Abstract
Cyclophilin D (CyPD), a mitochondrial matrix protein, has been widely studied for its role in mitochondrial-mediated cell death. Unexpectedly, we previously discovered that overexpression of CyPD in a stable cell line, increased mitochondrial membrane potentials and enhanced cell survival under conditions of oxidative stress. Here, we investigated the underlying mechanisms responsible for these findings. Spectrophotometric measurements in isolated mitochondria revealed that overexpression of CyPD in HEK293 cells increased respiratory chain activity, but only for Complex III (CIII). Acute treatment of mitochondria with the immumosupressant cyclosporine A did not affect CIII activity. Expression levels of the CIII subunits cytochrome b and Rieske-FeS were elevated in HEK293 cells overexpressing CyPD. However, CIII activity was still significantly higher compared to control mitochondria, even when normalized by protein expression. Blue native gel electrophoresis and Western blot assays revealed a molecular interaction of CyPD with CIII and increased levels of supercomplexes in mitochondrial protein extracts. Radiolabeled protein synthesis in mitochondria showed that CIII assembly and formation of supercomplexes containing CIII were significantly faster when CyPD was overexpressed. Taken together, these data indicate that CyPD regulates mitochondrial metabolism, and likely cell survival, by promoting more efficient electrons flow through the respiratory chain via increased supercomplex formation.
Keywords: mitochondrial permeability transition (MPT), mitochondrial respiratory chain complex, Chaperone, prolyl isomerase, metabolic regulation
1. Introduction
Mitochondria are major regulators of both cell death and cell survival [1–3]. The latter function is primarily attributed to the production of ATP, the final product of a coordinated action of five protein complexes in the mitochondrial respiratory chain (RC). As is well known, electrons are liberated by the oxidation of NADH and FADH2 within four consecutive complexes and two mobile carriers (coenzyme Q and cytochrome c), ultimately being transferred to oxygen. This generates the classic proton gradient across the inner mitochondrial membrane, which can then be used to drive ATP synthesis via the F0F1-ATP synthase (complex V). More recent work has shown that respiratory complexes can be organized, as functional entities, into supramolecular structures commonly referred to as “supercomplexes” [4]. The precise composition of complexes and supercomplexes appear to vary according to tissue type and temporal bioenergetic demands [5]. However, the overall impact of supercomplexes appears to be increased efficiency of electron transfer [6,7].
Complex III (coenzyme Q- cytochrome c reductase) anchors the central position in the RC, mediating electron transfer from ubiquinol to cytochrome c. It consists of 11 subunits, three of which constitute a functional core: cytochrome b (Cytb), cytochrome c1 (Cyt1) and the Rieske-FeS protein Rip1. Biogenesis of complex III (CIII) occurs in a modular step-wise assembly pathway. The subunit Cytb seeds the early core of the complex, which is subsequently stabilized by the incorporation of the Rip1 subunit [8].
A key event in necrotic cell death is the increased permeability of the mitochondrial inner membrane (MIM)[9], due to the functionally defined opening of the mitochondrial permeability transition pore (mPTP), a high conductance channel whose precise composition is unknown, but which includes, critically, cyclophilin-D (CyP-D) [10,11]. This is a mitochondrial isoform belonging to the highly conserved peptidyl-prolylcis-transisomerases (PPIases) family of proteins. Cyclophilins catalyze isomerizations around Xaa-Pro peptides, which are, in general, rate-limiting steps of protein folding [12]. Treatment of cells with the pseudo-CyP-D substrate cyclosporin A is widely known to inhibit cell death stimuli, which gives rise to the model that CyP-D sensitizes opening of the mPTP [13]. This model of cell death was constricted to necrosis based on the insensitivity of CyP-D null mice to high loads of mitochondrial calcium (Ca2+), which did not impact their ability to undergo apoptosis [14]. Unexpectedly, our group discovered that overexpression of CyP-D in HEK cells decreased their resistance to cell death stimuli [15]. We concluded that CyP-D, like mitochondria, played a role in both cell death and cell survival [15].
More recent work has implicated many CyPs as key players in larger protein complexes, although their functional significance is not yet completely understood [16,17]. The goal of the work presented here was to test the hypothesis that CyP-D is an important regulator of mitochondrial metabolism. We present evidence showing that overexpression of CyP-D in HEK293 cells significantly increases the efficiency of CIII activity itself. This increase in activity appears to be independent of PPIase enzymatic activity, at least acutely. Moreover, we find that the assembly of both CIII and supercomplexes are accelerated by CyPD overexpression. We note that others have reported CIII is present in all supercomplexes and is considered the main stabilizing complex [18–24]. Taken together, these findings suggest that CyP-D enhances cell survival by increasing the efficiency of mitochondrial respiration via CIII and its central role in supercomplex activity.
2. Material and methods
2.1 Cell culture and growth conditions
Cells were cultured according to Lin and Lechleiter [15]. Briefly, HEK293 cell were maintained at 37°C in Dulbecco’s modified Eagle’s medium/F-12 (Life Technology, Rockville, MD) supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 200 ug penicillin, and 100 ug/ml streptomycin in a humidified atmosphere of 5% CO2/95% air. The generation of a stable HEK293 cell line expressing CyPD was performed according to Lin and Lechleiter [15].
2.2 Mitochondrial isolation from HEK293 cells
Mitochondria from HEK293 cells were isolated according to the method described by Attardi et al. [25]. Briefly, HEK293 cells were scraped down in NKM solution (in mM: 130 NaCl, 5 KCl, 1 MgCl2) and spun down at 250 g for 5 min. Pellets were incubated in TKM buffer (in mM: 10 Tris-HCl pH 6.7, 10 KCl, 0.15 MgCl2). All the procedures were performed at 4 °C. The suspensions were manually homogenized (~50 strokes), transferred into a tube with 0.5ml of Sucr-TKM buffer (in mM:10 Tris-HCl pH 6.7, 10 KCl, 0.15 MgCl2, 2,000 Sucrose) and centrifuged at 1,000 g for 5 min. The subsequent supernatant was collected and centrifuged at 10,000 g for 10 min to obtain mitochondria pellets. The pellets were then resuspended with Suc-Tris-EDTA buffer (in mM: 10 Tris-HCl pH 6.7, 1 EDTA, 250 Sucrose) and centrifuged at 10,000 g for 10 min. TM-Suc buffer (in mM: 10 Tris-HCl pH 6.7, 0.15 MgCl2, 250 Sucrose) was added to the mitochondrial pellets and spun at 10,000 g for 10 min to obtain the final mitochondrial pellet.
2.3 DDM solubilization
1 mg of mitochondria protein from HEK293 cells were solubilized in 1% DDM (Sigma Aldrich, St. Louis, MO) for 45 min at 4°C on a rotator. Afterwards, the solubilization mix was spun at 72,000 g for 30 min. The supernatants were collected and benzonase treated (Sigma Aldrich, St. Louis, MO) for 30 min. The solubilized mitochondria supernatant was collected and aliquoted for Blue native gel electrophoresis or for SDS-PAGE analysis (12%) gels.
2.4 Activities of the respiratory chain complexes (ETCs) I–V
The activities of the electron transport complexes were measured by spectrophotometric assay. 10 μg of mitochondrial proteins in respiration buffer (in mM: 250 sucrose, 10 KH2PO4, 10 Tris-ClH, pH 7.4, 1 EGTA) were used to measure activity for complex I–IV. The activity for complex I was determined by monitoring the oxidation of NADH at 340 nm at 30°C, using ubiquinone-2 as an electron acceptor in the presence of 2, 6 -dichlorophenolindophenol (DPIP) [26]. CII activity was measured by the succinate-dependent reduction of DPIP. The reaction was monitored at 600 nm at 30°C using ubiquinone-2 as an electron acceptor [27]. CIII activity was measured by reduction of Cytochrome C Fe3+ using Decylubiquinol-2 as an electron donor at 550 nm. CIV activity was determined by monitoring the oxidation of cytochrome c Fe2+ at 550 nm. The data dimensions are μmol/min/mg protein.
2.5 SDS–PAGE and Western blotting
Electrophoresis was performed in SDS-polyacrylamide gels. Proteins were then either stained with Coomassie brilliant blue G-250 or transferred into PVDF membrane and probed with the following antibodies: anti-Core II, anti-Rieske (MitoProfile, Eugene, OR), polyclonal rabbit anti-CyPD antibody (custom made by Pocono Rabbit Farm & Laboratory, Inc. Canadensis, PA).
2.6 Blue native gel electrophoresis
Solubilized mitochondrial extracts (100 ug) were loaded onto a Native Page BisTris 4–12% gradient gel with Coomassie G250 (Life Technology, Carlsbad, CA) and run at 150 V using dark blue cathode buffer as directed by the manufacturer’s protocol with Native Mark standards (Bovine heart) in the cold room. Staining of the Native Page gel was performed using Colloidal Blue Staining kit (Life Technology, Carlsbad, CA) following the manufacturer’s protocol.
2.7 Blue Native Gel Transfer
Blue Native transfer was achieved by using the iBlot Dry Blotting transfer system (Life Technology, Carlsbad, Ca) on setting P3 for 7 min transfer time. Gels were immersed in 2X transfer buffer prior to transfer for 10 min on shaker at room temperature. During the last five minutes of shaking, a final 0.1% SDS was added to the 2X transfer buffer. The gel was then loaded onto a PVDF membrane, transferred and fixed with 8% acetic acid and rinsed with dH20 prior to blocking and immunodetection.
2.8 2-D Electrophoresis of Native Page Gel and transfer
After blue native gel electrophoresis and overnight distaining, the gel strips were treated following the manufacturer’s protocol (Life Technology, Carlsbad, CA). Briefly, the gel strips were incubated for 30 min in reduction solution. Next, the reducing agent was removed and incubated for 30 min in alkylating solution (1X Nupage LDS sample buffer and 50mM dimethylacrylamide). Afterwards, the alkylating solution was removed and a quenching solution was added (5mM DTT and 20% ethanol in the 1X Nupage LDS sample buffer). Lanes were loaded onto a 4–12% gradient 2D gel (Fife Technology, Carlsbad, CA) and run at 150V in the cold room. Afterwards, the gel was transferred to a nitrocellulose membrane for 1 hr at 100V. The membrane was probed for individual subunits of all five complexes with Total OXPHOS Human Antibody Cocktail (MitoProfile, Eugene, OR).
Briefly, the gel strips were sequentially incubated in reduction, alkylating and quenching solution. Lanes were loaded onto a 4–12% gradient 2D gel (Fife Technology, Carlsbad, CA). Afterwards, the gel was transferred to a nitrocellulose membrane and this was probed for individual subunits of all five complexes with Total OXPHOS Human Antibody Cocktail (MitoProfile, Eugene, OR).
2.9 Radiolabeling Mitochondria Proteins and Analysis
Pulse-labeling experiments with [35S]methionine/cysteine were performed according to a protocol described previously [28]. Briefly, cells were incubated for 20 h with 50 mg/ml chloramphenicol in complete medium before labeling. Next day, cells were washed with methionine-free DMEM 5 times for 1h. The same medium containing serum and 100 ug/ml of cycloheximide was added to cells for 20 min at 37°C. Then, 0.2 mCi of [35S]methionine/cysteine (PerkinElmer Life Sciences, Waltham, MA) was added, and the cells were incubated for 2 h at 37°C, followed by the addition of cold methionine to a final concentration of 1mM. After 15 min, the solution was removed and cells were washed 3 times by PBS. From this point, the cells were subjected to various time point chases in complete unlabeled medium in the absence of cycloheximide. Collected cell pellets were solubilized for 20 min on ice in 50 ul of 20 mM bis-Tris (pH 7.4), 40 mM NaCl, and 10% (v/v) glycerol containing 1% (w/v) digitonin, as described by Schagger and von Jagow [29]. 60 ug/lane of mitochondrial protein containing Coomassie Blue G was loaded to 3% to 11% Acrylamide BN gel. The gels were run for 30 min at 40V and then at 80V until the dye reached the end of the gel. The gels were dried and the radiolabeled proteins were analyzed by autoradiography. Quantification of the intensity of the bands was done by using a PhosphorImager (Molecular Dynamics) and the ImageQuant program.
3. Results
To investigate the underlying mechanism (s) responsible for enhanced cell survival in cells overexpressing CyPD [15], we measured O2 consumption and ATP production. We found, as expected, that both ATP and O2 were significantly higher in a stable cell line overexpressing CyPD (CyPD1) compared controls, consistent with an increase in metabolism (Fig 1a–c). Western blot analysis revealed that CyPD was significantly increased after over-expression (Fig 1d–e). We next determined the enzymatic activity of individual respiratory complexes in mitochondria isolated from the CyPD1 cell line as well as two additional stable cells lines differentially overexpressing CyPD (CyPD2 and CyPD3). Interestingly, we only observed a significant increase in CIII activity in each of the three cell lines overexpressing CyPD compared to controls. Surprisingly, the activity of the other respiratory chain complexes, I, II and IV, were not affected by overexpressing of CyPD under our assay conditions (Fig 2a–d).
Fig. 1. Overexpression of CyPD leads to enhanced cellular respiration and ATP production.
Oxygen consumption of HEK293 control cells (transfected with pcDNA3 vector) and stables cell lines overexpressing pcDNA3-CyPD were performed using StratKelvin oxygen consumption electrode. It is shown as a time course (a) and as absolute oxygen rate (b); representative traces of slope plots corresponding to control and oligomycin condition after control/vehicule (Water) and ATP addition (b1); absolute oxygen rate after control/vehicule (Water) and ATP addition (b2). HEK293 control cells and stables cell lines overexpressing CyPD were obtained. The ATPlite™ Luminescence Assay System was used to determine ATP concentrations (c). Western blot analysis of CyPD levels in HEK293 control cells (transfected with pcDNA3 vector) and overexpressed with pcDNA3-CyPD (CyPD) (d). Densitometry histogram is normalized with actin (e). All experiments were performed in triplicate. Oxygen consumption a representative experiment is shown of three performed separately, and ATP production data represent the mean of three independent experiments and are represented as mean ± SEM.
Fig. 2. Cyclophilin D over-expression increases CIII enzymatic activity.
Isolated mitochondria from HEK293 control cells (transfected with pcDNA3 vector) and stable cells lines differentially overexpressing CyPD (CyPD1, CyPD2 and CyPD3) were obtained. The enzymatic activity of individual respiratory complexes (I–IV) was measured by spectrophotometric assays as is indicated in Experimental Procedure (a–d). Data represent the mean either of two independent experiments (CI, CII and CIV enzymatic activities) or three (CIII enzymatic activity) and are represented as mean ± SEM.). Western blot analysis of CyPD levels in HEK293 control cells (transfected with pcDNA3 vector) and overexpressed with pcDNA3-CyPD (CyPD1, CyPD2 and CyPD3) and densitometry histogram normalized with actin (e). Western blot data are the mean of three independent experiment represented as mean ± SEM.
To test whether the stimulatory effect of CyPD on CIII was dependent on its PPIase activity, we utilized the immunosuppressant cyclosporine A (CsA), a prolyl isomerization catalysis inhibitor that forms a very tight stoichiometric complex with cyclophilins (Kd = 5 – 200 nM) [30] [31]. Mitochondria were isolated from control cells and from the CyPD3 cell line, then treated with CsA at 6 μM and 20 μM for 30 min prior to measuring CIII activity. We observed no effect of CsA treatments on CIII activity (Fig 3), despite the fact we used an inhibitor concentration that was 100-fold above its Kd [30] [31]. These data suggested that CyPD was not acutely affecting CIII activity via its PPIase activity.
Fig. 3. The effect of Cyclophilin D over-expression on enzymatic CIII activity is peptidyl-prolyl isomerase (PPIase) independent.

Isolated mitochondria from HEK293 control cells (transfected with pcDNA3 vector) and stable transfected cells line overexpressing CyPD (CyPD3) were obtained. The enzymatic activity of CIII was measured by spectrophotometric assay in the absence and presence of CsA at 6 μM and 20 μM incubated for 30 min. Data represent the mean of four independent experiments using different sets of isolated mitochondria and are represented as mean ± SEM.
To determine whether CyPD was affecting protein levels of the CIII subunits, mitochondria were isolated cell lines overexpressing CyPD (CyPD3) or not (control) and analyzed by Western blot analysis. Interestingly, we observed that cytochrome b (Cytb) and the iron-sulfur protein Rieske-FeS (Rip1), both part of the functional core of CIII, were expressed at significantly higher levels in mitochondria with overexpressed levels of CyPD (Fig 4a). A similar trend was observed for Core II expression (Fig 4b), another CIII subunit.
Fig 4. Cyclophilin D over-expression increases protein levels of CIII subunits and increases CIII specific enzymatic activity.
Mitochondrial protein extracts were obtained from HEK293 control cells (transfected with pcDNA3 vector) and stable transfected cells line overexpressing CyPD and analyzed by Western blot using antibodies against cytochrome b and the Rieske subunit of CIII (a), then the PVDF membrane was stripped and re-probed with antibodies against Core II and CyP-D (b). The PVDF membrane is shown stained with ponceau S, this was performed inmediately after transfer, before immunostainig (c). Values for CIII enzymatic activity were normalized either by total protein, ponceau S stained (d) or by levels of individual CIII subunits (e–g). Data represent the mean of four independent experiments and are represented as mean ± SEM.
We next tested whether or not the observed increase in CIII activity could be attributed to an increase in specific activity. Values for enzymatic activity were normalized either by expression levels of individual CIII subunits (Fig 4a and 4b) or by total protein (ponceau S staining, Fig 4c). We found that cells overexpressing CyPD exhibited higher CIII enzymatic activity when normalized not only with total protein, but also with expression levels of CIII subunits core II and Rieske (Fig 4d–g). Taken together, these data suggest that CyPD enhances both the expression of CIII and the amount of maturely folded, enzymatically active CIII.
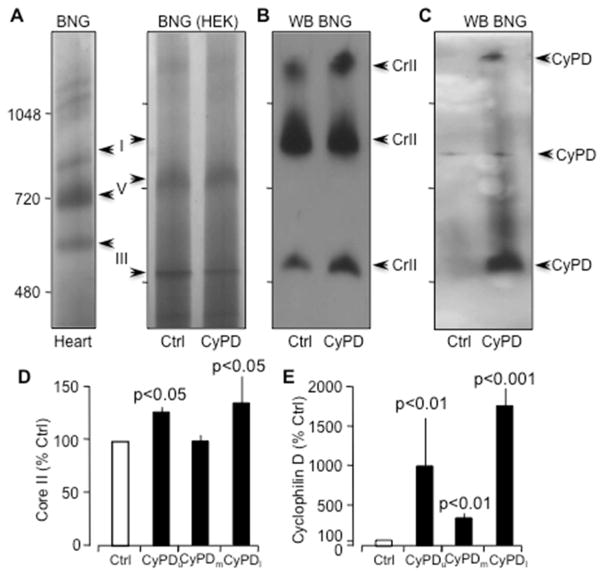
Supercomplexes are thought to increase the efficacy of electron flow through the respiratory chain [32]. Consequently, we investigated the potential interaction of CyPD with supercomplexes using blue native gel (BNG) electrophoresis. Mitochondrial protein extracts from control and CyPD-overexpressing cells were run on BNG’s and the native protein was immuno-blotted after transfer to polyvinylidene difluoride (PVDF) membrane for the CIII subunit Core II (Fig 5). Specific bands associated with respiratory chain complexes were identified by native protein masses according to Wittig et al. [33]. We observed an immunoreactive band just above molecular weight marker 480 KDa, corresponding to the expected size of individual CIII, and also bands near 900 and 1200 KDa, indicative of supercomplexes containing I and III or I, III and IV, respectively (Fig 5b). The native membrane was subsequently stripped and probed with an antibody against CyPD (Fig 5c). Immunoreactive bands matching those marked by core II were observed, suggesting CyPD remained associated with both individual CIII and fully assembled supercomplexes.
Fig 5. CyPD interacts with individual CIII and supercomplex.
Mitochondrial protein were obtained from HEK-293 control cells (transfected with pcDNA3 vector) and stable transfected cell line overexpressing CyPD were loaded onto BNGs; mitochondrial protein extracts from bovine heart were used as control (a). The mitochondrial extracts were loaded onto BNGs (100ug), run, transferred to PVDF membrane and probed using antibodies against Core II (b). The PVDF membrane was stripped and re-probed with antibodies against CyPD (c). Representative experiment is shown of three performed separately. (d) Densitometry histogram of Core II (upper, middle and lower bands) from CyPD transfected cell line are normalized with the corresponding value of control cells. (e) Densitometry histogram of CyPD (upper, middle and lower bands) from CyPD transfected cell line are normalized with the corresponding value of control cells.
To identify potential changes in individual complex and supercomplex stoichiometry, we performed a second-dimensional analysis by running excised BNG lanes, obtained from control and CyPD-overexpressing cells (CyPD3), on an SDS-PAGE. After protein transfer, the membranes were immune-blotted with an antibody cocktail probing for individual subunits of all five complexes (Abcam: ab110412). The CyPD3 cell line overexpressing CyPD showed significantly increased levels for all individual complexes as well as supercomplexes (Fig 6a and b).
Fig. 6. Cyclophilin D over-expression induces changes in supercomplex stoichiometry.
Mitochondrial protein extracts from control (a) and CyPD-overexpressing HEK293 cells (b) were loaded onto BNGs. Bands from each cell line were excised and ran in second dimension on SDS-PAGE, then transferred to PVDF membrane and analyzed by Western blot using an antibody cocktail which probes for individual subunits of all five complexes. Complex I (NDUFA9), complex II (subunit 70kDa), complex III (Core II), complex IV (subunit IV), and complex V (subunit alpha). Representative experiment is shown of four performed separately.
Finally, we performed in vivo labeling of mitochondrial translation products in order to determine whether the kinetics of supercomplex assembly or the rate of synthesis of individual complexes and supercomplexes was modulated by CyPD. Our strategy was to label newly synthesized subunits and follow their incorporation into, and later their dissociation from, the respiratory machinery. Control and overexpressing CyPD (CyPD3) cells were first pre-incubated in the presence of the mitochondrial protein synthesis inhibitor, chloramphenicol, for 20 h. This enriched nDNA-encoded subunits and facilitated the incorporation of the newly synthesized mtDNA-encoded subunits into complexes, which were labeled by incubating cells with [35S] methionine-cysteine for 2 h. This last step was performed in the presence of the reversible cytoplasmic protein synthesis inhibitor, cycloheximide, to ensure selective labeling of mtDNA-encoded protein [34,35]. Cells were then chased with unlabeled medium (in the absence of cycloheximide) for 1, 2, 4, 8, 24, 48 and 72 h (Fig 7a, d). Overall, this approach allowed us to follow labeled mtDNA encoded subunits present in intermediate assemblies at early time points (i.e. −1 and 0 h, Fig 7a, c) and permitted us to monitor the longer-term dynamics of respiratory complexes. Interestingly, we identified significantly higher intensity bands for CIII and supercomplex (V+III/IV) from the CyPD-overexpressing cells at earlier timepoints compared to controls. Densitometry analysis showed that CIII was assembled and incorporated into supercomplexes at a significantly faster rate in cells overexpressing CyPD (Fig 7b, d).
Fig. 7. Radiolabeling of mitochondrial proteins reveals complex III is assembled faster in cells overexpressing CyPD.
In vivo labeling of mitochondrial translation products was performed. Control HEK293 cells, transfected with pcDNA 3 vector (a) and stable transfected cells line overexpressing CyPD (b) were pre-incubated in the presence of chloramphenicol, for 20 h. Then the cells were subsequently washed and incubated with 0.2 mCi of [35S]methionine-cysteine for 2 h, in the presence of cycloheximide. Cells were then chased with unlabeled medium for 1, 2, 4, 8, 24, 48 and 72 h, in the absence of cycloheximide. Mitochondrial protein extracts were loaded onto BNGs; the gel was fixed, dried, and proteins were visualized by autoradiography. Quantification of bands from radiolabeling reveals relative CIII, CV+III/IV and supercomplexes amounts on multiple time points before and after 35S labeling (b, d). Data represent the mean of three independent experiments and are represented as mean ± SEM.
4. Discussion
We previously reported that overexpression of CyPD in HEK293 cells hyperpolarized mitochondria and increased their resistance to cell death stimuli [15]. The data presented here demonstrate that CyPD overexpression in HEK cells increases O2 consumption and ATP production. These observations are consistent with our previous work [15]. Unexpectedly, we also discovered that this increase in mitochondrial respiration was correlated with enhanced activity of CIII and not the other respiratory chain complexes. Enhanced CIII activity appeared to be due to both an increase in protein expression and an increase in the enzymatic efficiency of CIII, which may be due to an increase in the rate of supercomplex assembly in CyPD-overexpressing cells. Our data also indicate that CyP-D stably interacts with either CIII by itself or with CIII in supercomplexes.
Hafner et al. [36] showed that the acetylated CyPD form binds and positively regulates mPTP in a CsA sensitive manner, which promote mitochondria dysfunction. Although CyPD overexpression increases the ratio of CyPD (inactive)/acetylated CyPD (active), the metabolic effect that we observed was independent of CsA.
At present, we cannot precisely define the mechanism by which the expression of CyPD in HEK293 cells augments the level of expression of CIII subunits. CyPD could be increasing protein synthesis or facilitating the folding/stability of a specific subunit of CIII that is rate-limiting for its maturation and assembly. The possibility that an elevated level of CyPD enhances CIII stabilization suggests that this immunophilin may act as a co-chaperone under cell stress, consistent with reports of other PPIases in this family [37]. Similar functions for CIII have been reported for a conserved mitochondria protein named Mzm1, which was shown to be important for assembly/stability of CIII particularly when the cells grew in Zn-limited medium [8].
Our observations of CyPD function were obtained by overexpression studies. Thus, its possible that a similar increase under either physiological or pathological conditions could be used to enhanced electron transport efficiency according to the metabolic requirements of cell. These observations do not rule out the possibility that endogenous CyPD is also required for normal mitochondrial function, since our evidence showed CyPD interacts with CIII. In addition, the absence of any acute affect of cyclosporine A on CIII activity suggests that the mechanism of action of CyPD is not associated with moment-to-moment changes in protein folding, but rather, assembly and maturation of CIII, as discussed below. However, this does not rule out an enzymatic role in protein folding (PPIase activity) in the stabilization of assembled complexes and supercomplexes. This appears to be in contrast to the immunophilin Nina A, which is required for synthesis and localization of Drosophila melanogaster rhodopsin [16].
Native gel and Western blot analysis revealed that CyPD associated with CIII both individually and in supercomplexes, indicative of stable subunit binding (Fig 4). This approach also demonstrated that CyPD overexpression increased the levels of both individual CIII and in supercomplexes (Fig 5a and 5b). These data are again consistent with a model in which CyPD is important for stabilization rather than assembly. In this regard, CyPD could be included with the series of factors recently reported in yeast as being important for supercomplex stabilization, such as Rcf1 and Rcf2 (Respiratory supercomplex factor 1 and 2), and ACC2 (ADP/ATP carrier 2) [38] [39] [40]. In addition, Lapuente-Brun et al. [7] have reported that in mammalian cells, a protein named SCAFI (Supercomplex assembly factor I) stabilizes supercomplexes, specifically the interaction between CIII and IV.
Finally, we observed significant changes in the kinetics of supercomplex formation when we performed in vivo labeling of mitochondrial translation products (Fig 7). Note that all the respiratory supercomplexes involved in electron transfer (CICIII2, CICIII2CIVn, CIIICIV) contain CIII; evaluation of CIII integration into supercomplexes is essential to understand supercomplexes association. We think it unlikely that CyPD stimulated mitochondrial complex subunit synthesis, since the labeled radioactive bands at −1 and 0 h, in the presence of cycloheximide, are similar for control and CyPD-overexpressing cells. However, once cycloheximide was removed, we observed significant differences in the 35S-labeling pattern. Specifically, CyPD positively increased the appearance of individual CIII subunits and supercomplexes (V+III/IV), suggesting that CyPD was either accelerating stabilization or stimulating protein synthesis of nuclear-encoded mitochondrial subunits.
Irrespective of the precise underlying mechanisms, our data highlight a new level of regulation of mitochondrial respiratory activity by CyPD, which appears to act as an assembly/stabilization factor for CIII and supercomplexes. This new function of CyPD may provide novel insights into how other proteins could regulate mitochondrial function to meet the changing energetic requirements of a specific tissue or need. Additional studies will be required to better understand its pathological and/or physiological significance.
5. Conclusion
We report that CyPD overexpression selectively increases complex III activity and supercomplex formation in mitochondria, which increases the efficiency of electron transfer. We suggest that a likely mechanism for increased CIII activity in cells overexpressing CyPD is augmented expression of CIII, which, in turn, promotes faster assembly into supercomplexes. This data provide novel insights in the regulation of mitochondrial respiration in response to changing energetic needs.
Highlights.
We propose a new molecular mechanism that regulates mitochondrial metabolism.
The mechanism depends on increasing the efficiency of mitochondrial respiration.
Cyclophilin D promotes complex III activity and accelerates super-complexes formation.
Acknowledgments
We thank Dr. Ramaswamy Sharma for his thoughtful edits and critiques of the manuscript.
Funding: This work was supported by NIH R01 AG007218, 2016 to JDL and the Owens Foundation to JDL.
Abbreviations
- CyPD
Cyclophilin D
- CIII
Complex III
- RC
mitochondrial respiratory chain
- Cytb
cytochrome b
- Cyt1
cytochrome c1
- ROS
reactive oxygen species
- mPTP
mitochondrial permeability transition pore
- PPIases
peptidyl-prolylcis-transisomerases
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hansford RG. Physiological role of mitochondrial Ca2+ transport. J Bioenerg Biomembr. 1994;26(5):495–508. doi: 10.1007/BF00762734. [DOI] [PubMed] [Google Scholar]
- 2.Meier P, Finch A, Evan G. Apoptosis in development. Nature. 2000;407(6805):796–801. doi: 10.1038/35037734. [DOI] [PubMed] [Google Scholar]
- 3.Yuan J, Yankner BA. Apoptosis in the nervous system. Nature. 2000;407(6805):802–809. doi: 10.1038/35037739. [DOI] [PubMed] [Google Scholar]
- 4.Schagger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19(8):1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benard G, Faustin B, Passerieux E, Galinier A, Rocher C, Bellance N, Delage JP, Casteilla L, Letellier T, Rossignol R. Physiological diversity of mitochondrial oxidative phosphorylation. Am J Physiol Cell Physiol. 2006;291(6):C1172–1182. doi: 10.1152/ajpcell.00195.2006. [DOI] [PubMed] [Google Scholar]
- 6.Acin-Perez R, Fernandez-Silva P, Peleato ML, Perez-Martos A, Enriquez JA. Respiratory active mitochondrial supercomplexes. Mol Cell. 2008;32(4):529–539. doi: 10.1016/j.molcel.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 7.Lapuente-Brun E, Moreno-Loshuertos R, Acin-Perez R, Latorre-Pellicer A, Colas C, Balsa E, Perales-Clemente E, Quiros PM, Calvo E, Rodriguez-Hernandez MA, Navas P, Cruz R, Carracedo A, Lopez-Otin C, Perez-Martos A, Fernandez-Silva P, Fernandez-Vizarra E, Enriquez JA. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science. 2013;340(6140):1567–1570. doi: 10.1126/science.1230381. [DOI] [PubMed] [Google Scholar]
- 8.Atkinson A, Khalimonchuk O, Smith P, Sabic H, Eide D, Winge DR. Mzm1 influences a labile pool of mitochondrial zinc important for respiratory function. J Biol Chem. 2010;285(25):19450–19459. doi: 10.1074/jbc.M110.109793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kroemer G, Galluzzi L, Brenner C. Mitochondrial membrane permeabilization in cell death. Physiol Rev. 2007;87(1):99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- 10.Halestrap AP. The mitochondrial permeability transition: its molecular mechanism and role in reperfusion injury. Biochem Soc Symp. 1999;66:181–203. doi: 10.1042/bss0660181. [DOI] [PubMed] [Google Scholar]
- 11.Bernardi P, Petronilli V, Di Lisa F, Forte M. A mitochondrial perspective on cell death. Trends Biochem Sci. 2001;26(2):112–117. doi: 10.1016/s0968-0004(00)01745-x. [DOI] [PubMed] [Google Scholar]
- 12.Lu KP, Finn G, Lee TH, Nicholson LK. Prolyl cis-trans isomerization as a molecular timer. Nat Chem Biol. 2007;3(10):619–629. doi: 10.1038/nchembio.2007.35. [DOI] [PubMed] [Google Scholar]
- 13.Lemasters JJ, Qian T, Bradham CA, Brenner DA, Cascio WE, Trost LC, Nishimura Y, Nieminen AL, Herman B. Mitochondrial dysfunction in the pathogenesis of necrotic and apoptotic cell death. J Bioenerg Biomembr. 1999;31(4):305–319. doi: 10.1023/a:1005419617371. [DOI] [PubMed] [Google Scholar]
- 14.Baines CP, Kaiser RA, Purcell NH, Blair NS, Osinska H, Hambleton MA, Brunskill EW, Sayen MR, Gottlieb RA, Dorn GW, Robbins J, Molkentin JD. Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature. 2005;434(7033):658–662. doi: 10.1038/nature03434. [DOI] [PubMed] [Google Scholar]
- 15.Lin DT, Lechleiter JD. Mitochondrial targeted cyclophilin D protects cells from cell death by peptidyl prolyl isomerization. J Biol Chem. 2002;277(34):31134–31141. doi: 10.1074/jbc.M112035200. [DOI] [PubMed] [Google Scholar]
- 16.Baker EK, Colley NJ, Zuker CS. The cyclophilin homolog NinaA functions as a chaperone, forming a stable complex in vivo with its protein target rhodopsin. EMBO J. 1994;13(20):4886–4895. doi: 10.1002/j.1460-2075.1994.tb06816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giorgio V, Soriano ME, Basso E, Bisetto E, Lippe G, Forte MA, Bernardi P. Cyclophilin D in mitochondrial pathophysiology. Biochim Biophys Acta. 2010;1797(6–7):1113–1118. doi: 10.1016/j.bbabio.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenzie M, Lazarou M, Thorburn DR, Ryan MT. Mitochondrial respiratory chain supercomplexes are destabilized in Barth Syndrome patients. Journal of molecular biology. 2006;361(3):462–469. doi: 10.1016/j.jmb.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 19.Blakely EL, Mitchell AL, Fisher N, Meunier B, Nijtmans LG, Schaefer AM, Jackson MJ, Turnbull DM, Taylor RW. A mitochondrial cytochrome b mutation causing severe respiratory chain enzyme deficiency in humans and yeast. The FEBS journal. 2005;272(14):3583–3592. doi: 10.1111/j.1742-4658.2005.04779.x. [DOI] [PubMed] [Google Scholar]
- 20.Stroh A, Anderka O, Pfeiffer K, Yagi T, Finel M, Ludwig B, Schagger H. Assembly of respiratory complexes I, III, and IV into NADH oxidase supercomplex stabilizes complex I in Paracoccus denitrificans. The Journal of biological chemistry. 2004;279(6):5000–5007. doi: 10.1074/jbc.M309505200. [DOI] [PubMed] [Google Scholar]
- 21.D’Aurelio M, Gajewski CD, Lenaz G, Manfredi G. Respiratory chain supercomplexes set the threshold for respiration defects in human mtDNA mutant cybrids. Human molecular genetics. 2006;15(13):2157–2169. doi: 10.1093/hmg/ddl141. [DOI] [PubMed] [Google Scholar]
- 22.Abdrakhmanova A, Zickermann V, Bostina M, Radermacher M, Schagger H, Kerscher S, Brandt U. Subunit composition of mitochondrial complex I from the yeast Yarrowia lipolytica. Biochim Biophys Acta. 2004;1658(1–2):148–156. doi: 10.1016/j.bbabio.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 23.Diaz F, Fukui H, Garcia S, Moraes CT. Cytochrome c oxidase is required for the assembly/stability of respiratory complex I in mouse fibroblasts. Molecular and cellular biology. 2006;26(13):4872–4881. doi: 10.1128/MCB.01767-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Acin-Perez R, Bayona-Bafaluy MP, Fernandez-Silva P, Moreno-Loshuertos R, Perez-Martos A, Bruno C, Moraes CT, Enriquez JA. Respiratory complex III is required to maintain complex I in mammalian mitochondria. Molecular cell. 2004;13(6):805–815. doi: 10.1016/s1097-2765(04)00124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Attardi G, Costantino P, Lynch D, Mitchel C, Murphy W, Ojala D. Molecular and genetic approaches to the analysis of the informational content of the mitochondrial genome in mammalian cells. Mol Cell Biochem. 1977;14(1–3):151–164. doi: 10.1007/BF01734179. [DOI] [PubMed] [Google Scholar]
- 26.Birch-Machin MA, Turnbull DM. Assaying mitochondrial respiratory complex activity in mitochondria isolated from human cells and tissues. Methods Cell Biol. 2001;65:97–117. doi: 10.1016/s0091-679x(01)65006-4. [DOI] [PubMed] [Google Scholar]
- 27.Boveris A, Cadenas E. Mitochondrial production of superoxide anions and its relationship to the antimycin insensitive respiration. FEBS Lett. 1975;54(3):311–314. doi: 10.1016/0014-5793(75)80928-8. [DOI] [PubMed] [Google Scholar]
- 28.Chomyn A. In vivo labeling and analysis of human mitochondrial translation products. Methods in enzymology. 1996;264:197–211. doi: 10.1016/s0076-6879(96)64020-8. [DOI] [PubMed] [Google Scholar]
- 29.Schagger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199(2):223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 30.Handschumacher RE, Harding MW, Rice J, Drugge RJ, Speicher DW. Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science. 1984;226(4674):544–547. doi: 10.1126/science.6238408. [DOI] [PubMed] [Google Scholar]
- 31.Harding MW, Handschumacher RE, Speicher DW. Isolation and amino acid sequence of cyclophilin. J Biol Chem. 1986;261(18):8547–8555. [PubMed] [Google Scholar]
- 32.Schagger H, Pfeiffer K. The ratio of oxidative phosphorylation complexes I–V in bovine heart mitochondria and the composition of respiratory chain supercomplexes. J Biol Chem. 2001;276(41):37861–37867. doi: 10.1074/jbc.M106474200. [DOI] [PubMed] [Google Scholar]
- 33.Wittig I, Carrozzo R, Santorelli FM, Schagger H. Supercomplexes and subcomplexes of mitochondrial oxidative phosphorylation. Biochimica et biophysica acta. 2006;1757(9–10):1066–1072. doi: 10.1016/j.bbabio.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 34.Deng JH, Li Y, Park JS, Wu J, Hu P, Lechleiter J, Bai Y. Nuclear suppression of mitochondrial defects in cells without the ND6 subunit. Mol Cell Biol. 2006;26(3):1077–1086. doi: 10.1128/MCB.26.3.1077-1086.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vartak R, Deng J, Fang H, Bai Y. Redefining the roles of mitochondrial DNA-encoded subunits in respiratory Complex I assembly. Biochim Biophys Acta. 2015 doi: 10.1016/j.bbadis.2015.04.008. [DOI] [PMC free article] [PubMed]
- 36.Hafner AV, Dai J, Gomes AP, Xiao CY, Palmeira CM, Rosenzweig A, Sinclair DA. Regulation of the mPTP by SIRT3-mediated deacetylation of CypD at lysine 166 suppresses age-related cardiac hypertrophy. Aging. 2010;2(12):914–923. doi: 10.18632/aging.100252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang P, Heitman J. The cyclophilins. Genome Biol. 2005;6(7):226. doi: 10.1186/gb-2005-6-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen YC, Taylor EB, Dephoure N, Heo JM, Tonhato A, Papandreou I, Nath N, Denko NC, Gygi SP, Rutter J. Identification of a protein mediating respiratory supercomplex stability. Cell Metab. 2012;15(3):348–360. doi: 10.1016/j.cmet.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vukotic M, Oeljeklaus S, Wiese S, Vogtle FN, Meisinger C, Meyer HE, Zieseniss A, Katschinski DM, Jans DC, Jakobs S, Warscheid B, Rehling P, Deckers M. Rcf1 mediates cytochrome oxidase assembly and respirasome formation, revealing heterogeneity of the enzyme complex. Cell Metab. 2012;15(3):336–347. doi: 10.1016/j.cmet.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 40.Claypool SM, Oktay Y, Boontheung P, Loo JA, Koehler CM. Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. J Cell Biol. 2008;182(5):937–950. doi: 10.1083/jcb.200801152. [DOI] [PMC free article] [PubMed] [Google Scholar]