Abstract
Histone methylation is implicated in a number of biological and pathological processes, including cancer development. In this study, we investigated the molecular mechanism for the recruitment of Polycomb repressive complex-2 (PRC2) and its accessory component, JARID2, to chromatin, which regulates methylation of lysine 27 of histone H3 (H3K27), during epithelial-mesenchymal transition (EMT) of cancer cells. The expression of MEG3 long noncoding RNA (lncRNA), which could interact with JARID2, was clearly increased during transforming growth factor-β (TGF-β)-induced EMT of human lung cancer cell lines. Knockdown of MEG3 inhibited TGF-β-mediated changes in cell morphology and cell motility characteristic of EMT and counteracted TGF-β-dependent changes in the expression of EMT-related genes such as CDH1, ZEB family, and the microRNA-200 family. Overexpression of MEG3 influenced the expression of these genes and enhanced the effects of TGF-β in their expressions. Chromatin immunoprecipitation (ChIP) revealed that MEG3 regulated the recruitment of JARID2 and EZH2 and histone H3 methylation on the regulatory regions of CDH1 and microRNA-200 family genes for transcriptional repression. RNA immunoprecipitation and chromatin isolation by RNA purification assays indicated that MEG3 could associate with JARID2 and the regulatory regions of target genes to recruit the complex. This study demonstrated a crucial role of MEG3 lncRNA in the epigenetic regulation of the EMT process in lung cancer cells.
Keywords: cancer biology; epithelial-mesenchymal transition (EMT); histone methylation; long noncoding RNA (long ncRNA, lncRNA); transcription regulation
Introduction
The amino-terminal tail of histones can carry post-translational modifications such as methylation, acetylation, ubiquitination, and phosphorylation. These modifications regulate the structure of chromatin and influence many cellular processes, including DNA replication, DNA repair, transcription, and cell cycle progression (1). In particular, the importance of lysine methylation of histone H3 (Lys-4, Lys-9, Lys-27, and Lys-36) is highlighted because of its specific dynamics with respect to transcriptional regulation (2, 3). Methylation of H3K4 and H3K36 is closely associated with active transcription, whereas methylation of H3K27 and H3K9 is linked to transcriptional repression. These modifications are controlled by two classes of enzymes with opposing activities: histone lysine methyltransferases (KMTs)2 and lysine demethylases (KDMs). Deregulation of these enzymes has been reported to contribute to human diseases such as developmental disorders and many types of cancer (2–4).
Using retroviral insertional mutagenesis in mice, we have tried to identify novel genes involved in cancer development. As a result, we have isolated hundreds of candidate cancer genes, which include many genes encoding KMTs and KDMs (5, 6). We previously reported that KDM5B/PLU1 promoted cell invasion and epithelial-mesenchymal transition (EMT) of cancer cells (7, 8) and that DOT1L KMT enhanced cell migration and sphere formation potential of cancer cells (9). Thus our experiments demonstrated that histone KMTs and KDMs were implicated not only in the initiation but also in the malignant progression of cancer.
Aggressive cancer progression is often accompanied with the EMT program that is activated by extrinsic signals such as transforming growth factor-β (TGF-β) (10, 11). EMT is a paradigm of cell plasticity characterized by the reversible changes in epithelial and mesenchymal gene expression. Notably, transcriptional repression of CDH1/E-cadherin, a defining marker of epithelial cells, is important for EMT. Moreover, the plastic nature of EMT implies that epigenetic regulation such as histone modification, DNA methylation, and microRNA may be involved (12, 13). Thus the epigenetic analysis of the genes important for EMT has been carried out, and in particular, the functional connection between transcriptional repression of CDH1/E-cadherin and histone methyl-modifying enzymes has been investigated (8, 14, 15).
Previously, we showed that EED, a member of the Polycomb repressive complex-2 (PRC2) that catalyzes H3K27 methylation, was required for TGF-β-dependent repression of CDH1 and microRNA-200 (miR-200) family genes through histone H3 methylation, suggesting an essential role of PRC2 in the EMT of cancer cells (16). PRC2 is a complex of histone H3 methyltransferase essential for specific gene silencing during cancer and development (17–19). PRC2 consists of four core subunits, SUZ12, EED, RBBP4/7, and the catalytic EZH2. Proper recruitment of PRC2 to the target genes plays a crucial role in the epigenetic regulation of gene expression. JARID2, an interacting component of PRC2, has been reported as one of the essential factors for proper PRC2 recruitment in embryonic stem (ES) cells (20–23). We also found that JARID2 was indispensable for TGF-β-induced EMT of lung and colon cancer cell lines (24). Our ChIP experiments indicated that JARID2 might cause TGF-β-dependent repression of CDH1 and miR-200 family genes through EZH2 recruitment and H3K27 methylation on their regulatory regions. However, in the absence of TGF-β, JARID2 showed little effect on the levels of EZH2 occupancies and H3 methylation on these regions. Based on these results, we hypothesized that some additional factors and/or signals induced by TGF-β would be required for JARID2 function (24).
Long noncoding RNAs (lncRNAs) have been recognized as important regulatory factors in various cellular processes such as cell proliferation, differentiation, and establishment of cell identity (25). Expression of lncRNAs reveals highly developmental stage- or cell type-specific patterns and is frequently deregulated in cancer (26–28). Expression of lncRNAs reveals highly developmental stage- or cell type-specific patterns and is frequently deregulated in cancer (26–28). Functions of lncRNAs are largely unknown, but some lncRNAs were shown to interact with transcription factors and chromatin regulators to fine-tune the expression of specific genes (25). PRC2 is one of the most studied examples of chromatin-modifying factors that could be recruited and regulated by lncRNAs such as HOTAIR and RepA (29, 30). Thus we hypothesized that lncRNAs might be involved in the regulation of PRC2 and JARID2 during the EMT process. Because cells undergoing EMT are proposed to acquire stem cell-like properties (31), we focused on lncRNAs that were shown to be implicated in ES cells or induced pluripotent stem (iPS) cells (32, 33). Among them, MEG3 lncRNA was identified as a good candidate that might function in the TGF-β-induced EMT process based on its expression pattern (see Fig. 1, A and B). Moreover, during this study, it has been reported that JARID2 could bind to lncRNAs in mouse ES cells and that one of the candidates, Meg3, was implicated in PRC2 recruitment at a subset of target genes in pluripotent stem cells (34, 35). Therefore, we tried to examine functional interaction between MEG3, JARID2, and PRC2 during the EMT process of cancer cells.
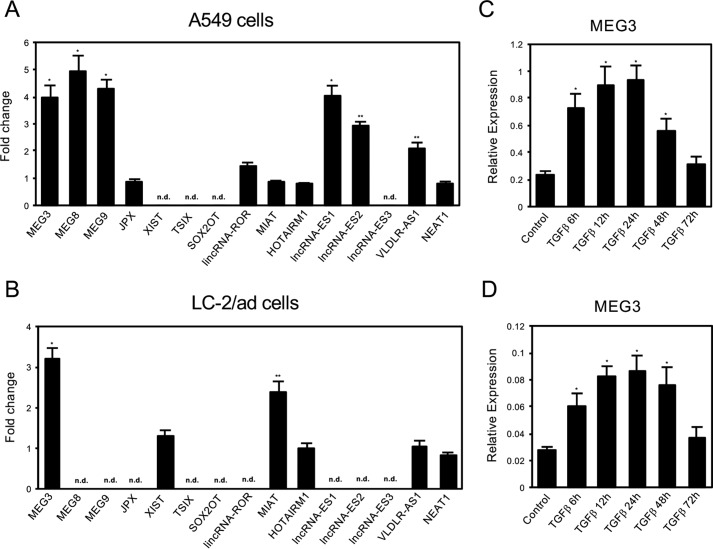
FIGURE 1.
Expression of long noncoding RNAs during TGF-β-induced EMT of A549 and LC-2/ad lung cancer cells. A and B, QRT-PCR analysis was performed to detect the expression of various lncRNAs, which were reported to be implicated in ES cells or iPS cells, in A549 cells (A), and LC-2/ad (B) cells before and after the treatment of 1 ng/ml TGF-β (24 h). Fold changes were calculated and presented. n.d. means not detected (*, p < 0.01 compared with control; **, p < 0.05 compared with control). C and D, QRT-PCR was performed to detect the expression of MEG3 lncRNA in A549 cells (C) and LC-2/ad (D) cells before and after the treatment of 1 ng/ml TGF-β (6, 12, 24, 48, and 72 h) (*, p < 0.01 compared with control).
In this study we found that MEG3 lncRNA was essential for the TGF-β-induced EMT process in A549 and LC-2/ad lung cancer cell lines. The gene expression program during EMT was disturbed by MEG3 knockdown and potentiated by MEG3 overexpression. MEG3 was directly involved in the epigenetic regulation of several EMT-related genes through the recruitment of JARID2 and EZH2 to the chromatin for histone H3 methylation.
Results
Expression of MEG3 Long Noncoding RNA Was Transiently Induced during TGF-β-induced EMT
To find the long noncoding RNAs (lncRNAs) involved in TGF-β-induced EMT of lung cancer cells, we have performed a candidate gene approach based on the previous studies (32, 33). Because cells undergoing EMT are thought to acquire stem cell-like properties (31), we picked up the candidate lncRNAs that were reported to be implicated in ES cells or iPS cells (32, 33). Then we examined the changes in the expression of these lncRNAs in the cells after TGF-β treatment (Fig. 1, A and B). We used the two lung cancer cell lines A549 and LC-2/ad because they are good model systems showing quick and clear changes for cell morphology and EMT-related gene expression during EMT caused by TGF-β (see Fig. 2 and supplemental Fig. S1 for the phenotypes of LC-2/ad cells) (38). Quantitative RT-PCR (QRT-PCR) was carried out to detect the expression of these lncRNAs in A549 and LC-2/ad cells with or without TGF-β treatment, and the fold changes were presented. We found that only MEG3 lncRNA was up-regulated by TGF-β in both A549 and LC-2/ad cells (Fig. 1, A and B). Then we examined the expression changes of MEG3 in TGF-β-induced EMT process of A549 and LC-2/ad cells (Fig. 1, C and D). The expression of MEG3 was immediately and transiently induced by TGF-β, suggesting its potential role in the induction of EMT. Therefore, we decided to focus on MEG3 lncRNA as a good candidate that might function during TGF-β-induced EMT.
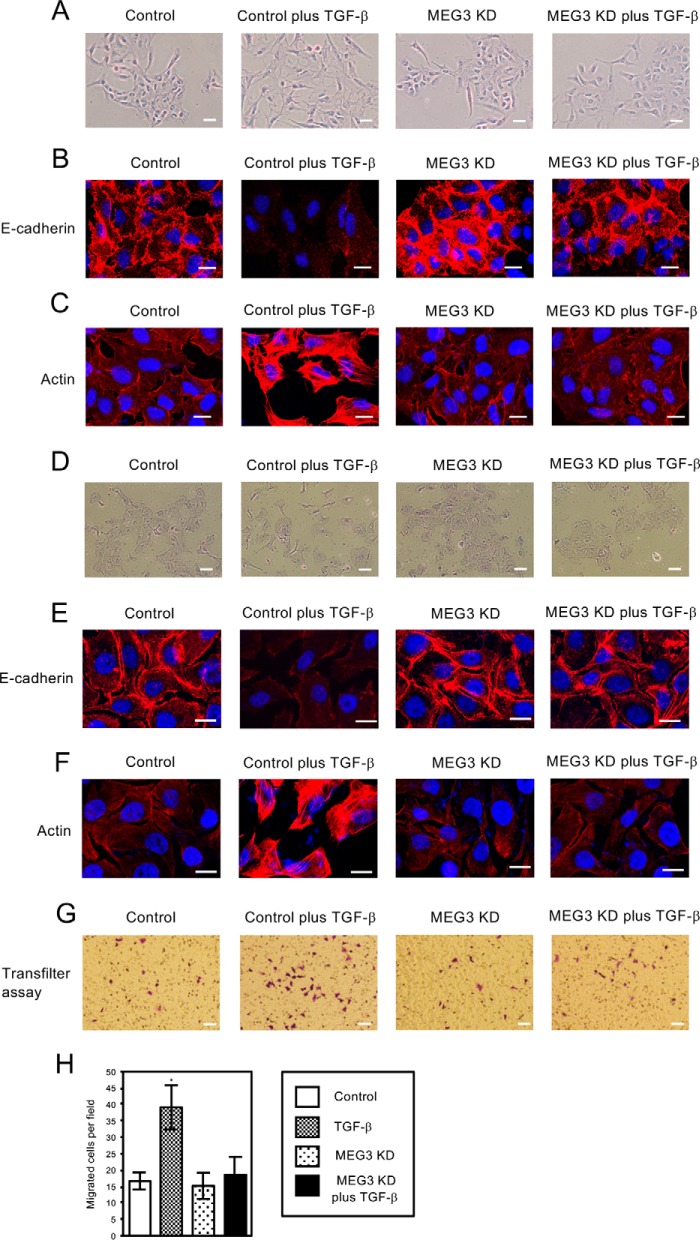
FIGURE 2.
Knockdown of MEG3 antagonized TGF-β-induced morphological changes of A549 and LC-2/ad cells and migratory activities of A549 cells. A and D, cell morphological changes of A549 (A) and LC-2/ad (D) cells after TGF-β treatment. A549 or LC-2/ad cells were infected with retroviruses expressing control shRNA or MEG3 shRNA#1 (described as MEG3 KD) without or with the treatment of 1 ng/ml TGF-β for 6 days. Cells were stained with 0.4% crystal violet. Scale bars, 20 μm. B and E, immunofluorescence images of cells showing the localization of E-cadherin. The cells were treated without or with TGF-β for 48 h. The panels of A549 (B) or LC-2/ad (E) cells with the same arrangement with A or D were stained with anti-E-cadherin antibody and DAPI. Scale bars, 10 μm. C and F, fluorescence images of A549 (C) and LC-2/ad (F) cells showing reorganization of actin cytoskeleton by staining with TRITC-phalloidin (Actin) and DAPI. The cells were treated without or with TGF-β for 48 h. Scale bars, 10 μm. G and H, MEG3 KD inhibited TGF-β-induced increase of migrated cells through the filter. Cells that migrated through the filter to the lower surface within 24 h were fixed and stained (G). The number of migrated cells was counted under a light microscope from at least five fields and three experiments, and the averages were calculated (H) (*, p < 0.01). Scale bars, 60 μm.
Knockdown of MEG3 Inhibited the Changes in Cell Morphologies Induced by TGF-β
To clarify the function of MEG3 lncRNA, we examined the effects of MEG3 knockdown in TGF-β-induced EMT. For knockdown of MEG3, we used two different shRNAs for MEG3 (MEG3 shRNA#1 and shRNA#2). MEG3 expression was clearly down-regulated in A549 and LC-2/ad cells with the infection of retroviruses expressing each MEG3 shRNA even in the presence of TGF-β (supplemental Fig. S2, A and B). We showed the data of MEG3 shRNA#1 as a representative result (indicated as MEG3 KD) because we confirmed that both MEG3 shRNAs led to the same effects in the expression of EMT-related genes in our EMT studies (Fig. 3, A and C). In the presence of TGF-β, control A549 cells showed dispersed and elongated cell phenotypes characteristic of EMT (Fig. 2A). MEG3 knockdown had little effect on cell shapes by itself, but it inhibited cell morphological changes mediated by TGF-β (Fig. 2A). We next examined the expression of E-cadherin, an epithelial cell marker, by immunofluorescence assay. Control A549 cells revealed heterogeneous E-cadherin staining, which had almost disappeared in TGF-β-treated cells (Fig. 2B) (38). MEG3 knockdown increased E-cadherin staining on the cell membrane compared with the control cells, which was still observed after TGF-β treatment (Fig. 2B). These results suggested that the epithelial cell characters might be enhanced by MEG3 knockdown and maintained in the presence of TGF-β. Because actin reorganization is associated with EMT, we observed the status of actin in the cells with TRITC-conjugated phalloidin staining (10). As shown in Fig. 2C, TGF-β treatment led to actin stress fiber formation, but in the MEG3 knockdown cells, we never detected actin fiber even after TGF-β treatment.
FIGURE 3.
Knockdown of MEG3 influenced the TGF-β-dependent changes in the expression of EMT-related genes in A549 or LC-2/ad cells. A and C, QRT-PCR analysis was performed to detect the expression of CDH1/E-cadherin, FN1/fibronectin, vimentin, ZEB1, ZEB2, miR-200a, miR-200c, JARID2, PMEPA1, SMAD7, ID1, and ID2 in A549 (A) and LC-2/ad (C) cells infected with retroviruses expressing control shRNA or each MEG3 shRNA (MEG3 sh#1 and sh#2) with or without the treatment of 1 ng/ml TGF-β for 24 h (*, p < 0.01 compared with control; **, p < 0.05 compared with control; ***, p < 0.01 compared with the sample without TGF-β treatment; ****, p < 0.05 compared with the sample without TGF-β treatment). B and D, immunoblotting was performed to detect the expression of E-cadherin, fibronectin, vimentin, ZEB1, ZEB2, phosphorylated SMAD3 (p-SMAD3), and GAPDH proteins using the corresponding antibodies in A549 (B) and LC-2/ad (D) cells. WB, Western blotting.
We further examined the effects of MEG3 knockdown in another cell line, LC-2/ad. After TGF-β treatment, control LC-2/ad cells became flat and scattered, accompanied by loss of E-cadherin staining and the appearance of actin fiber (Fig. 2, D–F). MEG3 knockdown strengthened E-cadherin staining and antagonized the TGF-β-induced phenotypes observed in the control cells (Fig. 2, D–F). Taken together, these results indicated that MEG3 knockdown antagonized TGF-β-induced EMT phenotypes of A549 and LC-2/ad lung cancer cells such as morphological changes and cytoskeletal rearrangements.
To see the effects of MEG3 knockdown in EMT-associated phenotypes, we performed the transfilter migration assay using Boyden modified chambers to measure cell migration activities. As shown in Fig. 2, G and H, the treatment of TGF-β significantly increased the number of migrated A549 cells per field. MEG3 knockdown had little effect on cell migration by itself but inhibited the TGF-β-dependent increase of migrated cells (Fig. 2, G and H). These results suggested that MEG3 knockdown inhibited TGF-β-induced EMT and also EMT-associated cell migration activity of lung cancer cells.
MEG3 Knockdown Antagonized the Changes in the Expression of EMT-related Genes Caused by TGF-β
EMT is associated with the changes in the expression of epithelial and mesenchymal marker genes (10). First, we examined the expression level of an epithelial cell marker, CDH1/E-cadherin, and mesenchymal markers, FN1/fibronectin and vimentin in the cells with MEG3 knockdown. QRT-PCR revealed that MEG3 knockdown increased CDH1 expression and inhibited transcriptional repression of CDH1 induced by TGF-β in A549 and LC-2/ad cells (Fig. 3, A and C), which was consistent with the results of E-cadherin staining (Fig. 2, B and E). In the cases of FN1/fibronectin and vimentin, MEG3 knockdown also counteracted the effect of TGF-β (Fig. 3, A and C). These findings suggested that MEG3 knockdown might inhibit the EMT-inducing gene expression program in A549 and LC-2/ad cells.
We further analyzed the expression of ZEB family transcription factors, important regulators of E-cadherin repression (39), and the miR-200 family of microRNAs, specific inhibitors of the ZEB family (40, 41), in the MEG3 knockdown cells. In A549 and LC-2/ad cells, TGF-β up-regulated the expression of ZEB1 and ZEB2 and decreased the expression of two representative miRNAs, miR-200a and miR-200c (Fig. 3, A and C). MEG3 knockdown itself slightly but significantly reduced the expression of ZEB1 and ZEB2 and increased the expression of miR-200a and miR-200c (Fig. 3, A and C). In the presence of TGF-β, MEG3 knockdown antagonized the TGF-β-dependent changes in their expressions (Fig. 3, A and C), confirming its inhibitory effects. We also examined the expression of JARID2 in the MEG3 knockdown cells, because JARID2 was induced by TGF-β and implicated in EMT (24). MEG3 knockdown did not have a significant effect on JARID2 expression and JARID2 induction by TGF-β in A549 and LC-2/ad cells (Fig. 3, A and C). JARID2 induction seemed to decrease slightly in the MEG3 knockdown cells compared with the TGF-β-treated control cells, but the decrease was not statistically significant (Fig. 3, A and C). To examine the specificity of MEG3 knockdown effects in gene regulation, we picked up several of the other known TGF-β-responsive genes (42, 43) and analyzed the changes in the expression of these genes. In A549 and LC-2/ad cells, TGF-β treatment increased the expression of TMEPA1 and SMAD7 and down-regulated the expression of ID1 and ID2 (Fig. 3, A and C). MEG3 knockdown did not have a significant effect in the expression of these genes and their expression changes induced by TGF-β. These results suggested that MEG3 was not involved in the expression of all of the TGF-β-responsive target genes, but might regulate a subset of genes, including some essential genes for EMT.
Next, we confirmed the changes in protein expression for several of the EMT-related gene products in A549 and LC-2/ad cells. The TGF-β-dependent decrease of E-cadherin protein and the increase of fibronectin, vimentin, ZEB1, and ZEB2 proteins were inhibited by MEG3 knockdown (Fig. 3, B and D), which was consistent with the QRT-PCR results (Fig. 3, A and C). To see whether MEG3 knockdown would influence the TGF-β signaling pathway in the cells or not, we analyzed the status of the phosphorylated SMAD3 protein (11). The phosphorylated SMAD3 proteins were detected after TGF-β treatment, and their levels were similar in the control cells and the MEG3 knockdown cells (Fig. 3B), indicating that MEG3 knockdown would not impair the activation of SMAD3 transcription factor by TGF-β. These results suggested that MEG3 might not influence TGF-β-induced activation of downstream transcription factors but could contribute to TGF-β-dependent transcriptional regulation of EMT-related genes in lung cancer cells.
MEG3 Is Involved in the Recruitment of EZH2 and Histone H3 Methylation on the Regulatory Regions of CDH1 and miR-200 Family Genes
MEG3 lncRNA has been reported to interact with JARID2 protein in mouse ES cells (34). JARID2 is a cofactor of PRC2 enzyme that controls histone H3K27 methylation and transcriptional repression in ES cells (20–23). Thus we performed chromatin immunoprecipitation (ChIP) to analyze the status of histone H3 methylation on the regulatory regions of CDH1 and miR-200 family genes, whose expressions were repressed in TGF-β-induced EMT. After immunoprecipitation, quantitative PCR was carried out using the primer pairs located around the transcription start sites of the CDH1 gene and two microRNA clusters as described previously (8, 44).
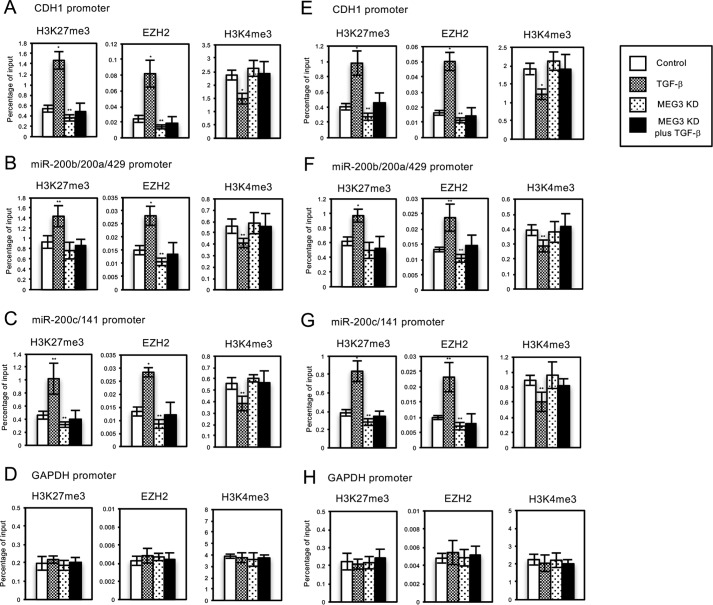
After TGF-β treatment, we could observe the significant increase of tri-methylated H3K27 (H3K27me3), the enhanced recruitment of EZH2, a catalytic subunit of PRC2, and the substantial decrease of transcriptionally active H3K4me3 marks on the regulatory regions of CDH1, miR-200b/200a/429, and miR-200c/141 genes in A549 and LC-2/ad cells (Fig. 4, A–C and E–G). These results strongly suggested that TGF-β-induced EZH2 recruitment on these regulatory regions might be responsible for the transcriptional repression. Knockdown of MEG3 caused the slight decrease of H3K27me3 and EZH2 signals on these regions compared with the control cells (Fig. 4, A–C and E–G), which was correlated with the increase of their expressions (Fig. 3). In addition, MEG3 knockdown counteracted TGF-β-mediated changes of histone methylation and EZH2 recruitment (Fig. 4, A–C and E–G). These findings suggested that endogenous MEG3 lncRNA was involved in EZH2 recruitment and histone H3 methylation on the specific target genes during the TGF-β-induced EMT process. Next, the ChIP assays on the regulatory region of the unrelated GAPDH gene were performed as a control experiment. We could not observe any changes in EZH2 signals and H3K27 and H3K4 methylation on the region of the GAPDH gene with MEG3 knockdown (Fig. 4, D and H). These results revealed that MEG3-mediated epigenetic changes would be specific to the target genes controlled by MEG3 and TGF-β, which was consistent with the specificity of transcriptional regulation.
FIGURE 4.
Knockdown of MEG3 affected the TGF-β-induced regulation of histone H3 methylation and EZH2 recruitment on the regulatory regions of CDH1 gene and miR-200 gene clusters in A549 or LC-2/ad cells. A549 or LC-2/ad cells were infected with retroviruses expressing control shRNA or MEG3 shRNA#1 without or with TGF-β treatment. ChIP analyses of H3K27me3, EZH2, and H3K4me3 on the regulatory regions of CDH1 (A and E), miR-200b/200a/429 (B and F), miR-200c/141 (C and G), and GAPDH genes (D and H) in A549 (A–D) or LC-2/ad (E–H) cells are shown. The occupancies of methylated histones or EZH2 protein on the regions were analyzed by quantitative PCR. Percentage enrichment over input chromatin DNA was presented (*, p < 0.01 compared with control; **, p < 0.05 compared with control).
Overexpression of MEG3 Partially Influenced the Gene Expression Program in EMT
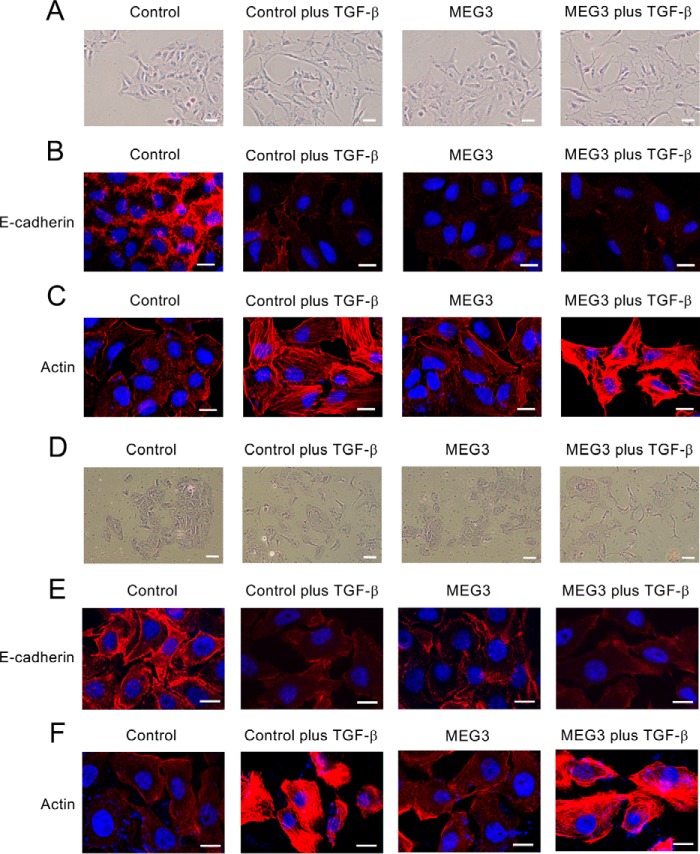
To deepen our understanding for MEG3 function in EMT regulation, we investigated the effects of MEG3 overexpression in A549 and LC-2/ad cells. First, we observed the cell morphologies and the status of E-cadherin and actin (Fig. 5). The cells with MEG3 overexpression looked slightly scattered compared with the control cells, but the effect was not significant (Fig. 5, A and D). More importantly, MEG3 overexpression itself could reduce E-cadherin staining of the cells but not induce actin stress fiber formation (Fig. 5, B, C, E, and F). In the presence of TGF-β, we confirmed morphological changes and cytoskeletal rearrangements of the cells associated with EMT (Fig. 5). These results suggested that MEG3 overexpression itself could down-regulate the expression of E-cadherin but might not proceed with the EMT process completely.
FIGURE 5.
Overexpression of MEG3 reduced the expression of E-cadherin but did not induce the reorganization of actin cytoskeleton of A549 and LC-2/ad cells. A and D, cell morphological changes of A549 (A) or LC-2/ad (D) cells after TGF-β treatment. The cells were infected with the control retrovirus or the retrovirus expressing MEG3 without or with the treatment of 1 ng/ml TGF-β for 6 days and were stained with 0.4% crystal violet. Scale bars, 20 μm. B and E, immunofluorescence images of cells showing the localization of E-cadherin. The panels of A549 (B) or LC-2/ad (E) cells were stained with anti-E-cadherin antibody and DAPI. The cells were treated without or with TGF-β for 48 h. Scale bars, 10 μm. C and F, fluorescence images of A549 (C) and LC-2/ad (F) cells showing reorganization of actin cytoskeleton by staining with TRITC-phalloidin (Actin) and DAPI. Scale bars, 10 μm.
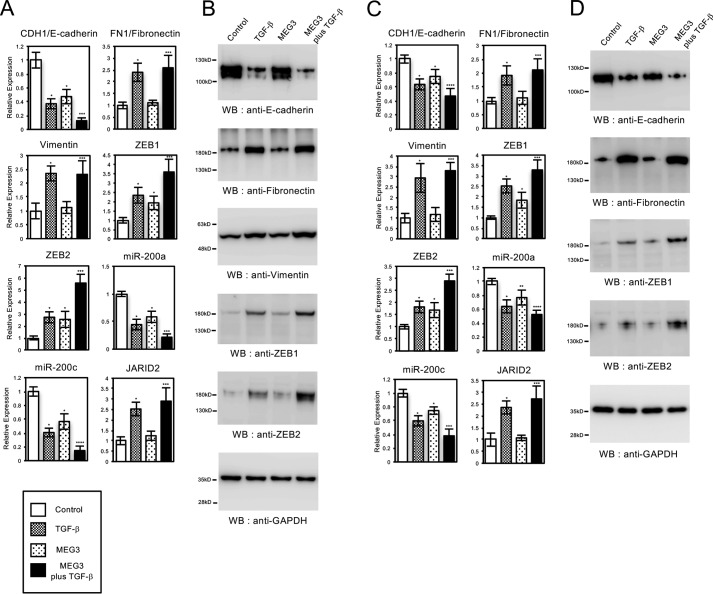
Next, we analyzed the expression of EMT-related genes in A549 and LC-2/ad cells with MEG3 overexpression. MEG3 overexpression caused significant changes on the expression of CDH1/E-cadherin, ZEB1, ZEB2, miR-200a, and miR-200c but had no effect on the expression of FN1/fibronectin, vimentin, and JARID2 (Fig. 6, A and C). The expression of CDH1, miR-200a, and miR-200c was repressed, and the expression of ZEB1 and ZEB2 was significantly up-regulated in the MEG3-overexpressing cells. In the presence of TGF-β, MEG3 overexpression potentiated the effects of TGF-β in the expression of CDH1/E-cadherin, ZEB1, ZEB2, miR-200a, and miR-200c but not significantly in the expression of FN1/fibronectin, vimentin, and JARID2 (Fig. 6, A and C). We also confirmed the effects of MEG3 overexpression in protein expression for several of the EMT-related genes. MEG3 reduced the expression of E-cadherin protein, increased ZEB1 and ZEB2 proteins, and enhanced the effects of TGF-β in their expressions (Fig. 6, B and D). These results were consistent with the QRT-PCR results (Fig. 6, A and C) and indicated that MEG3 overexpression itself partially influenced the gene expression program during the EMT process. This partial effect in gene expression might be related to the observed cell phenotypes caused by MEG3 overexpression (Fig. 5).
FIGURE 6.
Overexpression of MEG3 influenced the expression of part of EMT-related genes in A549 and LC-2/ad cells. QRT-PCR analysis was performed to detect the expression of CDH1/E-cadherin, FN1/fibronectin, vimentin, ZEB1, ZEB2, miR-200a, miR-200c, and JARID2 in A549 (A) and LC-2/ad (C) cells infected with the control retrovirus or the retrovirus expressing MEG3 with or without treatment of TGF-β for 24 h (*, p < 0.01 compared with control; **, p < 0.05 compared with control; ***, p < 0.01 compared with the sample without TGF-β treatment; ****, p < 0.05 compared with the sample without TGF-β treatment). B and D, immunoblotting was performed to detect the expression of E-cadherin, fibronectin, vimentin, ZEB1, ZEB2, and GAPDH proteins using the corresponding antibodies in A549 (B) and LC-2/ad (D) cells.
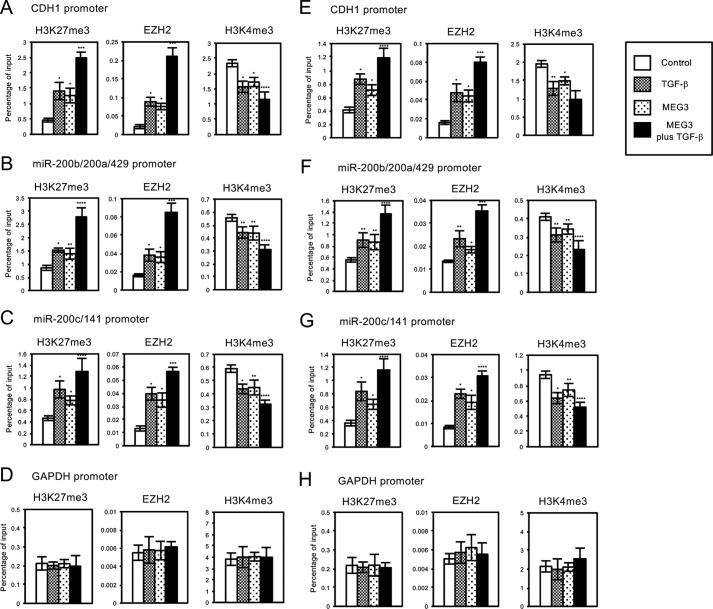
We also examined the status of histone H3 methylation and EZH2 occupancies on the regulatory regions of CDH1, miR-200b/200a/429, and miR-200c/141 genes in A549 and LC-2/ad cells with MEG3 overexpression. In these regions, MEG3 overexpression caused the significant increase of H3K27me3 and EZH2 recruitment and the decrease of H3K4me3 compared with the control cells (Fig. 7, A–C and E–G), which was correlated with the decrease of their expressions (Fig. 6). MEG3 overexpression also potentiated the effects of TGF-β in the EZH2 occupancies and histone H3 methylation on the target genes (Fig. 7, A–C and E–G). Again, we did not detect any changes in EZH2 signals and H3 methylation on the regulatory region of unrelated GAPDH gene by MEG3 overexpression (Fig. 7, D and H). These results suggested that MEG3 lncRNA could mediate EZH2 recruitment and histone H3 methylation on the specific target loci such as CDH1 and miR-200 family genes for transcriptional repression, which might be a critical step for the EMT process.
FIGURE 7.
Overexpression of MEG3 affected the regulation of histone H3 methylation and EZH2 recruitment on the regulatory regions of CDH1 gene and miR-200 gene clusters in A549 or LC-2/ad cells. A549 or LC-2/ad cells were infected with the control retrovirus or the retrovirus expressing MEG3 without or with TGF-β treatment. ChIP analyses of H3K27me3, EZH2, and H3K4me3 on the regulatory regions of CDH1 (A and E), miR-200b/200a/429 (B and F), miR-200c/141 (C and G), and GAPDH genes (D and H) in A549 (A–D) or LC-2/ad (E–H) cells are shown. The occupancies of methylated histones or EZH2 protein on the regions were analyzed by quantitative PCR (*, p < 0.01 compared with control; **, p < 0.05 compared with control; ***, p < 0.01 compared with the sample without TGF-β treatment; ****, p < 0.05 compared with the sample without TGF-β treatment). WB, Western blotting.
Effects of MEG3 Overexpression in EMT Were Cancelled by JARID2 Knockdown
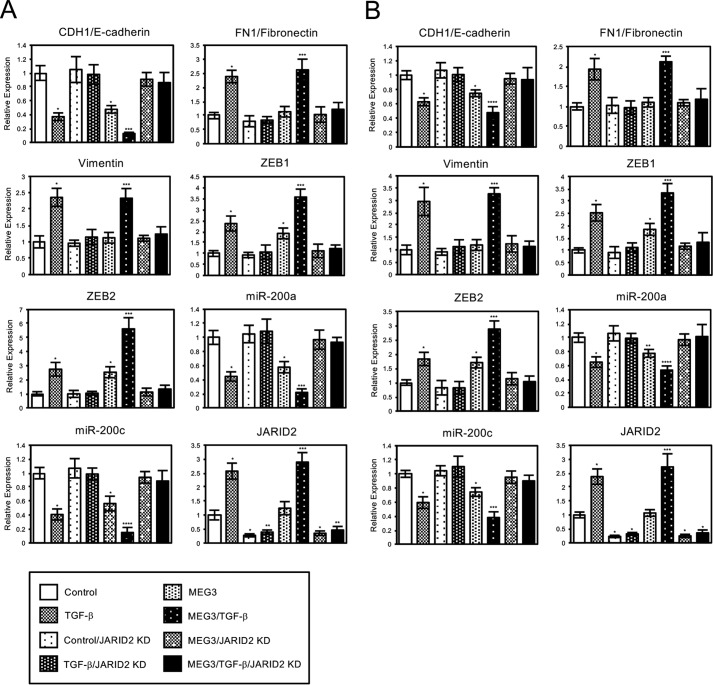
To understand the mechanism of how MEG3 regulates the expression of the target EMT-related genes, we examined the requirement of JARID2 in the process by the JARID2 knockdown experiment. A549 and LC2/ad cells with or without MEG3 overexpression were infected with the control retrovirus or JARID2 shRNA-expressing retrovirus. Then we analyzed the expression of EMT-related genes with or without TGF-β treatment (Fig. 8, A and B). In the case of CDH1/E-cadherin, JARID2 knockdown itself had no effect on its expression but inhibited the repression of CDH1 induced by TGF-β as reported previously (Fig. 8, A and B) (24). Moreover, JARID2 knockdown inhibited MEG3-induced repression of CDH1 in the absence or presence of TGF-β (Fig. 8, A and B). Similarly, JARID2 knockdown cancelled the effects of MEG3 overexpression and/or TGF-β in the expression of ZEB1, ZEB2, miR-200a, and miR-200c genes (Fig. 8, A and B). For the mesenchymal markers, FN1/fibronectin and vimentin, JARID2 knockdown inhibited only the effect of TGF-β, because MEG3 originally had little effect on them (Fig. 6). These results indicated that JARID2 knockdown prevented the ability of overexpressed MEG3 to regulate a subset of EMT-related genes. Therefore, JARID2 was suggested to be an essential factor for MEG3 function in EMT-related gene expression.
FIGURE 8.
Knockdown of JARID2 cancelled MEG3-mediated changes in the expression of part of EMT-related genes in A549 and LC-2/ad cells. QRT-PCR analysis was performed to detect the expression of CDH1/E-cadherin, FN1/fibronectin, vimentin, ZEB1, ZEB2, miR-200a, miR-200c, and JARID2 in A549 (A) and LC-2/ad (B) cells infected with the control retrovirus or the retrovirus expressing MEG3 and/or the retrovirus expressing JARID2 shRNA without or with treatment of TGF-β for 24 h (*, p < 0.01 compared with control; **, p < 0.05 compared with control; ***, p < 0.01 compared with the sample without TGF-β treatment; ****, p < 0.05 compared with the sample without TGF-β treatment).
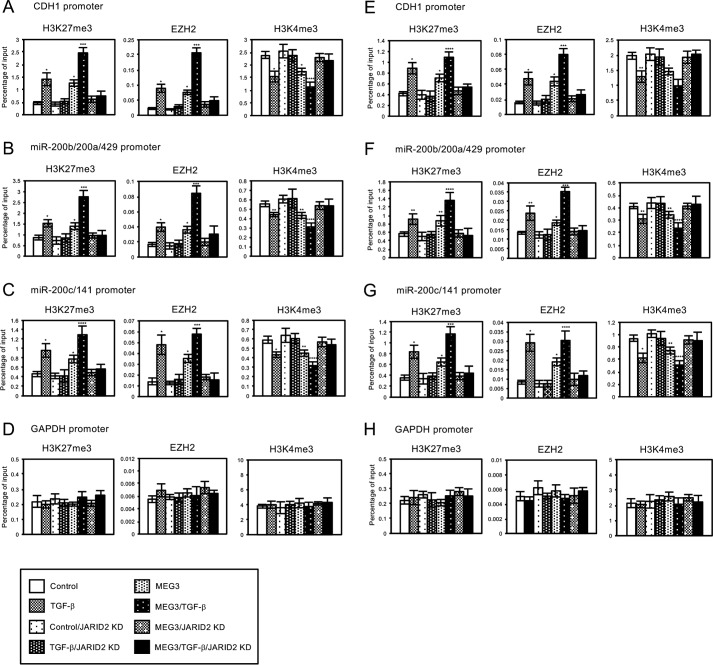
We also examined the effect of JARID2 knockdown in histone methylation and EZH2 occupancies on the regulatory regions of CDH1, miR-200b/200a/429, and miR-200c/141 genes in the cells with MEG3 overexpression. On these regions, JARID2 knockdown counteracted the TGF-β-mediated increase of H3K27me3 and EZH2 recruitment and the decrease of H3K4me3 as shown previously (Fig. 9, A and B) (24). In addition, JARID2 knockdown inhibited MEG3-induced changes in the EZH2 occupancies and histone H3 methylation on the target genes (Fig. 9, A and B). These epigenetic changes caused by JARID2 knockdown were well correlated with the changes in the expression of these genes. Again, we did not detect any changes in EZH2 signals and H3 methylation on the regulatory region of the unrelated GAPDH gene (Fig. 9, D and H). These results suggested that JARID2 was required for MEG3 lncRNA to induce EZH2 recruitment on the specific target loci, thereby regulating histone H3 methylation for transcriptional repression.
FIGURE 9.
Knockdown of JARID2 inhibited MEG3-induced changes in the regulation of histone H3 methylation and EZH2 recruitment on the regulatory regions of CDH1 gene and miR-200 gene clusters in A549 or LC-2/ad cells. A549 or LC-2/ad cells were infected with the control retrovirus or the retrovirus expressing MEG3 and/or the retrovirus expressing JARID2 shRNA without or with TGF-β treatment. ChIP analyses of H3K27me3, EZH2, and H3K4me3 on the regulatory regions of CDH1 (A and E), miR-200b/200a/429 (B and F), miR-200c/141 (C and G), and GAPDH genes (D and H) in A549 (A–D) or LC-2/ad (E–H) cells are shown. The occupancies of methylated histones or EZH2 protein on the regions were analyzed by quantitative PCR (*, p < 0.01 compared with control; **, p < 0.05 compared with control; ***, p < 0.01 compared with the sample without TGF-β treatment; ****, p < 0.05 compared with the sample without TGF-β treatment).
Overexpression of MEG3 Enhanced the Recruitment of JARID2 and EZH2 on the Regulatory Regions of CDH1 and miR-200 Family Genes
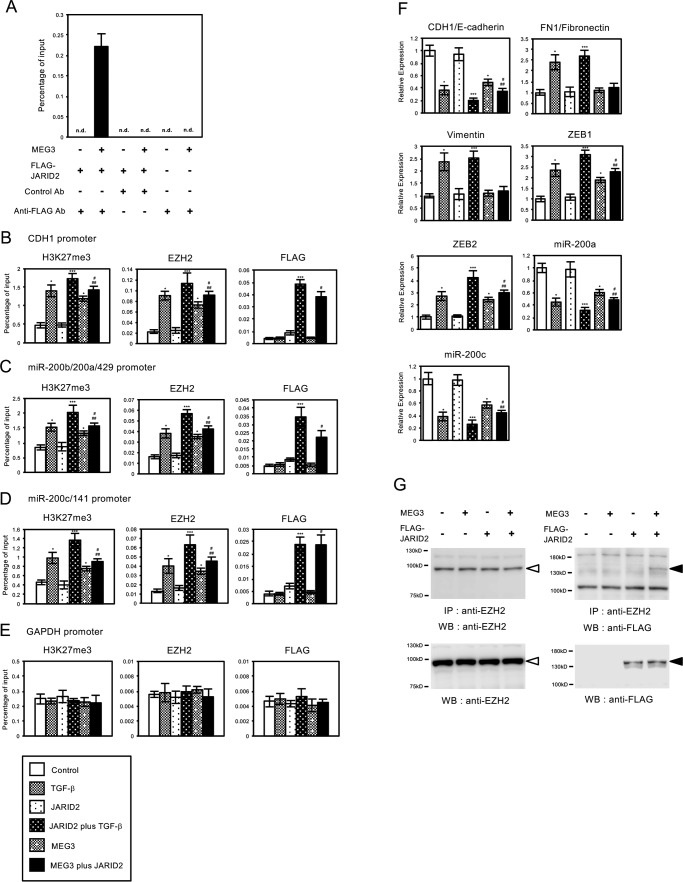
As the mechanism for the epigenetic regulation of MEG3, it was suggested that MEG3 could bind to JARID2 and recruit JARID2 and EZH2 to the specific target regions for histone methylation. However, in the above ChIP experiments, the recruitment of endogenous JARID2 protein was not detected on the regulatory regions of CDH1, miR-200b/200a/429, and miR-200c/141 genes with the anti-JARID2 antibody used in this study, possibly because of its low reactivity for immunoprecipitation. Thus we decided to examine the recruitment of JARID2 on the target genes when JARID2 was overexpressed in the cells. We first tried to confirm the interaction of MEG3 and JARID2, which was reported in mouse ES cells (34), by RNA immunoprecipitation (RIP) assay. A549 cells were infected with the various combinations of the retroviruses expressing MEG3 and FLAG-tagged JARID2, and the cross-linked cell lysates were prepared. The cell lysates were immunoprecipitated with control antibody (mouse IgG) or anti-FLAG antibody, and the co-precipitated RNA was quantified by QRT-PCR (Fig. 10A). The amplified signal for MEG3 lncRNA could be detected only when both MEG3 and JARID2 were overexpressed in the cells, and the cell lysate was immunoprecipitated with anti-FLAG antibody (Fig. 10A), confirming the specific interaction of overexpressed MEG3 and JARID2. We did not detect the signals derived from endogenous MEG3 lncRNA possibly due to the sensitivity of this assay (Fig. 10A).
FIGURE 10.
MEG3 interacted with JARID2 and stimulated the interaction and recruitment of JARID2 and EZH2 on the regulatory regions of CDH1, miR-200a, and miR-200c genes for histone H3K27 methylation in A549 cells. A, interaction of MEG3 and JARID2 detected by RIP. A549 cells were infected with the various combinations (as indicated) of the retroviruses expressing MEG3 and FLAG-tagged JARID2. The cross-linked cell lysates were immunoprecipitated with control antibody (mouse IgG) or anti-FLAG antibody, and the co-precipitated RNA was transcribed to cDNA. QPCR was performed to detect the enrichment of MEG3 in the precipitates. n.d. means not detected. B–E, A549 cells were infected with the various combinations (as indicated) of the retroviruses without or with TGF-β treatment. ChIP analyses of H3K27me3, EZH2, and FLAG-tagged JARID2 on the regulatory regions of CDH1 (B), miR-200b/200a/429 (C), miR-200c/141 (D), and GAPDH genes (E) are shown (*, p < 0.01 compared with control; ***, p < 0.01 compared with the sample without TGF-β treatment; #, p < 0.01 compared with the sample of JARID2 overexpressed cells; ##, p < 0.05 compared with the sample of MEG3 overexpressed cells). F, QRT-PCR analysis was performed to detect the expression of CDH1/E-cadherin, FN1/fibronectin, vimentin, ZEB1, ZEB2, miR-200a, and miR-200c in A549 cells infected with the various combinations of the retroviruses (*, p < 0.01 compared with control; ***, p < 0.01 compared with the sample without TGF-β treatment; #, p < 0.01 compared with the sample of JARID2 overexpressed cells; ##, p < 0.05 compared with the sample of MEG3 overexpressed cells). G, interaction of JARID2 and EZH2 was enhanced by MEG3. A549 cell lysates were immunoprecipitated (IP) with anti-EZH2 antibody and then subjected to immunoblotting with anti-EZH2 (left upper panel) and anti-FLAG antibodies (right upper panel). The cell lysates were also directly subjected to immunoblotting with anti-EZH2 and anti-FLAG antibodies to see the expression level of endogenous EZH2 and FLAG-tagged JARID2 proteins (lower panels). Open and closed arrowheads indicate endogenous EZH2 protein and FLAG-tagged JARID2 protein, respectively. WB, Western blotting.
Then we carried out ChIP analysis to see the effect of MEG3 overexpression in the recruitment of FLAG-tagged JARID2 on the regulatory regions of CDH1, miR-200b/200a/429, and miR-200c/141 genes (Fig. 10, B–D). As we reported previously (24), JARID2 overexpression itself did not alter H3K27 methylation, EZH2, and FLAG-tagged JARID2 recruitment on these regions compared with the control cells, but co-treatment of TGF-β enhanced the levels of methylation and their recruitment (Fig. 10, B–D). MEG3 overexpression itself caused the significant increase of H3K27me3 and EZH2 recruitment on these regions compared with the control cells similarly as shown in Fig. 7 (Fig. 10, B–D). Overexpression of both JARID2 and MEG3 caused the significant increase of H3K27me3 and the occupancies of EZH2 and FLAG-tagged JARID2 on these regions compared with JARID2-overexpressing cells and MEG3-overexpressing cells (Fig. 10, B–D). Again, we did not observe any changes in H3K27me3, EZH2, and JARID2 on the regulatory region of the unrelated GAPDH gene (Fig. 10E). These results indicated that MEG3 overexpression could increase the recruitment of JARID2 and EZH2 on the specific target regions for H3K27 methylation and that JARID2 overexpression also could enhance MEG3-induced EZH2 recruitment and histone methylation.
We also examined the effects of co-expression of JARID2 and MEG3 in the expression of EMT-related genes in A549 cells. JARID2 overexpression itself caused no significant changes in the expression of EMT-related genes but potentiated the effects of TGF-β as reported previously (Fig. 10F) (24). Again, MEG3 overexpression itself influenced the expression of part of the EMT-related genes such as CDH1/E-cadherin, ZEB1, ZEB2, miR-200a, and miR-200c. Co-expression of MEG3 and JARID2 induced a significant decrease of CDH1, miR-200a, and miR-200c expression and an increase of ZEB1 and ZEB2 expression compared with JARID2-overexpressing cells and MEG3-overexpressing cells (Fig. 10F). For the expression of FN1/fibronectin and vimentin, MEG3 and/or JARID2 had no effect. These results indicated that co-expression of JARID2 and MEG3 induced significant expression changes in several of EMT-related genes in the absence of TGF-β, suggesting the importance of cooperation of JARID2 and MEG3 in transcriptional regulation during EMT.
Next, we examined whether MEG3 would stimulate the interaction of JARID2 and EZH2 in the lung cancer cells. A549 cells were infected with the various combinations of the retroviruses expressing MEG3 and FLAG-tagged JARID2, and the cell lysates were prepared. We performed immunoprecipitation analysis with anti-EZH2 antibody followed by immunoblotting with anti-EZH2 and anti-FLAG antibodies. Immunoblot with anti-EZH2 revealed that endogenous EZH2 was detected at a similar level with or without expression of MEG3 and JARID2 (Fig. 10G, left panels, open arrowhead). In contrast, immunoblot with anti-FLAG showed that co-precipitated JARID2 was significantly detected when MEG3 was co-expressed in the cells (Fig. 10G, right upper panel, closed arrowhead). The expression level of FLAG-tagged JARID2 was not affected by MEG3 co-expression (Fig. 10G, right lower panel, closed arrowhead). Therefore, the complex of EZH2 and JARID2 was found to be increased in the presence of MEG3. These results strongly suggested that MEG3 enhanced the interaction of JARID2 and EZH2 in A549 lung cancer cells.
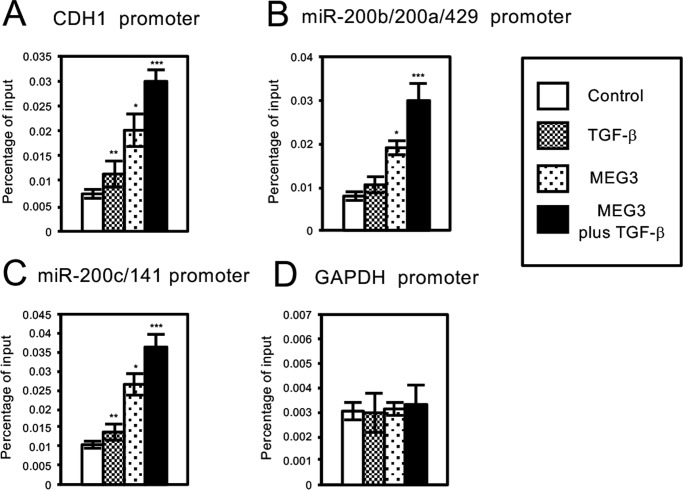
We also tried to examine whether MEG3 lncRNA could associate with the chromatin of the regulatory regions of CDH1, miR-200b/200a/429, and miR-200c/141 genes by chromatin isolation by RNA purification (ChIRP) assay (Fig. 11) (37). A549 cells were infected with the control retrovirus or the retrovirus expressing MEG3 with or without treatment of TGF-β. The cross-linked cell lysates were incubated with biotinylated DNA probes against MEG3 lncRNA, and the binding complexes were recovered by streptavidin-conjugated magnet beads. QPCR was performed to detect the enrichment of the specific regulatory regions that associated with MEG3 lncRNA. TGF-β treatment significantly increased the amplified signals for the regulatory regions of CDH1 and miR-200c/141 genes but not the regulatory region of the unrelated GAPDH gene (Fig. 11, A, C, and D). The signal for the regulatory region of miR-200b/200a/429 was slightly increased by TGF-β, but the difference was not statistically significant (Fig. 11B). These results suggested that endogenous MEG3 lncRNA might be recruited to the specific regulatory regions in the cells after TGF-β treatment. MEG3 overexpression caused a clear increase of the amplified signals for the regulatory regions of CDH1, miR-200b/200a/429, and miR-200c/141 genes, which was further enhanced by TGF-β but not of GAPDH gene (Fig. 11, A–D). These results indicated that the TGF-β signal was not necessary for the association of MEG3 lncRNA with the specific target regions but could enhance the interaction. Furthermore, the levels of MEG3 association with the target sites were well correlated with the recruitment of JARID2 and EZH2 on the chromatin, histone H3K27 methylation, and transcriptional repression of the target genes (Figs. 6, 7 and 10). Altogether, these results suggested that MEG3 lncRNA could bind to JARID2 and mediated the assembly of the JARID2- and EZH2-containing PRC2 complex on the regulatory regions of CDH1, miR-200b/200a/429, and miR-200c/141 genes for histone H3 methylation and transcriptional repression during EMT process of cancer cells.
FIGURE 11.
MEG3 could associate with the specific regulatory regions of CDH1, miR-200a, and miR-200c genes in A549 cells. ChIRP analyses of MEG3 on the regulatory regions of CDH1 (A), miR-200b/200a/429 (B), miR-200c/141 (C), and GAPDH genes (D) in A549 cells are shown. The cells were infected with the control retrovirus or the retrovirus expressing MEG3 with or without treatment of TGF-β. The cross-linked cell lysates were incubated with biotinylated DNA probes against MEG3 lncRNA, and the binding complexes were recovered by streptavidin-conjugated magnet beads. QPCR was performed to detect the enrichment of the specific regulatory regions that associated with MEG3 lncRNA. (*, p < 0.01 compared with control; **, p < 0.05 compared with control; ***, p < 0.01 comparing to the sample without TGF-β treatment).
Discussion
In this study, we found that MEG3 lncRNA was required for TGF-β-induced EMT of A549 and LC-2/ad lung cancer cell lines. MEG3 knockdown inhibited EMT by antagonizing TGF-β-dependent changes in the expression of EMT-related genes. In contrast, MEG3 overexpression partially influenced the expression of EMT-related genes, including CDH1, ZEB family, and miR-200 family, and contributed to E-cadherin down-regulation. Mechanistic investigations suggested that MEG3 could associate with the regulatory regions of CDH1 and miR-200 family genes and induce the recruitment of JARID2 and EZH2 and histone H3K27 methylation on these regions for transcriptional repression. This study demonstrated a novel role of MEG3 lncRNA in the epigenetic regulation of EMT process of cancer cells.
PRC2 plays an essential role in development, stem cell identity, and disease pathogenesis, including cancer (17, 18). The catalytic subunit, EZH2, regulates histone H3K27 methylation and establishes a chromatin structure for transcriptional repression. EZH2 overexpression is frequently observed in human solid tumors and is associated with a poor prognosis in several tumor types (19). Thus increased activity of EZH2 or PRC2 is proposed to be responsible for malignant phenotypes of cancer cells. It has been demonstrated that EZH2 could down-regulate the expression of CDH1 and miR-200 family genes through histone H3K27 methylation in cancer cells (14). We also found that EED, a core component of PRC2, and JARID2, an accessory factor of PRC2, were required for transcriptional repression of these genes through H3K27 methylation, which was essential for TGF-β-induced EMT (16, 24). Here we showed that MEG3 knockdown counteracted TGF-β-induced EMT of lung cancer cells by inhibiting EZH2 recruitment, H3K27 methylation, and transcriptional repression of these genes. Therefore, our studies validated the importance of PRC2 recruitment and activity during the EMT-inducing transcriptional program.
The core components of the mammalian PRC2 complex do not have sequence-specific DNA binding activities. Thus it would be an important issue how PRC2 is recruited to its specific target genes for transcriptional repression. So far, chromatin targeting mechanisms of PRC2 have been proposed, which include the interactions with various DNA-binding factors and long noncoding RNAs. During embryonic development, JARID2 is one of the essential factors for PRC2 recruitment (20–23). We also proposed that JARID2 was essential for PRC2 recruitment in TGF-β-induced EMT of lung cancer cells (24). Recently, a growing body of evidence has indicated that long noncoding RNAs play a crucial role in the recruitment of PRC2 and JARID2 to the target loci (34, 35, 45, 46). Although JARID2 and PRC2 could associate with many lncRNAs, Meg3 was shown to be one of the important regulators of JARID2 and PRC2 recruitment in mouse ES cells (34, 35). Meg3 bound to JARID2, stimulated the assembly of JARID2 and PRC2, and recruited them to the specific target genes in pluripotent stem cells. It was also emphasized that Meg3 affected JARID2 and PRC2 recruitment on only a subset of genes but not all of PRC2-regulated loci (34). Our previous studies revealed that transcriptional regulation of CDH1 and miR-200 family genes was dependent on JARID2 and PRC2, because knockdown of JARID2 or EED completely inhibited TGF-β-induced repression of these genes. In this study, we showed that MEG3 knockdown also inhibited EZH2 recruitment, H3K27 methylation, and transcriptional repression of these genes, and eventually EMT was induced by TGF-β (Figs. 2–4). In the MEG3 knockdown cells, elevated expression of endogenous JARID2 was observed after TGF-β treatment (Fig. 3, A and C), but JARID2 might not contribute to EZH2 recruitment and transcriptional repression of these genes without MEG3 expression (Figs. 3 and 4). These results suggested that transcriptional regulation of the critical target genes such as the CDH1 and miR-200 families, which were responsible for EMT progression in A549 and LC-2/ad lung cancer cells, was highly dependent on the function of MEG3, JARID2, and PRC2.
The experiments of MEG3 overexpression provide us another interesting clue for understanding the epigenetic regulation of EMT. MEG3 overexpression could induce the interaction and recruitment of JARID2 and EZH2, H3K27 methylation, and transcriptional repression on CDH1 and miR-200 family genes in the absence of TGF-β (Figs. 6, 7, and 10). This was in contrast to the previous observation that JARID2 overexpression itself had no effect (24). However, MEG3 itself could not induce the EMT phenotype completely because it could change the expression of only a subset of the EMT-related genes (Figs. 5 and 6). For example, the expression of mesenchymal markers, FN1/fibronectin and vimentin was not altered by MEG3 overexpression. These results suggested that the MEG3-dependent function of JARID2 and PRC2 on the specific target genes was required but not sufficient for the EMT-inducing program in lung cancer cells.
The presence of repressive H3K27me3 and active H3K4me3 marks on the gene promoter was not restricted to multipotent cells but was detected in more differentiated cells (47). For the genes that possessed these opposing methylation marks, it was reported that relative intensities of H3K27me3 and H3K4me3 determined their transcription levels (48). Our ChIP experiments were correlated with these previous studies; TGF-β resulted in the increase of EZH2 recruitment and K27me3 and the substantial decrease of K4me3 on the CDH1, miR-200b/200a/429, and miR-200c/141 genes, thereby causing transcriptional repression, but MEG3 knockdown inhibited TGF-β-dependent epigenetic changes, consequently relieving transcriptional repression (Figs. 3 and 4). In mammalian cells, H3K27 methylation is catalyzed by Polycomb family, and H3K4 methylation is mostly regulated by COMPASS family proteins (49). These families are well known for their opposing roles in balancing gene expression. In this study, the function of PRC2 in H3K27 methylation and transcriptional repression was mainly investigated, but the regulation of H3K4 methylation remained unknown. However, we and another group (8, 15) previously proposed a critical role of H3K4-methyl-modifying enzymes, including KDM5B and LSD1 in the promotion of EMT. Furthermore, it has been reported that HOTAIR lncRNA could mediate interaction of the PRC2- and LSD1-containing complex and coordinate the regulation of H3K27 and H3K4 methylation on the target genes (45). To understand the epigenetic regulation of EMT in detail, further studies will be required for the functional interaction of the H3K4-methyl-modifying complex and JARID2-containing PRC2 complex.
Rapidly accumulating evidence reveals that lncRNA is involved in the initiation and progression of human cancer (26–28). In addition to the observed deregulation of lncRNA expression in cancer, lncRNAs have been demonstrated to act as oncogenes or tumor suppressor genes. Although lncRNAs have been scarcely functionally characterized, some of them can serve as decoys to bind microRNAs, scaffolds to regulate protein-protein interaction, or guides to recruit epigenetic regulators on chromatin (25). The MEG3 gene encodes one of the lncRNAs and is expressed in many normal tissues. In many types of human tumors and cancer cell lines, loss of MEG3 expression has been detected (50). Re-introduction of MEG3 inhibited cell proliferation and colony formation of cancer cell lines, partly by MEG3-mediated induction of apoptosis (51, 52). MEG3 could also stabilize p53 protein and stimulated p53-mediated transcription of the genes (51, 53). Thus it is hypothesized that MEG3 is a tumor suppressor gene (50). In contrast to these results, our study indicated an important function of MEG3 in EMT, a hallmark of malignant tumor progression. In general, the roles of epigenetic enzymes and cofactors in cancer are sometimes controversial. Overexpression of EZH2 was detected in many types of tumors and correlated with poor prognosis (19), but its mutation was also detected in various myeloid and lymphoid neoplasms (54). JARID2 gene deletion was reported in leukemic transformation of chronic myeloid diseases (55), but a higher level expression of JARID2 was associated with metastasis in rhabdomyosarcomas (56). Thus the contribution of these epigenetic regulators to cancer development is highly dependent on many factors and cellular context. Therefore, careful analyses and discussions should be performed to understand the roles of the epigenetic regulatory factors.
We have found a novel function of MEG3 lncRNA during TGF-β-induced EMT of A549 and LC-2/ad lung cancer cell lines. MEG3 played an important role in the EMT process by regulating EMT-related gene expression through the modulation of PRC2 recruitment and histone H3 methylation. Our study provides novel insights into the involvement of long noncoding RNAs for epigenetic mechanism of cancer malignancies.
Experimental Procedures
Plasmids
The retrovirus vectors expressing small hairpin RNAs (shRNAs) were constructed as described previously (7). The oligonucleotide sequences for shRNAs are listed in supplemental Table S1. The sequences of control and JARID2 shRNAs were described previously (7, 24). For human MEG3 cDNA, the primer set described in supplemental Table S1 was designed based on the previous report (34) and was used for cloning. The amplified cDNA was cloned into pDON-5 Neo plasmid (Takara, Ohtsu, Japan) to produce retrovirus-expressing MEG3. pMXs-puro plasmid was kindly provided by Dr. C. Takahashi (Kanazawa University, Japan), and the plasmid that produces retrovirus expressing FLAG-tagged JARID2 was constructed as described previously (24).
Cell Culture and Transfections
LC-2/ad human lung cancer cell line was purchased from RIKEN BRC, Japan, and was maintained in the mixture of RPMI 1640 and Ham's F-12 medium with 10% FBS, 2 mm glutamine, and penicillin/streptomycin (Sigma) at 37 °C in 5% CO2. A549 human lung cancer cell line was kindly provided by Dr. Y. Endo (Kanazawa University, Japan) and maintained as described previously (16). For EMT induction, A549 and LC-2/ad cells were treated with 1 ng/ml TGF-β (R&D Systems, Minneapolis, MN) for 24 h to 6 days. The protocols for the production and infection of shRNA or cDNA-expressing retroviruses were essentially the same as described previously (7).
Quantitative PCR
Preparation of RNA and quantitative RT-PCR was performed as described previously (7). Quantitative PCR data were normalized with control human GAPDH expression. The averages from at least three independent experiments are shown with the standard deviations. p values were calculated between control and the samples using Student's t test. Primers used for the quantitative PCR were described previously (7, 8, 24) and are listed in supplemental Table S1. For the quantification of microRNAs, we used TaqMan microRNA assays (Applied Biosystems, Waltham, MA) for miR-200a (catalog no. 000502) and miR-200c (Applied Biosystems). All data were normalized with RNU6B (Applied Biosystems) expression.
Cell Staining, Immunofluorescence, and Immunoblotting
To observe the changes of cell morphologies, A549 or LC-2/ad cells were stained with 0.4% crystal violet. To visualize the actin cytoskeleton, the cells were stained with 0.25 μm TRITC-conjugated phalloidin (Sigma). For indirect immunofluorescence, the specimens were treated with anti-E-cadherin antibody (catalog no. 610181, BD Biosciences) and incubated with Alexa546-conjugated anti-mouse IgG antibody (Invitrogen). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Immunoblotting was done as described previously (8). We used anti-E-cadherin, anti-fibronectin (SAB4500974, Sigma), anti-vimentin (ab8069, Abcam, Cambridge, MA), anti-ZEB1 (catalog no. 3396, Cell Signaling, Danvers, MA), anti-ZEB2 (catalog no. 61096, Active Motif, Carlsbad, CA), anti-phosphorylated SMAD3 (ab51451, Abcam), and anti-GAPDH (6C5, Millipore) antibodies.
Cell Migration Assay
Cell migration abilities were measured in modified Boyden chambers consisting of Transwell membrane filter inserts (catalog no. 3422, Corning Costar). Serum-starved cells (2 × 105) suspended in 100 μl of DMEM containing 1 mg/ml BSA and 0.5% FBS were cultured in each Transwell chamber and allowed to migrate toward the underside of the membrane for 24 h for A549 cells. The lower chamber contained DMEM with 10% FBS. For TGF-β-treated cells, cells were pre-treated with 1 ng/ml TGF-β for 4 days and then allowed to migrate. Cells that had not penetrated the filter were wiped out, and cells on the lower surface of the filter were stained with 0.4% crystal violet. The number of migrated cells was counted under a light microscope from at least five fields and three experiments.
ChIP Assays
ChIP assays were carried out essentially as described previously (7, 36). The cross-linked cell lysates were immunoprecipitated with the following antibodies: anti-H3K27me3 and anti-H3K4me3 (36); anti-EZH2 (catalog no. 17-662, Millipore); anti-JARID2 (catalog no. NB100-2214, Novus Biologicals, Littleton, CO); and anti-FLAG (M2, catalog no. F1804, Sigma). Quantitative PCR was performed to detect the enrichment of a specific amplified region. Percentage enrichment over input chromatin was presented. Primers used for the QPCR correspond to region a of CDH1 gene, region b of miR-200b/a/429 gene, region b of miR-200c/141 gene; and region a of GAPDH gene, as described previously (8).
RIP
The preparation of cross-linked cell lysates was essentially the same with our ChIP experiments except that cells were lysed with RIP buffer (50 mm Tris-Cl, pH 7.5, 150 mm NaCl, 10 mm EDTA, 0.5% Nonidet P-40, protease inhibitors (Nakalai, Kyoto, Japan) and SUPERase-In (Thermo Fisher Scientific)). The lysates were treated with anti-FLAG antibody or normal mouse IgG bound to Dynabeads M-280 sheep anti-mouse IgG (Invitrogen). The co-precipitated RNAs were extracted with High Pure RNA tissue kit (Roche Applied Science, Basel, Switzerland) by following the manufacturer's instruction and were quantified by quantitative RT-PCR.
Immunoprecipitation Analysis
The infected cells were lysed in RIPA buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 0.1% SDS, 1 mm EDTA supplemented with 500 mm 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride, 150 nm aprotinin, 1 mm E-64, and 1 mm leupeptin), and the lysates were immunoprecipitated with anti-EZH2 antibody (catalog no. 17-662, Millipore) coupled with protein G-Sepharose 4 fast flow (GE Healthcare, South East England, UK). The precipitates were subjected to immunoblotting with anti-EZH2 and anti-FLAG antibodies (M2, catalog no. F1804, Sigma).
ChIRP Assays
The ChIRP experiment was performed as described by Chu et al. (37) with slight modifications. Briefly, antisense DNA probes against MEG3 lncRNA were designed by ChIRP Probe Designer and are listed in supplemental Table S1. The 3 ′-end Biotin-TEG-modified probes were synthesized by Rikaken Co. (Nagoya, Japan). A549 cells were infected with the control retrovirus or the retrovirus expressing MEG3 with or without treatment of TGF-β and were fixed with 1% glutaraldehyde for 10 min. The cross-linked cells were lysed with lysis buffer (50 mm Tris-Cl, pH 7.5, 10 mm EDTA, 1% SDS, protease inhibitors, and SUPERase-In). The lysates were sonicated by Bioruptor (Diagenode, Denville, NJ) at 4 °C at high setting with 30 s on and 45 s off pulse intervals for a total of 30 min. The sonicated cell lysates were hybridized with the mixture of biotinylated DNA probes against human MEG3 in hybridization buffer (50 mm Tris-Cl, pH 7.5, 750 mm NaCl, 1% SDS, 1 mm EDTA, 15% formamide, protease inhibitors and SUPERase-In) for overnight at 4 °C. Then the binding complexes were recovered by streptavidin-conjugated magnet beads C1 (Invitrogen), and DNA was eluted with elution buffer (50 mm NaHCO3, 1% SDS). Quantitative PCR was performed to detect the enrichment of specific regulatory regions that are associated with MEG3 lncRNA, and percentage enrichment of the locus over input DNA was presented.
Author Contributions
T. S. and M. T. designed the experiments. M. T. performed most of the experiments. S. T. and A. I. performed part of the experiments. T. S. wrote the manuscript with the help of M. T. and A. I.
Supplementary Material
Acknowledgments
We thank Drs. C. Takahashi and Y. Endo (Kanazawa University, Japan) for providing the plasmid and the cell line. We also thank Dr. H. Kimura (Tokyo Institute of Technology, Japan) and Dr. N. Nozaki (Monoclonal Antibody Institute, Japan) for providing the antibodies that specifically recognize methylated H3K residues.
Note Added in Proof
In the version that was published as a Paper in Press on November 16, 2016, the incorrect E-cadherin immunoblot was used in Fig. 3D. This error has now been corrected and does not affect the results or conclusions of this work.
This work was supported in part by Grants-in-aid for Scientific Research C 24590346 and 15K08263 (to T. S.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table S1 and Figs. S1 and S2.
- KMT
- lysine methyltransferase
- KDM
- lysine demethylase
- EMT
- epithelial-mesenchymal transition
- lncRNA
- long noncoding RNA
- QRT-PCR
- quantitative reverse transcription-PCR
- RIP
- RNA immunoprecipitation
- ChIRP
- chromatin isolation by RNA purification
- TRITC
- tetramethylrhodamine isothiocyanate
- KD
- knockdown
- iPS
- induced pluripotent stem.
References
- 1. Bannister A. J., and Kouzarides T. (2011) Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Greer E. L., and Shi Y. (2012) Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 13, 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kooistra S. M., and Helin K. (2012) Molecular mechanisms and potential functions of histone demethylases. Nat. Rev. Mol. Cell Biol. 13, 297–311 [DOI] [PubMed] [Google Scholar]
- 4. Suzuki T., Terashima M., Tange S., and Ishimura A. (2013) Roles of histone methyl-modifying enzymes in development and progression of cancer. Cancer Sci. 104, 795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Suzuki T., Shen H., Akagi K., Morse H. C., Malley J. D., Naiman D. Q., Jenkins N. A., and Copeland N. G. (2002) New genes involved in cancer identified by retroviral tagging. Nat. Genet. 32, 166–174 [DOI] [PubMed] [Google Scholar]
- 6. Suzuki T., Minehata K., Akagi K., Jenkins N. A., and Copeland N. G. (2006) Tumor suppressor gene identification using retroviral insertional mutagenesis in Blm-deficient mice. EMBO J. 25, 3422–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yoshida M., Ishimura A., Terashima M., Enkhbaatar Z., Nozaki N., Satou K., and Suzuki T. (2011) PLU1 histone demethylase decreases the expression of KAT5 and enhances the invasive activity of the cells. Biochem. J. 437, 555–564 [DOI] [PubMed] [Google Scholar]
- 8. Enkhbaatar Z., Terashima M., Oktyabri D., Tange S., Ishimura A., Yano S., and Suzuki T. (2013) KDM5B histone demethylase controls epithelial-mesenchymal transition of cancer cells by regulating the expression of the microRNA-200 family. Cell Cycle 12, 2100–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Oktyabri D., Ishimura A., Tange S., Terashima M., and Suzuki T. (2016) DOT1L histone methyltransferase regulates the expression of BCAT1 and is involved in sphere formation and cell migration of breast cancer cell lines. Biochimie 123, 20–31 [DOI] [PubMed] [Google Scholar]
- 10. Kalluri R., and Weinberg R. A. (2009) The basics of epithelial-mesenchymal transition. J. Clin. Invest. 119, 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Miyazono K., Ehata S., and Koinuma D. (2012) Tumor-promoting functions of transforming growth factor-β in progression of cancer. Uppsala J. Med. Sci. 117, 143–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tam W. L., and Weinberg R. A. (2013) The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat. Med. 19, 1438–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang Y., and Shang Y. (2013) Epigenetic control of epithelial-to-mesenchymal transition and cancer metastasis. Exp. Cell Res. 319, 160–169 [DOI] [PubMed] [Google Scholar]
- 14. Cao Q., Yu J., Dhanasekaran S. M., Kim J. H., Mani R. S., Tomlins S. A., Mehra R., Laxman B., Cao X., Yu J., Kleer C. G., Varambally S., and Chinnaiyan A. M. (2008) Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene 27, 7274–7284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lin T., Ponn A., Hu X., Law B. K., and Lu J. (2010) Requirement of the histone demethylase LSD1 in Snai1-mediated transcriptional repression during epithelial-mesenchymal transition. Oncogene 29, 4896–4904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oktyabri D., Tange S., Terashima M., Ishimura A., and Suzuki T. (2014) EED regulates epithelial-mesenchymal transition of cancer cells induced by TGF-β. Biochem. Biophys. Res. Commun. 453, 124–130 [DOI] [PubMed] [Google Scholar]
- 17. Margueron R., and Reinberg D. (2011) The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Di Croce L., and Helin K. (2013) Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 20, 1147–1155 [DOI] [PubMed] [Google Scholar]
- 19. Chase A., and Cross N. C. (2011) Aberrations of EZH2 in cancer. Clin. Cancer Res. 17, 2613–2618 [DOI] [PubMed] [Google Scholar]
- 20. Peng J. C., Valouev A., Swigut T., Zhang J., Zhao Y., Sidow A., and Wysocka J. (2009) Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 139, 1290–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shen X., Kim W., Fujiwara Y., Simon M. D., Liu Y., Mysliwiec M. R., Yuan G. C., Lee Y., and Orkin S. H. (2009) Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell 139, 1303–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pasini D., Cloos P. A., Walfridsson J., Olsson L., Bukowski J. P., Johansen J. V., Bak M., Tommerup N., Rappsilber J., and Helin K. (2010) JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 464, 306–310 [DOI] [PubMed] [Google Scholar]
- 23. Li G., Margueron R., Ku M., Chambon P., Bernstein B. E., and Reinberg D. (2010) Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 24, 368–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tange S., Oktyabri D., Terashima M., Ishimura A., and Suzuki T. (2014) JARID2 is involved in transforming growth factor-β-induced epithelial-mesenchymal transition of lung and colon cancer cell lines. PLoS One 9, e115684, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guttman M., and Rinn J. L. (2012) Modular regulatory principles of large non-coding RNAs. Nature 482, 339–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Iyer M. K., Niknafs Y. S., Malik R., Singhal U., Sahu A., Hosono Y., Barrette T. R., Prensner J. R., Evans J. R., Zhao S., Poliakov A., Cao X., Dhanasekaran S. M., Wu Y. M., Robinson D. R., et al. (2015) The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 47, 199–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yan X., Hu Z., Feng Y., Hu X., Yuan J., Zhao S. D., Zhang Y., Yang L., Shan W., He Q., Fan L., Kandalaft L. E., Tanyi J. L., Li C., Yuan C. X., et al. (2015) Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell 28, 529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bartonicek N., Maag J. L., and Dinger M. E. (2016) Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol. Cancer 15, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rinn J. L., Kertesz M., Wang J. K., Squazzo S. L., Xu X., Brugmann S. A., Goodnough L. H., Helms J. A., Farnham P. J., Segal E., and Chang H. Y. (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhao J., Sun B. K., Erwin J. A., Song J. J., and Lee J. T. (2008) Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322, 750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Scheel C., and Weinberg R. A. (2012) Cancer stem cells and epithelial-mesenchymal transition: concepts and molecular links. Semin. Cancer Biol. 22, 396–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hu W., Alvarez-Dominguez J. R., and Lodish H. F. (2012) Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep. 13, 971–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ghosal S., Das S., and Chakrabarti J. (2013) Long noncoding RNAs: new players in the molecular mechanism for maintenance and differentiation of pluripotent stem cells. Stem Cells Dev. 22, 2240–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kaneko S., Bonasio R., Saldaña-Meyer R., Yoshida T., Son J., Nishino K., Umezawa A., and Reinberg D. (2014) Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol. Cell 53, 290–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. da Rocha S. T., Boeva V., Escamilla-Del-Arenal M., Ancelin K., Granier C., Matias N. R., Sanulli S., Chow J., Schulz E., Picard C., Kaneko S., Helin K., Reinberg D., Stewart A. F., Wutz A., et al. (2014) Jarid2 is implicated in the initial xist-induced targeting of PRC2 to the inactive X chromosome. Mol. Cell 53, 301–316 [DOI] [PubMed] [Google Scholar]
- 36. Kimura H., Hayashi-Takanaka Y., Goto Y., Takizawa N., and Nozaki N. (2008) The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell Struct. Funct. 33, 61–73 [DOI] [PubMed] [Google Scholar]
- 37. Chu C., Quinn J., and Chang H. Y. (2012) Chromatin isolation by RNA purification (ChIRP). J. Vis. Exp. 61, 3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kasai H., Allen J. T., Mason R. M., Kamimura T., and Zhang Z. (2005) TGF-β1 induces human alveolar epithelial to mesenchymal cell transition (EMT). Respir. Res. 6, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peinado H., Olmeda D., and Cano A. (2007) Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat. Rev. Cancer 7, 415–428 [DOI] [PubMed] [Google Scholar]
- 40. Gregory P. A., Bert A. G., Paterson E. L., Barry S. C., Tsykin A., Farshid G., Vadas M. A., Khew-Goodall Y., and Goodall G. J. (2008) The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10, 593–601 [DOI] [PubMed] [Google Scholar]
- 41. Park S. M., Gaur A. B., Lengyel E., and Peter M. E. (2008) The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 22, 894–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Siegel P. M., and Massagué J. (2003) Cytostatic and apoptotic actions of TGF-β in homeostasis and cancer. Nat. Rev. Cancer 3, 807–821 [DOI] [PubMed] [Google Scholar]
- 43. Koinuma D., Tsutsumi S., Kamimura N., Taniguchi H., Miyazawa K., Sunamura M., Imamura T., Miyazono K., and Aburatani H. (2009) Chromatin immunoprecipitation on microarray analysis of Smad2/3 binding sites reveals roles of ETS1 and TFAP2A in transforming growth factor β signaling. Mol. Cell. Biol. 29, 172–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Davalos V., Moutinho C., Villanueva A., Boque R., Silva P., Carneiro F., and Esteller M. (2012) Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene 31, 2062–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tsai M. C., Manor O., Wan Y., Mosammaparast N., Wang J. K., Lan F., Shi Y., Segal E., and Chang H. Y. (2010) Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhao J., Ohsumi T. K., Kung J. T., Ogawa Y., Grau D. J., Sarma K., Song J. J., Kingston R. E., Borowsky M., and Lee J. T. (2010) Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell 40, 939–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mikkelsen T. S., Ku M., Jaffe D. B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T. K., Koche R. P., Lee W., Mendenhall E., O'Donovan A., Presser A., Russ C., et al. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ke X. S., Qu Y., Cheng Y., Li W. C., Rotter V., Øyan A. M., and Kalland K. H. (2010) Global profiling of histone and DNA methylation reveals epigenetic-based regulation of gene expression during epithelial to mesenchymal transition in prostate cells. BMC Genomics 11, 669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Piunti A., and Shilatifard A. (2016) Epigenetic balance of gene expression by Polycomb and COMPASS families. Science 352, aad9780. [DOI] [PubMed] [Google Scholar]
- 50. Zhou Y., Zhang X., and Klibanski A. (2012) MEG3 noncoding RNA: a tumor suppressor. J. Mol. Endocrinol. 48, R45–R53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang X., Gejman R., Mahta A., Zhong Y., Rice K. A., Zhou Y., Cheunsuchon P., Louis D. N., and Klibanski A. (2010) Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res. 70, 2350–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Braconi C., Kogure T., Valeri N., Huang N., Nuovo G., Costinean S., Negrini M., Miotto E., Croce C. M., and Patel T. (2011) microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene 30, 4750–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhou Y., Zhong Y., Wang Y., Zhang X., Batista D. L., Gejman R., Ansell P. J., Zhao J., Weng C., and Klibanski A. (2007) Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 282, 24731–24742 [DOI] [PubMed] [Google Scholar]
- 54. Ernst T., Chase A. J., Score J., Hidalgo-Curtis C. E., Bryant C., Jones A. V., Waghorn K., Zoi K., Ross F. M., Reiter A., Hochhaus A., Drexler H. G., Duncombe A., Cervantes F., Oscier D., et al. (2010) Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat. Genet. 42, 722–726 [DOI] [PubMed] [Google Scholar]
- 55. Puda A., Milosevic J. D., Berg T., Klampfl T., Harutyunyan A. S., Gisslinger B., Rumi E., Pietra D., Malcovati L., Elena C., Doubek M., Steurer M., Tosic N., Pavlovic S., Guglielmelli P., et al. (2012) Frequent deletions of JARID2 in leukemic transformation of chronic myeloid malignancies. Am. J. Hematol. 87, 245–250 [DOI] [PubMed] [Google Scholar]
- 56. Walters Z. S., Villarejo-Balcells B., Olmos D., Buist T. W., Missiaglia E., Allen R., Al-Lazikani B., Garrett M. D., Blagg J., and Shipley J. (2014) JARID2 is a direct target of the PAX3-FOXO1 fusion protein and inhibits myogenic differentiation of rhabdomyosarcoma cells. Oncogene 33, 1148–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.