Abstract
The Flavivirus genus encompasses several arboviruses of public health significance such as dengue, yellow fever, and Zika viruses. It also includes insect-specific flaviviruses (ISFs) that are only capable of infecting insect hosts. The vast majority of mosquito-infecting flaviviruses have been associated with mosquito species of the Aedes and Culex genera in the Culicinae subfamily, which also includes most arbovirus vectors. Mosquitoes of the Anophelinae subfamily are not considered significant arbovirus vectors; however, flaviviruses have occasionally been detected in field-caught Anopheles specimens. Whether such observations reflect occasional spillover or laboratory contamination or whether Anopheles mosquitoes are natural hosts of flaviviruses is unknown. Here, we provide in silico and in vivo evidence of transcriptionally active, flavivirus-derived endogenous viral elements (EVEs) in the genome of Anopheles minimus and Anopheles sinensis. Such non-retroviral endogenization of RNA viruses is consistent with a shared evolutionary history between flaviviruses and Anopheles mosquitoes. Phylogenetic analyses of the two newly described EVEs support the existence of a distinct clade of Anopheles-associated ISFs.
Keywords: Flavivirus, host range, Anopheles sinensis, Anopheles minimus, ISF, EVE
1. Introduction
Flaviviruses are positive-sense, single-stranded RNA viruses that infect a broad range of hosts including vertebrates (e.g. birds, primates) and arthropods (e.g. ticks, mosquitoes). In addition to major arboviruses of public health significance such as dengue, Zika, West Nile and yellow fever viruses, the Flavivirus genus also includes vertebrate-specific (not known vector; NKV) and insect-specific (insect-specific flaviviruses; ISFs) members (Moureau et al. 2015). The majority of mosquito-infecting flaviviruses have been associated with members of the Culicinae subfamily, mainly from the Culex and Aedes genera. Anopheles mosquitoes in the Anophelinae subfamily are well known for their role in the transmission of human malaria parasites, but they are not considered important vectors of arboviruses in general, and of flaviviruses in particular (Clements 2012). The only notable exception of Anopheles-transmitted arbovirus is the alphavirus O’nyong’nyong virus (Brault et al. 2004). Nevertheless, several studies have detected flaviviruses in field-caught Anopheles mosquitoes from different parts of the world. In North America, West Nile virus (WNV) was detected in Anopheles punctipennis (Bernard et al. 2001; Kulasekera et al. 2001). In Asia, Japanese encephalitis virus was detected in Anopheles sinensis (Feng et al. 2012; Liu et al. 2013). In Europe, Anopheles maculipennis was found positive for WNV (Filipe 1972; Kemenesi et al. 2014) and Usutu virus (Calzolari et al. 2013). Interestingly, some ISFs were also detected in An. sinensis (Zuo et al. 2014; Liang et al. 2015) and Anopheles atroparvus (Aranda et al. 2009). It is unknown whether these detections reflect occasional spillover or laboratory contamination, or whether Anopheles mosquitoes are in fact natural hosts of flaviviruses.
Endogenous viral elements (EVEs) are chromosomal integrations of partial or full viral genetic material into the genome of a host species. Not only retroviruses, whose replication cycle includes incorporation of a DNA form of the RNA viral genome into the host cell genome, but virtually all types of eukaryotic viruses can become endogenous (Feschotte and Gilbert 2012). Non-retroviral EVEs have been documented in the genome of a wide variety of host species, including vertebrates and arthropods (Feschotte and Gilbert 2012). Unlike detection of exogenous viruses, subject to possible laboratory or environmental contamination, EVEs are likely to reflect a long-lasting evolutionary relationship between an RNA virus and its natural host. This is because endogenization, for a single-stranded RNA virus, requires 1, reverse transcription; 2, integration of virus-derived DNA into the genome of germinal host cells; and 3, fixation of the integrated sequence in the host population (Holmes 2011; Aiewsakun and Katzourakis 2015). The low probability of this combination of events makes endogenization exceedingly unlikely to occur unless the viral infection is common in the host population over long evolutionary times. For example, flavivirus-derived EVEs have been reported in the genomes of Aedes aegypti (Crochu et al. 2004; Katzourakis and Gifford 2010) and Aedes albopictus (Crochu et al. 2004; Roiz et al. 2009; Chen et al. 2015). These EVEs are phylogenetically related to the clade of Aedes-associated ISFs (Crochu et al. 2004; Roiz et al. 2009; Katzourakis and Gifford 2010; Chen et al. 2015), which is consistent with the ancient evolutionary relationship between Aedes mosquitoes and ISFs.
Here, we report the discovery of two flavivirus-derived EVEs in the genomes of An. minimus and An. sinensis mosquitoes. We screened 24 publicly available Anopheles genomes (Holt et al. 2002; Zhou et al. 2014; Logue et al. 2015; Neafsey et al. 2015) for flaviviral sequences, and validated in silico hits both at the DNA and RNA levels in vivo. The two newly described flavivirus-derived EVEs are phylogenetically related to ISFs, and support the existence of a previously unsuspected Anopheles-associated clade of ISFs.
2. Material and Methods
2.1 In silico survey
2.1.1 Genome screen
Twenty-four Anopheles genomes (full list and accession numbers are provided in Table 1) were screened by tBLASTn (BLAST+ 2.2.28) (Camacho et al. 2009) for the presence of flavivirus-derived EVEs using a collection of 50 full flavivirus polyprotein queries (full list and accession numbers are provided in Supplementary Table S1) representing the currently known diversity of the Flavivirus genus. The sequences of hits whose E-value was < 10−4 were extracted using an in-house bash shell script. In order to reconstruct putative flavivirus-derived EVEs, BLAST hits were clustered using CD-HIT v.4.6.1 (Li and Godzik 2006) and aligned using MAFFT v7.017 (Katoh et al. 2002). Putative EVEs were extracted and used as query for a reciprocal tBLASTn (BLAST+ 2.2.30) against the National Center for Biotechnology Information (NCBI) nucleotide database (E-value < 10−4). Genetic features of identified EVE were analyzed using the NCBI Conserved Domain Database (Marchler-Bauer et al. 2015). Nucleotide sequence data reported are available in the Third Party Annotation Section of the DDBJ/ENA/GenBank databases under the accession numbers TPA: BK009978-BK009980.
Table 1.
Anopheles genomes screened in this study.
| Species | GenBank WGS project | Assembly | GenBank assembly ID |
|---|---|---|---|
| An. albimanus | APCK01 | AalbS1 | GCA_000349125.1 |
| An. arabiensis | APCN01 | AaraD1 | GCA_000349185.1 |
| An. atroparvus | AXCP01 | AatrE1 | GCA_000473505.1 |
| An. christyi | APCM01 | AchrA1 | GCA_000349165.1 |
| An. coluzzii | ABKP02 | AcolM1 | GCA_000150765.1 |
| An. culicifacies A | AXCM01 | AculA1 | GCA_000473375.1 |
| An. darlingi | ADMH02 | AdarC3 | GCA_000211455.3 |
| An. dirus A | APCL01 | AdirW1 | GCA_000349145.1 |
| An. epiroticus | APCJ01 | AepiE1 | GCA_000349105.1 |
| An. farauti | AXCN01 | AfarF1 | GCA_000473445.1 |
| An. funestus | APCI01 | AfunF1 | GCA_000349085.1 |
| An. gambiae | AAAB01 | AgamP4 | GCA_000005575.2 |
| An. koliensis | JXXB01 | AKwgs3 | GCA_000956275.1 |
| An. maculatus B | AXCL01 | AmacM1 | GCA_000473185.1 |
| An. melas | ACXO01 | AmelC1 | GCA_000473525.1 |
| An. merus | AXCQ01 | AmerM1 | GCA_000473845.1 |
| An. minimus A | APHL01 | AminM1 | GCA_000349025.1 |
| An. nili | ATLZ01 | Anili1 | GCA_000439205.1 |
| An. punctulatus | JXXA01 | APwgs2 | GCA_000956255.1 |
| An. quadriannulatus A | APCH01 | AquaS1 | GCA_000349065.1 |
| An. sinensis: Sinensis (Korean strain) | AXCK02 | AsinS2 | GCA_000472065.2 |
| An. sinensis: China (Chinese strain) | ATLV01 | AsinC2 | GCA_000441895.2 |
| An. stephensi: Indian | ALPR002 | AsteI2 | GCA_000300775.2 |
| An. stephensi: SDA-500 | APCG01 | AsteS1 | GCA_000349045.1 |
WGS, whole-genome shotgun sequencing.
2.1.2 Phylogenetic analyses
Translated EVE sequences were aligned to the corresponding sections of several flavivirus polyproteins (Supplementary Table S1) with MAFFT v7.017 and phylogenetically uninformative positions were trimmed using TrimAI v.1.3 (Capella-Gutiérrez et al. 2009) accessed through the webserver Phylemon 2 (Sánchez et al. 2011). The trimmed alignments were used to construct phylogenetic trees with PhyML Best AIC Tree (Sánchez et al. 2011). Best substitution models were Blosum62 + I + G+F for An. minimus (regardless of gene-coding domains) and Blosum62 + I + G for An. sinensis.
2.1.3 Transcriptome screen
Published RNA sequencing (RNA-seq) data were retrieved from NCBI Sequence Read Archive (Leinonen et al. 2011) and explored for the presence of the previously identified EVE sequences. Only one An. minimus transcriptome sequence read archive was found under accession number SRX265162. Six An. sinensis transcriptome sequence read archives were found under accession numbers SRX448985, SRX449003, SRX449006, and SRX277584 for experiments using Illumina sequencing technology, and SRX218691 and SRX218721 for experiments using Roche 454 sequencing technology. RNA-seq reads were mapped to the EVE nucleotide sequence using Bowtie2 v2.1.0 (Langmead and Salzberg 2012). The alignment file was converted, sorted, and indexed with Samtools v0.1.19 (Li et al. 2009). Coverage was assessed using bedtools v2.17.0 (Quinlan and Hall 2010).
2.2 In vivo validation
2.2.1 Mosquitoes
An. minimus and An. sinensis mosquitoes were obtained through BEI Resources (www.beiresources.org), National Institute of Allergy and Infectious Diseases, National Institutes of Health (An. minimus MINIMUS1, MRA-729; An. sinensis SINENSIS, MRA-1154). Anopheles minimus and An. sinensis specimens came from the 132nd and 65th generations of laboratory colonization, respectively. Eggs were hatched in filtered tap water, reared in 24 × 34 × 9 cm plastic trays and fed with fish food (TetraMin, Tetra, Melle, Germany). Adults were maintained in 30 × 30 × 30 cm screened cages under controlled insectary conditions (28 ± 1 °C, 75 ± 5% relative humidity, 12:12 h light–dark cycle). They were provided with cotton soaked in a 10% (m/v) sucrose solution ad libitum. An. stephensi nucleic acids, used as a reaction control, were kindly provided by the Genetics and Genomics of Insect Vectors unit, Institut Pasteur, Paris.
2.2.2 EVE genomic integration
Mosquitoes were homogenized in pools of 10 separated by sex in 300 µl of Dulbecco’s phosphate-buffered saline (DPBS) during two rounds of 30 s at 5,000 rpm in a mixer mill (Precellys 24, Bertin Technologies, Montigny le Bretonneux, France). DNA was extracted using All Prep DNA/RNA Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. EVE presence in genomic DNA was assessed by 35 cycles of PCR using Taq Polymerase (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) (Supplementary Table S2). PCR primers were designed to generate an amplicon spanning part of the EVE sequence and a section of the flanking host sequence. Identity of the EVE sequence was confirmed by Sanger sequencing of the PCR product.
2.2.3 EVE transcription level
Mosquitoes were homogenized in pools of five separated by sex or development stage in 300 µl of DPBS during two rounds of 30 s at 5,000 rpm in a mixer mill (Precellys 24). RNA was extracted from mosquito homogenates separated by sex using TRIzol Reagent (Life Technologies, Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s instructions. Samples were treated with Turbo DNA-free kit (Life Technologies) and reverse transcribed using random hexamers and M-MLV reverse transcriptase (Invitrogen). Complementary DNA was amplified with 35 cycles of PCR for An. minimus and 40 cycles of PCR for An. sinensis, respectively, using DreamTaq polymerase (Thermo Fisher Scientific) and primers located within the EVE sequence (Supplementary Table S2). To verify that RNA samples were free of DNA contamination, two sets of primers spanning exons 3 and 4 of the RPS7 gene of both Anopheles species (under VectorBase annotation number AMIN008193 and ASIC017918 for An. minimus and An. sinensis, respectively) were designed (Supplementary Table S2). Because the corresponding DNA sequence includes intron 3, DNA contamination is expected to result in a larger PCR product. The length of intron 3 is 252 base pairs (bp) for An. minimus and 295 bp for An. sinensis.
3. Results
The in silico screen of 24 Anopheles genomes identified two flavivirus-derived EVEs, one in the reference genome sequence of An. minimus and one in both genome sequences available for An. sinensis (Table 2).
Table 2.
Newly described flavivirus-derived EVEs in An. minimus and An. sinensis genomes.
| Element | GenBank accession no. | Protein identity | Supercontig accession no. | Supercontig length (bp) | Coordinates |
|||
|---|---|---|---|---|---|---|---|---|
| Supercontig |
Viral genomea |
|||||||
| Start | End | Start | End | |||||
| An. minimus EVE | BK009978 | NS4-NS5 | KB664005.1 | 2,043 | 107 | 1,987 | 6,567 | 8,432 |
| An. sinensis EVE (Korean strain) | BK009979 | NS3 | AXCK02024744 | 2,797 | 822 | 1,613 | 5,214 | 5,822 |
| An. sinensis EVE (Chinese strain) | BK009980 | NS3 | ATLV01019207.1 | 7,380 | 3,578 | 4,376 | 5,214 | 5,822 |
3.1 An. minimus EVE
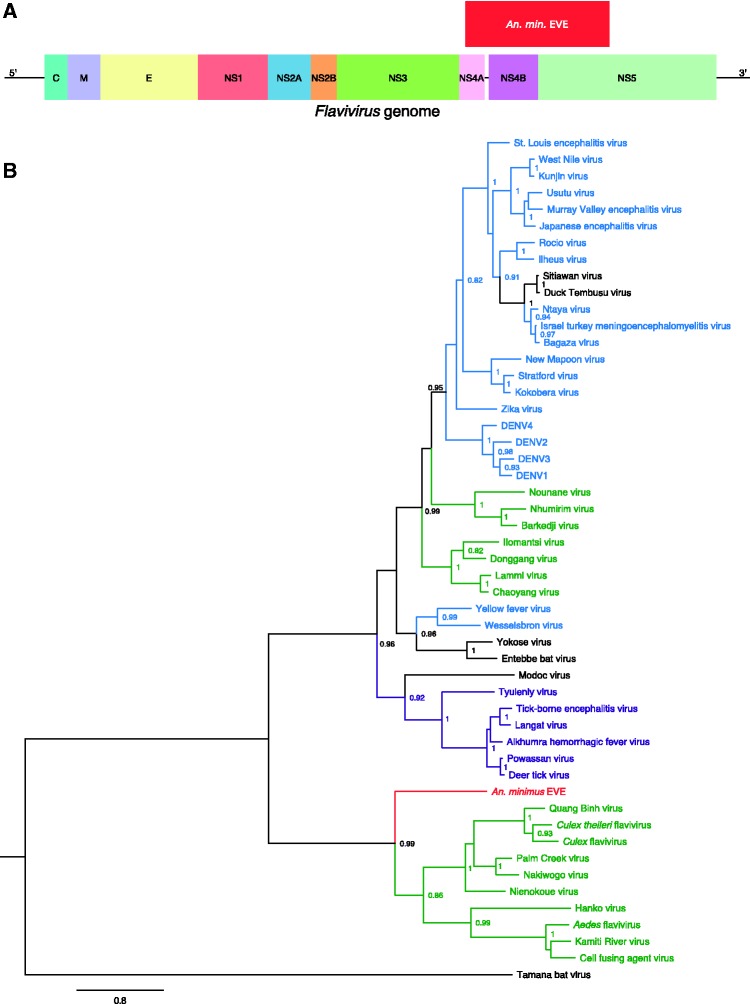
The An. minimus EVE is 1,881-bp long (627 amino-acid residues) with Nienokoue virus as the closest BLAST hit (44% amino-acid identity). The integrated sequence spans non-structural protein 4A (NS4A), NS4B and NS5 (Fig. 1A) without any stop codon. Conserved-domain search identified the NS5-methyltransferase domain involved in RNA capping and part of the RNA-directed RNA-polymerase domain. Phylogenetically, the An. minimus EVE is sister to the ISF clade (Fig. 1B). The An. minimus genome supercontig where the EVE was detected is 2,043-bp long and consists almost exclusively of the EVE sequence.
Figure 1.
Discovery of a flavivirus-derived EVE in An. minimus. (A) EVE location in a generic Flavivirus genome. Positioning is based on the genome sequence of Nienokoue virus (GenBank accession no. JQ957875). C, capsid protein; E, envelope glycoprotein; M, membrane glycoprotein; NS1, non-structural glycoprotein 1; NS2A, non-structural protein 2A; NS2B, non-structural protein 2B; NS3, non-structural protein 3 (protease/helicase); NS4A, non-structural protein 4A; NS4B, non-structural protein 4B; NS5, non-structural protein 5 (RNA-dependent RNA polymerase). (B) Phylogenetic relationships of the newly discovered An. minimus flavivirus-derived EVE with exogenous flaviviruses. Maximum likelihood trees were constructed based on the translated EVE sequence. Clades are color-coded according to known host specificity: green, ISFs; purple, tick-borne arboviruses; black, ‘NKV’ (vertebrate specific); blue, mosquito-borne arboviruses; red: EVEs. Scale bar indicates the number of substitutions. Node values represent Shimodaira-Hasegawa (SH)-like branch support (only values > 0.8 are shown).
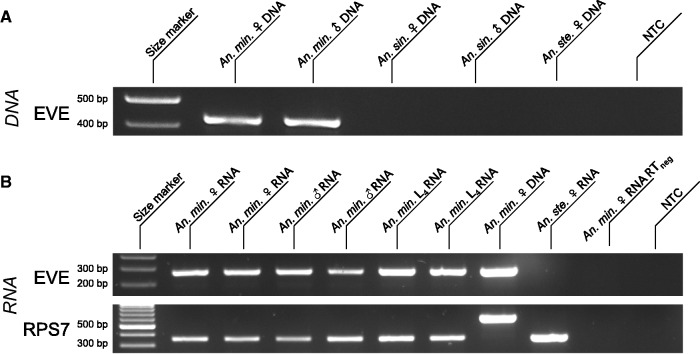
Presence of the EVE was verified in vivo by PCR on genomic DNA (Fig. 2A), followed by amplicon sequencing to confirm identity. The An. minimus EVE was found in both male and female genomic DNA, and was transcriptionally expressed for all combinations of sex and development stages tested (Fig. 2B). Evidence for transcriptional activity of the An. minimus EVE was confirmed in published RNA-seq data (Figure S1A).
Figure 2.
In vivo detection of the An. minimus flavivirus-derived EVE. (A) EVE detection in genomic DNA. Lane 1: size marker; lane 2: amplified genomic DNA from a pool of 10 An. minimus adult females; lane 3: amplified genomic DNA from a pool of 10 An. minimus adult males; lane 4: amplified genomic DNA from a pool of 10 An. sinensis adult females; lane 5: amplified genomic DNA from a pool of 10 An. sinensis adult males; lane 6: amplified DNA from a pool of 10 An. stephensi females; lane 7: no template control (NTC). (B) EVE detection in total RNA. Lane 1: size marker; lanes 2 and 3: amplified cDNA from pools of five adult females; lanes 4 and 5: amplified cDNA from pools of five adult males; lanes 6 and 7: amplified cDNA from pools of five L4 larvae; lane 8: amplified DNA from a pool of 10 females; lane 9: amplified cDNA from a pool of 5 An. stephensi females; lane 10: DNA contamination control (no reverse transcription) using the same pool of five adult females as lane 2; lane 11: NTC. First row: EVE; second row: RPS7 (control gene). The RPS7 target DNA sequence includes an intron, so that DNA contamination is expected to result in a larger PCR product.
3.2 An. sinensis EVE
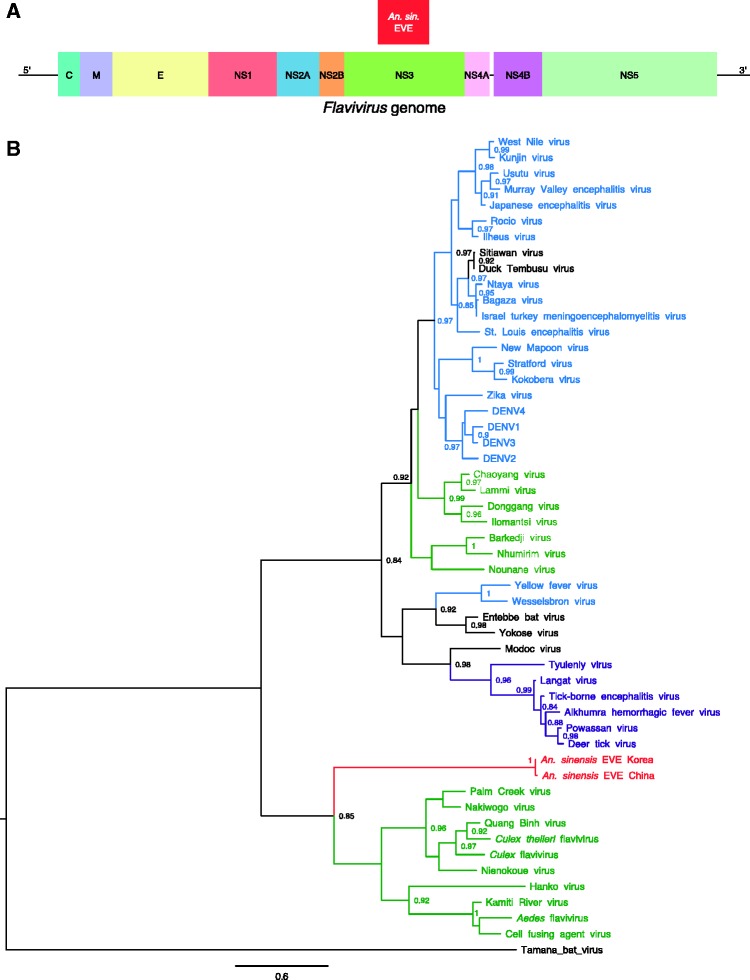
The An. sinensis EVE was detected in two distinct genome sequences that are available for this mosquito species, derived from a Korean strain and a Chinese strain, respectively. The EVE is 792-bp long (264 amino-acid residues) and 799-bp long (266 amino-acid residues) for the Korean and Chinese strains, respectively (Table 1). The closest BLAST hit is Culex flavivirus (45% amino-acid identity for the Korean strain, 44% amino-acid identity for the Chinese strain). The integrated sequence corresponds to the middle part of NS3 (Fig. 3A) and contains six and eight stop codons in the Korean and Chinese strains, respectively. Conserved-domain search identified the presence of a P-loop containing the nucleoside triphosphate hydrolase domain found in the NS3 protein of exogenous flaviviruses. Phylogenetically, the An. sinensis EVE is sister to the ISF clade (Fig. 3B).
Figure 3.
Discovery of a flavivirus-derived EVE in An. sinensis. (A) EVE location in a generic Flavivirus genome. Positioning is based on the genome sequence of Culex flavivirus (GenBank accession no. JQ308188). C, capsid protein; E, envelope glycoprotein; M, membrane glycoprotein; NS1, non-structural glycoprotein 1; NS2A, non-structural protein 2A; NS2B, non-structural protein 2B; NS3, non-structural protein 3 (protease/helicase); NS4A, non-structural protein 4A; NS4B, non-structural protein 4B; NS5, non-structural protein 5 (RNA-dependent-RNA polymerase). (B) Phylogenetic relationships of the newly discovered An. sinensis flavivirus-derived EVEs with exogenous flaviviruses. Maximum likelihood trees were constructed based on the translated EVE sequence. Clades are color-coded according to known host specificity: green, ISFs; purple, tick-borne arboviruses; black, ‘NKV’ (vertebrate specific); blue, mosquito-borne arboviruses; red, EVEs. Scale bar indicates the number of substitutions. Node values represent SH-like branch support (only values > 0.8 are shown).
The An. sinensis supercontig containing the EVE is 2,797-bp long for the Korean strain and 7,380-bp long for the Chinese strain. Analyses of flanking regions revealed the presence of another EVE in the same orientation, upstream of the flavivirus-derived EVE in both the Korean and the Chinese strains (Supplementary Table S3 and Supplementary Fig. S2). The closest BLAST hit of this additional EVE is Xincheng mosquito virus (43 and 42% amino-acid identity for the Korean and Chinese strains, respectively), an unclassified, negative-sense, single-stranded RNA virus detected in a pool of Chinese mosquitoes including An. sinensis specimens (Supplementary Table S3 and Supplementary Fig. S2). BLAST and conserved-domain search identified a class II Mariner-like transposase close to a mariner mos1 element, ∼1,000 bp downstream of the flavivirus-derived EVE (Supplementary Table S3 and Supplementary Fig. S2). This was only observed for the Chinese strain because the supercontig of the Korean strain did not span the downstream region far enough.
Presence of the An. sinensis EVE was verified in vivo by PCR on genomic DNA from the Korean strain (Fig. 4A), followed by amplicon sequencing to confirm identity. The An. sinensis EVE was found in both male and female genomic DNA, and was transcriptionally expressed, although less abundantly than the An. sinensis EVE, for all combinations of sex and development stages tested, especially in L4 larvae (Fig. 4B). Low expression observed for the An. sinensis EVE is consistent with barely detectable transcriptional activity in published RNA-seq data (Supplementary Fig. S1B).
Figure 4.
In vivo detection of the An. sinensis flavivirus-derived EVE. (A) EVE detection in genomic DNA from the Korean strain of An. sinensis. Lane 1, size marker; lane 2, amplified genomic DNA from a pool of 10 An. minimus adult females; lane 3, amplified genomic DNA from a pool of 10 An. minimus adult males; lane 4, amplified genomic DNA from a pool of 10 An. sinensis adult females; lane 5, amplified genomic DNA from a pool of 10 An. sinensis adult males; lane 6, amplified DNA from a pool of 10 An. stephensi females; lane 7, NTC. (B) EVE detection in total RNA from the Korean strain of An. sinensis. Lane 1: size marker; lanes 2 and 3, amplified cDNA from pools of five adult females; lanes 4 and 5, amplified cDNA from pools of five adult males; lanes 6 and 7, amplified cDNA from pools of five L4 larvae; lane 8, amplified DNA from a pool of ten females; lane 9, amplified cDNA from a pool of five An. stephensi females; lane 10: DNA contamination control (no reverse transcription) using the same pool of five adult females as lane 2; lane 11, NTC. First row, EVE; second row, RPS7 (control gene). The RPS7 target DNA sequence includes an intron, so that DNA contamination is expected to result in a larger PCR product.
4. Discussion
ISFs have attracted substantial interest in recent years after some of them were shown to enhance or suppress the replication of medically important flaviviruses in co-infected mosquito cells (Blitvich and Firth 2015). Over a dozen of ISFs have been formally identified to date, mainly in Aedes and Culex genera of the Culicinae subfamily (Blitvich and Firth 2015). ISFs were also reported in Anopheles mosquitoes of the Anophelinae subfamily (Aranda et al. 2009; Zuo et al. 2014; Liang et al. 2015). However, these Anopheles-associated ISFs are thought to infect a broad range of hosts including several mosquito species, mainly in the Culex genus, and are phylogenetically related to Culex-associated ISFs. Therefore, it is unclear whether Anopheles mosquitoes are true natural hosts of flaviviruses. Detection of ISFs in field-caught mosquitoes could result from incidental infection, or from a laboratory artifact. In this study, we discovered flavivirus-derived EVEs in the genomes of two Anopheles species. Phylogenetic analyses indicated that both EVEs are related to ISFs but belong to a clade that is distinct from Aedes-associated and Culex-associated ISFs.
Presence of flavivirus-derived EVEs in Anopheles genomes supports the hypothesis that Anopheles mosquitoes are, or were, natural hosts of flaviviruses. Endogenization of non-retroviral RNA viruses is unlikely to occur in the absence of recurrent host-virus interactions over a long evolutionary time scale. Endogenization requires reverse transcription, germ line integration and fixation in the host population, three steps whose combined probability is exceedingly low (Holmes 2011; Aiewsakun and Katzourakis 2015). The species-wide frequency of the An. minimus EVE is unknown because our in silico and in vivo analyses were based on the same mosquito strain. Presence of the An. sinensis EVE in two mosquito strains from different geographical locations, however, suggests that it could be fixed at the species level. Thus, our discovery of flavivirus-derived EVEs in Anopheles genomes is consistent with a long-lasting host-virus interaction between flaviviruses and mosquitoes of the Anophelinae subfamily. Although direct identification and characterization of exogenous Anopheles-associated flaviviruses are required to provide definite evidence that Anopheles are natural hosts of flaviviruses, our results provide strong indirect support to this hypothesis. Our finding emphasizes the need to include a broader range of mosquito genera and species than is typically screened in arbovirus surveillance programs.
ISFs are thought to be mainly maintained through vertical transmission from an infected female to its offspring (Blitvich and Firth 2015). Vertical transmission is likely to favor co-divergence of pathogens and hosts (Jackson and Charleston 2004), as illustrated by the existence of Aedes-associated and Culex-associated clades of ISFs (Moureau et al. 2015). Although extrapolation is limited by the scarcity of data on ISF host range and diversity, phylogenetic position of Anopheles-associated ISFs as sister to all other known ISFs is consistent with the co-divergence hypothesis. During the evolutionary history of mosquitoes, the Anophelinae diverged from the Culicinae prior to the separation of Culex and Aedes genera (Reidenbach et al. 2009). Further investigations will be necessary to determine whether an Anopheles-associated clade of exogenous ISFs exists, or existed.
Non-retroviral EVEs are thought to be generated by interaction of exogenous viruses with endogenous retro-elements, either with or without long terminal repeats (Holmes 2011). The short size of the supercontigs containing the An. minimus and An. sinensis EVEs limited our ability to investigate the integration mechanism(s). Another EVE sequence that we identified close to the flavivirus-derived EVE in An. sinensis may point to an EVE hotspot.
Sequence conservation of the An. minimus EVE (i.e. absence of stop codon across 1,881 bp) is consistent with a recent integration or a more ancient integration followed by non-neutral evolution. Our observation that the An. minimus EVE is abundantly transcribed may reflect a selective advantage for the host (Holmes 2011). Transcriptionally active EVEs have been suggested to confer protection or tolerance against related exogenous viruses (Flegel 2009; Holmes 2011; Aswad and Katzourakis 2012; Bell-Sakyi and Attoui 2013; Fujino et al. 2014). Despite the lack of empirical evidence so far, flavivirus-derived EVEs could contribute to antiviral immunity and arbovirus vector competence in mosquitoes.
Supplementary data
Supplementary data are available at Virus Evolution online.
Acknowledgements
The authors thank Inge Holm and Guillaume Carissimo for providing An. stephensi nucleic acids, Albin Fontaine for help with the in silico screen, Davy Jiolle and Elliott Miot for technical assistance, Clément Gilbert for advice and critical reading of an earlier version of the article, and members of the Lambrechts lab for insightful comments and discussions. We also thank Aris Katzourakis, Oliver Pybus and two anonymous reviewers for constructive comments on an earlier version of the article.
Funding
This work was supported by the French Government’s Investissement d’Avenir program Laboratoire d’Excellence Integrative Biology of Emerging Infectious Diseases grant ANR-10-LABX-62-IBEID, and the City of Paris Emergence(s) program in Biomedical Research. S.L. was supported by a doctoral fellowship from University Pierre and Marie Curie. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Conflict of interest: None declared.
References
- Aiewsakun P., Katzourakis A. (2015) ‘Endogenous Viruses: Connecting Recent and Ancient Viral Evolution’, Virology, 479–480, 26–37 [DOI] [PubMed] [Google Scholar]
- Aranda C. et al. (2009) ‘Detection and Monitoring of Mosquito Flaviviruses in Spain Between 2001 and 2005’, Vector Borne and Zoonotic Diseases, 9: 171–8 [DOI] [PubMed] [Google Scholar]
- Aswad A., Katzourakis A. (2012) ‘Paleovirology and Virally Derived Immunity’, Trends in Ecology and Evolution (Amsterdam), 27: 627–36 [DOI] [PubMed] [Google Scholar]
- Bell-Sakyi L., Attoui H. (2013) ‘Endogenous Tick Viruses and Modulation of Tick-Borne Pathogen Growth’, Frontiers in Cellular and Infection Microbiology, 3: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard K. A. et al. (2001) ‘West Nile Virus Infection in Birds and Mosquitoes, New York State, 2000’, Emerging Infectious Diseases, 7: 679–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitvich B. J., Firth A. E. (2015) ‘Insect-Specific Flaviviruses: A Systematic Review of Their Discovery, Host Range, Mode of Transmission, Superinfection Exclusion Potential and Genomic Organization’, Viruses, 7: 1927–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault A. C. et al. (2004) ‘Infection Patterns of O’nyong Nyong Virus in the Malaria-Transmitting Mosquito, Anopheles gambiae’, Insect Molecular Biology, 13: 625–35 [DOI] [PubMed] [Google Scholar]
- Calzolari M. et al. (2013) ‘Usutu Virus Persistence and West Nile Virus Inactivity in the Emilia-Romagna Region (Italy) in 2011’, PLoS One, 8: e63978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C. et al. (2009) ‘BLAST+: Architecture and Applications’, BMC Bioinformatics, 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T. (2009) ‘trimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses’, Bioinformatics, 25: 1972–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.-G. et al. (2015) ‘Genome sequence of the Asian Tiger mosquito, Aedes albopictus, Reveals Insights into its Biology, Genetics, and Evolution’, Proceedings of the National Academy of Sciences of the United States of America, 112: E5907–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements A. N. 2012. ‘Arboviruses - Characteristics and Concepts’, in The Biology of Mosquitoes. pp. 90–173. CABI. [Google Scholar]
- Crochu S. et al. (2004) ‘Sequences of Flavivirus-Related RNA Viruses Persist in DNA form Integrated in the Genome of Aedes spp. Mosquitoes’, Journal of General Virology, 85: 1971–80 [DOI] [PubMed] [Google Scholar]
- Feng Y. et al. (2012) ‘Distribution of Mosquitoes and Mosquito-Borne Viruses Along the China-Myanmar Border in Yunnan Province’, Japanese Journal of Infectious Diseases, 65: 215–21 [DOI] [PubMed] [Google Scholar]
- Feschotte C., Gilbert C. (2012) ‘Endogenous Viruses: Insights into Viral Evolution and Impact on Host Biology’, Nature Reviews Genetics, 13: 283–96 [DOI] [PubMed] [Google Scholar]
- Filipe A. R. (1972) ‘Isolation in Portugal of West Nile Virus from Anopheles maculipennis Mosquitoes’, Acta Virologica, 16: 361. [PubMed] [Google Scholar]
- Flegel T. W. (2009) ‘Hypothesis for Heritable, Anti-viral Immunity in Crustaceans and Insects’, Biology Direct, 4: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino K. et al. (2014) ‘Inhibition of Borna Disease Virus Replication by an Endogenous Bornavirus-Like Element in the Ground Squirrel Genome’, Proceedings of the National Academy of Sciences of the United States of America, 111: 13175–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E. C. (2011) ‘The Evolution of Endogenous Viral Elements’, Cell Host Microbe, 10: 368–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt R. A. et al. (2002) ‘The genome sequence of the malaria mosquito Anopheles gambiae’, Science, 298: 129–49 [DOI] [PubMed] [Google Scholar]
- Jackson A. P., Charleston M. A. (2004) ‘A Cophylogenetic Perspective of RNA-Virus Evolution’, Molecular Biology and Evolution, 21: 45–57 [DOI] [PubMed] [Google Scholar]
- Katoh K. et al. (2002) ‘MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform’, Nucleic Acids Research, 30: 3059–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzourakis A., Gifford R. J. (2010) ‘Endogenous Viral Elements in Animal Genomes’, PLoS Genetics, 6: e1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemenesi G. et al. (2014) ‘West Nile Virus Surveillance in Mosquitoes, April to October 2013, Vojvodina Province, Serbia: Implications for the 2014 Season’, Eurosurveillance, 19: 20779. [DOI] [PubMed] [Google Scholar]
- Kulasekera V. L. et al. (2001) ‘West Nile Virus Infection in Mosquitoes, Birds, Horses, and Humans, Staten Island, New York, 2000’, Emerging Infectious Diseases, 7: 722–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012) ‘Fast Gapped-Read Alignment with Bowtie 2’, Nature Methods, 9: 357–U54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen R., Sugawara H., Shumway M. International Nucleotide Sequence Database Collaboration. (2011) ‘The Sequence Read Archive’, Nucleic Acids Research, 39: D19–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., 1000 Genome Project Data Processing Subgroup. et al. (2009) ‘The Sequence Alignment/Map Format and SAMtools’, Bioinformatics, 25: 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Godzik A. (2006) ‘Cd-hit: A Fast Program for Clustering and Comparing Large Sets of Protein or Nucleotide Sequences’, Bioinformatics, 22: 1658–9 [DOI] [PubMed] [Google Scholar]
- Liang W. et al. (2015) ‘Distribution and Phylogenetic Analysis of Culex Flavivirus in Mosquitoes in China’, Archives in Virology, 160: 2259–68 [DOI] [PubMed] [Google Scholar]
- Liu H. et al. (2013) ‘Japanese Encephalitis Virus in Mosquitoes and Swine in Yunnan Province, China 2009-2010’, Vector Borne and Zoonotic Diseases, 13: 41–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue K. et al. (2015) ‘Whole-Genome Sequencing Reveals Absence of Recent Gene Flow and Separate Demographic Histories for Anopheles Punctulatus Mosquitoes in Papua New Guinea’, Molecular Ecology, 24: 1263–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A. et al. (2015) ‘CDD: NCBI’s Conserved Domain Database’, Nucleic Acids Research, 43: D222–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moureau G. et al. (2015) ‘New Insights into Flavivirus Evolution, Taxonomy and Biogeographic History, Extended by Analysis of Canonical and Alternative Coding Sequences’, PLoS One, 10: e0117849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neafsey D. E. et al. (2015) ‘Mosquito Genomics. Highly Evolvable Malaria Vectors: The Genomes of 16 Anopheles Mosquitoes’, Science, 347: 1258522–1258522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M. (2010) ‘BEDTools: A Flexible Suite of Utilities for Comparing Genomic Features’, Bioinformatics, 26: 841–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidenbach K. R. et al. (2009) ‘Phylogenetic Analysis and Temporal Diversification of Mosquitoes (Diptera: Culicidae) Based on Nuclear Genes and Morphology’, BMC Evolutionary Biology, 9: 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roiz D. et al. (2009) ‘Detection of Novel Insect Flavivirus Sequences Integrated in Aedes albopictus (Diptera: Culicidae) in Northern Italy’, Virology Journal, 6: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez R., Serra F., Tárraga J., Medina I., Carbonell J., Pulido L., de María A., Capella-Gutiérrez S., Huerta-Cepas J., Gabaldón T. et al. (2011) ‘Phylemon 2.0: A Suite of Web-Tools for Molecular Evolution, Phylogenetics, Phylogenomics and Hypotheses Testing’, Nucleic Acids Research, 39: W470–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D. et al. (2014) ‘Genome Sequence of Anopheles sinensis Provides Insight into Genetics Basis of Mosquito Competence for Malaria Parasites’, BMC Genomics, 15: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo S. et al. (2014) ‘Detection of Quang Binh Virus from mosquitoes in China’, Virus Research, 180: 31–8 [DOI] [PubMed] [Google Scholar]