden-Württemberg Hessen. Primary human monocytes were isolated from peripheral blood mononuclear cells after Ficoll density gradient centrifugation using either CD14-MicroBeads (Miltenyi Biotec; purity was >94%) for subsequent differentiation into MDMs or MDDCs or using a pan-monocyte isolation kit (Miltenyi Biotec; purity was > 90%) for purification of negative isolated monocytes for direct stimulation assays. Procedures were performed according to the manufacturer's guidelines, and separation from unlabeled cells was carried out in a MidiMACS or AutoMACS device (Miltenyi Biotec). Negatively isolated monocytes were cultured and treated with cytokines in 24-well cell culture plates (Falcon) with 7.5 × 105 cells/well in VLE-RPMI (Biochrom AG) supplemented with 2 mm l-glutamine, 1% FCS.
For differentiation of monocytes into MDMs, the cells were cultured in RPMI 1640 supplemented with 2 mm l-glutamine, 10% FCS, 1% (v/v) HEPES, 1 mm sodium pyruvate, and 100 units/ml GM-CSF (PeproTech). A second volume of medium including cytokines was added at day 3, and M1-like macrophages were used for subsequent experiments on day 5. MDDCs were differentiated similarly using RPMI 1640 supplemented with 2 mm l-glutamine, 10% FCS, 1% (v/v) HEPES, 1 mm sodium pyruvate, and 500 units/ml GM-CSF and 1,000 units/ml IL-4 (PeproTech). A second volume of medium including cytokines was added at day 3, and MDDCs were used for subsequent experiments on day 5. For IFN treatments, the cells were cultured in medium without differentiation cytokines but containing human interferon α2a (PBL Interferon Source), β (PBL Interferon Source), or γ (PeproTech) at the indicated concentrations.
Plasmids
SAMHD1 promoter reporter constructs were generated by cloning genomic sequences upstream of the SAMHD1 start codon ATG into a modified version of the pCpGfree-mcs plasmid (InvivoGen). mCMV enh, hEF1 prom, and SI 126 in pCpGfree-mcs plasmid had been replaced with a CpGfree firefly luciferase from InvivoGen, lacking a promoter (denoted as SAMHD1-Control). The earliest TSS found in our 5′-RACE at 200 nt upstream of the start codon was denoted as +1; upstream base pairs are indicated by negative numbers, whereas downstream bases have positive numbers.
Several SAMHD1 promoter reporter constructs were generated, reaching from +200 nt to −26 nt, −149 nt, −1500 nt, or −2000 nt. The sequence −2000 to +200 nt was amplified via PCR using the KOD Hot Start polymerase (Merck KGaA) with specific primers from THP-1 genomic DNA, whereas shorter sequences were again amplified from the −2000 to +200 nt sequence. A common reverse primer was used that introduced a 3′ NcoI cleavage site (CTCCATGGCTACACCTGGCGTCCGGC), whereas forward primers varied with every desired fragment length and included SbfI cleavage sites (SAMHD1(−2000), GACCTGCAGGTGGCCACCCAGCTAATTTTTG; SAMHD1(−1500), GACCTGCAGGTGAGTGCTCTGGCCTCCCAAAGTGCT; SAMHD1(−149), GACCTGCAGGGACGAGCCCCTAATTGGGGG; SAMHD1(−26), TTCCTGCAGGATTGATTTGAGGACGACTGG). PCR products were cleaved with SbfI and NcoI and cloned into the SAMHD1-empty plasmid upstream of the firefly luciferase.
For analysis of IFN stimulation on the SAMHD1 3′-UTR, a plasmid was obtained, which contained a destabilized luciferase and the SAMHD1 3′-UTR (SAMHD1-3′-UTR, Active Motif). SAMHD1-3′-UTR contains a 767-nt-long sequence that is composed of 29 nt from the end of the coding region of exon 16 followed by a 30–767-nt downstream genomic region.
To analyze the relevance of two putative miR-181, one miR-30, and one miR-155 target site in the 3′-UTR of SAMHD1 (indicated in Fig. 2B), a pmirGlo-SAMHD1-3′-UTR reporter plasmid was generated by transferring the 3′-UTR sequence of SAMHD1 from SAMHD1-3′-UTR into the pmirGlo-control plasmid (pmirGlo, Promega) using NheI and XhoI restriction enzymes. The pmirGlo plasmid encodes a firefly luciferase controlled by the human phosphoglycerate kinase promoter and may be post-transcriptionally modulated by insertion of a 3′-UTR sequence. In addition, a Renilla luciferase is constitutively expressed by an SV40 early enhancer/promoter. Double point mutations at positions 5 and 6 of one or both of the putative miR-181a seeding regions were introduced by site-directed mutagenesis of GA to AC to create plasmids pmirGlo_SAMHD1-3′-UTR_mut181a_1, pmirGlo_SAMHD1-3′-UTR_mut181a_2, and pmirGlo_SAMHD1-3′-UTR_mut181a_1+2, respectively. The putative miR-30a target site was mutated at positions 2 and 3 by TA to CG site-directed mutagenesis. The putative miR-155 target site was mutated at positions 2 and 3 by TT to CC site-directed mutagenesis. In brief, pmirGlo_SAMHD1-3′-UTR served as a template for a PCR using the KOD Hot Start polymerase (Merck KGaA) with specific primer and the following PCR cycling conditions: 2-h initial denaturation at 95 °C followed by 19 cycles of 20 min at 95 °C, 15 min at 50 or 55 °C, and 2 h 15 min at 70 °C. A final extension was performed for 5′ at 70 °C. (primers for miR-181a site 1, forward_mut181a_1 (GACCCAATGTACATGTCTGTAGTC) and reverse_mut181a:1 (GACTACAGACATGTACATTGGGTC); primers for miR-181a site 2, forward_mut181a_2 (TTTGTATTTTACATGTACACGCATGC) and reverse_mut181a_2 (GCATGCGTGTACATGTAAAATACAAA); primers for the miR-30a site, forward_mut_30a (GTCAGTTGTTCGCAAACTCCCT) and reverse_mut_30a (AGGGAGTTTGCGAACAACTGAC); primers for the miR-155 site, forward_mut_155 (ATGAAAAGCACCACCTTGCCTAT) and reverse_mut_155 (ATAGGCAAGGTGGTGCTTTTCAT)). Remaining template DNA was digested using DpnI. The construct pmirGlo_SAMHD1-3′-UTR_mut1+2 was generated the same way using primers for the second position using pmirGlo_SAMHD1-3′-UTR_mut1 as template. Signaling pathway reporters ISRE-Luc (based on pGL4.17 from Promega, ISG54 promoter was cloned into XhoI/HindIII) and pGAS-Luc (pGL3-GAS-Luc, Promega) were used.
Transfections of cDNA, siRNA, or miRNA
Plasmid DNA was transfected using FuGene HD (Promega) transfection reagent following the manufacturer's guidelines. FuGene HD/DNA ratios of 3:1 for adherent cell lines and 6:1 for suspension cell lines were used.
Transfection of siRNAs (siSP1 (Hs_SP1_1, Qiagen, caggtgcaaaccaacagatta), siSP3 (Hs_SP3_1, Qiagen, ctgcgcgagatgatactttga), siCtrl_1 (Allstar negative control siRNA, Qiagen), or siCtrl_2 (HP Custom siRNA-W/O Modification, 5′-AAGGTAATTGCGCGTGCAACT-3′, Qiagen)), miR mimics (miR-181a mimic (hsa-miR-181a-5p mimic, Dharmacon) or miR-181c (hsa-miR-181c-5p mimic, Dharmacon), or negative control mimic (Ctrl, miRIDIAN microRNA Mimic Negative Control 1, Dharmacon) was achieved using Lipofectamine RNAiMAX reagent (Invitrogen) for HEK293T and HeLa cells following the manufacturer's instructions. Co-transfection of plasmid DNA and miRNA was performed using DharmaFECT Duo transfection reagent (Dharmacon (Fisher)). Delivery of miRNAs either alone or in combination with reporter plasmids into U937 cells was conducted using an AmaxaTM NucleofectorTM 2b Device following the manufacturer's guidelines using the W01 program.
SAMHD1 Reporter Activity
To measure the transcriptional activity of SAMHD1 promoter sequences, the SAMHD1 reporter constructs were transfected in U937 and HEK293T cells using FuGene HD (Promega). For normalization, phRG-TK (Promega) encoding a Renilla luciferase was included and co-transfected in a 1:10 ratio. Transfections were conducted in duplicates or triplicates, and quantification of firefly and Renilla luciferase activity was carried out sequentially 48 h post-transfection using the Dual-Glo luciferase assay system (Promega). After knockdown of SP1 and SP3 in HeLa cells, firefly luciferase signals were normalized on protein content. For this, samples were lysed in 1× passive lysis buffer (Promega) and split to (i) determine the protein content and (ii) measure luciferase activities in technical triplicates.
SAMHD1 3′-UTR-mediated regulation was studied using the plasmid SAMHD1-3′-UTR transfected in HEK293T cells or U937 cells in triplicates as described above, and luciferase activities were quantified 24 h post-transfection using the LightSwitch luciferase assay kit (Active Motif). According to the manufacturer's instructions in well transfectable HEK293T cells, no additional normalization control was co-transfected, whereas in hard to transfect U937 cells, a firefly luciferase construct was co-transfected for normalization, and signals were normalized accordingly. The influence of cytokines on the transcriptional activities of the promoter/5′-UTR and 3′-UTR sequences was determined by treating transfected cells with 1000 or 2000 units/ml IFNα2a, IFNβ, or IFNγ 24 h post-transfection for an additional 24 h.
The relevance of two miR-181a, one miR-30a, and one miR-155 seed region as putative targets was investigated by co-transfection of plasmid pmirGlo-control, pmirGlo-SAMHD1-3′-UTR, pmirGlo_SAMHD1-3′-UTR_mut181a_1, pmirGlo_SAMHD1-3′-UTR_mut181a_2, pmirGlo_SAMHD1-3′-UTR_mut181a_1+2, pmirGlo_SAMHD1-3′-UTR_mut_30a, or pmirGlo_SAMHD1-3′-UTR_mut_155 together with either miR-181a, miR-155, miR-30a, or negative control miR mimics. Transfections were conducted in triplicates or quadruplicates, and quantification of firefly and Renilla luciferase activity was carried out sequentially 48 h post-transfection using the Dual-Glo luciferase assay system (Promega).
MicroRNA Quantification
To determine the relative levels of miR-181a, miR-155, and miR-30a expression in monocytes, we lysed cells at 4 and 24 h time points after stimulation with IFNα, -β, or -γ. Whole RNA samples were prepared with peqGOLD TriFast (Peqlab) as described by the manufacturer. RNA samples were then analyzed for miR-181a, miR-155, or miR-30a content using the TaqMan microRNA reverse transcription kit and TaqMan® microRNA assays for miR-181a/-155/-30a and RNU6B or U6 snRNA (all from Applied Biosystems) following the manufacturer's instructions. Briefly, miR-181a, miR-155, miR-30a, and RNU6B or U6 snRNA were reverse transcribed with specific primer sets generating elongated cDNA. Thereafter, relative levels of cDNA levels were quantified in 384-well format real-time quantitative RT-PCR on an ABI7900 cycler (Applied Biosystems). Levels of miRNAs were analyzed by the 2−ΔΔCT method (69) using RNU6B or U6 snRNA as a normalization control.
Real-time Quantitative RT-PCR
For quantification of cellular mRNA levels of SAMHD1, ISG-54, IRF1, TBP, or RPL13A, total cellular RNA was extracted using the RNeasy Plus minikit (Qiagen) or NucleoSpin® RNA kit (Macherey-Nagel) according to the manufacturer's guidelines. Relative expression of mRNAs was determined in 384-well format using the QuantiTect SYBR Green real-time quantitative RT-PCR kit (Qiagen) with the respective specific primers on an ABI7900 cycler (Applied Biosystems). The following primers were used for detection: hSAMHD1 fwd, TCACAGGCGCATTACTGCC; hSAMHD1 rev, GGATTTGAACCAATCGCTGGA; hISG-54 fwd, GGTGGCAGAAGAGGAAGATT; hISG-54 rev, TAGGCCAGTAGGTTGCACAT; hIRF1 fwd, CCTGGCTAGAGATGCAGATT; hIRF1 rev, AAGTTGGCCTTCCACGTCTT; hRPL13A fwd, CCTGGAGGAGAAGAGGAAAGAGA; hRPL13A rev, TTGAGGACCTCTGTGTATTTGTCAA. Data were normalized to reference gene RPL13A mRNA levels. Analysis for relative gene expression was performed using the 2−ΔΔCT method (69).
Cell Extract Preparation and Immunoblotting
Protein expression was assessed by immunoblotting. For this, whole cell extracts were prepared by lysing cells in radioimmunoprecipitation buffer (2 mm EDTA, 1% (v/v) glycerol, 137 mm NaCl, 1% (v/v) Nonidet P-40, 0.1% (w/v) SDS, 0.5% (w/v) sodium deoxycholate, 25 mm Tris (pH 8.0)) supplemented with protease inhibitors (Complete protease inhibitor mixture tablets, Roche Applied Science) and phosphatase inhibitors (PhosSTOP, Roche Applied Science) for 30 min on ice. Crude lysates were centrifuged to clear lysates from cell debris for 30 min at 17,000 × g and 4 °C. Protein concentrations of lysates were determined in a PierceTM BCA protein assay kit (Life Technologies, Inc.).
Protein samples were separated on SDS-PAGE using precast 4–12% NuPAGE® BisTris gradient gels (Life Technologies). Proteins were transferred to PVDF membranes (Hybond P 0.45, GE Healthcare) in 1× NuPAGE transfer buffer (Life Technologies). Membranes were blocked in 0.01% (v/v) Tris-buffered saline buffer with Tween 20 (TBST) + 5% (w/v) powdered milk or protease-free BSA (both from Carl Roth) for 2 h at 4 °C and probed with primary antibodies overnight at 4 °C. Primary antibodies used were rabbit anti-SAMHD1 (Proteintech; diluted 1:2000), rabbit anti-STAT1 (sc346, Santa Cruz Biotechnology, Inc.; diluted 1:5000), rabbit anti-SP1 (sc59, Santa Cruz Biotechnology; diluted 1:1500), rabbit anti-SP3 (sc644, Santa Cruz Biotechnology; diluted 1:3000), rabbit anti-GAPDH (catalog no. 2118, Cell Signaling Technology; diluted 1:4000), rabbit anti-ERK (catalog no. 9212, Cell Signaling Technology; diluted 1:4000), and mouse anti-β-actin (ACTB, A5441, Sigma-Aldrich; diluted 1:10,000). As secondary antibodies, the appropriate species-specific HRP-linked F(ab′)2 fragments (GE Healthcare) were incubated on the blot for 2 h at 4 °C. HRP activity was detected on autoradiography films (Fujifilm) after incubation with ECL Prime reagent (GE Healthcare) or Lumi-Light Western blotting substrate (Roche Applied Science).
Software
We used the earliest TSS found in our 5′-RACE at 200 nt upstream of the start codon as reference denoted as +1 for the analysis of the putative SAMHD1 promoter. Prediction and analysis of core promoter elements were carried out on the promoter sequence of SAMHD1 ranging from −2000 to +500 nt relative to this TSS. Predictions of promoter elements, such as the TATA-box, GC-boxes, or CCAAT-boxes, were performed using NNPP version 2.2 (70), FPROM (71), and McPromoter MM:II (72), GPMiner (73) or the eukaryotic promoter database (EPD) (74). An in silico analysis for CpG islands and C+G content was done using CpGPlot and GPMiner. AliBaba2.1 was used to predict SP1 transcription factor binding sites. GPMiner was used for prediction of high stringency human IFN signaling-responsive TF binding sites, and GAS consensus motifs (TT(C/A)NNN(G/T)AA) were searched with Vector NTI Advance® version 11.5.2 (Invitrogen).
miRNA target predictions on the 3′-UTR of SAMHD1 were performed using Miranda (50) (via microrna.org) (75), TargetScan version 7.1 (52), and miRcode (53) target predictions to find putative miRNAs and their target sites on the SAMHD1 mRNA (NM_015474). Search criteria were stringent, viewing only hits of conserved miRNAs. With Miranda, only hits with a mirSVR score of ≤−0.5, a PhastCons conservation score of ≥0.5, and at least 6-mer seed sequence complementarity were considered putative hits. The search by TargetScan version 7.1 was focused on miRNA families that are broadly conserved among vertebrates with either conserved or poorly conserved sites. MiRcode was used to search for sites with scores of ≥200.
Furthermore, miRcode and seed sequence comparison using Vector NTI Advance® version 11.5.2 (Invitrogen) were applied to predict miRNA target sites on the 200-nt 5′-UTR of SAMHD1 mRNA.
Quantification of immunoblotting data was carried out using Image Studio Lite version 5.0.21. Statistical analysis was performed with GraphPad Prism version 5.04 (GraphPad Software, La Jolla, CA). Data are represented as mean ± S.D. unless mentioned otherwise. Unpaired Student's t test was applied to assess statistical significance (ns, p > 0.05; *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001).
Author Contributions
R. K., N. V. F., and M. R. designed the study, analyzed and interpreted data, and wrote the paper. M. R. designed, performed, and analyzed the experiments shown in Figs. 1–7. E. F. and M. H. designed and constructed the initial promoter reporter construct. N. V. F. designed, performed, and analyzed the 5′- and 3′-RACE experiments. I. M. P. and A. I. performed and analyzed part of the experiments on miRNA in monocytes in Fig. 6. E. F. contributed reagents/material. I. M. P. contributed expertise and advice on miRNA experiments. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Prof. Carsten Münk for helpful discussions, Dr. Lise Rivière for proofreading the manuscript, Christiane Tondera and Heike Schmitz for excellent technical assistance, and Dr. Janna Seifried for advice on this project.
This work was supported by DZIF TTU 01.802 and TTU 04.802 (to R. K.). The authors declare that they have no conflicts of interest with the contents of this article.
- SAMHD1
- sterile α motif (SAM) and HD domain containing protein 1
- ISG
- interferon-stimulated gene
- MDDC
- monocyte-derived dendritic cell
- MDM
- monocyte-derived macrophage
- miRNA
- microRNA
- nt
- nucleotide(s)
- TF
- transcription factor
- TSS
- transcriptional start site
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- ISRE
- interferon-stimulated response element
- GAS
- IFNγ-activated sequence
- RRID
- research resource identifier.
References
- 1. Goldstone D. C., Ennis-Adeniran V., Hedden J. J., Groom H. C. T., Rice G. I., Christodoulou E., Walker P. A., Kelly G., Haire L. F., Yap M. W., de Carvalho L. P. S., Stoye J. P., Crow Y. J., Taylor I. A., and Webb M. (2011) HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382 [DOI] [PubMed] [Google Scholar]
- 2. Powell R. D., Holland P. J., Hollis T., and Perrino F. W. (2011) Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J. Biol. Chem. 286, 43596–43600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baldauf H.-M., Pan X., Erikson E., Schmidt S., Daddacha W., Burggraf M., Schenkova K., Ambiel I., Wabnitz G., Gramberg T., Panitz S., Flory E., Landau N. R., Sertel S., Rutsch F., et al. (2012) SAMHD1 restricts HIV-1 infection in resting CD4+ T cells. Nat. Med. 18, 1682–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim B., Nguyen L. A., Daddacha W., and Hollenbaugh J. A. (2012) Tight interplay among SAMHD1 protein level, cellular dNTP levels, and HIV-1 proviral DNA synthesis kinetics in human primary monocyte-derived macrophages. J. Biol. Chem. 287, 21570–21574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lahouassa H., Daddacha W., Hofmann H., Ayinde D., Logue E. C., Dragin L., Bloch N., Maudet C., Bertrand M., Gramberg T., Pancino G., Priet S., Canard B., Laguette N., Benkirane M., et al. (2012) SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 13, 223–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. St Gelais C., da Silva S., Amie S. M., Coleman C. M., Hoy H., Hollenbaugh J. A., Kim B., and Wu L. (2012) SAMHD1 restricts HIV-1 infection in dendritic cells (DCs) by dNTP depletion, but its expression in DCs and primary CD4+ T-lymphocytes cannot be upregulated by interferons. Retrovirology 9, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Franzolin E., Pontarin G., Rampazzo C., Miazzi C., Ferraro P., Palumbo E., Reichard P., and Bianchi V. (2013) The deoxynucleotide triphosphohydrolase SAMHD1 is a major regulator of DNA precursor pools in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 110, 14272–14277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bonifati S., Daly M. B., St Gelais C., Kim S. H., Hollenbaugh J. A., Shepard C., Kennedy E. M., Kim D.-H., Schinazi R. F., Kim B., and Wu L. (2016) SAMHD1 controls cell cycle status, apoptosis and HIV-1 infection in monocytic THP-1 cells. Virology 495, 92–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kretschmer S., Wolf C., König N., Staroske W., Guck J., Häusler M., Luksch H., Nguyen L. A., Kim B., Alexopoulou D., Dahl A., Rapp A., Cardoso M. C., Shevchenko A., and Lee-Kirsch M. A. (2015) SAMHD1 prevents autoimmunity by maintaining genome stability. Ann. Rheum. Dis. 74, e17–e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ballana E., and Esté J. A. (2015) SAMHD1: at the crossroads of cell proliferation, immune responses, and virus restriction. Trends Microbiol. 23, 680–692 [DOI] [PubMed] [Google Scholar]
- 11. Maelfait J., Bridgeman A., Benlahrech A., Cursi C., and Rehwinkel J. (2016) Restriction by SAMHD1 limits cGAS/STING-dependent innate and adaptive immune responses to HIV-1. Cell Rep. 16, 1492–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rice G. I., Bond J., Asipu A., Brunette R. L., Manfield I. W., Carr I. M., Fuller J. C., Jackson R. M., Lamb T., Briggs T. A., Ali M., Gornall H., Couthard L. R., Aeby A., Attard-Montalto S. P., et al. (2009) Mutations involved in Aicardi-Goutières syndrome implicate SAMHD1 as regulator of the innate immune response. Nat. Genet. 41, 829–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Crow Y. J., and Manel N. (2015) Aicardi-Goutieres syndrome and the type I interferonopathies. Nat. Rev. Immunol. 15, 429–440 [DOI] [PubMed] [Google Scholar]
- 14. Hrecka K., Hao C., Gierszewska M., Swanson S. K., Kesik-Brodacka M., Srivastava S., Florens L., Washburn M. P., and Skowronski J. (2011) Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Laguette N., Sobhian B., Casartelli N., Ringeard M., Chable-Bessia C., Ségéral E., Yatim A., Emiliani S., Schwartz O., and Benkirane M. (2011) SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Berger A., Sommer A. F. R., Zwarg J., Hamdorf M., Welzel K., Esly N., Panitz S., Reuter A., Ramos I., Jatiani A., Mulder L. C. F., Fernandez-Sesma A., Rutsch F., Simon V., König R., and Flory E. (2011) SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutières syndrome are highly susceptible to HIV-1 infection. PLoS Pathog. 7, e1002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Manel N., Hogstad B., Wang Y., Levy D. E., Unutmaz D., and Littman D. R. (2010) A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature 467, 214–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li N., Zhang W., and Cao X. (2000) Identification of human homologue of mouse IFN-γ induced protein from human dendritic cells. Immunol. Lett. 74, 221–224 [DOI] [PubMed] [Google Scholar]
- 19. Schmidt S., Schenkova K., Adam T., Erikson E., Lehmann-Koch J., Sertel S., Verhasselt B., Fackler O. T., Lasitschka F., and Keppler O. T. (2015) SAMHD1's protein expression profile in humans. J. Leukoc. Biol. 98, 5–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Préhaud C., Mégret F., Lafage M., and Lafon M. (2005) Virus infection switches TLR-3-positive human neurons to become strong producers of β interferon. J. Virol. 79, 12893–12904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhao D., Peng D., Li L., Zhang Q., and Zhang C. (2008) Inhibition of G1P3 expression found in the differential display study on respiratory syncytial virus infection. Virol. J. 5, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hartman Z. C., Kiang A., Everett R. S., Serra D., Yang X. Y., Clay T. M., and Amalfitano A. (2007) Adenovirus infection triggers a rapid, MyD88-regulated transcriptome response critical to acute-phase and adaptive immune responses in vivo. J. Virol. 81, 1796–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li S., Wang L., Berman M., Kong Y.-Y., and Dorf M. E. (2011) Mapping a dynamic innate immunity protein interaction network regulating type I interferon production. Immunity 35, 426–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Buchanan E. L., Espinoza D. A., McAlexander M. A., Myers S. L., Moyer A., and Witwer K. W. (2016) SAMHD1 transcript upregulation during SIV infection of the central nervous system does not associate with reduced viral load. Sci. Rep. 6, 22629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang S., Zhan Y., Zhou Y., Jiang Y., Zheng X., Yu L., Tong W., Gao F., Li L., Huang Q., Ma Z., and Tong G. (2016) Interferon regulatory factor 3 is a key regulation factor for inducing the expression of SAMHD1 in antiviral innate immunity. Sci. Rep. 6, 29665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liao W., Bao Z., Cheng C., Mok Y.-K., and Wong W. S. (2008) Dendritic cell-derived interferon-γ-induced protein mediates tumor necrosis factor-α stimulation of human lung fibroblasts. Proteomics 8, 2640–2650 [DOI] [PubMed] [Google Scholar]
- 27. Pauls E., Jimenez E., Ruiz A., Permanyer M., Ballana E., Costa H., Nascimiento R., Parkhouse R. M., Peña R., Riveiro-Muñoz E., Martinez M. A., Clotet B., Esté J. A., and Bofill M. (2013) Restriction of HIV-1 replication in primary macrophages by IL-12 and IL-18 through the upregulation of SAMHD1. J. Immunol. 190, 4736–4741 [DOI] [PubMed] [Google Scholar]
- 28. Sommer A. F. R., Rivière L., Qu B., Schott K., Riess M., Ni Y., Shepard C., Schnellbächer E., Finkernagel M., Himmelsbach K., Welzel K., Kettern N., Donnerhak C., Münk C., Flory E., Liese J., Kim B., Urban S., and König R. (2016) Restrictive influence of SAMHD1 on hepatitis B virus life cycle. Sci. Rep. 6, 26616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jin C., Peng X., Liu F., Cheng L., Xie T., Lu X., Wu H., and Wu N. (2016) Interferon-induced sterile α motif and histidine/aspartic acid domain-containing protein 1 expression in astrocytes and microglia is mediated by miR-181a. AIDS 30, 2053–2064 [DOI] [PubMed] [Google Scholar]
- 30. Goujon C., Schaller T., Galão R. P., Amie S. M., Kim B., Olivieri K., Neil S. J. D., and Malim M. H. (2013) Evidence for IFNα-induced, SAMHD1-independent inhibitors of early HIV-1 infection. Retrovirology 10, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cribier A., Descours B., Valadão A. L. C., Laguette N., and Benkirane M. (2013) Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep. 3, 1036–1043 [DOI] [PubMed] [Google Scholar]
- 32. Gottwein E., Mukherjee N., Sachse C., Frenzel C., Majoros W. H., Chi J.-T. A., Braich R., Manoharan M., Soutschek J., Ohler U., and Cullen B. R. (2007) A viral microRNA functions as an orthologue of cellular miR-155. Nature 450, 1096–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jin C., Peng X., Liu F., Cheng L., Lu X., Yao H., Wu H., and Wu N. (2014) MicroRNA-181 expression regulates specific post-transcriptional level of SAMHD1 expression in vitro. Biochem. Biophys. Res. Commun. 452, 760–767 [DOI] [PubMed] [Google Scholar]
- 34. Pilakka-Kanthikeel S., Raymond A., Atluri V. S. R., Sagar V., Saxena S. K., Diaz P., Chevelon S., Concepcion M., and Nair M. (2015) Sterile alpha motif and histidine/aspartic acid domain-containing protein 1 (SAMHD1)-facilitated HIV restriction in astrocytes is regulated by miRNA-181a. J. Neuroinflammation 12, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Filipowicz W., Bhattacharyya S. N., and Sonenberg N. (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature Rev. Genet. 9, 102–114 [DOI] [PubMed] [Google Scholar]
- 36. Bartel D. P. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moazed D. (2009) Small RNAs in transcriptional gene silencing and genome defence. Nature 457, 413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Miranda K. C., Huynh T., Tay Y., Ang Y.-S., Tam W.-L., Thomson A. M., Lim B., and Rigoutsos I. (2006) A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell 126, 1203–1217 [DOI] [PubMed] [Google Scholar]
- 39. Guo Z.-W., Xie C., Yang J.-R., Li J.-H., Yang J.-H., and Zheng L. (2015) MtiBase: a database for decoding microRNA target sites located within CDS and 5′UTR regions from CLIP-Seq and expression profile datasets. Database 10.1093/database/bav102: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lewis B. P., Burge C. B., and Bartel D. P. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 [DOI] [PubMed] [Google Scholar]
- 41. Liang Y., Ridzon D., Wong L., and Chen C. (2007) Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 8, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A. O., Landthaler M., Lin C., Socci N. D., Hermida L., Fulci V., Chiaretti S., et al. (2007) A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129, 1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pedersen I. M., Cheng G., Wieland S., Volinia S., Croce C. M., Chisari F. V., and David M. (2007) Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 449, 919–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sedger L. M. (2013) microRNA control of interferons and interferon induced anti-viral activity. Mol. Immunol. 56, 781–793 [DOI] [PubMed] [Google Scholar]
- 45. Abdel-Mohsen M., Deng X., Danesh A., Liegler T., Jacobs E. S., Rauch A., Ledergerber B., Norris P. J., Günthard H. F., Wong J. K., and Pillai S. K. (2014) Role of microRNA modulation in the interferon-α/ribavirin suppression of HIV-1 in vivo. PLoS One 9, e109220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Forster S. C., Tate M. D., and Hertzog P. J. (2015) MicroRNA as type I interferon-regulated transcripts and modulators of the innate immune response. Front. Immunol. 6, 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Danino Y. M., Even D., Ideses D., and Juven-Gershon T. (2015) The core promoter: at the heart of gene expression. Biochim. Biophys. Acta 1849, 1116–1131 [DOI] [PubMed] [Google Scholar]
- 48. Proudfoot N. J. (2011) Ending the message: poly(A) signals then and now. Genes Dev. 25, 1770–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tian B., Hu J., Zhang H., and Lutz C. S. (2005) A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 33, 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Enright A. J., John B., Gaul U., Tuschl T., Sander C., and Marks D. S. (2003) MicroRNA targets in Drosophila. Genome Biol. 5, R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Betel D., Koppal A., Agius P., Sander C., and Leslie C. (2010) Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 11, R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Agarwal V., Bell G. W., Nam J.-W., and Bartel D. P. (2015) Predicting effective microRNA target sites in mammalian mRNAs. eLife 4, e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jeggari A., Marks D. S., and Larsson E. (2012) miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics 28, 2062–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. St Gelais C., de Silva S, Hach J. C., White T. E., Diaz-Griffero F., Yount J. S., and Wu L. (2014) Identification of cellular proteins interacting with the retroviral restriction factor SAMHD1. J. Virol. 88, 5834–5844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kambara H., Gunawardane L., Zebrowski E., Kostadinova L., Jobava R., Krokowski D., Hatzoglou M., Anthony D. D., and Valadkhan S. (2014) Regulation of interferon-stimulated gene BST2 by a lncRNA transcribed from a shared bidirectional promoter. Front. Immunol. 5, 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Barriocanal M., Carnero E., Segura V., and Fortes P. (2014) Long non-coding RNA BST2/BISPR is induced by IFN and regulates the expression of the antiviral factor tetherin. Front. Immunol. 5, 655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schmitt M. J., Philippidou D., Reinsbach S. E., Margue C., Wienecke-Baldacchino A., Nashan D., Behrmann I., and Kreis S. (2012) Interferon-γ-induced activation of signal transducer and activator of transcription 1 (STAT1) up-regulates the tumor suppressing microRNA-29 family in melanoma cells. Cell Commun. Signal. 10, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schug J., Schuller W.-P., Kappen C., Salbaum J. M., Bucan M., and Stoeckert C. J. Jr. (2005) Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 6, R33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Juven-Gershon T., and Kadonaga J. T. (2010) Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 339, 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Saxonov S., Berg P., and Brutlag D. L. (2006) A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. U.S.A. 103, 1412–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shlyueva D., Stampfel G., and Stark A. (2014) Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 15, 272–286 [DOI] [PubMed] [Google Scholar]
- 62. Montgomery S. B., Griffith O. L., Sleumer M. C., Bergman C. M., Bilenky M., Pleasance E. D., Prychyna Y., Zhang X., and Jones S. J. M. (2006) ORegAnno: an open access database and curation system for literature-derived promoters, transcription factor binding sites and regulatory variation. Bioinformatics 22, 637–640 [DOI] [PubMed] [Google Scholar]
- 63. Griffith O. L., Montgomery S. B., Bernier B., Chu B., Kasaian K., Aerts S., Mahony S., Sleumer M. C., Bilenky M., Haeussler M., Griffith M., Gallo S. M., Giardine B., Hooghe B., van Loo P., et al. (2008) ORegAnno: an open-access community-driven resource for regulatory annotation. Nucleic Acids Res. 36, D107–D113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lesurf R., Cotto K. C., Wang G., Griffith M., Kasaian K., Jones S. J. M., Montgomery S. B., Griffith O. L., and Open Regulatory Annotation Consortium (2016) ORegAnno 3.0: a community-driven resource for curated regulatory annotation. Nucleic Acids Res. 44, D126–D132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hausser J., and Zavolan M. (2014) Identification and consequences of miRNA-target interactions: beyond repression of gene expression. Nat. Rev. Genet. 15, 599–612 [DOI] [PubMed] [Google Scholar]
- 66. Fang Z., and Rajewsky N. (2011) The impact of miRNA target sites in coding sequences and in 3′UTRs. PLoS One 6, e18067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li Q.-J., Chau J., Ebert P. J. R., Sylvester G., Min H., Liu G., Braich R., Manoharan M., Soutschek J., Skare P., Klein L. O., Davis M. M., and Chen C.-Z. (2007) miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell 129, 147–161 [DOI] [PubMed] [Google Scholar]
- 68. Martin-Gayo E., Buzon M. J., Ouyang Z., Hickman T., Cronin J., Pimenova D., Walker B. D., Lichterfeld M., and Yu X. G. (2015) Potent cell-intrinsic immune responses in dendritic cells facilitate HIV-1-specific T cell immunity in HIV-1 elite controllers. PLoS Pathog. 11, e1004930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 70. Reese M. G., and Eeckman F. (September 16–20, 1995) Novel neural network algorithms for improved eukaryotic promoter site recognition: the seventh international genome sequencing and analysis conference, Hilton Head Island, SC [Google Scholar]
- 71. Solovyev V. V., Shahmuradov I. A., and Salamov A. A. (2010) Identification of promoter regions and regulatory sites. Methods Mol. Biol. 674, 57–83 [DOI] [PubMed] [Google Scholar]
- 72. Ohler U., Stemmer G., Harbeck S., and Niemann H. (2000) Stochastic segment models of eukaryotic promoter regions. Pac. Symp. Biocomput. 2000, 380–91 [DOI] [PubMed] [Google Scholar]
- 73. Lee T.-Y., Chang W.-C., Hsu J. B.-K., Chang T.-H., and Shien D.-M. (2012) GPMiner: an integrated system for mining combinatorial cis-regulatory elements in mammalian gene group. BMC Genomics 13, S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Dreos R., Ambrosini G., Cavin Périer R., and Bucher P. (2013) EPD and EPDnew, high-quality promoter resources in the next-generation sequencing era. Nucleic Acids Res. 41, D157–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Betel D., Wilson M., Gabow A., Marks D. S., and Sander C. (2008) The microRNA.org resource: targets and expression. Nucleic Acids Res. 36, D149–D153 [DOI] [PMC free article] [PubMed] [Google Scholar]