Summary
Microarray of peripheral blood (PB) and synovial fluid mononuclear cells (PBMC, SFMC) of patients with juvenile idiopathic arthritis–enthesitis‐related arthritis (JIA‐ERA) has shown the involvement of monocytes. On the basis of CD14 and CD16 expression, monocytes are classified as classical, intermediate and non‐classical. In response to Toll‐like receptor (TLR) stimulation, intermediate monocytes produce proinflammatory cytokines and play a role in inflammatory diseases. Therefore, we have studied the microarray profile of monocytes, the frequency of their subsets and cytokine production. Monocyte‐specific microarray analysis was performed in six healthy controls' PBMC and six patients' PBMC and SFMC using Illumina chips WG12. Monocyte subsets were assessed in 46 patients with JIA‐ERA and 17 healthy controls and 17 disease controls by flow cytometry. Interleukin (IL)−23 and tumour necrosis factor (TNF) levels were measured in culture supernatants of eight controls and seven patients' PBMC/SFMC with/without lipopolysaccharide (LPS) stimulation. Cytokine‐producing intermediate monocytes were assessed by flow cytometry. Genes related to antigen presentation, cytokine signalling and TLR pathway were regulated differentially in PB and synovial monocytes of patients with JIA‐ERA. Key genes of intermediate monocytes, such as CLEC10A and MARCO, were expressed three‐ to fourfold more in JIA‐ERA. In PB, the frequency of intermediate monocytes was significantly higher in JIA‐ERA (4·90% ± 3·5) compared to controls (1·8% ± 1·06; P < 0·001). Patients' synovial cells also had more intermediate monocytes compared to PB (11·25% ± 11·32, 5·9% ± 4·8; P = 0.004). Intermediate monocytes are the major producers of IL‐23. Thus, intermediate monocytes may play an important role in JIA‐ERA, possibly by producing cytokines, and contribute to joint inflammation.
Keywords: gene expression, inflammation, intermediate monocytes, juvenile arthritis, synovial fluid
Introduction
Enthesitis‐related arthritis (ERA), a category of juvenile idiopathic arthritis (JIA), is characterized by lower limb asymmetrical arthritis and enthesitis 1. While the underlying causes of this disease are unknown, it has a strong association with human leucocyte antigen (HLA) B27. Later in life these children develop inflammatory back pain and a proportion of them progress to ankylosing spondylitis (AS). Thus it has been proposed to rename this category as juvenile spondyloarthropathy (JSpA). Studies aimed at understanding the pathogenic components suggest a role for innate immune cells. In particular, gene expression profiling of patient synovial fluid mononuclear cells (SFMCs) points to dysregulation of genes associated with natural killer cells and monocytes besides inflammatory mediators 2.
Monocytes have been classified broadly into three categories on the basis of CD14 and CD16 expression 3. The major population of monocytes (80–85%) with high CD14 expression and no CD16 expression are termed classical monocytes (CD14++CD16–). They have high phagocytic capabilities. The population with medium CD14 and CD16 expression are known as intermediate monocytes (CD14++CD16+; 5%). They have good antigen presentation and processing properties. The third subset is the non‐classical monocyte, which constitutes 10% of the monocytes. They have low CD14 and high CD16 expression (CD14+CD16++) and have patrolling behaviour in vivo 3, 4.
Intermediate monocytes express high levels of class MHC II marker (HLA‐DR) and antigen processing and presentation molecules such as CD74 (HLA‐DR‐associated invariant chain) and HLA‐DO. In addition, this monocyte subset has a higher cell surface expression of CD40 (co‐stimulatory molecule) and CD54 (intracellular adhesion molecule 1 or ICAM‐1), which induces T cell proliferation and stimulation 3, 4. Furthermore, in response to bacterial components such as lipopolysaccharide (LPS), these monocytes produce interleukin (IL)−6, IL‐1β and tumour necrosis factor (TNF)‐α 5 preferentially. CD14+CD16+ monocytes exhibit pro‐angiogenic behaviour and could form cell clusters upon vascular endothelial growth factor (VEGF) stimulation 6.
Patients with autoimmune diseases such as rheumatoid arthritis (RA) and systemic lupus erythematosus (SLE) have an increased frequency of intermediate monocytes 7, 8, 9. This suggests that they have role in immune‐mediated inflammation. Intermediate monocytes also skew the naive T cell population towards the pathogenic T helper type 17 (Th17) population in RA 5. Recently, even the non‐classical monocytes were found to help in the expansion of Th17 cells 10. JIA‐ERA is thought to be a Th17‐mediated disease 11, thus it would be interesting to determine these monocyte subsets in JIA‐ERA.
JIA‐ERA patients have increased synovial fluid levels of monocyte‐derived cytokines which possibly mediate joint inflammation 12. To dissect further the role of monocytes we performed gene expression analysis of monocytes isolated from patients' peripheral blood and synovial fluid. We studied the frequency of monocyte subsets in patients and controls and also their correlation with disease activity.
Methods
Patients and controls
Peripheral blood (PB) was collected from patients with JIA‐ERA and other JIA (enrolled as disease controls) who satisfied the International League of Association of Rheumatology (ILAR) criteria 1. Synovial fluid was obtained from those JIA‐ERA patients who required intra‐articular steroid injection as a part of treatment. Clinical assessments such as tender joint counts and swollen joint counts (SJC) were performed by a rheumatologist. The study was approved by the institutional ethics committee and consent was obtained from children/parents for their participation. Gender‐matched healthy controls were also included into the study.
Separation of mononuclear cells
Synovial fluid and peripheral blood mononuclear cells (SFMC and PBMC) were isolated by density gradient centrifugation using Histopaque 1077 (Sigma‐Aldrich, St Louis, MO, USA). Synovial fluid was first centrifuged at 700 g for 10 min. The cell‐free supernatant was stored in aliquots at −80°C. The cell pellet was suspended in 1× phosphate‐buffered saline (PBS). Blood was diluted with an equal volume of PBS. The diluted sample was layered carefully on Histopaque‐1077 at a ratio of 3 : 1 and the tube was centrifuged at 650 g for 30 min. The interface containing mononuclear cells was collected and washed thrice at 470 g for 15 min, at 300 g for 12 min and finally at 210 g for 10 min.
Separation of cell subsets
All steps henceforth were conducted at 4°C or in ice. Washing buffer, cell separation columns and magnetic separator were purchased from Miltenyi Biotech (Auburn, CA, USA). PBMCs/SFMCs were suspended in washing buffer (10 μl buffer per 106 total cells); 5 μl CD14 microbeads per 106 cells were added to the cell suspension and incubated for 15 min in the dark. Cells were washed with 2 ml buffer at 450 g for 5 min. Meanwhile, a ferromagnetic‐less sample depletion (LD) column was placed within the magnetic field of the magnetic affinity cell sorting (MACS) separator and washed with buffer.
The cells were suspended in 500 μl buffer with thorough mixing and then passed through the LD column. The column was never allowed to run dry and no more than 500 μl buffer was added at a time. Samples were eluted by flushing four to six times with buffer. The eluent was the CD14‐negative fraction. The column was removed subsequently from the magnetic field and filled with 3 ml buffer. Using a sterile plunger (provided with columns), the magnetically labelled cells were dislodged forcefully from the column into a fresh tube. This population comprised CD14+ cells. Purity was checked by flow cytometry.
RNA isolation
RNA was isolated by columns (RNeasy kit; Qiagen, Valencia, CA, USA); integrity was checked using the Agilent Bioanalyser (Agilent Technologies, Inc., Santa Clara, CA, USA). Samples with RNA integrity number (RIN) >8 were selected for further microarray procedures.
cDNA conversion and labelling
RNA isolation, cRNA conversion and labelling, array hybridization and scanning were performed at Sandor Proteomics, Hyderabad. mRNA was converted to cDNA by incubating at 42°C for 2 h. A second strand was synthesized at 16°C for 2 h. Double‐stranded cDNA was transcribed in vitro with T7 RNA polymerase and labelled with biotinylated uridine triphosphate (UTP).
Array hybridization and scanning
Human WG‐6 U3 Bead Chip Array were incubated overnight with streptavidin‐cyanin 3 (Cy3) and labelled cRNA. Data were scanned and imported into the bead study. Background correction was performed and data were normalized.
Data analysis
Genes were expressed differentially at a fold‐change cut‐off of ≥ 2·0 and a detection P‐value of <0·05 was chosen for further analysis. The online data analysis tool david was used to identify the significantly dysregulated pathways. A P‐value of <0·05 was considered significant after correcting for multiple testing using Benjamini–Hochberg correction.
Flow cytometry for monocyte subset
PB and SFMCs were stained with anti CD14‐fluorescein isothiocyanate (FITC), anti CD16‐allophycocyanin (APC) antibodies and isotype controls (BD Biosciences, San Jose, CA, USA). Cells were analysed in a Beckman Coulter Flow cytometer using Navios 6.1 software. CD14highCD16– cells were considered as classical monocytes, CD14highCD16low were considered as intermediate monocytes and CD14lowCD16high were taken as non‐classical monocytes (Supporting information, Fig. S1).
IL‐23p19 and TNF production by monocyte subsets
Intermediate monocytes skew naive T cells towards Th17 cells, thus we looked at IL‐23 production by intermediate monocytes. As intermediate monocytes comprise a small fraction of monocytes and a large volume of blood (approximately 200–250 ml) is needed to isolate adequate intermediate monocytes for in‐vitro experiment, we used flow cytometry to identify intermediate monocytes producing cytokines. Patients' PBMCs (2 × 106) were stimulated with 100 ng LPS for 5 h at 37°C and 5% CO2. Cells were then surface‐stained with anti‐CD14 FITC and anti‐CD16 peridinin chlorophyll (PerCP) (BD Biosciences). After fixation, cells were permeabilized and stained for intracellular cytokine anti‐IL‐23p19 phycoerythrin (PE) (R&D Systems, Minneapolis, MN, USA). IL‐23p19‐producing monocytes were analysed in all the monocyte subset gates. In addition, we measured the TNF‐producing monocytes in all the monocyte subset gates.
Expression of CLEC10A and HLA‐DR on monocytes
Whole blood from nine patients with JIA‐ERA and five gender‐matched healthy controls were stained for anti‐CD14 FITC, anti‐CD16 APC, anti‐CLEC10A PE and anti‐HLA‐DR PerCP. Mean fluorescence intensity (MFI) of CLEC10A and HLA‐DR were measured in total monocytes as well as in the monocyte subset gates.
Statistical analysis
The frequency of monocyte subsets is expressed as mean and standard deviation. Intergroup comparison and paired analysis between blood and synovial fluid was performed using non‐parametric tests. A P‐value less than 0·05 was taken as significant.
Results
Patients and controls
Monocytes from PBMCs and SFMCs from six JIA‐ERA patients were subjected to transcriptional profiling using microarrays. The median age of patients at the time of recruitment was 14 years and median duration of disease was 33 months. Five patients were HLA‐B27‐positive and all had active arthritis. The median age of gender‐matched healthy controls was 25 (range = 24–27) years.
Forty‐six JIA‐ERA patients, 17 patients with other forms of JIA and 17 healthy controls were studied for monocyte subset analysis. The median duration of disease was 48 months in JIA‐ERA and 24 months in other JIA subjects. Synovial fluid samples were available from 13 patients with JIA‐ERA. Demographic details are given in Table 1.
Table 1.
Clinical features of study population
| Demographic details |
JIA‐ERA (n = 46) |
Disease control (n = 17) [polyJIA (n = 9), oligoJIA (n = 4), SoJIA (n = 3), PsA (n = 1)] |
|---|---|---|
| Median age (range) in years | 16 (7–21) | 14 (7–26) |
| Median disease duration in months | 48 (3–192) | 24 (5–192) |
| Number with active arthritis | 41 | 14 |
| Number with enthesitis | 20 | 1 |
| Number with sacroiliitis | 13 | None |
| Number with uveitis | 02 | None |
| Numbers with inflammatory back pain | 18 | None |
| HLA B27‐positive | 39 | None |
| Median ESR | 65 (15–130) | 53 (10–142) |
| Patients on NSAIDS | 43 | 15 |
| Patients on methotrexate (MTX) | 8 | 4 |
| Patients with no drug | 2 | 2 |
JIA‐ERA = juvenile idiopathic arthritis–enthesitis‐related arthritis; ESR = erythrocyte sedimentation rate; HLA = human leucocyte antigen; NSAIDs = non‐steroidal anti‐inflammatory drugs.
The median erythrocyte sedimentation rate (ESR) of patients with JIA‐ERA was 65 mm (15–130) and of disease controls was 53 mm (10–142). Median swollen joint count and tender joint count of patients with JIA‐ERA was 2 (0–12) and 2 (0–20) and that of disease controls was 3 (0–21) and 4 (0–16), respectively. All patients except three in the JIA‐ERA group and two in the disease control group were on non‐steroidal anti‐inflammatory drugs (NSAIDs). Eight patients with JIA‐ERA and four with other JIA were on methotrexate.
Differential gene expression between JIA‐ERA and healthy controls
JIA‐ERA PB monocytes versus healthy control PB monocytes
A total of 2114 genes were dysregulated, 864 of which were up‐regulated and 1250 were down‐regulated (Supporting information, Data 1). When subjected to david analysis, a total of 27 pathways were found to be significantly dysregulated (Supporting information, Data 3). Among these, only eight pathways, i.e. immunoglobulin (IgA) production, lysosome, endocytosis, Toll‐like receptor, chemokine signalling, antigen processing and presentation, cytokine‐cytokine receptor interaction and Fcγ receptor‐mediated pathway had immunological relevance (Supporting information, Data 4). Some pathways were involved in cell metabolism, such as amino acid synthesis, glycolysis and galactose metabolism.
Further key genes related to intermediate monocytes, such as CLEC10A, GFRA2, MARCO and CD300C 13, were also found to be up‐regulated (Table 2). The functions of various genes specific to intermediate monocytes are described in Table 3. Similarly, in the genes related to non‐classical monocytes such as Siglec10 and CEACAM1, more than two fold changes were observed.
Table 2.
Fold changes of key genes of intermediate monocyte subset up‐regulated in patients' PB and SF
| JIA‐ERA PB monocytes versus control PB monocytes | JIA‐ERA SF monocytes versus ERA PB monocytes | ||
|---|---|---|---|
| Genes | Fold change | Genes | Fold change |
| CLEC10A | 4·3 | CLEC10A | 2·4 |
| MARCO | 2·94 | MARCO | 2·59 |
| HLA‐DRB1 | 6·4 | HLA‐DRB4 | 2·18 |
| HLA‐DRB5 | 5·4 | HLA‐DRB3 | 2·69 |
| GFRA2 | 3·03 | SCD | 2·57 |
| CD300C | 3·15 | FCGR3A | 4·74 |
| TGM2 | 4·64 | ||
| APOBEC3A | 2·9 | ||
PB = peripheral blood; SF = synovial fluid; JIA‐ERA = juvenile idiopathic arthritis–enthesitis‐related arthritis; CLEC10A = C‐type lectin domain family member 10 A; MARCO = macrophage receptor with collagenous structure; HLA‐DRB = major histocompatibility complex, class II, DR beta; GFRA2 = GDNF family receptor‐alpha2; CD300C = cluster of differentiation 300C antigen; = SCD = stearoyl CoA desaturase; FCGR3A = Fc fragment of immunoglobulin (Ig)G, low‐affinity receptor III a (CD16a); TGM2 = transglutaminase 2; APOBE3CA = apolipoprotein B mRNA editing enzyme.
Table 3.
Functions of the intermediate monocyte subset‐related genes
| Gene | Function | |
|---|---|---|
| APOBE3CA | Apolipoprotein B mRNA editing enzyme | Highly expressed in monocytes, DNA editing enzyme, restricts retroviruses |
| CD300C | CD300C antigen | Fc gamma‐dependent monocyte activation |
| CLEC10A | C‐type lectin domain family member 10 A | Binds carbohydrate, positively regulates cytokine production by NF‐κB pathway |
| FCGR3A | Fc fragment of IgG, low‐affinity receptor III a (CD16a) | Antibody‐mediated responses |
| GFRA2 | GDNF family receptor‐alpha2 | Monocyte differentiation |
| HLA‐DRB | Major histocompatibility complex, class II, DR beta | Antigen presentation |
| MARCO | Macrophage receptor with collagenous structure | Scavenger receptor, anti‐bacterial activity |
| SCD | Stearoyl CoA desaturase | Participates in inflammatory response |
| TGM2 | Transglutaminase 2 | Monocyte and macrophage activation |
APOBE3CA = apolipoprotein B mRNA editing enzyme; CD300C = cluster of differentiation 300C antigen; CLEC10A = C‐type lectin domain family member 10 A; FCGR3A = Fc fragment of immunoglobulin (Ig)G, low‐affinity receptor III a (CD16a); GFRA2 = GDNF family receptor‐alpha2; HLA‐DRB = major histocompatibility complex, class II, DR beta; MARCO = macrophage receptor with collagenous structure; SCD = stearoyl CoA desaturase; TGM2 = transglutaminase 2; NF‐κB = nuclear factor kappa B; IgG = immunoglobulin G.
JIA‐ERA SF monocytes versus JIA‐ ERA PB monocytes
A total of 1129 genes were dysregulated, 618 of which were up‐regulated and 511 were down‐regulated (Supporting information, Data 2). When subjected to david analysis, a total of 18 pathways were found to be regulated significantly differentially (Supporting information, Data 3). Among these, only five pathways were involved in immune regulation and activation, i.e. complement activity, chemokine signalling, IgA production, antigen presentation and proteasome pathway (Supporting information, Data 4). Other pathways were related to glycolysis and steroid biosynthesis, and some were also involved in other autoimmune disease such as SLE and type I diabetes.
Synovial monocytes of JIA‐ERA patients also had increased expression (two‐ to fourfold) of genes representing intermediate monocyte subsets, such as FCGR3A, CLEC10A, MARCO and TGM2 genes (Table 2).
Monocyte subset analysis
Intermediate monocytes
The frequency of intermediate monocytes (4·90% ± 3·5) was increased in the JIA‐ERA group compared to disease controls (2·20% ± 1·58, P = 0·002) and healthy controls (1·8% ± 1·06, P < 0·001) (Fig. 1a). Further, in paired analysis, intermediate monocytes were more frequent in SF (11·25% ± 11·32) compared to PB (5·9% ± 4·8, P = 0·004) (Fig. 1d).
Figure 1.
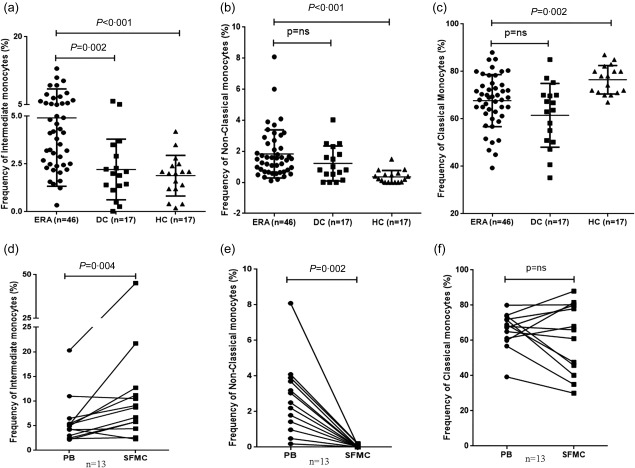
Frequency of monocyte subsets. Scatter‐plots representing the frequency of peripheral blood. (a) Intermediate monocytes (CD14++CD16+); (b) non‐classical monocytes (CD14+CD16++); and (c) classical monocytes (CD14++CD16–) in juvenile idiopathic arthritis–enthesitis‐related arthritis (JIA‐ERA) (ERA, n = 46), disease controls (DC, n = 17) and healthy controls (HC, n = 17). Paired frequency (n = 13) of (d) intermediate monocytes; (e) non‐classical monocytes; and (f) classical monocytes in peripheral blood and synovial fluid of JIA‐ERA patients.
Non‐classical monocytes
Non‐classical monocytes were more frequent in JIA‐ERA (1·83% ± 1·55) compared to healthy individuals [0·35% ± 0·4, P = not significant (n.s.)] (Fig. 1b). In contrast, the non‐classical subset of monocytes was decreased in synovial fluid (0·03% ± 0·07) compared to PB (2·7% ± 2·05, P = 0·002) (Fig. 1e).
Classical monocytes
The frequency of classical monocytes, i.e. CD14high and CD16–, was slightly less in JIA‐ERA patients (67·7% ± 10·9) compared to healthy controls (76·01% ± 6·0, P = 0·002), while no difference was observed between JIA‐ERA and disease controls (61·50% ± 13·4, P = n.s.) (Fig. 1c). The frequency of classical monocytes (61·65% ± 19·80) in SF was similar to PB (66·04% ± 10·20) (Fig. 1f).
Expression of CLEC10A and HLA‐DR on monocytes
Patients with JIA‐ERA had a higher median expression of CLEC10A [36·72 (26·02–70·35)] on total monocytes compared to controls [16·79 (16·36–32·53, P = 0·007)]. However, median expression of HLA‐DR was similar on the total monocytes of patients and controls.
Among monocyte subsets, intermediate monocytes of patients had more expression of CLEC10A [103·7 (35·14–209·2) versus 40·0 (36·5–63·0), P = 0·019] and HLA‐DR [48·14 (28·96–89·00) versus 19·4 (13·70‐24·17), P = 0·001] compared to controls.
Correlation of intermediate monocyte subsets
PB intermediate monocytes had a modest correlation with swollen joint count (r = 0·333, P = 0·03) and a high correlation with SF intermediate monocytes (r = 0·721, P = 0·006). However, no correlation was found with ESR and C‐reactive protein (CRP).
IL‐23p19‐producing intermediate monocytes
In JIA‐ERA, intermediate monocytes contributed 72·5% (56–94) of IL‐23p19‐producing monocytes. However, classical monocytes contributed 78% (35–90) of TNF‐producing monocytes (Table 4).
Table 4.
Median (range) frequency of different monocyte subsets among monocytes producing that cytokine (n = 6)
| Cytokine | Classical | Intermediate | Non‐classical |
|---|---|---|---|
| IL‐23 | 4·35% (1·2–23) | 72·5% (56–94) | 3·15% (0–6) |
| TNF‐α | 78% (35–90) | 4·2% (0·2–14) | 12·25% (0·1–35) |
IL = interleukin; TNF‐α = tumour necrosis factor alpha.
Discussion
JIA‐ERA is an HLA‐B27‐related disorder. In HLA‐B27‐related diseases innate immune cells, including monocytes, are thought to play a major role in pathogenesis. Monocytes have multiple functions, such as antigen presentation and cytokine secretion. In HLA‐B27‐related SpA, monocytes produce cytokines in response to pattern‐associated molecular patterns (PAMPs), as well as via unfolded protein response or autophagy. To our knowledge, this is the first monocyte‐specific gene expression profiling study in JIA‐ERA. Microarray analysis of monocytes of JIA‐ERA patients revealed an increased expression of genes related to intermediate monocytes. Further, the frequency of intermediate monocytes was increased in PB of patients compared to healthy controls. In paired samples, SF had a higher frequency of intermediate monocytes. PB intermediate monocytes had a positive correlation with swollen joint count.
Most of the gene expression studies available in rheumatic disease, including juvenile arthritis, are on whole blood or PBMCs. Increased expression of inflammation‐related genes which correlate with an increased PB monocyte count was found in PB gene expression profiling in RA 14. PB gene expression profiling in oligoarticular JIA (a subcategory of JIA) has revealed dysregulation of the genes involved in inflammation and monocyte/macrophage activation 15. A microarray study in different subgroups of JIA, which also included patients with JIA‐ERA, has reported dysregulation of the genes involved in complement cascade 16. We have also found up‐regulation of the complement pathway genes such as C1Q, C2, C3 and CDBPA in PB monocytes. This increase in complement pathway in JIA‐ERA is probably the consequence of increased inflammation, which leads to increased secretion of complement protein by monocytes. Gene expression profile of patients' PBMCs with jSpA has shown dysregulation of the genes related to antigen presentation, immune activation and migration of inflammatory cells, namely TLR4, NLPR3 and CXCR4 17. Taken together, these reports suggest a strong case for the use of gene expression profiling to identify potential pathogenic components in ERA. Similar to these studies, our data also revealed dysregulation of genes related to inflammation and immune activation.
Data relating to monocyte‐specific gene expression in rheumatic diseases are sparse. In type 1 diabetes and primary Sjögren's syndrome gene expression profiling of CD14+ monocytes identified cytokine signalling, chemotaxis, adhesion, motility and metabolic pathways 18. Our microarray results also identified differentially regulated pathways related to cytokines and cytokine receptors, such as IL‐15, IL‐15RA, IL‐17RA, IL‐18R and TNFA and chemokine signalling such as CCL2, CCL3, CCL5, CCL8 and CCL13 [involved in chemotaxis of monocytes, natural killer (NK) cells, T cells and neutrophils], in addition to the TLR and antigen presentation pathways. This is also similar to our previous gene expression profiling data on PBMCs and SFMCs of JIA‐ERA patients, in which genes related to monocyte functions such as CD163 scavenger receptor, CD1b, CD1d and MHC class II molecules were over‐expressed 2.
Further, the gene expression profile showed higher expression of genes related to intermediate monocytes. HLA‐DRB1, HLA‐DRB5 and FCGR3A (CD16) represent genes for antigen presentation and were found to be up‐regulated highly. Genes related to intermediate monocyte activation, i.e. GFRA2, TGM2 and CD300C, were also expressed highly in PB. CLEC10A, highly specific for intermediate monocytes and known to induce cytokine production, was over‐expressed in both PB and SF of JIA‐ERA patients. A recent gene profiling study on monocyte subsets in obesity has revealed the enhanced expression of intermediate monocytes 19. This suggests that the intermediate monocyte subset may have a role in inflammation in JIA‐ERA.
Our data of expansion of the intermediate monocyte population in patients with JIA‐ERA is in agreement with the data in other immune‐mediated diseases such as RA, SLE, Crohn's disease and ulcerative colitis 7, 8, 9, 20, 21. This population has been widely reported to expand during chronic inflammation and also has a major role in infection 3. A study in RA has reported the skewing of naive T cells towards pathogenic Th17 cells by co‐culturing with CD14+CD16+ monocytes, and this is mediated by IL‐23 production 5. However, we have not found any correlation between this subset and Th17 cell frequency in PB (data not shown), but we have not performed in‐vitro co‐culture experiments. Intermediate monocytes are more frequent in JIA‐ERA compared to disease controls; this could be explained by other subtypes of JIA having a different pathogenesis. For example, SoJIA is more similar to autoinflammatory disease and is driven by cytokines such as IL‐1b, IL‐18 and IL‐6 as major players, and oligo‐JIA and polyarticular JIA are more driven by the adaptive immune system, as they have autoantibodies. However, no data are available on intermediate monocytes in different JIA subgroups.
Similar to intermediate monocytes, non‐classical monocytes were also increased in JIA‐ERA PB, suggesting that the CD16+ monocytes are involved in systemic inflammation. CD16 is a low‐affinity FcγRIII receptor, which is considered mainly as an activation marker. However, studies in RA, SLE and Crohn's disease have reported a similar frequency of CD14+CD16++ monocytes in patients and controls 8, 9, 21. Recently, non‐classical monocytes were also found to induce the expansion of Th17 cells, suggesting their inflammatory nature 10.
In SF of JIA‐ERA, intermediate monocytes were increased further compared to PB. Previous studies have also reported the expansion of intermediate monocytes at the site of inflammation, such as at the synovial site in RA 22, salivary glands in Sjögren's syndrome 23 and intestinal mucosa in Crohn's disease 20. This was validated further as increased expression of genes related to intermediate monocytes (HLA‐DR, CLEC10A, MARCO) in SF monocytes. This enrichment of intermediate monocytes with increased antigen presentation and cytokine production properties at the local site suggests their important role in mediating local inflammation. A decrease in the SF non‐classical monocyte subset may suggest selective recruitment of intermediate monocytes at the synovial site. Another possible explanation is the down‐regulation of CD16 expression on monocytes as reported in RA, when PB monocytes were stimulated with LPS 22. Our limited data on six patients suggest that intermediate monocytes are the major producers of IL‐23, a key cytokine in the pathogenesis of JIA‐ERA. IL‐23 acts on CD4 cells, NK cells and gamma‐delta T cells and induces IL‐17 production. We have recently shown an increased frequency of IL‐17‐producing CD4, NK and gamma‐delta T cells in patients with JIA‐ERA 24.
To summarize our findings, transcriptome analysis of monocytes of JIA‐ERA patients had increased expression of genes related to antigen presentation, the TLR pathway and cytokine and chemokine signalling. Further key genes related to intermediate monocytes (CLEC10A, GFRA2, MARCO) were up‐regulated in patients' PB and SF. This was supported further by the increased frequency of intermediate monocytes in PB and SF of patients compared to controls. These observations thus indicate that the intermediate monocyte subset may play an important role in JIA‐ERA pathogenesis by producing proinflammatory cytokines and altering the immune environment.
Disclosure
The authors declare no disclosures.
Supporting information
Additional Supporting information may be found in the online version of this article at the publisher's web‐site:
Fig. S1. (a) Forward‐ and side‐scatter plot of lysed peripheral blood (PB) of patient samples, monocyte population gated. (b) Dot‐plot representing the CD14 and CD16 population in the monocyte gate; cells expressing CD14++\CD16– are classical monocytes, CD14++CD16+ are intermediate monocytes and CD14+CD16++ are non‐classical monocytes (++high expression, +medium expression, –no expression).
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Acknowledgement
This work was supported by grants to AA from Indian council of Medical Research and Department of Biotechnology. PG was supported by a senior research fellowship from university grants commission.
References
- 1. Petty RE, Southwood TR, Manners P et al International League of Associations for Rheumatology classification of juvenile idiopathic arthritis: second revision, Edmonton 2001. J Rheumatol 2004; 31:390–2. [PubMed] [Google Scholar]
- 2. Myles A, Tuteja A, Aggarwal A. Synovial fluid mononuclear cell gene expression profiling suggests dysregulation of innate immune genes in enthesitis‐related arthritis patients. Rheumatology 2012; 51:1785–9. [DOI] [PubMed] [Google Scholar]
- 3. Wong KL, Yeap WH, Tai JJ, Ong SM, Dang TM, Wong SC. The three human monocyte subsets: implications for health and disease. Immunol Res 2012; 53:41–57. [DOI] [PubMed] [Google Scholar]
- 4. Ziegler‐Heitbrock L, Hofer TP. Toward a refined definition of monocyte subsets. Front Immunol 2013; 4:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rossol M, Kraus S, Pierer M, Baerwald C, Wagner U. The CD14 (bright) CD16+ monocyte subset is expanded in rheumatoid arthritis and promotes Th17 expansion. Arthritis Rheum 2012; 64:671–7. [DOI] [PubMed] [Google Scholar]
- 6. Zawada AM, Rogacev KS, Rotter B et al SuperSAGE evidence for CD14++CD16+ monocytes as a third monocyte subset. Blood 2011; 118:e50–61. [DOI] [PubMed] [Google Scholar]
- 7. Kawanaka N, Yamamura M, Aita T et al CD14+CD16+ blood monocytes and joint inflammation in rheumatoid arthritis. Arthritis Rheum 2002; 46:2578–86. [DOI] [PubMed] [Google Scholar]
- 8. Henriques A, Inês L, Carvalheiro T et al Functional characterization of peripheral blood dendritic cells and monocytes in systemic lupus erythematosus. Rheumatol Int 2012; 32:863–9. [DOI] [PubMed] [Google Scholar]
- 9. Cairns AP, Crockard AD, Bell AL. The CD14+ CD16+ monocyte subset in rheumatoid arthritis and systemic lupus erythematosus. Rheumatol Int 2002; 21:189–92. [DOI] [PubMed] [Google Scholar]
- 10. Traunecker E, Gardner R, Fonseca JE et al Blocking of LFA‐1 enhances expansion of Th17 cells induced by human CD14(+) CD16(++) nonclassical monocytes. Eur J Immunol 2015; 45:1414–25. [DOI] [PubMed] [Google Scholar]
- 11. Mahendra A, Misra R, Aggarwal A. Th1 and Th17 predominance in the enthesitis‐related arthritis form of juvenile idiopathic arthritis. J Rheumatol 2009; 36:1730–6. [DOI] [PubMed] [Google Scholar]
- 12. Saxena N, Aggarwal A, Misra R. Elevated concentrations of monocyte derived cytokines in synovial fluid of children with enthesitis related arthritis and polyarticular types of juvenile idiopathic arthritis. J Rheumatol 2005; 32:1349–53. [PubMed] [Google Scholar]
- 13. Wong KL, Tai JJ, Wong WC, Han H, Sem X, Yeap WH. Gene expression profiling reveals the defining features of the classical, intermediate and non‐classical human monocyte subsets. Blood 2011; 118:e16–31. [DOI] [PubMed] [Google Scholar]
- 14. Batliwalla FM, Baechler EC, Xiao X et al Peripheral blood gene expression profiling in rheumatoid arthritis. Genes Immun 2005; 6:388–97. [DOI] [PubMed] [Google Scholar]
- 15. Hunter PJ, Nistala K, Jina N et al Biologic predictors of extension of oligoarticular juvenile idiopathic arthritis as determined from synovial fluid cellular composition and gene expression. Arthritis Rheum 2010; 62:896–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barnes MG, Grom AA, Thompson SD et al Subtype‐specific peripheral blood gene expression profiles in recent onset juvenile idiopathic arthritis. Arthritis Rheum 2009; 60:2102–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lamot L, Borovecki F, Bukovac LT et al Aberrant expression of shared master‐key genes contributes to the immunopathogenesis in patients with juvenile spondyloarthritis. PLOS ONE 2014; 9:e115416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Padmos RC, Schloot NC, Beyan H et al Distinct monocyte gene‐expression profiles in autoimmune diabetes. Diabetes 2008; 57:2768–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Devêvre EF, Renovato‐Martins M, Clément K, Sautès‐Fridman C, Cremer I, Poitou C. Profiling of the three circulating monocyte subpopulations in human obesity. J Immunol 2015; 194:3917–23. [DOI] [PubMed] [Google Scholar]
- 20. Grip O, Bredberg A, Lindgren S, Henriksson G. Increased subpopulations of CD16(+) and CD56(+) blood monocytes in patients with active Crohn's disease. Inflamm Bowel Dis 2007; 13:566–72. [DOI] [PubMed] [Google Scholar]
- 21. Koch S, Kucharzik T, Heidemann J, Nusrat A, Luegering A. Investigating the role of proinflammatory CD16+ monocytes in the pathogenesis of inflammatory bowel disease. Clin Exp Immunol 2010; 161:332–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yoon BR, Yoo SJ, Choi Y et al Functional phenotype of synovial monocytes modulating inflammatory T‐cell responses in rheumatoid arthritis(RA). PLOS ONE 2014; 9:e109775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wildenberg ME, Welzen‐Coppens JM, van Helden‐Meeuwsen CG et al Increased frequency of CD16+ monocytes and the presence of activated dendritic cells in salivary glands in primary Sjögren syndrome. Ann Rheum Dis 2009; 68:420–6. [DOI] [PubMed] [Google Scholar]
- 24. Gaur P, Misra R, Aggarwal A. Natural killer cell and gamma delta T cell alterations in enthesitis related arthritis category of juvenile idiopathic arthritis. Clin Immunol 2015; 161:163–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting information may be found in the online version of this article at the publisher's web‐site:
Fig. S1. (a) Forward‐ and side‐scatter plot of lysed peripheral blood (PB) of patient samples, monocyte population gated. (b) Dot‐plot representing the CD14 and CD16 population in the monocyte gate; cells expressing CD14++\CD16– are classical monocytes, CD14++CD16+ are intermediate monocytes and CD14+CD16++ are non‐classical monocytes (++high expression, +medium expression, –no expression).
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information


