Abstract
For several decades, many bacteria, among which A. baumannii, have shown their ability to colonize the upper surface of static liquids, forming a biofilm at the air-liquid interface named pellicle. Despite the ubiquity of these pellicles in both natural and artificial environments, few studies have investigated this biofilm type. The present data set provides the first description of the whole proteome of A. baumannii cells grown as pellicle, using a label-free mass spectrometry approach. Results are in accord with the general findings reporting that sessile bacteria are far more resistant to detrimental conditions than their planktonic counterparts, by the accumulation of stress proteins. The present investigation also confirmed previous studies suggesting a correlation between the pellicle forming ability and the bacterial virulence. Indeed, we showed the up-regulation of numerous virulence factors during the pellicle growth, e.g. phospholipases, adhesion factors, as well as those of the GacAS Two-Component System (TCS) and Type 6 Secretion System (T6SS). We also highlighted that Bam and Tam systems, both related to the OM insertion machinery, play a critical role during pellicle biogenesis. Moreover, sessile bacteria activate several pathways, e.g. iron, magnesium, phosphate pathways, which allows for increasing the panel of nutrient sources.
Acinetobacter baumannii is a member of the ESKAPE group of bacterial pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, A. baumannii, Pseudomonas aeruginosa, and Enterobacter sp.) (1), responsible for a majority of hospital-acquired infections. This Gram-negative pathogenic bacterium has become an important human pathogen owing to both an increasing number of infections and an emergence of multidrug-resistant (MDR) strains (2). A. baumannii infections occur mostly in intensive care units and cause severe nosocomial infections including pneumonia, bacteremia, endocarditis, skin and soft tissue infections, urinary tract infections, or meningitis (3–5). A. baumannii is known for its long-time survival in hospital settings because of its great ability to survive desiccation (6), oxidative stress (7), or treatment with disinfectants (8). This persistence is mostly linked to its capacity to form biofilms (9–11). Indeed, MDR clinical isolates showed a high ability to form biofilm, which was positively associated with a capability to adhere to human bronchial epithelial cells (12, 13).
Biofilms are structured communities of bacteria encapsulated within a polymeric matrix formed by extracellular polymeric substances (EPS)1, e.g. exopolysaccharides, proteins, nucleic acids, and other substances (14). This matrix acts as a protective layer and creates an optimal environment for genetic material exchange between the microorganisms (15). The dynamics of the transition between planktonic and biofilm growth modes occur in response to different hostile environmental signals (16). In the biofilm state, bacteria are generally more tolerant to antibiotics than in planktonic state (17). The mechanism of bacterial biofilm resistance to antibiotics is still under investigation but several explanations have been proposed: (1) the biofilm EPS matrix acts as penetration barriers to antibiotics, (2) the biofilm bacteria up-regulate efflux pumps decreasing intracellular antibiotic concentration, and (3) finally the presence of a nondividing dormant bacterial subpopulation that is protected from antibiotics (18, 19).
Biofilms can develop on a wide variety of interfaces. The best studied biofilms are those formed at the solid-liquid (S-L) interface in which bacteria adhere to biotic or abiotic surfaces (17, 20). Some bacteria, like A. baumannii, also form biofilms at the air-liquid (A-L) interface, usually named pellicles. This interface is an ideal environment for strictly aerobic bacteria to obtain oxygen from the air and nutrients from the liquid media (9, 21–23). A-L biofilms form more complex structures and require a higher level of organization, compared with S-L biofilms, owing to the lack of a solid surface on which the growth can be initiated (24). A recent study in our laboratory (8) showed that the Acinetobacter species forming pellicles are those mainly involved in nosocomial infections such as A. baumannii and A. nosocomialis (23). This obviousness suggested a correlation between this feature and the bacterial virulence.
Today, transcriptomic (25) and proteomic (26, 27) studies are performed to investigate the physiology of S-L A. baumannii biofilms. Only a proteomic work focused on the membrane compartment of pellicle bacteria (28) and to our knowledge, no comparative study was performed to give information on the protein expression dynamics during the pellicle formation.
In this context, the goal of the present work was to monitor the proteomic alterations during the pellicle formation in order to give insight into the biological processes that are involved. To this aim, we investigated the differential protein expression by bacteria grown in the planktonic and pellicle modes, with a particular focus on the protein dynamics of pellicle formation (1- and 4-day pellicles). Both cytoplasmic and membrane proteins were here analyzed, using a label-free approach. We thus successfully highlighted 620 proteins that were differentially expressed in young and/or mature pellicles. Data suggest a higher virulence of A. baumannii pellicle cells compared with planktonic counterparts.
EXPERIMENTAL PROCEDURES
Planktonic and Pellicle Growth Conditions
An overnight culture of A. baumannii strain ATCC 17978, performed in Mueller-Hinton Broth (MHB, Difco), was used to inoculate 500 ml of MHB at a final concentration of 107 Colony Forming Unit (CFU)/ml. For the planktonic growth, the culture was performed at 37 °C for 24 h, under constant shaking. Cells were harvested at the stationary phase of growth (i.e. OD600 = 1.9), by centrifugation (2700 × g, 20 min, 4 °C), and washed 3 times with deionized water. For the pellicle formation, bacterial growth was performed in glass Erlenmeyer containing 500 ml of MHB during 1-day or 4-day, at 37 °C, without shaking. Pellicles (1-day and 4-day) were recovered from the surface of the culture, and resuspended in 5 ml of a sterile phosphate buffered saline solution (PBS, 10 mm, pH 7.4). Cell recovery from pellicles was performed by sonication (twice during 15 min) and then by centrifugation (after each sonication at 4000 × g, 15 min, 4 °C). For each growth condition, the bacterial culture was performed in triplicate.
Pellicle Growth in Minimal Medium Supplemented with Aromatic Amino Acids, Magnesium, and Phosphate Ions
To measure the impact of nutrients on the 1-day pellicle growth, a minimal media M9 (Fluka) was supplemented with glucose (Glc) and/or amino acids (Ser, Tyr, His, or Phe, Sigma-Aldrich) at 2.5 mm. Magnesium or phosphate enrichment effects on 1-day and 4-day pellicle formation were also assessed at 2 and 10 mm (MgSO4 or H2PO4−/HPO42−), using M9 and MMG minimal media, respectively. After an overnight MHB culture, minimal media supplemented or not with the appropriate nutrients, were inoculated and incubated at 37 °C in 5 ml polystyrene tubes. After removing subphase under pellicles, biomass quantification was achieved by staining with a crystal violet assay, as described by O'Toole and Kolter (29). After rinsing pellicles with water, staining was performed by incubation for 20 min in 0.5% crystal violet. Crystal violet was then solubilized by adding 2 ml of ethanol in each tube, and the OD590 was finally measured. Assays were performed in triplicates and data were statistically analyzed by ANOVA one-way test, using the GraphPad Prism software.
Protein Extraction
Protein extraction from pellicle and planktonic bacteria were performed as described by Marti et al. (28) and Ouidir et al. (30), respectively. Briefly, after centrifugation (2700 × g, 10 min, 4 °C) the resulting cell pellets were resuspended in 10 ml of Tris-HCl buffer (20 mm, pH 7.4). The mixtures were sonicated twice, each for 3 min (Vibra Cell 75115, Bioblock Scientific, Illkirch, France). Unbroken cells were eliminated by centrifugation (10,000 × g, 10 min, 4 °C). An ultracentrifugation (60,000 × g, 45 min, 4 °C) of supernatants allowed to separate cytoplasmic and membrane protein fractions for further analyses. The protein concentration in each fraction was evaluated by the Bradford assay (Bio-Rad).
Enzymatic Digestion of Protein Extracts
Twenty-five micrograms of proteins were mixed with SDS loading buffer (63 mm Tris-HCl, pH 6.8, 10 mm DTT, 2% SDS, 0.02% bromphenol blue, 10% glycerol), then loaded onto a SDS-PAGE stacking gel (7%). A short electrophoresis was performed (10 mA, 20 min) in order to concentrate proteins. After migration, gels were stained with Coomassie Blue and destained (50% ethanol, 10% acetic acid, 40% deionized water). The revealed protein band from each fraction was excised, washed with water, and then immersed in a reductive medium (5 mm DTT). Cysteines were irreversibly alkylated with 25 mm iodoacetamide in the dark. Following washing steps in water, gel bands were submitted to protein digestion with trypsin (2 μg per band), overnight at 37 °C, in ammonium bicarbonate buffer (10 mm and pH 8). Peptide were extracted with H2O/CH3CN/TFA mixtures (49.5/49.5/1) and then dried. For each growth conditions, three biological replicates were carried and two technical replicates were realized for each of them (in total 6 samples per conditions were analyzed).
Tandem Mass Spectrometry
Cytoplasmic and membrane fractions were separately analyzed by mass spectrometry. All experiments were performed on a LTQ-Orbitrap Elite (Thermo Scientific) coupled to an Easy nLC II system (Thermo Scientific). One microliter of sample (1 μg) was injected onto an enrichment column (C18 PepMap100, Thermo Scientific). The separation was performed with an analytical column needle (NTCC-360/internal diameter: 100 μm; particle size: 5 μm; length: 153 mm, NikkyoTechnos, Tokyo, Japan). The mobile phase consisted of H2O/0.1% formic acid (FA) (buffer A) and CH3CN/FA 0.1% (buffer B). Tryptic peptides were eluted at a flow rate of 300 nL/min using a three-step linear gradient: from 2 to 40% B over 75 min, from 40 to 80% B in 4 min and 11 min at 80% B. The mass spectrometer was operated in positive ionization mode with capillary voltage and source temperature set at 1.5 kV and 275 °C, respectively. The samples were analyzed using CID (collision induced dissociation) method. The first scan (MS spectra) was recorded in the Orbitrap analyzer (r = 60,000) with the mass range m/z 400–1800. Then, the 20 most intense ions were selected for tandem mass spectrometry (MS2) experiments. Singly charged species were excluded for MS2 experiments. Dynamic exclusion of already fragmented precursor ions was applied for 30 s, with a repeat count of 1, a repeat duration of 30 s and an exclusion mass width of ±10 ppm. Fragmentation occurred in the linear ion trap analyzer with collision energy of 35 eV. All measurements in the Orbitrap analyzer were performed with on-the-fly internal recalibration (lock mass) at m/z 445.12002 (polydimethylcyclosiloxane).
Protein Quantification
For protein quantification, a label-free experiment was performed as previously described by Obry et al. (31). Two independent analyses were achieved on cytoplasmic and membrane samples. Briefly, after MS analysis, raw data were imported in Progenesis LC-MS software (Nonlinear Dynamics, version 4.0.4441.29989, Newcastle, UK). For comparison, one sample was set as a reference and the retention times of all other samples within the experiment were aligned. After alignment and normalization, statistical analysis was performed for one-way analysis of variance (ANOVA) calculations. For quantitation, peptide features presenting a p value and a q-value less than 0.05, and a power greater than 0.8_were retained. MS/MS spectra from selected peptides were exported for peptide identification with Mascot (Matrix Science, version 2.2.04) against the database restricted to A. baumannii ATCC 17978 containing 4071 protein sequences (http://www.genoscope.cns.fr/) (32). Database searches were performed with the following parameters: 1 missed trypsin cleavage site allowed; variable modifications: carbamidomethylation of cysteine and oxidation of methionine. Mass tolerances for precursor and fragment ions were set at 10 ppm and 0.5 Da, respectively. False discovery rates (FDR) were calculated using a decoy-fusion approach in Mascot (version 2.2.04). Identified peptide-spectrum-matches with −10logP value of 20 or higher were kept, at a FDR threshold of 5%. Mascot search results were imported into Progenesis. For each growth condition, the total cumulative abundance of the protein was calculated by summing the abundances of peptides. Proteins identified with less than 3 peptides were discarded. Only the proteins that varied by 2-fold in their average normalized abundances between growth conditions were retained. In this study, protein expressions in 1-day and 4-day pellicle states were systematically compared with those in planktonic state. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD004167.
Bioinformatic Analyses
Both protein cellular localization and biological function were determined from the Genoscope database (http://www.genoscope.cns.fr) (32). For unknown function and localization, the Kyoto Encyclopedia of Genes and Genomes (KEGG) and National Center for Biotechnology Information (NCBI) databases were used. InterPro database (http://www.ebi.ac.uk/interpro/) (33) was used for protein sequence analysis. STRING database were used to investigate protein-protein network interactions (http://string91.embl.de) (34).
Experimental Design and Statistical Rationale
We investigated the protein contents of bacteria grown in three conditions i.e. planktonic, 1-day-, and 4-day-old pellicles. For each condition, three biological replicates were performed. Two technical replicates per biological replicate were achieved (i.e. a total of 6 samples per condition was analyzed). Growth conditions were closely matched for planktonic and pellicle samples (same broth composition, same volumes, and same vessel material (glass)).
R software (35) was used for the statistical analyses to determine protein markers. To compare protein expression variations between the three conditions, a one-way analysis of variance (ANOVA) test with post hoc pairwise was used. The univariate associations between selected proteins and the different groups were assessed by simple regression. Additionally, a principal coordinate analysis (PCoA) was conducted, to visualize the relationships among groups at each stage. Proteins with a similar expression pattern were presented as heat map, visualized using the hclust function. Proteins with the same expression variation were grouped, showing strong correlation. Relationships between the marker proteins were visualized with a correlation matrix with corrplots (Pearson's r correlation coefficient, corrplot package) (36). Hierarchical clustering of each growth condition was performed using Euclidean distance metric and Average linkage method (unsupervised clustering).
RESULTS AND DISCUSSION
Proteome Expression Profile in Planktonic and Pellicle States
In the present study, we investigated the protein differential expression of the fully sequenced clinical isolate A. baumannii ATCC 17978 (35) in three growth conditions: planktonic, i.e. 1-day and 4 days-old pellicles, to access to the protein dynamics during the pellicle formation. Both cytoplasmic and membrane proteomes were separately investigated by a label-free approach (Fig. 1). A total of 1668 proteins were identified in the whole cell lysates with a minimum of 2 peptides (supplemental Table S1). The quantitative analysis using the Progenesis software pointed out 620 proteins (37% of the proteins) that exhibited a significant fold change (≥2) according to the culture conditions (supplemental Table S2). Different statistical parameters (see Materials and Methods section) were used both at the peptide and protein levels, to select specifically proteins with a difference of expression profile in one condition. A PCoA analysis, clustering the data of membrane and cytoplasmic proteins analyses, showed a good homogeneity of the biological replicates because three distinct protein populations were discriminated (supplemental Fig. S1).
Fig. 1.
Experimental design used to determine protein expression variations between planktonic, 1-day pellicle and 4-day pellicle states in A. baumannii.
The study pointed out some specific accumulated proteins in each growth condition, i.e. 93, 95, and 97 proteins in planktonic, 1-day and 4-day pellicle states, respectively (supplemental Fig. S2). Some proteins were commonly more abundant in two different growth conditions. For example, 147 polypeptides were up-regulated in both 1-day and 4-day pellicle cells; 119 in planktonic and 1-day pellicle cells and 69 in planktonic and 4-day pellicle states (supplemental Fig. S2). Protein profiles in mature pellicle (4-day pellicle) underwent the greatest modifications compared with planktonic state, as previously demonstrated for S-L biofilms (20).
The biological processes of these 620 differentially produced proteins were inventoried from different databases (see Material and Methods section). As previously observed in a P. aeruginosa planktonic/S-L biofilm comparative study (36), overproduced proteins in planktonic state was mainly involved in carbohydrate catabolism, protein maturation, DNA transcription and replication, and cell division (Fig. 2A). Translation process was particularly well represented with 55 proteins (mainly ribosomal). All these processes are in accordance with a planktonic growth mode, an ideal environment for bacterial biomass production (20, 36). The classification of biological functions for pellicle proteomes highlighted different categories like amino acid metabolism, biofilm, drug resistance, ions acquisition and metabolism, lipid metabolism, quorum sensing and secretion systems.
Fig. 2.
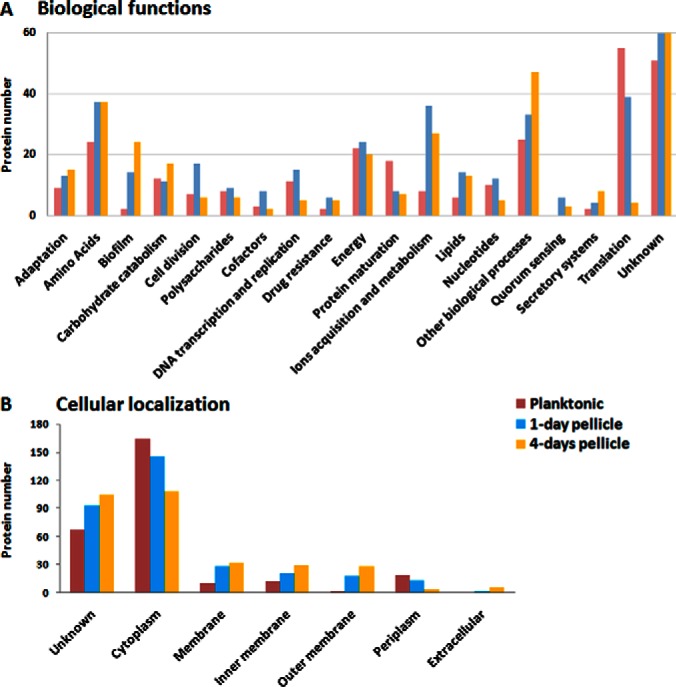
Protein distribution in the 3 experimental culture conditions according to their (A) biological process, and (B) cellular localization.
The cellular localization was also investigated. Membrane proteins were obviously overexpressed in pellicle states (20 and 28 membrane proteins in 1-day and 4-day pellicles, respectively) whereas only 10 up-regulated proteins were found in planktonic samples (Fig. 2B). This observation points out that biological processes involved in bacterial membrane trafficking or membrane structure are crucial during pellicle formation. Next sections will focus on accumulated proteins in (1) 1-day and 4-day pellicles, (2) 1-day pellicle, and (3) 4-day pellicle.
Accumulated Proteins in 1-Day and 4-Day-Old Pellicle Bacteria
Outer Membrane Proteins (OMPs) in Pellicles
We observed, in 1-day and 4-day pellicle cells, the accumulation of several proteins involved in the process of outer membrane (OM) β-barrel proteins assembly into outer membrane, i.e. three proteins of the Sec system (SecA (A1S_2862), SecD (A1S_2914), and SecY (A1S_3061)), as well as the chaperonin Skp (A1S_1968), and all the proteins of the Bam system which is related to the OM insertion machinery: BamA (A1S_1969), BamB (A1S_0505), BamC (A1S_3424), BamD (A1S_0840), and BamE (A1S_0894) (Fig. 3). Moreover, we observed the accumulation of a protein (A1S_2132), sharing 34% of identity with TamA of E. coli. The TamAB system (Translocation and Assembly Module) also participates to the β-barrel protein assembly and might be more specifically required for efficient translocation of the autotransporters cell surface domain (37).
Fig. 3.
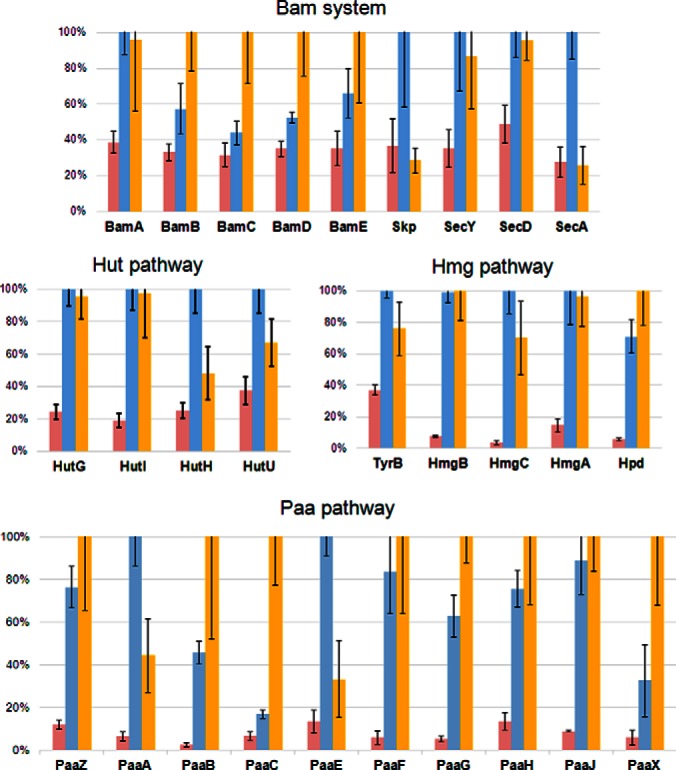
Examples of protein expression variations in the 3 culture conditions.
All these data demonstrate that the overall pathway required for OMP assemblies was mobilized. It is well known that newly synthesized OMPs have to cross the inner membrane via the Sec-dependent pathway. Once in the periplasm, periplasmic chaperone proteins, e.g. Skp, SurA and DegP assist the delivery of the unfolded proteins to the Bam system. Final folding and insertion of OMPs into the membrane is then performed via a protein-lipid interphase provided by BamA, assisted by four lipoproteins: BamB and BamC:D:E module (38, 39). In accordance with an overexpression of this pathway by pellicle bacteria, we observed that OMPs underwent an accumulation in pellicle cells compared with planktonic counterparts (Fig. 2B). This phenomenon could be involved in the pellicle establishment, EPS matrix production and/or adhesion (40) but, also, could be a response to particular environments such as a nutrient starvation (17, 27).
Thus, several porins, that may use the Bam pathway for OM insertion, were significantly accumulated in pellicle cells, probably to improve bacterial nutrients assimilation, i.e. OprB (A1S_2849), OprC (A1S_0170), OprD-like (A1S_0201), DcaP-like (A1S_2753), and CarO (A1S_2538). Apart from OprC, these proteins were already described as up-regulated in S-L biofilm-forming A. baumannii (27). It has been shown that loss of most of these OMPs in mutant strains induces the formation of small cell aggregates as compared with a wild-type strain that forms a dense biofilm (27). The accumulation of CarO and OprD-like porins, that are respectively involved in the uptake of ornithine (precursor of siderophore biosynthesis from hydroxamate family (41, 42)), and uptakes of Fe3+ and Mg2+, could contribute to the bacterial adaptation to magnesium- and/or iron-depleted environments (41, 43, 44). An overexpression of the DcaP-like protein has already been observed in an A. baumannii S-L biofilms; however, the biological function of this protein remains unclear (22, 23, 28, 21). Lastly, the abundance of the copper-regulated protein OprC in A-L biofilms might highlight oxygen limited conditions (28).
Increase of Drug Resistance and Stress Adaptation Proteins by Pellicle Microorganisms
Several proteins belonging to resistance-nodulation-cell division type efflux pumps (RND pumps) were accumulated in pellicle states. It is the case of AdeI (A1S_2735) and AdeJ (A1S_2736) that belong to the constitutive AdeIJK efflux pump, as well as an inner membrane protein (A1S_3446) and a membrane fusion protein (A1S_3447) that are, to our knowledge, described for the first time. These two last efflux pumps are part of a RND efflux system and are different from those found as up-regulated in S-L biofilms (25, 27) and so highlight the specificity of A-L biofilms. The AdeIJK efflux pump has a high broad substrate specificity; its overexpression in A. baumannii would confer a low bacterium susceptibility to major antibiotic classes, including β-lactams, fluoroquinolones, tetracyclines, phenicols, antifolates, and fusidic acid (45). This feature is reminiscent of the bacterial biofilms, sessile bacteria being able to be up to 1000 times more tolerant to antibiotics than their planktonic counterparts (46). Rao et al. (47) investigated the correlation between biofilm production and multidrug resistance of clinical A. baumannii isolates. They showed that high biofilm producer strains were more resistance to amikacin, cefotaxime, ciprofloxacin and aztreonam than low biofilm producers. On the other hand, we observed a downregulation of the cephalosporinase AmpC (A1S_2367) in pellicule bacteria, suggesting a higher sensibility to cephalothin, cefazolin, cefoxitin, and most penicillins (48).
It has been advanced that the matrix contributes not only to the biofilm cohesion but also participates to the drug resistance by limiting the antibiotics penetration (18). To date, few pathways involved in EPS matrix synthesis have been described in A. baumannii (27, 49). It has been shown that the synthesis and export of cell-associated poly-β-(1–6)-N-acetylglucosamine (PNAG) is ensured by the pgaABCD locus, that was demonstrated as essential for the biofilm growth at the S-L interface under high shear forces (49). The present study points out a strong accumulation of PgaA (A1S_2162), a porin allowing PNAG translocation across the OM (49), in 1-day and 4-day pellicle cells (by 3.5- and 4.5-fold, respectively). Neither transcriptomic (25) nor proteomic studies performed on S-L biofilms (26, 27) had revealed this overexpression up to now, suggesting that the PNAG production in static conditions is specific to biofilms at the A-L interface. It has been shown on E. coli biofilms containing PNAG that the increased tolerance to cationic peptides, e.g. polymyxin B or colistin, is because of an electrostatic repulsion between the positively charged of PNAG and those of the cationic peptides (50). Consequently, we can advance that the same phenomenon might confer the same colistin resistance within the A. baumannii pellicle biofilm. Though many other antimicrobial resistance mechanisms exist in A. baumannii, the overproduction of efflux pumps and EPS matrix establishment might be crucial for antibiotic resistance of A. baumannii pellicles.
It is also well known that biofilm cells often exhibited an oxidative stress (51, 53). In agreement with data reported by Cabral et al. on S-L biofilms (27), we showed here an up-regulation of the catalase hydroperoxidase II (KatE, A1S_1386) and the alkyl hydroperoxide reductase F (AhpF, A1S_1458) in pellicles. The paraquat inducible protein (PqiB, A1S_2204) was also accumulated. Paraquat is a superoxide radical-generating agent that, in E. coli (52), induces the transcription of the pqiAB cluster, via a control by the soxRS system which is involved in the oxidative stress response. Little information has been reported on PqiB, and its role remains unknown. We observed however, that the superoxide dismutase B (SodB, A1S_2343) amount decreased in pellicles, suggesting that this protein is differently regulated. Reactive oxygen species like H2O2 that are generated may cause the bacterial lysis (53) leading to chromosomal DNA releasing and providing extracellular DNA (eDNA) which contributes to the matrix cohesion (53). Many proteins involved in adaptation to several stress conditions (nutrient, osmotic, heavy metals, xenobiotic, or heat shock stresses) were accumulated (supplemental Table S2). All these data demonstrate that the alterations in the proteome of pellicle bacteria are widely involved in the antibiotic resistance and stress adaptation.
Iron Uptake and Pellicle Formation
Iron availability plays a key role in a lot of bacterial mechanisms, like adhesion (54), virulence (55–57), quorum sensing (58, 59), drug resistance (59), and biofilm formation (60). We observed here that numerous proteins belonging to the 5 principal iron uptake systems (54, 61–65), were significantly up-regulated in both 1-day and 4-day pellicles, as compared with planktonic culture: (1) the ferrous iron uptake system (FeoB A1S_0243), (2) the heme or acinetobactin uptake system (TonB A1S_0452 and ExbB A1S_0453 of the ExbB:ExbD:TonB complex), (3) the acinetobactin synthesis and transport system (BarB A1S_2375, BasF A1S_2380, BasC A1S_2384, BauA A1S_2385, BauB A1S_2386 and BasB A1S_2390), and (4) the hydroxamate siderophore synthesis and transport system (siderophore synthetase A1S_1650, TonB-dependent OM receptor A1S_1655 and acetyltransferase A1S_1657). The two components of the enterobactin receptor FepA, i.e., A1S_0980 and A1S_0981, were also accumulated, as expected (28). Some other ferric siderophore receptor proteins, never described up to now (proteins A1S_0474, A1S_1063, A1S_1667, A1S_2080, and A1S_3324), were also overexpressed by pellicle cells. This accumulation of iron uptake systems in pellicles agrees well with previous data by Marti et al. (28), which pointed out the importance of iron in A. baumannii pellicle formation.
Surprisingly, the present study shows that most of these proteins (17/20) were more abundant in 1-day pellicle than in 4-day one (supplemental Table S2) though we might imagine that nutrient-starved environment prevail in mature biofilms. The decrease of iron scavenging capability were also reported in P. aeruginosa 4-day S-L biofilm compared with early stages (i.e. one and 2 day S-L biofilm) (36). This discrepancy might be explained by a higher cellular activity in young biofilms.
An overexpression of the iron uptake has never been described in A. baumannii biofilms (25, 27). Some works even concluded that iron availability did not have a significant role in the ability of the bacterium to form a biofilm at the S-L interface (54, 66), or on the contrary generated an inhibitory effect (10, 58). The present results may suggest 2 hypotheses: (1) in contrast to S-L biofilm, iron is an essential inducer for A. baumannii pellicle formation; (2) as suggested for static culture growth conditions, a gradient of nutrient from the bottom to the top, including iron, was established (67) and A-L biofilms, because of their location, might be more starved.
Histidine and Aromatic Amino Acids Metabolism During Pellicle Biogenesis
Amino acids availability can also modulate bacterial biofilm development (27, 68, 69). Several proteins involved in the aromatic amino acids metabolism were here found accumulated in pellicles. Thus, proteins involved in the histidine degradation, i.e. HutG (A1S_3402), HutH (A1S_3405), HutI (A1S_3403) and HutU (A1S_3409), were up-regulated in 1-day pellicle cells (Fig. 3). The overproduction of the hut cluster products is in agreement with previous investigations on young (1-day) S-L biofilms (25, 27). This cluster leads to the production of purines and pyrimidines that may be required for eDNA production in matrix (70). We also showed, for the first time, a high abundance of proteins related to histidine biosynthesis, i.e. HisA (A1S_3238), HisC (A1S_0688) and HisZ (A1S_1178) in 4-day pellicle (supplemental Table S2). Histidine may serve as a precursor for various metabolites synthesis. Here, it might be necessary for histamine production that is required for acinetobactin siderophore biosynthesis (71).
Furthermore, two pathways (hmg and paa clusters) involved in the degradation of phenylalanine and tyrosine, were overexpressed in pellicles. Thus, five proteins belonging to the homogentisate (Hmg) degradation pathway controlled by the hmg cluster (72) were accumulated: TyrB (A1S_0071), HmgA (A1S_3416), HmgB (A1S_3415), HmgC (A1S_3414), and Hpd (A1S_3418,) (Fig. 3). Ten proteins of the phenylacetic acid (paa) degradation cluster i.e. A1S_1335–1338, A1S_1340–1344, and A1S_1347, were highly up-regulated in pellicle cells compared with planktonic counterparts (Fig. 3) (e.g. 18- and 41-fold for PaaB, respectively, supplemental Table S2). Acetoacetate (cleaved in acetyl-CoA) and fumarate are the final products of the hmg pathway, whereas the degradation paa pathway essentially generates acetyl-CoA and succinyl-CoA. These metabolites then integrate TCA cycle to generate energy, which is required for pellicle development (73–76).
Histidine metabolism is likely linked to that of aromatic amino acids (77, 78). Thus, protein-protein interaction network performed on the differentially expressed proteins showed that HisC protein was predicted to interact with the accumulated AroA (A1S_2276, supplemental Table S2, supplemental Fig. S3). AroA (also designated as EPSP synthase) is involved in the shikimate pathway leading to aromatic amino acids biosynthesis (79, 80). HisC also participates in the final steps of tyrosine and phenylalanine biosynthesis, by converting 4-hydroxyphenylpyruvate to tyrosine and phenylpyruvate to phenylalanine by transamination (77, 78).
To investigate the role of these aromatic amino acids in pellicles, we assessed the capacity of A. baumannii to form a pellicle in a minimum culture medium M9 containing Tyr or Phe as a sole carbon source. His and Ser were used as positive and negative controls, respectively (27). As expected, His promoted the pellicle formation as described for S-L biofilm (27). Moreover, Tyr and Phe significantly enhanced the pellicle development (supplemental Fig. S4A). These results corroborate those previously observed for P. aeruginosa in which Phe and Tyr robustly promoted biofilm formation (69) and support the hypothesis suggesting the important role of the aromatic acids metabolism in the pellicle formation.
Proteins Overrepresented in 1-Day Pellicle Cells
The analysis of the protein expressions dynamic revealed a group of proteins mostly overexpressed in 1-day pellicle cells (supplemental Table S2). Some biological processes in which these proteins are involved are discussed below.
Quorum Sensing
A cluster of proteins (A1S_0112 to A1S_0118, except A1S_0114), that was described as an operon related to the quorum sensing (QS) and motility (supplemental Fig. S5) (81, 82), was accumulated in 1-day pellicles. This operon could be involved in the synthesis of a biosurfactant lipopeptide, or an N-Acyl homoserine lactone (AHL) (25, 81). It is independently regulated by three different factors: (1) by the AHL, N-(3-hydroxydodecanoyl)- homoserine lactone (3-OH-C12-HSL), itself in a abaI-dependent manner (AbaI, A1S_0109, being the AHL synthase (81, 83), (2) by the cAMP via the CpdA regulator (82), and (3) via an iron limitation (58). A1S_0112 and A1S_0115 proteins are an Acyl-CoA synthetase and an AMP-dependent synthetase, respectively. Loss of these proteins in mutant strains induces an alteration of the pellicle formation, motility and hydrophobicity (82). In S-L biofilm, the A1S_0112–0118 genes have been shown overexpressed after 24 h of growth and an A1S_0114 deletion mutant (the corresponding protein being an acyl carrier protein ACP), also exhibited a reduced S-L biofilm formation compared with the wild-type (25). The relationship between the QS and the biofilm formation dynamics, has been widely investigated in P. aeruginosa (84). These studies, in particular, showed that lasI (abaI-homolog) expression decreased over the course of the biofilm development. The QS might so have a leading function during the early stages of the biofilm development, notably in attachment and microcolonies formation (84). The accumulation of autoinducers at this early stage of biofilm might play a crucial role to trigger the differentiation process (85).
Three of the four gene products from A1S_0104–0107 cluster, located upstream to abaI HSL-synthase, were also accumulated in 1-day pellicle organisms. These proteins, i.e., the acetyl-CoA synthetase/AMP-fatty acid ligase (A1S_0104), the acyl-CoA dehydrogenase (A1S_0105), and the enoyl-CoA hydratase/isomerase (A1S_0106), are predicted to be involved in fatty acids metabolism. Interestingly, it seems that biological functions of A1S_0104 and A1S_0105 are similar to those of A1S_0112 and A1S_0113 respectively. A1S_0106 and A1S_0107 may be required for the maturation of the acyl part during AHL biosynthesis, conducting to speculate that A1S_0104–0107 cluster could be also related to QS (supplemental Fig. S5). Recently, a transcriptomic analysis on imipenem resistant mutants derivative from ATCC 17978 strain, demonstrated a correlation between a decrease of the biofilm formation and a decrease of the expression of genes belonging to both clusters A1S_0104–0107 and A1S_0112–0118 (86) (supplemental Fig. S5).
Finally, it is known that QS may also affect lipopolysaccharide (LPS) synthesis (87). In E. coli, a knock-out mutant of luxS induced a 2-fold reduction in LPS amount when compared with the wild-type strain (87). Here, LPS-synthesis-related proteins, i.e., the UDP-acetylglucosamine acyltransferase LpxA (A1S_1965), the LPS glycosyltransferase (A1S_3841), and the phosphoglucosamine mutase GlmM (A1S_3320) were accumulated in 1-day pellicle. LPS has been suspected to be a component of the pellicle extracellular matrix in A. baumannii (22). Consequently, the accumulation of proteins involved in its synthesis might favor the pellicle establishment and facilitate, via its amphiphilic nature, the initial cells attachment (88).
Magnesium and Phosphate Enhanced Pellicle Formation
During biofilm formation, bacteria are in a constant interaction with their surrounding environment, in particularly with ions (as discussed above for iron). It has been described that magnesium ions may influence biofilm formation according to the bacterial species (89–91). The present work points out that the Mg2+ ATPase transporter MgtA (A1S_2070) was overexpressed in 1-day pellicle by 15- and 9-fold, compared with planktonic and 4-day pellicle cells, respectively. Mg2+ seems so to be required for the A. baumannii initial pellicle formation, as described for P. fluorescens for S-L biofilms (89), probably by limiting electrostatic repulsions or/and as a co-factor for some enzymes (90).
Two proteins, i.e. PstB (A1S_2445) and PstS-like (A1S_2448), related to the assimilation of inorganic phosphate (Pi), were also accumulated. Pi was shown to modulate S-L biofilm formation via PstSCAB-PhoU complex (92, 93). In non-limited Pi conditions, Pi assimilation is mainly provided by PstSCAB transporter to drive biofilm formation.
To confirm the importance of magnesium and phosphate on the pellicle formation, biofilm kinetics were performed in the presence of increasing ion concentrations. The positive effect of Mg2+ and phosphate on the pellicle formation was clearly observed, particularly after 1-day of growth (supplemental Fig. 4B and 4C).
Acetylation and Pellicle Development
Data analysis shows that the acetyltransferase A1S_1657 was specifically overexpressed in 1-day pellicle cells (by 17-fold). This result might suggest an important role of acetylation in young pellicle cells. Thus, the highest protein change fold (i.e., 51-fold) that we observed here, concerns an uncharacterized protein (A1S_1281), which has no homolog in other A. baumannii strains, but shares 51% of identity with the Sir2-like protein EMW58725.1 of E. coli strain 2762100. Sir-2 proteins are NAD+-dependent deacetylases and are broadly conserved from bacteria to higher eukaryotes (94). In M. tuberculosis, the NAD+-dependent deacetylase (CobB) mutant forms less biofilm compared with the wild-type strain (95, 96). Moreover, we also recently demonstrated that several adhesin/pili involved in biofilm formation in A. baumannii, e.g., Ata, Bap, LysM, CsuA/B and CsuC, were acetylated (article under minor revisions in Journal of Proteomics, 2016).
Proteins Overexpressed in 4-Day Pellicle State
Pellicle Adaptation to Nutrient Starvation
Bacterial cultures were here carried out in batch conditions, characterized by no nutrients renewing. After 4 days of growth, nutrients e.g., carbohydrates, are probably scarcer in the culture medium. Consequently, bacteria have to adapt their metabolism to a nutrient starvation. It is well known that matrix constituents (EPS, proteins, and lipids), can be recycled and utilized as carbon and energy sources (97). Thus, in A. baumannii, it has been reported that 5-day pellicle accumulated a lipase and a long chain fatty acid transporter, two proteins that could be used for fatty acids recycling (28). In accordance with this hypothesis, we observed here the 5-fold overproduction of both the phospholipase PLD1 (A1S_1919) (98) and the uncharacterized protein A1S_2738 which shares 75% of identity with the phospholipase C (PlcB) of Psychrobacter sp. Phospholipases are known as virulence factors in A. baumannii, they allow its growth in serum and promote host cell invasion (98, 99). The expression of extracellular phospholipases, allowing degradation of phospholipids as one nutrient source, has already been associated with the expression of fatty acids transporters (100). Here, we observed that the long-chain fatty acid transporter FadL-like (A1S_2773) was accumulated by 5-fold. These FadL transporters may be involved in the first steps of bacterial adherence, leading to host colonization (101). In E. coli, FadL transporter works in association with FadB, FadD, FadG, and FadI to convert fatty acids to acetyl-CoA in the cytoplasm to finally generate energy in TCA cycle (102). In accordance with this observation, FadG (A1S_0818) and FadB (A1S_0863) were up-regulated in 4-day cells and FadD (A1S_0817) and FadI (A1S_0534) in both 1-day and 4-day cells. All these data suggest that pellicle bacteria are highly adapted to nutrient limitation and more virulent compared than planktonic counterparts.
Importance of the TCS GacSA-regulated Proteins in Mature Pellicle
Bacterial TCS are designed to sense diverse stimuli and enact an appropriate and rapid adaptive physiological response. Recently, a novel global virulence regulator TCS has been described in A. baumannii, the GacSA system, which is composed by the inner membrane sensory kinase GacS (A1S_0574) and its response regulator GacA (A1S_0236) (73). In our data, GacA was found to be accumulated (4-fold) in 4-day pellicle cells. This overexpression is of particular importance. Indeed, this TCS regulates various virulence aspects, including the control of pili synthesis, the bacterial motility, the biofilm resistance and the metabolism of aromatic compounds. GacSA is also relevant for A. baumannii to persist and to cause infection in healthcare environments (73). We also observed that different gacA-regulated genes products varied in the same way: OmpA/MotB (A1S_1193), the paa cluster products (A1S_1335–1349), the ast operon product (A1S_3128–3132, a cluster involved in the arginine metabolism), as well as CsuA/B (A1S_2218), CsuC (A1S_2215) and CsuD (A1S_2214) (Table SI-2), which belong to the csu chaperone-usher pili operon which is associated in A. baumannii to adhesion on abiotic surfaces (15, 101).
Pili and other adhesion factors to maintain mature pellicle cohesion
The activation of the csu operon is mediated by the BfmRS TCS in A. baumannii strain ATCC 19606 (103). Indeed, we observed here an accumulation of the BfmR response regulator (A1S_0748) in mature biofilm cells. We also identified an accumulation of FilF (A1S_0695), shown to be involved in the structure of filamentous type III pili in Burkholderia cepacia (104). The overproduction of these appendices in A. baumannii pellicles has been already described (28). In Klebsiella pneumoniae, a type III pili deficient strain exhibited a reduced biofilm thickness and a higher rigidity of capsular polysaccharides. This type III pili may play a key role in capsular polysaccharide organization, improving the mechanical properties of biofilms (105). In the present study, among proteins involved in capsular biosynthesis, Wzi (A1S_0999) and Amylovoran glycosyltransferase AmsE (A1S_0060) were overexpressed both in 1-day and 4-day, Wzc and Wza were overexpressed at 1- and 4-day, respectively. Of note, it was recently demonstrated that the K locus genes (A1S_0049-A1S_0066,) expression, which is responsible of the capsular exopolysaccharide production, is dependent of the BfmRS TCS (106).
Some other adhesion factors were found overexpressed in mature biofilms: (1) OmpA (A1S_2840) which is an A. baumannii virulence factor facilitating adherence to eukaryotic cells and the cell invasion (103, 104). Two proteins presenting an OmpA/MotB peptidoglycan-associated domain (A1S_0884 and A1S_1193) were also accumulated in 4-day pellicle. These proteins share, 37 and 28% of identity with OmpA, respectively. (2) The LysM protein (A1S_0820), that belongs to the class of staphylococcal adhesins (109–111), presented a 2-fold amount increase in 4-day pellicle cells. It has been shown that a ΔLysM mutant strain presented a reduced S-L biofilm forming ability in A. baumannii (27). (3) Finally, the protein Bap (A1S_2724) was overexpressed in mature pellicle cells (supplemental Table S2). This protein is a surface adhesin that mediates primary attachment and which is also involved in intercellular adhesion in the mature biofilm (112, 113).
All these data show that mature pellicle cells mobilize an arsenal of adhesion factors to probably circumvent the lack of solid surface and to maintain the pellicle cohesion in improving cell-to-cell adherence.
Type VI Secretion System
Type VI secretion systems (T6SS) have been identified in a large number of pathogens. They are involved in cell invasion and competition among bacteria. T6SS have been implicated in numerous processes, including toxin delivery, bacterial fitness in chronic infection and biofilm formation (114–116). Recently, a genomic analysis of different Acinetobacter strains revealed the existence of T6SS related genes (117). In the present study, some T6SS-related proteins were mainly overexpressed in 4-day pellicle cells: TssC (A1S_1295), type VI secretion-associated protein (A1S_1301), TssM (A1S_1303), TssA (A1S_1308), and TssK (A1S_1309). TssC forms a tubular complex with TssB, participating to T6SS biogenesis (118). TssM is actively required for T6SS function in strain ATCC 17978 and controls the expression of Hcp and VgrG proteins, thought to form the needle of the secretion system (112, 114). A1S_1301, TssA and TssK proteins, presenting a high degree of conservation across bacterial T6SSs, are required for T6SS function, but their exact roles remain unclear (115). However, T6SS seems less operating in P. aeruginosa 4-day S-L biofilm than planktonic counterpart (36). T6SS is used by A. baumannii for interbacterial competition but may also play an important role in its virulence (120–122). In A. baumannii strain ATCC 17978, Weber et al. (2015) showed that the negative regulators of T6SS were harbored in a large self-transmissible resistance plasmid, pAB3. When A. baumannii is in competition with other bacteria, a part of the population undergoes plasmid loss, resulting in the activation of the T6SS (122). We might consequently suggest that after 4 days of growth in pellicle, a subset of the cellular population lost their pAB3 plasmid, leading to the T6SS activation, in response to environmental conditions. The T6SS has also been described as essential in the biofilm-specific drug resistance in A-L biofilms. Thus, the loss of tssC1 in P. aeruginosa PA14 resulted in a reduction of biofilm resistance to antibiotics such as tobramycin, gentamicin and ciprofloxacin, compared with the wild type whereas the mutation had no impact on the susceptibility of planktonic cells (123). TssC1 shared 47% of identity with A. baumannii TssC, overexpressed in A-L biofilm.
Protein Markers of Planktonic and Pellicle States
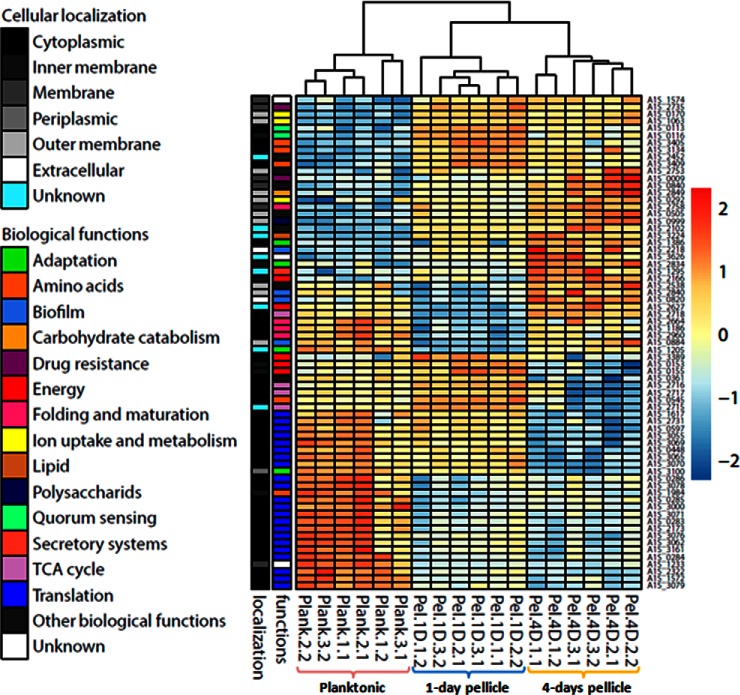
We then look for some protein markers specific of each growth condition (planktonic, 1- and 4-day pellicles). To this aim, an ANOVA-one way test was performed on the 620 identified proteins; 69 of them allowed to discriminate the 3 growth conditions (supplemental Table S3 and supplemental Fig. S1B). They were classified according to their correlation in abundance variation (Fig. 4). Their localization and their function are also given. This representation clearly separated the three culture conditions (planktonic, 1- and 4-day pellicles). The degree of positive correlation between these 69 proteins is described in supplemental Fig. S6. From this analysis, different groups could be discriminated (Fig. 4 and supplemental Fig. S6A). For the planktonic group (group G), proteins involved in translation were overrepresented with 12 ribosomal proteins (RplW (A1S_3078), RplK (A1S_0283), RplA (A1S_0284), RplJ (A1S_0285), RplL (A1S_0286), RpsA (A1S_1572), RplI (A1S_2173), RplM (A1S_3000), RplN (A1S_3071), RplV (A1S_3076), RplD (A1S_3079) and RplS (A1S_3161)) and the elongation factor EF-Ts (A1S_2322). For 1-day pellicle, two groups were discriminated (groups A and E), which clustered proteins involved in histidine catabolism (HutH, A1S_3405 and HutU, A1S_3409), the energy (AtpD, A1S_0155) or QS (A1S_0113 and A1S_0116). The group B and C concern the markers of 4-day pellicles. The corresponding proteins are involved in (1) bacterial attachment (CsuA/B A1S_2218) (2) virulence (LysM A1S_0820, OmpA A1S_2840, and TssC A1S_1295), (3) nutrient assimilation (CarO, A1S_2538), or (4) stress adaptation (KatE, A1S_1386) (supplemental Fig. S6B, supplemental Table S3). Therefore, this analysis provides the first biomarkers associated to the culture growth condition, and the pellicle development stage of A. baumannii ATCC 17978. Further experiments are required to validate these indicators at the species level.
Fig. 4.
Heatmap hierarchical cluster representing the correlation in abundance variation of the 69 protein markers.
CONCLUSION
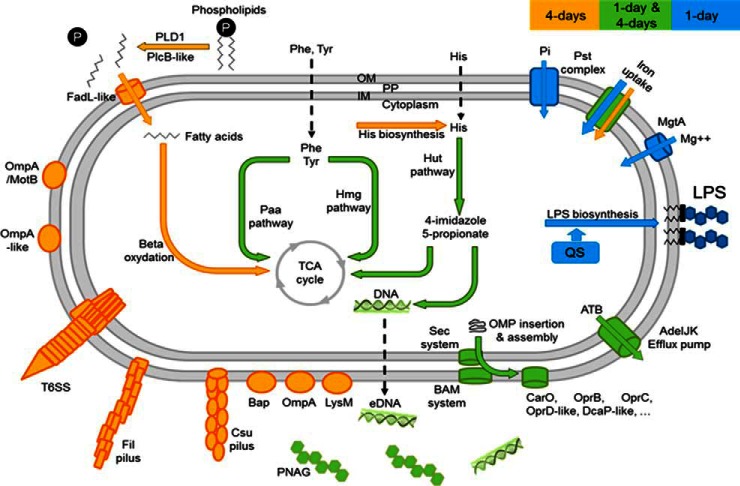
Despite the ubiquity of pellicles in both natural and artificial environments (124), few studies have investigated this biofilm type. This study provides the widest proteomic investigation devoted to the dynamic protein expression of A. baumannii cells grown as pellicle. Here, different mechanisms seem to be involved in pellicle development, summarized in Fig. 5. An outstanding result is the confirmation that A. baumanni cells enhance the expression of virulence factors in pellicle, confirming clinical observations (26). The bacterial adaptation to this growth mode also requires drastic changes at the membrane level, highlighted by the accumulation of adhesion factors and proteins involved in the nutrient assimilation. According to the high number of iron uptake systems overexpressed in pellicles, iron seems also essential for this growth mode. Multiple unknown proteins were also pointed out as playing a role in the pellicle phenotype. To identify their function constitutes yet a future challenge.
Fig. 5.
Schematic representation of biological processes during A. baumannii pellicle development at the cellular level. Intervening proposed metabolic and assimilation pathways are shown by arrows. OM: outer membrane, IM: inner membrane, PP: periplasm, Pi: phosphate ion, ATB: antibiotics.
Supplementary Material
Acknowledgments
T.K. thanks the Région Haute-Normandie for and the IRIB institute for the PhD financial support.
Footnotes
Author contributions: E.D. and J.H. designed research; T.K. performed research; T.K. and J.H. analyzed data; T.K., T.J., E.D., and J.H. wrote the paper; A.B. statistical analysis.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- EPS
- extracellular polymeric substances
- S-L
- solid-liquid
- A-L
- air-liquid
- MS2
- tandem MS
- OMP
- outer membrane proteins.
REFERENCES
- 1. Boucher H. W., Talbot G. H., Bradley J. S., Edwards J. E., Gilbert D., Rice L. B., Scheld M., Spellberg B., and Bartlett J. (2009) Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48, 1–12 [DOI] [PubMed] [Google Scholar]
- 2. Tien H. C., Battad A., Bryce E. A., Fuller J., Mulvey M., Bernard K., Brisebois R., Doucet J. J., Rizoli S. B., Fowler R., and Simor A. (2007) Multi-drug resistant Acinetobacter infections in critically injured Canadian forces soldiers. BMC Infect. Dis. 7, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bergogne-Bérézin E., and Towner K. J. (1996) Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9, 148–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sengstock D. M., Thyagarajan R., Apalara J., Mira A., Chopra T., and Kaye K. S. (2010) Multidrug-resistant Acinetobacter baumannii: an emerging pathogen among older adults in community hospitals and nursing homes. Clin. Infect. Dis. 50, 1611–1616 [DOI] [PubMed] [Google Scholar]
- 5. Dijkshoorn L., Nemec A., and Seifert H. (2007) An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5, 939–951 [DOI] [PubMed] [Google Scholar]
- 6. Gayoso C. M., Mateos J., Méndez J. A., Fernández-Puente P., Rumbo C., Tomás M., Martínez de Ilarduya O., and Bou G. (2014) Molecular mechanisms involved in the response to desiccation stress and persistence in Acinetobacter baumannii. J. Proteome Res. 13, 460–476 [DOI] [PubMed] [Google Scholar]
- 7. Soares N. C., Cabral M. P., Gayoso C., Mallo S., Rodriguez-Velo P., Fernandez-Moreira E., and Bou G. (2010) Associating Growth-Phase-Related Changes in the Proteome of Acinetobacter baumannii with Increased Resistance to Oxidative Stress. J. Proteome Res. 9, 1951–1964 [DOI] [PubMed] [Google Scholar]
- 8. Peleg A. Y., Seifert H., and Paterson D. L. (2008) Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21, 538–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Davey M. E., and O'toole G. A. (2000) Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. MMBR 64, 847–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tomaras A. P., Dorsey C. W., Edelmann R. E., and Actis L. A. (2003) Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149, 3473–3484 [DOI] [PubMed] [Google Scholar]
- 11. Rodríguez-Baño J., Martí S., Soto S., Fernández-Cuenca F., Cisneros J. M., Pachón J., Pascual A., Martínez-Martínez L., McQueary C., Actis L. A., Vila J., and the Spanish Group for the Study of Nosocomial Infections (GEIH). (2008) Biofilm formation in Acinetobacter baumannii: associated features and clinical implications. Clin. Microbiol. Infect. 14, 276–278 [DOI] [PubMed] [Google Scholar]
- 12. Lee H. W., Koh Y. M., Kim J., Lee J. C., Lee Y.-C., Seol S. Y., Cho D. T., and Kim J. (2008) Capacity of multidrug-resistant clinical isolates of Acinetobacter baumannii to form biofilm and adhere to epithelial cell surfaces. Clin. Microbiol. Infect. 14, 49–54 [DOI] [PubMed] [Google Scholar]
- 13. Greene C., Vadlamudi G., Newton D., Foxman B., and Xi C. (2016) The influence of biofilm formation and multidrug resistance on environmental survival of clinical and environmental isolates of Acinetobacter baumannii. Am. J. Infect. Control 44, e65–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Monds R. D., and O'Toole G. A. (2009) The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 17, 73–87 [DOI] [PubMed] [Google Scholar]
- 15. Donlan R. M. (2002) Biofilms: Microbial Life on Surfaces. Emerg. Infect. Dis. 8, 881–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hall-Stoodley L., and Stoodley P. (2009) Evolving concepts in biofilm infections. Cell. Microbiol. 11, 1034–1043 [DOI] [PubMed] [Google Scholar]
- 17. Gaddy J. A., and Actis L. A. (2009) Regulation of Acinetobacter baumannii biofilm formation. Future Microbiol. 4, 273–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stewart P. S., and William Costerton J. (2001) Antibiotic resistance of bacteria in biofilms. The Lancet 358, 135–138 [DOI] [PubMed] [Google Scholar]
- 19. Mah T. F., and O'Toole G. A. (2001) Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9, 34–39 [DOI] [PubMed] [Google Scholar]
- 20. Sauer K., Camper A. K., Ehrlich G. D., Costerton J. W., and Davies D. G. (2002) Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184, 1140–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koza A., Hallett P. D., Moon C. D., and Spiers A. J. (2009) Characterization of a novel air–liquid interface biofilm of Pseudomonas fluorescens SBW25. Microbiology 155, 1397–1406 [DOI] [PubMed] [Google Scholar]
- 22. Nait Chabane Y., Marti S., Rihouey C., Alexandre S., Hardouin J., Lesouhaitier O., Vila J., Kaplan J. B., Jouenne T., and De E. (2014) Characterisation of pellicles formed by Acinetobacter baumannii at the air-liquid interface. PLoS ONE 9, e111660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martí S., Rodríguez-Baño J., Catel-Ferreira M., Jouenne T., Vila J., Seifert H., and Dé E. (2011) Biofilm formation at the solid-liquid and air-liquid interfaces by Acinetobacter species. BMC Res. Notes 4, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Branda S. S., Vik Å., Friedman L., and Kolter R. (2005) Biofilms: the matrix revisited. Trends Microbiol. 13, 20–26 [DOI] [PubMed] [Google Scholar]
- 25. Rumbo-Feal S., Gómez M. J., Gayoso C., Alvarez-Fraga L., Cabral M. P., Aransay A. M., Rodríguez-Ezpeleta N., Fullaondo A., Valle J., Tomás M., Bou G., and Poza M. (2013) Whole Transcriptome Analysis of Acinetobacter baumannii Assessed by RNA-Sequencing reveals different mRNA expression profiles in biofilm compared to planktonic cells. PLoS ONE 8, e72968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shin J.-H., Lee H.-W., Kim S.-M., and Kim J. (2009) Proteomic analysis of Acinetobacter baumannii in biofilm and planktonic growth mode. J. Microbiol. 47, 728–735 [DOI] [PubMed] [Google Scholar]
- 27. Cabral M. P., Soares N. C., Aranda J., Parreira J. R., Rumbo C., Poza M., Valle J., Calamia V., Lasa I., and Bou G. (2011) Proteomic and functional analyses reveal a unique lifestyle for Acinetobacter baumannii biofilms and a key role for histidine metabolism. J. Proteome Res. 10, 3399–3417 [DOI] [PubMed] [Google Scholar]
- 28. Marti S., Nait Chabane Y., Alexandre S., Coquet L., Vila J., Jouenne T., and Dé E. (2011) Growth of Acinetobacter baumannii in pellicle enhanced the expression of potential virulence factors. PLoS ONE 6, e26030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. O'Toole G. A., and Kolter R. (1998) Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28, 449–461 [DOI] [PubMed] [Google Scholar]
- 30. Ouidir T., Jarnier F., Cosette P., Jouenne T., and Hardouin J. (2015) Characterization of N-terminal protein modifications in Pseudomonas aeruginosa PA14. J. Proteomics 114, 214–225 [DOI] [PubMed] [Google Scholar]
- 31. Obry P. C. (2014) AB0161 Identification and validation of a protein combination including s100a9 able to predict the response to the mtx/etanercept association in rheumatoid arthritis patients. Ann. Rheum. Dis. 72, A834–A835 [Google Scholar]
- 32. Vallenet D., Belda E., Calteau A., Cruveiller S., Engelen S., Lajus A., Le Fèvre F., Longin C., Mornico D., Roche D., Rouy Z., Salvignol G., Scarpelli C., Thil Smith A. A., Weiman M., and Médigue C. (2013) MicroScope–an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res. 41, D636–D647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mitchell A., Chang H. Y., Daugherty L., Fraser M., Hunter S., Lopez R., McAnulla C., McMenamin C., Nuka G., Pesseat S., Sangrador-Vegas A., Scheremetjew M., Rato C., Yong S. Y., Bateman A., Punta M., Attwood T. K., Sigrist C. J., Redaschi N., Rivoire C., Xenarios I., Kahn D., Guyot D., Bork P., Letunic I., Gough J., Oates M., Haft D., Huang H., Natale D. A., Wu C. H., Orengo C., Sillitoe I., Mi H., Thomas P. D., and Finn R. D. (2015) The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 43, D213–D221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., Lin J., Minguez P., Bork P., von Mering C., and Jensen L. J. (2013) STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–D815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smith M. G., Gianoulis T. A., Pukatzki S., Mekalanos J. J., Ornston L. N., Gerstein M., and Snyder M. (2007) New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 21, 601–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Park A. J., Murphy K., Krieger J. R., Brewer D., Taylor P., Habash M., and Khursigara C. M. (2014) A temporal examination of the planktonic and biofilm proteome of whole cell Pseudomonas aeruginosa PAO1 using quantitative mass spectrometry. Mol. Cell. Proteomics MCP 13, 1095–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Selkrig J., Mosbahi K., Webb C. T., Belousoff M. J., Perry A. J., Wells T. J., Morris F., Leyton D. L., Totsika M., Phan M.-D., Celik N., Kelly M., Oates C., Hartland E. L., Robins-Browne R. M., Ramarathinam S. H., Purcell A. W., Schembri M. A., Strugnell R. A., Henderson I. R., Walker D., and Lithgow T. (2012) Discovery of an archetypal protein transport system in bacterial outer membranes. Nat. Struct. Mol. Biol. 19, 506–510, S1 [DOI] [PubMed] [Google Scholar]
- 38. Voulhoux R., Bos M. P., Geurtsen J., Mols M., and Tommassen J. (2003) Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299, 262–265 [DOI] [PubMed] [Google Scholar]
- 39. Selkrig J., Leyton D. L., Webb C. T., and Lithgow T. (2014) Assembly of β-barrel proteins into bacterial outer membranes. Biochim. Biophys. Acta 1843, 1542–1550 [DOI] [PubMed] [Google Scholar]
- 40. Dallo S. F., Denno J., Hong S., and Weitao T. (2010) Adhesion of Acinetobacter baumannii to extracellular proteins detected by a live cell-protein binding assay. Ethn. Dis. 20, S1–7–11 [PubMed] [Google Scholar]
- 41. Li H., Luo Y.-F., Williams B. J., Blackwell T. S., and Xie C.-M. (2012) Structure and function of OprD protein in Pseudomonas aeruginosa: From antibiotic resistance to novel therapies. Int. J. Med. Microbiol. IJMM 302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mussi M. A., Relling V. M., Limansky A. S., and Viale A. M. (2007) CarO, an Acinetobacter baumannii outer membrane protein involved in carbapenem resistance, is essential for L-ornithine uptake. FEBS Lett. 581, 5573–5578 [DOI] [PubMed] [Google Scholar]
- 43. Mussi M. A., Limansky A. S., and Viale A. M. (2005) Acquisition of Resistance to Carbapenems in Multidrug-Resistant Clinical Strains of Acinetobacter baumannii: Natural Insertional Inactivation of a Gene Encoding a Member of a Novel Family of β-Barrel Outer Membrane Proteins. Antimicrob. Agents Chemother. 49, 1432–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Catel-Ferreira M., Nehmé R., Molle V., Aranda J., Bouffartigues E., Chevalier S., Bou G., Jouenne T., and Dé E. (2012) Deciphering the function of the outer membrane protein OprD homolog of Acinetobacter baumannii. Antimicrob. Agents Chemother. 56, 3826–3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yoon E. J., Chabane Y. N., Goussard S., Snesrud E., Courvalin P., Dé E., and Grillot-Courvalin C. (2015) Contribution of resistance-nodulation-cell division efflux systems to antibiotic resistance and biofilm formation in Acinetobacter baumannii. mBio 6, e00309–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Costerton J. W., Lewandowski Z., Caldwell D. E., Korber D. R., and Lappin-Scott H. M. (1995) Microbial biofilms. Annu. Rev. Microbiol. 49, 711–745 [DOI] [PubMed] [Google Scholar]
- 47. Rao R. S., Karthika R. U., Singh S. P., Shashikala P., Kanungo R., Jayachandran S., and Prashanth K. (2008) Correlation between biofilm production and multiple drug resistance in imipenem resistant clinical isolates of Acinetobacter baumannii. Indian J. Med. Microbiol. 26, 333–337 [DOI] [PubMed] [Google Scholar]
- 48. Jacoby G. A. (2009) AmpC β-Lactamases. Clin. Microbiol. Rev. 22, 161–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Choi A. H., Slamti L., Avci F. Y., Pier G. B., and Maira-Litrán T. (2009) The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-β-1–6-N-acetylglucosamine, which is critical for biofilm formation. J. Bacteriol. 191, 5953–5963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Amini S., Goodarzi H., and Tavazoie S. (2009) Genetic dissection of an exogenously induced biofilm in laboratory and clinical isolates of E. coli. PLoS Pathog. 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hassett D. J., Cuppoletti J., Trapnell B., Lymar S. V., Rowe J. J., Yoon S. S., Hilliard G. M., Parvatiyar K., Kamani M. C., Wozniak D. J., Hwang S. H., McDermott T. R., and Ochsner U. A. (2002) Anaerobic metabolism and quorum sensing by Pseudomonas aeruginosa biofilms in chronically infected cystic fibrosis airways: rethinking antibiotic treatment strategies and drug targets. Adv. Drug Deliv. Rev. 54, 1425–1443 [DOI] [PubMed] [Google Scholar]
- 52. King T., Lucchini S., Hinton J. C., and Gobius K. (2010) Transcriptomic Analysis of Escherichia coli O157:H7 and K-12 cultures exposed to inorganic and organic acids in stationary phase reveals acidulant- and strain-specific acid tolerance responses. Appl. Environ. Microbiol. 76, 6514–6528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Das T., and Manefield M. (2012) Pyocyanin promotes extracellular DNA release in Pseudomonas aeruginosa. PloS One 7, e46718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Eijkelkamp B. A., Hassan K. A., Paulsen I. T., and Brown M. H. (2011) Investigation of the human pathogen Acinetobacter baumannii under iron limiting conditions. BMC Genomics 12, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rhodes E. R., Menke S., Shoemaker C., Tomaras A. P., McGillivary G., and Actis L. A. (2007) Iron acquisition in the dental pathogen Actinobacillus actinomycetemcomitans: what does it use as a source and how does it get this essential metal? Biometals Int. J. Role Met. Ions Biol. Biochem. Med. 20, 365–377 [DOI] [PubMed] [Google Scholar]
- 56. Antunes L. C., Imperi F., Carattoli A., and Visca P. (2011) Deciphering the multifactorial nature of Acinetobacter baumannii pathogenicity. PloS One 6, e22674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gaddy J. A., Arivett B. A., McConnell M. J., López-Rojas R., Pachón J., and Actis L. A. (2012) Role of acinetobactin-mediated iron acquisition functions in the interaction of Acinetobacter baumannii strain ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infect. Immun. 80, 1015–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Modarresi F., Azizi O., Shakibaie M. R., Motamedifar M., Mosadegh E., and Mansouri S. (2015) Iron limitation enhances acyl homoserine lactone (AHL) production and biofilm formation in clinical isolates of Acinetobacter baumannii. Virulence 6, 152–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Modarresi F., Azizi O., Shakibaie M. R., Motamedifar M., Valibeigi B., and Mansouri S. (2015) Effect of iron on expression of efflux pump (adeABC) and quorum sensing (luxI, luxR) genes in clinical isolates of Acinetobacter baumannii. APMIS Acta Pathol. Microbiol. Immunol. Scand. 123, 959–968 [DOI] [PubMed] [Google Scholar]
- 60. Banin E., Vasil M. L., and Greenberg E. P. (2005) Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 102, 11076–11081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Antunes L. C., Imperi F., Towner K. J., and Visca P. (2011) Genome-assisted identification of putative iron-utilization genes in Acinetobacter baumannii and their distribution among a genotypically diverse collection of clinical isolates. Res. Microbiol. 162, 279–284 [DOI] [PubMed] [Google Scholar]
- 62. Zimbler D. L., Penwell W. F., Gaddy J. A., Menke S. M., Tomaras A. P., Connerly P. L., and Actis L. A. (2009) Iron acquisition functions expressed by the human pathogen Acinetobacter baumannii. Biometals Int. J. Role Met. Ions Biol. Biochem. Med. 22, 23–32 [DOI] [PubMed] [Google Scholar]
- 63. Nwugo C. C., Gaddy J. A., Zimbler D. L., and Actis L. A. (2011) Deciphering the iron response in Acinetobacter baumannii: A proteomics approach. J. Proteomics 74, 44–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mihara K., Tanabe T., Yamakawa Y., Funahashi T., Nakao H., Narimatsu S., and Yamamoto S. (2004) Identification and transcriptional organization of a gene cluster involved in biosynthesis and transport of acinetobactin, a siderophore produced by Acinetobacter baumannii ATCC 19606T. Microbiol. Read. Engl. 150, 2587–2597 [DOI] [PubMed] [Google Scholar]
- 65. Cartron M. L., Maddocks S., Gillingham P., Craven C. J., and Andrews S. C. (2006) Feo–transport of ferrous iron into bacteria. Biometals Int. J. Role Met. Ions Biol. Biochem. Med. 19, 143–157 [DOI] [PubMed] [Google Scholar]
- 66. Gentile V., Frangipani E., Bonchi C., Minandri F., Runci F., and Visca P. (2014) Iron and Acinetobacter baumannii biofilm formation. Pathogens 3, 704–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Carpentier B., and Cerf O. (1993) Biofilms and their consequences, with particular reference to hygiene in the food industry. J. Appl. Bacteriol. 75, 499–511 [DOI] [PubMed] [Google Scholar]
- 68. Goh S.-N., Fernandez A., Ang S.-Z., Lau W.-Y., Ng D.-L., and Cheah E. S. G. (2013) Effects of different amino acids on biofilm growth, swimming motility and twitching motility in Escherichia Coli BL21. J. Biol. Life Sci. 4 [Google Scholar]
- 69. Bernier S. P., Ha D. G., Khan W., Merritt J. H., and O'Toole G. A. (2011) Modulation of Pseudomonas aeruginosa surface-associated group behaviors by individual amino acids through c-di-GMP signaling. Res. Microbiol. 162, 680–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kapatral V., Campbell J. W., Minnich S. A., Thomson N. R., Matsumura P., and Prüss B. M. (2004) Gene array analysis of Yersinia enterocolitica FlhD and FlhC: regulation of enzymes affecting synthesis and degradation of carbamoylphosphate. Microbiol. Read. Engl. 150, 2289–2300 [DOI] [PubMed] [Google Scholar]
- 71. Hasan T., Choi C. H., and Oh M. H. (2015) Genes involved in the biosynthesis and transport of acinetobactin in Acinetobacter baumannii. Genomics Inform. 13, 2–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Arias-Barrau E., Olivera E. R., Luengo J. M., Fernández C., Galán B., García J. L., Díaz E., and Miñambres B. (2004) The homogentisate pathway: a central catabolic pathway involved in the degradation of L-phenylalanine, L-tyrosine, and 3-hydroxyphenylacetate in Pseudomonas putida. J. Bacteriol. 186, 5062–5077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cerqueira G. M., Kostoulias X., Khoo C., Aibinu I., Qu Y., Traven A., and Peleg A. Y. (2014) A global virulence regulator in Acinetobacter baumannii and its control of the phenylacetic acid catabolic pathway. J. Infect. Dis. 210, 46–55 [DOI] [PubMed] [Google Scholar]
- 74. Fernández C., Díaz E., and García J. L. (2014) Insights on the regulation of the phenylacetate degradation pathway from Escherichia coli. Environ. Microbiol. Rep. 6, 239–250 [DOI] [PubMed] [Google Scholar]
- 75. Hamlin J. N., Bloodworth R. A., and Cardona S. T. (2009) Regulation of phenylacetic acid degradation genes of Burkholderia cenocepacia K56–2. BMC Microbiol. 9, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Olivera E. R., Minambres B., Garcia B., Muniz C., Moreno M. A., Ferrandez A., Diaz E., Garcia J. L., and Luengo J. M. (1998) Molecular characterization of the phenylacetic acid catabolic pathway in Pseudomonas putida U: The phenylacetyl-CoA catabolon. Proc. Natl. Acad. Sci. U.S.A. 95, 6419–6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Fernandez F. J., Vega M. C., Lehmann F., Sandmeier E., Gehring H., Christen P., and Wilmanns M. (2004) Structural studies of the catalytic reaction pathway of a hyperthermophilic histidinol-phosphate aminotransferase. J. Biol. Chem. 279, 21478–21488 [DOI] [PubMed] [Google Scholar]
- 78. Nester E. W., and Montoya A. L. (1976) An enzyme common to histidine and aromatic amino acid biosynthesis in Bacillus subtilis. J. Bacteriol. 126, 699–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sutton K. A., Breen J., Russo T. A., Schultz L. W., and Umland T. C. (2016) Crystal structure of 5-enolpyruvylshikimate-3-phosphate (EPSP) synthase from the ESKAPE pathogen Acinetobacter baumannii. Acta Crystallogr. Sect. F Struct. Biol. Commun. 72, 179–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Umland T. C., Schultz L. W., MacDonald U., Beanan J. M., Olson R., and Russo T. A. (2012) In vivo-validated essential genes identified in Acinetobacter baumannii by using human ascites overlap poorly with essential genes detected on laboratory media. mBio 3, e00113–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Clemmer K. M., Bonomo R. A., and Rather P. N. (2011) Genetic analysis of surface motility in Acinetobacter baumannii. Microbiology 157, 2534–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Giles S. K., Stroeher U. H., Eijkelkamp B. A., and Brown M. H. (2015) Identification of genes essential for pellicle formation in Acinetobacter baumannii. BMC Microbiol. 15, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Niu C., Clemmer K. M., Bonomo R. A., and Rather P. N. (2008) Isolation and characterization of an autoinducer synthase from Acinetobacter baumannii. J. Bacteriol. 190, 3386–3392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. De Kievit T. R., Gillis R., Marx S., Brown C., and Iglewski B. H. (2001) Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl. Environ. Microbiol. 67, 1865–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Davies D. G., Parsek M. R., Pearson J. P., Iglewski B. H., Costerton J. W., and Greenberg E. P. (1998) The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280, 295–298 [DOI] [PubMed] [Google Scholar]
- 86. Chang K.-C., Kuo H.-Y., Tang C. Y., Chang C.-W., Lu C.-W., Liu C.-C., Lin H.-R., Chen K.-H., and Liou M.-L. (2014) Transcriptome profiling in imipenem-selected Acinetobacter baumannii. BMC Genomics 15, 815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Palaniyandi S., Mitra A., Herren C. D., Zhu X., and Mukhopadhyay S. (2013) LuxS contributes to virulence in avian pathogenic Escherichia coli O78:K80:H9. Vet. Microbiol. 166, 567–575 [DOI] [PubMed] [Google Scholar]
- 88. Uppu D. S., and Haldar J. (2016) Lipopolysaccharide neutralization by cationic-amphiphilic polymers through pseudoaggregate formation. Biomacromolecules 17, 862–873 [DOI] [PubMed] [Google Scholar]
- 89. Song B., and Leff L. G. (2006) Influence of magnesium ions on biofilm formation by Pseudomonas fluorescens. Microbiol. Res. 161, 355–361 [DOI] [PubMed] [Google Scholar]
- 90. Simoni S. F., Bosma T. N. P., Harms H., and Zehnder A. J. B. (2000) Bivalent cations increase both the subpopulation of adhering bacteria and their adhesion efficiency in sand columns. Environ. Sci. Technol. 34, 1011–1017 [Google Scholar]
- 91. Marcus H., Austria A., and Baker N. R. (1989) Adherence of Pseudomonas aeruginosa to tracheal epithelium. Infect. Immun. 57, 1050–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Monds R. D., Silby M. W., and Mahanty H. K. (2001) Expression of the Pho regulon negatively regulates biofilm formation by Pseudomonas aureofaciens PA147–2. Mol. Microbiol. 42, 415–426 [DOI] [PubMed] [Google Scholar]
- 93. Danhorn T., Hentzer M., Givskov M., Parsek M. R., and Fuqua C. (2004) Phosphorus limitation enhances biofilm formation of the plant pathogen Agrobacterium tumefaciens through the PhoR-PhoB regulatory system. J. Bacteriol. 186, 4492–4501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Blander G., and Guarente L. (2004) The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 73, 417–435 [DOI] [PubMed] [Google Scholar]
- 95. Liu F., Yang M., Wang X., Yang S., Gu J., Zhou J., Zhang X.-E., Deng J., and Ge F. (2014) Acetylome analysis reveals diverse functions of lysine acetylation in Mycobacterium tuberculosis. Mol. Cell. Proteomics 13, 3352–3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ouidir T., Kentache T., and Hardouin J. (2016) Protein lysine acetylation in bacteria: Current state of the art. Proteomics 16, 301–309 [DOI] [PubMed] [Google Scholar]
- 97. Flemming H. C., and Wingender J. (2010) The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633 [DOI] [PubMed] [Google Scholar]
- 98. Stahl J., Bergmann H., Göttig S., Ebersberger I., and Averhoff B. (2015) Acinetobacter baumannii virulence is mediated by the concerted action of three phospholipases D. PLOS ONE 10, e0138360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Jacobs A. C., Hood I., Boyd K. L., Olson P. D., Morrison J. M., Carson S., Sayood K., Iwen P. C., Skaar E. P., and Dunman P. M. (2010) Inactivation of phospholipase D diminishes Acinetobacter baumannii pathogenesis. Infect. Immun. 78, 1952–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Miller R. M., Tomaras A. P., Barker A. P., Voelker D. R., Chan E. D., Vasil A. I., and Vasil M. L. (2008) Pseudomonas aeruginosa twitching motility-mediated chemotaxis towards phospholipids and fatty acids: specificity and metabolic requirements. J. Bacteriol. 190, 4038–4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. de Lima Pimenta A., Di Martino P., Le Bouder E., Hulen C., and Blight M. A. (2003) In vitro identification of two adherence factors required for in vivo virulence of Pseudomonas fluorescens. Microbes Infect. Inst. Pasteur 5, 1177–1187 [DOI] [PubMed] [Google Scholar]
- 102. van and den Berg B. (2010) Going forward laterally: transmembrane passage of hydrophobic molecules through protein channel walls. Chembiochem Eur. J. Chem. Biol. 11, 1339–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Tomaras A. P., Flagler M. J., Dorsey C. W., Gaddy J. A., and Actis L. A. (2008) Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology 154, 3398–3409 [DOI] [PubMed] [Google Scholar]
- 104. Goldstein R., Sun L., Jiang R. Z., Sajjan U., Forstner J. F., and Campanelli C. (1995) Structurally variant classes of pilus appendage fibers coexpressed from Burkholderia (Pseudomonas) cepacia. J. Bacteriol. 177, 1039–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wang H., Wilksch J. J., Strugnell R. A., and Gee M. L. (2015) Role of capsular polysaccharides in biofilm formation: An AFM Nanomechanics Study. ACS Appl. Mater. Interfaces 7, 13007–13013 [DOI] [PubMed] [Google Scholar]
- 106. Geisinger E., and Isberg R. R. (2015) Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii. PLOS Pathog 11, e1004691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Ofori-Darko E., Zavros Y., Rieder G., Tarlé S. A., Van Antwerp M., and Merchant J. L. (2000) An OmpA-like protein from Acinetobacter spp. stimulates gastrin and interleukin-8 promoterse. Infect. Immun. 68, 3657–3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Gaddy J. A., Tomaras A. P., and Actis L. A. (2009) The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect. Immun. 77, 3150–3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Downer R., Roche F., Park P. W., Mecham R. P., and Foster T. J. (2002) The elastin-binding protein of Staphylococcus aureus (EbpS) is expressed at the cell surface as an integral membrane protein and not as a cell wall-associated protein. J. Biol. Chem. 277, 243–250 [DOI] [PubMed] [Google Scholar]
- 110. Heilmann C., Hartleib J., Hussain M. S., and Peters G. (2005) The multifunctional Staphylococcus aureus autolysin aaa mediates adherence to immobilized fibrinogen and fibronectin. Infect. Immun. 73, 4793–4802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Heilmann C., Thumm G., Chhatwal G. S., Hartleib J., Uekötter A., and Peters G. (2003) Identification and characterization of a novel autolysin (Aae) with adhesive properties from Staphylococcus epidermidis. Microbiol. Read. Engl. 149, 2769–2778 [DOI] [PubMed] [Google Scholar]
- 112. Loehfelm T. W., Luke N. R., and Campagnari A. A. (2008) Identification and characterization of an Acinetobacter baumannii biofilm-associated protein. J. Bacteriol. 190, 1036–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. De Gregorio E., Del Franco M., Martinucci M., Roscetto E., Zarrilli R., and Di Nocera P. P. (2015) Biofilm-associated proteins: news from Acinetobacter. BMC Genomics 16, 933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Tian Y., Zhao Y., Wu X., Liu F., Hu B., and Walcott R. R. (2015) The type VI protein secretion system contributes to biofilm formation and seed-to-seedling transmission of Acidovorax citrulli on melon. Mol. Plant Pathol. 16, 38–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Silverman J. M., Brunet Y. R., Cascales E., and Mougous J. D. (2012) Structure and regulation of the type VI secretion system. Annu. Rev. Microbiol. 66, 453–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Filloux A., Hachani A., and Bleves S. (2008) The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiol. Read. Engl. 154, 1570–1583 [DOI] [PubMed] [Google Scholar]
- 117. Weber B. S., Miyata S. T., Iwashkiw J. A., Mortensen B. L., Skaar E. P., Pukatzki S., and Feldman M. F. (2013) Genomic and functional analysis of the type VI secretion system in Acinetobacter. PLoS ONE 8, e55142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Bönemann G., Pietrosiuk A., Diemand A., Zentgraf H., and Mogk A. (2009) Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 28, 315–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Ruiz F. M., Santillana E., Spínola-Amilibia M., Torreira E., Culebras E., and Romero A. (2015) Crystal structure of Hcp from Acinetobacter baumannii: A component of the type VI secretion system. PLoS ONE 10, e0129691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Carruthers M. D., Nicholson P. A., Tracy E. N., and Munson R. S. Jr. (2013) Acinetobacter baumannii utilizes a Type VI secretion system for bacterial competition. PLoS ONE 8, e59388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Repizo G. D., Gagné S., Foucault-Grunenwald M.-L., Borges V., Charpentier X., Limansky A. S., Gomes J. P., Viale A. M., and Salcedo S. P. (2015) Differential role of the T6SS in Acinetobacter baumannii virulence. PLoS ONE 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Weber B. S., Ly P. M., Irwin J. N., Pukatzki S., and Feldman M. F. (2015) A multidrug resistance plasmid contains the molecular switch for type VI secretion in Acinetobacter baumannii. Proc. Natl. Acad. Sci. U.S.A. 112, 9442–9447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Zhang L., Hinz A. J., Nadeau J. P., and Mah T. F. (2011) Pseudomonas aeruginosa tssC1 links type VI secretion and biofilm-specific antibiotic resistance▿. J. Bacteriol. 193, 5510–5513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Armitano J., Méjean V., and Jourlin-Castelli C. (2014) Gram-negative bacteria can also form pellicles. Environ. Microbiol. Rep. 6, 534–544 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.