Abstract
The network of activator protein-protein interactions (PPIs) that underpin transcription initiation is poorly defined, particularly in the cellular context. The transient nature of these contacts and the often low abundance of the participants present significant experimental hurdles. Through the coupling of in vivo covalent chemical capture and shotgun LC-MS/MS (MuDPIT) analysis, we can trap the PPIs of transcriptional activators in a cellular setting and identify the binding partners in an unbiased fashion. Using this approach, we discover that the prototypical activators Gal4 and VP16 target the Snf1 (AMPK) kinase complex via direct interactions with both the core enzymatic subunit Snf1 and the exchangeable subunit Gal83. Further, we use a tandem reversible formaldehyde and irreversible covalent chemical capture approach (TRIC) to capture the Gal4-Snf1 interaction at the Gal1 promoter in live yeast. Together, these data support a critical role for activator PPIs in both the recruitment and positioning of important enzymatic complexes at a gene promoter and represent a technical advancement in the discovery of new cellular binding targets of transcriptional activators.
Graphical abstract
INTRODUCTION
Executive orders within a cell are implemented by multi-component complexes whose functional state and subcellular location are often influenced by association with exchangeable subunits.1–3 In the process of transcription initiation, for example, several lines of evidence indicate that the Snf1/AMPK kinase complex arrives at the promoters of galactose catabolism genes only when the exchangeable subunit Gal83 is present within the complex.4 Although the molecular mechanism by which enzymatic complexes such as Snf1/AMPK localize to promoters has been the subject of debate, biochemical and genetic data suggest that the most likely suspects performing the recruitment function are one or more transcriptional activators via protein-protein interactions (PPIs).5–9 Distinguishing which of the components of an enzymatic (or coactivator) complex is the binding target of a promoter-bound activator is, however, quite difficult using conventional strategies. This is particularly true in cellular settings, where the transient nature of the activator-PPI network and the structural plasticity of the individual binding partners prove especially challenging.10–12 For this reason, the PPI network of transcriptional activators that underpins transcriptional activation is limited to a handful of transcriptional machinery proteins.13
The prototypical transcriptional activator Gal4 stimulates the colocalization of a long list of enzymatic complexes at the promoters of yeast galactose catabolism genes.14 In the presence of glucose, Gal4 is tightly repressed by its masking protein Gal80; however, when galactose becomes the primary carbon source, repression of Gal4 is relieved, enabling recruitment of complexes involved in galactose sensing and catabolism.15 To define the PPI network that Gal4 engages to recruit these functions to promoters, we sought an approach that would mitigate false positives caused by the often promiscuous in vitro binding profile of transcriptional activators. Thus, we focused on the interrogation of activator PPIs in the native cellular environment.
Here we describe the combination of in vivo covalent chemical capture with the photo-cross-linking amino acid p-benzoyl-L-phenylalanine (Bpa) and label-free shotgun liquid chromatography mass spectrometry analysis using the multidimensional protein identification technology (MuDPIT) to identify novel binding targets of the yeast activator Gal4 in live Saccharomyces cerevisiae that accompany gene activation (Figure 1). Both Gal4 and the viral activator VP16 associate directly with two components within the central metabolic regulator Snf1/AMPK: the kinase Snf1 and an exchangeable subunit Gal83. Additionally, the combined use of reversible formaldehyde-based cross-linking and covalent chemical capture pinpoints the location of the Gal4-Snf1 PPI directly within the Gal1 promoter region. These data support a central role for amphipathic activators in the functional response of Snf1/AMPK to changes in cellular environment. As PPI networks emerge as viable therapeutic targets, covalent chemical capture in vivo will be a powerful tool for the discovery of molecular interaction points, particularly in dynamic assemblies.
Figure 1.
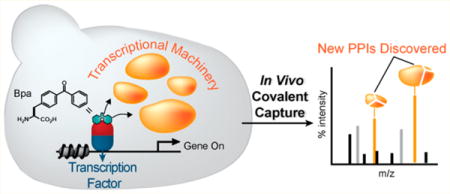
An in vivo covalent chemical capture and mass spectrometric-based approach for the identification of the cellular targets of transcriptional activators. Live yeast expressing Bpa-containing activators (blue and red) are irradiated with UV light to covalently capture the spectrum of protein binding partners that activators directly contact in cells. Following cell lysis, affinity chromatography and immunoprecipitation are used to enrich for cross-linked complexes. The purified products are then subjected to mass spectrometric-based analysis such as Mud-PIT, thus allowing for the concurrent verification of previously identified activator targets (green) and the discovery of novel binding partners (orange).
RESULTS AND DISCUSSION
Combined in Vivo Covalent Chemical Capture and MuDPIT Identifies Gal4-Gal80 Interaction
For in vivo covalent chemical capture, the strategy of unnatural amino acid incorporation using nonsense suppression was employed.16–18 Incorporation of Bpa at position 849 in Gal4 results in the robust capture of several unidentified protein partners, illustrating the complexity of activator-PPI networks at a single position. Therefore, Bpa incorporation at position Phe849 in Gal4 was selected for the combined covalent chemical capture-MuDPIT experiments.16 Initial optimization studies with this approach examined the sugar-dependent changes of the complex between Gal4 and Gal80 at a genomically integrated Gal1-Lacz reporter gene.19 Importantly, earlier work by our group demonstrated that Bpa-containing Gal4 mutants retain responsiveness to changes in nutrient availability (glucose, raffinose, and raffinose-galactose).16 Thus, the Bpa-containing Gal4 mutants in our yeast expression system behave in accordance with the findings of others who have studied activation and repression at the Gal1 promoter under various nutrient contexts.15,20 As shown in Figure 2a, Gal4 Phe849bpa is functionally active in galactose but inhibited in glucose, indicating a change in the Gal4-Gal80 interaction that is reflected in the in vivo covalent chemical capture-MuDPIT data in which Gal80 was significantly enriched under glucose growth conditions (Figure 2b and Supplementary Table 1). Together, these data indicate that the Gal4 mutant retains the responsiveness of its wild-type counterpart and additionally that Bpa cross-linking coupled with MuDPIT serves as a reliable platform upon which to examine activator PPIs in vivo.
Figure 2.
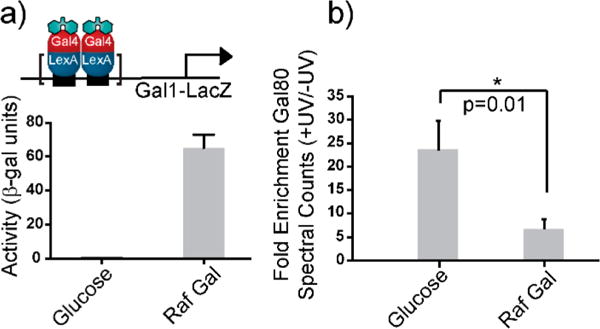
In vivo covalent chemical capture and MuDPIT validate the masking protein Gal80 as a direct target of Gal4 in live yeast and reveal novel direct targets within covalently captured complexes. (a) Incorporation of the unnatural amino acid Bpa at position 849 in the Gal4 transcriptional activation domain yields a functional activator that retains its ability to sense changes in carbon source availability, as determined via β-galactosidase assays on an integrated pGal1-LacZ reporter gene. In the presence of glucose, Gal4 is transcriptionally repressed but is activated in the presence of the inducing sugar, galactose, indicating that Gal4 remains responsive to repression by its masking protein Gal80. (b) In line with the activity data, in vivo covalent chemical capture followed by MuDPIT analysis of purified Gal4 covalent adducts shows significant enrichment of the Gal4-Gal80 interaction under repressive glucose conditions (N = 3) biological replicates.
Identification of Novel Targets of Gal4 Via in Vivo Covalent Chemical Capture and MuDPIT
Large scale (10L) transcriptionally active yeast cultures were subjected to the covalent chemical capture-MuDPIT workflow (see Supplementary Figure 5 and Supplementary Methods for detailed procedure). The resulting data sets for non UV-treated (N = 2 biological replicates) and UV-treated (N = 3 biological replicates) samples were compiled, and the spectral counts detected for each protein under each condition were averaged (See Supplementary Table 2 for complete MS data set and analysis). Proteins demonstrating a maximal mean of <2 spectral counts were removed from further analysis. The remaining 638 proteins were subjected to stringent filter cutoffs according to the following criteria (Figure 3a): (i) Proteins must display a minimum average of 5 spectral counts; (ii) proteins must show a fold enrichment (+UV/−UV) ≥5; and (iii) a t test between UV-treated and non-UV control data sets for a protein must generate a p-value of ≤0.05.21 A selection of protein targets meeting this criteria is displayed in Figure 3a.
Figure 3.
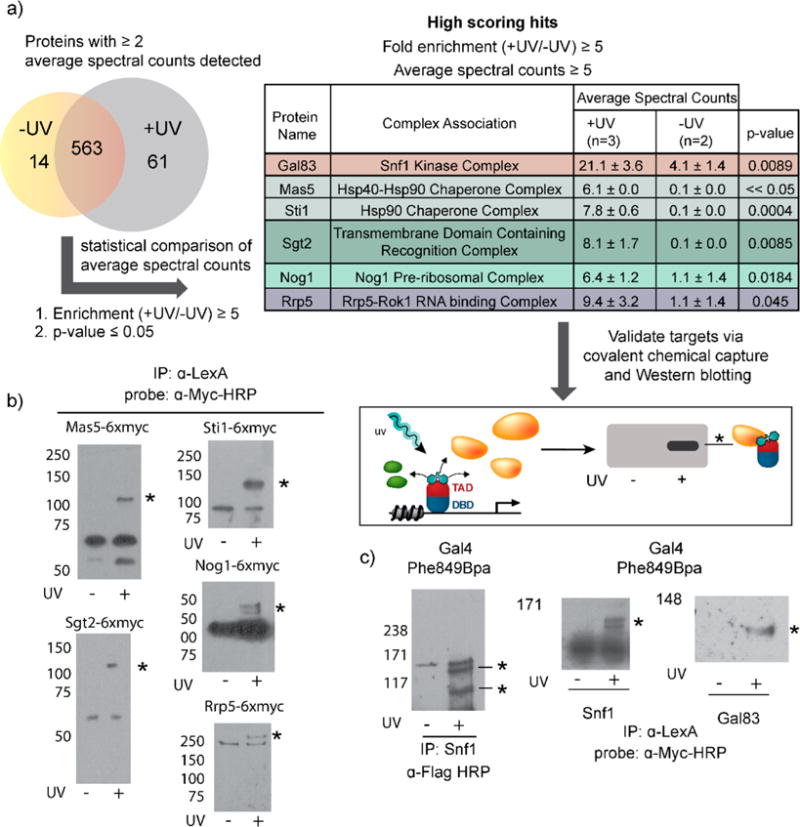
Covalent chemical capture and MuDPIT reveals several novel targets of the transcriptional activator Gal4 in vivo. (a) Data sets from UV treated (n = 3) and non-UV control (n = 2) data sets were compiled and filtered to remove proteins with less than two average spectral counts detected. Of the proteins remaining, 14 were unique to the non-UV treated samples, 61 proteins were unique to the UV-treated condition, and 563 proteins were detected under both conditions. Stringent filtering criteria were applied to generate a list of proteins that were significantly enriched in the UV treated sample set. Proteins that were <5-fold enriched (+UV/−UV) and with p-values >0.05 were removed from further analysis. A selection of previously unidentified direct targets of Gal4 Phe849Bpa are displayed as the number of spectral counts detected in UV treated and non-UV treated conditions. Novel targets detected via MuDPIT should be validated using covalent chemical capture Western blotting. See Supporting Information for additional details and complete MS results. (b) Covalent chemical capture Western blotting validates the proteins identified from MuDPIT analysis are genuine targets of Gal4 within their respective complexes. Proteins identified from MuDPIT analysis were co-expressed with Gal4 Phe849Bpa in yeast and then subjected to the covalent chemical capture workflow. Yeast lysates were immunoprecipitated with a LexA antibody to enrich for covalent activator species, and the subsequent Westerns were probed with a Myc-HRP antibody to detect the presence of covalently bound myc-tagged proteins. (c) Gal4 directly contacts the catalytic subunit Snf1 and the exchangeable subunit Gal83 in live yeast. In vivo covalent chemical capture was carried out in yeast expressing Gal4 Phe849Bpa. Immunoprecipitation of yeast lysates with a Snf1 antibody and Western detection with a Flag-HRP antibody indicated that Gal4 contacts two proteins within the Snf1 complex. Myc-tagged versions of the Snf1 and Gal83 subunits were co-expressed alongside Gal4 Phe849Bpa in yeast. The resulting yeast lysates were immunoprecipitated with a LexA antibody as indicated, and the immunoprecipitated complexes were analyzed by Western blot with a myc-HRP antibody to detect covalently bound Gal4 complexes.
These cross-linking-MS studies revealed several previously unidentified targets of Gal4, all of which participate in larger complexes (Figure 3a). To validate a direct interaction between Gal4 and each of the proteins, myc-tagged versions of each identified target were co-expressed alongside LexA+Gal4 F849Bpa, and covalent chemical capture experiments were carried out. Following irradiation of live cells, yeast lysates were enriched with a LexA antibody, and enriched proteins were analyzed via Western blot with a myc-antibody to detect the presence of a covalently bound activator/target complex (Figure 3b). Two chaperones, Mas5 and Sti1, directly interact with Gal4; consistent with this result, deletion of these chaperone proteins in yeast has been previously demonstrated to significantly delay or abrogate induction of Gal1.22 We additionally observe proteins involved in kinase complexes (Gal83),23,24 RNA binding complexes (Rrp5),25 and those that are regulated by the TOR pathway (Nog1) directly interact, suggesting that previously identified crosstalk between the TOR and Snf1 nutrient sensing pathways extends to the level of transcription.26–29
GAL4 Directly Contacts the SNF1 Kinase and GAL83 Exchangeable Subunit of the SNF1/AMPK Kinase Complex
Of the targets identified in this study, one protein of particular note belongs to the Snf1 kinase complex. The Snf1 complex is a heterotrimeric complex containing a catalytic alpha subunit (Snf1), a regulatory gamma subunit (Snf4), and a third beta subunit that exchanges between Sip1, Sip2, and Gal83 to regulate subcellular localization of the complex. Importantly, both Gal4 and the Snf1 complexes regulate galactose-inducible genes such as Gal1.14,30 The Snf1 complex plays a critical role in the response to environmental changes in carbon source.4,24 Several lines of evidence suggest that Snf1 phosphorylation of the Mig1 repressor contributes to the derepression of carbon source-regulated genes.24,31,32 The Gal83-containing isoform has been shown to be the complex that predominantly localizes to the nucleus, and, consistent with this model, our MuDPIT data shows a 5-fold increase in spectral counts over background, thus supporting a nuclear Gal4-Gal83 interaction when cells are grown in galactose.4 The Snf1 subunit of the complex is also present in the MuDPIT data set, but it appeared robustly under both conditions. A closer examination of the Snf1 sequence revealed the presence of a polyhistidine stretch on its amino terminus, and we thus postulated that it was likely enriched during the denaturing Ni-affinity column purification and additionally retained throughout the milder FLAG affinity purification, presumably via an interaction with Flag-tagged Gal4 (Supplementary Table 2 and Supplementary Figure 5). In support of Gal4 targeting multiple proteins within the Snf1 complex, enrichment of yeast lysates with a Snf1 antibody revealed two proteins that Gal4 directly contacts within the complex under irradiating conditions (Figure 3c). We were able to confirm Snf1 and Gal83 as direct targets of Gal4 by co-expressing Gal4 Phe849Bpa with either myc-Snf1 or myc-Gal83 and following our established covalent chemical capture workflow (Figure 3c). Furthermore, a functionally inactive mutant, Gal4 867–869Ala, was unable to cross-link to Snf1, demonstrating that this interaction is specific and is not mediated by the presence of Bpa (Supplementary Figure 3).
To further refine the Snf1 model, we performed chromatin immunoprecipitation (ChIP) to determine if Gal4 and Snf1 colocalize at the Gal1-LacZ gene used in our experiments. Consistent with earlier studies, both proteins showed significant enrichment, supporting a model where Gal4 and the Snf1 complex work collaboratively at Gal1 to drive reporter gene expression (Figure 4a).6 Additional ChIP experiments with the Gal4 triple alanine mutant show a substantial loss in Snf1 localization to Gal1, supporting the importance of Gal4 in coordinating Snf1 localization to the promoter (Supplementary Figure 2 and Supplementary Table 3). β-Galactosidase experiments with WT Gal4 in a Snf1 delete strain complemented with either WT Snf1 or a kinase dead T210A Snf1 mutant additionally support this model (Supplementary Figure 4).
Figure 4.
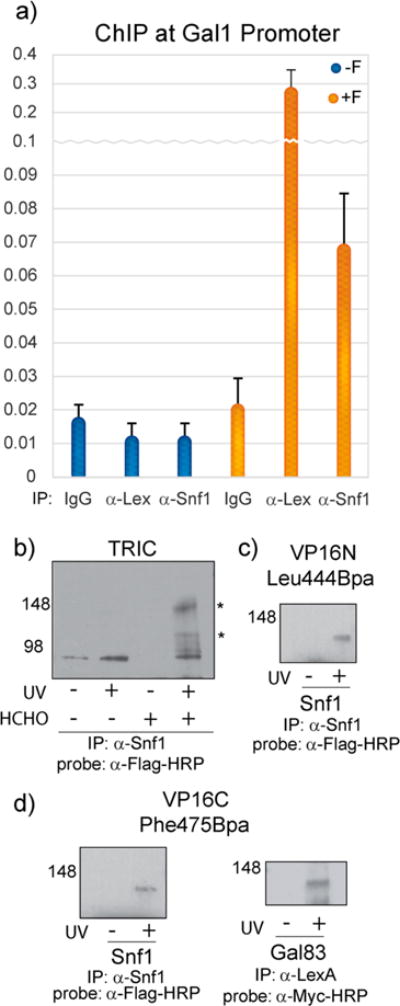
In vivo photo-cross-linking with Bpa captures the direct targets of amphipathic activators within the Snf1 kinase complex. (a) Chromatin immunoprecipitation with Snf1 and LexA antibodies verify that Gal4 and the Snf1 kinase complex colocalize to the Gal1-LacZ reporter gene used in these studies. (b) The Gal4-Snf1 interaction is maintained on DNA. TRIC was performed to investigate the interaction of DNA bound Gal4 with Snf1 at the Gal1 promoter. (c,d) The Snf1 complex is a common target of amphipathic activators. The amino- and carboxy-terminal subdomains of the amphipathic activator VP16 were tested for cross-linking to the catalytic subunit of the Snf1 kinase complex as well as a myc-Gal83 construct in yeast. Covalent complexes were analyzed by Western blot with a Flag or Myc antibody to detect covalently bound Snf1-VP16 and Gal83-VP16, respectively.
The GAL4-SNF1 PPI Is Sustained at the Gal1 Promoter
ChIP assays are suitable for determining the colocalization of proteins at the same promoter over a defined period of time, but they lack the resolution to determine if these proteins are in direct contact with one another when bound to DNA.33 In the absence of methods to accomplish this goal and determine if the Gal4-Snf1 interaction is sustained at Gal1, we used a new strategy which we have coined tandem reversible and irreversible cross-linking (TRIC) that combines the irreversible cross-link arising from UV-activated Bpa and the reversible protein-protein and protein-DNA cross-links formed upon formaldehyde (HCHO) treatment.34 In this experiment, cells bearing Gal4 modified to have Bpa at position 849 were first treated with formaldehyde to stabilize transient protein-DNA and PPIs. The cells were then subjected to our standard UV-irradiation protocol to activate the Bpa moiety and generate an irreversible covalent cross-link between Gal4 and its directly interacting protein partners within the immobilized complexes. The treated cells were then lysed, the chromatin fraction isolated and sheared, and then immunoprecipitation with α-Snf1 antibody was carried out. Next, the formaldehyde cross-links were reversed, thus leaving only the irreversible Bpa-cross-links intact, and a Western blot of the Bpa cross-linked species with α-FLAG antibody conclusively demonstrated that Gal4 directly interacts with the Snf1 complex on the DNA (Figure 4b). Taken together, these data support a model of recruitment for the Snf1 complex in which Gal4 directly contacts the catalytic Snf1 subunit and forms additional contacts with the exchangeable regulatory subunit Gal83.
SNF1 and GAL83 Are Shared Targets of Amphipathic Activators
Given the high homology between yeast Snf1 and mammalian AMPK, the interactions of Snf1 and VP16, an amphipathic viral activator that functions in the HSV-1 infection of mammalian cells, were examined.35 The VP16 transcriptional activation domain is large (77 amino acids) and consists of two subdomains that can function independently. As has been done previously, we examined the binding behavior of each subdomain individually.36 In vivo covalent chemical capture with VP16N (413–456) bearing Bpa at position 444 resulted in an observable cross-link with Snf1 (Figure 4c), but no interaction with Gal83 was identified for this subdomain under any conditions examined. In contrast, VP16C (446–490) with a Phe475Bpa mutation interacts with both of the subunits (Figure 4d). These data suggest that the Snf1 and Gal83 subunits of Snf1/AMPK are a common target for these two activators specifically and perhaps for amphipathic activators as a class.
DISCUSSION
Snf1/AMPK and Gal4 each play a central role in the ability of yeast to rapidly respond to changes in nutrient context.14,37 Upon the switch to galactose, the Gal83-containing isoform of Snf1/AMPK transits to the nucleus where it phosphorylates targets at galactose-catabolism genes, contributing to derepression of nutrient response genes.4,24 Galactose similarly signals a change in the Gal4-Gal80 complex, leading to rapid up-regulation of the GAL genes.14 The covalent chemical capture data with Gal4 reveal two direct PPIs that connect these two critical nutrient response pathways. Our results show that the activator Gal4 directly contacts not only the kinase Snf1 within the Snf1/AMPK complex but also the exchangeable subunit Gal83. It is conceivable that by contacting two components of a single complex, Gal4 is not only able to cooperatively recruit the Snf1/AMPK complex and its associated kinase activities but also to orient the complex in its appropriate configuration, ultimately providing a rapid transcriptional response to process the external stimulus. In retrospect, we observed the same phenomenon with the Swi/Snf chromatin modifying complex; in that case, two regions of VP16 contacted the enzymatic subunit and a second scaffolding component.36 An open question is what role these two distinct types of interactions play. Certainly, proper positioning at the promoter is critical, and, in the case of the VP16-Swi/Snf interaction, EM studies support this role.38 An additional possibility is that the PPIs alter the activity and/or substrate profile of the enzymatic subunit through allosteric effects. Studies investigating this latter possibility are currently underway.
The use of in vivo covalent chemical capture followed by adduct analysis via shotgun liquid chromatography-mass spectrometry (LC-MS) methods (e.g., MuDPIT) represents a significant step toward the development of a complete in vivo interaction map of transcriptional activators. While a powerful approach, it should be noted that issues of relative abundance, stability, and ease of digestion may contribute to uneven coverage of proteins across the proteome.39 Indeed, many of the “typical” targets of transcriptional activators are of lower abundance including, for example, Gal11/Med15.40,41 In the case of Gal11/Med15, even in an in vitro cross-linking experiment in which an abundance of cross-linked products (Gal11/Med15-activator) were isolated and purified prior to MS analysis, it was necessary to use two different digestion strategies and two different ionization techniques in order to achieve sufficient sequence coverage.42 Thus, continued advances in MS methods coupled with fine-tuning cross-linker reactivities will undoubtedly enhance the utility of tandem covalent chemical capture-MS approaches.43–46 Additionally, this study captured binding partners of Gal4 that rely upon the sequence surrounding position 849 for interaction. As there is considerable evidence that transcriptional activators use distinct but overlapping sequences within their activation domains to contact an array of binding partners, future experiments with a cross-linking amino acid placed at distinct positions will likely lead to the identification of additional coactivator targets.15,16,36,47
In this study, this strategy resulted in the identification of two subunits of the Snf1 kinase complex as novel, direct targets of the amphipathic activators Gal4 and VP16 in vivo. The Snf1 complex is highly conserved among eukaryotes, and the mammalian counterpart of the yeast Snf1 kinase complex, AMPK, plays a significant role in maintaining cellular homeostasis by functioning in some cases as a tumor suppressor and additionally as a regulator of energy response.37,48 Given its important role in the cell, AMPK is a critical target in the treatment of diseases that exhibit abnormal metabolic profiles including certain cancers and diabetes.49,50 The establishment of a direct link between the Snf1/AMPK complex and transcriptional activators suggests a novel intervention point for extrinsic regulation of Snf1/AMPK promoter activity. This will be the focus of future efforts.
EXPERIMENTAL SECTION
Yeast Strains and Plasmid Construction
Yeast strains used in this study were LS41 [JPY9::pZZ41, Matαhis3Δ200 leu2Δ1 trp1Δ63 ura3-52 lys2Δ385 gal4 URA::pZZ41] and ΔSnf1 LS41 [JPY9::pZZ41, Matαhis3Δ200 leu2Δ1 trp1Δ63 ura3-52 lys2Δ385 gal4 URA::pZZ41 SNF1::TRP1].
All plasmids were generated using standard restriction enzyme digestion and ligation reactions, and the sequences of all the isolated plasmids were verified by sequencing at the University of Michigan Core Facility (Ann Arbor, MI). T4 DNA ligase and all restriction enzymes were all purchased from New England Biolabs. Mutagenesis of Snf1 and Gal4 was carried out using a two-step site directed mutagenesis protocol (Quikchange, Qiagen).
In Vivo Covalent Chemical Capture
In vivo covalent chemical capture was carried out as previously described, but on a larger scale (1–10 L).16 A detailed procedure regarding culture growth, cryolysis, and protein purification conditions can be found in the Supplementary Methods.
β-Galactosidase Assays
To evaluate the ability of each LexA+Gal4 F849Bpa-Flag-6HIS to activate transcription in the presence or absence of glucose, saturated cultures (SC media + 2% raffinose) of yeast expressing Gal4 Phe849Bpa were used to inoculate 5 mL SC media containing either 2% glucose or 2% raffinose and 2% galactose but lacking histidine and tryptophan for selection. The cells were grown to an OD660 of 0.8–1 and harvested. The activity of each construct was monitored using β-galactosidase assays as previously described.16 Additional activity data can be found Supplementary Figures 3 and 4 and Supplementary Methods.
MuDPIT Analysis
All MuDPIT analyses were performed at the Center for Physiological Proteomics in La Jolla, California using a biphasic strong cation exchange/reverse phase capillary column for 2D liquid chromatography and an 11 step gradient on an LTQ mass spectrometer for tandem MS. The data were analyzed against the entire yeast genome using the SEQUEST algorithm, and the analysis was performed using the DTASelect software package. The complete MS data sets and analysis can be found in Supplementary Tables 1 and 2 and additional experimental details regarding sample runs and data analysis can be found in the Supplementary Methods.
Preparation and Western Blotting of Snf1 and Gal83 Cross-Linked Samples
In vivo covalent chemical capture was carried out as previously described.16 Following lysis, yeast whole cell lysates were immunoprecipitated with either a LexA antibody (sc-1725, Santa Cruz Biotechnologies) or a Snf1 antibody (sc-15621, Santa Cruz Biotechnologies), as indicated in figure legends. Immunoprecipitated proteins were immobilized to 40 μL Dynabeads Protein G magnetic bead slurry (Life Technologies) and washed. Western blots were probed with 1:1000 dilutions of either α-Myc-HRP antibody (sc-40 HRP, Santa Cruz Biotechnologies) or α-Flag-HRP antibody (M2, Sigma) in 5% milk PBST. Full blots can be found in Supplementary Figure 1, and additional procedural details can be found in Supplementary Methods.
Chromatin Immunoprecipitation (ChIP) at Gal1-LacZ
All ChIP experiments were conducted in ΔSnf1 LS41 strain with pGADT7-Snf1-6xmyc complemented back in. Cultures were grown in 2% glucose to mid-log phase and then induced for 3 h in 2% galactose before treating with formaldehyde. Solubilized chromatin was split equally among three clean tubes to which 2 μg Snf1 antibody (sc-15621, Santa Cruz Biotechnologies), LexA antibody (sc-1725, Santa Cruz Biotechnologies), or control IgG (sc-2027, Santa Cruz Biotechnologies) was added. Immunoprecipitated samples were immobilized to magnetic Dynabeads Protein G (Life Technologies) and washed. Samples were eluted at 65 °C for 30 min, the eluate transferred to a new tube, and the formaldehyde cross-links were reversed overnight in a 65 °C water bath. Samples were purified using a Qiagen PCR clean up kit prior to qPCR analysis. qPCR reactions were set up in triplicate for each sample using Promega GoTaq qPCR Master Mix reagents. All samples were run on an Applied Biosystems StepOne Plus, and a minimum of three independent experiments were conducted to generate average % input values for each immunoprecipitation condition. Detailed experimental procedure and data analysis can be found in the Supplementary Methods.
Tandem Reversible and Irreversible Cross-Linking (TRIC) at Gal1-LacZ
Cultures expressing pLexA+Gal4 Phe849Bpa-6HIS-3xFLAG were grown to mid log phase in SC media containing 2% raffinose and 2% galactose and 1 mM Bpa. The cultures receiving only UV treatment were treated as previously described.16 Cultures receiving formaldehyde treatment were equipped with a stirbar and cross-linked with 1% formaldehyde for 5 min prior to quenching with 250 mM glycine for 5 min. Cells were then centrifuged, and cell pellets were washed with media and recentrifuged. Samples intended to additionally receive UV cross-linking were then resuspended in 2 mL H-W-U media containing 2% raffinose and 2% galactose, transferred to a small cell culture dish, and subjected to UV irradiation at 365 nm UV light (Eurosolar 15 W UV lamp) with cooling for 0.5 h.
Cell lysis was carried out with glass bead mechanical shearing as previously described.16,36 Subsequent lysates were removed, and the remaining pellets washed and sonicated to solubilize the chromatin fraction. Samples were then centrifuged for 20 min at max speed, and the soluble chromatin was then immunoprecipitated with Snf1 antibody (sc-15621, Santa Cruz Biotechnologies) and immobilized to protein G magnetic Dynabeads (Life Technologies). After immunoprecipitation, the beads were washed, and the cross-linked sample was eluted from the beads. Formaldehyde cross-links were reversed by heating at 95 °C for 20 min in NuPAGE 4x LDS sample buffer (Life Technologies) containing 250 mM DTT. Samples were run on a 4–20% Tris-glycine TGX gel (Bio-Rad) and transferred to a PVDF membrane. Western Blot analysis was carried out using a 1:1000 dilution of anti-FLAG (M2) antibody (Sigma) in 5% PBST.
Supplementary Material
Acknowledgments
We would like to thank B. Martin and J.P. Carolan for helpful discussions and the Skiniotis Lab for use of their cryolysis equipment. We thank NIH-NIGMS 2R0106553 for funding in addition to NSF CHE 1412759. R.P. was supported by the Biology Interface Training Program 5T32GM008597-17.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.6b07680.
Supporting figures and methods (PDF)
Complete MuDPIT data set (XLSX)
Complete MuDPIT data set (XLSX)
Notes
The authors declare no competing financial interest.
References
- 1.Thompson AD, Dugan A, Gestwicki JE, Mapp AK. ACS Chem Biol. 2012;7:1311. doi: 10.1021/cb300255p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paoletti A, Parmely T, Tomomori-Sato C, Sato S, Zhu D, Conaway R, Conaway J, Florens L, Washburn M. Proc Natl Acad Sci U S A. 2006;103:18928. doi: 10.1073/pnas.0606379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sikorski TW, Buratowski S. Curr Opin Cell Biol. 2009;21:344. doi: 10.1016/j.ceb.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vincent O, Townley R, Kuchin S, Carlson M. Genes Dev. 2001;15:1104. doi: 10.1101/gad.879301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X-F, Lehmann L, Lin JJ, Vashisht A, Schmidt R, Ferrari R, Huang C, McKee R, Mosley A, Plath K, Kurdistani SK, Wohlschlegel J, Carey M. Cell Rep. 2012;2:1061. doi: 10.1016/j.celrep.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lo W-S, Gamache ER, Henry KW, Yang D, Pillus L, Berger SL. EMBO J. 2005;24:997. doi: 10.1038/sj.emboj.7600577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ptashne M, Gann A. Nature. 1997;386:569. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 8.Vincent O, Carlson M. EMBO J. 1999;18:6672. doi: 10.1093/emboj/18.23.6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sikorski TW, Joo YJ, Ficarro SB, Askenazi M, Buratowski S, Marto JA. J Biol Chem. 2012;287:35397. doi: 10.1074/jbc.M112.391581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai K-L, Tomomori-Sato C, Sato S, Conaway RC, Conaway JW, Asturias FJ. Cell. 2014;157:1430. doi: 10.1016/j.cell.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wands AM, Wang N, Lum JK, Hsieh J, Fierke CA, Mapp AK. J Biol Chem. 2011;286:16238. doi: 10.1074/jbc.M110.207589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka Y, Bond MR, Kohler JJ. Mol BioSyst. 2008;4:473. doi: 10.1039/b803218a. [DOI] [PubMed] [Google Scholar]
- 13.Mapp AK, Ansari AZ. ACS Chem Biol. 2007;2:62. doi: 10.1021/cb600463w. [DOI] [PubMed] [Google Scholar]
- 14.Traven A, Jelicic B, Sopta M. EMBO Rep. 2006;7:496. doi: 10.1038/sj.embor.7400679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ansari AZ, Reece RJ, Ptashne M. Proc Natl Acad Sci U S A. 1998;95:13543. doi: 10.1073/pnas.95.23.13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Majmudar CY, Lee LW, Lancia JK, Nwokoye A, Wang Q, Wands AM, Wang L, Mapp AK. J Am Chem Soc. 2009;131:14240. doi: 10.1021/ja904378z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG. Science. 2003;301:964. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt MJ, Summerer D. Front Chem. 2014;2:1. doi: 10.3389/fchem.2014.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaman Z, Ansari AZ, Koh SS, Young R, Ptashne M. Proc Natl Acad Sci U S A. 2001;98:2550. doi: 10.1073/pnas.041611198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnston M, Flick JS, Pexton T. Mol Cell Biol. 1994;14:3834. doi: 10.1128/mcb.14.6.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin BR, Cravatt BF. Nat Methods. 2009;6:135. doi: 10.1038/nmeth.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Floer M, Bryant GO, Ptashne M. Proc Natl Acad Sci U S A. 2008;105:2975. doi: 10.1073/pnas.0800053105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young ET, Zhang C, Shokat KM, Parua PK, Braun KA. J Biol Chem. 2012;287:29021. doi: 10.1074/jbc.M112.380147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papamichos-Chronakis M, Gligoris T, Tzamarias D. EMBO Rep. 2004;5:368. doi: 10.1038/sj.embor.7400120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hierlmeier T, Merl J, Sauert M, Perez-Fernandez J, Schultz P, Bruckmann A, Hamperi S, Ohmayer U, Rachel R, Jacob A, Hergert K, Deutzmann R, Griesenbeck J, Hurt E, Milkereit P, Bassler J, Tschochner H. Nucleic Acids Res. 2013;41:1191. doi: 10.1093/nar/gks1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hedbacker K, Carlson M. Eukaryotic Cell. 2006;5:1950. doi: 10.1128/EC.00256-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertram PG, Choi JH, Carvalho J, Chan TF, Ai W, Zheng XF. Mol Cell Biol. 2002;22:1246. doi: 10.1128/MCB.22.4.1246-1252.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitchell SF, Jain S, She M, Parker R. Nat Struct Mol Biol. 2012;20:127. doi: 10.1038/nsmb.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shashkova S, Welkenhuysen N, Hohmann S. FEMS Yeast Res. 2015;15(4):fov026. doi: 10.1093/femsyr/fov026. [DOI] [PubMed] [Google Scholar]
- 30.Flick JS, Johnston M. Mol Cell Biol. 1990;10:4757. doi: 10.1128/mcb.10.9.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Treitel MA, Kuchin S, Carlson M. Mol Cell Biol. 1998;18:6273. doi: 10.1128/mcb.18.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ostling J, Ronne H. Eur J Biochem. 1998;252:162. doi: 10.1046/j.1432-1327.1998.2520162.x. [DOI] [PubMed] [Google Scholar]
- 33.Hall DB, Struhl K. J Biol Chem. 2002;277:46043. doi: 10.1074/jbc.M208911200. [DOI] [PubMed] [Google Scholar]
- 34.Dugan A, Pricer R, Katz M, Mapp AK. Protein Sci. 2016;25:1371. doi: 10.1002/pro.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woods A, Munday MR, Scott J, Yang X, Calson M, Carling D. J Biol Chem. 1994;269:19509. [PubMed] [Google Scholar]
- 36.Krishnamurthy M, Dugan A, Nwokoye A, Fung Y, Lancia J, Majmudar CY, Mapp AK. ACS Chem Biol. 2011;6:1321. doi: 10.1021/cb200308e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hardie DG. Nat Rev Mol Cell Biol. 2007;8:774. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 38.Dechassa ML, Zhang B, Horowitz-Scherer R, Persinger J, Woodcock CL, Peterson CL, Bartholomew B. Mol Cell Biol. 2008;28:6010. doi: 10.1128/MCB.00693-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Falsone SF, Gesslbauer B, Tirk F, Piccinini AM, Kungl AJ. FEBS Lett. 2005;579:6350. doi: 10.1016/j.febslet.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 40.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Nature. 2003;425:737. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 41.Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Nat Methods. 2014;11:319. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 42.Majmudar CY, Wang B, Lum JK, Hakansson K, Mapp AK. Angew Chem Int Ed. 2009;48:7021. doi: 10.1002/anie.200902669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lancia JK, Nwokoye A, Dugan A, Joiner C, Pricer R, Mapp AK. Biopolymers. 2014;101:391. doi: 10.1002/bip.22395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hulce JJ, Cognetta AB, Niphakis MJ, Tully SE, Cravatt BF. Nat Methods. 2013;10:259. doi: 10.1038/nmeth.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mann M. Nat Rev Mol Cell Biol. 2006;7:952. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 46.Preston GW, Radford SE, Ashcroft AE, Wilson AJ. ACS Chem Biol. 2014;9:761. doi: 10.1021/cb400731s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herbig E, Warfield L, Fish L, Fishburn J, Knutson BA, Moorefield B, Pacheco D, Hahn S. Mol Cell Biol. 2010;30:2376. doi: 10.1128/MCB.01046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Faubert B, Boily G, Izreig S, Griss T, Samborska B, Dong Z, Dupuy F, Chambers C, Fuerth BJ, Viollet B, Mamer OA, Avizonis D, DeBerardinis RJ, Siegel PM, Jones RG. Cell Metab. 2013;17:113. doi: 10.1016/j.cmet.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liang J, Mills GB. Cancer Res. 2013;73:2929. doi: 10.1158/0008-5472.CAN-12-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viollet B, Lantier L, Devin-Leclerc J, Hebrard S, Amouyal C, Mounier R, Foretz M, Andreelli F. Front Biosci Landmark Ed. 2009;14:3380. doi: 10.2741/3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


