Abstract
A major goal for HIV-1 vaccine development is an ability to elicit strong and durable broadly neutralizing antibody (bNAb) responses. The trimeric envelope glycoprotein (Env) spikes on HIV-1 are known to contain multiple epitopes that are susceptible to bNAbs isolated from infected individuals. Nonetheless, all trimeric and monomeric Env immunogens designed to date have failed to elicit such antibodies. We report the structure-guided design of HIV-1 cyclically permuted gp120 that forms homogeneous, stable trimers, and displays enhanced binding to multiple bNAbs, including VRC01, VRC03, VRC-PG04, PGT128, and the quaternary epitope-specific bNAbs PGT145 and PGDM1400. Constructs that were cyclically permuted in the V1 loop region and contained an N-terminal trimerization domain to stabilize V1V2-mediated quaternary interactions, showed the highest homogeneity and the best antigenic characteristics. In guinea pigs, a DNA prime-protein boost regimen with these new gp120 trimer immunogens elicited potent neutralizing antibody responses against highly sensitive Tier 1A isolates and weaker neutralizing antibody responses with an average titer of about 115 against a panel of heterologous Tier 2 isolates. A modest fraction of the Tier 2 virus neutralizing activity appeared to target the CD4 binding site on gp120. These results suggest that cyclically permuted HIV-1 gp120 trimers represent a viable platform in which further modifications may be made to eventually achieve protective bNAb responses.
Keywords: human immunodeficiency virus (HIV); immunogenicity; protein conformation; protein engineering; protein stability; CD4 binding site; Env trimers; cyclic permutation; immunogen design; protein stabilization, dynamics
Introduction
The HIV-1 Env displayed on the virion surface mediates entry of the virion into target CD4+T cells to establish infection. Due to its surface accessibility, Env is the predominant target for neutralizing antibody responses (1, 2). Env is synthesized as a gp160 precursor protein, which is further cleaved into surface proximal gp120 and membrane anchored gp41 subunits. A functional HIV-1 Env spike consists of a trimer of gp120 and gp41 heterodimers. The base of the trimer is proximal to the viral membrane (3) and is held together by trimerization motifs within the N-heptad repeat of gp41 (4–6). The trimer apex is stabilized by interactions between the V1V2 loops of adjacent gp120 monomers (3). Binding of Env to the CD4 receptor on T cells causes extensive conformational changes in the Env trimer and leads to the formation of an open CD4-bound conformation in which the cryptic, co-receptor binding site becomes accessible. This facilitates interaction of the co-receptor (CCR5 or CXCR4) with Env, further driving the fusion of viral and host cell membranes (3, 7).
In natural HIV-1 infection, most of the B cell response is elicited against the Env protein, due to its location on the virion surface and relatively high immunogenicity. The antibody response induced during natural HIV-1 infection is predominantly non-neutralizing and strain specific, due to the shed gp120 (immunodominant decoys) and various other immune evasive mechanisms (8). However, ∼20% of HIV-1-infected patients develop bNAbs during the course of 1–3 years of infection (9). For the past few years, with the help of advanced technologies for isolating antibody genes from single memory B cells, a large number of neutralizing antibodies (NAbs)4 have been isolated from HIV patients that define potential conserved, neutralizing epitopes on HIV-1 Env (10). The bNAbs described to date target the CD4 binding site (CD4bs) on the gp120 subunit (NAbs b12, VRC01–03, NIH45–46, 3BNC117), quaternary epitopes in the V1V2 region on the gp120 trimer (PG9, PG16, PGT141–145, PGDM1400, and CAP256-VRC26.25), glycan-dependent epitopes in the V3 loop (PGT series of NAbs), the membrane proximal external region of the gp41 subunit (NAbs 2F5, 4E10, and 10E8), and a cleavage-dependent epitope at the gp41-gp120 interface of the Env trimer (PGT151 and PGT152) (11–14). The neutralizing epitopes on gp41 are less accessible to bNAbs in the context of native HIV-1 Env (15, 16). Thus, designing immunogens that efficiently display the more accessible neutralizing epitopes present on gp120 in its native trimeric conformation may elicit bNAb responses.
A recent RV144 clinical trial based on the ALVAC vector and clade B/E gp120 regimen showed ∼31.2% efficacy, appreciably higher than other vaccines tested in earlier clinical trials (17). The vaccine efficacy in the RV144 trial was ∼59.9% within the first year but the efficacy diminished drastically over the course of the study indicating low antibody persistence or weak memory B cell responses (17, 18). The antibodies elicited in the RV144 clinical trial did not have broad neutralizing activity suggesting that some other immunological mechanisms conferred protection (19). The results of this clinical study (17, 20) were consistent with earlier evidence (21) indicating that monomeric gp120 does not trigger broadly neutralizing responses. Recent evidence suggests that functional antibodies to Env are necessary and sufficient to significantly reduce the probability of infection by SIV in non-human primates. Although the non-human primate sera against Env exhibited mild in vitro neutralization, they showed enhanced antibody effector functions in non-human primates (22, 23).
Various approaches have been employed to design Env immunogens to mimic the native, pre-fusion trimeric conformation. These include the design of cleavage-deficient gp140, cleavage-competent gp140 stabilized by a disulfide linkage between gp120 and gp41 (SOSIP-gp140), and trimeric gp140 and trimeric gp120 immunogens where a heterologous trimerization domain has been fused at the C terminus (24–30). Although these immunogens are trimeric and some of the sera displayed robust Tier 1 and strong autologous Tier 2 neutralization titers, in most cases the resulting antisera show only a marginal improvement in the breadth of neutralization response relative to those elicited by monomeric gp120 (31–33).
In a prior study (34) we showed that sera capable of neutralizing multiple Tier 2 isolates could be elicited by priming with a fragment containing a part of the epitope for the bNAb b12 and boosting with full-length gp120. However, this study employed a year long immunization schedule. In the present study, we explore an alternative approach with a shorter, 20-week schedule, immunizing with trimeric, cyclically permuted gp120 derivatives.
In the native Env trimer (3–6, 35) V1V2 loops from gp120 monomers are oriented at the apex of the trimer, forming quaternary epitopes. The V1V2 and V3 loops in the native trimer restrict the angle at which antibodies can approach the CD4bs. However, the loop regions are highly dynamic (35, 36). To minimize conformational dynamics in these loops, based on available structural information, we inserted a heterologous trimerization domain into the V1 loop region to trimerize the gp120 molecule at the apex instead of at the C terminus of gp120 as had been done previously. We have previously designed cyclically permuted gp120 by making new N and C termini at the V1 loop and by connecting the native N and C termini with an appropriate flexible linker (37). Here we describe the antigenicity and immunogenicity of a new generation of cyclically permuted gp120s that exhibit improved homogeneity and trimer stability.
Results
Structure Guided HIV-1 Trimeric gp120 Immunogen Design
Several cryo-EM and crystallographic studies indicate that the three monomers of gp120 in pre-fusion, native Env interact though quaternary contacts at the apex of the trimer that are largely mediated by the V1V2 regions of the gp120 subunits (3–6, 35). We hypothesized that insertion of a heterologous trimerization sequence in the V1V2 region might help in the formation of native-like gp120 trimers. We anticipated that the long distances between the N and C termini of commonly used coiled-coil trimerization domains would make them unsuitable for insertion into V1V2 loop regions of gp120 protein. To overcome this problem, we designed gp120 derivatives that are cyclically permuted at the V1 loop region of gp120 at sites where mutations and insertions are tolerated (supplemental Fig. S1) (37). To this end, the native N and C termini of the gp120 molecule were connected with a 20-amino acid linker and new N and C termini were created within the V1 loop region at specific positions in the gp120 sequence. When a heterologous, disulfide-linked, coiled-coil trimerization domain from human cartilage matrix protein (hCMP) was inserted at the V1 loop region of cyclically permuted gp120, it facilitated the formation of homogeneous trimers. In the present study we characterized the antigenicity and immunogenicity of cyclically permuted gp120 trimers made from both JRFL and JRCSF gp120 sequences. The flexible C1 region of gp120 (residues 31–83) was removed from all the constructs to restrict conformational flexibility (Fig. 1A and supplemental Fig. S2). We also used JRFL gp120 (residues 83–511) and JRFLgp120-L6-hCMP (JRFL gp120 with a C-terminal hCMP domain connected with a 6-residue linker) for comparison, to assess the biochemical properties, antigenicity, and immunogenicity of cyclically permuted gp120 trimers (Fig. 1 and supplemental Fig. S2).
FIGURE 1.
Engineered cyclically permuted gp120 immunogens form homogeneous, stable trimers. A, schematic representation of the four different immunogens characterized in this study. Cyclically permuted gp120 trimers (JRFL-hCMP-V1cyc and JRCSF-hCMP-V1cyc) were designed by joining native N and C termini of gp120 with a flexible, soluble 20-residue linker and new N and C termini were created by opening the V1 loop followed by fusion of an hCMP trimerization domain to the new N terminus. A more conventional gp120 trimer (JRFLgp120-L6-hCMP) was made by connecting the hCMP trimerization domain to the C terminus of JRFL gp120 with a 6-residue flexible linker. L6, 6-residue linker; L20, 20-residue linker. All constructs were secreted from HEK 293T cells under control of the CMV promoter with a tpa leader. B, purified proteins were subjected to BN-PAGE followed by Western blotting with rabbit anti-gp120 polyclonal sera to characterize the oligomeric state of the proteins. Cyclically permuted gp120 trimers appeared as trimers, whereas JRFL gp120 and JRFLgp120-L6-hCMP appeared as a mixture of oligomers. C, analytical gel-filtration chromatography of cyclically permuted constructs on a Superdex-200 column in PBS buffer at room temperature. The absorbance at 220 nm is shown as a function of the elution volume. Cyclically permuted gp120 trimers showed a single peak corresponding to a trimer, whereas JRFL gp120 and JRFLgp120-L6-hCMP showed two major peaks corresponding to a dimer and a monomer. D, the mass fractions and corresponding molecular weights of each peak for the proteins analyzed by SEC-MALS. E, the functional stability of all proteins was tested by monitoring binding to bNAbs, VRC01 (CD4bs specific), and PGT128 (V3 loop glycan specific) before and after heating. Proteins were incubated at 60 or 100 °C for 1 h and cooled down to room temperature to monitor binding to bNAbs VRC01 (left panel) and PGT128 (right panel) immobilized on the surface of a CM5 chip. Biacore response units were plotted after incubation at different temperatures to compare stabilities. Cyclically permuted gp120 trimers are stable at 60 °C showing similar binding before and after heating to VRC01 and PGT128 antibodies. They retained detectable binding to these antibodies even after incubation at 100 °C for 1 h. F, temperature-induced equilibrium unfolding transitions of monomeric JRFL gp120 (black open circles, Tm = 337.1 K (64.0 °C)), and trimeric JRFL-hCMP-V1cycgp120 (red open circles, Tm = 341.8 K (68.7 °C)). The lines through the symbols represent fits using a two-state model.
Purification and Characterization of gp120 Monomer and Cyclically Permuted gp120 Trimers
All constructs were expressed in HEK 293T cells and proteins were purified from cell supernatants by lentil lectin affinity chromatography. Both cyclically permuted gp120 constructs (JRFL-hCMP-V1cyc and JRCSF-hCMP-V1cyc) appeared as homogeneous trimers as determined by blue native-PAGE (BN-PAGE) (Fig. 1B), analytical gel filtration chromatography (Fig. 1C), and multiangle light scattering (SEC-MALS) experiments (Fig. 1D and supplemental Fig. S3), whereas JRFL gp120 appeared as mixed oligomers with predominantly monomeric species and JRFLgp120-L6-hCMP appeared as a mixture of oligomers. The JRFL-hCMP-V1cyc and JRCSF-hCMP-V1cyc were not dissociated into dimers or monomers after purification and they were stable at 4 °C for 40 days without any degradation. In addition, we monitored the stability of all constructs after heating them to 60 and 100 °C for 1 h and subsequently monitoring their interaction with bNAbs using surface plasmon resonance (SPR). Heating at 60 °C did not affect the binding of cyclically permuted gp120 trimers and JRFLgp120-L6-hCMP to surface-immobilized bNAbs, VRC01, and PGT128 (Fig. 1E). In contrast, binding of JRFL gp120 to VRC01 and PGT128 was significantly decreased upon heating at 60 °C. When all proteins were heated at 100 °C, cyclically permuted gp120 trimers retained detectable binding to VRC01 and PGT128 bNAbs, in contrast to JRFL gp120 and JRFLgp120-L6-hCMP, which lost binding (Fig. 1E). The thermal stability was also monitored by CD spectroscopy. Data were analyzed assuming two-state thermal unfolding and yielded apparent melting temperatures (Tm) of 341.8 (68.7 °C) and 337.1 K (64.0 °C) for JRFL trimeric cyclic permutant and monomeric gp120, respectively (Fig. 1F).
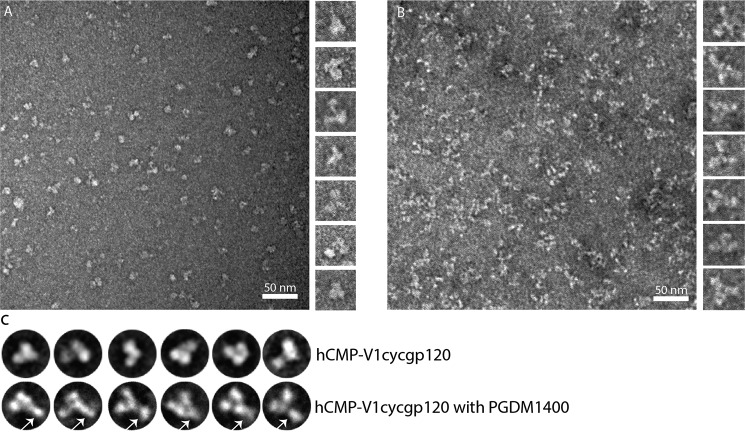
To further understand the conformation and trimeric nature of cyclically permuted gp120 trimers we subjected purified protein to negative stain EM. Visual inspection of the negative stained EM images indicates that cyclically permuted gp120 attained trimeric structures, consistent with our SEC-MALS data (Fig. 2A and supplemental Fig. S4), albeit with some degree of conformational heterogeneity. In addition, we performed negative stain EM on the cyclically permuted gp120 trimers complexed with PGDM1400 (38), a gp120 trimer-specific antibody. We observed a prominent density for the PGDM1400 antibody at the apex of the cyclically permuted trimer and one PGDM1400 antibody molecule bound to one gp120 subunit of the trimer (Fig. 2, B and C, and supplemental Fig. S4).
FIGURE 2.
Raw EM images of negative stained gp120 and gp120 complexed with mAb PGDM1400. A, raw EM image of cyclically permuted gp120 particles embedded in negative stain. Panel on the right shows projections of excised gp120 particles. B, raw EM image of particles of gp120 complexed with PGDM1400 embedded in negative stain. Panel on the right shows projections of excised particles. C, representative class averages of gp120 and gp120 complexed with mAb PGDM1400. PGDM1400 antibodies are marked with white arrows.
Cyclically Permuted gp120 Trimers Show High Affinity Binding by bNAbs
The antigenic properties of the cyclically permuted gp120 trimers were assessed by SPR using the CD4bs bNAbs (supplemental Fig. S5) VRC01, VRC03, and VRC-PG04 (39), the V3-glycan bNAb PGT128 and the quaternary epitope-specific bNAbs PGT145, PGDM1400, and CAP256-VRC26.25, which bind glycan-dependent epitopes in the V1V2 loop of gp120 (40–42). bNAbs were immobilized on the chip surface and protein immunogens were used as analytes. The cyclically permuted gp120 trimers showed high affinity binding to all three CD4bs mAbs often with undetectable dissociation rates, whereas JRFL gp120 and JRFLgp120-L6-hCMP showed measurable dissociation from the same mAbs (Table 1 and supplemental Fig. S5A). Cyclically permuted gp120 trimers showed no dissociation even after ∼200 min from VRC01 (supplemental Fig. S5B). We have previously reported that cyclically permuted gp120 trimers show improved affinity to neutralizing antibody b12 and lower affinity for non-neutralizing CD4bs antibody F105 relative to monomeric gp120 (37). The F105 binding studies were repeated (Table 1, supplemental Fig. S6A) with similar results. Binding to another CD4bs non-neutralizing mAb b6 was also characterized. b6 bound tightly to all four constructs with negligible dissociation. However, the overall extent of binding signal appeared to be measurably lower for the hCMP-V1cyc gp120 constructs (supplemental Fig. S6B).
TABLE 1.
Cyclically permuted gp120 trimers bind bNAbs with high affinity
Kinetic parameters for binding of immunogens to bNAbs VRC01, VRC03, VRCPG-04, PGT128, PGT145, PGDM1400, F105, and b6 by surface plasmon resonance. ∼1000 RUs of each IgG (ligand) was immobilized on the surface of a CM5 chip at 25 °C.
| Immunogen | Ligand | kon | koff | KD |
|---|---|---|---|---|
| m−1.s−1 | s−1 | m | ||
| JRFL gp120 | VRC01 | 1.2 ± 2.3 × 104 | 1.2 ± 0.42 × 10−3 | 1.0 ± 1.4 × 10−7 |
| VRC03 | 1.2 ± 0.1 × 104 | 3.7 ± 0.2 × 10−3 | 3.5 ± 0.3 × 10−7 | |
| VRC-PG04 | 2.6 ± 0.5 × 103 | 1.2 ± 0.1 × 10−3 | 5.6 ± 0.2 × 10−7 | |
| PGT128 | 8.9 ± 3.1 × 103 | 1.1 ± 0.4 × 10−3 | 1.2 ± 1.3 × 10−7 | |
| PGT145 | NBa | NB | NB | |
| PGDM1400 | NB | NB | NB | |
| F105 | 1.0 ± 0.1 × 105 | 3.8 ± 0.3 × 10−4 | 3.7 ± 0.3 × 10−9 | |
| b6 | 1.8 ± 1.5 × 104 | NDb | ND | |
| JRFL-hCMP-V1cyc | VRC01 | 1.2 ± 0.1 × 104 | ND | ND |
| VRC03 | 3.9 ± 0.2 × 104 | 6.0 ± 0.4 × 10−4 | 1.6 ± 0.2 × 10−8 | |
| VRC-PG04 | 4.4 ± 0.3 × 104 | ND | ND | |
| PGT128 | 4.1 ± 3.6 × 104 | 2.8 ± 0.6 × 10−4 | 6.8 ± 1.8 × 10−9 | |
| PGT145 | 1.5 ± 0.3 × 104 | 7.2 ± 2.8 × 10−3 | 4.7 ± 2.4 × 10−7 | |
| PGDM1400 | 1.5 ± 0.3 × 105 | 4.1 ± 0.4 × 10−3 | 3.7 ± 1.2 × 10−8 | |
| F105 | 1.4 ± 0.1 × 104 | 8.4 ± 1.4 × 10−4 | 6.4 ± 1.6 × 10−8 | |
| b6 | 1.1 ± 1.6 × 105 | ND | ND | |
| JRCSF-hCMP-V1cyc | VRC01 | 1.8 ± 0.8 × 104 | ND | ND |
| VRC03 | 4.0 ± 0.5 × 104 | ND | ND | |
| VRC-PG04 | 6.6 ± 0.7 × 104 | ND | ND | |
| PGT128 | 7.8 ± 2.1 × 104 | 2.2 ± 1.1 × 10−4 | 2.8 ± 1.5 × 10−9 | |
| PGT145 | 6.3 ± 1.9 × 103 | 1.0 ± 0.2 × 10−2 | 2.1 ± 0.7 × 10−6 | |
| 1.9 ± 4.2 × 104 | 7.5 ± 2.1 × 10−3 | 4.0 ± 1.6 × 10−7 | ||
| PGDM1400 | 3.3 ± 1.5 × 105 | 4.5 ± 0.2 × 10−3 | 4.0 ± 1.5 × 10−8 | |
| F105 | 10.0 ± 0.4 × 104 | 2.1 ± 0.1 × 10−3 | 2.0 ± 0.1 × 10−8 | |
| b6 | 3.4 ± 1.8 × 104 | ND | ND | |
| JRFLgp120-L6-hCMP | VRC01 | 7.7 ± 0.1 × 103 | 4.3 ± 0.5 × 10−4 | 5.6 ± 0.7 × 10−8 |
| VRC03 | 4.4 ± 0.4 × 103 | 3.6 ± 0.5 × 10−3 | 1.1 ± 1.0 × 10−6 | |
| VRC-PG04 | 8.3 ± 0.8 × 103 | 5.5 ± 0.9 × 10−4 | 5.3 ± 1.4 × 10−8 | |
| PGT128 | 2.2 ± 0.7 × 104 | 2.4 ± 0.3 × 10−3 | 1.1 ± 0.8 × 10−7 | |
| 1.6 ± 0.7 × 104 | 5.5 ± 0.2 × 10−4 | 3.4 ± 1.1 × 10−8 | ||
| PGT145 | NB | NB | NB | |
| PGDM1400 | NB | NB | NB | |
| F105 | 1.5 ± 0.1 × 104 | 4.0 ± 1.3 × 10−3 | 2.8 ± 1.1 × 10−7 | |
| b6 | 3.9 ± 0.8 × 104 | ND | ND | |
a NB, no measurable binding.
b ND, no measurable dissociation.
Cyclically permuted gp120 trimers exhibited measurable binding to PGT145, whereas JRFL gp120 and JRFLgp120-L6-hCMP did not bind to PGT145 (Table 1 and supplemental Fig. S5C). Both cyclic permutants showed a KD of ∼400 nm with biphasic dissociation rates. The trimers also bound preferentially to the trimer-specific antibodies PGDM1400 (Table 1 and supplemental Fig. S5E) as well as PG9 and PG16 (37). This data indicates that trimerization of cyclically permuted gp120 at the V1 loop retains native-like quaternary epitopes at the V1V2 region. No binding was seen to the V1V2-specific mAb CAP256-VRC26.25 (42) consistent with its known lack of ability to neutralize JRFL and JRCSF viral strains. Cyclically permuted gp120 trimers showed a 20–40-fold increase in affinity to PGT128 relative to JRFL gp120 and JRFLgp120-L6-hCMP (Table 1 and supplemental Fig. S5C). The increase in affinity was driven by both increased association rates to and decreased dissociation rates from PGT128.
Binding of CD4 receptor to Env induces conformational changes in the gp120 subunit, which exposes a cryptic co-receptor binding site and CD4-induced (CD4i) epitopes. In general, CD4i epitopes are thought to be buried in the native Env trimer and become accessible only after CD4 interaction (43). To examine whether these trimers behave like gp120 in the context of native Env, we studied the interaction of the CD4i epitope-specific antibody, 17b, to cyclically permuted gp120 trimers in the presence and absence of soluble CD4 (sCD4) (supplemental Fig. S7). In the absence of sCD4, cyclically permuted gp120 trimers show low but marginally higher binding to the 17b antibody as compared with JRFL gp120. This might be due to local conformational changes at the base of the V1V2 region (which is proximal to CD4i epitopes) caused by insertion of the hCMP trimerization insertion (supplemental Fig. S7). Upon addition of sCD4, all the gp120 derivatives expectedly show significantly enhanced binding to 17b.
It is known that the epitope for VRC01 does not overlap with quaternary epitopes in the V1V2 region targeted by bNAbs such as PGT145 (44). Because our cyclically permuted gp120 trimers are trimeric and retain binding to PGT145, we monitored sequential binding of the designed gp120 derivatives to surface-immobilized VRC01, followed by binding of PGT145 to the resulting complex (supplemental Fig. S8). Cyclically permuted gp120 trimers bound well to surface-immobilized VRC01, and the resulting complex retained binding to the PGT145 antibody (supplemental Fig. S8). In contrast, interaction of JRFL gp120 or JRFLgp120-L6-hCMP to VRC01 did not allow subsequent binding to PGT145. These results indicate that the cyclically permuted gp120 trimers adopt a native-like trimer conformation.
Immunogenicity of Cyclically Permuted gp120 Trimers in Guinea Pigs
To investigate the immunogenicity of cyclically permuted gp120 trimers relative to JRFLgp120 and JRFLgp120-L6-hCMP, a DNA prime-protein boost guinea pig immunization study was executed. We have previously shown that a similar protocol results in good antigen-specific antibody responses (45). Guinea pigs (n = 4 per group) were immunized with 2 mg of plasmid DNA encoding the respective immunogen with Adju-Phos adjuvant at weeks 0 and 4 followed by a boost with 45 μg of protein in MAA/IMX adjuvant at weeks 10.5 and 18 (Fig. 3A). Serum samples were collected 2 weeks after each immunization, and the animals were terminated 16.5 weeks after the last protein boost (Fig. 3A). The elicitation of gp120-specific antibodies was assessed by ELISA.
FIGURE 3.
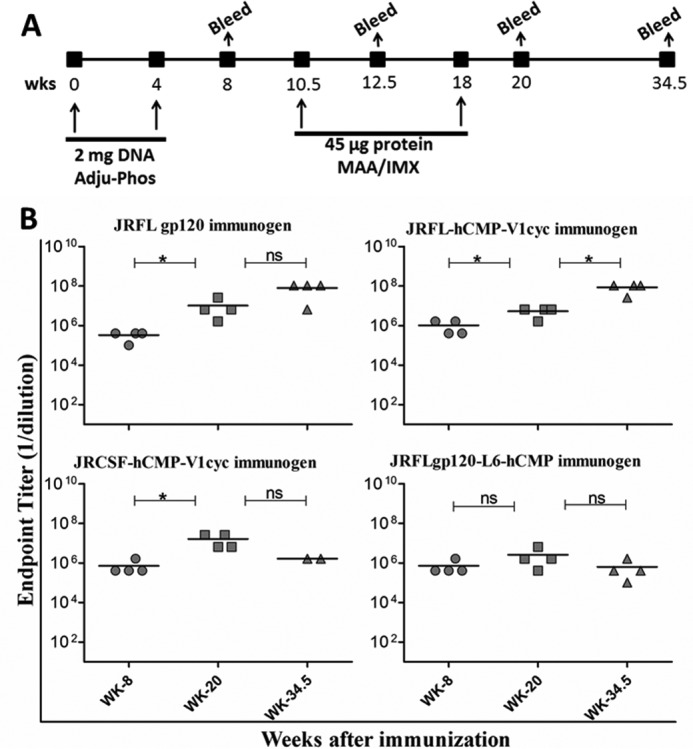
Guinea pig immunizations. A, immunization parameters and time lines of DNA prime and protein boost in guinea pigs. Immunization and bleed time points are indicated in weeks. B, gp120-specific ELISA end point titers of sera collected from immunized animals. Horizontal lines represent mean titers. Preimmune sera obtained before immunization had no detectable anti-gp120 titer. A Mann-Whitney test was performed to compare end point titers, between different time points. *, p < 0.05; ns, no significant difference (p > 0.05) between two compared groups.
Guinea pigs from all four groups generated robust IgG titers to JRFL gp120 (full-length JRFL gp120) after DNA priming and boosting with protein (Fig. 3B). Terminal bleed sera (collected at week 34.5) also maintained high end point IgG titers to JRFL gp120, in the range of ∼106 to 107 (Fig. 3B). The antisera titers were increased substantially following a protein boost (p < 0.05, Mann-Whitney test) in all animals except those immunized with JRFLgp120-L6-hCMP (Fig. 3B). Overall, there are no statistically significant differences in ELISA titers among the four different groups at any time point and the DNA prime-protein boost regimen elicited robust anti-gp120 IgG titers irrespective of the type of immunogen used.
Neutralization Studies
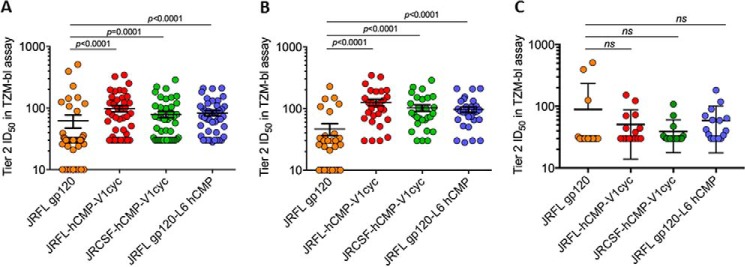
Next we assessed neutralizing activity against a panel of highly sensitive Tier 1 isolates and a multiclade global panel of Tier 2 isolates (46) in the TZM-bl assay using peak immune serum samples (week 20). All four immunogens generated high neutralizing titers (ID50 = 25,000–50,000) to the Tier 1A clade B virus MN.3, and low titers of neutralizing antibodies against the Tier 1A clade C virus, MW965.26 (Fig. 4). Notably, weak neutralizing activity against multiple Tier 2 viruses from clades B and C was detectable in all four groups but was significantly stronger in the groups immunized with cyclically permuted gp120 with mean ID50 values of 126 and 103 for groups 2 and 3, respectively (Figs. 4 and 5). We tested additional non-clade B and C Tier 2 viruses (246_F3, CNE_55, Ce703010217, and CNE8) but did not observe detectable neutralization for these (Figs. 4 and 5). To examine the persistence of the neutralization, we measured neutralization titers for sera from individual animals against two Tier 2 viruses (supplemental Fig. S9) in the week 34.5 terminal bleed sera. The neutralization titers are 2–3-fold lower than at week 20. To confirm whether the observed neutralization was Env directed, sera were adsorbed onto immobilized gp120 or trimeric cyclic permutant protein essentially as described previously (34, 47). The data (Table 2) clearly show that the observed neutralization is gp120 directed and could be adsorbed out by incubation with either immobilized gp120 or immobilized trimeric cyclic permutant.
FIGURE 4.
Antisera from cyclically permuted gp120 trimers elicit broad cross-reactive neutralizing responses. TZM-bl ID50 values (neutralizing titers) for week 20 antisera from guinea pigs in TZM-bl cell-based neutralization assays. The neutralizing activity of antisera from each group of guinea pigs was tested against Tier 1 and Tier 2 HIV-1 isolates from a global panel and additional Tier-2 isolates, which included viruses from clades A, B, C, and CRFs. The neutralization titers are corrected for their respective preimmune backgrounds. The color code indicates the following: ID50 ≤ 60 or ≤ 2-fold of MuLV neutralizing titers are in white, ID50 from 60 to 90 are in yellow, ID50 from 90 to 500 are in orange, and ID50 >500 are highlighted in red. Group number and corresponding immunogen used for vaccination are indicated in the first column.
FIGURE 5.
Relative to the other two immunogens, cyclically permuted gp120 trimers elicited significantly higher neutralizing titers against Tier 2 HIV-1 isolates. Scatter plot of neutralization ID50 values of week 20 antisera from JRFL gp120 (orange), JRFL-hCMP-V1cyc (red), JRCSF-hCMP-V1cyc (green), and JRFLgp120-L6-hCMP (blue). Error bars in the diagram represents mean ± S.E. Neutralization ID50 values of: A, all Tier 2; B, clade B and C viruses from Tier 2; and C, non-clade B and C Tier 2 viruses are represented after subtracting preimmune titers. Neutralizing ID50 titers elicited by JRFL gp120 were compared with corresponding ID50 titers elicited by other immunogens by a Mann-Whitney test.
TABLE 2.
Neutralization ID50 values obtained with pooled terminal bleed sera (week 34.5) before and after depletion
Animals in Group 1 were immunized with JRFL gp120 and those in Group 2 with JRFL-hCMP-V1cyc gp120.
| Group | ID50 in TZM-bl cells |
||
|---|---|---|---|
| JRFL | REJO | DU422 | |
| Group 1 undepleted | 48 | 81 | <30 |
| Group 2 undepleted | 156 | 167 | 123 |
| Group 1 depleted on JRFL gp120 | <30 | <30 | <30 |
| Group 2 depleted on JRFL-hCMP-V1cyc | 38 | <30 | <30 |
| Group 1 depleted on BSA | 52 | 76 | <30 |
| Group 2 depleted on BSA | 149 | 171 | 118 |
Epitope Mapping
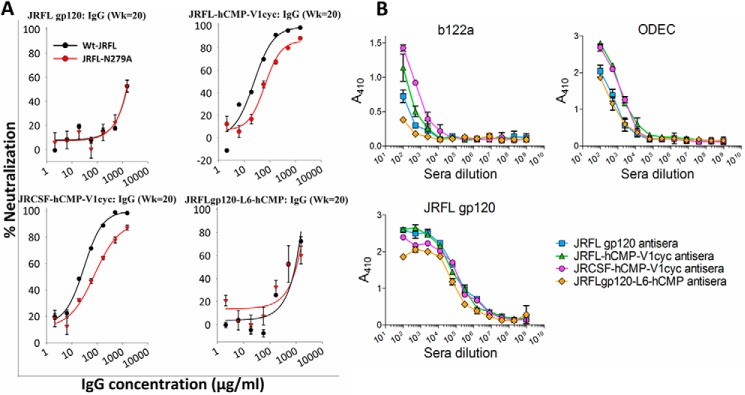
We next probed whether the antisera to cyclically permuted gp120 trimers harbor neutralizing specificities similar to known bNAbs targeting the CD4bs, quaternary epitopes in V1V2, and V3 glycan epitopes. These three neutralizing epitopes are highly conserved in the panel of HIV-1 isolates we tested. To map the neutralizing antibody specificities, we mutated important amino acid residues to abolish bNAb binding. We incorporated the N279A mutation into HIV-1 JRFL pseudotyped virus to monitor CD4bs-specific neutralizing specificities, as it was previously reported that mutating this residue diminishes the binding of CD4bs-specific neutralizing antibodies like VRC01 (40, 48) (supplemental Fig. S10). Results of TZM-bl neutralization assays indicate that introduction of this mutation led to a modest decrease in the neutralizing activity induced by cyclically permuted gp120 trimers, suggesting the presence of a fraction of CD4bs specificities (Fig. 6A). To further validate these results we tested the binding of sera from all four groups to fragment immunogens of the gp120 outer domain that harbor most of the CD4 binding site of gp120 (34, 49). Antibodies induced by cyclically permuted gp120 trimers showed substantially higher affinity to these fragment immunogens, indicating the presence of a substantial fraction of CD4bs-specific antibodies (Fig. 6B). Introducing a D368R mutation in gp120 prevents the binding of CD4 and CD4bs bNAbs to gp120 (50). Thus, we tested the binding of sera from all four groups to JRFL gp120-D368R. Only sera from cyclically permuted gp120 trimer-immunized animals showed marginally lower binding to JRFL gp120 in the presence of the D368R mutation (supplemental Fig. S11).
FIGURE 6.
Mapping of CD4 binding site specificities in antisera from cyclically permuted gp120 trimers. A, neutralization activity of IgG purified from pooled week 20 sera against JRFL and JRFL-N279A pseudoviruses. Introduction of the N279A mutation in Env makes JRFL pseudovirus insensitive to neutralization by the CD4 binding site specific, VRC01-like antibodies. The percent neutralization as a function of IgG concentration is shown. Error bars represent the S.D. from two independent experiments. Only IgG from antisera of cyclically permuted gp120 trimers showed decreased neutralization against JRFL-N279A pseudovirus indicating the presence of CD4 binding site specific neutralizing antibodies. B, binding of pooled antisera to CD4 binding site focused outer domain fragments (b122a and ODEC) and JRFL gp120. Antigens were coated directly onto ELISA plate wells and probed with pooled antisera from week 20. The absorbance at 410 nm is proportional to the amount of bound antiserum. Antisera from cyclically permuted gp120 trimers show higher binding to the CD4 binding site focused outer domain immunogens than antisera from JRFL gp20 and JRFLgp120-L6-hCMP.
We also individually mutated glycan sites in the V1V2 region (N156K and N160K) that are essential for binding of V1V2 quaternary epitope-specific bNAbs (51), and we mutated a glycan position at the base of the V3 loop region (N332T) essential for binding of V3 glycan-specific bNAbs (40, 41). These residues were individually replaced with the second most frequent residues occurring at that particular position (based on a multiple sequence alignment) to prevent the loss of infectivity of JRFL pseudoviruses (supplemental Fig. S10). Results of TZM-bl neutralization assays indicate that mutating glycans at positions 156 and 160 did not decrease the neutralizing activity of antibodies elicited by cyclically permuted gp120 trimers (supplemental Fig. S12). Unexpectedly, removal of glycan at position 156 increased the neutralizing activity of sera from all four groups, perhaps due to unmasking epitopes that remain to be defined (supplemental Fig. S12). Mutating the glycosylation site at the V3 loop base (N332T) had no effect on neutralization. These results suggest that sera from cyclically permuted gp120 trimer-immunized animals do not have neutralizing specificities against V2-glycan or V3-glycan quaternary epitopes.
Discussion
Despite considerable effort, there has been limited success in designing immunogens that elicit potent and bNAbs against HIV-1. The best trimeric HIV-1 Env immunogens designed to date elicit potent Tier 1 and autologous Tier 2 neutralizing antibody responses (33, 52) but the antibodies exhibit little activity against heterologous Tier 2 HIV-1 isolates. These trimeric HIV-1 Env immunogens (33, 52) were derived from the ectodomain of the BG505 subtype A isolate, contain an engineered disulfide bond between gp120 and gp41 subunits, and lack the membrane proximal external region of gp41. In the case of other trimeric gp120 and trimeric gp140 immunogens, a heterologous trimerization domain was fused at the C terminus of gp120 or gp140 (8, 24, 29, 31). Relative to monomeric gp120, these trimeric immunogens showed a marginal improvement in eliciting neutralizing responses, but also failed to neutralize Tier 2 viruses from heterologous strains. During acute infection, it was shown that the initial antibody responses are gp41 directed, non-neutralizing (53) and derived from polyreactive B cells that bind both Env and intestinal microbiota (54). In a recent study (14) it was shown that vaccination with Env ectodomain (gp140) (HVTN clinical trials) also led to an antibody response dominated by gp41 reactive antibodies that were derived from pre-existing pools of intestinal microbiota cross-reactive B-cells (14). These results indicate that Env-based vaccines containing gp41 primarily induce non-neutralizing gp41 reactive antibodies (14, 20, 55). In the present study, we therefore omitted the gp41 subunit from the immunogen.
We further rationally engineered a trimerized gp120 molecule by inserting a heterologous trimerization domain in the V1 loop region to mimic native gp120 trimers in the pre-fusion, native state of HIV-1 Env. Initial attempts to incorporate an hCMP trimerization domain into the V1 loop region of monomeric gp120 were unsuccessful although the resulting constructs were soluble. We circumvented this problem by creating new chain termini within the V1 loop region of gp120 by cyclically permuting the gp120 molecule, followed by fusion of the hCMP domain to the new N terminus in the V1 loop. This resulted in the formation of soluble, homogeneous, stable gp120 trimers. The fraction of monomers and other species was small. Hence, apart from lectin affinity purification, no additional purification steps were required. These trimers display high affinity for bNAbs against CD4bs, V2-glycan, and V3-glycan epitopes on gp120 (Table 1). Moreover, the cyclically permuted gp120 trimers induced neutralizing activity against multiple isolates in a global panel of heterologous Tier 2 viruses that, although relatively weak, was clearly detectable and significantly stronger than the response induced by monomeric gp120. We attempted to trimerize gp120 that was cyclically permuted in both the V1 loop (142–144) and the V2 loop (185–186) regions where mutations and insertions are tolerated (supplemental Fig. S1). Both constructs yielded soluble trimers but the V1 loop cyclically permuted gp120 trimers display superior antigenic properties to the V2 loop trimer. In addition, we attempted to add trimerization domains to cyclically permuted gp120 at both the V1 loop and within the linker connecting the original C termini of gp120 (hCMP domain at V1 loop and foldon domain at C terminus). However, this construct did not yield stable trimers.
In the past few years, multiple cryo-EM and X-ray crystallographic structures have been solved for disulfide-stabilized Env trimers (4–6, 35). The 142–144 (V1cyc) and 185–186 (V2cyc) positions lie within loops in the V1V2 region (Fig. 7A). These constructs were designed (37) before crystal structures of the gp140 trimer were solved. Recent structures of the BG505 SOSIP.664 gp140 trimer (4, 5, 35) show that the region from 135 to 153 forms a flexible loop with the apex at residue 140. In the BG505 isolate there is a deletion of residues 142–149 relative to JRFL and HXBc2. In an extended conformation the 10-residue stretch from 144 to 154 will span ∼36 Å and fusion of hCMP to the residue 144, which is the new N terminus in V1cyc, should allow enough conformational freedom for the hCMP trimerization domains to align with the Env trimer axis. The 142–144 position of the V1 loop, where we inserted the hCMP trimerization motif, is a conformationally flexible region and tolerates insertions (supplemental Fig. S1). We therefore anticipated that the insertion of trimerization motifs at this region would not prevent binding of the trimer-specific V1V2 NAbs, consistent with our SPR data. In addition, the negative stain EM data clearly confirms the presence of PGDM1400 bound to the cyclically permuted gp120 trimer (Fig. 2 and supplemental Fig. S4). It is worth noting that the glycosylation sites (Asn-156 and Asn-160) that constitute a major part of V1V2 trimer-specific epitopes are intact in these cyclically permuted gp120 trimers.
FIGURE 7.
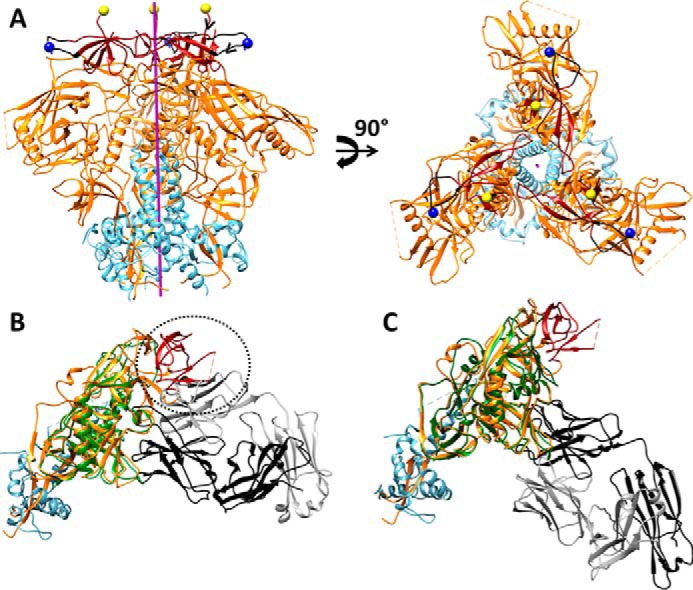
A, approximate positions of new N termini of gp120 in V1cyc and V2cyc, respectively, mapped onto the structure of the BG505 SOSIP.664 HIV-1 Env trimer (Protein Data Bank code 4TVP). The color scheme of the structures is as follows: gp120 (orange), gp41 (cyan), V1V2 region (dark red), residue 141 of V1 region (blue spheres), residue 187 of V2 region (yellow spheres), and region 136–154 of V1 loop (black). The trimer axis is represented as a purple rod. The hCMP trimerization domain was fused to the above N-terminal positions to trimerize gp120. The chain direction at the N termini of V1cyc and V2cyc are indicated by black arrowheads. Residue 144 (the actual N terminus of V1cyc) in JRFL is absent in BG505 because of a deletion of residues 142–149. Hence residue 141 is shown as the N terminus. B, superimposition of core gp120 bound to CD4bs non-neutralizing F105 Fab (PDB code 3HI1) onto gp140 from the BG505 Env trimer (PDB code 4TVP). The color scheme of the structure is as follows: F105 bound gp120 (green), V1V2 region (dark red), and the remaining gp120 subunit from BG505 Env trimer (orange), gp41 from BG505 Env trimer (cyan), F105 Fab light chain (gray), and heavy chain (black) of F105 antigen binding fragment Fab. Inability of F105 to bind gp120 in the presence of the V1V2 region is indicated with a dotted circle demonstrating steric overlap between the F105 light chain and V1V2 in the superimposed structure. C, superimposition of core gp120 bound to CD4bs neutralizing VRC01 Fab (PDB code 3NGB), onto gp140 from the BG505 Env trimer (PDB code 4TVP) shows the absence of any steric clash between the Fab and native trimer. The color scheme of the structure is as follows: VRC01 bound gp120 (green), V1V2 region (dark red), and the gp120 subunit (orange) from BG505 Env trimer, gp41 from BG505 Env trimer (cyan), VRC01 Fab light chain (gray), and heavy chain (black).
A recent study shows that native, pre-fusion Env on the HIV-1 virion surface is dynamic with significant motions occurring in variable loop (V1V2V3) regions (35). Conformational fluctuations in such a dynamic structure likely result in transient exposure of immunodominant, non-neutralizing epitopes on the Env trimer. The cyclically permuted gp120 trimers designed in the present study have a trimerization domain in the V1V2 loop that should reduce the conformational fluctuations both in these loops and the spatially proximal V3 loop and reduce access to the CD4bs by non-neutralizing antibodies like F105 and b13 (Fig. 7B), but not the neutralizing antibodies like VRC01 and PGV04 (Fig. 7C). This effect would not occur in conventional gp140 derivatives where trimerization is largely mediated by interactions between gp41 subunits, distal from V1 and V3 loops.
V1 loop cyclically permuted gp120 trimers based on JRFL and JRCSF retain significant binding to native, quaternary epitopes in the V1V2 loop, and bind to bNAbs PGT145 and PGDM1400 (Table 1) as well as PG9 and PG16 (37). They also showed high affinity to the CD4b-specific bNAbs, VRC01, VRC03, and VRC-PG04 (Table 1) and decreased binding to the non-neutralizing CD4bs antibody, F105 (Table 1). In addition, in contrast to the JRFLgp120-L6-hCMP construct, they also retained binding to PGT145 antibody after VRC01 interaction, implying that these soluble gp120 trimers appeared to attain a native-like conformation. The trimeric cyclic permutants bind PGT145 with poorer affinity than the BG505-SOSIP-664 trimers possibly due to the lack of the gp41 subunit, or due to the presence of the hCMP domain proximal to the V1V2 epitope regions. Moreover, we observed that PGT145 binding is enhanced in the presence of the CD4bs bNAb, VRC01.
The cyclically permuted gp120 trimers were thermostable and retained binding to bNAbs even after incubation at 60 °C for 1 h. The designed gp120 trimers were used to immunize guinea pigs. Four different antigens were tested, namely monomeric gp120, JRFL-hCMP-V1cyc, JRCSF-hCMP-V1cyc, and JRFLgp120-L6-hCMP. Following two DNA primes and two protein boosts, high titers of antigen-specific antibodies were obtained in all four groups. Previous studies demonstrated high gp120 binding titers (45, 56, 57) when a DNA prime-protein boost strategy was adopted.
Neutralization assays with an HIV-1 global panel, representing Tier 2 HIV-1 isolates from diverse geographic regions, demonstrated that antisera from cyclically permuted gp120 trimers neutralized several heterologous Tier 2 HIV-1 isolates, which are genetically diverse. The neutralization profiles of these antisera were superior to those elicited by the other two immunogens tested (JRFL gp120 and JRFLgp120-L6-hCMP). Immunization with clade C trimeric gp140 immunogens in guinea pigs was shown to elicit antisera with significant neutralizing activity to homologous clade C Tier 2 HIV-1 isolates in a highly sensitive A3R5 neutralization assay (31). Immunization with a JRFL-Env DNA prime and JRFL gp140 trimeric protein boost in primates elicited antisera with robust neutralizing activity to Tier-1 isolates, and sporadic neutralizing titers to Tier-2 isolates in a TZM-bl assay. The sera showed higher neutralization of clade B and C Tier 2 isolates in more sensitive A3R5 assays with neutralizing specificities directed predominantly to the CD4bs (32). Very recently, it was shown that immunization of rabbits with structurally well characterized, native-like BG505 SOSIP gp140 trimers elicited robust, cross-clade Tier-1 and autologous Tier 2 neutralizing titers. Most of the neutralizing activity was specific to the CD4bs and trimer apex (V1V2 regions) of gp120 (33). Consistent with these studies, our results demonstrate that it is possible to elicit CD4bs-specific bNAbs by using trimeric Env immunogens.
Mapping of serum neutralization specificities of antisera from cyclically permuted gp120 trimers indicate that these antisera harbor a moderate fraction of CD4bs-specific neutralizing responses, but not quaternary epitope and glycan-specific neutralizing titers. Consistent with the above mentioned studies, our results clearly suggest that the CD4bs are potentially immunogenic on native-like gp120 trimers. Although V1V2 glycan quaternary and V3 glycan-based epitopes are surface accessible on the gp120 trimer, we could not detect neutralizing antibody responses targeting known bNAb epitopes in these regions. The Tier 2 HIV-1 isolates, used for determining the breadth of neutralizing activity, contain conserved linear epitopes in the V2 and V3 regions of gp120 (supplemental Fig. S13), which are surface accessible and immunodominant in nature. It is possible that antibodies against these epitopes might have contributed to the neutralization breadth of cyclically permuted gp120 antisera. The conserved V2 region in these Tier 2 HIV-1 isolates harbor a high number of positively charged residues, which were shown to be essential for binding to V2-specific antibodies in the RV144 clinical trial (58, 59).
In conclusion, this study describes the antigenicity and immunogenicity of native-like cyclically permuted gp120 trimers in small animals. Encouragingly, neutralizing responses could be elicited with a relatively short duration immunization schedule. A measurable fraction of the Tier 2 virus neutralizing activity appears to be directed to the CD4bs. Although these data are encouraging, there are several avenues to explore in future studies. The present trimers still bind the non-neutralizing CD4bs antibodies F105 and b6 albeit more weakly than more monomeric gp120 (37) and show conformational heterogeneity in EM. In addition, the immunogenicity studies employed a modest number of four animals per group, and with the guinea pig model, monoclonal antibody isolation is not possible.
Although encouraging heterologous Tier 2 neutralization is observed, the breadth needs to be improved. In natural infection broad neutralization is observed after years of infection (2) and ongoing replication and viral evolution drive this process. It is possible that sequential immunizations with cyclic permutant constructs designed to engage appropriate B-cell precursors (60) and with diverse sequences from multiple clades might improve the neutralization breadth.
Future directions will include antigen optimization, employing alternative sites for cyclic permutation, using sequences from other subtypes, varying the nature and locations of trimerization domains used, and incorporation of other stabilizing mutations to elicit a more potent and broad NAb response to circulating HIV-1 isolates. Future immunization studies will also involve non-human primates to better validate the utility of the cyclic permutant approach for HIV-1 Env immunogen design.
Experimental Procedures
Construct Descriptions
All constructs were codon optimized for expression in mammalian cell lines and were cloned into the V1Jns vector between BglII and KpnI sites (45). In the V1Jns vector, the protein coding genes were cloned under control of the CMV promoter with a tpa leader sequence. The gp120 sequences used to design constructs are derived from JRFL or JRCSF gp120 sequences. The E168K mutation was introduced in the sequence to facilitate recognition of PG9/PG16 and PGT145 NAbs (51). The hCMP trimerization domain used in this study is a 43-residue coiled-coil domain derived from human cartilage matrix protein (61) that contains two cysteine residues N-terminal to the coiled-coil, which form inter-chain disulfide bonds. We designed four different immunogens (i) JRFL gp120, (ii) JRFL-hCMP-V1cyc, (iii) JRCSF-hCMP-V1cyc, and (iv) JRFLgp120-L6-hCMP. All of these constructs lack the C1 region of gp120 (amino acids 31–82). JRFL-hCMP-V1cyc and JRCSF-hCMP-V1cyc were made by connecting the native N and C termini of gp120 with a 20-residue flexible linker and by creating new N and C termini in the V1 loop region at residues 144 and 142, respectively, followed by fusion of the hCMP domain to the N terminus. In the JRFLgp120-L6-hCMP construct, a 6-residue linker and the hCMP domain were fused to the C terminus of JRFL gp120.
To map neutralizing specificities of antisera from immunized animals, we introduced N279A, N156K, N160K, and N332T mutations individually by site-directed mutagenesis into the pSVIII-JRFL gp160dCT plasmid, which encodes for envelope glycoprotein (62). The mutant plasmids were used for making pseudoviruses as described in a later section.
Purification of Proteins and Analysis by BN-PAGE and Western Blot
All constructs were transiently transfected into HEK 293T cells using polyethyleneimine transfection reagent. After 6–8 h of transfection, media was replaced with serum-free medium. At 72 h post-transfection, supernatants were collected and protein was purified by affinity chromatography using lentil lectin-Sepharose 4B (GE Healthcare). Bound protein was eluted with 600 mm α-d-methylmannopyranoside in phosphate-buffered saline (PBS) (pH 7.4). The eluted fractions were pooled and dialyzed against PBS (pH 7.4). The purified protein concentrations were estimated by using a BCA assay (Sigma). Concentrations were confirmed using both quantitative Western blotting with gp120 of known concentration as a standard and by far-UV CD spectroscopy as described (45). The proteins were stored at −80 °C until further use.
BN-PAGE was performed to determine the oligomeric state of proteins. In BN-PAGE, protein samples were mixed with sample buffer (250 mm MOPS, 250 mm Tris-HCl, 0.1% Coomassie Brilliant Blue G-250, and 40% glycerol). Samples were loaded onto a 4–12% gradient BisTris NuPAGE gel (Invitrogen). Electrophoresis was done at 4 ºC for 4 h at 100 V. The cathode buffer was comprised of 50 mm MOPS, 50 mm Tris (pH 7.7), and 0.002% Coomassie Brilliant Blue G-250. The anode buffer contained 50 mm MOPS and 50 mm Tris (pH 7.7). The BN-PAGE gels were stained with Coomassie-colloidal blue staining reagent (Invitrogen). For Western blot analysis, following BN-PAGE, proteins were electrophoretically transferred onto a PVDF (polyvinylidene difluoride) membrane. After transfer, the PVDF membrane was blocked with 5% nonfat milk. The membrane was washed with PBST (PBS with 0.05% Tween 20) and incubated with anti-JRCSF gp120 polyclonal sera generated in rabbits, at 1:1000 dilution. The membranes were washed again with PBST and then incubated with anti-rabbit IgG conjugated to HRP (horseradish peroxidase) (Sigma). After washing with PBST, the membrane was incubated the ClarityTM Western ECL substrate (Bio-Rad) at room temperature for ∼2 min.
Gel Filtration Analysis
The oligomeric states of purified proteins were analyzed using analytical gel filtration chromatography. 60 μg of native protein in PBS (pH 7.4) at room temperature was subjected to analytical gel filtration chromatography on a Superdex-200 column. Standard calibration markers, namely conalbumin (75 kDa), aldolase (158 kDa), β-amylase (200 kDa), ferritin (440 kDa), and thyroglobulin (669 kDa) were used to determine the expected position of the trimeric, dimeric, and monomeric peaks. JRFL gp120 (obtained from the Neutralizing Antibody Consortium) having a mass of 120 kDa was also used as a marker.
SEC-MALS
All the proteins were subjected to HPLC (Shimadzu liquid chromatography) using a Superdex-200 analytical gel filtration column (GE Healthcare) equilibrated in PBS (pH 7.4) buffer coupled to UV (Shimadzu), MALS (miniDAWN TREOS, Wyatt Technology), and refractive index detectors (Waters) for molecular mass determination. For each run, 100 μg of each protein was injected. UV, MALS, and RI data were collected and analyzed using ASTRATM software (Wyatt Technology).
SPR Using Biacore-2000
SPR experiments were performed with a Biacore-2000 optical biosensor (Biacore, Uppsala, Sweden) at 25 ºC. Binding kinetics of designed proteins were assessed with VRC01, VRC03 (63), VRC-PG04 (64), PGT128, PGT145, PGDM1400 (38), CAP256-VRC26.25 (42), F105, b6, and 17b antibodies. Antibodies were immobilized on a CM5 sensor chip at a surface density of ∼1000 RUs. Various concentrations of gp120 constructs were passed at a flow rate of 30 μl/min over the sensor chip. The sensor chip was regenerated after each binding reaction with 4 m MgCl2. The kinetic parameters for interactions with different antibodies were obtained by fitting the data to a 1:1 Langmuir interaction model using Bia Evaluation software. Because the binding of some of the proteins to PGT128 and PGT145 are biphasic, the dissociation rates were obtained by fitting the data to a double exponential decay equation in SigmaPlot 10.0.
For monitoring stability, the purified proteins were heated to 60 and 100 °C for 1 h in a PCR cycler (Bio-Rad) with a heated lid to prevent evaporation. The protein samples were cooled to room temperature and binding affinity to VRC01 and PGT128 bNAbs was determined by SPR experiments. In addition, the thermal stability was characterized using CD spectroscopy by monitoring the ellipticity at 230 nm as a function of temperature essentially as described previously (65, 66).
For sequential injection experiments, VRC01 antibody was immobilized on a CM5 sensor chip. The individual proteins were sequentially injected along with PGT145 antibody over the VRC01 surface to monitor the sequential interaction of protein immunogens and PGT145 antibody with the VRC01 antibody.
Immunization Studies
Four groups, each having 4 guinea pigs were used for immunization studies. Immunogens for groups 1–4 were JRFL gp120, JRFL-hCMP-V1cyc, JRCSF-hCMP-V1cyc, and JRFLgp120-L6-hCMP, respectively. Immunogens were given as a DNA prime and protein boost. For priming, all guinea pigs were injected intramuscularly at weeks 0 and 4 with 2 mg of DNA in Adju-Phos adjuvant by using a Biojector-2000 (Bioject). For boosting, all guinea pigs were intramuscularly injected with 45 μg of protein adjuvanted with MAA (Merck amorphous aluminum hydroxylphosphate sulfate adjuvant) and IMX (ISCOMATRIX®, CSL) at weeks 10.5 and 18. At 34.5 weeks after the first prime, the guinea pigs were terminated. Serum samples were collected at weeks 0, 8, 12.5, 20, and 34.5, heat-inactivated, and stored in aliquots for further analysis.
Sample Preparation and Negative-stained EM Imaging of Samples
The JRFL-hCMP-V1cyc and JRFL-hCMP-V1cyc with PGDM1400 samples were prepared for EM using the conventional negative staining protocol. Briefly, carbon-coated copper grids were glow-discharged for 1 min and negative staining was carried out using 2% uranyl acetate. All samples were visualized and imaged at room temperature with a Tecnai T12 electron microscope operated at 120 kV. Images were collected at a magnification of ×87,000 and a defocus value of ∼1.3 μm on a side-mounted Olympus VELITA (2Kx2K) CCD camera. All images were collected at a pixel size of 2.97 Å on the specimen level.
Two-dimensional Classifications of JRFL-hCMP-V1cyc gp120 and JRFL-hCMP-V1cyc gp120 with PGDM1400
Raw particle projections were interactively selected and excised using e2boxer.py and e2projectmanager.py (EMAN2.1 software (67)). The two-dimensional reference-free alignment and classification of particle projections were performed using e2refine2d.py.
ELISA (Enzyme-linked Immunosorbent Assay) to Determine Antisera Titers Against JRFL gp120 and CD4 Binding Site, Stabilized Outer Domain Immunogens
To determine serum IgG titers elicited by different gp120 constructs, 2 μg/ml of HIV-1 gp120 (JRFL) in PBS (pH 7.4) was coated at 4 °C overnight onto Immulon 4HBX plates (50 μl/well). gp120-coated wells were washed 6 times with PBS containing 0.05% Tween 20 (0.05% PBST) and blocked with a solution of PBS containing 0.05% Tween 20 and 3% nonfat dried milk (blocking buffer). Guinea pig serum samples collected at weeks 8, 12.5, 20, or 34.5 were 4-fold serially diluted from 1:100 to 1:1.05 × 108, and diluted sera was added to antigen-coated wells. Following a 2–4-h incubation at room temperature, plates were washed 6 times with 0.05% PBST, and HRP goat anti-guinea pig IgG Fc secondary antibody (Jackson ImmunoResearch) was added at 1:5000 dilution in blocking buffer. Following a 1-h room temperature incubation, plates were washed 6 times with 0.05% PBST and developed with TMB substrate solution (Thermo Scientific). The reaction was stopped by addition of 2 n sulfuric acid, and absorbance of each well was measured at 450 nm.
To screen binding of the CD4 binding site-stabilized outer domain immunogens (b122a, ODEC) (34, 49), 96-well plates were coated overnight at 4 °C with 25 μl of PBS containing 300 ng of protein per well. The wells were washed four times with PBS containing 0.025% Tween 20 (PBST) and blocked with 3% BSA at room temperature for 1 h. Serial dilutions of pooled antisera of each group of animals from week 20 were added to antigen-coated wells, incubated for 2 h at room temperature, and washed 4 times with PBST. Binding was probed with alkaline phosphatase-conjugated goat anti-guinea pig IgG (Sigma) at 1:10,000 dilution. The plate was incubated at room temperature for 2 h, washed four times with PBST, and developed by adding 40 μl of alkaline phosphatase substrate. The absorbance (OD) was measured at 410 nm.
Serum Depletion Studies
12.5 mg of Dynabeads (Invitrogen) were coupled to 0.5 mg of JRFL gp120 or JRFL-hCMP-V1cyc protein. FACS was used to check the antigenic integrity of the bound protein as reported (34, 47). 500 μl of pooled terminal bleed sera (diluted 1:5 in DMEM) were incubated with 12.5 mg of JRFL gp120-coupled magnetic beads for 2 h at room temperature with gentle rocking. The flow-through was incubated with another 12.5 mg of gp120-coupled magnetic beads. The flow-through collected after the second round of binding is the depleted serum. The same protocol was used for depletion on JRFL-hCMP-V1cyc protein. Sera before and after depletions were used for neutralization assays using Group 1 (monomeric) and Group 2 (trimeric) terminal bleed sera.
Neutralization Assays
Neutralization assays of antisera from guinea pigs were performed in TZM-bl cells as described (68). Pseudoviruses capable of a single round of infection were produced by co-transfection of HEK 293T cells with a genomic plasmid, pSG3Δenv, and an Env expression plasmid. Following transfection, supernatant containing pseudovirus was harvested and used to infect TZM-bl cells in a neutralization assay. TZM-bl cells are genetically engineered HeLa cell lines that constitutively express the CD4 receptor and the CCR5 and CXCR4 co-receptors. TZM-bl cells also contain a Tat-regulated reporter gene for firefly luciferase under the regulatory control of an HIV-1 long terminal repeat sequence. 100,000 to 150,000 RLU equivalents of virus were incubated with serial dilutions of serum for 1 h at 37 °C in a total volume of 150 μl of DMEM growth medium. TZM-bl cells were harvested by trypsinization and 10,000 cells in 100 μl of growth medium containing 30 μg/ml of DEAE-dextran were added to each well containing a pseudovirus pre-mixed with serum. After 48 h of incubation at 37 ºC, 150 μl of culture volume from each well was removed and 100 μl of Bright Glo reagent (Promega or PerkinElmer Life Sciences) was added. After a 2-min incubation at room temperature for cell lysis, 150 μl of the cell lysate was transferred to a 96-well black plate for luminescence measurements. Neutralization titers (ID50) represent the sample dilution at which the relative luminescence units (RLUs) are reduced by 50% compared with RLU in virus control wells after subtraction of background RLU in cell control wells.
Statistical Analysis
The p values for differences in antibody binding and neutralization titers were generated by analyzing the data with a two-tailed Mann-Whitney test using the GraphPad Prism software.
Author Contributions
S. K. and R. V. designed the immunogens; J. G. J. and J. A. F. planned the immunization experiments; S. K. and R. Das carried out immunogen expression, biochemical and biophysical characterization, and epitope mapping; M. C., R. Das, L. E., and D. D. S. carried out immunizations and serum titer estimations; R. S. carried out the gp120 ELISAs; C. C. L. and D. C. M. were responsible for neutralization assays with undepleted sera. R. Datta carried out neutralization assays with depleted sera. N. S. contributed to SPR data collection. S. D. carried out EM data acquisition and analysis. S. K. and R. V. wrote the original draft; S. K., R. V., J. A. F., C. C. L, and D. C. M. edited the draft.
Supplementary Material
Acknowledgments
We thank the Neutralizing Antibody Consortium and the National Institutes of Health AIDS Research and Reference Reagent Program for providing sCD4, HIV-1 envelope-directed monoclonal antibodies b12, b6, F105, 17b, PGT128, and PGT145 and Standard Reference Panels of Subtype B and Subtype C HIV-1 Env clones. We thank members of the R. V. laboratory, Drs. V. Vamsee Aditya Mallajosyula, Ujjwal Rathore, Nilesh Aghera, Tariq Najar, and Chetana Baliga for helpful discussions. We thank Aparna Asok, Devanarayanan Sivasankar, and Uddipan Kar for help with thermal stability measurements. We thank Satyavati of the Department of Microbiology and Cell Biology and Govindswamy of the Advanced Facility for Microscopy and Microanalysis, IISc, for assistance with electron microscopic data acquisition. We are grateful to Dr. Dennis Burton for providing the broadly neutralizing antibody PGDM1400, and Dr. John Mascola for providing the antibodies VRC01, VRC03, VRC-PG04, and CAP256-VRC26.25.
This work was supported in part by the Departments of Biotechnology and Science and Technology Government of India, the International AIDS Vaccine Initiative, and National Institutes of Health NIAID Grant R01AI118366 (to R. V.), and National Institutes of Health NIAID Contract HHSN27201100016C (to D. C. M.) for the neutralizing antibody assays. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Figs. S1–S13.
- NAb
- neutralizing antibodies
- CD4bs
- CD4 binding site
- hCMP
- human cartilage matrix protein
- SEC
- size exclusion chromatography
- MALS
- multiangle light scattering
- SPR
- surface plasmon resonance
- sCD4
- soluble CD4
- RLU
- relative luminescence unit
- RU
- response unit
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
References
- 1. Burton D. R., Ahmed R., Barouch D. H., Butera S. T., Crotty S., Godzik A., Kaufmann D. E., McElrath M. J., Nussenzweig M. C., Pulendran B., Scanlan C. N., Schief W. R., Silvestri G., Streeck H., Walker B. D., et al. (2012) A blueprint for HIV vaccine discovery. Cell Host Microbe 12, 396–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Burton D. R., and Mascola J. R. (2015) Antibody responses to envelope glycoproteins in HIV-1 infection. Nat. Immunol. 16, 571–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu J., Bartesaghi A., Borgnia M. J., Sapiro G., and Subramaniam S. (2008) Molecular architecture of native HIV-1 gp120 trimers. Nature 455, 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Julien J. P., Cupo A., Sok D., Stanfield R. L., Lyumkis D., Deller M. C., Klasse P. J., Burton D. R., Sanders R. W., Moore J. P., Ward A. B., and Wilson I. A. (2013) Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342, 1477–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lyumkis D., Julien J. P., de Val N., Cupo A., Potter C. S., Klasse P. J., Burton D. R., Sanders R. W., Moore J. P., Carragher B., Wilson I. A., and Ward A. B. (2013) Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342, 1484–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bartesaghi A., Merk A., Borgnia M. J., Milne J. L., and Subramaniam S. (2013) Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat. Struct. Mol. Biol. 20, 1352–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Merk A., and Subramaniam S. (2013) HIV-1 envelope glycoprotein structure. Curr. Opin. Struct. Biol. 23, 268–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rathore U., Kesavardhana S., Mallajosyula V. V., and Varadarajan R. (2014) Immunogen design for HIV-1 and influenza. Biochim. Biophys. Acta 1844, 1891–1906 [DOI] [PubMed] [Google Scholar]
- 9. Stamatatos L., Morris L., Burton D. R., and Mascola J. R. (2009) Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat. Med. 15, 866–870 [DOI] [PubMed] [Google Scholar]
- 10. Mascola J. R., and Haynes B. F. (2013) HIV-1 neutralizing antibodies: understanding nature's pathways. Immunol. Rev. 254, 225–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kwong P. D., Mascola J. R., and Nabel G. J. (2013) Broadly neutralizing antibodies and the search for an HIV-1 vaccine: the end of the beginning. Nat. Rev. Immunol. 13, 693–701 [DOI] [PubMed] [Google Scholar]
- 12. Blattner C., Lee J. H., Sliepen K., Derking R., Falkowska E., de la Peña A. T., Cupo A., Julien J. P., van Gils M., Lee P. S., Peng W., Paulson J. C., Poignard P., Burton D. R., Moore J. P., et al. (2014) Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity 40, 669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van Gils M. J., and Sanders R. W. (2013) Broadly neutralizing antibodies against HIV-1: templates for a vaccine. Virology 435, 46–56 [DOI] [PubMed] [Google Scholar]
- 14. Williams W. B., Liao H. X., Moody M. A., Kepler T. B., Alam S. M., Gao F., Wiehe K., Trama A. M., Jones K., Zhang R., Song H., Marshall D. J., Whitesides J. F., Sawatzki K., Hua A., et al. (2015) HIV-1 VACCINES. Diversion of HIV-1 vaccine-induced immunity by gp41-microbiota cross-reactive antibodies. Science 349, aab1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ruprecht C. R., Krarup A., Reynell L., Mann A. M., Brandenberg O. F., Berlinger L., Abela I. A., Regoes R. R., Günthard H. F., Rusert P., and Trkola A. (2011) MPER-specific antibodies induce gp120 shedding and irreversibly neutralize HIV-1. J. Exp. Med. 208, 439–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kesavardhana S., and Varadarajan R. (2014) Stabilizing the native trimer of HIV-1 Env by destabilizing the heterodimeric interface of the gp41 postfusion six-helix bundle. J. Virol. 88, 9590–9604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rerks-Ngarm S., Pitisuttithum P., Nitayaphan S., Kaewkungwal J., Chiu J., Paris R., Premsri N., Namwat C., de Souza M., Adams E., Benenson M., Gurunathan S., Tartaglia J., McNeil J. G., Francis D. P., et al. (2009) Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361, 2209–2220 [DOI] [PubMed] [Google Scholar]
- 18. Lewis G. K., DeVico A. L., and Gallo R. C. (2014) Antibody persistence and T-cell balance: two key factors confronting HIV vaccine development. Proc. Natl. Acad. Sci. U.S.A. 111, 15614–15621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Montefiori D. C., Karnasuta C., Huang Y., Ahmed H., Gilbert P., de Souza M. S., McLinden R., Tovanabutra S., Laurence-Chenine A., Sanders-Buell E., Moody M. A., Bonsignori M., Ochsenbauer C., Kappes J., Tang H., et al. (2012) Magnitude and breadth of the neutralizing antibody response in the RV144 and Vax003 HIV-1 vaccine efficacy trials. J. Infect. Dis. 206, 431–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haynes B. F., Gilbert P. B., McElrath M. J., Zolla-Pazner S., Tomaras G. D., Alam S. M., Evans D. T., Montefiori D. C., Karnasuta C., Sutthent R., Liao H. X., DeVico A. L., Lewis G. K., Williams C., et al. (2012) Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 366, 1275–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Johnston M. I., and Fauci A. S. (2007) An HIV vaccine: evolving concepts. N. Engl. J. Med. 356, 2073–2081 [DOI] [PubMed] [Google Scholar]
- 22. Roederer M., Keele B. F., Schmidt S. D., Mason R. D., Welles H. C., Fischer W., Labranche C., Foulds K. E., Louder M. K., Yang Z. Y., Todd J. P., Buzby A. P., Mach L. V., Shen L., Seaton K. E., et al. (2014) Immunological and virological mechanisms of vaccine-mediated protection against SIV and HIV. Nature 505, 502–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Barouch D. H., Alter G., Broge T., Linde C., Ackerman M. E., Brown E. P., Borducchi E. N., Smith K. M., Nkolola J. P., Liu J., Shields J., Parenteau L., Whitney J. B., Abbink P., Ng'ang'a D. M., et al. (2015) HIV-1 vaccines: protective efficacy of adenovirus/protein vaccines against SIV challenges in rhesus monkeys. Science 349, 320–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang X., Farzan M., Wyatt R., and Sodroski J. (2000) Characterization of stable, soluble trimers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 74, 5716–5725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sanders R. W., Schiffner L., Master A., Kajumo F., Guo Y., Dragic T., Moore J. P., and Binley J. M. (2000) Variable-loop-deleted variants of the human immunodeficiency virus type 1 envelope glycoprotein can be stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits. J. Virol. 74, 5091–5100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Binley J. M., Sanders R. W., Clas B., Schuelke N., Master A., Guo Y., Kajumo F., Anselma D. J., Maddon P. J., Olson W. C., and Moore J. P. (2000) A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74, 627–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sanders R. W., Derking R., Cupo A., Julien J. P., Yasmeen A., de Val N., Kim H. J., Blattner C., de la Peña A. T., Korzun J., Golabek M., de Los Reyes K., Ketas T. J., van Gils M. J., King C. R., et al. (2013) A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 9, e1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang X., Lee J., Mahony E. M., Kwong P. D., Wyatt R., and Sodroski J. (2002) Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J. Virol. 76, 4634–4642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pancera M., Lebowitz J., Schön A., Zhu P., Freire E., Kwong P. D., Roux K. H., Sodroski J., and Wyatt R. (2005) Soluble mimetics of human immunodeficiency virus type 1 viral spikes produced by replacement of the native trimerization domain with a heterologous trimerization motif: characterization and ligand binding analysis. J. Virol. 79, 9954–9969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sneha Priya R., Veena M., Kalisz I., Whitney S., Priyanka D., LaBranche C. C., Sri Teja M., Montefiori D. C., Pal R., Mahalingam S., and Kalyanaraman V. S. (2015) Antigenicity and immunogenicity of a trimeric envelope protein from an Indian clade C HIV-1 isolate. J. Biol. Chem. 290, 9195–9208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kovacs J. M., Nkolola J. P., Peng H., Cheung A., Perry J., Miller C. A., Seaman M. S., Barouch D. H., and Chen B. (2012) HIV-1 envelope trimer elicits more potent neutralizing antibody responses than monomeric gp120. Proc. Natl. Acad. Sci. U.S.A. 109, 12111–12116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chakrabarti B. K., Feng Y., Sharma S. K., McKee K., Karlsson Hedestam G. B., Labranche C. C., Montefiori D. C., Mascola J. R., and Wyatt R. T. (2013) Robust neutralizing antibodies elicited by HIV-1 JRFL envelope glycoprotein trimers in non-human primates. J. Virol. 87, 13239–13251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sanders R. W., van Gils M. J., Derking R., Sok D., Ketas T. J., Burger J. A., Ozorowski G., Cupo A., Simonich C., Goo L., Arendt H., Kim H. J., Lee J. H., Pugach P., Williams M., et al. (2015) HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers Science 349, aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bhattacharyya S., Singh P., Rathore U., Purwar M., Wagner D., Arendt H., DeStefano J., LaBranche C. C., Montefiori D. C., Phogat S., and Varadarajan R. (2013) Design of an Escherichia coli expressed HIV-1 gp120 fragment immunogen that binds to b12 and induces broad and potent neutralizing antibodies. J. Biol. Chem. 288, 9815–9825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pancera M., Zhou T., Druz A., Georgiev I. S., Soto C., Gorman J., Huang J., Acharya P., Chuang G. Y., Ofek G., Stewart-Jones G. B., Stuckey J., Bailer R. T., Joyce M. G., Louder M. K., et al. (2014) Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zolla-Pazner S., Cohen S. S., Boyd D., Kong X. P., Seaman M., Nussenzweig M., Klein F., Overbaugh J., and Totrov M. (2015) Structure/function studies involving the V3 region of the HIV-1 envelope delineate multiple factors that affect neutralization sensitivity. J. Virol. 90, 636–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Saha P., Bhattacharyya S., Kesavardhana S., Miranda E. R., Ali P. S., Sharma D., and Varadarajan R. (2012) Designed cyclic permutants of HIV-1 gp120: implications for envelope trimer structure and immunogen design. Biochemistry 51, 1836–1847 [DOI] [PubMed] [Google Scholar]
- 38. Sok D., van Gils M. J., Pauthner M., Julien J. P., Saye-Francisco K. L., Hsueh J., Briney B., Lee J. H., Le K. M., Lee P. S., Hua Y., Seaman M. S., Moore J. P., Ward A. B., Wilson I. A., Sanders R. W., and Burton D. R. (2014) Recombinant HIV envelope trimer selects for quaternary-dependent antibodies targeting the trimer apex. Proc. Natl. Acad. Sci. U.S.A. 111, 17624–17629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou T., Georgiev I., Wu X., Yang Z. Y., Dai K., Finzi A., Kwon Y. D., Scheid J. F., Shi W., Xu L., Yang Y., Zhu J., Nussenzweig M. C., Sodroski J., Shapiro L., Nabel G. J., Mascola J. R., and Kwong P. D. (2010) Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329, 811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pejchal R., Doores K. J., Walker L. M., Khayat R., Huang P. S., Wang S. K., Stanfield R. L., Julien J. P., Ramos A., Crispin M., Depetris R., Katpally U., Marozsan A., Cupo A., Maloveste S., et al. (2011) A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334, 1097–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Walker L. M., Huber M., Doores K. J., Falkowska E., Pejchal R., Julien J. P., Wang S. K., Ramos A., Chan-Hui P. Y., Moyle M., Mitcham J. L., Hammond P. W., Olsen O. A., Phung P., Fling S., et al. (2011) Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477, 466–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Doria-Rose N. A., Bhiman J. N., Roark R. S., Schramm C. A., Gorman J., Chuang G. Y., Pancera M., Cale E. M., Ernandes M. J., Louder M. K., Asokan M., Bailer R. T., Druz A., Fraschilla I. R., Garrett N. J., et al. (2015) New member of the V1V2-directed CAP256-VRC26 lineage that shows increased breadth and exceptional potency. J. Virol. 90, 76–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tran E. E., Borgnia M. J., Kuybeda O., Schauder D. M., Bartesaghi A., Frank G. A., Sapiro G., Milne J. L., and Subramaniam S. (2012) Structural mechanism of trimeric HIV-1 envelope glycoprotein activation PLoS Pathog. 8, e1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Derking R., Ozorowski G., Sliepen K., Yasmeen A., Cupo A., Torres J. L., Julien J. P., Lee J. H., van Montfort T., de Taeye S. W., Connors M., Burton D. R., Wilson I. A., Klasse P. J., Ward A. B., Moore J. P., and Sanders R. W. (2015) Comprehensive antigenic map of a cleaved soluble HIV-1 envelope trimer. PLoS Pathog. 11, e1004767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Varadarajan R., Sharma D., Chakraborty K., Patel M., Citron M., Sinha P., Yadav R., Rashid U., Kennedy S., Eckert D., Geleziunas R., Bramhill D., Schleif W., Liang X., and Shiver J. (2005) Characterization of gp120 and its single-chain derivatives, gp120-CD4D12 and gp120-M9: implications for targeting the CD4i epitope in human immunodeficiency virus vaccine design. J. Virol. 79, 1713–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. deCamp A., Hraber P., Bailer R. T., Seaman M. S., Ochsenbauer C., Kappes J., Gottardo R., Edlefsen P., Self S., Tang H., Greene K., Gao H., Daniell X., Sarzotti-Kelsoe M., Gorny M. K., et al. (2014) Global panel of HIV-1 Env reference strains for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 88, 2489–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li Y., Svehla K., Louder M. K., Wycuff D., Phogat S., Tang M., Migueles S. A., Wu X., Phogat A., Shaw G. M., Connors M., Hoxie J., Mascola J. R., and Wyatt R. (2009) Analysis of neutralization specificities in polyclonal sera derived from human immunodeficiency virus type 1-infected individuals. J. Virol. 83, 1045–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Falkowska E., Ramos A., Feng Y., Zhou T., Moquin S., Walker L. M., Wu X., Seaman M. S., Wrin T., Kwong P. D., Wyatt R. T., Mascola J. R., Poignard P., and Burton D. R. (2012) PGV04, an HIV-1 gp120 CD4 binding site antibody, is broad and potent in neutralization but does not induce conformational changes characteristic of CD4. J. Virol. 86, 4394–4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bhattacharyya S., Rajan R. E., Swarupa Y., Rathore U., Verma A., Udaykumar R., and Varadarajan R. (2010) Design of a non-glycosylated outer domain-derived HIV-1 gp120 immunogen that binds to CD4 and induces neutralizing antibodies. J. Biol. Chem. 285, 27100–27110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li Y., Migueles S. A., Welcher B., Svehla K., Phogat A., Louder M. K., Wu X., Shaw G. M., Connors M., Wyatt R. T., and Mascola J. R. (2007) Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat. Med. 13, 1032–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Walker L. M., Phogat S. K., Chan-Hui P. Y., Wagner D., Phung P., Goss J. L., Wrin T., Simek M. D., Fling S., Mitcham J. L., Lehrman J. K., Priddy F. H., Olsen O. A., Frey S. M., Hammond P. W., et al. (2009) Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326, 285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. de Taeye S. W., Ozorowski G., Torrents de la Peña A., Guttman M., Julien J. P., van den Kerkhof T. L., Burger J. A., Pritchard L. K., Pugach P., Yasmeen A., Crampton J., Hu J., Bontjer I., Torres J. L., Arendt H., et al. (2015) Immunogenicity of stabilized HIV-1 envelope trimers with reduced exposure of non-neutralizing epitopes. Cell 163, 1702–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tomaras G. D., Yates N. L., Liu P., Qin L., Fouda G. G., Chavez L. L., Decamp A. C., Parks R. J., Ashley V. C., Lucas J. T., Cohen M., Eron J., Hicks C. B., Liao H. X., Self S. G., et al. (2008) Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J. Virol. 82, 12449–12463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Trama A. M., Moody M. A., Alam S. M., Jaeger F. H., Lockwood B., Parks R., Lloyd K. E., Stolarchuk C., Scearce R., Foulger A., Marshall D. J., Whitesides J. F., Jeffries T. L. Jr, Wiehe K., Morris L., et al. (2014) HIV-1 envelope gp41 antibodies can originate from terminal ileum B cells that share cross-reactivity with commensal bacteria. Cell Host Microbe 16, 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Moody M. A., Yates N. L., Amos J. D., Drinker M. S., Eudailey J. A., Gurley T. C., Marshall D. J., Whitesides J. F., Chen X., Foulger A., Yu J. S., Zhang R., Meyerhoff R. R., Parks R., Scull J. C., et al. (2012) HIV-1 gp120 vaccine induces affinity maturation in both new and persistent antibody clonal lineages. J. Virol. 86, 7496–7507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vaine M., Wang S., Crooks E. T., Jiang P., Montefiori D. C., Binley J., and Lu S. (2008) Improved induction of antibodies against key neutralizing epitopes by human immunodeficiency virus type 1 gp120 DNA prime-protein boost vaccination compared to gp120 protein-only vaccination. J. Virol. 82, 7369–7378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hoffenberg S., Powell R., Carpov A., Wagner D., Wilson A., Kosakovsky Pond S., Lindsay R., Arendt H., Destefano J., Phogat S., Poignard P., Fling S. P., Simek M., Labranche C., Montefiori D., et al. (2013) Identification of an HIV-1 clade A envelope that exhibits broad antigenicity and neutralization sensitivity and elicits antibodies targeting three distinct epitopes. J. Virol. 87, 5372–5383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liao H. X., Bonsignori M., Alam S. M., McLellan J. S., Tomaras G. D., Moody M. A., Kozink D. M., Hwang K. K., Chen X., Tsao C. Y., Liu P., Lu X., Parks R. J., Montefiori D. C., Ferrari G., et al. (2013) Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity 38, 176–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nicely N. I., Wiehe K., Kepler T. B., Jaeger F. H., Dennison S. M., Rerks-Ngarm S., Nitayaphan S., Pitisuttithum P., Kaewkungwal J., Robb M. L., O'Connell R. J., Michael N. L., Kim J. H., Liao H. X., Munir Alam S., et al. (2015) Structural analysis of the unmutated ancestor of the HIV-1 envelope V2 region antibody CH58 isolated from an RV144 vaccine efficacy trial vaccinee. EBioMedicine. 2, 713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. McGuire A. T., Gray M. D., Dosenovic P., Gitlin A. D., Freund N. T., Petersen J., Correnti C., Johnsen W., Kegel R., Stuart A. B., Glenn J., Seaman M. S., Schief W. R., Strong R. K., Nussenzweig M. C., and Stamatatos L. (2016) Specifically modified Env immunogens activate B-cell precursors of broadly neutralizing HIV-1 antibodies in transgenic mice. Nat. Commun. 7, 10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dames S. A., Kammerer R. A., Wiltscheck R., Engel J., and Alexandrescu A. T. (1998) NMR structure of a parallel homotrimeric coiled coil. Nat. Struct. Biol. 5, 687–691 [DOI] [PubMed] [Google Scholar]
- 62. Pancera M., and Wyatt R. (2005) Selective recognition of oligomeric HIV-1 primary isolate envelope glycoproteins by potently neutralizing ligands requires efficient precursor cleavage. Virology 332, 145–156 [DOI] [PubMed] [Google Scholar]
- 63. Wu X., Yang Z. Y., Li Y., Hogerkorp C. M., Schief W. R., Seaman M. S., Zhou T., Schmidt S. D., Wu L., Xu L., Longo N. S., McKee K., O'Dell S., Louder M. K., Wycuff D. L., Feng Y., et al. (2010) Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wu X., Zhou T., Zhu J., Zhang B., Georgiev I., Wang C., Chen X., Longo N. S., Louder M., McKee K., O'Dell S., Perfetto S., Schmidt S. D., Shi W., Wu L., et al. (2011) Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333, 1593–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Prajapati R. S., Indu S., and Varadarajan R. (2007) Identification and thermodynamic characterization of molten globule states of periplasmic binding proteins. Biochemistry 46, 10339–10352 [DOI] [PubMed] [Google Scholar]
- 66. Greenfield N. J. (2006) Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc. 1, 2527–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., and Ludtke S. J. (2007) EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 [DOI] [PubMed] [Google Scholar]
- 68. Li M., Gao F., Mascola J. R., Stamatatos L., Polonis V. R., Koutsoukos M., Voss G., Goepfert P., Gilbert P., Greene K. M., Bilska M., Kothe D. L., Salazar-Gonzalez J. F., Wei X., Decker J. M., Hahn B. H., and Montefiori D. C. (2005) Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79, 10108–10125 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.