Abstract
Pathophysiologies of cancer-associated syndromes such as cachexia are poorly understood and no routine biomarkers have been established, yet. Using shotgun proteomics, known marker molecules including PMEL, CRP, SAA, and CSPG4 were found deregulated in patients with metastatic melanoma. Targeted analysis of 58 selected proteins with multiple reaction monitoring was applied for independent data verification. In three patients, two of which suffered from cachexia, a tissue damage signature was determined, consisting of nine proteins, PLTP, CD14, TIMP1, S10A8, S10A9, GP1BA, PTPRJ, CD44, and C4A, as well as increased levels of glycine and asparagine, and decreased levels of polyunsaturated phosphatidylcholine concentrations, as determined by targeted metabolomics. Remarkably, these molecules are known to be involved in key processes of cancer cachexia. Based on these results, we propose a model how metastatic melanoma may lead to reprogramming of organ functions via formation of platelet activating factors from long-chain polyunsaturated phosphatidylcholines under oxidative conditions and via systemic induction of intracellular calcium mobilization. Calcium mobilization in platelets was demonstrated to alter levels of several of these marker molecules. Additionally, platelets from melanoma patients proved to be in a rather exhausted state, and platelet-derived eicosanoids implicated in tumor growth were found massively increased in blood from three melanoma patients. Platelets were thus identified as important source of serum protein and lipid alterations in late stage melanoma patients. As a result, the proposed model describes the crosstalk between lipolysis of fat tissue and muscle wasting mediated by oxidative stress, resulting in the metabolic deregulations characteristic for cachexia.
Serum is the most important diagnostic sample type because of its minimal invasive access, a relatively high stability and its comprehensive representation of the physiological state of an individual. The latest technological development of mass spectrometric instruments, such as high resolution orbitrap instruments, improved substantially the quality and reliability of serum proteomics data (1). Shotgun proteomics analysis using the orbitrap technology is mainly applied for protein screening purposes (2). This method allows performing untargeted analyses, applicable for hypothesis-generating clinical applications. Targeted proteomics using triple-quadrupole mass spectrometric instruments, represents a complementary approach which is applied for protein quantification, hypotheses verification and validation (3). Enormous efforts have been invested in the last decades focusing on serum biomarker discovery (4). However, mass spectrometry-based proteomics analyses hardly managed to cope with biological variation. As a consequence, almost no clinically validated biomarker has emerged from these investigations yet, and a lot of questions about pathophysiological mechanisms remain unanswered.
Using mass spectrometry-based proteomics, we have previously investigated melanoma and neighboring stroma cells, focusing on the identification of biomarkers associated with intrinsic and extrinsic drug resistance (5). In the present article, we followed the question how metastatic melanoma may reprogram distant organ functions, and how these changes may be depicted in serum signatures taken from patients treated in a phase II trial on metastatic melanoma. Such studies are directed on uncovering tumor promoting communication-mediated loops, drug resistance and novel approaches for targeting for example cachexia associated syndromes. The research questions emphasize the relevance of blood - as circulating fluid - as promising target for studying biomarkers characteristic for melanoma progression. Actually, in melanoma, PMEL is most widely used as melanoma marker in serum samples (6). Chondroitin sulfate proteoglycan 4 and S100A8 and S100A9 have been reported to be upregulated during melanoma progression (7).
In the present study an orbitrap shotgun screening experiment was complemented with a newly established targeted assay using multiple reaction monitoring. For data evaluation, free software tools such as MaxQuant (8), Skyline (9) and MSStats (10) were applied. Furthermore, a targeted metabolomics and a shotgun lipidomics approach were applied. All obtained results pointed to a systemic calcium deregulation as most apparent characteristic in patients with metastatic melanoma. A proposed novel pathomechanism on melanoma-associated reprogramming of distant organ functions, potentially related to cachexia, was tested and confirmed by the analysis of platelets isolated from healthy donors and patients with metastatic melanoma.
EXPERIMENTAL PROCEDURES
Preparation of Serum Samples
Using VACUETTE® serum tubes (Greiner Bio One, Germany), serum samples were routinely collected from six patients with metastatic melanoma treated within a phase I/II trial (ClinicalTrials.gov Identifier: NCT01614301) after giving written informed consent. All serum samples were stored at −80 °C immediately after collection. These serum samples were used for proteome profiling, targeted proteomics and targeted metabolomics analyses.
Serum Depletion
Serum samples were depleted employing Pierce™ Top 12 Abundant Protein Depletion Spin Columns (Thermo Fisher Scientific). The depletion was performed according to the protocol of the manufacturer, using 7 μl of serum per depletion. In the following, 250 μl of depleted serum eluate containing not more than 25 μg total protein amount was used for digestion.
In-solution Digestion
In-solution digestion was performed using 3 kDa MW cut-off filters (Nanosep with Omega membrane, Pall Austria Filter GmbH, Vienna, Austria) After washing with 50 mm ammonium bicarbonate (Sigma Aldrich), proteins were reduced with 32 mm dithiothreitol (Gerbu Biotechnik GmbH, Heidelberg, Germany) for 30 min at 35 °C on a thermal shaker (1000 rpm) and alkylated with 54 mm 2-iodoacetamide solution (Sigma Aldrich) at 30 °C for 45 min. Both, dithiothreitol and 2-iodoacetamide solutions were prepared with 8 m guanidinium hydrochloride (Sigma Aldrich) solution in 50 nm ammonium bicarbonate. Afterward, proteins were digested on the filter using a Trypsin/Lys-C Mix (MS grade; Promega Corporation, Madison, WI) with a total enzyme to protein ratio of 1:20. For clean-up of resulting peptide samples C18 spin columns (Pierce™ C18 Spin Columns, Thermo Fisher Scientific) were used and, after drying via vacuum centrifugation, the samples were stored at −20 °C. Upon LC-MS analysis the dried peptides were reconstituted in 5 μl of the equimolar 10 fmol standard peptide mix and 40 μl of mobile phase A (98% H2O, 2% acetonitrile (ACN)1, 0.1% formic acid (FA)) for shotgun analysis, and for targeted analysis in 30 μl of the 10 fmol/μl standard peptide mix solution containing 30% formic acid.
Shotgun LC-MS Analysis
Digested peptides were separated by UltiMate 3000 RSLC nano System (Pre-column: Acclaim PepMap 100, C18 100 μm × 2 cm; Analytical column: Acclaim PepMap RSLC C18 75 μm × 50 cm; Dionex, California). Per injection, 1 μl of the sample was loaded on the precolumn with a flow rate of 10 μl/min. For separation, peptides were eluted to the analytical column applying a gradient from 8 to 40% mobile phase B (80% ACN, 2% H2O, 0.1% FA) over 95 min and operating at a flow rate of 300 nL/min. Data acquisition was conducted on a QExactive mass spectrometer (Thermo Fischer Scientific) using a top 8 data dependent method described previously (11). The full scan MS was acquired at a resolution of 70,000, the MS2 scan at 17,500, both at m/z 200. HCD fragmentation was applied at 30% normalized collision energy.
Peptide and Protein Identification
Protein identification was achieved using the MaxQuant 1.5.2.8 software (8) employing the Andromeda search engine (12) and searching against the UniProt database for human proteins (version 102014 with 20,195 entries). For statistical analysis, data obtained from both biological as well as technical replicates were included and the Perseus statistical analysis package was used (13). Search criteria included a maximum of two missed cleavages with trypsin/Lys-C as proteases and a maximal mass deviation of 5 ppm for peptide ions and of 20 ppm for fragment ions. Additionally, a minimum of two peptide identifications per protein (including one unique) was requested and an FDR of less than 0.01 was applied at both peptide and protein level. Carbamidomethylation of cysteines was set as fixed modification and methionine oxidation as well as N-terminal protein acetylation as variable modifications. The match-between-runs feature was used to ensure as much identifications as possible. Over all samples, we were able to identify 485 different proteins (supplemental Table S2). The mass spectrometry-based proteomics data (including raw files, result files and peak list files, peptide sequences, precursor charges, mass to charge ratios, amino acid modifications, peptide identification scores, protein accession numbers, number of distinct peptides assigned for each identified protein, percent coverage of each identified protein in each individual experiment and annotated MS2 spectra for each peptide spectrum match) have been deposited to the ProteomeXchange Consortium (14) via the PRIDE partner repository and are freely available with accession numbers PXD004624–26.
Protein Quantification of Shotgun LC-MS Data
A MS1-based label-free quantification approach and statistical analysis was applied to quantify the identified proteins based on label-free quantification (LFQ) values using Perseus (supplemental Table S2) (13). Known contaminants were excluded from analysis. Proteins with a minimum fold change of two (p value ≤ 0.05) of LFQ values between high TL and low TL, high TL and control, or low TL and control samples were taken into consideration for MRM assay development. Additionally, nonsignificant but highly regulated (> 4-fold change) proteins and proteins with high biological relevance were also included, finally resulting in a panel of 96 candidate proteins.
LC-MRM Analysis
Targeted analysis was performed on an Agilent 6490 triple quadrupole mass spectrometer coupled with a nano-Chip-LC Agilent Infinity Series HPLC1290 system, as described recently (15). Solvent compositions were 97.8% H2O, 2% ACN and 0.2% FA for solvent A, and 97.8% ACN, 2% H2O and 0.2% FA for solvent B.
MRM Assay Development
The selection of targeted peptides was based on the same criteria as recently published (15). Briefly, only prototypic peptides with a length of 8 to 25 amino acids and without any methionine residue or missed cleavages were accepted. Additionally, peptide extracted ion chromatograms (XICs) on MS1 and MS2 level were manually checked. At the MS1 level, this involved selection of the right peak, considering coelution of all precursors. Identification triggers over all samples and technical replicates had to align within a certain time window. Peptides had to be identified only once over the whole chromatographic run (multiple identifications were only accepted for very broad peaks) and had to be identified in at least two of the three different sample types (low TL, high TL, control). Thereby, 940 transitions for 188 peptides (93 proteins) were used for unscheduled MRM measurements, applying different dwell times depending on the intensities achieved for peptide precursors in shotgun MS: 100ms for those with the lowest intensities (<106), 50ms for those with moderate intensities (106-107), and 20ms for those with highest signal intensities (>107). One biological sample, each of the low TL, high TL and control group, previously used for shotgun MS, was injected multiple times for measuring the 940 selected transitions in unscheduled MRM mode.
The peak picking for the scheduled MRM assay was done based on the same criteria as recently published (15), which were: selection of only co-eluting peaks with consistent peak shape for at least three out of the five investigated product ions and a dot-product (dotp) value of at least 0.8. Additionally, measured and calculated (based on the Skyline SSRcalc 3.0) retention times had to match with a correlation greater r = 0.9. Finally, three most intense and interference-free transitions were selected per peptide for the scheduled MRM method, resulting in 372 transitions for 126 peptides interfered from 88 proteins. The interference free properties of transitions corresponding to peptides derived from proteins, which were found significantly regulated in the targeted proteomics analysis (Fig. 2), are shown in the supplemental Fig. S1. The unchanged peak area percentages over 90 injections indicate the interference-free character of selected transitions. Although trem-like transcript 1 protein (TREML1) fulfil the threshold criteria for significant regulation, it was excluded from the panel of biomarker candidates because selected peptides show strong variation of the transition peak area percentage (supplemental Fig. S1). In order to maintain a minimum dwell time of 20 ms, an additional and final revision was performed, limiting the number of peptides per protein to two, removing peptides with insufficient signal intensities in all biological groups and removing proteins with low biological significance (based on literature and shotgun data). In the end, a final scheduled MRM method for 92 peptides (276 transitions) derived from 58 target proteins and using a 3 min time window was established (supplemental Table S3). Transitions of standard peptides spiked in each samples were included in the final MRM method. All clinical samples were measured as technical triplicates. Skyline software (9) was used to implement this MRM method, allowing data analysis and data preparation for statistical analysis.
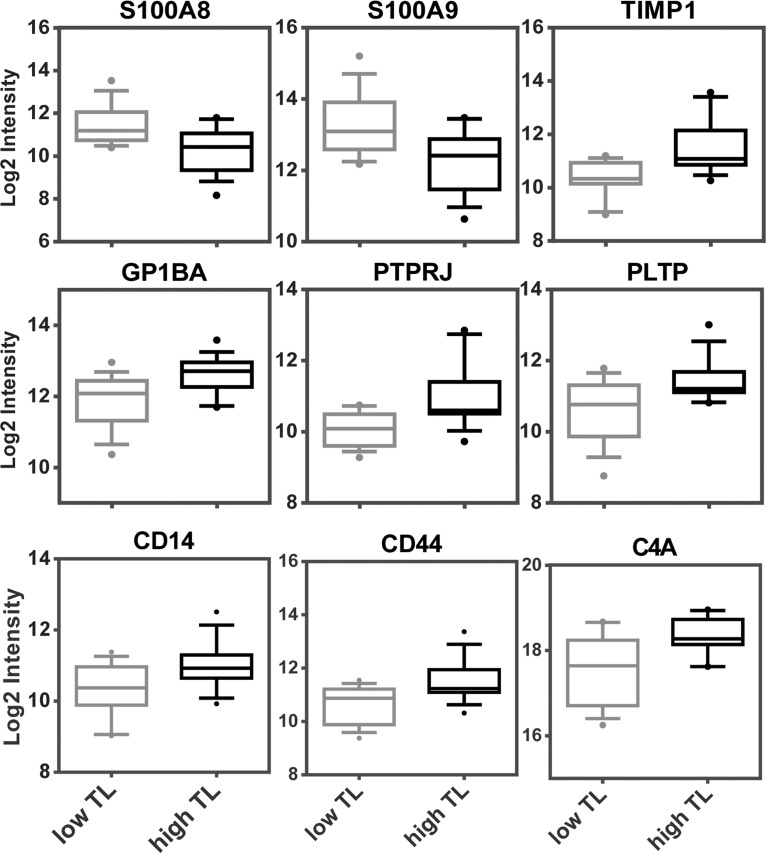
Fig. 2.
Results of targeted proteomics analyses. The MRM analysis revealed nine proteins which were significantly up- or downregulated in serum samples from patients with with high TL or low TL. For statistical analysis, the MSStats software was used and the significance threshold was set to a fold change higher than 1.5 with a p value lower than 0.05. Sample quantification data from MSStats are represented as log2 intensities.
Statistical Analysis of MRM Data
The statistical analysis of the data was performed with MSStats (v. 2.3.5) (10) employing the linear mixed effects model. The manual inspection regarding correct peak selection, interferences, and integration boundaries of the data was done with Skyline. All other steps were conducted in R using MSStats package. Data pre-processing and quality control steps consisted on log2 transformation and normalization to the four spiked standard peptides. The missing peaks were replaced with NA value. Furthermore, in order to remove the very low abundant transitions and to consider only the most informative features for further analysis, the Feature Selection was set to TRUE by applying the default setting. Both, for comparison of the abundances between sample groups and for abundance calculation of each protein sample, missed values were subjected to imputation using the average minimum intensity across all runs. Additionally, p values were adjusted according to Benjamini and Hochberg and a significant threshold of 0.05 was applied. The statistical analysis was performed with expanded scope of conclusions for biological replication.
Targeted Metabolomics Analysis
Targeted metabolomics was performed using the AbsoluteIDQ p180 kit (Biocrates Life Sciences AG, Innsbruck, Austria). The kit allows the identification and (semi-) quantitation of metabolites including acylcarnitines, amino acids, and biogenic amines, the sum of hexoses, sphingolipids, and glycerophospholipids by LC- and flow injection analysis (FIA)-MRM. The samples were analyzed on an AB SCIEX QTrap 4000 mass spectrometer using an Agilent 1200 RR HPLC system (Agilent, Santa Clara, CA), which were operated with Analyst 1.6.2 (AB SCIEX, Redwood City, CA). The chromatographic column was purchased from Biocrates. The serum samples and additional blanks, calibration standards and quality controls were prepared according to the user manual. All amino acids and biogenic amines were derivatized with phenylisothiocyanate. The experiments were validated with the supplied software (MetIDQ, Version 5–4-8-DB100-Boron-2607, Biocrates Life Sciences, Innsbruck, Austria). Correlation coefficients of the calibration curves of the amino acids and biogenic amines were between 0.9636 (ADMA) and 0.9938 (Met-SO). The quality controls (analyzed every 20th samples) revealed that leucine/isoleucine, acylcarnitine-D0, total DMA, t4-hydroxyproline, spermine, SDMA, histamine, Dopa and dopamine were not valid; these analytes were thus excluded from data evaluation. Finally, 178 metabolites were analyzed.
Platelets Isolation and Activation
Peripheral venous blood was drawn from two healthy volunteers and three metastatic melanoma patients with high tumor load, with their written consent and ethical approval (ClinicalTrials.gov Identifier: NCT01614301). As the melanoma patients recruited for the first part of the study were not available, other patients were chosen. Two blood collection tubes containing 3.2% sodium citrate (Vacuette system; Greiner, Kremsmuenster, Austria) were used per donor. To avoid premature activation of platelets, blood was collected into open tubes, letting the blood drop freely into the tubes after discarding the first few drops. The tubes were closed and gently inverted four times. Platelet rich plasma (PRP) was obtained by centrifugation of blood samples for 20min at 100g, yielding 1 ml of PRP per tube. To obtain a purified platelet suspension, PRP was further filtered through a Sepharose column. Firstly, 10 ml of Sepharose 2B (Sigma, Germany) were mixed with 10 ml RPMI medium (RPMI medium 1640 with glutamine, Gibco, Paisley, UK), containing no FCS, and filled in plastic columns plugged with cotton wool. Then, 1 ml of PRP was applied on top of each column. Elution was performed with RPMI medium, and, after discarding 2.5 ml, 1.5 ml platelet suspension was collected in a 15-ml tube. The two platelet fractions of a same donor were pooled and kept for 1 h at room temperature in a dark place, turning the tubes occasionally. Two fractions each of 1 ml platelet suspension were used for controls and for platelet activation. Activation was performed for 15 min by adding 100 μl of 10 μm ionomycin (Sigma, Germany) dissolved in PBS to a final concentration of 1 μm.
Protein Extraction of Whole Cell Lysates and Secretomes
Platelet samples were centrifuged at 1500 × g for 10min. Afterward, pellets were suspended in 100 μl sample buffer (7.5 m urea, 1.5 m thiourea, 4% CHAPS, 0.05% SDS, 100 mm dithiothreitol) and lysed by sonication with an ultrasonic homogenizer to obtain proteins from whole platelet lysates. For lipidomics analysis (see below), internal lipid standards (PGE2-d4 and PGF2a-d4, Cayman Europe) were added to the platelet supernatants, each at an end-concentration of 100 nm. Proteins were precipitated from these supernatants by adding 4 ml of ice cold absolute ethanol (Merck, Darmstadt, Germany) and incubated at −20 °C overnight. After centrifugation for 20min at 4750g and 4 °C, proteins were dissolved in 100 μl sample buffer. Resulting supernatant were used for lipidomics analysis.
Proteomics Analysis of Platelet Samples
The identical digestion protocol used for serum samples was applied also to platelet supernatants and lysates. The shotgun analysis was performed injecting 5 μl of digested sample, otherwise using the same instrumentation equipment and settings as used for the analysis of serum samples.
Lipid Extraction
Ethanol from platelet supernatants was reduced by vacuum centrifugation at 35 °C for 25 min to a volume of 1 ml. Samples were then diluted 1:3 with LC-MS grade water (Merck Millipore, Germany), vortexed and ultra-sonicated in a water bath at room temperature for 5 min. Lipids were extracted using solid phase extraction columns (StrataX 30 mg/ml, Phenomenex, Torrance, CA) as described recently (16). Columns were conditioned and equilibrated with HPLC gradient methanol (MeOH) (VWR International, Vienna, Austria) and water, respectively. After sample loading, the columns were washed with 10% MeOH solution, and eicosanoids were eluted into a glass vial with 1 ml of solution containing 49% ACN, 49% MeOH and 2% FA. Eluents were evaporated by vacuum centrifugation and samples were reconstituted in 200 μl mobile phase A (30% ACN, 70% H2O, 0.02% FA). The same extraction protocol was applied also to 200 μl of serum samples after overnight protein precipitation with ice cold ethanol. To 8 ml of whole blood sample, immediately after blood collection, 8 ml of ice cold methanol was added und incubated over night at −20 °C. On the next day, additionally 8 ml dichloromethane (VWR International, Austria) were added and vortexed. After centrifugation for 30 min at 1132 g, the methanol phase was isolated and the volume was reduced to 2.5 ml by evaporation in a vacuum centrifuge. Afterward, samples were diluted to 50 ml with LC-MS grade water, vortexed and ultra-sonicated. Again, lipid extraction using solid phase extraction was performed as described above.
Lipidomics LC-MS Analysis
Samples were pipetted into 200 μl glass inserts. All eluents were degassed prior to usage. Using Infinity 1290 UHPLC (Agilent Technologies Austria GmbH, Vienna, Austria), a 40 min gradient flow method was established using a Kinetex™ 2.1 mm × 15 cm, 2.6 μm, C18, 100 Å reversed phase column (Phenomenex); the flow rate was set to 250 μl/min and a gradient was applied (0–2 min: 5% mobile phase B (90% ACN, 10%, MeOH, 0.02% FA), 2–28 min: 5–85% mobile phase B, 28–35 min: 95% mobile phase B and finally re-equilibration with 5% mobile phase B). The injection needle was washed extensively after each injection with ACN: MeOH: 2-Propanol: H2O, 25% each). Columns oven was set to 45 °C and auto sampler was set to 4 °C. All samples (20 μl injected) were analyzed in technical duplicates. Mass spectrometric detection was performed with a Q-Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific) using the HESI-source to achieve negative ion mode ionization. MS scans were performed with an m/z range from 250 to 750 and a resolution of 35 000 (at m/z = 300). A lock mass was set; drift was ± 5ppm over all experiments. MS/MS scans of the six most abundant ions were achieved through HCD fragmentation at 30% normalized collision energy and analyzed in the orbitrap at a resolution of 17,500 (at m/z = 300). Raw files generated by the Q-Exactive Orbitrap were analyzed and scored manually using Thermo Xcalibur 2.2 Sp1.48 (Qual browser). Libraries from Lipid Maps depository were used and implemented as references.
Experimental Design and Statistical Rationale
For the screening study with shotgun proteomics, serum samples of six melanoma patients and three control samples collected from heathy donors were analyzed. The shotgun analysis was performed in technical duplicates for all samples. Based on obtained abundance values for selected marker proteins, melanoma patients were grouped into patient with high tumor load (high TL) and patients with low tumor load (low TL). In the next step, MRM analysis was applied to serum samples collected from these melanoma patients. Five serum samples collected within four months from each patient, thus in total 15 serum samples per patient group were analyzed. The criteria applied for the development of label-free MRM analyses are described in the above section of MRM method development. Additionally, the analytic parameters of MRM analysis were evaluated including sample preparation steps and LC-MS measurement (supplemental Fig. S3). All MRM measurements were performed in technical triplicates. In order to assess potential metabolomics alteration, targeted metabolomics analysis using a validated kit was applied to the three serum samples per sample group, high TL and low TL melanoma patients as well as control serum samples. The shotgun analysis of lipids was performed to assess the PAF abundance in serum samples of six melanoma patients included in the study and grouped in patients with high TL and low TL. The lipidomics shotgun analysis was performed in technical duplicates for all samples. Platelets isolated form two healthy donors were treated with ionomycin and compared with corresponding untreated platelets for confirmation of calcium dependent release of biomarker candidates. The exhausted state of platelets freshly isolated from three melanoma patients were assessed by their comparison with platelets isolated from three healthy donors. Shotgun proteomics was applied to lysates and supernatants of isolated platelets. Moreover, shotgun analysis of eicosanoids was performed with the supernatant of platelets. In order to overcome potential sampling issues regarding platelets activation potentially influencing the obtained results, whole blood of two healthy donors and three metastatic melanoma patients with high TL were analyzed as well. Results of shotgun analysis of eicosanoids supported the hypothesis of exhausted state of platelets and their contribution on tumor metastasis. The LC-MS analysis of eicosanoids was performed in technical duplicates for all samples measured in this study. Student t-tests were applied to assess the significance of obtained differences in abundance values.
RESULTS
Serum Proteome Profiling of Patients with Metastatic Melanoma
Based on an ongoing study performed at the University Hospital of Regensburg evaluating novel therapeutic options for patients with metastatic melanoma (ClinicalTrials.gov Identifier: NCT01614301) (17), we analyzed serum samples from six study participants. Assessment of these patients according to clinical evaluation is provided in supplemental Table S1. Comparative analysis with respect to healthy donors revealed statistically significant proteome alterations between the six patients and normal volunteers (Fig. 1A). Although standard serum proteins such as vitamin d-binding protein (GC), clusterin (CLU) and fibronectin (FN1) were detected at similar levels, typical inflammation marker molecules such as C-reactive protein (CRP), serum amyloid-A1 protein (SAA1) and SAA2, as well as the melanoma marker proteins melanocyte protein PMEL (PMEL) were significantly upregulated (Fig. 1A, supplemental Table S2). Several other proteins such as the liver-specific enzymes hydroxymethylglutaryl-CoA synthase (HMGCS2) and carbamoyl-phosphate synthase (CPS1) were found significantly downregulated (Fig. 1A).
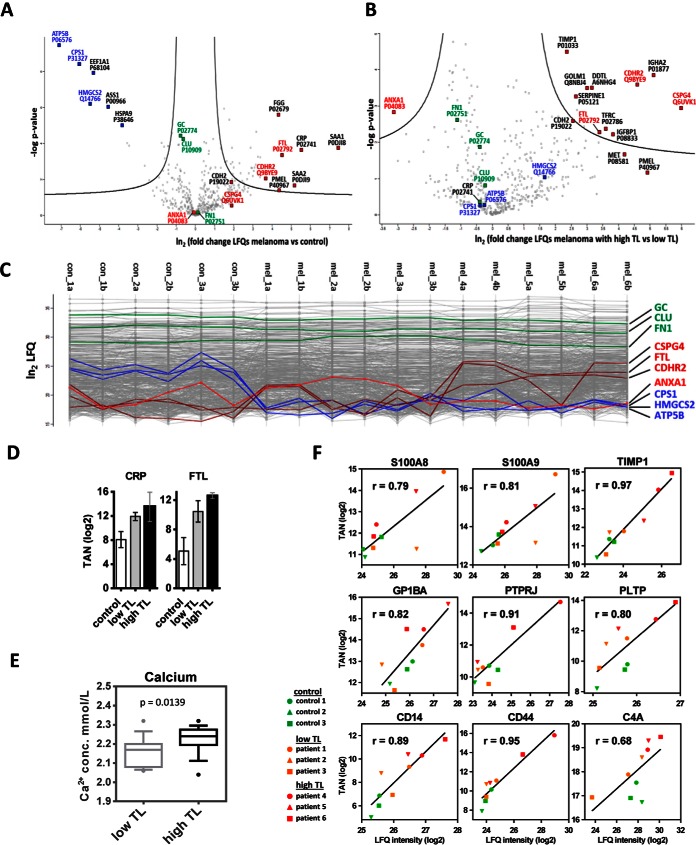
Fig. 1.
Regulation of proteins in serum from melanoma patients. Differences of LFQ values (logarithmic scale base two) of proteins determined in serum samples from melanoma patients versus controls (A), as well as from melanoma patients with high tumor load (high TL) or low tumor load (low TL) (B), with corresponding p values (logarithmic scale base 10), are represented as volcano plots. The area above the two black lines encompasses at least 2-fold significantly regulated proteins with a global FDR<0.05 as determined by a permutation-based method. C, Profile plots showing LFQ values (logarithmic scale base two) obtained from each individual measurement. Values for non-regulated proteins (green), proteins upregulated upon escalation of melanoma (dark red), proteins which were downregulated in melanoma versus control serum samples (blue), and one protein (ANXA1) which was upregulated in low TL, but downregulated in high TL (light red) are highlighted. Patients annotated as mel_4a to mel_6b showed increased levels of melanoma marker molecules D, Results from targeted analyses showing that CRP levels were similar in serum samples from patients with high TL or low TL, but elevated in patients with metastatic melanoma in comparison to controls. Progressive FTL increase associated with high TL melanoma was also determined with targeted analysis; TAN, log2 of normalized total peak area for a selected peptide. E, Calcium levels, as determined in clinics, were higher in serum samples from patients with high TL in comparison to low TL melanoma. Boxplots represent data collected in a four month period of time and consist of 15 samples per group. F, Correlation of quantification data obtained fom MRM and shotgun analyses. TAN, log2 of the normalized total peak area for proteins obtained by MRM; LFQ intensity, log2 of the label-free quantification value obtained by shotgun analysis; r, Pearson correlation coefficient.
In a second step, proteins were classified according to the most relevant source of origin in order to facilitate data interpretation. Thus, three important sources of the affected proteins were distinguished: liver, melanoma cells and platelets.
Assuming that high levels of the melanoma-cell derived proteins PMEL, chondroitin sulfate proteoglycan 4 (CSPG4) and cadherin-2 (CDH2) are indicative for increased tumor load (TL), we distinguished three patients with high levels of these serum proteins indicating high TL (Fig. 1B). Interestingly, two of the three patients suffered from cachexia (weight loss, muscle wasting and fatigue) at study inclusion. Independent clinical assessment of the patients mainly based on the presentation of metastases perfectly matched the discrimination based on the proteome analysis data.
Reprogramming of Distant Organ Functions May be Associated with Changes in Liver Function Plus Deregulation of Iron and Calcium Homoeostasis
In order to investigate proteome alterations potentially associated with the extent of TL according to the defined serum parameters, we compared serum analysis data from three patients with high and three patients with low TL according to the defined serum parameters (Fig. 1B and 1C). Several proteins were found at significantly different levels in the two patient groups. An increase in Golgi membrane protein 1 (GOLM1) and insulin-like growth factor-binding protein 1 (IGFBP1) is typically associated with altered liver function (18, 19). Increased levels of the tumor-promoting molecules transferrin receptor protein 1 (TFRC) and ferritin light chain (FTL) indicate aberrant iron metabolism. Deregulation of the proto-oncogene hepatocyte growth factor receptor (MET) as well as FTL are known to contribute to metastasis (20, 21). Targeted proteomics analyses confirmed upregulation of FTL in the high TL group (Fig. 1D). According to our data, the group with high TL was not necessarily associated with increased CRP indicative for tumor-associated inflammation (Fig. 1D). Downregulation of the anti-inflammatory molecule annexin A1 (ANXA1) as presently observed (Fig. 1B) has also been associated with advanced cancer stages (22). Intracellular calcium actually regulates ANXA1 secretion (23). Decreased levels of ANXA1 may thus indicate calcium release. Interestingly, free serum calcium levels were found significantly increased in melanoma patients with high versus low TL (Fig. 1E).
Targeted Analysis Characterizes Molecular Tissue Damage Signature
The shotgun data indicated that the abundance of several serum proteins not derived from tumor cells were altered in the patients with high TL, pointing to tumor-associated reprogramming of functions in other organs. Although shotgun analysis is best-suited for exploring unknown samples, the variance of quantitative determinations is not optimal. However, the high-resolution orbitrap MS-data can be used to set up an independent multiple reaction monitoring assay (15) best suited for independent data validation (3). Thus, a targeted proteomics method including 276 transitions for 58 selected proteins was developed and evaluated (supplemental Fig. S1, supplemental Table S3). Here, the median coefficient of variation (CV) was determined to be 6.1% (supplemental Fig. S2A) and the coefficient of determination was typically better than 0.99 (supplemental Fig. S2B). Sample preparation included depletion of the 12 most abundant serum proteins to enrich low-abundant proteins (Materials and Methods). The depletion efficiency was determined for nine proteins and varied between 42 and 99% (supplemental Fig. S2C). In average, the median CV of the entire assay including depletion, digestion and variations of LC-MS measurements was less than 20% (supplemental Fig. S2D). To avoid random effects caused by sampling or other individual variations, serum samples were taken from five different time points from each patient. This resulted in fifteen independent serum samples, digested separately and analyzed in technical duplicates, each from high TL and low TL melanoma patients. The targeted analysis revealed nine proteins significantly regulated between the two groups. (Fig. 2; supplemental Table S4 and S5).
In order to allow a comparison of quantification data obtained from MRM and shotgun analyses, normalized total peak areas for proteins obtained by MRM were plotted against respective LFQ quantification values obtained by shotgun analysis of the same sample. Pearson correlation coefficients were determined and proved good correlation between MRM and shotgun data (Fig. 1F).
Among the nine proteins, S100-A8 (S100A8) and S100-A9 (S100A9) proteins were found downregulated in high TL melanoma. These proteins form heterodimers binding two calcium ions and are well-known calcium sensors (24). Platelet glycoprotein Ib alpha chain (GP1BA) and receptor-type tyrosine-protein phosphatase eta (PTPRJ) are platelet membrane proteins known to be released upon platelets activation (25, 26). Increased serum levels of monocyte differentiation antigen CD14, intercellular adhesion molecule 1 (ICAM) and phospholipid transfer protein (PTLP) have been correlated with high endotoxin load (27–29). ICAM1 and CD136 were also found almost 2-fold upregulated, with p values just missing the significance threshold of 0.05 (supplemental Fig. S3; supplemental Table S4 and S5). CD44 antigen is also a transmembrane receptor protein and elevated levels of its soluble form in circulating fluids are known to promote tumor metastasis (30). Complement C4-A (C4A), secreted by macrophages, is part of the innate immune system. Intriguingly, the regulation of six of these proteins (S100A8, S100A9, GP1BA, PTPRJ, CD44, and C4A) is linked to intracellular calcium mobilization. The occurrence of cell stress-associated membrane protein shedding is evidenced by the increased serum levels of CD14, CD44, ICAM1, MET, CDH2, TFRC, GP1BA, and PTPRJ. Together with increased metalloproteinase inhibitor 1 (TIMP1) and FTL as well as decreased ANXA1 these proteome alterations may be considered as molecular tissue damage signature determined in blood serum samples.
Metabolomics Analyses of Serum Samples Revealed Downregulation of Selected Amino Acids and PCs
In addition to the proteomics analyses, we also applied a targeted metabolomics approach to the serum samples. After protein precipitation, the soluble fraction containing the metabolites was derivatized with phenylisothiocyanate. The p180 kit returned 178 validated metabolites for data evaluation. The increased concentration of several amino acids such as glycine and asparagine in high TL melanoma, most plausibly caused by increased proteolytic activity, is in accordance with descriptions of processes occurring in tumor escalation (31). All 39 acylcarnitines were found upregulated likewise, pointing to extensive lipolysis, which is typical for the wasting syndrome cachexia (31). Additionally, the targeted metabolomics analysis revealed significant decreased concentrations of several long-chain and highly unsaturated phosphatidylcholines (PC) (Fig. 3), which has been related to dysfunction of lipid synthesis in liver, as well as to altered properties of cell membranes (32, 33). Under oxidative conditions, typically developing upon tumor progression, PCs may be converted into platelet activating factor (PAF) and other platelet activating factor receptor (PAF-R) agonists (34, 35). Actually, increased abundance of PAF in samples from patients with high TL were demonstrated by a mass spectrometry-based serum analysis (Fig. 3).
Fig. 3.
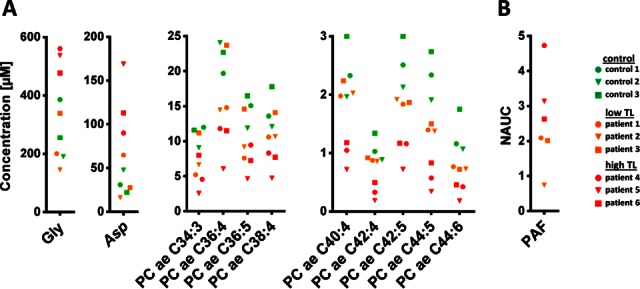
Metabolomics analysis. A, The concentrations of glycine (Gly), aspartic acid (Asp) and selected phospohatidylcholine acyl alkyl (PC ae) derivatives determined with targeted analysis in samples from healthy donors as well as from melanoma patients are represented. B, The abundance of platelet activating factor (PAF) obtained by shotgun analysis was found increased in high TL melanoma patients; NAUC; normalized area under the curve.
Systemic Intracellular Calcium Mobilization May be Responsible for Disease Escalation
Activation of PAF-R by binding of PAF or other PAF-R agonists induces intracellular calcium mobilization (36). Considering the above-described alterations of several calcium-regulated proteins, the implication of calcium mobilization in the group of melanoma patients with damage signature becomes evident. Thus, we here propose a model implicating intracellular calcium mobilization in manifest or probably impending melanoma-associated cachexia. (Fig. 4). The fact that PAF-R is expressed in many tissues and cells (37, 38), together with the increased abundance of PAF in the blood of patients with melanoma damage signature, suggests that PAF-R activation may occur in a systemic fashion. In order to test this model, we investigated whether calcium mobilization was capable of inducing proteome alterations compatible with the presently described serum proteome alterations. For this purpose, platelets appeared to be a good choice as they can be easily stimulated in vitro by inducing calcium mobilization (39).
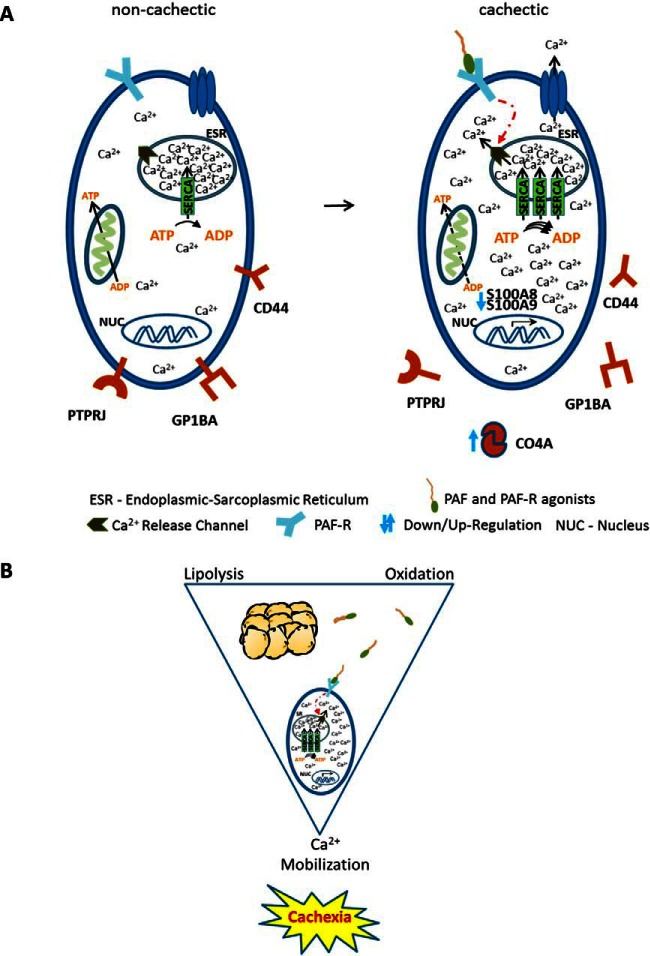
Fig. 4.
Proposed mechanism for cachexia development. A, From long-chain highly unsaturated phosphatidylcholines acyl alkyl metabolites (PC ae) sourcing from adipose tissue platelet activating factor (PAF) and platelet activating factor-receptor (PAF-R) agonists are produced under oxidative conditions. These molecules induce calcium release from ESR in patients with high TL upon binding to PAF-R (two of three with cachexia). To transfer calcium back into the ESR and maintain cytosolic calcium concentrations tolerable, cells may upregulate SERCA pumps, which is an energy consuming process. At increased calcium concentration, S100 proteins may form insoluble complexes which can explain the reduced levels of these proteins. C4A levels are increased as protection mechanism of the cells as response to the elevated levels of calcium concentration. Additionally, membrane protein-receptors are released and calcium dependent signaling pathways may be activated as a consequence of the sustained calcium influx in the cytosol. Disruption of ESR-mitochondria bidirectional communication may also be induced by incorporation of PC ae into membranes. B, The combination of the three processes lipolysis, oxidation and intracellular calcium mobilization may be a critical combination resulting in melanoma damage signature and probably in cachexia establishment. Phosphatidylcholines sourcing from lipolysis of adipose tissues can be converted into PAF and PAF-R agonists upon oxidation which induce intracellular calcium mobilization by activating the PAF-R. Subsequently, systemic intracellular calcium mobilization may eventually result in cachexia.
Proteome Alterations Induced by Calcium Mobilization in Platelets Correlate with Serum Proteome Alterations
Platelets were isolated from two healthy donors and treated in vitro with the calcium ionophore ionomycin. Shotgun proteomics analysis of the resulting platelet supernatants showed that S100 proteins were significantly downregulated, whereas the platelet receptor proteins GP1BA and PTPRJ were upregulated (Fig. 5A). This was consistent with the MRM data obtained with serum samples (Fig. 2). Additionally, the analysis of whole platelet lysates revealed that sarcoplasmic/endoplasmic reticulum calcium ATPases (SERCA) pumps sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (ATP2A2), sarcoplasmic/endoplasmic reticulum calcium ATPase 3 (ATP2A3) and calcium-transporting ATPase type 2C member 1 (ATP2C1) were significantly upregulated in platelets upon ionomycin treatment (Fig. 5A). These pumps translocate calcium from the cytosol back to the sarcoplasmic reticulum lumen, a process which is coupled to the hydrolysis of ATP (40). Increased activity of SERCA pumps potentially required to control otherwise critical levels of intracellular calcium is clearly associated with high energy consumption (40, 41). As one of the typical symptoms of cachexia is actually energy wasting, such biochemical activity may be related to this clinical feature (31, 42).
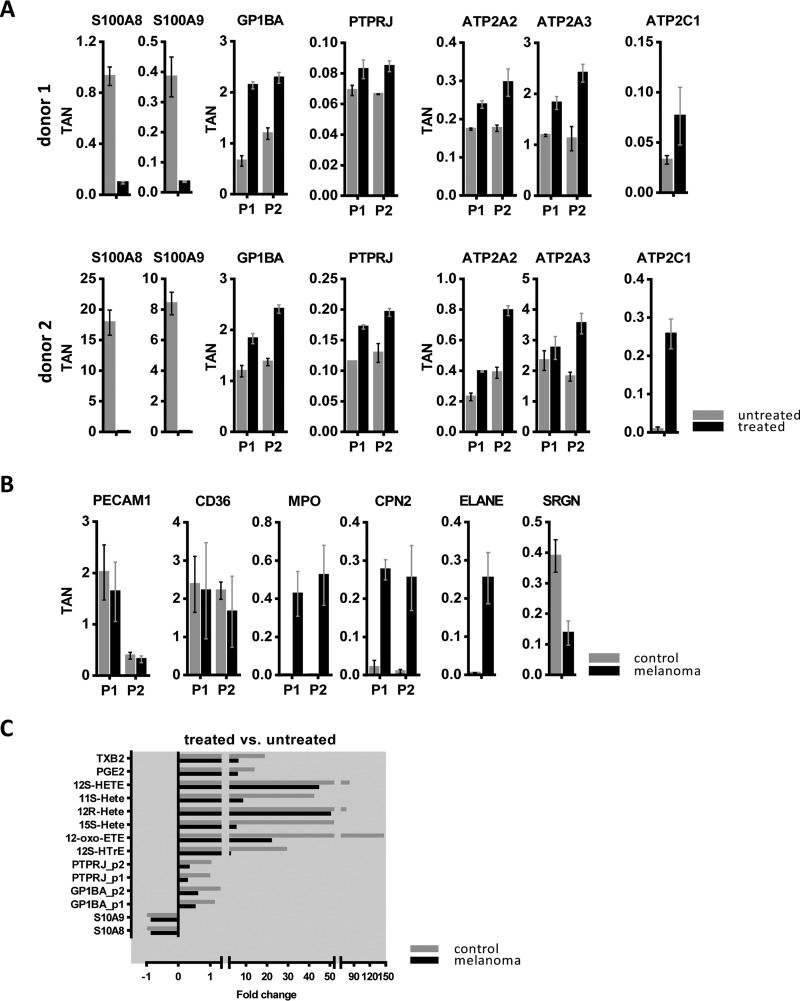
Fig. 5.
Shotgun analysis of platelets at protein and eicosanoid levels. A, Results for selected proteins included in the mechanistic model of cachexia development, obtained from analysis of untreated and ionomycin-treated platelets isolated from two healthy donors, are shown. Each protein is represented with one or two peptides; TAN, normalized total peak area for a selected peptide. B, Results for selected proteins obtained from the analysis of whole lysates of platelets obtained from healthy donors and melanoma patients. P1 and P2 refer to independent peptides of the same protein. C, The fold change in abundance of proteins and eicosanoids secreted from treated versus untreated platelets are shown. Platelets were isolated from three healthy donors as well as from three melanoma patients. The isolated platelets were treated with ionomycin for 15 min.
Platelets Derived from Melanoma Patients Display an Exhausted State
The present data indicated that platelet activation may be involved in the pathogenesis of melanoma. Thus, we investigated whether platelets from a newly chosen set of metastatic melanoma patients with high TL (n = 3) were distinguishable from those of healthy donors. Shotgun proteomics analyses revealed that two healthy donors showed similar regulation patterns of platelet proteins (Fig. 5A), whereas several proteins were regulated differentially in platelets from melanoma patients. Fig. 5B shows proteins upregulated in platelets derived from patients with metastatic melanoma relative to healthy controls. Interestingly, myeloperoxidase (MPO) was described to induce priming of platelets, however without leading to platelet aggregation (43). Circulating neutrophil elastase (ELANE) has been described to be released from platelets upon hypercoagulable conditions (44). Carboxypeptidase N subunit 2 (CPN2) is able to act on stromal cell-derived factor 1, reducing its activity as a pre-B-cell growth factor and chemoattractant (45). Serglycin (SRGN) was on the contrary downregulated in platelets from melanoma patients relative to controls (Fig. 5B). This protein is involved in regulating the half-life of cytokines and apoptosis induction (46); the downregulation of this protein may thus also contribute to pathophysiological processes resulting in tumor escalation. Other proteins such as platelet endothelial cell adhesion molecule (PECAM1) and platelet glycoprotein 4 (CD36) were detected with similar levels in all platelets, demonstrating that differential regulation was only affecting a few selected proteins.
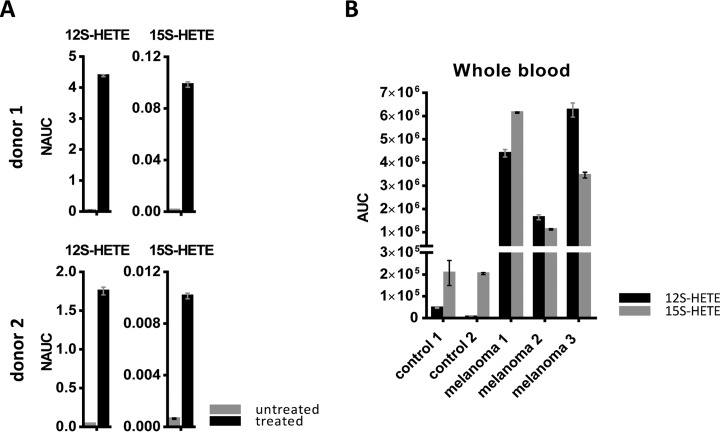
Platelets are also important producers of eicosanoids, which play an essential role in physiological as well as pathophysiological processes including tumor progression (47). Therefore we also analyzed 12S-hydroxyeicosatetraenoic acid (12S-HETE) and 15S-HETE formation by platelets from healthy donors upon ionomycin treatment (Fig. 6A).
Fig. 6.
Eicosanoid analysis in platelets and whole blood samples. A, Results for 12S-HETE and 15S-HETE, obtained from analysis of untreated and ionomycin-treated platelets isolated from two healthy donors, are shown NAUC; normalized area under the curve. B, The shotgun analysis of eicosanoids was applied to 8 ml blood collected from melanoma patients and healthy donors. The results obtained for 12S-HETE and 15S-HETE are represented; AUC, area under the curve of MS1 peak.
Finally, we investigated whether ionomycin might have different effects when applied to platelets of healthy donors or those of patients with metastatic melanoma. Indeed, activation-induced protein secretion was much higher in platelets isolated from healthy donors than in those isolated from patients. This trend was observed for all selected protein biomarker candidates as well as the two HETEs (Fig. 5C). Assuming that platelets may lose secretion capacity in the in vitro assay when some activation had already occurred in vivo before the blood donation, the present observations suggest that platelets from melanoma patients were already somewhat exhausted. Such exhausted state of platelets was already suggested by Mannucci et al. in case of malignant melanoma (48).
Increased Blood Levels of 12S-HETEand 15-HETE in Melanoma Patients May be Caused by Systemic Stimulation of Platelets Because of Calcium Mobilization
A melanoma-associated exhausted state of platelets would suggest that molecules released upon platelet stimulation should be present in increased concentrations in the blood of patients. This is of great relevance, as both proteins and lipids released from activated platelets may have strong tumor-promoting capabilities (47, 49). Indeed, 12S-HETE and 15S-HETE were found strongly increased in whole blood of melanoma patients when compared with whole blood of healthy donors (Fig. 6B). Our findings indicate that increased blood levels of 12S-HETE and 15S-HETE in patients with metastatic melanoma may be caused by platelet activation because of systemic calcium mobilization. Both, 15S-HETE and 12S-HETE are known to increase protein kinase C (PKC) activity which plays an important role for tumor promotion; 12S-HETE has been demonstrated to regulate PKC-alpha in melanoma cells (50). As a consequence, we are going to further investigate HETEs as potential biomarkers for patients with metastatic melanoma to monitor the on-set of the damage signature, presumably related to cachexia and systemic tumor-promoting features.
DISCUSSION
Cancer treatment mainly relies on the removal of cancer cells by surgery and/or pharmacological interventions designed to eradicate cancer cells in the advanced stage of tumor disease. Amazing results have been obtained with modern targeted therapies in metastatic melanoma, demonstrating complete remission with combined B-raf and MEK inhibition, anti-PD1 and/or CTLA4 antibodies. Complete remissions may be now translated into long-term improved overall survival (51). By putting the tumor disease in a holistic communicative context, thereby considering tumor mediated functional changes at distant organ sites (52), we may better meet the diagnostic and therapeutic requirements of late stage tumor diseases with their typical syndromes, B-symptoms (weight loss and night sweats), or cachexia and fatigue (53).
The current study was designed to investigate melanoma-induced functional changes at distant organ sites in patients with metastatic melanoma (17). Proteome profiling from blood serum samples was the method of choice for the screening of any kind of melanoma-associated reprogramming of organ functions at distant sites.
As expected, known clinical markers for melanoma such as PMEL, CSPG4 and CDH2 were readily identified in patients with metastatic melanoma by mass spectrometry, as well as inflammation markers such as CRP, SAA1 and SAA2 (Fig. 1). Statistical analysis of the shotgun data allowed us to evaluate 485 different proteins (supplemental Table S2). Patients with high TL had a functional impairment of the liver as well as deregulation of iron and calcium homeostasis (Fig. 1B). In order to improve the accuracy of serum protein determinations by mass spectrometry, we developed a targeted method based on multiple reaction monitoring. The present strategy of combining high-resolution mass spectrometry-based screening with targeted quantitative analysis of selected candidate proteins proved to be very successful, and was based, among others, on the good correlation between quantification data obtained from shotgun and MRM experiments (Fig. 1F). Evaluation of the MRM data set, including five independent time points for each melanoma patient, strongly indicated that systemic calcium mobilization might be a common reason for most of the observed proteome alterations in the subgroup of melanoma patients with high TL. The identification of a large number of membrane proteins in serum, most probably derived from ectodomain shedding of those proteins (54), indicated that these molecules may represent a molecular signature for tissue damage. Ongoing clinical studies performed by us will demonstrate the potential use of such a signature for improving diagnosis, melanoma systemic therapy and therapy monitoring.
Actually, the biomedical implications of the observed proteome alterations are simply stunning: Calcium mobilization turned out to be the common triggering event for the release of most marker molecules found in the presently described tissue damage signature. TIMP, CD14, CD44, and ICAM1 (Fig. 2, supplemental Fig. S3) have already been identified by us in the secretome of mesenchymal cells in the context of tumor-associated inflammation (55). CD44 shedding has directly been linked to calcium mobilization and contributes to metastasis (56). The interrelation between inflammation, calcium, reactive oxygen species and specific stress signatures like endoplasmic reticulum stress is well established (57). GP1BA (Fig. 2) is also known to be released from platelets upon calcium mobilization and associated with reactive oxygen species (58). The present metabolomics study demonstrated significant downregulation of several long-chain and highly unsaturated phosphatidylcholines (PCs) which are precursor molecules for the formation of PAF in inflammatory cells (59). The contribution of PAF and PAF receptors for melanoma progression and chemotherapy resistance has already been suggested (60).
All these results prompted us to investigate the status of platelets in melanoma patients as potential surrogate markers for systemic calcium mobilization (Fig. 5). Indeed, platelets derived from melanoma patients showed dramatic differences when compared with platelets of healthy individuals (Fig. 5). Platelets have been implicated with metastasis for decades (49). However, to the best of our knowledge this is the first study analyzing and comparing platelets isolated from melanoma patients and platelets from healthy donors as well as the corresponding serum samples. The present data suggest that indeed major proteome and lipidome alterations evidenced in the blood of melanoma patients are related to tumor-associated altered platelet functions (Figs. 5, 6). The identified molecules may not only serve as disease markers, but are evidently eminent contributors to disease progression. To give examples, GP1BA and 12S-HETE are known as powerful tumor promoters (47, 61). Most importantly, the altered platelet functions may be considered as surrogate markers for manifest systemic reprogramming of distant organ sites in metastatic melanoma, mainly resulting from calcium mobilization.
Existing literature strongly supports the notion that the described damage signature directly results in melanoma-associated cachexia, still a dreadful and essentially incurable disease state. It remains to be studied whether other tumor types may express similar signatures in the metastatic tumor stage.
Lipolysis associated with oxidative stress is capable of producing PAF which may affect all PAF-R positive cells, causing calcium mobilization. PAF-R are mainly expressed by mesenchymal cells, which may explain the implication of adipose, muscle, liver, and heart tissue, as well as gut barrier and brain dysfunctions in cachexia, supporting our proposed model (31, 32, 62–64). Also, the described crosstalk between wasting of fat tissue and skeletal muscle is compatible with the proposed mechanism (65–67).
Several important conclusions may be drawn from the present study. First, here we present analytical methods and tools to determine marker molecules building a potential tissue damage signature directly in blood serum. Second, the interpretation of the present data suggests a comprehensive model for a tumor-associated damage signature related to melanoma-mediated cachexia. Third, understanding the origin and effects of powerful tumor promoters provides us with means to establish novel therapeutic options for both cancer in general as well as cancer-associated cachexia (52, 66). Evidently, many anti-cancer strategies already exist. Understanding how metastatic tumors may reprogram distant organ sites, as demonstrated for metastatic melanoma via calcium mobilization with lipid processing and oxidative stress conditions (Fig. 4), gives the opportunity for developing more comprehensive diagnostic instruments and therapy strategies (52) aimed at improving quality of life and overall survival. To sum up, the present application of a combined proteomics, metabolomics and lipidomics approach clearly connected widespread fragment knowledge of melanoma biology (53), helping us to build a simple, clear and comprehensive model for systemic tumor-promoting activities. Currently, we are establishing clinical studies with appropriate statistical power for the evaluation of the here presented model. Both, improved diagnosis and improved therapeutic options will hopefully be supported by the present new insights.
Supplementary Material
Acknowledgments
We thank Lisa M. Boehm for practical support.
Footnotes
Author contributions: B.M. and C.G. designed research; B.M., M.E., S.M.M., and A.T. performed research; T.P., S.H., and A.R. contributed new reagents or analytic tools; B.M., A.S., and C.G. analyzed data; B.M., A.R., A.S., and C.G. wrote the paper.
* This study was supported by the Faculty of Chemistry of the University of Vienna.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- ACN
- acetonitrile
- ANXA1
- annexin A1
- C4A
- Complement C4-A
- CD14
- monocyte differentiation antigen CD14
- CD44
- CD44 antigen
- CDH2
- cadherin-2
- CLU
- clusterin
- CRP
- C-reactive protein
- CSPG4
- chondroitin sulfate proteoglycan 4
- CPS1
- carbamoyl-phosphate synthase
- CV
- coefficient of variation
- FA
- formic acid
- FDR
- false discovery rate
- LFQ
- label-free quantification
- FTL
- ferritin light chain
- GP1BA
- Platelet glycoprotein Ib alpha chain
- ICAM1
- intercellular adhesion molecule 1
- MET
- hepatocyte growth factor receptor
- MRM
- multiple reaction monitoring
- PAF
- platelet activating factor
- PAF-R
- platelet activating factor receptor
- PC
- phosphatidylcholine
- TFRC
- transferrin receptor protein 1
- PGE2
- prostaglandin E2
- PGF2a
- prostaglandin F2 alpha
- PKC
- protein kinase C
- PMEL
- proteins melanocyte protein
- PRP
- platelet rich plasma
- PTLP
- phospholipid transfer protein
- PTPRJ
- receptor-type tyrosine-protein phosphatase eta
- S100A8
- protein S100-A8
- S100A9
- protein S100-A9
- SAA1
- serum amyloid-A1 protein
- SERCA
- sarcoplasmic/endoplasmic reticulum calcium ATPase
- TIMP1
- metalloproteinase inhibitor 1
- TL
- tumor load
- 12S-HETE
- 12S-hydroxyeicosatetraenoic acid
- 15S-HETE
- 15S-hydroxyeicosatetraenoic acid.
REFERENCES
- 1. Adkins J. N., Varnum S. M., Auberry K. J., Moore R. J., Angell N. H., Smith R. D., Springer D. L., and Pounds J. G. (2002) Toward a human blood serum proteome: analysis by multidimensional separation coupled with mass spectrometry. Mol. Cell. Proteomics 1, 947–955 [DOI] [PubMed] [Google Scholar]
- 2. Michalski A., Damoc E., Hauschild J. P., Lange O., Wieghaus A., Makarov A., Nagaraj N., Cox J., Mann M., and Horning S. (2011) Mass Spectrometry-based Proteomics Using Q Exactive, a High-performance Benchtop Quadrupole Orbitrap Mass Spectrometer. Mol. Cell. Proteomics 10, M111.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aebersold R., Burlingame A. L., and Bradshaw R. A. (2013) Western blots versus selected reaction monitoring assays: time to turn the tables? Mol. Cell. Proteomics 12, 2381–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Abbatiello S. E., Schilling B., Mani D. R., Zimmerman L. J., Hall S. C., MacLean B., Albertolle M., Allen S., Burgess M., Cusack M. P., Gosh M., Hedrick V., Held J. M., Inerowicz H. D., Jackson A., Keshishian H., Kinsinger C. R., Lyssand J., Makowski L., Mesri M., Rodriguez H., Rudnick P., Sadowski P., Sedransk N., Shaddox K., Skates S. J., Kuhn E., Smith D., Whiteaker J. R., Whitwell C., Zhang S., Borchers C. H., Fisher S. J., Gibson B. W., Liebler D. C., MacCoss M. J., Neubert T. A., Paulovich A. G., Regnier F. E., Tempst P., and Carr S. A. (2015) Large-Scale Interlaboratory Study to Develop, Analytically Validate and Apply Highly Multiplexed, Quantitative Peptide Assays to Measure Cancer-Relevant Proteins in Plasma. Mol. Cell. Proteomics 14, 2357–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Paulitschke V., Berger W., Paulitschke P., Hofstatter E., Knapp B., Dingelmaier-Hovorka R., Fodinger D., Jager W., Szekeres T., Meshcheryakova A., Bileck A., Pirker C., Pehamberger H., Gerner C., and Kunstfeld R. (2015) Vemurafenib resistance signature by proteome analysis offers new strategies and rational therapeutic concepts. Mol. Cancer Therap. 14, 757–768 [DOI] [PubMed] [Google Scholar]
- 6. Weinstein D., Leininger J., Hamby C., and Safai B. (2014) Diagnostic and prognostic biomarkers in melanoma. J. Clin. Aesthet. Dermatol. 7, 13–24 [PMC free article] [PubMed] [Google Scholar]
- 7. Ruma I. M., Putranto E. W., Kondo E., Murata H., Watanabe M., Huang P., Kinoshita R., Futami J., Inoue Y., Yamauchi A., Sumardika I. W., Youyi C., Yamamoto K. I., Nasu Y., Nishibori M., Hibino T., and Sakaguchi M. (2016) MCAM, as a novel receptor for S100A8/A9, mediates progression of malignant melanoma through prominent activation of NF-kappaB and ROS formation upon ligand binding. Clin. Exp. Metastasis 33, 609–627 [DOI] [PubMed] [Google Scholar]
- 8. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 9. MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., and MacCoss M. J. (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Choi M., Chang C. Y., Clough T., Broudy D., Killeen T., MacLean B., and Vitek O. (2014) MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics 30, 2524–2526 [DOI] [PubMed] [Google Scholar]
- 11. Slany A., Bileck A., Kreutz D., Mayer R. L., Muqaku B., and Gerner C. (2016) Contribution of human fibroblasts and endothelial cells to the hallmarks of inflammation as determined by proteome profiling. Mol. Cell. Proteomics 15, 1982–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 13. Cox J., and Mann M. (2012) 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics 13, S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolome S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Muqaku B., Slany A., Bileck A., Kreutz D., and Gerner C. (2015) Quantification of cytokines secreted by primary human cells using multiple reaction monitoring: evaluation of analytical parameters. Anal. Bioanal. Chem. 407, 6525–6536 [DOI] [PubMed] [Google Scholar]
- 16. Muqaku B., Tahir A., Klepeisz P., Bileck A., Kreutz D., Mayer R. L., Meier S. M., Gerner M., Schmetterer K., and Gerner C. (2016) Coffee consumption modulates inflammatory processes in an individual fashion. Mol. Nutrition Food Res. 60, 2529–2541 [DOI] [PubMed] [Google Scholar]
- 17. Hart C., Vogelhuber M., Hafner C., Landthaler M., Berneburg M., Haferkamp S., Herr W., and Reichle A. (2015) Biomodulatory metronomic therapy in stage IV melanoma is well-tolerated and may induce prolonged progression-free survival, a phase I trial. J Eur. Acad. Dermatol. Venereol. 30, e119–e121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tsuchiya N., Sawada Y., Endo I., Saito K., Uemura Y., and Nakatsura T. (2015) Biomarkers for the early diagnosis of hepatocellular carcinoma. World J. Gastroenterol. 21, 10573–10583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li H. H., Doiron K., Patterson A. D., Gonzalez F. J., and Fornace A. J. Jr. (2013) Identification of serum insulin-like growth factor binding protein 1 as diagnostic biomarker for early-stage alcohol-induced liver disease. J. Translational Med. 11, 266–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peschard P., and Park M. (2007) From Tpr-Met to Met, tumorigenesis and tubes. Oncogene 26, 1276–1285 [DOI] [PubMed] [Google Scholar]
- 21. Baldi A., Lombardi D., Russo P., Palescandolo E., De Luca A., Santini D., Baldi F., Rossiello L., Dell'Anna M. L., Mastrofrancesco A., Maresca V., Flori E., Natali P. G., Picardo M., and Paggi M. G. (2005) Ferritin contributes to melanoma progression by modulating cell growth and sensitivity to oxidative stress. Clin. Cancer Res. 11, 3175–3183 [DOI] [PubMed] [Google Scholar]
- 22. Wang L. P., Bi J., Yao C., Xu X. D., Li X. X., Wang S. M., Li Z. L., Zhang D. Y., Wang M., and Chang G. Q. (2010) Annexin A1 expression and its prognostic significance in human breast cancer. Neoplasma 57, 253–259 [DOI] [PubMed] [Google Scholar]
- 23. Sato-Matsumura K. C., Matsumura T., Nakamura H., Sawa H., Nagashima K., and Koizumi H. (2000) Membrane expression of annexin I is enhanced by calcium and TPA in cultured human keratinocytes. Arch. Dermatol. Res. 292, 496–499 [DOI] [PubMed] [Google Scholar]
- 24. Bresnick A. R., Weber D. J., and Zimmer D. B. (2015) S100 proteins in cancer. Nat. Rev. Cancer 15, 96–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Senis Y. A., Tomlinson M. G., Ellison S., Mazharian A., Lim J., Zhao Y., Kornerup K. N., Auger J. M., Thomas S. G., Dhanjal T., Kalia N., Zhu J. W., Weiss A., and Watson S. P. (2009) The tyrosine phosphatase CD148 is an essential positive regulator of platelet activation and thrombosis. Blood 113, 4942–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Savage B., Saldivar E., and Ruggeri Z. M. (1996) Initiation of platelet adhesion by arrest onto fibrinogen or translocation on von Willebrand factor. Cell 84, 289–297 [DOI] [PubMed] [Google Scholar]
- 27. Anas A., van der Poll T., and de Vos A. F. (2010) Role of CD14 in lung inflammation and infection. Crit. Care 14, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zonneveld R., Martinelli R., Shapiro N. I., Kuijpers T. W., Plotz F. B., and Carman C. V. (2014) Soluble adhesion molecules as markers for sepsis and the potential pathophysiological discrepancy in neonates, children and adults. Crit. Care 18, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Albers J. J., Wolfbauer G., Cheung M. C., Day J. R., Ching A. F., Lok S., and Tu A. Y. (1995) Functional expression of human and mouse plasma phospholipid transfer protein: effect of recombinant and plasma PLTP on HDL subspecies. Biochim. Biophys. Acta 1258, 27–34 [DOI] [PubMed] [Google Scholar]
- 30. Misra S., Hascall V. C., Markwald R. R., and Ghatak S. (2015) Interactions between Hyaluronan and Its Receptors (CD44, RHAMM) Regulate the Activities of Inflammation and Cancer. Front. Immunol. 6, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Argiles J. M., Busquets S., Stemmler B., and Lopez-Soriano F. J. (2014) Cancer cachexia: understanding the molecular basis. Nat. Rev. Cancer 14, 754–762 [DOI] [PubMed] [Google Scholar]
- 32. Masri S., Papagiannakopoulos T., Kinouchi K., Liu Y., Cervantes M., Baldi P., Jacks T., and Sassone-Corsi P. (2016) Lung adenocarcinoma distally rewires hepatic circadian homeostasis. Cell 165, 896–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Antunes D., Padrao A. I., Maciel E., Santinha D., Oliveira P., Vitorino R., Moreira-Goncalves D., Colaco B., Pires M. J., Nunes C., Santos L. L., Amado F., Duarte J. A., Domingues M. R., and Ferreira R. (2014) Molecular insights into mitochondrial dysfunction in cancer-related muscle wasting. Biochim. Biophys. Acta 1841, 896–905 [DOI] [PubMed] [Google Scholar]
- 34. Sahu R. P., Ocana J. A., Harrison K. A., Ferracini M., Touloukian C. E., Al-Hassani M., Sun L., Loesch M., Murphy R. C., Althouse S. K., Perkins S. M., Speicher P. J., Tyler D. S., Konger R. L., and Travers J. B. (2014) Chemotherapeutic agents subvert tumor immunity by generating agonists of platelet-activating factor. Cancer Res. 74, 7069–7078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marathe G. K., Johnson C., Billings S. D., Southall M. D., Pei Y., Spandau D., Murphy R. C., Zimmerman G. A., McIntyre T. M., and Travers J. B. (2005) Ultraviolet B radiation generates platelet-activating factor-like phospholipids underlying cutaneous damage. J. Biol. Chem. 280, 35448–35457 [DOI] [PubMed] [Google Scholar]
- 36. Chao W., Liu H., Hanahan D. J., and Olson M. S. (1992) Platelet-Activating Factor-Stimulated Protein Tyrosine Phosphorylation and Eicosanoid Synthesis in Rat Kupffer Cells - Evidence for Calcium-Dependent and Protein-Kinase C-Dependent and C-Independent Pathways. J. Biol. Chem. 267, 6725–6735 [PubMed] [Google Scholar]
- 37. Gill P., Jindal N. L., Jagdis A., and Vadas P. (2015) Platelets in the immune response: Revisiting platelet-activating factor in anaphylaxis. J. Allergy Clin. Immun. 135, 1424–1432 [DOI] [PubMed] [Google Scholar]
- 38. Fillon S., Soulis K., Rajasekaran S., Benedict-Hamilton H., Radin J. N., Orihuela C. J., El Kasmi K. C., Murti G., Kaushal D., Gaber M. W., Weber J. R., Murray P. J., and Tuomanen E. I. (2006) Platelet-activating factor receptor and innate immunity: uptake of gram-positive bacterial cell wall into host cells and cell-specific pathophysiology. J. Immunol. 177, 6182–6191 [DOI] [PubMed] [Google Scholar]
- 39. Massini P., and Naf U. (1980) Ca2+ Ionophores and the Activation of Human-Blood Platelets - the Effects of Ionomycin, Beauvericin, Lysocellin, Virginiamycin-S, Lasalocid-Derivatives and Mcn-4308. Biochim. Biophys. Acta 598, 575–587 [DOI] [PubMed] [Google Scholar]
- 40. Fontes-Oliveira C. C., Busquets S., Toledo M., Penna F., Paz Aylwin M., Sirisi S., Silva A. P., Orpi M., Garcia A., Sette A., Ines Genovese M., Olivan M., Lopez-Soriano F. J., and Argiles J. M. (2013) Mitochondrial and sarcoplasmic reticulum abnormalities in cancer cachexia: altered energetic efficiency? Biochim. Biophys. Acta 1830, 2770–2778 [DOI] [PubMed] [Google Scholar]
- 41. Smith I. C., Bombardier E., Vigna C., and Tupling A. R. (2013) ATP Consumption by Sarcoplasmic Reticulum Ca2+ Pumps Accounts for 40–50% of Resting Metabolic Rate in Mouse Fast and Slow Twitch Skeletal Muscle. PloS one 8, e68924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lok C. (2015) The last illness. Nature 528, 182–183 [DOI] [PubMed] [Google Scholar]
- 43. Kolarova H., Klinke A., Kremserova S., Adam M., Pekarova M., Baldus S., Eiserich J. P., and Kubala L. (2013) Myeloperoxidase induces the priming of platelets. Free Radic. Biol. Med. 61, 357–369 [DOI] [PubMed] [Google Scholar]
- 44. Joo S. Y., Kim J. E., Kim J. Y., Han K. S., and Kim H. K. (2010) Usefulness of circulating vascular endothelial growth factor and neutrophil elastase as diagnostic markers of disseminated intravascular coagulation in non-cancer patients. Kor. J. Hematol. 45, 23–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Davis D. A., Singer K. E., De La Luz Sierra M., Narazaki M., Yang F., Fales H. M., Yarchoan R., and Tosato G. (2005) Identification of carboxypeptidase N as an enzyme responsible for C-terminal cleavage of stromal cell-derived factor-1alpha in the circulation. Blood 105, 4561–4568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Waern I., Karlsson I., Pejler G., and Wernersson S. (2016) IL-6 and IL-17A degradation by mast cells is mediated by a serglycin:serine protease axis. Immun., Inflamm. Dis. 4, 70–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Powell W. S., and Rokach J. (2015) Biosynthesis, biological effects, and receptors of hydroxyeicosatetraenoic acids (HETEs) and oxoeicosatetraenoic acids (oxo-ETEs) derived from arachidonic acid. Biochim. Biophys. Acta 1851, 340–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mannucci P. M., Cattaneo M., Canciani M. T., Maniezzo M., Vaglini M., and Cascinelli N. (1989) Early presence of activated (exhausted) platelets in malignant-tumors (Breast Adenocarcinoma and Malignant-Melanoma). Eur. J. Cancer Clin. Oncol. 25, 1413–1417 [DOI] [PubMed] [Google Scholar]
- 49. Gay L. J., and Felding-Habermann B. (2011) Contribution of platelets to tumour metastasis. Nat. Rev. Cancer 11, 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu B., Khan W. A., Hannun Y. A., Timar J., Taylor J. D., Lundy S., Butovich I., and Honn K. V. (1995) 12(S)-Hydroxyeicosatetraenoic acid and 13(S)-hydroxyoctadecadienoic acid regulation of protein-kinase C-alpha in melanoma-cells - role of receptor-mediated hydrolysis of inositol phospholipids (Vol 92, Pg 9323, 1995). Proc. Natl. Acad. Sci. U.S.A. 92, 11322–11322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tang T., Eldabaje R., and Yang L. (2016) Current status of biological therapies for the treatment of metastatic melanoma. Anticancer Res. 36, 3229–3241 [PubMed] [Google Scholar]
- 52. Hart C., Vogelhuber M., Wolff D., Klobuch S., Ghibelli L., Foell J., Corbacioglu S., Rehe K., Haegeman G., Thomas S., Herr W., and Reichle A. (2015) Anakoinosis: Communicative reprogramming of tumor systems - for rescuing chemorefractory neoplasia. Cancer Microenviron. 8, 75–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Miyamoto Y., Hanna D. L., Zhang W., Baba H., and Lenz H. J. (2016) Molecular Pathways: Cachexia Signaling-A Targeted Approach to Cancer Treatment. Clin. Cancer Res. 22, 3999–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Garton K. J., Gough P. J., and Raines E. W. (2006) Emerging roles for ectodomain shedding in the regulation of inflammatory responses. J. Leukocyte Biol. 79, 1105–1116 [DOI] [PubMed] [Google Scholar]
- 55. Slany A., Meshcheryakova A., Beer A., Ankersmit H. J., Paulitschke V., and Gerner C. (2014) Plasticity of fibroblasts demonstrated by tissue-specific and function-related proteome profiling. Clin. Proteomics 11, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee S. B., Schramme A., Doberstein K., Dummer R., Abdel-Bakky M. S., Keller S., Altevogt P., Oh S. T., Reichrath J., Oxmann D., Pfeilschifter J., Mihic-Probst D., and Gutwein P. (2010) ADAM10 is upregulated in melanoma metastasis compared with primary melanoma. J. Investig. Dermatol. 130, 763–773 [DOI] [PubMed] [Google Scholar]
- 57. Chaudhari N., Talwar P., Parimisetty A., Lefebvre d'Hellencourt C., and Ravanan P. (2014) A molecular web: endoplasmic reticulum stress, inflammation, and oxidative stress. Front. Cell. Neurosci. 8, 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang P., Du J., Zhao L., Wang X., Zhang Y., Yan R., Dai J., Liu G., Zhang F., and Dai K. (2013) The role of intraplatelet reactive oxygen species in the regulation of platelet glycoprotein Ibalpha ectodomain shedding. Thromb. Res. 132, 696–701 [DOI] [PubMed] [Google Scholar]
- 59. Shindou H., Hishikawa D., Nakanishi H., Harayama T., Ishii S., Taguchi R., and Shimizu T. (2007) A single enzyme catalyzes both platelet-activating factor production and membrane biogenesis of inflammatory cells. Cloning and characterization of acetyl-CoA:LYSO-PAF acetyltransferase. J. Biol. Chem. 282, 6532–6539 [DOI] [PubMed] [Google Scholar]
- 60. Melnikova V., and Bar-Eli M. (2007) Inflammation and melanoma growth and metastasis: the role of platelet-activating factor (PAF) and its receptor. Cancer Metastasis Rev. 26, 359–371 [DOI] [PubMed] [Google Scholar]
- 61. Jain S., Zuka M., Liu J., Russell S., Dent J., Guerrero J. A., Forsyth J., Maruszak B., Gartner T. K., Felding-Habermann B., and Ware J. (2007) Platelet glycoprotein Ib alpha supports experimental lung metastasis. Proc. Natl. Acad. Sci. U.S.A. 104, 9024–9028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Das S. K., Eder S., Schauer S., Diwoky C., Temmel H., Guertl B., Gorkiewicz G., Tamilarasan K. P., Kumari P., Trauner M., Zimmermann R., Vesely P., Haemmerle G., Zechner R., and Hoefler G. (2011) Adipose triglyceride lipase contributes to cancer-associated cachexia. Science 333, 233–238 [DOI] [PubMed] [Google Scholar]
- 63. Stephens N. A., Skipworth R. J., Macdonald A. J., Greig C. A., Ross J. A., and Fearon K. C. (2011) Intramyocellular lipid droplets increase with progression of cachexia in cancer patients. J. Cachexia, Sarcopenia Muscle 2, 111–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Klein G. L., Petschow B. W., Shaw A. L., and Weaver E. (2013) Gut barrier dysfunction and microbial translocation in cancer cachexia: a new therapeutic target. Curr. Opin. Support Pa 7, 361–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kir S., Komaba H., Garcia A. P., Economopoulos K. P., Liu W., Lanske B., Hodin R. A., and Spiegelman B. M. (2016) PTH/PTHrP Receptor Mediates Cachexia in Models of Kidney Failure and Cancer. Cell Metabolism 23, 315–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Reichle A., and Vogt T. (2008) Systems biology: a therapeutic target for tumor therapy. Cancer Microenvironment 1, 159–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fukawa T., Yan-Jiang B. C., Min-Wen J. C., Jun-Hao E. T., Huang D., Qian C. N., Ong P., Li Z., Chen S., Mak S. Y., Lim W. J., Kanayama H. O., Mohan R. E., Wang R. R., Lai J. H., Chua C., Ong H. S., Tan K. K., Ho Y. S., Tan I. B., Teh B. T., and Shyh-Chang N. (2016) Excessive fatty acid oxidation induces muscle atrophy in cancer cachexia. Nature Med. 22, 666–671 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.