Abstract
Objectives
To determine whether peak blood PCT measured within 48 hours of pediatric intensive care unit (PICU) admission can differentiate severe bacterial infections from sterile inflammation and viral infection and identify potential subgroups of PICU patients for whom PCT may not have clinical utility.
Study design
This was a retrospective, observational study of 646 critically ill children who had PCT measured within 48 hours of admission to an urban, academic PICU. Patients were stratified into 6 categories by infection status. We compared test characteristics for peak PCT, C-reactive protein (CRP), white blood cell count (WBC), absolute neutrophil count (ANC), and percentage immature neutrophils (% Imm). The area under the receiver operating characteristic curve (AUROC) was determined for each biomarker to discriminate bacterial infection.
Results
The AUROC was similar for PCT (0.73, 95% CI 0.69, 0.77) and CRP (0.75, 95% CI 0.71, 0.79; p=0.36), but both outperformed WBC, ANC, and % immature neutrophils (p<0.01 for all pairwise comparisons). The combination of PCT and CRP was no better than either PCT or CRP alone. Diagnostic patterns prone to false-positive and false-negative PCT values were identified.
Conclusions
Peak blood PCT measured close to PICU admission was not superior to CRP in differentiating severe bacterial infection from viral illness and sterile inflammation; both PCT and CRP outperformed WBC, ANC, and % immature neutrophils. PCT appeared especially prone to inaccuracies in detecting localized bacterial central nervous system infections or bacterial co-infection in acute viral illness causing respiratory failure.
Keywords: PICU, biomarker, sepsis
Difficulty in distinguishing bacterial infections from non-infectious systemic inflammatory illness exposes many patients to unnecessary antibiotic therapy in the intensive care unit.1–6 There remains an unmet need to identify early biomarkers of severe bacterial infections in critically ill pediatric patients that can help to optimize antibiotic utilization. Procalcitonin (PCT) is an emerging biomarker with demonstrable utility to guide antibiotic utilization in adults.7–12 Several trials in adults have shown that serum PCT level is higher with invasive bacterial infections than with viral or sterile inflammatory conditions and can help to optimize antibiotic utilization without increasing morbidity or mortality.13–15
In critically ill children, however, the utility of PCT to augment early recognition of severe bacterial infections compared with routinely available laboratory tests remains unclear. Prior pediatric studies have reported mixed results, and few studies have specifically examined the use of PCT in the pediatric intensive care unit (PICU).16–19 In some cases, PCT has yielded superior test characteristics than routinely used laboratory tests, such as measurement of C-reactive protein (CRP), white blood cell count (WBC), and percentage immature neutrophils (% Imm), but the optimal cut-point reported for PCT to guide clinical decision-making remains highly variable across studies.20–24 One common limitation of prior studies has been the relatively small sample size of subjects analyzed. Additionally, although few diagnostic tests perform universally well in all patient subgroups, prior PICU-based studies of PCT have not attempted to consider diagnostic patterns for which PCT testing may have more or less clinical utility. Along these lines, one recently published prospective study suggested that there may be subgroups of patients in the PICU for whom PCT measurement is less useful, but the study but had too few patients to draw firm conclusions.19
We sought to determine if peak blood PCT measured within 48 hours of PICU admission could differentiate severe bacterial infections from severe viral illness and systemic sterile inflammation and identify potential subgroups of critically ill children for whom PCT may not have clinical utility. We hypothesized that a low PCT cut-point may perform as well as or better than routinely available laboratory tests to identify PICU patients with a low likelihood of bacterial infection who required prolonged treatment with antibiotics, and there are identifiable diagnostic patterns of PICU disease that are prone to false-positive and false-negative PCT results for whom PCT testing may be less useful.
METHODS
We performed a retrospective, observational study of all patients age 29 days to 21 years admitted to a 55-bed PICU at an academic medical center between August 1, 2012, and February 15, 2014. Patients were included if blood PCT was sent as part of routine care within 48 hours of PICU admission, and the maximum measured PCT within this timeframe was utilized. For patients with multiple PICU admissions, only data from the first episode were included. We also excluded patients with superficial (i.e., non-invasive) bacterial infections, those transferred from another unit or hospital with established antibiotic therapy for >48 hours, or those whose final infection status could not be determined because of transfer out to another institution before all diagnostic testing was complete. This study was approved by the Institutional Review Board at The Children’s Hospital of Philadelphia, and a waiver of consent was granted.
Study design and data collection followed published guidelines for chart reviews.25 A review of the electronic medical record was completed for all eligible patients. Demographics, comorbid conditions, duration of hospitalization, and laboratory and microbiologic data were collected, and any missing data were noted. Recognizing that patients may come to attention at different timepoints in their courses of illness, the maximum values of PCT, CRP, and WBC within 48 hours prior to and 48 hours after PICU admission were recorded as (measured) biomarker peaks. The absolute neutrophil count (ANC) and % Imm corresponding to the highest WBC also were recorded. Severity of illness was determined by the Pediatric Risk of Mortality (PRISM)-III and Pediatric Index of Mortality (PIM)-2 scores.26,27 Definitions of types of infections were adapted from guidelines by the Centers for Disease Control and Prevention (CDC) and the National Healthcare Safety Network (NHSN).28 All data were recorded onto a standardized case report form using the web-based Research Electronic Data Capture (REDCap) system.29 The case-report form, a glossary of terms, and a coding sheet for infection categorization were developed with collaborative input from all study group members. Four abstractors (AJL, ACD, ARD, KAO) were trained to collect data and categorize patients in a similar manner.
Patients were classified into one of six mutually exclusive categories of infection (Table I; available at www.jpeds.com): (1) no infection; (2) viral infection; (3) suspected bacterial infection without shock; (4) documented bacterial infection without shock; (5) bacterial infection with shock (bacterial septic shock); and (6) septic shock without definitive microbiologic evidence of bacterial infection (“culture-negative septic shock”). Patients categorized as having no infection had no pathogenic organisms identified and no imaging suggestive of infection. Patients with viral infection had either an identified viral pathogen or a documented strong suspicion of viral infection without concurrent bacterial infection. Criteria for bacterial infection without shock included a clinical syndrome consistent with a likely bacterial infection, with (for documented infection) or without (for suspected infection) isolation of a bacterial or fungal pathogen from a sterile site.28 For example, most patients with pneumonia who did not have shock were categorized as suspected bacterial infection without shock. Patients with bacterial septic shock had a documented bacterial or fungal pathogen and met criteria for severe sepsis or septic shock.30 Culture-negative septic shock included patients with suspected infection (including documented viral infection) without isolation of a bacterial or fungal pathogen but who met criteria for severe sepsis or septic shock. Although culture-negative septic shock likely included some patients with undocumented bacterial infection, we a priori determined to analyze this group separately from documented bacterial septic shock because it was not possible to differentiate these patients from non-bacterial (eg, viral) septic shock and because their severity of illness justified empiric antibiotic administration regardless of pathogen.7,20
Table 1.
Criteria for categorization
| Infection Category | Definition |
|---|---|
| No infection |
|
| Viral infection |
|
| Suspected bacterial infection without shock |
|
| Documented bacterial infection without shock |
|
| Bacterial infection with shock (bacterial septic shock) |
|
| Culture-negative septic shock |
|
CDC=Centers for Disease Control and Prevention, NHSN=National Healthcare Safety Network, CXR=chest x-ray, UTI=urinary tract infection, IPSCC=International Pediatric Sepsis Consensus Conference
Horan TC, Andrus M, Dudeck MA. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. American journal of infection control. 2008;36(5):309–332.
Goldstein B, Giroir B, Randolph A, International Consensus Conference on Pediatric S. International pediatric sepsis consensus conference: definitions for sepsis and organ dysfunction in pediatrics. Pediatric critical care medicine: a journal of the Society of Critical Care Medicine and the World Federation of Pediatric Intensive and Critical Care Societies. 2005;6(1):2–8.
Inter-rater reliability testing was undertaken to ensure congruent classification of infection. Fifteen charts were randomly selected for all abstractors to review. The mean percent agreement across all abstractors to determine the infection category was 83% (Kappa 0.71). When categories of infection were conservatively grouped by presence or absence of bacterial infection (i.e., no infection and viral infection versus bacterial with/without shock and culture-negative septic shock), the mean percent agreement increased to 87%. Following consensus review, agreement of the final assigned infection category reached 100%. Because inter-rater reliability for infection category did not reach 100% until after consensus review, abstractors continued to flag any cases for which the category of infection was not clear during the remainder of the chart review process. Regular meetings were held to monitor overall performance and to establish final categorization by consensus agreement for all cases with uncertainty. In total, 24% of patients were reviewed for consensus agreement. Abstractors were blinded to PCT values during chart abstraction, categorization, and consensus review. PCT values were separately provided by the institution’s Department of Biomedical and Health Informatics. Other laboratory values, including CRP and WBC, were directly abstracted from the medical record by the chart reviewer after determination of infectious categorization.
PCT was measured at the discretion of the clinical team, using the VIDAS B.R.A.H.M.S. PCT assay (Biomerieux) in the hospital’s clinical laboratory.
Statistical Analyses
Analysis was performed using Stata Version 13.1 (StataCorp, College Station, TX). Summary statistics are reported as medians with interquartile ranges (IQR) for continuous variables and compared with the Wilcoxon rank-sum, test of trend, or Kruskal-Wallis tests. Categorical variables are reported as proportions and analyzed using chi-squared or Fisher’s exact tests. Receiver operating characteristic (ROC) curves were constructed by comparing patients with bacterial infection (suspected or documented bacterial infection with/without shock and culture-negative septic shock) with those with no infection or viral infection. Comparison of the area under the ROC curves (AUROC) was performed by generating linear predictions following separate logistic regression models with bacterial infection as the outcome and either a single biomarker alone or several biomarkers as independent variables.31,32 The sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), positive likelihood ratio, and negative likelihood ratio at various cut-points were determined, with the best cut-points a priori defined to maximize sensitivity and NPV in order to minimize the number of false-negative results (ie, patients for whom antibiotics could be incorrectly withheld). P-values ≤0.05 were significant.
Because the clinical utility of PCT could be optimized if subgroups were identified for which PCT testing was prone to false-positive or -negative results, we performed a qualitative exploration of patients with outlier PCT values in each infection category. This was a post-hoc exploratory analysis done after infection category was established and PCT values were unblinded. For patients with no infection or viral infection, false-positive PCT values greater than 1.5 times the upper quartile were considered extreme outliers. For patients with bacterial infection with or without shock or culture-negative shock, false-negative PCT values less than the identified optimal cut-point of 0.1 ng/mL were considered outliers. We used a more strict definition of outliers for false-negatives because we considered stopping antibiotics for a patient with a bacterial infection to be a more substantial error than continuing empiric antibiotics in a patient without a bacterial infection.
RESULTS
Of the 5,521 PICU admissions within the study period, 667 patients met initial inclusion criteria. Twenty-one patients underwent full chart review but subsequently were excluded following determination of non-invasive (superficial) bacterial infections18, leaving 646 patients for the final analysis (Figure 1; available at www.jpeds.com).
Figure 1.
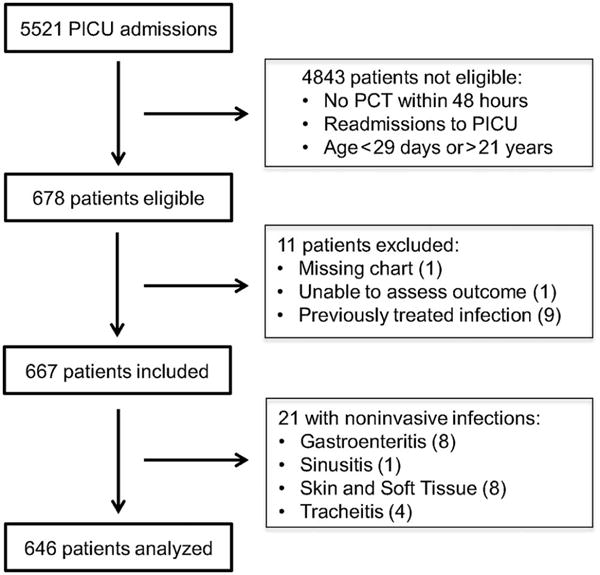
(online). Flow diagram of patient selection for analysis.
Patients were categorized as having no infection (n=188), viral infection (n=162), suspected bacterial infection without shock (n=89), documented bacterial infection without shock (n=48), bacterial septic shock (n=61), and culture-negative septic shock (n=98). Patient characteristics by infection category are shown in Table II. Patients in each group had similar pre-PICU hospital length of stays, suggesting that infections were predominantly community-acquired.
Table 2.
Patient characteristics by category of infection
| Variable | No infection | Viral | Suspected bacterial without shock | Documented bacterial without shock | Bacterial with shock | Culture-negative septic shock | p-value |
|---|---|---|---|---|---|---|---|
| n=188 | n=162 | n=89 | n=48 | n=61 | n=98 | ||
| Age (years) | 8.1 (2.5–13.1) | 2.2 (0.9–5.2) | 4.1 (1.6–9.2) | 6.2 (1.2–12.4) | 10.5 (3.5–16.6) | 7.3 (1.3–14.5) | <0.001 |
| Sex | 0.63 | ||||||
| Female | 80 (43) | 68 (42) | 39 (44) | 16 (33) | 31 (51) | 41 (42) | |
| Male | 108 (57) | 94 (58) | 50 (56) | 32 (67) | 30 (49) | 57 (58) | |
| Race | 0.14 | ||||||
| White | 79 (42) | 64 (40) | 38 (43) | 26 (54) | 31 (51) | 49 (50) | |
| Black | 66 (35) | 56 (35) | 32 (36) | 8 (17) | 19 (31) | 21 (21) | |
| Other | 43 (23) | 42 (26) | 19 (21) | 14 (29) | 11 (18) | 28 (29) | |
| Comorbidities | |||||||
| None | 56 (30) | 54 (33) | 32 (36) | 17 (35) | 11 (18) | 22 (22) | 0.07 |
| Respiratory | 38 (20) | 52 (32) | 26 (29) | 7 (15) | 13 (21) | 25 (26) | 0.06 |
| Cardiovascular | 15 (8) | 18 (11) | 5 (6) | 4 (8) | 3 (5) | 7 (7) | 0.66 |
| Neurologic | 54 (29) | 43 (27) | 18 (20) | 13 (27) | 15 (25) | 31 (32) | 0.61 |
| Hepatic | 4 (2) | 0 (0) | 1 (1) | 0 (0) | 1 (2) | 2 (2) | 0.36 |
| Renal | 4 (2) | 6 (4) | 1 (1) | 1 (2) | 4 (7) | 1 (1) | 0.31 |
| Oncologic | 29 (15) | 3 (2) | 1 (1) | 6 (12) | 19 (31) | 13 (13) | <0.001 |
| Hematologic | 3 (2) | 1 (1) | 3 (3) | 0 (0) | 1 (2) | 2 (2) | 0.58 |
| Genetic | 27 (14) | 34 (21) | 16 (18) | 8 (17) | 8 (13) | 16 (16) | 0.64 |
| Other | 42 (22) | 27 (17) | 13 (15) | 14 (29) | 15 (25) | 27 (28) | 0.10 |
| Reason for PICU admissiona | <0.001 | ||||||
| Sepsis | 21 (11) | 12 (7) | 9 (10) | 16 (33) | 53 (87) | 48 (49) | |
| Shock (not septic) | 15 (8) | 4 (2) | 3 (3) | 0 (0) | 1 (2) | 11 (11) | |
| Respiratory distress | 33 (18) | 106 (65) | 63 (71) | 11 (23) | 7 (11) | 29 (30) | |
| Status epilepticus | 28 (15) | 21 (13) | 4 (4) | 3 (6) | 0 (0) | 4 (4) | |
| Altered mental status | 22 (12) | 11 (7) | 2 (2) | 1 (2) | 0 (0) | 1 (1) | |
| Post-op management | |||||||
| General | 21 (11) | 4 (2) | 2 (2) | 1 (2) | 0 (0) | 3 (3) | |
| Neurosurgical | 25 (13) | 2 (1) | 2 (2) | 12 (25) | 0 (0) | 1 (1) | |
| Trauma | 8 (4) | 1 (1) | 0 (0) | 0 (0) | 0 (0) | 1 (1) | |
| Acute kidney injury | 2 (1) | 0 (0) | 1 (1) | 0 (0) | 0 (0) | 0 (0) | |
| Other | 13 (7) | 1 (1) | 3 (3) | 4 (8) | 0 (0) | 0 (0) | |
| PIM-2 | 1.3 (0.8–3.9) | 0.9 (0.2–3.3) | 1.0 (0.8–3.4) | 0.9 (0.8–2.7) | 1.3 (1.1–4.4) | 2.9 (1.0–4.6) | <0.001 |
| PRISM-III | 4 (0–9) | 2 (0–5) | 1 (0–4) | 2 (0–5) | 9 (3–14)b | 9 (6–15) | <0.001 |
| PICU mortality | 11 (6) | 4 (2) | 0 (0) | 2 (4) | 4 (7) | 5 (5) | 0.09 |
| LOS >48 hrs before PICU | 39 (21) | 20 (12) | 9 (10) | 9 (19) | 15 (25) | 16 (16) | 0.07 |
| Source of bacterial infection | |||||||
| Bloodstream | 0 (0) | 10 (21) | 31 (51) | 0 (0) | |||
| Cardiacd | 0 (0) | 0 (0) | 2 (3) | 2 (2) | |||
| Mediastinitis | 0 (0) | 0 (0) | 0 (0) | 2 (2) | |||
| Lower respiratory | 71 (80) | 6 (13) | 4 (7) | 29 (30) | |||
| Upper respiratory | 0 (0) | 5 (10) | 5 (8) | 1 (1) | |||
| Central nervous system | 6 (7) | 11 (23) | 1 (2) | 1 (1) | |||
| Abdominal | 6 (7) | 1 (2) | 4 (7) | 9 (9) | |||
| Genitourinary | 2 (2) | 10 (21) | 9 (15) | 1 (1) | |||
| Musculoskeletal | 1 (1) | 2 (4) | 1 (2) | 1 (1) | |||
| Othere | 3 (3) | 3 (6) | 4 (7) | 5 (5) | |||
| Unknown | 0 (0) | 0 (0) | 0 (0) | 47 (48) |
PICU=Pediatric Intensive Care Unit, PIM=Pediatric Index of Mortality, PRISM=Pediatric Risk of Mortality, LOS=length of stay, hrs=hours, BCx=blood culture, PCT=procalcitonin, abx=antibiotics
Data presented as n (%) or median (IQR) unless otherwise indicated.
As determined at time of PICU admission
PRISM-III not calculated for one patient because PICU length of stay was less than 2 hours
IV antibiotics administered within 24 hours prior to procalcitonin level
Includes endocarditis, myocarditis, and pericarditis
Includes rickettsial infections and toxic shock syndrome
The maximum (peak) PCT was the first available value for 596 patients (92.3%), and in 545 patients (84.4%) the peak PCT was sent within the 12 hours before through 24 hours after PICU admission. The median PCT for patients with no infection (0.22 [IQR 0.05–1.70] ng/mL) was not different from those with viral infection (0.33 [0.07–1.45] ng/mL; p=0.16). Patients with suspected and documented bacterial infection without shock had slightly higher median PCT levels (1.51 [0.41–4.04] and 0.91 [0.10–10.80] ng/mL, respectively; p<0.05 for comparisons with both no infection and viral infection), and those in shock had the highest median PCT values (7.16 [2.21–42.28] and 3.22 [0.36–24.93] ng/mL, respectively for bacterial septic shock and culture-negative septic shock; p<0.05 for pairwise comparisons with no infection, viral infection, and suspected and documented bacterial infection without shock). Values for PCT, CRP, WBC, ANC, and % Imm are shown in Table III. Only CRP and % Imm demonstrated a similar stepwise increase across infection category as PCT. PCT values by site and category of infection are shown in Figure 2.
Table 3.
Test characteristics by category of infection
| Biomarker | No infection | Viral | Suspected bacterial without shock | Documented bacterial without shock | Bacterial with shock | Culture-negative septic shock | p-valuea |
|---|---|---|---|---|---|---|---|
| PCT (ng/mL)b | 0.22 (0.05–1.70) | 0.33 (0.07–1.45) | 1.51 (0.41–4.04) | 0.91 (0.10–10.80) | 7.16 (2.21–42.28) | 3.22 (0.36–24.93) | <0.001 |
| CRP (mg/dL)c | 1.9 (0.5–6.8) | 1.4 (0.6–3.9) | 5.2 (2.4–15.5) | 5.0 (2.3–15.8) | 18.3 (7.4–31.7) | 6.2 (1.8–21.5) | <0.001 |
| WBC (thou/μL)d | 12.3 (8.1–16.0) | 11.6 (8.6–15.0) | 13.7 (9.5–18.6) | 14.8 (10.6–18.5) | 11.6 (2.8–18.8) | 12.7 (8.3–20.4) | 0.25 |
| ANC (thou/μL)e | 8.3 (4.1–12.0) | 7.6 (5.1–12.1) | 9.4 (6.3–14.7) | 9.8 (5.9–14.0) | 7.6 (0.4–15.4) | 9.0 (4.6–13.7) | 0.08 |
| % Immaturef | 0 (0–1.0) | 0 (0–4.0) | 0.5 (0–8.2) | 0 (0–0.3) | 0 (0–14.0) | 2 (0–14.0) | <0.001 |
Data presented as median (IQR).
p-value calculated as test of trend across categories. For the purposes of test of trend calculation, suspected bacterial infection without shock and documented bacterial infection without shock were combined into a single category (bacterial infection without shock), and bacterial infection with shock and culture-negative septic shock were combined into a single category (shock).
PCT available from all 646 patients
CRP available from 546 patients
WBC available from 633 patients
ANC available from 609 patients
% Immature available from 605 patients
Figure 2.
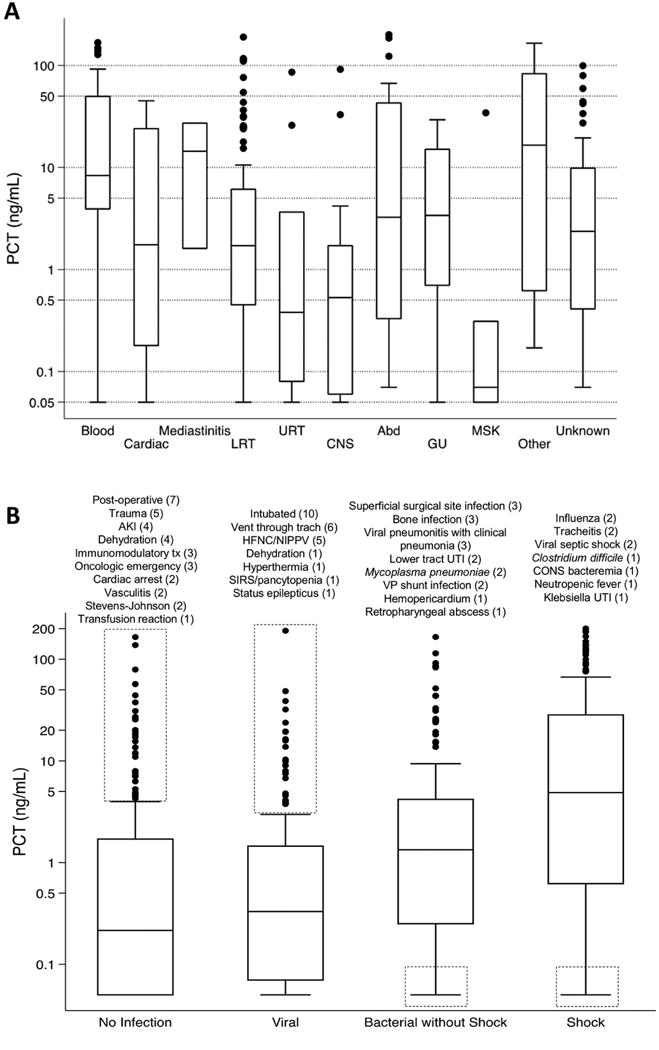
A, PCT level (on logarithmic scale) by site of bacterial infection. Cardiac includes endocarditis, myocarditis, and pericarditis. Upper respiratory tract infection includes tracheitis. Other infection includes rickettsial infections and toxic shock syndrome. B, PCT by category of infection, with outliers noted in dashed boxes. Bacterial without shock includes suspected and documented infections. Shock includes bacterial and culture-negative septic shock. Description and number (n) of outliers are noted above each box plot. Abd, abdominal; AKI, acute kidney injury; CNS, central nervous system; CONS, coagulase-negative Staphylococcus; GU, genitourinary; HFNC, high flow nasal cannula; LRT, lower respiratory tract; MSK, musculoskeletal; NIPPV, noninvasive positive pressure ventilation; SIRS, system inflammatory response syndrome; Tx, treatment; URT, upper respiratory tract; UTI, urinary tract infection; VP, ventriculoperitoneal.
Figure 3 shows the ROC curves for each individual biomarker to discriminate between patients with no infection and viral infection versus suspected or documented bacterial infection with/without shock and culture-negative septic shock. The AUROC was similar for PCT (0.73, 95% CI 0.69, 0.77) and CRP (0.75, 95% CI 0.71, 0.79; p=0.36). The AUROC for WBC, ANC, and % Imm was lower than either PCT or CRP (p<0.01 for all pairwise comparisons between PCT or CRP and WBC, ANC, and % Imm). The combination of PCT and CRP offered no additional benefit, with an AUROC of 0.76 (95% CI 0.72, 0.80; p=0.12 compared with PCT alone, p=0.07 compared to CRP alone). Table IV (available at www.jpeds.com) shows the test characteristics of select cut-points for PCT, CRP, and PCT+CRP to discriminate bacterial infection. The highest sensitivity and NPV for PCT occurred at a cut-point of 0.1 ng/dL. Although PPVs and positive likelihood ratios are overall low for PCT (in isolation and in combination with CRP), a PCT cut-point of 0.1 ng/mL yielded a clinically-relevant negative likelihood ratio of 0.3 as a solitary biomarker and of 0.1 when used in combination with CRP <0.8 mg/dL to exclude bacterial infection. Table V (available at www.jpeds.com) demonstrates the differential ability of PCT, CRP, WBC, ANC, and % Imm to discriminate no/viral infection from documented and suspected bacterial infections. PCT had an AUROC of 0.73 (95% CI 0.67, 0.79) for documented and 0.73 (95% CI 0.68, 0.77) for suspected bacterial infections.
Figure 3.
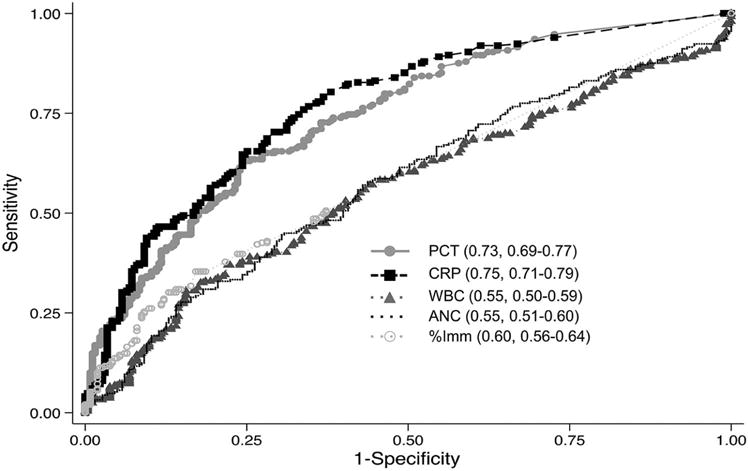
ROC curves for biomarkers values within 48 hours of PICU admission to predict need for antibiotics. Data are presented as AUROC, 95% CI. There was no difference in the AUROC between PCT and CRP (p=0.36). p>0.01 for all pairwise comparisons of PCT or CRP with WBC, ANC, and %Imm.
Table 4.
Test characteristics for PCT and CRP to identify need for antibiotic therapy
| PCT (ng/mL) | CRP (mg/dL) | Sensitivity | Specificity | PPV | NPV | LR+ | LR− |
|---|---|---|---|---|---|---|---|
| 0.1 | – | 91 (87–94) | 36 (31–41) | 54 (50–59) | 82 (75–88) | 1.4 | 0.3 |
| 0.25 | – | 81 (76–85) | 48 (43–54) | 57 (52–62) | 75 (69–81) | 1.6 | 0.4 |
| 0.5 | – | 74 (69–79) | 60 (55–65) | 61 (56–66) | 73 (68–78) | 1.9 | 0.4 |
| – | 0.5 | 95 (91–97) | 21 (17–25) | 50 (46–55) | 82 (72–89) | 1.2 | 0.2 |
| – | 0.8 | 93 (89–95) | 30 (25–35) | 53 (48–57) | 83 (75–89) | 1.3 | 0.2 |
| – | 1.0 | 91 (87–94) | 34 (29–39) | 54 (49–58) | 81 (73–87) | 1.4 | 0.3 |
| 0.1 | 0.5 | 98 (96–99) | 12 (9–16) | 49 (45–53) | 90 (77–97) | 1.1 | 0.2 |
| 0.1 | 0.8 | 98 (96–99) | 18 (14–22) | 50 (46–54) | 91 (82–97) | 1.2 | 0.1 |
| 0.1 | 1.0 | 97 (95–99) | 19 (15–23) | 50 (46–55) | 89 (80–95) | 1.2 | 0.2 |
| 0.25 | 0.5 | 98 (95–99) | 15 (11–19) | 49 (45–53) | 88 (77–95) | 1.2 | 0.1 |
| 0.25 | 1.0 | 95 (92–97) | 23 (19–28) | 51 (47–55) | 85 (76–92) | 1.2 | 0.2 |
| 0.5 | 0.5 | 97 (94–98) | 17 (13–22) | 50 (45–54) | 86 (75–93) | 1.2 | 0.2 |
| 0.5 | 1.0 | 94 (91–97) | 27 (22–32) | 52 (48–56) | 85 (76–91) | 1.3 | 0.2 |
PCT=procalcitonin, CRP=C-reactive protein, PPV=positive predictive value, NPV=negative predictive value, LR+=positive likelihood ratio, LR−=negative likelihood ratio
Data presented as % (95% CI). True positive (sensitivity) is defined as a patient with a bacterial infection and test value > cut-point.
Table 5.
Test characteristics for suspected versus definite bacterial infection
| Groups Compared | AUROC (95% CI) | ||||
|---|---|---|---|---|---|
| PCT | CRP | WBC | ANC | % Imm | |
| A vs B | 0.73 (0.67,0.79) |
0.82 (0.77, 0.86) |
0.52 (0.45, 0.59) |
0.53 (0.46, 0.60) |
0.54 (0.48, 0.60) |
| A vs C | 0.73 (0.68, 0.77) |
0.72 (0.67, 0.76) |
0.56 (0.51, 0.61) |
0.57 (0.52, 0.62) |
0.63 (0.58, 0.68) |
A: No infection or viral infection
B: Documented bacterial with or without shock
C: Suspected bacterial without shock and culture-negative septic shock
In a post-hoc exploratory analysis, outliers were examined to identify potential subgroups for whom PCT may have limited clinical utility (Figure 2, B). For patients with no infection, PICU admission following surgery, trauma, cardiac arrest, immunomodulatory therapy with chimeric antigen T lymphocytes33 or for acute kidney injury (AKI) or dehydration accounted for the 25 of the 33 (76%) false-positive PCT outliers. Ten of the 25 false-positive PCT outliers with viral infection were intubated due to respiratory failure, and an additional five patients received high-flow nasal cannula (HFNC) or non-invasive positive pressure ventilation. Patients with surgical site infections, bone infections, viral pneumonitis with clinical suspicion of bacterial superinfection, and ventriculoperitoneal (VP) shunt infections accounted for 11 of the 17 (65%) false-negative PCT outliers for bacterial infection without shock.
DISCUSSION
PCT in critically ill patients is more likely to be used as a guide to discontinue unnecessary empiric antibiotics in the absence of a microbiologically-proven bacterial infection than as a diagnostic biomarker for to initiate antibiotic therapy. Nonetheless, a clear understanding of the test characteristics of PCT and scenarios prone to false interpretation of results is necessary to optimize use of PCT in critically ill children. In this relatively large study of PCT in critically ill children, we found that maximum measured PCT level drawn close to PICU admission was not superior to CRP measurement in differentiating severe bacterial infection from viral illness and sterile inflammation, but was better than WBC, ANC, and % Imm for this purpose. Overall, PCT yielded moderately useful test characteristics to rule out bacterial infection with a clinically-relevant negative likelihood ratio using a cut-point of 0.1 ng/mL. However, recurring patterns of false-negative and false-positive PCT values suggest that test characteristics of PCT could be optimized with a more selective use of this biomarker in the PICU.
There has been notable heterogeneity of PCT test characteristics among prior studies of critically ill children,. In 175 patients, Hatherill et al reported a higher AUROC for PCT (0.96) than CRP or WBC (0.83 and 0.51, respectively) for the identification of septic shock. Similarly, in 94 PICU patients, Rey et al found PCT yielded a higher AUROC (0.91) than CRP (AUROC 0.75) or WBC (0.53) to diagnose septic shock. However, neither of these studies analyzed PCT for differentiation of bacterial from non-bacterial infection.20,24 In our study, PCT outperformed several routinely available laboratory tests to identify PICU patients with a low likelihood of bacterial infection, but there was no a clear benefit of PCT over CRP to differentiate bacterial infection from viral illness or sterile inflammation.
Mandell et al reported that PCT may be even less useful than CRP at PICU admission for early identification of culture-positive bacterial infection.19 Although that study was performed prospectively, only 33% (n=107) of eligible patients were included in the primary analysis, and most of the patients with incorrect PCT classification were false-positive rather than false-negative results (ie, PCT was more useful to “rule-out” rather than “rule-in” bacterial infection). Also, patients were categorized without regard to shock by Mandell et al and 25.7% of patients without bacterial infection required inotropic support. As PCT is elevated in culture-negative septic shock (as reported in our study and by Anand et al)7, this approach likely contributed to higher PCT levels in many patients categorized as low suspicion for bacterial infection. Finally, 50% of patients in the “no bacterial infection” group required mechanical ventilation, a subgroup for whom we also found PCT to have a high rate of false-positive results in our study. Notably, the only two patients reported with bacterial infection with false-negative PCT <0.05 ng/mL both had brain abscesses without shock, similar to the low PCT values observed in our study in patients with localized central nervous system (CNS) infections. Taken together, the prior study by Mandell et al supports our findings that PCT may not be superior to CRP in critically ill children and that PCT in isolation may be inaccurate to diagnose bacterial co-infection in patients in the PICU with acute viral illness causing respiratory failure or to rule out localized CNS infections.
The optimal cut-point identified in our study was PCT ≤0.1 ng/mL to rule out bacterial infection. This cut-point was selected to optimize sensitivity, NPV, and negative likelihood ratio to minimize false-negative results that could lead to antibiotics being discontinued inappropriately. Notably, although a proposed PCT cut-point of 0.1 ng/mL is lower than the commonly suggested value of 0.5 ng/mL, much of the literature supporting this higher PCT cut-point for bacterial infections relied on an early-generation PCT assay with a lower limit of detection of 0.5 ng/mL.16 Other studies using a more sensitive assay similar to our study also have identified lower cut-points to rule-out bacterial infection.14,34,36 Although combining PCT with CRP did not offer a statistical advantage over either biomarker alone, using both PCT <0.1 ng/mL and CRP <0.8 mg/dL did yield slightly more favorable test characteristics to rule out bacterial infection. However, one would expect sensitivity to increase with the application of two serially performed tests. Ultimately, to determine whether the low PCT cut-point suggested by our data (with or without CRP) can truly help to guide antibiotic utilization, specific investigation in prospective studies is required.
In a post-hoc qualitative exploration of patients with outlier PCT values, we identified several diagnostic patterns for which PCT may have limited clinical utility including patients with viral respiratory failure and localized CNS infections. It is important to note that, given the post-hoc exploratory nature of this analysis, these findings are preliminary and require validation in subsequent targeted studies. However, identification of subgroups for whom PCT may have limited utility does not necessarily diminish the overall value of PCT as a biomarker of severe bacterial infection. Rather, we believe that such limitations of PCT should be incorporated into the design of future prospective studies to optimize PCT in PICU patients.
Our study has several limitations. First, because there is no “gold standard” for identifying bacterial infections, some patients may have been categorized incorrectly. This has been a common criticism of prior studies, and we thus took several measures to limit potential misclassification bias, including detailed chart review, inter-rater reliability testing across chart abstractors, and liberal use of a consensus review process that required universal agreement for final infectious categorization. Any remaining misclassification should have biased our results toward the null and diminished the overall performance of PCT in differentiating true bacterial infection from non-bacterial illness. Second, in this retrospective study only 12% of PICU admissions had PCT testing available during the study period, raising concern for potential selection bias. It is possible that PCT may have been used to confirm bacterial infections more often than to assist with diagnostic uncertainty. Consequently, prospective validation of our proposed PCT cut-points therefore is necessary. Third, the differential kinetics of PCT and CRP relative to infection onset was not considered in our study. Prior studies have shown that PCT rises faster and peaks earlier than CRP following bacterial infections, which may be important when considering the differential utility of these two biomarkers at PICU admission.16 That the initial PCT value was the maximum recorded in over 92% of patients supports a clinically pragmatic role for this biomarker early in the course of diagnostic evaluation. Finally, our data reflect the experience of a single institution, and we acknowledge the need for a multicenter study to provide a more broadly generalizable relationship between PCT and bacterial infection in subgroups of critically ill children.
PCT was more useful to “rule out” rather than to identify infection, but caution should be used in interpreting PCT in critically ill children with acute viral illness causing respiratory failure or with localized central nervous system (CNS) infections because high PCT did not consistently indicate bacterial co-infection in critical viral illnesses nor did low PCT rule out all CNS infections.
Acknowledgments
We would like to thank Sherri Kubis, RN, for her assistance with PIM-2 and PRISM-III data collection.
Supported by the Division of Critical Care Medicine at The Children’s Hospital of Philadelphia. S.W. is supported by National Institute of General Medical Sciences (K23GM110496).
List of Abbreviations and Acronyms
- % Imm
Percentage immature neutrophils
- ANC
Absolute Neutrophil Count
- AUROC
Area under the ROC curve
- CRP
C-reactive protein
- NPV
Negative Predictive Value
- PCT
Procalcitonin
- PICU
Pediatric Intensive Care Unit
- ROC
Receiver operating characteristic
- WBC
White blood cell count
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest.
References
- 1.Cantey JB, Patel SJ. Antimicrobial stewardship in the NICU. Infect Dis Clin North Am. 2014;28:247–261. doi: 10.1016/j.idc.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Cantey JB, Wozniak PS, Sanchez PJ. Prospective Surveillance of Antibiotic Use in the Neonatal Intensive Care Unit: Results From the SCOUT Study. Pediatr Infect Dis J. 2015;34:267–272. doi: 10.1097/INF.0000000000000542. [DOI] [PubMed] [Google Scholar]
- 3.Garnacho-Montero J, Escoresca-Ortega A, Fernandez-Delgado E. Antibiotic deescalation in the ICU: how is it best done? Curr Opin Infect Dis. 2015;28:193–198. doi: 10.1097/QCO.0000000000000141. [DOI] [PubMed] [Google Scholar]
- 4.Nobre V, Harbarth S, Graf JD, Rohner P, Pugin J. Use of procalcitonin to shorten antibiotic treatment duration in septic patients: a randomized trial. Am J Respir Crit Care Med. 2008;177:498–505. doi: 10.1164/rccm.200708-1238OC. [DOI] [PubMed] [Google Scholar]
- 5.Shlaes DM, Gerding DN, John JJF, Craig WA, Bornstein DL, Duncan RA, et al. Society for Healthcare Epidemiology of America and Infectious Diseases Society of America Joint Committee on the Prevention of Antimicrobial Resistance: Guidelines for the Prevention of Antimicrobial Resistance in Hospitals. Clin Infect Dis. 1997;25:584–599. doi: 10.1086/513766. [DOI] [PubMed] [Google Scholar]
- 6.Dellit TH, Owens RC, McGowan JE, Jr, Gerding DN, Weinstein RA, Burke JP, et al. Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America guidelines for developing an institutional program to enhance antimicrobial stewardship. Clin Infect Dis. 2007;44:159–177. doi: 10.1086/510393. [DOI] [PubMed] [Google Scholar]
- 7.Anand D, Das S, Bhargava S, Srivastava LM, Garg A, Tyagi N, et al. Procalcitonin as a rapid diagnostic biomarker to differentiate between culture-negative bacterial sepsis and systemic inflammatory response syndrome: a prospective, observational, cohort study. J Crit Care. 2015;30:218 e217–212. doi: 10.1016/j.jcrc.2014.08.017. [DOI] [PubMed] [Google Scholar]
- 8.Charles PE, Tinel C, Barbar S, Aho S, Prin S, Doise JM, et al. Procalcitonin kinetics within the first days of sepsis: relationship with the appropriateness of antibiotic therapy and the outcome. Crit Care. 2009;13:R38. doi: 10.1186/cc7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harbarth S, Holeckova K, Froidevaux C, Pittet D, Ricou B, Grau GE, et al. Diagnostic value of procalcitonin, interleukin-6, and interleukin-8 in critically ill patients admitted with suspected sepsis. Am J Respir Crit Care Med. 2001;164:396–402. doi: 10.1164/ajrccm.164.3.2009052. [DOI] [PubMed] [Google Scholar]
- 10.Chirouze C, Schuhmacher H, Rabaud C, Gil H, Khayat N, Estavoyer JM, et al. Low serum procalcitonin level accurately predicts the absence of bacteremia in adult patients with acute fever. Clin Infect Dis. 2002;35:156–161. doi: 10.1086/341023. [DOI] [PubMed] [Google Scholar]
- 11.Assicot M, Bohuon C, Gendrel D, Raymond J, Carsin H, Guilbaud J. High serum procalcitonin concentrations in patients with sepsis and infection. Lancet. 1993;341:515–518. doi: 10.1016/0140-6736(93)90277-N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gendrel D, Raymond J, Coste J, Moulin F, Lorrot M, Guérin S, et al. Comparison of procalcitonin with C-reactive protein, interleukin 6 and interferon-alpha for differentiation of bacterial vs. viral infections. Pediatr Infect Dis J. 1999;18:875–881. doi: 10.1097/00006454-199910000-00008. [DOI] [PubMed] [Google Scholar]
- 13.Prkno A, Wacker C, Brunkhorst FM, Schlattmann P. Procalcitonin-guided therapy in intensive care unit patients with severe sepsis and septic shock–a systematic review and meta-analysis. Crit Care. 2013;17:R291. doi: 10.1186/cc13157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuetz P, Albrich W, Mueller B. Procalcitonin for diagnosis of infection and guide to antibiotic decisions: past, present and future. BMC Med. 2011;9:107. doi: 10.1186/1741-7015-9-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuetz P, Chiappa V, Briel M, Greenwald JL. Procalcitonin algorithms for antibiotic therapy decisions: a systematic review of randomized controlled trials and recommendations for clinical algorithms. Arch Intern Med. 2011;171:1322–1331. doi: 10.1001/archinternmed.2011.318. [DOI] [PubMed] [Google Scholar]
- 16.Becker KL, Snider R, Nylen ES. Procalcitonin assay in systemic inflammation, infection, and sepsis: clinical utility and limitations. Crit Care Med. 2008;36:941–952. doi: 10.1097/CCM.0B013E318165BABB. [DOI] [PubMed] [Google Scholar]
- 17.Deis JN, Creech CB, Estrada CM, Abramo TJ. Procalcitonin as a marker of severe bacterial infection in children in the emergency department. Pediatr Emerg Care. 2010;26:51–60. doi: 10.1097/PEC.0b013e3181c399df. quiz 61–53. [DOI] [PubMed] [Google Scholar]
- 18.Fernandez Lopez A, Luaces Cubells C, Garcia Garcia JJ, Fernandez Pou J, Spanish Society of Pediatric Emergencies Procalcitonin in pediatric emergency departments for the early diagnosis of invasive bacterial infections in febrile infants: results of a multicenter study and utility of a rapid qualitative test for this marker. Pediatr Infect Dis J. 2003;22:895–903. doi: 10.1097/01.inf.0000091360.11784.21. [DOI] [PubMed] [Google Scholar]
- 19.Mandell IM, Aghamohammadi S, Deakers T, Khemani RG. Procalcitonin to Detect Suspected Bacterial Infections in the PICU. Pediatr Crit Care Med. 2016;17:e4–e12. doi: 10.1097/PCC.0000000000000571. [DOI] [PubMed] [Google Scholar]
- 20.Hatherill M, Tibby SM, Sykes K, Turner C, Murdoch IA. Diagnostic markers of infection: comparison of procalcitonin with C reactive protein and leucocyte count. Arch Dis Child. 1999;81:417–421. doi: 10.1136/adc.81.5.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simon L, Saint-Louis P, Amre DK, Lacroix J, Gauvin F. Procalcitonin and C-reactive protein as markers of bacterial infection in critically ill children at onset of systemic inflammatory response syndrome. Pediatr Crit Care Med. 2008;9:407–413. doi: 10.1097/PCC.0b013e31817285a6. [DOI] [PubMed] [Google Scholar]
- 22.Casado-Flores J, Blanco-Quiros A, Asensio J, Arranz E, Garrote JA, Nieto M. Serum procalcitonin in children with suspected sepsis: a comparison with C-reactive protein and neutrophil count. Pediatr Crit Care Med. 2003;4:190–195. doi: 10.1097/01.PCC.0000059420.15811.2D. [DOI] [PubMed] [Google Scholar]
- 23.Cies JJ, Chopra A. Procalcitonin use in a pediatric intensive care unit. Pediatr Infect Dis J. 2014;33:984–986. doi: 10.1097/INF.0000000000000370. [DOI] [PubMed] [Google Scholar]
- 24.Rey C, Los Arcos M, Concha A, Medina A, Prieto S, Martinez P, et al. Procalcitonin and C-reactive protein as markers of systemic inflammatory response syndrome severity in critically ill children. Intensive Care Med. 2007;33:477–484. doi: 10.1007/s00134-006-0509-7. [DOI] [PubMed] [Google Scholar]
- 25.Gilbert EH, Lowenstein SR, Koziol-McLain J, Barta DC, Steiner J. Chart reviews in emergency medicine research: Where are the methods? Ann Emerg Med. 1996;27:305–308. doi: 10.1016/s0196-0644(96)70264-0. [DOI] [PubMed] [Google Scholar]
- 26.Pollack MM, Patel KM, Ruttimann UE. Prism III: An updated pediatric risk of mortality score. Crit Care Med. 1996;24:743–752. doi: 10.1097/00003246-199605000-00004. [DOI] [PubMed] [Google Scholar]
- 27.Slater A, Shann F, Pearson G, Paediatric Index of Mortality Study Group PIM2: a revised version of the Paediatric Index of Mortality. Intensive Care Med. 2003;29:278–285. doi: 10.1007/s00134-002-1601-2. [DOI] [PubMed] [Google Scholar]
- 28.Horan TC, Andrus M, Dudeck MA. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control. 2008;36:309–332. doi: 10.1016/j.ajic.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 29.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)–a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldstein B, Giroir B, Randolph A, International Consensus Conference on Pediatric Sepsis International pediatric sepsis consensus conference: definitions for sepsis and organ dysfunction in pediatrics. Pediatr Crit Care Med. 2005;6:2–8. doi: 10.1097/01.PCC.0000149131.72248.E6. [DOI] [PubMed] [Google Scholar]
- 31.Ramby AL, Goodman DM, Wald EL, Weiss SL. Red Blood Cell Distribution Width as a Pragmatic Marker for Outcome in Pediatric Critical Illness. PLoS One. 2015;10:e0129258. doi: 10.1371/journal.pone.0129258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pregibon D. Logistic regression diagnostics. Ann Stat. 1981;9:705–724. [Google Scholar]
- 33.Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. 2014;371:1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maniaci V, Dauber A, Weiss S, Nylen E, Becker KL, Bachur R. Procalcitonin in young febrile infants for the detection of serious bacterial infections. Pediatrics. 2008;122:701–710. doi: 10.1542/peds.2007-3503. [DOI] [PubMed] [Google Scholar]
- 35.Milcent K, Faesch S, Gras-Le Guen C, Dubos F, Poulalhon C, Badier I, et al. Use of Procalcitonin Assays to Predict Serious Bacterial Infection in Young Febrile Infants. JAMA Pediatr. 2016;170:62–69. doi: 10.1001/jamapediatrics.2015.3210. [DOI] [PubMed] [Google Scholar]
- 36.Kutz A, Briel M, Christ-Crain M, Stolz D, Bouadma L, Wolff M, et al. Prognostic value of procalcitonin in respiratory tract infections across clinical settings. Crit Care. 2015;19:74. doi: 10.1186/s13054-015-0792-1. [DOI] [PMC free article] [PubMed] [Google Scholar]


