SUMMARY
Chronic rhinosinusitis (CRS) encompasses a heterogeneous group of debilitating chronic inflammatory sinonasal diseases. Despite considerable research, the etiology of CRS remains poorly understood, and debate on potential roles of microbial communities is unresolved. Modern culture-independent (molecular) techniques have vastly improved our understanding of the microbiology of the human body. Recent studies that better capture the full complexity of the microbial communities associated with CRS reintroduce the possible importance of the microbiota either as a direct driver of disease or as being potentially involved in its exacerbation. This review presents a comprehensive discussion of the current understanding of bacterial, fungal, and viral associations with CRS, with a specific focus on the transition to the new perspective offered in recent years by modern technology in microbiological research. Clinical implications of this new perspective, including the role of antimicrobials, are discussed in depth. While principally framed within the context of CRS, this discussion also provides an analogue for reframing our understanding of many similarly complex and poorly understood chronic inflammatory diseases for which roles of microbes have been suggested but specific mechanisms of disease remain unclear. Finally, further technological advancements on the horizon, and current pressing questions for CRS microbiological research, are considered.
INTRODUCTION
Rhinosinusitis is a heterogeneous group of diseases affecting 5 to 15% of people (1–3). It is characterized by inflammation of the nasal and sinus mucosa (2, 4) and may include nasal congestion or discharge, facial pain or pressure, loss of the sense of smell, polyposis, mucopurulent discharge, and edema or obstruction of the sinuses and nasal cavity (2, 5). While acute rhinosinusitis (ARS) and subacute rhinosinusitis involve the resolution of symptoms within 4 and 12 weeks, respectively, progression to chronic rhinosinusitis (CRS), characterized by symptoms persisting for longer than 12 weeks, occurs in up to 5% of the general population (2, 5–7). CRS is a debilitating condition, with effects on quality of life being equal to or greater than those with chronic bronchitis, asthma, peptic ulcer disease, chronic obstructive pulmonary disease, congestive heart failure, and angina (2, 8–10). In addition to the physical burden on the individual, the annual economic burden of CRS is enormous (estimated to be $8.6 billion in health care costs in the United States) (6, 11, 12).
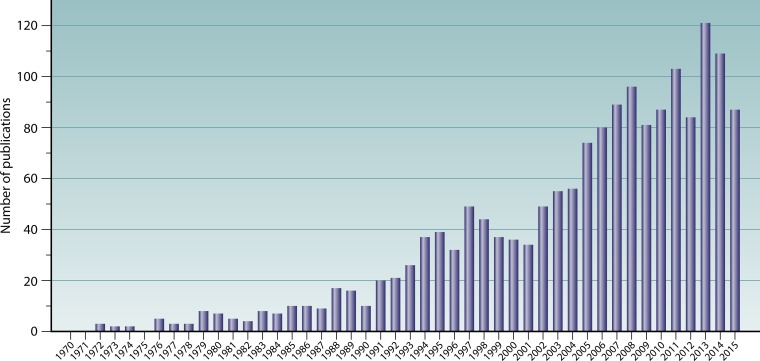
Despite considerable research, the etiology of CRS remains poorly understood. A diverse range of factors have been investigated as potential causative agents, including infectious agents; aberrant inflammatory patterns; anatomic variations affecting the ostiomeatal complex (such as nasal septal deviation and concha bullosa) or sinonasal drainage abnormalities caused by aberrant frontal sinus cells; genetics underlying the innate immune system, such as epithelial barrier integrity and mucociliary clearance; and acquired disorders such as secondary ciliary dyskinesia, hypersensitivities associated with asthma, hormonal imbalance, autoimmune disorders, and immunodeficiency (5, 7, 13, 14). Early research on CRS was principally based on an infection model of disease, with a focus on identifying a pathogenic microbial driver of the inflammatory response. Viruses, fungi, and bacteria have each been considered potentially central to the development of CRS. In recent years, however, an infection-based model of CRS has largely given way to an inflammation-based model, with the general focus shifting from possible causative pathogens to immunological factors that might underlie the disease process. Limited specific findings, together with a wealth of contradictions in the research to date, have led some to conclude that CRS may not be related to specific microbial taxa (or even microbial communities) (7, 15) but rather that CRS is a purely immune-mediated disease in which microbial involvement may be limited to causing exacerbations. Nonetheless, interest in the microbiology of CRS continues to increase (Fig. 1), and recent technological advances are now enabling new light to be shed on the unresolved role of microbes in CRS. These advances have facilitated a shift in the perspective of microbial pathogenesis from the effects of single organisms to more complex models allowing for interacting effects of both microbial communities and the environment which they inhabit, in this case the host tissues of the sinonasal tract.
FIG 1.
Publication output between 1970 and 2015 in the field of microbiology and CRS. Data for the number of publications per year were generated by using the following Scopus database search string: TITLE-ABS (“chronic rhinosinusitis” OR “chronic sinusitis”) AND (TITLE-ABS [bacteria OR bacterial OR virus OR viral OR fungus OR fungi OR fungal OR microbe OR microbial OR microbiota OR microbiome OR infectious OR infection]) AND NOT (TITLE-ABS [review]) AND (PUBYEAR > 1970) AND (EXCLUDE [PUBYEAR, 2016]) (search date, 13 May 2016).
While the discussion here is limited to the context of CRS, this new perspective is equally applicable to the disease processes of a number of complex chronic inflammatory mucosal diseases irrespective of a relation to CRS. At present, the list of disorders in which mucosal inflammation and degradation may play a role is rapidly expanding and includes such varied disorders as inflammatory bowel disease, type 2 diabetes, cardiac diseases, and autism (16, 17). In many of these cases, a role of microbes has been hypothesized, but as in CRS, specific associations remain unclear.
Until recently, reviews of the microbiology of CRS were limited largely to culture-based data. However, the complex picture offered by sequencing-era studies is now receiving increased attention (18–20). Here we present a comprehensive review of the current understanding of bacterial, fungal, and viral associations with CRS together with a detailed summary of the potential mechanisms of action driving the inflammatory process in CRS. We present this review with a specific focus on the transition to the new perspective offered in recent years by modern technology in microbiological research. Importantly, this recent shift in perspective will have far-reaching implications for both the understanding and clinical management of many chronic inflammatory diseases. Clinical implications of this new perspective are discussed. Finally, future directions, including new technological advancements and current pressing questions for research into the role of microbes in CRS, are considered. This review specifically aims to further the understanding of CRS and focus research going forward but also provides a new perspective for approaches to clinical management of the microbiology of diseases like CRS.
BACTERIA
The Culture-Based Perspective
As recently as 2009, healthy sinuses were still viewed by some to be predominantly sterile (21). Culture results from the nasal cavity included species of the genera Corynebacterium, Staphylococcus (in particular S. epidermidis), and Rhodococcus (22, 23). Pathogenicity was attributed largely to the detection of common known potential pathogens such as Staphylococcus aureus, Haemophilus influenzae, and Pseudomonas aeruginosa. However, more recent studies have questioned the role of these putative pathogens. Recent clinical culture-based studies have extended the view of the sinonasal tract in both healthy and disease states to include S. aureus, coagulase-negative staphylococci, Propionibacterium acnes, Corynebacterium diphtheriae and other Corynebacterium spp., Streptococcus viridans, Streptococcus pneumoniae, Streptococcus milleri, Haemophilus spp., Escherichia coli, and Pseudomonas aeruginosa and other Pseudomonas spp. (24, 25). Highlighting the limitations of routine culture assessment of the resident sinonasal microbial communities, a more intensive culture-based study recently identified 4- to 5-fold more bacterial species per individual than those identified by standard clinical culture methods (26). The core community within most subjects, regardless of disease status, included S. epidermidis, P. acnes, Corynebacterium accolens, Corynebacterium tuberculostearicum, S. aureus, Propionibacterium avidum, Propionibacterium granulosum, and Finegoldia magna. Other common taxa included Staphylococcus haemolyticus, Staphylococcus capitis, Staphylococcus hominis, Staphylococcus warneri, Staphylococcus lugdunensis, Streptococcus mitis-S. oralis, Streptococcus parasanguinis, as well as strict anaerobes from the genera Clostridium, Anaerococcus, Finegoldia, Parvimonas, Peptoniphilus, Veillonella, and Fusobacterium. In total, 139 distinct bacterial species were identified across the entire cohort (26).
Much of the research into the microbiology of CRS has focused on identifying and isolating candidate microbial species for pathogenicity related to the development or exacerbation of the condition. These culture-based studies have yielded much information about the potential roles of individual organisms and the causative mechanisms that they may initiate within the host. However, the clinical relevance of these organisms within the context of the polymicrobial communities of the sinonasal mucosa, many members of which are recalcitrant to standard culture efforts, remains unclear, and such an approach may be a constrained representation of the in vivo processes involved in disease. Standard culture approaches offer only a limited range of defined conditions for microbial growth and thus oftentimes omit taxa that require alternative or more niche conditions, such as slower-growing organisms that are outcompeted for a limited range of nutrient sources or those that depend upon cooperation to survive in vivo (such as cross-feeding relationships). The disparity between the identification of microbes by culture and those (viable but nonculturable or difficult to culture) identified by molecular methods has been termed “the great plate count anomaly,” with estimates of the nonculturable portion of microbial communities ranging between 25% and 99% (27, 28). Culture-independent sequencing-based studies regularly identify up to an order of magnitude more distinct taxa per individual (24, 29, 30). In direct comparisons between results of culture and sequencing approaches, the dominant bacteria identified in most patients by sequencing were identified by culture only approximately half of the time. Of those taxa present at a relative sequence abundance of <1%, only about 5% were also identified by culture (24), revealing the extent of information on lower-abundance taxa missed by standard clinical culturing. Next-generation sequencing-based studies of both healthy and CRS-associated microbiota provide a new perspective, and only now is the complexity of human-associated microbial communities becoming more fully appreciated (Fig. 2).
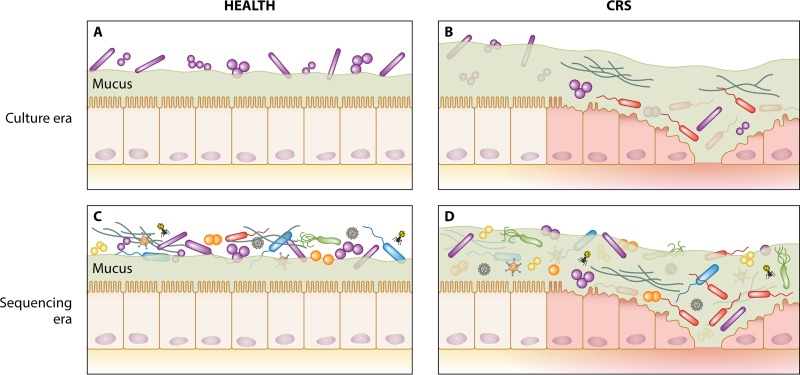
FIG 2.
Shift in the perspective of mucosa-associated microbial communities from the culture era to the sequencing era in healthy and CRS states. (A) Previous culture-based assessments of sinonasal-associated microbiology in healthy subjects led to the view of the sinuses as being a predominantly sterile site and identified a relatively simple bacterial community associated with the nasal cavity. (B) Early culture-based assessment of CRS patients highlighted the possible role of putative bacterial and fungal pathogens in the development or progressive course of CRS. (C and D) Sequencing-era assessment of the microbiota subsequently identified markedly more diverse microbial communities associated with both healthy (C) and CRS (D) states, with little consensus emerging on unique associations with CRS.
The Sequencing Era
Modern culture-independent (molecular) techniques have vastly improved our understanding of the complex microbial communities associated with the human body. Recent technological advances in next-generation sequencing allow the cost-effective assessment of microbial communities (microbiota) from complex environmental samples without the need for prior culturing of organisms. Specifically, targeted sequencing of taxonomically informative regions of the genome (such as the 16S rRNA gene and internal transcribed spacer regions) can enable the reliable identification of the majority of bacteria and fungi down to the taxonomic resolution of genus.
Complex, and often relatively comparable, microbial communities have been identified in healthy and diseased sinonasal mucosae by molecular techniques. Various Corynebacterium spp., Staphylococcus spp. (including, among others, both S. epidermidis and S. aureus), as well as Propionibacterium spp. have been identified as typical constituents of the sinonasal microbiota. Other regular, albeit less common, bacterial community members include species within the genera Anaerococcus, Streptococcus, Pseudomonas, Haemophilus, Moraxella, Peptoniphilus, Dolosigranulum, Finegoldia, Lactobacillus, Stenotrophomonas, Prevotella, Clostridium, Ralstonia, Veillonella, Neisseria, and Klebsiella, with up to 20 or 30 distinct genera commonly being identified within any given individual (24, 29–43). Furthermore, a number of particular species commonly viewed as being pathogenic, including P. aeruginosa and Haemophilus spp., have been observed in both healthy and CRS subjects.
Given the complexities of both the definition of CRS (4, 13) and the extent of natural interpatient microbiota variability (40), larger numbers of patients need to be studied. Until recently, most studies involved a limited number of subjects; however, larger-cohort community sequencing-based studies are finally beginning to emerge, with one recent study including 101 cases (41). The presence of purulent secretions, asthma, and a history of smoking has each been associated with the enrichment or depletion of certain taxa within the community (29, 41). In both healthy and CRS subjects, a history of smoking is also associated with reduced diversity of the bacterial community (41). Less bacterial diversity is also a feature of CRS-associated bacterial communities generally (34, 40) and is associated with postoperative outcomes (29); however, the nature of the relationship between less bacterial community diversity and pathophysiology of CRS remains unclear, as does the direction of the effect between these two observed phenomena. Interestingly, none of the other patient variables considered in these studies, including patient age, allergies, diabetes, ethnicity, gender, polyposis, antibiotic usage in the 6 months prior to surgery, saline washing routines, intranasal steroid use, and previous surgery, were associated with observed changes in bacterial communities in this cohort (29). Given the high degree of intersubject variability seen in both healthy and CRS individuals, numbers of patients in each substratified group may yet remain underpowered to detect subtle differences, and thus, further investigation is warranted. Notably, a history of smoking appears to have a stronger association with effects on the bacterial community in healthy subjects than in CRS subjects (41). While these results are yet to directly implicate smoking in the CRS disease process itself, smoking may be an additional exacerbating factor in driving aberrant microbiota changes that are commonly seen in CRS.
Even though these larger-cohort studies are beginning to garner important insights into clinical associations with changes in the microbiota, they also highlight the inadequacy of many studies in discerning subtle associations due to an insufficient study cohort size. Recent studies by Ramakrishnan and colleagues (29, 41) highlight this particularly well: in an initial cohort of 56 CRS and 26 control subjects, no association between the microbiota and patient smoking history was found (29). A subsequent follow-up study that included an additional 14 CRS and 5 healthy control subjects to bolster statistical power (bringing the total to 70 CRS subjects and 31 controls, respectively) found an association between microbiota differences and a history of smoking (41). Of note, this study involved the largest cohort for sequencing-based studies of CRS thus far. The difference in the patterns of association with smoking seen here indicate that many studies of CRS likely remain underpowered to discern genuine patterns of association between clinical parameters and the microbiota, from the noise of the high interpatient microbiota variability seen even in healthy controls.
While the composition of the microbiota in healthy controls and CRS patients is increasingly well described, the role of the microbiota in the pathogenesis of CRS remains unclear. In part, this is likely on account of the substantial technical variation in the methodological approaches employed by different studies: differences in sampling sites and techniques, target gene regions, sequencing platforms, bioinformatics pipelines, and taxonomic assignment databases can render meaningful comparisons difficult and may obscure overall trends. This is further compounded by difficulties in the definition of CRS itself, which is a heterogeneous group of conditions that share symptomatic expression but not necessarily underlying causative factors (Fig. 3) (2, 4, 7, 13, 44–53). This is particularly evident when we consider the range of comorbidities and complications associated with CRS, including polyposis, asthma, allergy, immunodeficiency, primary and secondary ciliary dyskinesia, gastroesophageal reflux disease, cystic fibrosis, aspirin hypersensitivity, and dental disease (2, 7, 13). If these associated comorbidities reflect the heterogeneity of the etiology of CRS, diagnosis of CRS subtypes may have important treatment implications. Furthermore, genuine patterns associated with each disease type may have been obscured by the inclusion of subtypes that involve distinct etiologies. An overview of subclassification schemas for delineating possible subtypes of CRS is provided in Table 1.
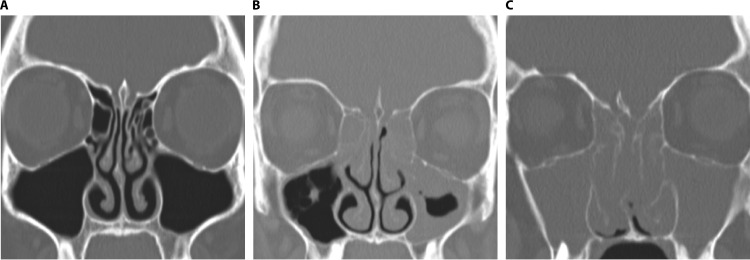
FIG 3.
Phenotypic heterogeneity of CRS on the basis of polyposis. Computed tomography scans show the differential extent of inflammation and sinonasal blockage in healthy subjects (A), patients with chronic rhinosinusitis without nasal polyps (B), and patients with chronic rhinosinusitis with nasal polyps (C).
TABLE 1.
Suggested subclassification schemas for subtypes of CRSa
| Subtype(s) | Definition | Distinguishing characteristic(s) | Clinical finding(s) | Reference(s) |
|---|---|---|---|---|
| CRSsNP | Chronic rhinosinusitis without nasal polyps | Polyposis | Tends to be Th1-skewed neutrophilic disease with fibrosis and often high levels of collagen deposition, basement membrane thickening, and goblet cell hyperplasia | 2, 45–47 |
| CRSwNP | Chronic rhinosinusitis with nasal polyps | Polyposis | Tends to be eosinophilic disease associated with edema and elevated inflammatory signaling; generally Th2-skewed immune response in Caucasian populations but Th1 in Chinese populations and Th17 in cystic fibrosis patients; presence of polyposis in patients tends to be associated with worse preoperative CT and both pre- and postoperative symptom scores | 2, 4, 7, 45–51 |
| eCRSsNP | Eosinophilic chronic rhinosinusitis without nasal polyps | Polyposis, eosinophilia | Appears to be uncommon | |
| neCRSsNP | Noneosinophilic chronic rhinosinusitis without nasal polyps | Polyposis, eosinophilia | Tends to be representative of most CRSsNP patients | 46, 51 |
| eCRSwNP | Eosinophilic chronic rhinosinusitis with nasal polyps | Polyposis, eosinophilia | Tends to be representative of most cases of CRSwNP in Caucasian populations; eosinophilia correlates with both frequency of polyps and comorbid asthma and tends to be associated with worse preoperative CT and both pre- and postoperative symptom scores than neCRSwNP | 2, 4, 7, 45–51 |
| neCRSwNP | Noneosinophilic chronic rhinosinusitis with nasal polyps | Polyposis, eosinophilia | Tends to be the most common form of CRSwNP in Chinese populations (Th1) and cystic fibrosis patients (Th17) | 2, 4, 7, 45, 47–51 |
| CHES, ECHRS | Chronic hyperplastic eosinophilic sinusitis, eosinophilic chronic hyperplastic rhinosinusitis | Hyperplasia | Comparable to eCRSwNP | 49, 52 |
| CIS, NECHRS | Chronic inflammatory sinusitis, noneosinophilic chronic hyperplastic rhinosinusitis | Hyperplasia | Comparable to neCRSwNP | 49, 52 |
| eCRSsNP, neCRSsNP, eCRSwNP, neCRSwNP, CHES, ECHRS, CIS, and NECHRS with or without polyposis, eosinophilia, neutrophilia, asthma, atopy, allergic fungal rhinosinusitis, aspirin-exacerbated respiratory disease, cystic fibrosis | Polyposis, eosinophilia, neutrophilia, asthma, atopy, allergic fungal rhinosinusitis, aspirin-exacerbated respiratory disease, cystic fibrosis | Asthmatic, aspirin-exacerbated respiratory disease, and allergic fungal rhinosinusitis patients tend to have higher CT and endoscopy scores and higher rates of polyposis and eosinophilia; nonasthmatic patients, in contrast, tend to have disease more associated with purulence but with lower CT scores than those of their asthmatic counterparts; cystic fibrosis patients represent a mix of the above-described patterns, associated with both purulence and higher CT scores | 53 | |
| CRS, delineated into 10 distinct subclusters of underlying inflammatory profiles | Distinct profiles of tissue inflammatory markers | 44 |
CT, computed tomography.
Role of Bacteria in CRS
In spite of the more complete view emerging of the sinonasal microbiota, there remains a relative paucity of comprehensive microbiological studies of CRS, and to date, there is no real consensus on the microbial ecology associated with this disease. Changes in S. aureus abundance or activity, a decreased abundance Prevotella spp., an increased abundance C. accolens, less microbial diversity, and an increased abundance of microbes have all been suggested to be associated with CRS, with little consensus between individual studies (33–35, 40, 54, 55). Below, we present various hypotheses that have been considered for potential roles of bacteria or bacterial assemblages in the etiology or exacerbation of CRS. For the purposes of introduction, each of the sections below is considered in relative isolation; however, the reality is much more complex. Each hypothesis and mechanism of action are not mutually exclusive and are likely interlinked and interactive, representing the multiple layers of changes in the microbiota that are associated with differences between healthy and disease states.
Single-pathogen hypothesis.
Much research has focused on the possible role of S. aureus in the etiology of CRS. However, recent studies have not identified a significant difference between subjects with CRS and controls in S. aureus carriage, the presence of known S. aureus exotoxins (SAEs), or correlations with inflammatory cells (56, 57).
While rates of S. aureus identification are comparable in healthy controls and CRS patients (∼30% to 40% [56, 58]), alterations in the virulence and activity of S. aureus remain possible etiological or exacerbating factors in the subset of CRS patients colonized by S. aureus. The main mechanisms by which S. aureus can influence inflammatory disease include the release of exotoxins that degrade epithelial barrier integrity or drive inflammation via superantigenic activation of lymphocytes (59, 60) and anti-inflammatory activity (including affecting complement, antimicrobial peptide production, and adhesion and chemotactic processes) (61). Exposure of cultured epithelial and polyp cells to either S. aureus or its secretory proteins can induce apoptosis and alter cytokine signaling and nitric oxide production (an important endogenous antimicrobial) in mucosal tissue from both healthy and CRS subjects (62–64). These mechanisms offer potential S. aureus-mediated immune-modulating roles, together with several viable mechanisms for the mucosal inflammation and epithelial cell damage seen in diseases such as CRS. Elsewhere, S. aureus delta-toxin production has been found to induce mast cell degranulation, suggesting another possible mechanistic link between S. aureus and allergic inflammatory disease (65). Direct evidence of S. aureus promoting CRS via these mechanisms remains to be identified.
In one recent study, changes in the abundances of S. aureus, Prevotella spp., and C. accolens and increased abundances of microbes overall were not associated with CRS. Rather, the increased abundance of a single species, C. tuberculostearicum, was found to correlate with the severity of CRS and also with a relative decrease in the abundance of Lactobacillus spp. (34). This finding was reinforced with data from a murine model in which C. tuberculostearicum inoculation resulted in the development of CRS-like symptoms, while coinoculation of Lactobacillus sakei mitigated this effect. However, subsequent studies have failed to replicate the observed negative correlation between Corynebacterium and Lactobacillus spp.
Intramucosal bacteria.
S. aureus has a remarkable ability to live within epithelial cells or submucosally in the interstitium while evading the host immune response (66–70). Submucosal S. aureus has been identified in 40% to 75% of all CRS subjects, compared to 0% to 12.5% of control subjects (68–70), and is particularly prevalent in subjects with aspirin-exacerbated respiratory disease-related CRS (66). In one study, intracellular S. aureus induced interleukin-6 (IL-6) production in nasal epithelial cells in vitro (67). An alternative ex vivo study of CRS subjects, however, found that submucosal S. aureus elicited no immune reaction at all (69), supporting a pathogen immune evasion hypothesis. Identification of submucosal S. aureus in CRS subjects has been significantly associated with an increased risk of relapse of disease following surgical intervention (70).
The specific etiology of the increased abundance of intramucosal S. aureus in CRS and the role that it might play in the disease process (or, indeed, whether it plays a direct role or is merely a bystander) remains unclear. Loss of epithelial barrier integrity is a hallmark of CRS and thus may provide an opportunity for intramucosal invasion. Furthermore, S. aureus has been shown to bind to Candida albicans hyphae in vitro, with epithelial invasion by C. albicans enabling coinvasion by S. aureus (71, 72); however, a clear association or role for either S. aureus or C. albicans in CRS remains to be seen. One mechanism by which S. aureus might achieve submucosal immune evasion is through phenotype switching to metabolically less active small-colony variants (SCVs). Submucosal or intracellular SCVs could act as a reservoir for S. aureus, evading the host immune response and antimicrobial efforts, enabling a seed bank of putative pathogens, and potentially explaining the recalcitrance of CRS to antibiotic treatment. S. aureus bacteria from experimentally infected airway epithelial cell cultures exhibit decreased enterotoxin secretion and alterations of phenotypes and growth patterns (73). However, in a recent study, no association between S. aureus SCVs and CRS was found, with comparable rates of both S. aureus and S. aureus SCVs in CRS and control subjects. Furthermore, putative S. aureus SCVs were later identified via 16S rRNA gene sequencing to in fact be bacterial taxa that were unrelated to Staphylococcus altogether (74), calling into question both the general role of S. aureus in CRS pathogenesis as well as the specific role of S. aureus SCVs. Submucosal bacteria have been found at comparable rates across all patient groups assessed, including controls and subjects with CRS without nasal polyps (CRSsNP), subjects with CRS with nasal polyps (CRSwNP), and CRS subjects with underlying cystic fibrosis (75). Accordingly, the pathogenic role of intramucosal bacteria is far from clear.
Cooccurrence of bacteria.
The cooccurrence of particular bacterial taxa of interest in a range of human mucosal inflammatory conditions, including CRS, has received growing attention. A comprehensive analysis of bacterial networks within the Human Microbiome Project data set identified 3,005 significant cooccurrence and coexclusion relationships (76). Metabolic modeling also identified a plethora of interspecies metabolic exchanges that can shape and maintain interdependent relationships between particular microbes (77). In CRS, S. aureus, for example, is most frequently associated with S. epidermidis and P. acnes (26, 37). Alternatively, in a predominantly sequencing-based study coupled with follow-up coculture experiments, C. accolens and S. aureus mutually facilitated the growth of each other, while Corynebacterium pseudodiphtheriticum and S. aureus were mutually inhibitory (37). A recent landmark study that moved beyond observational patterns alone and investigated underlying molecular mechanisms of interactions between specific microbes identified an antagonistic relationship between a putative pathogen and a common commensal of the sinonasal tract: C. accolens metabolism of human skin surface triacylglycerols led to the release of the free fatty acid oleic acid, inhibiting the growth of pneumococcus (78). Similarly, numerous antimicrobial small molecules that are produced by bacteria in the skin microbiota (including S. epidermidis, which is also a common member of the sinonasal microbiota), altering cooccurrence and exclusion patterns, have now been identified (79, 80). This suggests that species-level differences and interactions within the community may drive important changes of the community structure as a whole. Analyzing species-level community differences is not always accurate in next-generation sequencing-based studies, which reliably identify taxa only to the genus level. In a comparative study by Kaspar et al. (26), sequencing identified a higher total number of phylotypes, but due to the limited resolution of 16S rRNA gene fragment sequencing identification, their culture-based approach identified more taxa down to the species level (26). Microbial genetic potential and activity, virulence, and involvement in important disease-associated processes can vary markedly on the scale of species or even strain differences between bacteria of the same genus (81). This offers a compelling argument for studies that combine molecular and culture-based approaches to more fully understand the complexity of bacterial community interactions both overall and at the bacterial species or strain level.
Biofilms.
Biofilms are complex, multicellular assemblages that are largely comprised of a polysaccharide matrix that serves as a structural basis for microbial clusters and as a barrier to the surrounding environment (82, 83). When free-swimming planktonic cells come into contact with a surface, rapid phenotypic changes and exogenous small-molecule signaling drive the adhesion and coaggregation of microbes together with the secretion of the polysaccharide matrix that ultimately encapsulates the microbial cluster (84). Living within biofilms offers protection from a wide array of possible threats, including phagocytic cells, predatory amoebae, bacteriophages, surfactants, and antimicrobials (83, 85). Biofilms can lead to a reduction in complement activation and susceptibility to phagocytosis by host immune cells as well as resistance to humoral aspects of the immune system such as endogenous antimicrobial peptides (83) and may by some measure be the preferred state of bacteria in the human body (86).
In the context of CRS, specific taxa identified in bacterial biofilms include P. aeruginosa, S. aureus, coagulase-negative staphylococci, H. influenzae, Enterobacter spp., Proteus mirabilis, Hafnia spp., Acinetobacter spp., and Streptococcus pneumoniae (87–90). In a number of these studies, several taxa were concurrently associated with biofilms, highlighting the likelihood of polymicrobial biofilm formation. Furthermore, fungal biofilms have also been identified in CRS patients (91), as have increased rates of fungal elements in bacterial biofilms in patients with allergic fungal rhinosinusitis (AFRS) and eosinophilic mucin CRS (92), suggesting a multidomain dimension to biofilm activity in CRS. Importantly, both multispecies and multidomain polymicrobial biofilms can exhibit properties and interactions distinct from those of single species alone (93). These factors can include changes in morphology, the production of antimicrobials, protection from antibiotics, improved attachment due to coaggregation, competitive exclusion for binding sites, effects on the production of virulence factors, and changes in nutrient availability (93). Each of these modulating factors could markedly influence microbial community dynamics.
However, comparable rates of biofilm identification in CRS patients and healthy controls bring into question the role of biofilms in CRS (87, 88, 90–92, 94–97). As noted above, it is also increasingly understood that biofilms likely represent the predominant mode of existence of microbes in the environment. This suggests that the presence of a biofilm or biofilm-forming capacity does not necessarily imply a direct association with the disease process. A mechanistic link that distinguishes the role of biofilms in CRS from their comparable presence in healthy controls has been sought. Increased rates of S. aureus biofilms in particular, together with their superantigens, have been identified in CRS subjects compared to controls (98). In CRS patients, biofilms tend to colocate with a disruption of the epithelial barrier and associate with an increased presence of T cells and macrophages (97). This offers a compelling explanation for why the presence of biofilms alone is not closely associated with inflammation in the sinonasal tract: a disrupted epithelial barrier may also be required for biofilms to drive the increased inflammatory response. An alternative argument for a role of biofilm formation in CRS suggests that the first point of formation is binding to mucin. MUC gene expression levels were significantly elevated in CRS subjects who had biofilm formation in comparison to those who did not (99). Importantly, it remains to be seen whether biofilms in CRS directly drive disease and an aberrant inflammatory response or whether CRS disease processes drive the inflammatory response, epithelial barrier breakdown, and increased MUC expression in parallel.
While the direct role of biofilms in CRS remains unclear, subjects who have biofilm formation also tend to have a worse prognosis, are more likely to have poorer outcomes postsurgery, and exhibit an increased immune response when biofilms are associated with the disrupted epithelium that is ubiquitous in CRS-afflicted mucosa (87, 89, 95, 97, 100). The search for an effective treatment modality addressing the possible involvement of biofilms in CRS remains an ongoing effort (100).
The dysbiosis hypothesis.
While a few associations of microbes with CRS are beginning to emerge, intersubject variation, regardless of health status, is often more prominent than specific associations with disease (26, 29, 40, 42). Increased variability has been observed, as is particularly true of CRS subjects, suggesting a general process of imbalance, or dysbiosis, in their bacterial community structures. This dysbiosis may be an important mediator of the disease process (40). Furthermore, as noted above, each of the above-described hypotheses likely involves intertwined processes that reflect overall changes in the microbiota, which may arguably be best understood under the umbrella of the dysbiosis hypothesis. Dysbiosis and instability in the community as a whole are likely reflected in the dominance of particular microbes (single-pathogen hypothesis) or clusters of microbes (cooccurrence hypothesis), intramucosal invasion and persistence (intramucosal hypothesis), or changes in the membership and activity of the biofilms present.
In contrast to standard single-agent models of pathogenicity, a shift away from the normal “recognized” microbial and molecular milieu in the sinonasal tract toward an unbalanced community may in itself drive an exacerbated and ongoing inflammatory response. It remains unclear whether community shifts and increasing dysbiosis might directly drive the initial etiology or subsequent exacerbation of the disease or if they are merely benign reflections of the changing microbial habitat as a result of the disease. Nevertheless, the loss of epithelial barrier integrity that results from progressive inflammatory disease facilitates further exposure to the environmental stimuli within the sinonasal tract, including fungi, viruses, bacteria, and their metabolic products, which may further drive the inflammatory disease (16, 101). Thus, even in the event that CRS proves to be a chiefly immune-mediated disease, the microbial milieu may be important in exacerbations and the progression of the condition, even if it is not primarily responsible for the initial etiology.
FUNGI
The involvement of fungi in rhinosinusitis is divided into invasive forms, including variants of acute, chronic, and granulomatous invasive fungal rhinosinusitis, and noninvasive forms. Acute invasive forms tend to be found in immunocompromised patients, while chronic and granulomatous forms are usually found in a background of immunocompetence. While the latter two are chiefly treated with systemic antifungals, acute invasive fungal rhinosinusitis additionally requires the recovery of immunocompetence (5). Invasive forms are considered independent entities of rhinosinusitis and are thus typically excluded from considerations of fungal involvement in idiopathic CRS more generally. However, there is a long history of research into the putative noninvasive roles of fungi in idiopathic CRS.
Early Studies and the Ponikau Hypothesis
The earliest reports of fungal rhinosinusitis described an infectious disease process associated with Aspergillus and identified by the presence of eosinophilic mucin together with fungal hyphae (102, 103). Initially known as aspergillosis or allergic aspergillus sinusitis, this disease included cases of benign fungal balls as well as debilitating cases of necrotizing cavitary granulomatosis (102). Subsequent diagnostic criteria specified AFRS in much the same terms as other forms of eosinophilic rhinosinusitis—sinusitis of one or more paranasal sinuses; eosinophil-rich mucin; and the absence of invasive fungal disease, immunodeficiency, or diabetes—with the addition of a positive fungal stain or culture (104). Comparable criteria largely stand to this day (105). Following the establishment of diagnostic criteria for AFRS by DeShazo and Swain (104), early culture methodologies identified fungi in approximately 7% of CRS subjects (106). Using highly sensitive fungal collection and culturing techniques, Ponikau and colleagues (107) subsequently identified fungi in 93% of CRS patients in a large cohort. This led to the proposal that most cases of CRS are mediated by fungi (107). However, similarly comprehensive culturing techniques later showed comparable rates of the presence of fungi in both CRS subjects and healthy controls (106). Furthermore, in a neonatal study, nasal mucus specimens from 94% of subjects were positive for fungal cultures within the first 4 months of life, suggesting that fungi are a normal constituent of the sinonasal tract (108). Together, these findings challenge the suggested key role for fungi in the majority of CRS cases.
The Aberrant Immune Response Hypothesis
Given similar rates of detection of fungi in CRS patients and healthy subjects, it was proposed that hypersensitivity to fungi due to the state of the host's immune system may distinguish individuals who develop disease from those who do not (109–112). One supportive study found significantly increased immune and inflammatory responses in peripheral blood mononuclear cells cocultured with Alternaria antigens in the majority of CRS subjects compared with the responses of healthy controls (109). Other research efforts and reviews have in turn both implicated and ruled out fungus-specific immune responses as uniquely defining AFRS as being distinct from CRS (107, 113–119).
Proposed Mechanisms of Action
Fungal protease activation of epithelial cells can elicit increased inflammatory cytokine production and the subsequent migration of inflammatory cells (120, 121). Similarly, coculture of human eosinophils from healthy individuals with the common environmental airborne fungi Alternaria and Penicillium can induce exocytosis in cells from normal individuals and general activation of eosinophils (122). This highlights one possible mechanism by which fungi could drive eosinophilic inflammatory cascades such as those seen in cases of eosinophilic CRS. An effect of fungi on epithelial barrier integrity has also been investigated. Epithelial cell cultures from AFRS cases show increased permeability and alterations of the expression of intercellular junction proteins in comparison to controls (123). However, increased epithelial barrier permeability is a feature of CRS regardless of a suspected role of fungi (124).
An interactive role of fungi and bacteria has also been suggested. Synergistic interactions between C. albicans and a diverse range of bacteria, including Streptococcus sanguinis, Streptococcus salivarius, Streptococcus mutans, S. mitis, Fusobacterium nucleatum, and Actinomyces viscosus, have been noted (125). S. aureus has been identified significantly more often in AFRS subjects than in non-AFRS CRS subjects (126). The presence of fungi may aid in the establishment and growth of this bacterium in AFRS patients: adhesion of S. aureus to C. albicans hyphae may enable intramucosal invasion (71, 72, 93, 125, 127). In contrast, antagonistic interactions between P. aeruginosa and fungi, including C. albicans, Scedosporium aurantiacum, and Aspergillus fumigatus, have been well documented (128–131) and reflect a further aspect in which interactions between particular fungi and bacteria present in the microbiota can shape the microbiota as a whole. Additionally, other factors that also affect cooccurrence patterns might be considered. Antibiotic treatment has previously been described as a risk factor for fungal outbreaks and disease (132). In CRS specifically, patients who are most at risk for AFRS include those patients with a recent history of corticosteroid or antibiotic treatment (102). Antibiotic-resistant strains of S. aureus may also flourish in a community under the effects of antibiotics that are otherwise effective against many other bacterial community members.
A New Perspective: the Sequencing Era
Few studies have investigated the fungal communities of the sinonasal tract by using molecular techniques. This highlights an important dearth in our microbial understanding of CRS. Two studies have confirmed that fungi are ubiquitous in both CRS patients and control subjects and that the fungal portion of the sinonasal microbiota is markedly more diverse than previously appreciated (35, 133). Interestingly, both of these studies found little meaningful difference between fungal communities of CRS patients and those of control subjects. However, the dominant fungi in each study were different. Cryptococcus neoformans var. neoformans dominated fungal communities in a study by Aurora et al. (35). Malassezia spp. were also ubiquitous in CRS and control subjects but at a much lower relative abundance. In contrast, the genus Malassezia was both the most prevalent, found in all subjects, including controls and CRS patients, and the most abundant fungus in a study by Cleland et al. (133). The prevalences and relative abundances of other fungi were also markedly different between the two studies. Predominant fungal taxa in CRS patients and controls in the former study included C. neoformans, Rhodosporidium diobovatum, Davidiella tassiana, and two Malassezia species (35). In contrast, in the latter study, the genera Malassezia, Calicium, Neocosmospora, Fusarium, Saccharomyces, Aspergillus, and Scutellospora were predominant (133). It remains unclear whether the contrast in these findings is due to differences in the technological approaches used or whether it is instead reflective of the confounding geographic influence of fungal dispersal and human exposure. Of note, Alternaria and Aspergillus (the two fungal taxa previously most commonly associated with CRS on the basis of data from culture-based studies) were found only at low relative abundances when they were present in either of these culture-independent studies, questioning their putative significance in the pathogenesis of CRS.
Ultimately, the specific allergic role, and the more general role, of fungi in CRS has more recently been viewed as being secondary to the primary disease process (134, 135). Nonetheless, with few sequencing-based studies to date, a great deal more needs to be understood about the presence and possible role of fungi in CRS. For example, it remains unclear whether detected fungi are active, colonizing members of the microbiota or merely transient environmental stimuli. Fungal spores are ubiquitous in the air that we breathe, and so the upper respiratory tract is continually exposed to these spores. Gene-targeted sequencing is unable to distinguish between fungal DNA present as transiently passing spores and that present as an actual colonizing member of the mucosal microbiota. Metatranscriptomics (discussed below) is a more appropriate technique for answering the question of whether fungi in the upper respiratory tract are passive (but nonetheless still potential environmental stimuli) or active members of the microbiota.
VIRUSES
Viruses are a key causative factor of acute rhinosinusitis (ARS), while acute bacterial rhinosinusitis (ABRS) is a complication of ARS in <2% of patients (2). Viral ARS and ABRS are clinically indistinguishable, hampering differential diagnosis efforts (with delineation generally being done solely on the basis of the time course of symptom presentation) (5). Thus, it remains unclear whether ABRS or CRS represents a temporal continuation of viral ARS or a distinct condition that presents similarly to ARS in the initial stages.
The potential means by which viral activity might influence the development or progressive course of CRS are manifold. Ex vivo studies of mucosal tissue have shown that once an individual is infected, rhinovirus infection can be linked with exacerbations of CRS, including increased bacterial adhesion; increased susceptibility to secondary microbial infection (by bacteria, fungi, or other viruses) (136); reduction of epithelial barrier integrity (137); respiratory exacerbations (as in asthma, cystic fibrosis, and bronchiectasis subjects) (138–140); epithelial damage, including necrosis; ciliary dysfunction; and impairment of mucociliary clearance (141) and mucus overproduction (142–144).
Current Knowledge on Viruses and CRS
The roles of a relatively small number of respiratory viruses have been the focus of research to date, and the limited number of studies investigating specific virus associations with CRS present contradictory results. One of these studies found that human rhinovirus was the only virus that differed significantly between CRS patients and controls (145). While an additional study identified an increased presence of virus in CRS patients compared to controls, this was not associated with one particular virus, and there was no association with disease severity scores (146). Another study identified 8 distinct human rhinovirus strains in CRS, 4-fold more than those seen in controls (147), suggesting that strain-level variation may play a role in either the risk of the initial development of CRS or exacerbation events. Rhinovirus infection can alter both inflammatory signaling and gene and protein expression of remodeling factors (148, 149), indicating a potential link with the ongoing disease processes of CRS. Alternatively, cell cultures from CRS subjects with nasal polyps have been found to be no more susceptible to rhinovirus infection than those from controls (150), calling into question the hypothesis that increased rhinovirus susceptibility and subsequent infection may be linked with the initial development of CRS. While targeted assessments of common respiratory viruses have identified viruses in CRS subjects with various frequencies, these viruses have also been observed in nonrhinosinusitis controls (145, 151, 152), leading to the question of whether the viruses studied play any more of a significant role in CRS disease processes than in the general population.
The Missing Viral Diversity
As with the transition from culture-based to culture-independent assessments of bacterial and fungal communities, technological developments mean that we are on the brink of a new, and increasingly complex, understanding of the virus diversity associated with the sinonasal tract. Bacteria and fungi each contain taxonomically useful genetic regions (16S rRNA and 18S rRNA, 26S rRNA, and internal transcribed spacer regions, respectively), enabling gene-targeted sequencing. Viruses, however, lack such a prospective panviral genetic target. Instead, our understanding of virus associations with the human body has been based largely on techniques that require a priori knowledge of genetic targets of specific viruses of interest. Our understanding of the viral component of the human microbiome in many cases remains akin to the culture-era view of the bacterial and fungal components of the community, likely a significant underestimation.
Metagenome studies do not target specific genes but instead indiscriminately sequence all DNA or RNA in a sample and represent an equivalent transition for our understanding of viral communities as the advent of next-generation sequencing techniques has done for bacterial and fungal communities. Early studies indicate that the virus diversity associated with all environments, including the human body, is vastly more complex than previously appreciated (153, 154). For example, a diverse range of viruses, including many strains across 39 different species, have been identified in nasopharyngeal aspirate samples (155). As with bacteria and fungi, it is increasingly understood that viruses are likely to be ubiquitously present in mucosal sites and also to play important commensal roles as resident members of our microbiota (153, 154). Thus, the true complexity and role of viruses likely remain markedly underappreciated. Beyond simply infecting host cells and driving cytolysis, both favorable and pathogenic interactions may result from the variety of ways in which viruses can influence not only our own cells but also the other members of our microbiota. Bacterium-infecting viruses (bacteriophages) are thought to comprise the vast majority of viruses associated with the human body (156, 157). The incorporation of viral genetic elements into both our own chromosomes and those of our resident microbiota can influence gene and protein expression, with both negative and positive potential roles in human health and disease. Community dynamics can be markedly influenced via conferring a competitive advantage or disadvantage through direct killing and encoding of toxin production as well as through altering infected-cell cellular mechanisms, gene and protein expression, microbial resilience to disturbance, and immune evasion. Furthermore, viruses play an important mediating role in horizontal gene transfer of genetic elements, including antibiotic resistance genes (153, 154, 158–160). The potential for viruses to influence our microbial community is of great interest for the understanding of the ecology and dynamics of our resident microbes more generally and is also garnering increasing interest as a therapeutic option for targeting specific pathogens or modifying the community structure via the administration of specific bacteriophages (161–163).
CLINICAL IMPLICATIONS
In standard practice, CRS is initially treated medically, usually with combinations of systemic or topical corticosteroids and antibiotics, which in many cases can go on for years (6). When medical management fails to bring about sufficient improvement, endoscopic sinus surgery is performed to remove inflamed tissue and open up sinus ostia, reestablishing the airway, relieving sinus obstruction, as well as allowing in-office debridement and improved postoperative delivery of topical medications and irrigations (5). With the microbiological picture of CRS becoming more complicated in recent years, we consider the clinical implications of where our knowledge currently stands. In particular, we discuss the effectiveness of current treatment approaches and their impact on mitigating or exacerbating CRS (Fig. 4). We also explore the available alternatives.
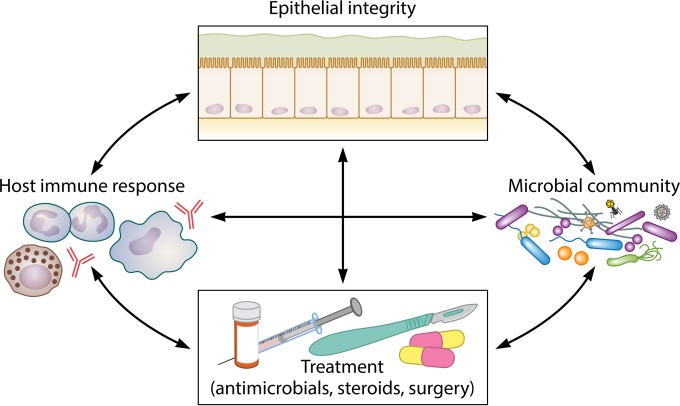
FIG 4.
Multifaceted interactions that may influence our understanding of CRS and the progressive course of chronic mucosal inflammation. The interacting effects of epithelial barrier integrity, the host immune response, the resident microbiota (including bacteria, fungi, viruses, and archaea), and treatment courses on one another are likely multidirectional and complex and remain poorly understood.
Antibiotic Use in CRS
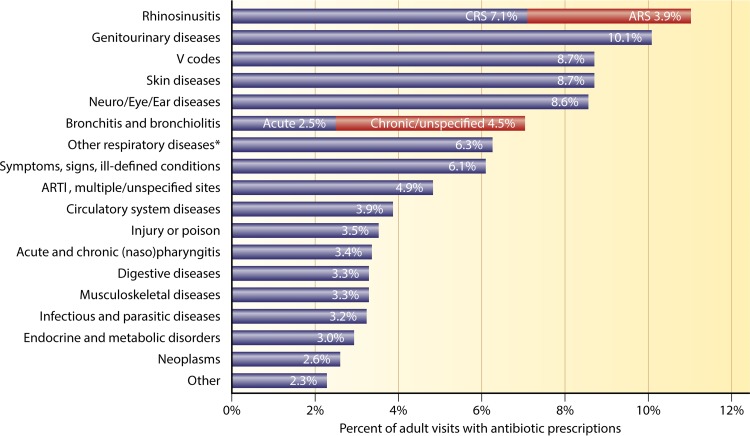
In 2003, rhinosinusitis was the single most common indication for antibiotic use in the United States, accounting for 15% of all broad-spectrum antibiotic prescriptions (approximately 8 million prescriptions) (164). Furthermore, antibiotics accounted for the greatest proportion of medical expenses associated with the treatment of CRS (12). There remains little indication that this usage has changed: in 2013, CRS remained the primary diagnosis that accounted for the most outpatient prescriptions for antibiotics (Fig. 5) (165).
FIG 5.
Primary diagnoses for adult outpatient visits resulting in antibiotic prescriptions. ARTI, acute respiratory tract infection. *, other respiratory diseases include tonsillitis/adenoiditis, laryngitis/tracheitis, deviated septum, peritonsillar abscess, allergic rhinitis, adenoid/tonsil hypertrophy/vegetations, pneumonia, influenza, emphysema, asthma, bronchiectasis, extrinsic allergic alveolitis, chronic airway obstruction not otherwise specified, pneumoconiosis, pleurisy, pneumothorax, lung/mediastinal abscess, pulmonary congestion/hypostasis, postinflammatory pulmonary fibrosis, other alveolar diseases, other parieto/pneumopathies, sclerotic lung disease, acute chest syndrome, lung involvement in other disorders, and other respiratory diseases. (Adapted from reference 165 with permission of Elsevier.)
While antibiotics remain the most common treatment for CRS, there is surprisingly little evidence to support their efficacy, and antibiotics are not routinely recommended by several clinical guidelines (2, 7, 12, 15, 166–168). A small number of studies have suggested that long-term treatment (3 months) with macrolide antibiotics may be effective, particularly in patients with normal serum IgE levels (169). However, macrolides also possess anti-inflammatory properties, including suppression of cytokine signaling, downregulation of prolonged inflammation, alteration of chemotaxis, activation of neutrophils, and reduction of reactive oxygen species production (2, 170). Thus, the observed effect on CRS may be due to the anti-inflammatory rather than antibiotic activity of macrolides. Due to a paucity of evidence supporting the efficacy of antibiotics for the treatment of CRS and concerns about the increasing development of antibiotic resistance, antibiotics are ideally reserved for cases in which known pathogens have been linked with symptomatic signs of bacterial infection, such as purulent mucus discharge, localized pressure or pain, or fever (2, 7). Even in these cases, such symptoms are not reliable predictors of either a specific bacterial infection or a likely benefit of antibiotic treatment (167).
Nonetheless, antibiotic administration remains widespread practice for the management of CRS. For those patients who ultimately progress to surgery, almost all of them will have been prescribed multiple courses of antimicrobials. More than 90% of the courses of medical therapy given to CRS patients before surgery include courses of systemic antibiotics and topical corticosteroids (171, 172). Furthermore, at the time of CRS diagnosis, patients have increased rates of associated premorbid diagnoses, including anxiety, headaches, gastroesophageal reflux disease, sleep apnea, and acute infections of both the respiratory tract and other sites (173). Often, this results in an increased use of antibiotics and steroids. In the case of antibiotics, this translates to roughly twice as many prescriptions in comparison to those of the general population prior to the initial diagnosis of CRS (173).
Efficacy of Antibiotics for Treatment of CRS: Clinical Trials and Systematic Reviews
One systematic review of clinical trials of antimicrobials for the treatment of CRS found limited evidence for the efficacy of topical antibacterials, and those authors recommend against their general use (174). Of note, this review found that the best evidence in support of antibacterial use was limited to postsurgical patients and culture-directed therapy, suggesting a possible beneficial role in targeting aberrant bacterial community structures postsurgery. Similarly, a more recent evidence-based review of topical therapies for the treatment of CRS supported the use of saline irrigation and nasal steroid therapy but specifically recommended against topical antifungals and topical antibiotics delivered via nebulizers and spray techniques (175). Research on topical antibiotics delivered by other means remained insufficient for a meaningful assessment. Another comprehensive review found only low-level evidence for an improved quality of life with antibiotic use, particularly in the case of culture-directed antibiotics. Studies with higher-level evidence found no difference between antibiotics and saline rinsing alone (176). Of note, in the 50 years preceding the 2008 review by Lim et al. (174), only 7 controlled trials (5 double blind and randomized) of topical antimicrobial use for the treatment of CRS had been conducted. Most recently, a further systematic review by Orlandi and colleagues (5) again noted a lack of enough robust studies to determine the efficacy of many delivery forms of nonmacrolide antibiotics in the treatment of CRS (while macrolide antibiotics are given the recommendation of optional). Where there was adequate evidence to establish recommendations (topical antibiotics for CRSsNP and oral nonmacrolide antibiotic courses of <3 weeks for CRSwNP), the evidence supported a recommendation against these therapeutic options.
Antifungals, Antivirals, and Bacteriophage Therapy
Antifungals.
Antifungals are now recognized as being ineffective treatments for the management of CRS, whether or not fungi are thought to play a role in the case being treated (105, 135, 175, 176). A meta-analysis of data from double-blind randomized control trials of antifungals for the treatment of CRS found no significant benefit of either systemic or topical antifungals (177). On account of these findings, together with reported side effects of systemic antifungals, such as renal and hepatic toxicity, antifungals are not advocated for the general treatment of CRS outside known cases of AFRS (5, 178).
Antivirals.
On account of inconclusive findings in studies on the role of viruses in CRS to date, there remains little research on the usage or potential benefit of antivirals for the management of CRS. Given the above-noted limitations of our understanding of the true complexity of the viral members of our associated microbiota, this remains an open discussion and may yet become a viable area of therapeutic interest should a clearer role of viruses in CRS emerge.
Bacteriophage.
While not yet applied to the context of CRS, interest in the potential applications of bacteriophage therapy has recently reemerged and may soon represent an additional option for targeted treatment of putatively pathogenic bacteria associated with disease. Bacteriophage therapy involves the administration of highly selective bacterium-targeting viruses (often targeting specific bacterial strains) with the aim of attacking only the disease-causing bacteria of interest (161). With more selective targeting of particular bacteria than with traditional antibacterials and reduced concerns of the development of antimicrobial resistance, bacteriophage therapy represents an attractive alternative as a bactericidal treatment for disease. This has been of increasing interest for P. aeruginosa-associated exacerbations in cystic fibrosis patients, for example (162). Furthermore, a promising initial trial in a murine model showed reductions in bacterial loads and inflammation in treated mice (163). However, progress toward possible widespread clinical applications of bacteriophage therapy remains in its early stages and also includes regulatory hurdles that are not inconsiderable (179, 180).
Are Antibiotics Appropriate for General Treatment of CRS?
It is important to clarify the effectiveness of antibiotic treatment for CRS in order to limit unnecessary or unhelpful prescribing patterns. Importantly, many microbially oriented treatment approaches for CRS have failed to consider the microbial component of CRS at the complex community ecology level. Standard antimicrobial treatment modalities address specific microbial infections and pathogenicity. Their role and effect may be quite different in complex diseases where no specific pathogenic agent has been identified but where imbalance, instability, or broader alterations in the normal commensal microbiota more generally may play a role in ongoing disease (181).
A recurring theme in recent studies is the observation that shifts in the microbial (particularly bacterial) community structure are associated with CRS or subsets of CRS delineated on the basis of associated clinical parameters (29, 34, 40, 41). These findings support the argument that changes in the microbiota in CRS patients are driven largely by the disease process and associated clinical factors. However, it remains unclear whether these associations play a role in the initial etiology or ongoing exacerbations and progression of CRS or whether they are driven by the environmental changes associated with the underlying disease process itself. Whether or not the initial etiology of CRS is microbiological in nature, the resultant changes to the microbial community structure may also feed back as an ongoing disease modifier.
Where there might be an antimicrobial effect, it also remains unclear what the direction of this effect may be. Antibiotic treatment that destabilizes the normal commensal microbiota may facilitate the dominance of antibiotic-resistant taxa. For example, one study showed that following antibiotic therapy, communities were often dominated by taxa that tended to be less susceptible to the prescribed antibiotics (39). In an animal model, antibiotic treatment exacerbated, and to a large extent was required for, the CRS-mediating effect of C. tuberculostearicum (34). Exacerbation following antibiotic treatment has also been observed elsewhere: in the cystic fibrosis lung, dominance by the bacterium Stenotrophomonas maltophilia was reduced in one patient, and diversity was increased with medical treatment, but following antibiotic treatment, dominance by S. maltophilia was restored (182). Similarly, in both the cystic fibrosis lung and the sinonasal microbiota in CRS patients, antibiotic treatment has been identified as perhaps being the most powerful driver of microbial community change and has been associated with a decrease in community diversity. Both of these factors may predispose a patient to susceptibility to secondary infection (33, 183, 184). For CRS, while one study found that bacterial diversity was significantly reduced following treatment, the shifts in the microbiota were highly individualized from patient to patient, and no single microbiota profile was apparent (39). A similar variability was seen in a subsequent study: communities were different posttreatment but not in any consistent manner. In contrast to the study described above, culture-directed antibiotics led to increased diversity in the posttreatment group (185). The cause of this disparity remains unclear. Culture-directed antibiotics may better target the aberrant aspects of a diversity-depleted microbiota during disease, allowing the more complex normal community structure to reestablish. Notably, both studies consist of only a few cases. Given the highly variable nature of the microbiota in CRS patients (29, 40, 41), it is likely that many more subjects are needed to tease apart genuine associations between the dynamics of the microbiota, antibiotic use, and the underlying disease process.
Ultimately, if it is indeed the destabilizing nature of the disease that allows aberrant communities to take hold at the expense of the normal microbiota, we might reconsider the extent to which current treatment modalities could equally promote the dysregulation of the existing communities as much as they might inhibit disease-associated ones.
The Future of Targeted Treatment of the Microbiota in CRS
The controversial role of antibacterial treatment in CRS warrants investigations of alternative treatments and therapies for the management of aberrant microbiota. Therapies that reconstruct and stabilize the healthy microbiota may prove more effective. For example, the pattern observed by Abreu et al. (34) suggests a promising new approach to the treatment of CRS via probiotic manipulation of the microbiota: following prior antimicrobial bacterial depletion in a murine model, inoculation with C. tuberculostearicum via application to the nares and inhalation led to the development of CRS-like symptoms (goblet cell hyperplasia and mucin hypersecretion), while Lactobacillus sakei coinoculation mitigated this effect. Other studies have similarly entertained the idea of microbial community rehabilitation as a viable avenue for the treatment of CRS, identifying the inhibitory effects of both S. epidermidis and Corynebacterium sp. against S. aureus (23, 186). Efforts aimed toward proactive restoration of the healthy microbiota remain promising avenues to explore. The potential of probiotic approaches for the treatment of CRS was extensively reviewed recently (187).
A further understanding of the complexities of microbial communities associated with inflammatory diseases such as CRS is vital for clarifying the extent to which antibiotics are appropriate as a treatment recourse or whether other strategies of modulating microbial communities, such as targeted probiotics, will prove more fruitful.
Anti-Inflammatory Drugs and Biologics
While the role of antimicrobials in the management of CRS remains a contentious issue, the use and effectiveness of anti-inflammatory drugs continue to be a mainstay. Intranasal corticosteroids administered via standard delivery methods have been shown to improve subjective and objective disease measures in both CRSsNP and CRSwNP patients, and a recent systematic evidence-based review recommended their continued use as a key therapeutic option for the medical management of CRS (5). Additionally, biologic therapies such as monoclonal antibodies (MAbs) that target specific inflammatory signaling molecules (for example, anti-IL-5 or anti-immunoglobulin E MAb) represent a further area of interest in the treatment of the inflammatory process underlying conditions such as CRS (188–190). The complex heterogeneity of inflammatory processes underlying CRS (likely including mixed inflammatory types) remains a complication for such specific targeted approaches to treating underlying inflammatory processes (5). However, an initial study of CRS highlighted anti-IL-5 MAb as a promising therapeutic option in cases of severe eosinophilic nasal polyposis (191).
Importantly, for our purposes here, where the microbiology of CRS is the chief focus, if microbes play a role in the ongoing disease process, novel approaches aimed at modifying the microbiota to return to a healthy state may better address underlying causative mechanisms, while anti-inflammatory drugs may be limited to mitigating symptoms. However, given the broad changes in the microenvironment within the inflamed sinonasal tract, anti-inflammatory drugs and/or inflammation-mediating biologics may be equally fundamental, even in attempts explicitly aimed at remedying the microbiota, by enabling environmental conditions that are more amenable to a healthy microbial community. Ultimately, the inherent complexities of both the inflammatory processes underlying the ongoing pathophysiology of the disease as well as the dynamics of the associated microbial communities will likely demand a multifaceted approach to the clinical management of complex chronic inflammatory mucosal conditions such as CRS.
FUTURE DIRECTIONS
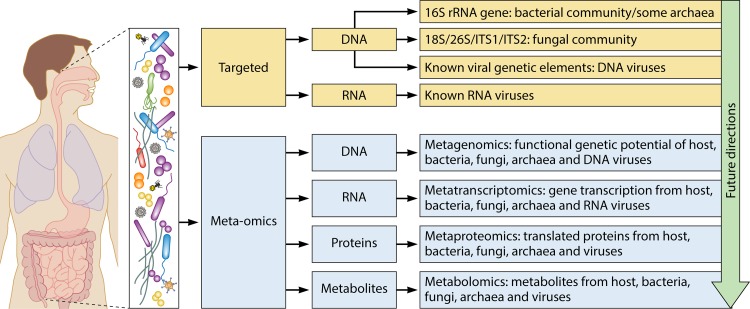
The advent of molecular microbiological techniques has enabled researchers to study in unprecedented depth the complex microbial communities, and their functional capabilities, that inhabit the human body. Large-scale projects, such as the Human Microbiome Project, describe both microbial diversity and function across a wide range of body habitats and provide attractive avenues for potential research and clinical studies (192). In what follows, we introduce the wider context within which CRS research fits, among other human studies regarding the role of host-associated microbial communities in health and disease, and offer prospective ideas for future CRS research (Table 2). An overview of current approaches to assessing microbial community structure, capability, and function is presented in Fig. 6 and is described in detail below.
TABLE 2.
Outstanding questions in CRS microbiology research
| Question |
|---|
| What are the dynamics of the sinonasal microbiota over time in both healthy and CRS subjects? |
| Do changes in the microbiota during standard medical management of CRS predict improvement or deterioration of disease progression? |
| What is the effect of systemic antibiotics on the sinonasal microbiota in both healthy and CRS subjects? |
| What is the efficacy of standard or culture-directed antibiotics in treatment of CRS? |
| Can virus-targeted DNA and RNA (metagenome and metatranscriptome) sequencing approaches furnish a more complete picture of the role of viruses in the microbiota? |
| Can improved subclassification of variants of disease better delineate distinct microbial associations? |
| Can routine assessment of “personalized microbial genomes”a via gene-targeted or metagenomics analyses identify biomarkers of disease and inform personalized treatment approaches? |
See reference 197.
FIG 6.
Future directions in microbiological research on inflammatory mucosal disease. CRS research thus far remains limited to gene-targeted assessments of community membership and structure, including bacterial 16S rRNA gene sequencing, fungal internal transcribed spacer (ITS) region sequencing, and known viral genetic elements. Approaches that sequence all DNA (metagenomics) or all transcribed RNA (metatranscriptomics) or that identify proteins (metaproteomics) or metabolites (metabolomics) will provide greater insights into the true diversity and structure, as well as the full genetic potential and in situ activity, of the mucosa-associated microbiota.
The Next Technological Frontier in Microbiological Research of CRS
Extensions of gene-targeted sequencing approaches.
While the single-gene approaches discussed above offer insights into the taxonomic diversity of a community, they are relatively uninformative with respect to in situ functional capacity. Furthermore, highly variable taxonomic profiles in both CRS-affected and unaffected sinonasal microbiota make it difficult to distinguish microbial biomarkers of disease. Community-wide functional capability can be indirectly investigated by using computational approaches such as PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) (193). The combined use of gene-targeted data and PICRUSt functional prediction software offers a preliminary look into the genetic potential of microbial communities and has successfully been applied in several gut microbiome studies to date (194–196).
Metagenomics.
In contrast to the above-mentioned gene-targeted studies, metagenomic studies sequence total DNA within a sample, identifying the full genetic potential of the microbiota associated with health and disease and providing increased resolution of all microbial members of a sample (bacterial, fungal, viral, and archaeal), potentially distinguishing between individual strains (197). Metagenomics has been successfully applied to a wide variety of host-associated environments, especially gut microbial communities (192, 198–200), and non-host-associated environments, including soil and water (201, 202). However, to date, there are no published metagenomic studies of the sinonasal microbiota. Recent pilot metagenomic studies have identified the difficulty in applying this technique to the system of CRS, where sampled DNA is overwhelmingly dominated by contaminating host DNA (>99%), leaving little sequencing information on the associated microbial community (B. Wagner Mackenzie, unpublished data). Nonetheless, the application of metagenomic sequencing to identify functional capacity, strain-level differences in composition, and the contribution of microbes to the pathogenesis of CRS represents the next logical step for research and may provide invaluable insight into the disease process of CRS and avenues for the development of novel approaches to treatment.
Metatranscriptomics.
Where gene-targeted sequencing elucidates microbial community composition and metagenomics identifies community-wide functional potential, metatranscriptomics offers a picture of in situ microbial activity. Sequencing of community RNA, in contrast to DNA, provides information regarding transcription levels of genes and a snapshot of total community activity (203–207). Metatranscriptomics, in conjunction with community structure data, can track whether shifts in community composition are associated with the differential expression of microbial genes in healthy and diseased individuals, which may better represent and explain functional differences related to disease. In one study, the application of metatranscriptomics to periodontal disease identified conserved changes in microbial community metabolism associated with this disease, against a background of high interpatient variability in community composition (205). Furthermore, metatranscriptomics can be used to track the microbial response to antibiotics and host-targeted therapies (207). Metatranscriptomics has already been successfully applied to other human microbiome fields, including gut, oral, and vaginal microbiomes (203, 205–208). Additionally, recent studies have begun to highlight the importance of distinguishing between live, active members of the microbiota and the remains of dead microbes that may still be identified by gene-targeted approaches (209, 210). While both are relevant to the dynamics of the microbial community and interactions with the host system, each will play distinct ecological roles. Metatranscriptomics studies can offer further insight into this largely unknown factor of microbiota community dynamics and its potential roles in inflammatory disease.
Metaproteomics.
While examination of transcribed RNA can elucidate microbial activity in situ, actual interactions within the microbiota, and between the microbiota and host tissues, take place on the level of the proteins subsequently translated. Metaproteomics typically employs mass spectrometry to examine the proteins present at any given time within a microbial community (211–214). This provides a direct picture of microbial activity and interactions as well as the potential for the identification of protein biomarkers of disease. In combination with metatranscriptomics, disparities between transcribed RNA and proteins present in a sample can also provide valuable insight into posttranslational processes in microbial communities. Metaproteomics has been successfully applied to a diverse range of microbial communities, including those associated with the human gut (215, 216), irritable bowel syndrome and Crohn's disease (217, 218), human saliva (219), and the open ocean (220), shedding light on biomarker protein profiles in disease, translational regulation patterns, bacterial interactions, and bacterial metabolism, including changes in nutrient utilization and energy transduction.
Metametabolomics.
Representing the final layer in the biological system, metabolomics examines both endogenous and exogenous small-molecule metabolites. Subsequently, metametabolomics involves the study of all metabolites within a given sample (including microbial communities) and can provide insight into finer-scale metabolic interactions within the microbiota as well as molecular modifiers important in both healthy and disease states (196, 221–223). Metametabolomics has already been applied in studies examining the influence of the microbiota on host metabolism (224, 225), the presence and importance of immunomodulatory microbiota-derived metabolites such as short-chain fatty acids in circulation within the host (226), and changes in xenobiotic metabolism (196, 227) and represents a logical next step in efforts to better understand the finest-scale processes taking place within the microbiota of individuals with inflammatory mucosal conditions such as CRS.
Ultimately, accurate modeling of human-associated microbial communities requires a cross-disciplinary approach that includes longitudinal studies of genetic, immunological, environmental, and microbiological facets. The use of gene-targeted sequencing, metagenomic, metatranscriptomic, metaproteomic, and metametabolomic techniques together offers an integrated approach to help elucidate the role of microbial communities in inflammatory diseases such as CRS. High-throughput next-generation sequencing techniques have dramatically improved our understanding of the role of microbes in other sites of the human body. These studies can help to serve as a guide for future CRS research with the overall aim of establishing predictive models of CRS-associated microbiomes and individualized, targeted treatments (228–230). Identifying trends in the structure and function of CRS-associated microbiota may provide further targets for therapies and treatments that address the causative mechanisms of disease rather than secondary symptoms and comorbidities alone.
SUMMARY AND CONCLUDING REMARKS
Despite considerable research over the last few decades, the role of microbes in CRS remains unclear. In part, this has arisen due to a lack of consensus between studies, with numerous examples of either opposing findings or a complete lack of evidence to implicate specific bacterial associations with CRS. Additionally, results from next-generation sequencing studies suggest that numerous bacteria commonly thought of as being pathogens and previously thought to be associated with the inflammatory process of CRS are also characteristic constituents of the complex polymicrobial communities associated with the healthy human mucosa. In contrast, recent studies that better capture the full complexity of the microbial communities associated with CRS reintroduce the possible importance of the microbiota either as a direct driver of the disease or as potentially being involved in its exacerbation. Nonetheless, a great deal remains to be understood.
The clinical implications of clarifying the role of the microbiota in CRS are manifold. Antibiotics remain a mainstay of the treatment of CRS at a significant cost to the health care system and with a risk for the further development of antimicrobial resistance, and yet the nature of the involvement of microbes in CRS remains unclear. Moreover, the efficacy of antibiotic treatment of CRS remains unknown, and antibiotic treatment could in principle act as an exacerbating factor if microbial dysbiosis proves to be an important facet of the pathophysiology of CRS.
It is increasingly apparent that there is a need to progress beyond simplistic models of traditional single-agent pathogenicity and instead embrace multifaceted perspectives that retain the inherent complexities of both the microbial and host systems and examine the dynamic ecological interactions within and between these two systems across space and time. Untangling the web of interactions between microbial communities and the host mucosa in the inflammatory disease process is vital for a better understanding of the progression of the pathophysiology of this disease. In this respect, recent technological advances in molecular microbiology will continue to offer new perspectives on many poorly understood diseases. An interdisciplinary approach is vital to further our understanding of diseases as multifaceted as CRS and may highlight key facets toward which targeted therapeutic efforts can be focused.
ACKNOWLEDGMENTS
Research on CRS in our laboratories is supported by the Garnett Passe and Rodney Williams Memorial Foundation, the Green Lane Research and Educational Fund, the Maurice and Phyllis Paykel Trust, and the University of Auckland.
We declare no conflict of interest.
Biographies

Michael Hoggard is a Ph.D. candidate in the School of Biological Sciences at the University of Auckland, New Zealand, with a research focus on processes and interactions in both the human and microbial aspects of the chronic inflammatory mucosal condition chronic rhinosinusitis. He received his bachelor's degrees in arts (B.A.) and science (B.Sc. [hons]) from the University of Auckland, New Zealand, with majors in philosophy, ecology, and biomedical science. He has been involved in research into the microbial ecology associated with a diverse range of host environments, including the human sinonasal tract in health and CRS, the ear in health and chronic suppurative otitis media, and marine invertebrates. More generally, his research interests focus on host-microbe interactions at the ecosystem level and the application of principles of ecological theory to better understand the nature of microbial roles in both health and disease.

Brett Wagner Mackenzie, M.Sc., is a Ph.D. candidate at the School of Medicine, Department of Surgery, at the University of Auckland in Auckland, New Zealand. Her thesis focuses on the role of the human sinonasal microbiome in chronic rhinosinusitis and how to better understand the role of the sinonasal microbiome during health and disease. She completed her undergraduate degree at Gonzaga University in Spokane, WA, and her master's degree at the School of Biological Sciences at the University of Auckland, New Zealand. Her research interests include host-microbiome interactions and microbial community network stability and response to disturbances.

Ravi Jain graduated from the University of Otago, New Zealand, with a bachelor's degree in medicine and surgery in 2008. He is currently an advanced trainee of the Royal Australasian College of Surgeons in Otolaryngology, Head and Neck Surgery, and a Ph.D. candidate at the University of Auckland, New Zealand. Over the last four years, his research has investigated the effects of perioperative therapies used in treating chronic rhinosinusitis.

Michael W. Taylor is an Associate Professor of Microbiology at the University of Auckland, New Zealand. He received his B.Sc. (Zoology) from the University of Otago, M.Sc. (Biology) from the University of Auckland, and Ph.D. (Marine Microbiology) from the University of New South Wales. Following postdoctoral research in microbial ecology at the University of Vienna, he started his own research group in Auckland in 2007. His research applies molecular biology approaches and advanced bioinformatics techniques to study the interactions between microbes and their animal (including human) “hosts.” He has recently focused on the human microbiome, particularly the sinonasal microbiota and the role of the gut microbiome in type 2 diabetes and autism spectrum disorder. Professor Taylor is currently Vice President of the New Zealand Microbiological Society, Co-Convenor of the New Zealand Microbial Ecology Consortium, and Board Member of the International Society for Microbial Ecology.

Kristi Biswas is currently a postdoctoral fellow at the Department of Surgery, University of Auckland, New Zealand. She obtained her Ph.D. in molecular microbiology in 2014 from the University of Auckland and completed her M.Sc. in 2007 at the University of Otago, New Zealand. The focus of her research has been on microbes associated with humans. More specifically, her research has moved towards investigating the interplay between the host immune responses and the associated microbes in the inflammatory disease chronic rhinosinusitis.

Richard G. Douglas, M.D., F.R.A.C.S., F.R.A.C.P., attended Medical School at the University of Auckland and trained in Otorhinolaryngology in New Zealand. He has completed fellowships in Clinical Immunology and Allergy at the Alfred Hospital in Melbourne, Australia, and in Rhinology at the Queen Elizabeth Hospital, Adelaide, Australia. He is currently an Associate Professor in the Department of Surgery of the University of Auckland and a rhinologist at Auckland Hospital. His main areas of research are the pathogenesis and treatment of chronic rhinosinusitis. Our understanding of the complex etiology of this condition is incomplete, and currently available treatments are less than perfect. CRS is very often treated with antibiotics, but evidence for their efficacy is poor. There is a strong research imperative to clarify the role of the microbiome and treatment options for this condition, which have been central themes in his involvement in CRS research for more than a decade.
REFERENCES
- 1.Hastan D, Fokkens WJ, Bachert C, Newson RB, Bislimovska J, Bockelbrink A, Bousquet PJ, Brozek G, Bruno A, Dahlen SE, Forsberg B, Gunnbjornsdottir M, Kasper L, Kramer U, Kowalski ML, Lange B, Lundback B, Salagean E, Todo-Bom A, Tomassen P, Toskala E, Van Drunen CM, Bousquet J, Zuberbier T, Jarvis D, Burney P. 2011. Chronic rhinosinusitis in Europe—an underestimated disease. A GA2LEN study. Allergy 66:1216–1223. doi: 10.1111/j.1398-9995.2011.02646.x. [DOI] [PubMed] [Google Scholar]
- 2.Fokkens WJ, Lund VJ, Mullol J, Bachert C, Alobid I, Baroody F, Cohen N, Cervin A, Douglas R, Gevaert P, Georgalas C, Goossens H, Harvey R, Hellings P, Hopkins C, Jones N, Joos G, Kalogjera L, Kern B, Kowalski M, Price D, Riechelmann H, Schlosser R, Senior B, Thomas M, Toskalaw E, Voegels R, Wang DY, Wormald PJ. 2012. EPOS 2012: European position paper on rhinosinusitis and nasal polyps 2012. A summary for otorhinolaryngologists. Rhinology 50:1–12. doi: 10.4193/Rhino50E2. [DOI] [PubMed] [Google Scholar]
- 3.Pleis J, Benson V, Schiller J. 2003. Summary health statistics for U.S. adults: National Health Interview Survey, 2000. Vital Heal Stat 10:1–132. [PubMed] [Google Scholar]
- 4.Van Crombruggen K, Zhang N, Gevaert P, Tomassen P, Bachert C. 2011. Pathogenesis of chronic rhinosinusitis: inflammation. J Allergy Clin Immunol 128:728–732. doi: 10.1016/j.jaci.2011.07.049. [DOI] [PubMed] [Google Scholar]
- 5.Orlandi RR, Kingdom TT, Hwang PH, Smith TL, Alt JA, Baroody FM, Batra PS, Bernal-Sprekelsen M, Bhattacharyya N, Chandra RK, Chiu A, Citardi MJ, Cohen NA, Delgaudio J, Desrosiers M, Dhong HJ, Douglas R, Ferguson B, Fokkens WJ, Georgalas C, Goldberg A, Gosepath J, Hamilos DL, Han JK, Harvey R, Hellings P, Hopkins C, Jankowski R, Javer AR, Kern R, Kountakis S, Kowalski ML, Lane A, Lanza DC, Lebowitz R, Lee HM, Lin SY, Lund V, Luong A, Mann W, Marple BF, Mcmains KC, Metson R, Naclerio R, Nayak JV, Otori N, Palmer JN, Parikh SR, Passali D, Peters A, et al. . 2016. International consensus statement on allergy and rhinology: rhinosinusitis. Int Forum Allergy Rhinol 6:S22–S209. doi: 10.1002/alr.21695. [DOI] [PubMed] [Google Scholar]
- 6.Bhattacharyya N. 2011. Incremental health care utilization and expenditures for chronic rhinosinusitis in the United States. Ann Otol Rhinol Laryngol 120:423–427. doi: 10.1177/000348941112000701. [DOI] [PubMed] [Google Scholar]
- 7.Cain RB, Lal D. 2013. Update on the management of chronic rhinosinusitis. Infect Drug Resist 6:1–14. doi: 10.2147/IDR.S26134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gliklich RE, Metson R. 1995. The health impact of chronic sinusitis in patients seeking otolaryngologic care. Otolaryngol Head Neck Surg 113:104–109. doi: 10.1016/S0194-5998(95)70152-4. [DOI] [PubMed] [Google Scholar]
- 9.Bhattacharyya N. 2009. Contemporary assessment of the disease burden of sinusitis. Am J Rhinol Allergy 23:392–395. doi: 10.2500/ajra.2009.23.3355. [DOI] [PubMed] [Google Scholar]
- 10.Soler ZM, Wittenberg E, Schlosser RJ, Mace JC, Smith TL. 2011. Health state utility values in patients undergoing endoscopic sinus surgery. Laryngoscope 121:2672–2678. doi: 10.1002/lary.21847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ray NF, Baraniuk JN, Thamer M, Rinehart CS, Gergen PJ, Kaliner M, Josephs S, Pung YH. 1999. Healthcare expenditures for sinusitis in 1996: contributions of asthma, rhinitis, and other airway disorders. J Allergy Clin Immunol 103:408–414. doi: 10.1016/S0091-6749(99)70464-1. [DOI] [PubMed] [Google Scholar]
- 12.Bhattacharyya N. 2003. The economic burden and symptom manifestations of chronic rhinosinusitis. Am J Rhinol 17:27–32. [PubMed] [Google Scholar]
- 13.Benninger MS, Ferguson BJ, Hadley JA, Hamilos DL, Jacobs M, Kennedy DW, Lanza DC, Marple BF, Osguthorpe JD, Stankiewicz JA, Anon J, Denneny J, Emanuel I, Levine H. 2003. Adult chronic rhinosinusitis: definitions, diagnosis, epidemiology, and pathophysiology. Otolaryngol Head Neck Surg 129:S1–S32. doi: 10.1016/S0194-5998(03)01397-4. [DOI] [PubMed] [Google Scholar]
- 14.Hsu J, Avila PC, Kern RC, Hayes MG, Schleimer RP, Pinto JM. 2013. Genetics of chronic rhinosinusitis: state of the field and directions forward. J Allergy Clin Immunol 131:977–993. doi: 10.1016/j.jaci.2013.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kennedy JL, Borish L. 2013. Chronic rhinosinusitis and antibiotics: the good, the bad, and the ugly. Am J Rhinol Allergy 27:467–472. doi: 10.2500/ajra.2013.27.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho I, Blaser MJ. 2012. The human microbiome: at the interface of health and disease. Nat Rev Genet 13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. 2012. Host-gut microbiota metabolic interactions. Science 336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 18.Lam K, Schleimer R, Kern RC. 2015. The etiology and pathogenesis of chronic rhinosinusitis: a review of current hypotheses. Curr Allergy Asthma Rep 15:41. doi: 10.1007/s11882-015-0540-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahdavinia M, Keshavarzian A, Tobin MC, Landay A, Schleimer RP. 2016. A comprehensive review of the nasal microbiome in chronic rhinosinusitis (CRS). Clin Exp Allergy 46:21–41. doi: 10.1111/cea.12666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JT, Frank DN, Ramakrishnan V. 2016. Microbiome of the paranasal sinuses: update and literature review. Am J Rhinol Allergy 30:3–16. doi: 10.2500/ajra.2016.30.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamad WA, Matar N, Elias M, Nasr M, Sarkis-Karam D, Hokayem N, Haddad A. 2009. Bacterial flora in normal adult maxillary sinuses. Am J Rhinol Allergy 23:261–263. doi: 10.2500/ajra.2009.23.3317. [DOI] [PubMed] [Google Scholar]
- 22.Rasmussen TT, Kirkeby LP, Poulsen K, Reinholdt J, Kilian M. 2000. Resident aerobic microbiota of the adult human nasal cavity. APMIS 108:663–675. doi: 10.1034/j.1600-0463.2000.d01-13.x. [DOI] [PubMed] [Google Scholar]
- 23.Uehara Y, Nakama H, Agematsu K, Uchida M, Kawakami Y, Abdul Fattah ASM, Maruchi N. 2000. Bacterial interference among nasal inhabitants: eradication of Staphylococcus aureus from nasal cavities by artificial implantation of Corynebacterium sp. J Hosp Infect 44:127–133. doi: 10.1053/jhin.1999.0680. [DOI] [PubMed] [Google Scholar]
- 24.Hauser LJ, Feazel LM, Ir D, Fang R, Wagner BD, Robertson CE, Frank DN, Ramakrishnan VR. 2015. Sinus culture poorly predicts resident microbiota. Int Forum Allergy Rhinol 5:3–9. doi: 10.1002/alr.21428. [DOI] [PubMed] [Google Scholar]
- 25.Tabet P, Endam LM, Boisvert P, Boulet L-P, Desrosiers M. 2015. Gram-negative bacterial carriage in chronic rhinosinusitis with nasal polyposis is not associated with more severe inflammation. Int Forum Allergy Rhinol 5:289–293. doi: 10.1002/alr.21481. [DOI] [PubMed] [Google Scholar]
- 26.Kaspar U, Kriegeskorte A, Schubert T, Peters G, Rudack C, Pieper DH, Wos-Oxley M, Becker K. 5 June 2015. The culturome of the human nose habitats reveals individual bacterial fingerprint patterns. Environ Microbiol doi: 10.1111/1462-2920.12891. [DOI] [PubMed] [Google Scholar]
- 27.Browne HP, Forster SC, Anonye BO, Kumar N, Neville BA, Stares MD, Goulding D, Lawley TD. 2016. Culturing of “unculturable” human microbiota reveals novel taxa and extensive sporulation. Nature 533:543–546. doi: 10.1038/nature17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hugenholtz P. 2002. Exploring prokaryotic diversity in the genomic era. Genome Biol 3:REVIEWS0003. doi: 10.1186/gb-2002-3-2-reviews0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramakrishnan VR, Hauser LJ, Feazel LM, Ir D, Robertson CE, Frank DN. 2015. Sinus microbiota varies among chronic rhinosinusitis phenotypes and predicts surgical outcome. J Allergy Clin Immunol 136:334–342. doi: 10.1016/j.jaci.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 30.Bassiouni A, Cleland EJ, Psaltis AJ, Vreugde S, Wormald PJ. 2015. Sinonasal microbiome sampling: a comparison of techniques. PLoS One 10:e0123216. doi: 10.1371/journal.pone.0123216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wos-Oxley ML, Plumeier I, von Eiff C, Taudien S, Platzer M, Vilchez-Vargas R, Becker K, Pieper DH. 2010. A poke into the diversity and associations within human anterior nare microbial communities. ISME J 4:839–851. doi: 10.1038/ismej.2010.15. [DOI] [PubMed] [Google Scholar]
- 32.Stressmann FA, Rogers GB, Chan SW, Howarth PH, Harries PG, Bruce KD, Salib RJ. 2011. Characterization of bacterial community diversity in chronic rhinosinusitis infections using novel culture-independent techniques. Am J Rhinol Allergy 25:133–140. doi: 10.2500/ajra.2011.25.3628. [DOI] [PubMed] [Google Scholar]
- 33.Feazel LM, Robertson CE, Ramakrishnan VR, Frank DN. 2012. Microbiome complexity and Staphylococcus aureus in chronic rhinosinusitis. Laryngoscope 122:467–472. doi: 10.1002/lary.22398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abreu NA, Nagalingam NA, Song Y, Roediger FC, Pletcher SD, Goldberg AN, Lynch S. 2012. Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis. Sci Transl Med 4:151ra124. doi: 10.1126/scitranslmed.3003783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aurora R, Chatterjee D, Hentzleman J, Prasad G, Sindwani R, Sanford T. 2013. Contrasting the microbiomes from healthy volunteers and patients with chronic rhinosinusitis. JAMA Otolaryngol Head Neck Surg 139:1328–1338. doi: 10.1001/jamaoto.2013.5465. [DOI] [PubMed] [Google Scholar]
- 36.Ramakrishnan VR, Feaze LM, Gitomer SA, Ir D, Robertson CE, Frank DN. 2013. The microbiome of the middle meatus in healthy adults. PLoS One 8:e85507. doi: 10.1371/journal.pone.0085507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan M, Pamp SJ, Fukuyama J, Hwang PH, Cho DY, Holmes S, Relman DA. 2013. Nasal microenvironments and interspecific interactions influence nasal microbiota complexity and S. aureus carriage. Cell Host Microbe 14:631–640. doi: 10.1016/j.chom.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bassis CM, Tang AL, Young VB, Pynnonen MA. 2014. The nasal cavity microbiota of healthy adults. Microbiome 2:27. doi: 10.1186/2049-2618-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu CM, Soldanova K, Nordstrom L, Dwan MG, Moss OL, Contente-Cuomo TL, Keim P, Price LB, Lane AP. 2013. Medical therapy reduces microbiota diversity and evenness in surgically recalcitrant chronic rhinosinusitis. Int Forum Allergy Rhinol 3:775–781. doi: 10.1002/alr.21195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biswas K, Hoggard M, Jain R, Taylor MW, Douglas RG. 2015. The nasal microbiota in health and disease: variation within and between subjects. Front Microbiol 9:134. doi: 10.3389/fmicb.2015.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramakrishnan VR, Frank DN. 2015. Impact of cigarette smoking on the middle meatus microbiome in health and chronic rhinosinusitis. Int Forum Allergy Rhinol 5:981–989. doi: 10.1002/alr.21626. [DOI] [PubMed] [Google Scholar]
- 42.Kim RJT, Biswas K, Hoggard M, Taylor MW, Douglas RG. 2015. Paired analysis of the microbiota of surface mucus and whole-tissue specimens in patients with chronic rhinosinusitis. Int Forum Allergy Rhinol 5:877–883. doi: 10.1002/alr.21600. [DOI] [PubMed] [Google Scholar]
- 43.Cleland EJ, Bassiouni A, Vreugde S, Wormald P-J. 2016. The bacterial microbiome in chronic rhinosinusitis: richness, diversity, postoperative changes, and patient outcomes. Am J Rhinol Allergy 30:37–43. doi: 10.2500/ajra.2016.30.4261. [DOI] [PubMed] [Google Scholar]
- 44.Tomassen P, Vandeplas G, Van Zele T, Cardell L-O, Arebro J, Olze H, Förster-Ruhrmann U, Kowalski ML, Olszewska-Ziąber A, Holtappels G, De Ruyck N, Wang X, Van Drunen C, Mullol J, Hellings P, Hox V, Toskala E, Scadding G, Lund V, Zhang L, Fokkens W, Bachert C. 2016. Inflammatory endotypes of chronic rhinosinusitis based on cluster analysis of biomarkers. J Allergy Clin Immunol 137:1457–1459. doi: 10.1016/j.jaci.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 45.Van Zele T, Claeys S, Gevaert P, Van Maele G, Holtappels G, Van Cauwenberge P, Bachert C. 2006. Differentiation of chronic sinus diseases by measurement of inflammatory mediators. Allergy 61:1280–1289. doi: 10.1111/j.1398-9995.2006.01225.x. [DOI] [PubMed] [Google Scholar]
- 46.Bachert C, Zhang N, Van Zele T, Gevaert P. 2012. Chronic rhinosinusitis: from one disease to different phenotypes. Pediatr Allergy Immunol 23(Suppl 22):2–4. doi: 10.1111/j.1399-3038.2012.01318.x. [DOI] [PubMed] [Google Scholar]
- 47.Park SJ, Kim TH, Jun YJ, Lee SH, Ryu HY, Jung KJ, Jung JY, Hwang GH, Lee SH. 2013. Chronic rhinosinusitis with polyps and without polyps is associated with increased expression of suppressors of cytokine signaling 1 and 3. J Allergy Clin Immunol 131:772–780. doi: 10.1016/j.jaci.2012.12.671. [DOI] [PubMed] [Google Scholar]
- 48.Bachert C, Gevaert P, Holtappels G, Johansson SGO, Van Cauwenberge P. 2001. Total and specific IgE in nasal polyps is related to local eosinophilic inflammation. J Allergy Clin Immunol 107:607–614. doi: 10.1067/mai.2001.112374. [DOI] [PubMed] [Google Scholar]
- 49.Kountakis SE, Arango P, Bradley D, Wade ZK, Borish L. 2004. Molecular and cellular staging for the severity of chronic rhinosinusitis. Laryngoscope 114:1895–1905. doi: 10.1097/01.mlg.0000147917.43615.c0. [DOI] [PubMed] [Google Scholar]
- 50.Bachert C, Zhang N, Holtappels G, De Lobel L, Van Cauwenberge P, Liu S, Lin P, Bousquet J, Van Steen K. 2010. Presence of IL-5 protein and IgE antibodies to staphylococcal enterotoxins in nasal polyps is associated with comorbid asthma. J Allergy Clin Immunol 126:962–968. doi: 10.1016/j.jaci.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 51.Chaaban MR, Walsh EM, Woodworth BA. 2013. Epidemiology and differential diagnosis of nasal polyps. Am J Rhinol Allergy 27:473–478. doi: 10.2500/ajra.2013.27.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Alarcón A, Steinke JW, Caughey R, Barekzi E, Hise K, Gross CW, Han JK, Borish L. 2006. Expression of leukotriene C4 synthase and plasminogen activator inhibitor 1 gene promoter polymorphisms in sinusitis. Am J Rhinol 20:545–549. doi: 10.2500/ajr.2006.20.2934. [DOI] [PubMed] [Google Scholar]
- 53.Han JK. 2013. Subclassification of chronic rhinosinusitis. Laryngoscope 123(Suppl 2):S15–S27. doi: 10.1002/lary.23979. [DOI] [PubMed] [Google Scholar]
- 54.Boase S, Foreman A, Cleland E, Tan L, Melton-Kreft R, Pant H, Hu FZ, Ehrlich GD, Wormald P-J. 2013. The microbiome of chronic rhinosinusitis: culture, molecular diagnostics and biofilm detection. BMC Infect Dis 13:210. doi: 10.1186/1471-2334-13-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choi EB, Hong SW, Kim DK, Jeon SG, Kim KR, Cho SH, Gho YS, Jee YK, Kim YK. 2014. Decreased diversity of nasal microbiota and their secreted extracellular vesicles in patients with chronic rhinosinusitis based on a metagenomic analysis. Allergy 69:517–526. doi: 10.1111/all.12374. [DOI] [PubMed] [Google Scholar]
- 56.Heymans F, Fischer A, Stow NW, Girard M, Vourexakis Z, Des Courtis A, Renzi G, Huggler E, Vlaminck S, Bonfils P, Mladina R, Lund V, Schrenzel J, Francois P, Lacroix JS. 2010. Screening for staphylococcal superantigen genes shows no correlation with the presence or the severity of chronic rhinosinusitis and nasal polyposis. PLoS One 5:e9525. doi: 10.1371/journal.pone.0009525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rom D, Snidvongs K, Sacks P-L, Dalgorf D, Pratt E, Earls P, Sacks R, Harvey RJ. 2014. The impact of culturable bacterial community on histopathology in chronic rhinosinusitis. Int Forum Allergy Rhinol 4:29–33. doi: 10.1002/alr.21220. [DOI] [PubMed] [Google Scholar]
- 58.Plouin-Gaudon I, Clement S, Huggler E, Chaponnier C, François P, Lew D, Schrenzei J, Vaudaux P, Lacroix JS. 2006. Intracellular residency is frequently associated with recurrent Staphylococcus aureus rhinosinusitis. Rhinology 44:249–254. [PubMed] [Google Scholar]
- 59.Stow NW, Douglas R, Tantilipikorn P, Lacroix JS. 2010. Superantigens. Otolaryngol Clin North Am 43:489–502. doi: 10.1016/j.otc.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 60.Seiberling KA, Conley DB, Tripathi A, Grammer LC, Shuh L, Haines GK, Schleimer R, Kern RC. 2005. Superantigens and chronic rhinosinusitis: detection of staphylococcal exotoxins in nasal polyps. Laryngoscope 115:1580–1585. doi: 10.1097/01.mlg.0000168111.11802.9c. [DOI] [PubMed] [Google Scholar]
- 61.Yu R-L, Dong Z. 2009. Proinflammatory impact of Staphylococcus aureus enterotoxin B on human nasal epithelial cells and inhibition by dexamethasone. Am J Rhinol Allergy 23:15–20. doi: 10.2500/ajra.2009.23.3252. [DOI] [PubMed] [Google Scholar]
- 62.Okano M, Fujiwara T, Kariya S, Haruna T, Higaki T, Noyama Y, Makihara SI, Kanai K, Nishizaki K. 2015. Staphylococcal protein A-formulated immune complexes suppress enterotoxin-induced cellular responses in nasal polyps. J Allergy Clin Immunol 136:343–350. doi: 10.1016/j.jaci.2014.10.058. [DOI] [PubMed] [Google Scholar]
- 63.Cantero D, Cooksley C, Bassiouni A, Tran HB, Roscioli E, Wormald PJ, Vreugde S. 2015. Staphylococcus aureus biofilms induce apoptosis and expression of interferon-γ, interleukin-10, and interleukin-17A on human sinonasal explants. Am J Rhinol Allergy 29:23–28. doi: 10.2500/ajra.2015.29.4130. [DOI] [PubMed] [Google Scholar]
- 64.Carey RM, Workman AD, Chen B, Adappa ND, Palmer JN, Kennedy DW, Lee RJ, Cohen NA. 2015. Staphylococcus aureus triggers nitric oxide production in human upper airway epithelium. Int Forum Allergy Rhinol 5:808–813. doi: 10.1002/alr.21568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakamura Y, Oscherwitz J, Cease KB, Chan SM, Muñoz-Planillo R, Hasegawa M, Villaruz AE, Cheung GYC, McGavin MJ, Travers JB, Otto M, Inohara N, Núñez G. 2013. Staphylococcus δ-toxin induces allergic skin disease by activating mast cells. Nature 503:397–401. doi: 10.1038/nature12655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Corriveau MN, Zhang N, Holtappels G, Van Roy N, Bachert C. 2009. Detection of Staphylococcus aureus in nasal tissue with peptide nucleic acid-fluorescence in situ hybridization. Am J Rhinol Allergy 23:461–465. doi: 10.2500/ajra.2009.23.3367. [DOI] [PubMed] [Google Scholar]
- 67.Sachse F, Becker K, Von Eiff C, Metze D, Rudack C. 2010. Staphylococcus aureus invades the epithelium in nasal polyposis and induces IL-6 in nasal epithelial cells in vitro. Allergy 65:1430–1437. doi: 10.1111/j.1398-9995.2010.02381.x. [DOI] [PubMed] [Google Scholar]
- 68.Tan NCW, Foreman A, Jardeleza C, Douglas R, Tran H, Wormald PJ. 2012. The multiplicity of Staphylococcus aureus in chronic rhinosinusitis: correlating surface biofilm and intracellular residence. Laryngoscope 122:1655–1660. doi: 10.1002/lary.23317. [DOI] [PubMed] [Google Scholar]
- 69.Wood AJ, Fraser JD, Swift S, Patterson-Emanuelson EAC, Amirapu S, Douglas RG. 2012. Intramucosal bacterial microcolonies exist in chronic rhinosinusitis without inducing a local immune response. Am J Rhinol Allergy 26:265–270. doi: 10.2500/ajra.2012.26.3779. [DOI] [PubMed] [Google Scholar]
- 70.Tan NCW, Foreman A, Jardeleza C, Douglas R, Vreugde S, Wormald PJ. 2013. Intracellular Staphylococcus aureus: the Trojan horse of recalcitrant chronic rhinosinusitis? Int Forum Allergy Rhinol 3:261–266. doi: 10.1002/alr.21154. [DOI] [PubMed] [Google Scholar]
- 71.Peters BM, Jabra-Rizk MA, Scheper MA, Leid JG, Costerton JW, Shirtliff ME. 2010. Microbial interactions and differential protein expression in Staphylococcus aureus-Candida albicans dual-species biofilms. FEMS Immunol Med Microbiol 59:493–508. doi: 10.1111/j.1574-695X.2010.00710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schlecht LM, Peters BM, Krom BP, Freiberg JA, Hänsch GM, Filler SG, Jabra-Rizk MA, Shirtliff ME. 2015. Systemic Staphylococcus aureus infection mediated by Candida albicans hyphal invasion of mucosal tissue. Microbiology 161:168–181. doi: 10.1099/mic.0.083485-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tan NCW, Cooksley CM, Roscioli E, Drilling AJ, Douglas R, Wormald PJ, Vreugde S. 2014. Small-colony variants and phenotype switching of intracellular Staphylococcus aureus in chronic rhinosinusitis. Allergy 69:1364–1371. doi: 10.1111/all.12457. [DOI] [PubMed] [Google Scholar]
- 74.Gitomer SA, Ramakrishnan VR, Malcolm KC, Kofonow JM, Ir D, Frank DN. 2015. Initial investigation of small colony variants of Staphylococcus aureus in chronic rhinosinusitis. Am J Rhinol Allergy 29:29–34. doi: 10.2500/ajra.2015.29.4133. [DOI] [PubMed] [Google Scholar]
- 75.Chalermwatanachai T, Zhang N, Holtappels G, Bachert C. 2015. Association of mucosal organisms with patterns of inflammation in chronic rhinosinusitis. PLoS One 10:e0136068. doi: 10.1371/journal.pone.0136068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, Huttenhower C. 2012. Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol 8:e1002606. doi: 10.1371/journal.pcbi.1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zelezniak A, Andrejev S, Ponomarova O, Mende DR, Bork P, Patil KR. 2015. Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc Natl Acad Sci U S A 112:6449–6454. doi: 10.1073/pnas.1421834112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bomar L, Brugger SD, Yost BH, Davies SS, Lemon KP. 2016. Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. mBio 7:e01725-15. doi: 10.1128/mBio.01725-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fyhrquist N, Salava A, Auvinen P, Lauerma A. 2016. Skin biomes. Curr Allergy Asthma Rep 16:40. doi: 10.1007/s11882-016-0618-5. [DOI] [PubMed] [Google Scholar]
- 80.Zipperer A, Konnerth MC, Laux C, Berscheid A, Janek D, Weidenmaier C, Burian M, Schilling NA, Slavetinsky C, Marschal M, Willmann M, Kalbacher H, Schittek B, Brötz-Oesterhelt H, Grond S, Peschel A, Krismer B. 2016. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 535:511–516. doi: 10.1038/nature18634. [DOI] [PubMed] [Google Scholar]
- 81.Boucher Y, Nesbø CL, Doolittle WF. 2001. Microbial genomes: dealing with diversity. Curr Opin Microbiol 4:285–289. doi: 10.1016/S1369-5274(00)00204-6. [DOI] [PubMed] [Google Scholar]
- 82.Nadell CD, Drescher K, Wingreen NS, Bassler BL. 2015. Extracellular matrix structure governs invasion resistance in bacterial biofilms. ISME J 9:1700–1709. doi: 10.1038/ismej.2014.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Davies D. 2003. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov 2:114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 84.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 85.Costerton WJ, Cheng K-J, Greesy GG, Ladd TI, Nickel JC, Dasgupta M, Marrie TJ. 1987. Bacterial biofilms in nature and disease. Annu Rev Microbiol 41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 86.Al-Mutairi D, Kilty SJ. 2011. Bacterial biofilms and the pathophysiology of chronic rhinosinusitis. Curr Opin Allergy Clin Immunol 11:18–23. doi: 10.1097/ACI.0b013e3283423376. [DOI] [PubMed] [Google Scholar]
- 87.Ferguson BJ, Stolz DB. 2005. Demonstration of biofilm in human bacterial chronic rhinosinusitis. Am J Rhinol 19:452–457. [PubMed] [Google Scholar]
- 88.Sanderson AR, Leid JG, Hunsaker D. 2006. Bacterial biofilms on the sinus mucosa of human subjects with chronic rhinosinusitis. Laryngoscope 116:1121–1126. doi: 10.1097/01.mlg.0000221954.05467.54. [DOI] [PubMed] [Google Scholar]
- 89.Prince AA, Steiger JD, Khalid AN, Dogrhamji L, Reger C, Eau Claire S, Chiu AG, Kennedy DW, Palmer JN, Cohen NA. 2008. Prevalence of biofilm-forming bacteria in chronic rhinosinusitis. Am J Rhinol 22:239–245. doi: 10.2500/ajr.2008.22.3180. [DOI] [PubMed] [Google Scholar]
- 90.Singh P, Mehta R, Agarwal S, Mishra P. 2015. Bacterial biofilm on the sinus mucosa of healthy subjects and patients with chronic rhinosinusitis (with or without nasal polyposis). J Laryngol Otol 129:46–49. doi: 10.1017/S002221511400303X. [DOI] [PubMed] [Google Scholar]
- 91.Foreman A, Psaltis AJ, Tan LW, Wormald P-J. 2009. Characterization of bacterial and fungal biofilms in chronic rhinosinusitis. Am J Rhinol Allergy 23:556–561. doi: 10.2500/ajra.2009.23.3413. [DOI] [PubMed] [Google Scholar]
- 92.Healy DY, Leid JG, Sanderson AR, Hunsaker DH. 2008. Biofilms with fungi in chronic rhinosinusitis. Otolaryngol Head Neck Surg 138:641–647. doi: 10.1016/j.otohns.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 93.Wargo MJ, Hogan DA. 2006. Fungal-bacterial interactions: a mixed bag of mingling microbes. Curr Opin Microbiol 9:359–364. doi: 10.1016/j.mib.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 94.Sanclement JA, Webster P, Thomas J, Ramadan HH. 2005. Bacterial biofilms in surgical specimens of patients with chronic rhinosinusitis. Laryngoscope 115:578–582. doi: 10.1097/01.mlg.0000161346.30752.18. [DOI] [PubMed] [Google Scholar]
- 95.Bendouah Z, Barbeau J, Hamad WA, Desrosiers M. 2006. Biofilm formation by Staphylococcus aureus and Pseudomonas aeruginosa is associated with an unfavorable evolution after surgery for chronic sinusitis and nasal polyposis. Otolaryngol Head Neck Surg 134:991–996. doi: 10.1016/j.otohns.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 96.Psaltis AJ, Ha KR, Beule AG, Tan LW, Wormald P-J. 2007. Confocal scanning laser microscopy evidence of biofilms in patients with chronic rhinosinusitis. Laryngoscope 117:1302–1306. doi: 10.1097/MLG.0b013e31806009b0. [DOI] [PubMed] [Google Scholar]
- 97.Wood AJ, Fraser J, Swift S, Amirapu S, Douglas RG. 2011. Are biofilms associated with an inflammatory response in chronic rhinosinusitis? Int Forum Allergy Rhinol 1:335–339. doi: 10.1002/alr.20060. [DOI] [PubMed] [Google Scholar]
- 98.Foreman A, Holtappels G, Psaltis AJ, Jervis-Bardy J, Field J, Wormald P-J, Bachert C. 2011. Adaptive immune responses in Staphylococcus aureus biofilm-associated chronic rhinosinusitis. Allergy 66:1449–1456. doi: 10.1111/j.1398-9995.2011.02678.x. [DOI] [PubMed] [Google Scholar]
- 99.Mao YJ, Chen HH, Wang B, Liu X, Xiong GY. 2015. Increased expression of MUC5AC and MUC5B promoting bacterial biofilm formation in chronic rhinosinusitis patients. Auris Nasus Larynx 42:294–298. doi: 10.1016/j.anl.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 100.Jain R, Douglas R. 2014. When and how should we treat biofilms in chronic sinusitis? Curr Opin Otolaryngol Head Neck Surg 22:16–21. doi: 10.1097/MOO.0000000000000010. [DOI] [PubMed] [Google Scholar]
- 101.Macia L, Thorburn AN, Binge LC, Marino E, Rogers KE, Maslowski KM, Vieira AT, Kranich J, Mackay CR. 2012. Microbial influences on epithelial integrity and immune function as a basis for inflammatory diseases. Immunol Rev 245:164–176. doi: 10.1111/j.1600-065X.2011.01080.x. [DOI] [PubMed] [Google Scholar]
- 102.Jahrsdoerfer R, Ejercito V, Johns M, Cantrell R, Sydnor J. 1979. Aspergillosis of the nose and paranasal sinuses. Am J Otolaryngol 1:6–14. doi: 10.1016/S0196-0709(79)80003-4. [DOI] [PubMed] [Google Scholar]
- 103.Katzenstein A-LA, Sale SR, Greenberger PA. 1983. Allergic Aspergillus sinusitis: a newly recognized form of sinusitis. J Allergy Clin Immunol 72:89–93. doi: 10.1016/0091-6749(83)90057-X. [DOI] [PubMed] [Google Scholar]
- 104.DeShazo RD, Swain RE. 1995. Diagnostic criteria for allergic fungal sinusitis. J Allergy Clin Immunol 96:24–35. doi: 10.1016/S0091-6749(95)70029-3. [DOI] [PubMed] [Google Scholar]
- 105.Laury AM, Wise SK. 2013. Allergic fungal rhinosinusitis. Am J Rhinol Allergy 27(Suppl 1):S26–S27. doi: 10.2500/ajra.2013.27.3891. [DOI] [PubMed] [Google Scholar]
- 106.Braun H, Buzina W, Freudenschuss K, Beham A, Stammberger H. 2003. “Eosinophilic fungal rhinosinusitis”: a common disorder in Europe? Laryngoscope 113:264–269. doi: 10.1097/00005537-200302000-00013. [DOI] [PubMed] [Google Scholar]
- 107.Ponikau JU, Sherris DA, Kern EB, Homburger HA, Frigas E, Gaffey TA, Roberts GD. 1999. The diagnosis and incidence of allergic fungal sinusitis. Mayo Clin Proc 74:877–884. doi: 10.4065/74.9.877. [DOI] [PubMed] [Google Scholar]
- 108.Lackner A, Stammberger H, Buzina W, Freudenschuss K, Panzitt T, Schosteritsch S, Braun H. 2005. Fungi: a normal content of human nasal mucus. Am J Rhinol 19:125–129. [PubMed] [Google Scholar]
- 109.Shin S-H, Ponikau JU, Sherris DA, Congdon D, Frigas E, Homburger HA, Swanson MC, Gleich GJ, Kita H. 2004. Chronic rhinosinusitis: an enhanced immune response to ubiquitous airborne fungi. J Allergy Clin Immunol 114:1369–1375. doi: 10.1016/j.jaci.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 110.Sasama J, Sherris DA, Shin S-H, Kephart GM, Kern EB, Ponikau JU. 2005. New paradigm for the roles of fungi and eosinophils in chronic rhinosinusitis. Curr Opin Otolaryngol Head Neck Surg 13:2–8. doi: 10.1097/00020840-200502000-00003. [DOI] [PubMed] [Google Scholar]
- 111.Ponikau JU, Sherris DA, Kephart GM, Adolphson C, Kita H. 2006. The role of ubiquitous airborne fungi in chronic rhinosinusitis. Clin Rev Allergy Immunol 30:187–194. doi: 10.1385/CRIAI:30:3:187. [DOI] [PubMed] [Google Scholar]
- 112.Soler ZM, Schlosser RJ. 2012. The role of fungi in diseases of the nose and sinuses. Am J Rhinol Allergy 26:351–358. doi: 10.2500/ajra.2012.26.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Collins M, Nair S, Smith W, Kette F, Gillis D, Wormald PJ. 2004. Role of local immunoglobulin E production in the pathophysiology of noninvasive fungal sinusitis. Laryngoscope 114:1242–1246. doi: 10.1097/00005537-200407000-00019. [DOI] [PubMed] [Google Scholar]
- 114.Pant H, Kette FE, Smith WB, Wormald PJ, Macardle PJ. 2005. Fungal-specific humoral response in eosinophilic mucus chronic rhinosinusitis. Laryngoscope 115:601–606. doi: 10.1097/01.mlg.0000161341.00258.54. [DOI] [PubMed] [Google Scholar]
- 115.Ryan MW, Marple BF. 2007. Allergic fungal rhinosinusitis: diagnosis and management. Curr Opin Otolaryngol Head Neck Surg 15:18–22. doi: 10.1097/MOO.0b013e328013dbd9. [DOI] [PubMed] [Google Scholar]
- 116.Ebbens FA, Georgalas C, Rinia AB, van Drunen CM, Lund VJ, Fokkens WJ. 2007. The fungal debate: where do we stand today? Rhinology 45:178–189. [PubMed] [Google Scholar]
- 117.Wise SK, Ahn CN, Lathers DMR, Mulligan RM, Schlosser RJ. 2008. Antigen-specific IgE in sinus mucosa of allergic fungal rhinosinusitis patients. Am J Rhinol 22:451–456. doi: 10.2500/ajr.2008.22.3227. [DOI] [PubMed] [Google Scholar]
- 118.Ebbens FA, Georgalas C, Fokkens WJ. 2009. Fungus as the cause of chronic rhinosinusitis: the case remains unproven. Curr Opin Otolaryngol Head Neck Surg 17:43–49. doi: 10.1097/MOO.0b013e32831de91e. [DOI] [PubMed] [Google Scholar]
- 119.Hutcheson PS, Schubert MS, Slavin RG. 2010. Distinctions between allergic fungal rhinosinusitis and chronic rhinosinusitis. Am J Rhinol Allergy 24:405–408. doi: 10.2500/ajra.2010.24.3533. [DOI] [PubMed] [Google Scholar]
- 120.Shin S-H, Lee Y-H, Jeon C. 2006. Protease-dependent activation of nasal polyp epithelial cells by airborne fungi leads to migration of eosinophils and neutrophils. Acta Otolaryngol 126:1286–1294. doi: 10.1080/00016480500395179. [DOI] [PubMed] [Google Scholar]
- 121.Ebert CS, McKinney KA, Urrutia G, Wu M, Rose AS, Fleischman GM, Thorp B, Senior BA, Zanation AM. 2014. Expression of protease-activated receptors in allergic fungal rhinosinusitis. Int Forum Allergy Rhinol 4:266–271. doi: 10.1002/alr.21295. [DOI] [PubMed] [Google Scholar]
- 122.Inoue Y, Matsuwaki Y, Shin S-H, Ponikau JU, Kita H. 2005. Nonpathogenic, environmental fungi induce activation and degranulation of human eosinophils. J Immunol 175:5439–5447. doi: 10.4049/jimmunol.175.8.5439. [DOI] [PubMed] [Google Scholar]
- 123.Den Beste KA, Hoddeson EK, Parkos CA, Nusrat A, Wise SK. 2013. Epithelial permeability alterations in an in vitro air-liquid interface model of allergic fungal rhinosinusitis. Int Forum Allergy Rhinol 3:19–25. doi: 10.1002/alr.21077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tieu DD, Kern RC, Schleimer RP. 2009. Alterations in epithelial barrier function and host defense responses in chronic rhinosinusitis. J Allergy Clin Immunol 124:37–42. doi: 10.1016/j.jaci.2009.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Allison DL, Willems HME, Jayatilake JAMS, Bruno VM, Peters BM, Shirtliff ME. 13 May 2016. Candida-bacteria interactions: their impact on human disease. Microbiol Spectr doi: 10.1128/microbiolspec.VMBF-0030-2016. [DOI] [PubMed] [Google Scholar]
- 126.Clark DW, Wenaas A, Luong A, Citardi MJ, Fakhri S. 2013. Staphylococcus aureus prevalence in allergic fungal rhinosinusitis vs other subsets of chronic rhinosinusitis with nasal polyps. Int Forum Allergy Rhinol 3:89–93. doi: 10.1002/alr.21090. [DOI] [PubMed] [Google Scholar]
- 127.Shirtliff ME, Peters BM, Jabra-Rizk MA. 2009. Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol Lett 299:1–8. doi: 10.1111/j.1574-6968.2009.01668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hogan DA, Kolter R. 2002. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science 296:2229–2232. doi: 10.1126/science.1070784. [DOI] [PubMed] [Google Scholar]
- 129.Hogan DA, Vik Å, Kolter R. 2004. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol Microbiol 54:1212–1223. doi: 10.1111/j.1365-2958.2004.04349.x. [DOI] [PubMed] [Google Scholar]
- 130.Kaur J, Pethani BP, Kumar S, Kim M, Sunna A, Kautto L, Penesyan A, Paulsen IT, Nevalainen H. 2015. Pseudomonas aeruginosa inhibits the growth of Scedosporium aurantiacum, an opportunistic fungal pathogen isolated from the lungs of cystic fibrosis patients. Front Microbiol 6:866. doi: 10.3389/fmicb.2015.00866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shirazi F, Ferreira JAG, Stevens DA, Clemons KV, Kontoyiannis DP. 2016. Biofilm filtrates of Pseudomonas aeruginosa strains isolated from cystic fibrosis patients inhibit preformed Aspergillus fumigatus biofilms via apoptosis. PLoS One 11:e0150155. doi: 10.1371/journal.pone.0150155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Huffnagle GB, Noverr MC. 2013. The emerging world of the fungal microbiome. Trends Microbiol 21:334–341. doi: 10.1016/j.tim.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cleland EJ, Bassioni A, Boase S, Dowd S, Vreugde S, Wormald PJ. 2014. The fungal microbiome in chronic rhinosinusitis: richness, diversity, postoperative changes and patient outcomes. Int Forum Allergy Rhinol 4:259–265. doi: 10.1002/alr.21297. [DOI] [PubMed] [Google Scholar]
- 134.Chakrabarti A, Denning DW, Ferguson BJ, Ponikau J, Buzina W, Kita H, Marple B, Panda N, Vlaminck S, Kauffmann-Lacroix C, Das A, Singh P, Taj-Aldeen SJ, Kantarcioglu AS, Handa KK, Gupta A, Thungabathra M, Shivaprakash MR, Bal A, Fothergill A, Radotra BD. 2009. Fungal rhinosinusitis: a categorization and definitional schema addressing current controversies. Laryngoscope 119:1809–1818. doi: 10.1002/lary.20520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fokkens WJ, van Drunen C, Georgalas C, Ebbens F. 2012. Role of fungi in pathogenesis of chronic rhinosinusitis: the hypothesis rejected. Curr Opin Otolaryngol Head Neck Surg 20:19–23. doi: 10.1097/MOO.0b013e32834e9084. [DOI] [PubMed] [Google Scholar]
- 136.Wang JH, Kwon HJ, Jang YJ. 2009. Rhinovirus enhances various bacterial adhesions to nasal epithelial cells simultaneously. Laryngoscope 119:1406–1411. doi: 10.1002/lary.20498. [DOI] [PubMed] [Google Scholar]
- 137.Yeo N-K, Jang YJ. 2010. Rhinovirus infection-induced alteration of tight junction and adherens junction components in human nasal epithelial cells. Laryngoscope 120:346–352. doi: 10.1002/lary.20764. [DOI] [PubMed] [Google Scholar]
- 138.Busse WW, Lemanske RF, Gern JE. 2010. Role of viral respiratory infections in asthma and asthma exacerbations. Lancet 376:826–834. doi: 10.1016/S0140-6736(10)61380-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Goffard A, Lambert V, Salleron J, Herwegh S, Engelmann I, Pinel C, Pin I, Perrez T, Prevotat A, Dewilde A, Delhaes L. 2014. Virus and cystic fibrosis: rhinoviruses are associated with exacerbations in adult patients. J Clin Virol 60:147–153. doi: 10.1016/j.jcv.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kapur N, Mackay IM, Sloots TP, Masters IB, Chang AB. 2014. Respiratory viruses in exacerbations of non-cystic fibrosis bronchiectasis in children. Arch Dis Child 99:749–753. doi: 10.1136/archdischild-2013-305147. [DOI] [PubMed] [Google Scholar]
- 141.Jackson DJ, Johnston SL. 2010. The role of viruses in acute exacerbations of asthma. J Allergy Clin Immunol 125:1178–1187. doi: 10.1016/j.jaci.2010.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kuyper LM, Pare PD, Hogg JC, Lambert RK, Ionescu D, Woods R, Bai TR. 2003. Characterization of airway plugging in fatal asthma. Am J Med 115:6–11. doi: 10.1016/S0002-9343(03)00241-9. [DOI] [PubMed] [Google Scholar]
- 143.Rogers DF. 2003. The airway goblet cell. Int J Biochem Cell Biol 35:1–6. doi: 10.1016/S1357-2725(02)00083-3. [DOI] [PubMed] [Google Scholar]
- 144.He S-H, Zheng J, Duan M-K. 2004. Induction of mucin secretion from human bronchial tissue and epithelial cells by rhinovirus and lipopolysaccharide. Acta Pharmacol Sin 25:1176–1181. [PubMed] [Google Scholar]
- 145.Cho GS, Moon BJ, Lee BJ, Gong CH, Kim NH, Kim YS, Kim HS, Jang YJ. 2013. High rates of detection of respiratory viruses in the nasal washes and mucosae of patients with chronic rhinosinusitis. J Clin Microbiol 51:979–984. doi: 10.1128/JCM.02806-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rowan NR, Lee S, Sahu N, Kanaan A, Cox S, Phillips CD, Wang EW. 2015. The role of viruses in the clinical presentation of chronic rhinosinusitis. Am J Rhinol Allergy 29:197–200. doi: 10.2500/ajra.2015.29.4242. [DOI] [PubMed] [Google Scholar]
- 147.Lee SB, Yi JS, Lee BJ, Gong CH, Kim NH, Joo CH, Jang YJ. 2015. Human rhinovirus serotypes in the nasal washes and mucosa of patients with chronic rhinosinusitis. Int Forum Allergy Rhinol 5:197–203. doi: 10.1002/alr.21472. [DOI] [PubMed] [Google Scholar]
- 148.Yuta A, Doyle WJ, Gaumond E, Ali M, Tamarkin L, Baraniuk JN, Van Deusen M, Cohen S, Skoner DP. 1998. Rhinovirus infection induces mucus hypersecretion. Am J Physiol 274:L1017–L1023. [DOI] [PubMed] [Google Scholar]
- 149.Wang JH, Kwon HJ, Jang YJ. 2009. Rhinovirus upregulates matrix metalloproteinase-2, matrix metalloproteinase-9, and vascular endothelial growth factor expression in nasal polyp fibroblasts. Laryngoscope 119:1834–1838. doi: 10.1002/lary.20574. [DOI] [PubMed] [Google Scholar]
- 150.Wang JH, Kwon HJ, Chung Y-S, Lee B-J, Jang YJ. 2008. Infection rate and virus-induced cytokine secretion in experimental rhinovirus infection in mucosal organ culture. Arch Otolaryngol Head Neck Surg 134:424–427. doi: 10.1001/archotol.134.4.424. [DOI] [PubMed] [Google Scholar]
- 151.Ramadan HH, Farr RW, Wetmore SJ. 1997. Adenovirus and respiratory syncytial virus in chronic sinusitis using polymerase chain reaction. Laryngoscope 107:923–925. doi: 10.1097/00005537-199707000-00017. [DOI] [PubMed] [Google Scholar]
- 152.Jang YJ, Kwon H-J, Park H-W, Lee B-J. 2006. Detection of rhinovirus in turbinate epithelial cells of chronic sinusitis. Am J Rhinol 20:634–636. doi: 10.2500/ajr.2006.20.2899. [DOI] [PubMed] [Google Scholar]
- 153.Abeles SR, Pride DT. 2014. Molecular bases and role of viruses in the human microbiome. J Mol Biol 426:3892–3906. doi: 10.1016/j.jmb.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Virgin HW. 2014. The virome in mammalian physiology and disease. Cell 157:142–150. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lysholm F, Wetterbom A, Lindau C, Darban H, Bjerkner A, Fahlander K, Lindberg AM, Persson B, Allander T, Andersson B. 2012. Characterization of the viral microbiome in patients with severe lower respiratory tract infections, using metagenomic sequencing. PLoS One 7:e30875. doi: 10.1371/journal.pone.0030875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Breitbart M, Hewson I, Felts B, Mahaffy JM, Nulton J, Salamon P, Rohwer F. 2003. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol 185:6220–6223. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Pride DT, Salzman J, Haynes M, Rohwer F, Davis-Long C, White RA, Loomer P, Armitage GC, Relman DA. 2012. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J 6:915–926. doi: 10.1038/ismej.2011.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brüssow H. 2003. Phage as agents of lateral gene transfer. Curr Opin Microbiol 6:417–424. doi: 10.1016/S1369-5274(03)00086-9. [DOI] [PubMed] [Google Scholar]
- 159.Quiros P, Colomer-Lluch M, Martinez-Castillo A, Miro E, Argente M, Jofre J, Navarro F, Muniesa M. 2014. Antibiotic resistance genes in the bacteriophage DNA fraction of human fecal samples. Antimicrob Agents Chemother 58:606–609. doi: 10.1128/AAC.01684-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Muniesa M, Colomer-Lluch M, Jofre J. 2013. Could bacteriophages transfer antibiotic resistance genes from environmental bacteria to human-body associated bacterial populations? Mob Genet Elements 3:e25847. doi: 10.4161/mge.25847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kingwell K. 2015. Bacteriophage therapies re-enter clinical trials. Nat Rev Drug Discov 14:515–516. doi: 10.1038/nrd4695. [DOI] [PubMed] [Google Scholar]
- 162.Hraiech S, Brégeon F, Rolain J-M. 2015. Bacteriophage-based therapy in cystic fibrosis-associated Pseudomonas aeruginosa infections: rationale and current status. Drug Des Devel Ther 9:3653–3663. doi: 10.2147/DDDT.S53123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Pabary R, Singh C, Morales S, Bush A, Alshafi K, Bilton D, Alton EWFW, Smithyman A, Davies JC. 2016. Antipseudomonal bacteriophage reduces infective burden and inflammatory response in murine lung. Antimicrob Agents Chemother 60:744–751. doi: 10.1128/AAC.01426-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Steinman MA, Gonzales R, Linder JA, Landefeld CS. 2003. Changing use of antibiotics in community-based outpatient practice, 1991-1999. Ann Intern Med 138:525–533. doi: 10.7326/0003-4819-138-7-200304010-00008. [DOI] [PubMed] [Google Scholar]
- 165.Smith SS, Evans CT, Tan BK, Chandra RK, Smith SB, Kern RC. 2013. National burden of antibiotic use for adult rhinosinusitis. J Allergy Clin Immunol 132:1230–1232. doi: 10.1016/j.jaci.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Van Buchem F, Knottnerus J, Schrijnemaekers V, Peeters M. 1997. Primary-care-based randomised placebo-controlled trial of antibiotic treatment in acute maxillary sinusitis. Lancet 349:683–687. doi: 10.1016/S0140-6736(96)07585-X. [DOI] [PubMed] [Google Scholar]
- 167.Gonzales R, Bartlett JG, Besser RE, Hickner JM, Hoffman JR, Sande MA. 2001. Principles of appropriate antibiotic use for treatment of nonspecific upper respiratory tract infections in adults: background. Ann Emerg Med 37:698–702. doi: 10.1067/S0196-0644(01)70088-1. [DOI] [PubMed] [Google Scholar]
- 168.Young J, De Sutter A, Merenstein D, van Essen GA, Kaiser L, Varonen H, Williamson I, Bucher HC. 2008. Antibiotics for adults with clinically diagnosed acute rhinosinusitis: a meta-analysis of individual patient data. Lancet 371:908–914. doi: 10.1016/S0140-6736(08)60416-X. [DOI] [PubMed] [Google Scholar]
- 169.Wallwork B, Coman W. 2004. Chronic rhinosinusitis and eosinophils: do macrolides have an effect? Curr Opin Otolaryngol Head Neck Surg 12:14–17. doi: 10.1097/00020840-200402000-00005. [DOI] [PubMed] [Google Scholar]
- 170.Zarogoulidis P, Papanas N, Kioumis I, Chatzaki E, Maltezos E, Zarogoulidis K. 2012. Macrolides: from in vitro anti-inflammatory and immunomodulatory properties to clinical practice in respiratory diseases. Eur J Clin Pharmacol 68:479–503. doi: 10.1007/s00228-011-1161-x. [DOI] [PubMed] [Google Scholar]
- 171.Dubin MG, Liu C, Lin SY, Senior BA. 2007. American Rhinologic Society member survey on “maximal medical therapy” for chronic rhinosinusitis. Am J Rhinol 21:483–488. doi: 10.2500/ajr.2007.21.3047. [DOI] [PubMed] [Google Scholar]
- 172.Schwartz JS, Tajudeen BA, Cohen NA. 2016. Medical management of chronic rhinosinusitis—an update. Expert Rev Clin Pharmacol 9:695–704. doi: 10.1586/17512433.2016.1150780. [DOI] [PubMed] [Google Scholar]
- 173.Tan BK, Chandra RK, Pollak J, Kato A, Conley DB, Peters AT, Grammer LC, Avila PC, Kern RC, Stewart WF, Schleimer RP, Schwartz BS. 2013. Incidence and associated premorbid diagnoses of patients with chronic rhinosinusitis. J Allergy Clin Immunol 131:1350–1360. doi: 10.1016/j.jaci.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lim M, Citardi MJ, Leong J-L. 2008. Topical antimicrobials in the management of chronic rhinosinusitis: a systematic review. Am J Rhinol 22:381–389. doi: 10.2500/ajr.2008.22.3189. [DOI] [PubMed] [Google Scholar]
- 175.Rudmik L, Hoy M, Schlosser RJ, Harvey RJ, Welch KC, Lund V, Smith TL. 2013. Topical therapies in the management of chronic rhinosinusitis: an evidence-based review with recommendations. Int Forum Allergy Rhinol 3:281–298. doi: 10.1002/alr.21096. [DOI] [PubMed] [Google Scholar]
- 176.Huang A, Govindaraj S. 2013. Topical therapy in the management of chronic rhinosinusitis. Curr Opin Otolaryngol Head Neck Surg 21:31–38. doi: 10.1097/MOO.0b013e32835bc4ab. [DOI] [PubMed] [Google Scholar]
- 177.Sacks P-L, Harvey RJ, Rimmer J, Gallagher RM, Sacks R. 10 August 2011. Topical and systemic antifungal therapy for the symptomatic treatment of chronic rhinosinusitis. Cochrane Database Syst Rev doi: 10.1002/14651858.CD008263.pub2. [DOI] [PubMed] [Google Scholar]
- 178.Sacks P-L, Harvey RJ, Rimmer J, Gallagher RM, Sacks R. 2012. Antifungal therapy in the treatment of chronic rhinosinusitis: a meta-analysis. Am J Rhinol Allergy 26:141–147. doi: 10.2500/ajra.2012.26.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Pelfrene E, Willebrand E, Sanches AC, Sebris Z, Cavaleri M. 10 April 2016. Bacteriophage therapy: a regulatory perspective. J Antimicrob Chemother doi: 10.1093/jac/dkw083. [DOI] [PubMed] [Google Scholar]
- 180.Cooper CJ, Khan Mirzaei M, Nilsson AS. 2016. Adapting drug approval pathways for bacteriophage-based therapeutics. Front Microbiol 7:1209. doi: 10.3389/fmicb.2016.01209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Willing BP, Russell SL, Finlay BB. 2011. Shifting the balance: antibiotic effects on host-microbiota mutualism. Nat Rev Microbiol 9:233–243. doi: 10.1038/nrmicro2536. [DOI] [PubMed] [Google Scholar]
- 182.Lim YW, Evangelista JS, Schmieder R, Bailey B, Haynes M, Furlan M, Maughan H, Edwards R, Rohwer F, Conrad D. 2014. Clinical insights from metagenomic analysis of sputum samples from patients with cystic fibrosis. J Clin Microbiol 52:425–437. doi: 10.1128/JCM.02204-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Fodor AA, Klem ER, Gilpin DF, Elborn JS, Boucher RC, Tunney MM, Wolfgang MC. 2012. The adult cystic fibrosis airway microbiota is stable over time and infection type, and highly resilient to antibiotic treatment of exacerbations. PLoS One 7:e45001. doi: 10.1371/journal.pone.0045001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Zhao J, Schloss PD, Kalikin LM, Carmody LA, Foster BK, Petrosino JF, Cavalcoli JD, VanDevanter DR, Murray S, Li JZ, Young VB, LiPuma JJ. 2012. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc Natl Acad Sci U S A 109:5809–5814. doi: 10.1073/pnas.1120577109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Merkley MA, Bice TC, Grier A, Strohl AM, Man L-X, Gill SR. 2015. The effect of antibiotics on the microbiome in acute exacerbations of chronic rhinosinusitis. Int Forum Allergy Rhinol 5:884–893. doi: 10.1002/alr.21591. [DOI] [PubMed] [Google Scholar]
- 186.Cleland EJ, Drilling A, Bassiouni A, James C, Vreugde S, Wormald PJ. 2014. Probiotic manipulation of the chronic rhinosinusitis microbiome. Int Forum Allergy Rhinol 4:309–314. doi: 10.1002/alr.21279. [DOI] [PubMed] [Google Scholar]
- 187.Cope EK, Lynch SV. 2015. Novel microbiome-based therapeutics for chronic rhinosinusitis. Curr Allergy Asthma Rep 15:504. doi: 10.1007/s11882-014-0504-y. [DOI] [PubMed] [Google Scholar]
- 188.Landolina N, Levi-Schaffer F. 2016. Monoclonal antibodies: the new magic bullets for allergy. IUPHAR review 17. Br J Pharmacol 173:793–803. doi: 10.1111/bph.13396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Rodgers KR, Chou RC. 25 July 2016. Therapeutic monoclonal antibodies and derivatives: historical perspectives and future directions. Biotechnol Adv doi: 10.1016/j.biotechadv.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 190.Khanna R, Chande N, Vermeire S, Sandborn WJ, Parker CE, Feagan BG. 2016. The next wave of biological agents for the treatment of IBD: evidence from Cochrane Reviews. Inflamm Bowel Dis 22:1737–1743. doi: 10.1097/MIB.0000000000000808. [DOI] [PubMed] [Google Scholar]
- 191.Gevaert P, Van Bruaene N, Cattaert T, Van Steen K, Van Zele T, Acke F, De Ruyck N, Blomme K, Sousa AR, Marshall RP, Bachert C. 2011. Mepolizumab, a humanized anti-IL-5 mAb, as a treatment option for severe nasal polyposis. J Allergy Clin Immunol 128:989–995. doi: 10.1016/j.jaci.2011.07.056. [DOI] [PubMed] [Google Scholar]
- 192.Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, Spector TD, Clark AG, Ley RE. 2014. Human genetics shape the gut microbiome. Cell 159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, Morgan XC, Kostic AD, Luo C, González A, McDonald D, Haberman Y, Walters T, Baker S, Rosh J, Stephens M, Heyman M, Markowitz J, Baldassano R, Griffiths A, Sylvester F, Mack D, Kim S, Crandall W, Hyams J, Huttenhower C, Knight R, Xavier RJ. 2014. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe 15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ursell LK, Haiser HJ, Van Treuren W, Garg N, Reddivari L, Vanamala J, Dorrestein PC, Turnbaugh PJ, Knight R. 2014. The intestinal metabolome: an intersection between microbiota and host. Gastroenterology 146:1470–1476. doi: 10.1053/j.gastro.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Sharon I, Banfield JF. 2013. Genomes from metagenomics. Science 342:1057–1058. doi: 10.1126/science.1247023. [DOI] [PubMed] [Google Scholar]
- 198.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. 2009. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto J-M, Hansen T, Le Paslier D, Linneberg A, Nielsen B, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, et al. . 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Korem T, Zeevi D, Suez J, Weinberger A, Avnit-Sagi T, Pompan-Lotan M, Matot E, Jona G, Harmelin A, Cohen N, Sirota-Madi A, Thaiss CA, Pevsner-Fischer M, Sorek R, Xavier R, Elinav E, Segal E. 2015. Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science 349:1101–1106. doi: 10.1126/science.aac4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Fierer N, Leff JW, Adams BJ, Nielsen UN, Bates ST, Lauber CL, Owens S, Gilbert JA, Wall DH, Caporaso JG. 2012. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc Natl Acad Sci U S A 109:21390–21395. doi: 10.1073/pnas.1215210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Yutin N, Suzuki MT, Teeling H, Weber M, Venter JC, Rusch DB, Béjà O. 2007. Assessing diversity and biogeography of aerobic anoxygenic phototrophic bacteria in surface waters of the Atlantic and Pacific Oceans using the Global Ocean Sampling expedition metagenomes. Environ Microbiol 9:1464–1475. doi: 10.1111/j.1462-2920.2007.01265.x. [DOI] [PubMed] [Google Scholar]
- 203.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Jiang Y, Xiong X, Danska J, Parkinson J. 2016. Metatranscriptomic analysis of diverse microbial communities reveals core metabolic pathways and microbiome-specific functionality. Microbiome 4:2. doi: 10.1186/s40168-015-0146-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Jorth P, Turner KH, Gumus P, Nizam N, Buduneli N, Whiteley M. 2014. Metatranscriptomics of the human oral microbiome during health and disease. mBio 5:e01012-14. doi: 10.1128/mBio.01012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Macklaim JM, Fernandes AD, Di Bella JM, Hammond J-A, Reid G, Gloor GB. 2013. Comparative meta-RNA-seq of the vaginal microbiota and differential expression by Lactobacillus iners in health and dysbiosis. Microbiome 1:12. doi: 10.1186/2049-2618-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Maurice CF, Haiser HJ, Turnbaugh PJ. 2013. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152:39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Booijink CCGM, Boekhorst J, Zoetendal EG, Smidt H, Kleerebezem M, De Vos WM. 2010. Metatranscriptome analysis of the human fecal microbiota reveals subject-specific expression profiles, with genes encoding proteins involved in carbohydrate metabolism being dominantly expressed. Appl Environ Microbiol 76:5533–5540. doi: 10.1128/AEM.00502-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Willis AL, Calton JB, Carr TF, Chiu AG, Chang EH. 2016. Dead or alive: deoxyribonuclease I sensitive bacteria and implications for the sinus microbiome. Am J Rhinol Allergy 30:94–98. doi: 10.2500/ajra.2016.30.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Carini P, Marsden PJ, Leff JW, Morgan EE, Strickland MS, Fierer N. 2016. Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. bioRxiv doi: 10.1101/043372. [DOI] [PubMed] [Google Scholar]
- 211.Hettich RL, Sharma R, Chourey K, Giannone RJ. 2012. Microbial metaproteomics: identifying the repertoire of proteins that microorganisms use to compete and cooperate in complex environmental communities. Curr Opin Microbiol 15:373–380. doi: 10.1016/j.mib.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 212.Siggins A, Gunnigle E, Abram F. 2012. Exploring mixed microbial community functioning: recent advances in metaproteomics. FEMS Microbiol Ecol 80:265–280. doi: 10.1111/j.1574-6941.2011.01284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wilmes P, Bond PL. 2006. Metaproteomics: studying functional gene expression in microbial ecosystems. Trends Microbiol 14:92–97. doi: 10.1016/j.tim.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 214.Wilmes P, Heintz-Buschart A, Bond PL. 2015. A decade of metaproteomics: where we stand and what the future holds. Proteomics 15:3409–3417. doi: 10.1002/pmic.201500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Verberkmoes NC, Russell AL, Shah M, Godzik A, Rosenquist M, Halfvarson J, Lefsrud MG, Apajalahti J, Tysk C, Hettich RL, Jansson JK. 2009. Shotgun metaproteomics of the human distal gut microbiota. ISME J 3:179–189. doi: 10.1038/ismej.2008.108. [DOI] [PubMed] [Google Scholar]
- 216.Xiong W, Abraham PE, Li Z, Pan C, Hettich RL. 2015. Microbial metaproteomics for characterizing the range of metabolic functions and activities of human gut microbiota. Proteomics 15:3424–3438. doi: 10.1002/pmic.201400571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Erickson AR, Cantarel BL, Lamendella R, Darzi Y, Mongodin EF, Pan C, Shah M, Halfvarson J, Tysk C, Henrissat B, Raes J, Verberkmoes NC, Fraser CM, Hettich RL, Jansson JK. 2012. Integrated metagenomics/metaproteomics reveals human host-microbiota signatures of Crohn's disease. PLoS One 7:e49138. doi: 10.1371/journal.pone.0049138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Presley LL, Ye J, Li X, Leblanc J, Zhang Z, Ruegger PM, Allard J, McGovern D, Ippoliti A, Roth B, Cui X, Jeske DR, Elashoff D, Goodglick L, Braun J, Borneman J. 2012. Host-microbe relationships in inflammatory bowel disease detected by bacterial and metaproteomic analysis of the mucosal-luminal interface. Inflamm Bowel Dis 18:409–417. doi: 10.1002/ibd.21793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Jagtap P, McGowan T, Bandhakavi S, Tu ZJ, Seymour S, Griffin TJ, Rudney JD. 2012. Deep metaproteomic analysis of human salivary supernatant. Proteomics 12:992–1001. doi: 10.1002/pmic.201100503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Morris RM, Nunn BL, Frazar C, Goodlett DR, Ting YS, Rocap G. 2010. Comparative metaproteomics reveals ocean-scale shifts in microbial nutrient utilization and energy transduction. ISME J 4:673–685. doi: 10.1038/ismej.2010.4. [DOI] [PubMed] [Google Scholar]
- 221.Bundy JG, Davey MP, Viant MR. 2009. Environmental metabolomics: a critical review and future perspectives. Metabolomics 5:3–21. doi: 10.1007/s11306-008-0152-0. [DOI] [Google Scholar]
- 222.Heinken A, Thiele I. 2015. Systems biology of host-microbe metabolomics. Wiley Interdiscip Rev Syst Biol Med 7:195–219. doi: 10.1002/wsbm.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Turnbaugh PJ, Gordon JI. 2008. An invitation to the marriage of metagenomics and metabolomics. Cell 134:708–713. doi: 10.1016/j.cell.2008.08.025. [DOI] [PubMed] [Google Scholar]
- 224.Marcobal A, Kashyap P, Nelson T, Aronov P, Donia M, Spormann A, Fischbach M, Sonnenburg J. 2013. A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. ISME J 7:1933–1943. doi: 10.1038/ismej.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. 2009. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A 106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. 2013. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Carmody RN, Turnbaugh PJ. 2014. Host-microbial interactions in the metabolism of therapeutic and diet-derived xenobiotics. J Clin Invest 124:4173–4181. doi: 10.1172/JCI72335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Bucci V, Xavier JB. 2014. Towards predictive models of the human gut microbiome. J Mol Biol 426:3907–3916. doi: 10.1016/j.jmb.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Nagalingam NA, Cope EK, Lynch SV. 2013. Probiotic strategies for treatment of respiratory diseases. Trends Microbiol 21:485–492. doi: 10.1016/j.tim.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 230.D'Haens GR, Sartor RB, Silverberg MS, Petersson J, Rutgeerts P. 2014. Future directions in inflammatory bowel disease management. J Crohns Colitis 8:726–734. doi: 10.1016/j.crohns.2014.02.025. [DOI] [PubMed] [Google Scholar]