Abstract
Cranberry contains high levels of nutrients and bioactive molecules that have health‐promoting properties. The purpose of the present studies was to determine if cranberry extracts (CEs) contain phytochemicals that exert anti‐inflammatory effects. The human monocytic cell line THP‐1 was treated with two CEs (CE and 90MX) and subsequently challenged with Lipopolysaccharides (LPS). Tumor necrosis factor α (TNF α) expression was decreased in the CE‐treated cells, indicative of an anti‐inflammatory effect. Gene expression microarrays identified several immune‐related genes that were responsive to CEs including interferon‐induced protein with tetratricopeptide repeats 1 and 3 (IFIT 1 and 3), macrophage scavenger receptor 1 (MSR1) and colony‐stimulating factor 2 (CSF2). In addition, in the CE‐treated cells, metallothionein 1F and other metal‐responsive genes were induced. Taken together, this data indicates that CEs contain bioactive components that have anti‐inflammatory effects and may protect cells from oxidative damage.
Keywords: Gene expression, macrophage, nutrigenomics, proanthocyanidins, reactive oxygen species
Introduction
Epidemiological, human intervention, and mechanistic trials have shown that polyphenol‐rich foods, often found in a diet rich in fruits and vegetables, may have protective effects against chronic degenerative diseases linked to oxidative stress and reactive oxygen species (ROS)‐mediated cell damage (Cassidy et al. 2011; Wedick et al. 2012; Del Rio et al. 2013). Flavonoids and other phenolic compounds in these foods are bioactive health components that are likely responsible for these effects. The North American cranberry (Vaccinium macrocarpon Ait. Ericaceae) is of growing public interest as a functional food because of potential health benefits linked to the high content of bioactive flavonoids in the fruit, including proanthocyanidins, flavonols such as quercetin and myricetin and anthocyanins. In fact, cranberry ranks high among fruit in both antioxidant quality and quantity (Vinson et al. 2001; Gu et al. 2004; Côté et al. 2010), and contain a higher amount of total phenols per serving (507–709 mg gallic acid equivalents/100 g) than other common fruits including blueberries (258–531 mg/100 g), apples (185–347 mg/100 g), and red grapes (175–370 mg/100 g) (Wu et al. 2004). Cranberries, in powder, extract or juice, have been reported to be beneficial for helping prevent urinary tract infections (Wang et al. 2012; Blumberg et al. 2013; Foxman et al. 2015), oxidative stress and cardiometabolic risk factors such as reducing C‐reactive protein and lowering blood pressure (Basu et al. 2011; Dohadwala et al. 2011; Novotny et al. 2015). As cranberry is a natural food product and not a drug, the effect seen is often mild but that is consistent with the recommendation for a food with health components that is consumed as part of a healthy balanced lifestyle.
In vitro evidence has shown the antioxidant potential of cranberry phenolic compounds including protection of human microvascular endothelial cells against ROS, and HepG2 cells from inflammatory insults and reduced glycation (Caton et al. 2010; Crozier et al. 2010; Liu et al. 2011; Watson et al. 2014; Martín et al. 2015). A recent publication also showed the bioavailability and bioactivity of cranberry phenolics and metabolites from consuming cranberry juice beverage (McKay et al. 2015). The protective effect of these compounds may be related to their function in sequestering ROS and/or maintaining the cell components in their correct redox state, but emerging findings indicate that natural compounds may act by increasing the endogenous antioxidant defense potential through regulation of cell signaling pathways (Seeram et al. 2004; Stevenson and Hurst 2007; Martín et al. 2015). The objective of this study is to examine the anti‐inflammatory activity of cranberry phenolic compounds in two different cranberry powders (90 MX and CE) by looking at changes in gene expression using human monocytic cells lines.
Material and Methods
Chemicals
Cranberry juice powder and extract were obtained from Ocean Spray Cranberries (Lakeville‐Middleboro, MA; for chemical analysis see Table 1 and reference [Martín et al. 2015]). Cranberry juice powder (90MX) was prepared from the juice of mature berries of the commonly cultivated cranberry plant (Vaccinium macrocarpon). Cranberry juice processing consists of the milling and pressing of the berries after a hot (50°C for 1 h) commercial pectinase maceration. 90MX was then prepared by spray drying cranberry concentrate with magnesium hydroxide as the carrier and tri‐calcium phosphate as an anti‐caking agent. The powder is fine, free‐flowing and rosy red in color, and contains approximately 90% cranberry solids. The cranberry extract (CE) powder is a water‐soluble, phenolic‐rich extract of cranberry utilizing a proprietary resin separation process. CE was standardized to 55% proanthocyanidin (PAC) content on a dry weight basis as analyzed by a modified 4‐dimethylaminocinnamaldehyde (DMAC) method (Martín et al. 2015). Finally, 100 mg/mL stock solutions of 90MX and CE were prepared in Dimethylsulfoxide (DMSO) for cell treatment. All media components and fetal bovine serum (FBS) were purchased from Gibco BRL/Life Technologies (Carlsbad, CA). Phorbol 12‐myristate 13‐acetate (PMA), used to differentiate THP‐1 cells, was purchased from Sigma Chemical Company (St. Louis, MO). All other chemicals and reagents were of highest quality available.
Table 1.
Content of phenolic compounds and other major components in cranberry juice and extract powders
| Cranberry juice powder (90MX, mg/100 g) | Cranberry extract powder (CE, mg/100 g) | Enrichment (CE/90MX)c | |
|---|---|---|---|
| Proanthocyanidins | |||
| By OS‐DMAC methoda | 2800 | 51,600 | 18.4 |
| By BL‐DMAC methodb | 653 | 15,000 | 23.0 |
| Organic acids | |||
| Citric acid | 14,200 | 2850 | 0.2 |
| Malic acid | 8510 | 1660 | 0.2 |
| Quinic acid | 13,500 | 2040 | 0.2 |
| Galacturonic acid | 3300 | 78.4 | 0.0 |
| Sugars | |||
| Glucose | 31,300 | 4700 | 0.2 |
| Fructose | 8120 | 1100 | 0.1 |
| Anthocyanins | |||
| Cyanidin‐3‐arabinoside | 33.8 | 420 | 12.4 |
| Cyanidin‐3‐galacoside | 41.7 | 508 | 12.2 |
| Cyanidin‐3‐glucose | 1.3 | 25.4 | 19.5 |
| Peonidin‐3‐arabinoside | 17.3 | 239 | 13.8 |
| Peonidin‐3‐galactose | 41.9 | 530 | 12.6 |
| Peonidin‐3‐glucoside | 3.0 | 36.6 | 12.2 |
| Phenolic acids | |||
| Benzoic acid | 84.5 | 2550 | 30.2 |
| Salicylic acid | 0.6 | 13.4 | 22.3 |
| Protocatechuic acid | 15.6 | 120 | 7.7 |
| Gallic acid | 4.2 | 25.8 | 6.1 |
| Vanilic acid | 4.9 | 85.4 | 17.4 |
| t‐Cinnamic acid | 7.7 | 72.8 | 9.5 |
| p‐Coumaric acid (4‐hydroxycinnamic acid) | 30.2 | 436 | 14.4 |
| Caffeic acid (3,4‐dihydroxycinnamic acid) | 3.0 | 50.5 | 16.8 |
| Ferulic acid (3‐methoxy‐4‐hydroxycinnamic acid | 0.9 | 54.7 | 60.8 |
| Chlorogenic acid | 22.8 | 793 | 34.8 |
| Ellagic acid | 1.3 | 2.2 | 1.7 |
| Flavon‐3‐ols | |||
| Catechin | 8.0 | 43.3 | 5.4 |
| Epicatechin | 31.7 | 2.2 | 0.1 |
| Flavonols | |||
| Quercetin | 37.3 | 404 | 10.8 |
| Quercitrin (quercetin‐3‐O‐rhamnoside | 20.8 | 527 | 25.3 |
| Hyperoside (quercitin‐3‐O‐galactoside) | 53.1 | 2720 | 51.2 |
| Myricetin | 40.5 | 657 | 16.2 |
| Myricetrin (myrocetin‐3‐O‐rhamnoside) | 7.6 | 125 | 16.4 |
| Total phenolic by Folin‐Ciocalteu method (GAE) | 2600 | 45,000 | 17.3 |
From Martín et al. (2015).
Expressed in milligram (mg) of cranberry‐specific PACs equivalents per 100 g of powder.
Expressed in milligram (mg) of procyanidine A‐2 equivalents per 100 g of powder.
Enrichment is the ratio of each component in the CE extract relative to that of 90MX.
Cell culture and treatment
THP‐1 (Homo sapiens monocyte) cells were obtained from the American Type Culture Collection (ATCC; Rockville, MD) and cultured in RPMI 1640 with 10% heat inactivated FBS, 50 μmol/L 2‐mercaptoethanol, 1 mmol/L sodium pyruvate, 100 U/mL penicillin and 100 μg/mL streptomycin. These cells were seeded in 24‐well plates at a density of 3 × 105/well and differentiated into macrophages with 100 nmol/L PMA (Sigma) for 48 h. For in vitro treatment experiments, THP‐1 cells were grown to 75% confluency and treated with 90MX or CE (0, 25, 50, 100 μg dry weight/mL media, heretofore listed as μg/mL). Sixteen hrs after treatment, the cells will be stimulated with 10 ng/mL LPS for 6 h.
RNA extraction, reverse transcription, real‐time PCR
Total RNA was isolated by Qiagen RNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. The total RNA was reverse transcribed, using the ABI High Capacity cDNA archive kit (Applied Biosystems, Foster City, CA). Standard curves were made using serial dilutions from pooled cDNA samples. Real‐time polymerase chain reaction (PCR) was performed with the use of the SYBR Green PCR Master Mix (Applied Biosystems) according to the manufacturer's protocol and amplified on the ABI Prism 7000 Sequence Detection System. Primers are listed in Table 2.
Table 2.
Primer sequences used for quantitative real‐time PCR
| Gene | Forward primer | Reverse primer |
|---|---|---|
| 18S | ACCCGTTGAACCCCATTCGTGA | GCCTCACTAAACCATCCAATCGG |
| ABCG2 | ACGAACGGATTAACAGGGTCA | CTCCAGACACACCACGGAT |
| CAT | TGTTGCTGGAGAATCGGGTTC | TCCCAGTTACCATCTTCTGTGTA |
| CCNL2 | GTACTCCGGGGTGCTCATC | GAGGTCGGTCTCTGTGTCG |
| COX2 | CGGTGTTGAGCAGTTTTCTCC | AAGTGCGATTGTACCCGGAC |
| CSF2 | TCCTGAACCTGAGTAGAGACAC | TGCTGCTTGTAGTGGCTGG |
| HERC5 | GGTGAGCTTTTTGCCTGGG | TTCTCCGGCAGAAATCTGAGC |
| IFIT1 | GCGCTGGGTATGCGATCTC | CAGCCTGCCTTAGGGGAAG |
| IFIT3 | AGAAAAGGTGACCTAGACAAAGC | CCTTGTAGCAGCACCCAATCT |
| IL‐1α | CTCCCAATCTCCATTCCCAA | CGTAAGGCCTCAGCCAGAAG |
| IL6 | GCCACTCACCTCTTCAGAACG | CCGTCGAGGATGTACCGAATT |
| MSR1 | GCAGTGGGATCACTTTCACAA | AGCTGTCATTGAGCGAGCATC |
| MT1F | TCCTGCAAGAAGAGCTGCTG | ACTTCTCTGACGCCCCTTTG |
| OAS1 | TGTCCAAGGTGGTAAAGGGTG | CCGGCGATTTAACTGATCCTG |
| SLC7A11 | TCTCCAAAGGAGGTTACCTGC | AGACTCCCCTCAGTAAAGTGAC |
| SOD1 | GGTGTGGCCGATGTGTCTAT | CCTTTGCCCAAGTCATCTGC |
| TNFA | ATCAATCGGCCCGACTATCTC | TGGATGTTCGTCCTCCTCACA |
Microarray experiments and statistical analysis
THP‐1 cells were treated with CE or 90MX at 100 μg/mL or control (DMSO, 0.1% v/v) for 16 h followed by stimulation with LPS for 6 h. RNA was extracted, using Qiagen RNeasy and quality assessed by RNA Nano Chips on the Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA). Each sample was labeled using the Affymetrix IVT Express Kit according to the manufacturer's protocol. The GeneChip Human Genome U133A 2.0 (Affymterix, Santa Clara, CA), representing 14,500 well‐characterized genes, was hybridized with the labeled RNA using GeneChip Hybridization Wash and Stain Kit (#702232) in the Affymetrix GeneChip Hybridization Oven 640, according to the manufacturer's instructions. Following hybridization, the arrays were washed and stained using the Affymetrix GeneChip Fluidics Station 450 according to the manufacturer's protocol and scanned using the GeneChip Scanner 3000 7G. The scanned image file (DAT) and the intensity data (CEL) was imported into ArrayStar (DNASTAR, Inc., Madison, WI). The Robust Multi‐array Average (RMA) was used to normalize the data. The nine slides were grouped based on treatment and Student t‐test with asymptotic P‐value and Benjamini–Hochberg multiple corrections was performed comparing CE versus DMSO and 90MX versus DMSO. At a P‐value of 0.05 and a twofold change, a total of 236 entities were significantly regulated by CE while there were no genes that met the criteria for 90MX. The group of genes was examined by hierarchical clustering using complete linkage analysis of the normalized data (JMP 7.0; SAS Institute, Cary, NC). Gene Ontology (GO) and pathway analysis was performed, using Ingenuity Pathway Analysis (Qiagen, Redwood City, CA).
Statistical analysis
One‐way analysis of variance, followed by Dunnett's post hoc test, was used to test the difference between treatments (P < 0.05). The values were expressed as mean ± SEM. All data analyses were performed by JMP 7.0 (SAS Institute) and data plotted by Prism 5.01 (GraphPad Software, Inc., San Diego, CA).
Results
Regulation of inflammation‐related genes in THP‐1 cells
The ability of CEs to regulate proinflammatory gene expression in THP‐1 cells was examined using an LPS‐challenge model (Lee et al. 2009; Zhang et al. 2010). A single concentration of each extract (100 μg/mL) was examined; there was no sign of overt toxicity at this dose (data not shown). The 90MX powder had no effect on expression of interleukin‐1α (IL‐1α), interleukin‐6 (IL6), tumor necrosis factor‐α (TNFα), catalase (CAT), cyclooxygenase‐2 (COX2) or superoxide dismutase 1 (SOD1) mRNA (Fig. 1). The CE extract at this dose resulted in a significant increase in IL‐6 mRNA (2.5‐fold) with decreased expression of TNFα, CAT and SOD1 mRNA (1.6, 1.5 and 1.8‐fold, respectively) relative to the vehicle control. The pattern of gene expression, with several LPS‐induced genes being repressed while others are either unaffected or augmented, suggests an atypical mechanism of action and prompted a more comprehensive examination of mRNA concentration.
Figure 1.
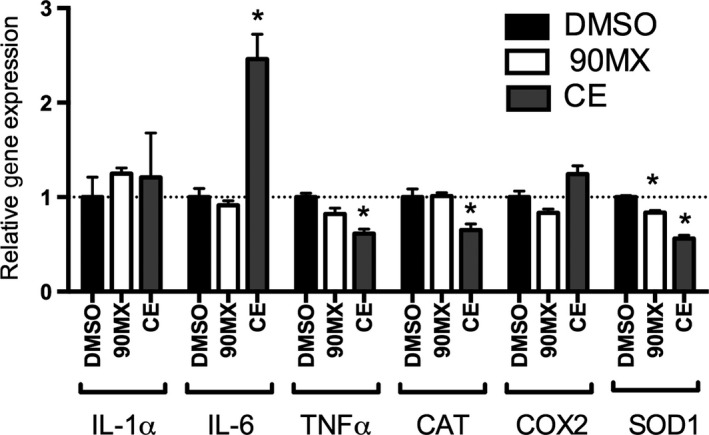
Regulation of inflammation‐related genes by cranberry extracts in THP‐1 cells. Cells were treated and RNA extracted as described in Materials and Methods. Gene expression following treatment with DMSO (0.1% v/v),CE or 90MX (100 μg/mL) is expressed relative to a housekeeping gene (18S) and normalized to vehicle control (DMSO). Asterisks denote significantly different than control (P < 0.05, n = 3, bars represent mean and SEM).
Comprehensive analysis of gene expression
The design of the present experiments was aimed to understand the anti‐inflammatory effects of cranberry polyphenol extracts to identify sensitive biomarkers for subsequent studies and to begin understanding their potential mechanism of action. The THP‐1 monocytes were treated with CE or 90MX at 100 μg/mL for 16 h, followed by stimulation with LPS and the RNA was used to examine gene expression by high‐density microarray. A total of 236 genes were significantly regulated by CE with a twofold change (Table S1 and Fig. 2) with 64 genes reaching a threefold threshold (Table 3). There were no significant changes using these same criteria with the 90MX extract. A majority of the genes (78%) were repressed by CE; Metallothionein 1F (MT1F) mRNA was increased fivefold while interferon‐induced protein with tetratricopeptide repeats 5 (IFIT5) was reduced 10‐fold relative to the DMSO‐treated group.
Figure 2.
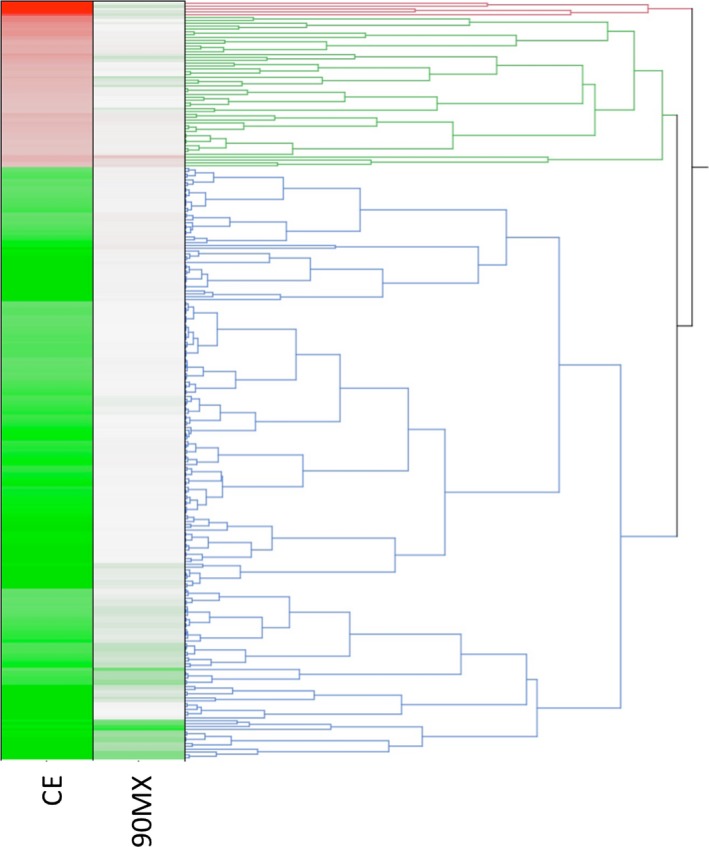
Comprehensive analysis of gene expression. Analysis of gene expression by high‐density microarrays was performed by microarrays as described in Material and Methods. Average expression values (n = 3 arrays per group) were exported to JMP (SAS Institute, Cary NC) and hierarchical clustering performed. Data were expressed relative to the DMSO‐treated group with green representing a decrease and red an increase relative to control (depicted as white).
Table 3.
Genes significantly regulated by cranberry extracts (threefold, P < 0.05)
| Probe set ID | Gene symbol | Gene title | 90MX | CE |
|---|---|---|---|---|
| 213629_x_at | MT1F | Metallothionein 1F | 0.80 | 4.84 |
| 211906_s_at | SERPINB4 | Serpin peptidase inhibitor, clade B (ovalbumin), member 4 | 0.70 | 4.47 |
| 210524_x_at | MT1F | Metallothionein 1F | 0.81 | 4.10 |
| 212859_x_at | MT1E | Metallothionein 1E | 0.75 | 3.87 |
| 220322_at | IL36G | Interleukin 36, gamma | 0.72 | 3.56 |
| 203213_at | CDK1 | Cyclin‐dependent kinase 1 | 0.89 | 0.33 |
| 210764_s_at | CYR61 | Cysteine‐rich, angiogenic inducer, 61 | 0.87 | 0.33 |
| 203725_at | GADD45A | Growth arrest and DNA‐damage‐inducible, alpha | 0.94 | 0.32 |
| 207386_at | CYP7B1 | Cytochrome P450, family 7, subfamily B, polypeptide 1 | 1.05 | 0.32 |
| 211668_s_at | PLAU | Plasminogen activator, urokinase | 1.04 | 0.32 |
| 218662_s_at | NCAPG | Non‐SMC condensin I complex, subunit G | 0.91 | 0.32 |
| 214453_s_at | IFI44 | Interferon‐induced protein 44 | 0.57 | 0.31 |
| 204747_at | IFIT3 | Interferon‐induced protein with tetratricopeptide repeats 3 | 0.63 | 0.31 |
| 203362_s_at | MAD2L1 | MAD2 mitotic arrest deficient‐like 1 (yeast) | 0.94 | 0.31 |
| 219209_at | IFIH1 | Interferon induced with helicase C domain 1 | 0.80 | 0.31 |
| 202127_at | PRPF4B | Pre‐mRNA processing factor 4B | 0.56 | 0.30 |
| 201291_s_at | TOP2A | Topoisomerase (DNA) II alpha 170 kDa | 0.96 | 0.30 |
| 218883_s_at | CENPU | Centromere protein U | 0.74 | 0.30 |
| 213226_at | CCNA2 | Cyclin A2 | 0.80 | 0.29 |
| 202508_s_at | SNAP25 | Synaptosomal‐associated protein, 25 kDa | 0.51 | 0.29 |
| 209969_s_at | STAT1 | Signal transducer and activator of transcription 1, 91 kDa | 0.80 | 0.29 |
| 210559_s_at | CDK1 | Cyclin‐dependent kinase 1 | 0.90 | 0.29 |
| 211597_s_at | HOPX | HOP homeobox | 0.99 | 0.29 |
| 216442_x_at | FN1 | Fibronectin 1 | 1.00 | 0.28 |
| 206513_at | AIM2 | Absent in melanoma 2 | 0.85 | 0.28 |
| 203967_at | CDC6 | Cell division cycle 6 | 0.77 | 0.28 |
| 204127_at | RFC3 | Replication factor C (activator 1) 3, 38 kDa | 0.81 | 0.28 |
| 204994_at | MX2 | Myxovirus (influenza virus) resistance 2 (mouse) | 0.63 | 0.28 |
| 210495_x_at | FN1 | Fibronectin 1 | 0.98 | 0.28 |
| 212464_s_at | FN1 | Fibronectin 1 | 0.95 | 0.27 |
| 203708_at | PDE4B | Phosphodiesterase 4B, cAMP‐specific | 0.78 | 0.27 |
| 211719_x_at | FN1 | Fibronectin 1 | 0.98 | 0.27 |
| 202833_s_at | SERPINA1 | Serpin peptidase inhibitor, clade A (alpha‐1 antiproteinase, antitrypsin), member 1 | 1.03 | 0.27 |
| 205479_s_at | PLAU | Plasminogen activator, urokinase | 1.02 | 0.27 |
| 201506_at | TGFBI | Transforming growth factor, beta‐induced, 68 kDa | 1.12 | 0.26 |
| 204823_at | NAV3 | Neuron navigator 3 | 0.94 | 0.26 |
| 220104_at | ZC3HAV1 | Zinc finger CCCH‐type, antiviral 1 | 0.79 | 0.26 |
| 213293_s_at | TRIM22 | Tripartite motif containing 22 | 0.41 | 0.26 |
| 222162_s_at | ADAMTS1 | ADAM metallopeptidase with thrombospondin type 1 motif, 1 | 0.90 | 0.26 |
| 201340_s_at | ENC1 | Ectodermal‐neural cortex 1 (with BTB domain) | 0.99 | 0.25 |
| 201341_at | ENC1 | Ectodermal‐neural cortex 1 (with BTB domain) | 0.93 | 0.25 |
| 202628_s_at | SERPINE1 | Serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 | 0.99 | 0.24 |
| 219148_at | PBK | PDZ‐binding kinase | 0.99 | 0.24 |
| 202086_at | MX1 | Myxovirus (influenza virus) resistance 1, interferon‐inducible protein p78 (mouse) | 0.70 | 0.23 |
| 218039_at | NUSAP1 | Nucleolar and spindle‐associated protein 1 | 0.85 | 0.23 |
| 201890_at | RRM2 | Ribonucleotide reductase M2 | 0.89 | 0.22 |
| 202589_at | TYMS | Thymidylate synthetase | 0.85 | 0.21 |
| 219691_at | SAMD9 | Sterile alpha motif domain containing 9 | 0.62 | 0.21 |
| 205552_s_at | OAS1 | 2′‐5′‐oligoadenylate synthetase 1, 40/46 kDa | 0.54 | 0.20 |
| 202627_s_at | SERPINE1 | Serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 | 1.01 | 0.20 |
| 214146_s_at | PPBP | Proplatelet basic protein (chemokine (C‐X‐C motif) ligand 7) | 1.26 | 0.20 |
| 219684_at | RTP4 | Receptor (chemosensory) transporter protein 4 | 0.72 | 0.20 |
| 219908_at | DKK2 | Dickkopf WNT signaling pathway inhibitor 2 | 0.88 | 0.19 |
| 218585_s_at | DTL | Denticleless E3 ubiquitin protein ligase homolog (Drosophila) | 0.95 | 0.19 |
| 203596_s_at | IFIT5 | Interferon‐induced protein with tetratricopeptide repeats 5 | 0.77 | 0.19 |
| 213294_at | EIF2AK2 | Eukaryotic translation initiation factor 2‐alpha kinase 2 | 0.75 | 0.18 |
| 209773_s_at | RRM2 | Ribonucleotide reductase M2 | 0.86 | 0.18 |
| 202869_at | OAS1 | 2′‐5′‐oligoadenylate synthetase 1, 40/46 kDa | 0.54 | 0.16 |
| 205239_at | AREG | Amphiregulin | 1.00 | 0.15 |
| 212977_at | ACKR3 | Atypical chemokine receptor 3 | 1.06 | 0.15 |
| 203153_at | IFIT1 | Interferon‐induced protein with tetratricopeptide repeats 1 | 0.54 | 0.14 |
| 201289_at | CYR61 | Cysteine‐rich, angiogenic inducer, 61 | 0.83 | 0.13 |
| 218943_s_at | DDX58 | DEAD (Asp‐Glu‐Ala‐Asp) box polypeptide 58 | 0.69 | 0.11 |
| 203595_s_at | IFIT5 | Interferon‐induced protein with tetratricopeptide repeats 5 | 0.75 | 0.09 |
Quantitative real‐time PCR was used to confirm a subset of transcripts identified by the microarray experiments. Care was taken to choose genes that were both increased and decreased by CE treatment as well as those with known biological functions. THP‐1 cells were treated as described above, with the exception that three doses of each extract was examined. As shown in Figure 3 (top panel), unlike the microarray experiment, the 90MX extract significantly regulated colony‐stimulating factor 2 (CSF2) and cyclin L2 (CCNL2) mRNA accumulation. All genes studied, with the exception of solute carrier family 7A11 (SLC7A11), were significantly regulated by CE, albeit to a different extent and with varying potency (Fig. 3, bottom panel). However, in the ATP‐binding cassette, the sub‐family G, member 2 (ABCG2) mRNA was affected only at the highest dose, CSF2, 2′‐5′‐oligoadenylate synthetase 1 (OAS1), metallothionein 1F (MT1F), and CCNL2 mRNA were regulated at the 25 μg/mL dose.
Figure 3.
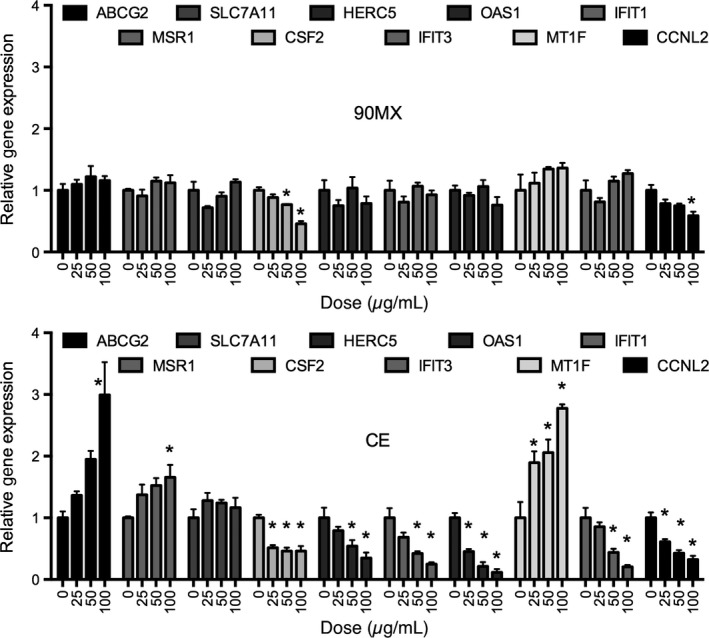
Expression of selected genes identified as responsive to cranberry extracts in THP‐1 cells. Genes were selected from Table 2 to Figure 2 for validation by real‐time PCR. Cells were treated and RNA extracted as described in Material and Methods. Gene expression following treatment with 90MX or CE (0, 25, 50, 100 μg/mL) is expressed relative to a housekeeping gene (18S) and normalized to vehicle control (DMSO). Asterisks denote significantly different than control (P < 0.05, n = 3, bars represent mean and SEM).
GO and pathway analysis
The list of genes that were significantly regulated by the CE extract in THP‐1 cells (twofold and P < 0.05, 236 genes) were classified according to their functional ontology terms (GO). Several GO biological processes were significantly enriched in these CE‐regulated genes (http://pantherdb.org, see Table S2), including immune effector process, response to interferon, cell cycle checkpoint, cell cycle phase, and response to metal ion. The gene expression patterns were further examined to determine if they were consistent with certain diseases and functions (Ingenuity Pathway Analysis; Table 4 and Figures S1, S2). Genes regulated by CEs, such as ABCG2, CCNA2, CCNB1, and CDC25A were associated with cell cycle control and DNA repair processes. The majority of the genes in this network were decreased in expression relative to the DMSO‐treated cells. Similarly, several CE‐regulated genes were enriched in the inflammatory response network and most genes, with the exception of IL6, were repressed. Potential mechanisms by which CE regulates genes within these networks is shown in Table 5. Several transcription factors and signaling molecules are implicated, including HGF signaling, PPAR, LXR, or VDR activation and importantly inhibition of inflammatory signaling such as interferon, MAPK, or NFAT activity.
Table 4.
Gene ontology analysis: top diseases and functions
| ID | Top diseases and functions | Molecules in network |
|---|---|---|
| 1 | Cell cycle, cellular assembly and organization, DNA replication, recombination, and repair | ABCG2, alcohol group acceptor phosphotransferase, ALT, ASAH1, AURKA, BUB1, BUB1B, CCNA2, CCNB1, CD163, Cdc2, CDC25A, CDK1, Cyclin B, ENO2, GADD45A, GINS1, HISTONE, Histone H1, Histone h3, HOPX, IL6, MCM4, MT1G, NASP, NCAPG, PRC1, PRPF4B, PTPase, RNA polymerase II, SFPQ, SMC4, SNX10, Sod, TTK |
| 2 | Cell cycle, embryonic development, organismal survival | AIM2, APC (complex), ATM/ATR, ATR, CDC6, CDC20, Cdk, Cyclin A, Cyclin D, Cyclin E, DTL, Dynein, E2f, FANCI, GMNN, HERC5, LUC7L3, MAD2L1, MCM10, MELK, NBN, NFkB (complex), NUSAP1, PCNA, PPP4R4, Rb, Rfc, RFC3, RFC4, RPA, RRM2, SKP2, TRIM22, TYMS, UNG |
| 3 | Antimicrobial response, inflammatory response, cell signaling | 2′ 5′ oas, DDX58, DDX60, EIF2AK2, ERK, IFI16, IFI44, IFIH1, IFIT1, IFIT3, IFIT5, Ifn, IFN alpha/beta, IFN Beta, IFN type 1, Ifnar, IRF, ISG15, ISG20, ISGF3, JAK, MT1M, MX1, Oas, OAS1, OAS2, PARP12, RSAD2, STAT4, Stat1‐Stat2, THEMIS2, USP18, XAF1, ZC3HAV1 |
| 4 | Nervous system development and function, cancer, organismal injury and abnormalities | ACKR3, ACPP, ADAMTS1, ADCY, ADRB, Alpha catenin, AREG, BNIP3, CAP2, Cg, Creb, DUSP2, EMR2, FGF13, FSH, Gpcr, GPR56, GPR65, Hdac, HRH1, Lh, Mapk, Metalloprotease, MT1F, MT1X, NMDA Receptor, OXTR, Pdgfr, PLC, PVR, SLC7A11, SNAP25, STEAP1, TCF, Vegf |
| 5 | Infectious disease, cell morphology, hair and skin development, and function | AIF1, Akt, c‐Src, Calcineurin A, Collagen(s), CYR61, Fascin, FCGR2B, FERMT1, Fgf, Fibrin, Fibrinogen, GDF15, Integrin, Integrin alpha 3 beta 1, Integrin alpha 4 beta 1, ITGA1, JINK1/2, KIAA0101, Laminin, Lfa‐1, LRIG1, MAP2K1/2, MT1E, Notch, PBK, PMAIP1, PNISR, PNN, PPIG, SLC4A7, TLR2/TLR4, TRIB3, TRIM5, ZWINT |
Table 5.
Gene ontology analysis: top pathways
| Canonical pathways | z‐score | Molecules |
|---|---|---|
| Cell cycle: G2/M DNA damage checkpoint regulation | 1.134 | GADD45A, TOP2A, ATR, AURKA, CDK1, SKP2, CCNB1 |
| HGF signaling | 1 | MET, FOS, IL6, ELK3, PRKCB |
| PPAR signaling | 1 | IL33, FOS, IL36G, IL1A |
| LXR/RXR activation | 0.447 | IL33, IL36G, IL1A, SERPINA1, IL6 |
| Activation of IRF by cytosolic pattern recognition receptors | −0.447 | IFIH1, IL10, DDX58, IL6, STAT1, ISG15 |
| PDGF signaling | −0.447 | FOS, CAV1, EIF2AK2, STAT1, PRKCB |
| IL‐6 signaling | −0.447 | IL33, FOS, IL36G, IL1A, IL6 |
| NF‐κB signaling | −0.447 | IL33, IL36G, IL1A, EIF2AK2, PRKCB |
| ERK/MAPK signaling | −0.447 | FOS, STAT1, ELK3, DUSP2, PRKCB |
| Cholecystokinin/gastrin‐mediated signaling | −0.816 | IL33, FOS, IL36G, IL1A, MEF2C, PRKCB |
| Toll‐like receptor signaling | −1 | IL33, FOS, IL36G, IL1A, EIF2AK2 |
| Growth hormone signaling | −1 | FOS, IGFBP3, STAT1, PRKCB |
| JAK/stat signaling | −1 | STAT4, FOS, IL6, STAT1 |
| TREM1 signaling | −1 | IL10, IL6, FCGR2B, CSF2 |
| Tec kinase signaling | −1 | STAT4, FOS, STAT1, PRKCB |
| Production of nitric oxide and reactive oxygen species in macrophages | −1 | FOS, SERPINA1, STAT1, PRKCB |
| Colorectal cancer metastasis signaling | −1 | FOS, IL6, STAT1, MMP19 |
| Role of CHK proteins in cell cycle checkpoint control | −1.342 | PCNA, RFC4, ATR, CDK1, CDC25A, RFC3, NBN |
| HMGB1 signaling | −1.342 | FOS, IL1A, IL6, SERPINE1, CSF2, PLAT |
| p38 MAPK signaling | −1.342 | IL33, IL36G, IL1A, MEF2C, STAT1 |
| Aryl hydrocarbon receptor signaling | −1.342 | CCNA2, FOS, IL1A, ATR, IL6 |
| Acute phase response signaling | −1.414 | IL33, FOS, IL36G, IL1A, FN1, SERPINA1, IL6, SERPINE1 |
| VDR/RXR activation | −2 | GADD45A, IGFBP3, CYP27B1, CSF2, PRKCB |
| UVA‐induced MAPK signaling | −2 | FOS, ZC3HAV1, PARP12, STAT1 |
| Role of NFAT in regulation of the immune response | −2 | FOS, MEF2C, FCGR2B, RCAN2 |
| Interferon signaling | −2.236 | IFIT3, IFIT1, OAS1, MX1, STAT1 |
Discussion
Several berry fruits, including the American cranberry (Vaccinium macrocarpon), have recently received attention as a result of their effects in vitro and their associations with observational studies with lowered risk of some chronic diseases (Zafra‐Stone et al. 2007; Blumberg et al. 2013). Cranberries, consumed in sauces, juices, dried fruit, or taken as a dietary supplement in powders or extract form, are a rich source of polyphenols that are associated with a variety of biological effects. In vitro studies have demonstrated antibacterial, antiviral, antimutagenic, anticarcinogenic, antitumorigenic, antiangiogenic, anti‐inflammatory, and antioxidant properties (Yan et al. 2002; Bagchi et al. 2004; Ferguson et al. 2004; Seeram et al. 2004; Neto 2007; Zafra‐Stone et al. 2007) (3,7,8). In vivo, animal models reveal that CEs: reduce proinflammatory interleukins (Kim et al. 2011); suppress Helicobacter pylori infection (Blumberg et al. 2013), and; improve pancreatic β‐cell glucose responsiveness and β‐cell mass (Zhu et al. 2011). The clinical effects of cranberry and their bioactives include: lowering LDL cholesterol and total cholesterol (Basu and Lyons 2012); improving markers of endothelial function (Bagchi et al. 2004; Dohadwala et al. 2011), lowering glycemic responses (Wilson et al. 2010), elevating plasma antioxidant capacity (McKay et al. 2015); modulation of gastric Helicobacter pylori infection (Shmuely et al. 2012); reducing biomarkers of metabolic syndrome (Wilson et al. 2010; Basu and Lyons 2012; Simão et al. 2013), and; protecting against urinary tract infections (UTIs) (Foxman et al. 2015).
Cranberries are a particularly rich source of phenolic phytochemicals, including phenolic acids (benzoic, hydroxycinnamic, and ellagic acids) and flavonoids (anthocyanins, flavonols, and flavanols), and these bioactive molecules are believed to be associated with the health benefits described above. The major anthocyanins in cranberry are galactosides and arabinosides of cyanidin and peonidin (Neto 2007). Anthocyanin content varies widely among cranberry cultivars with averages between 25 and 65 mg per 100 g of ripe fruit at harvest. The primary flavonol in cranberries is quercetin, which exists in several glycosidic forms, and the total flavonol content of cranberry fruit typically in the range of 20–30 mg per 100 g of fresh fruit weight. In comparing the two CEs used in the present study (Martín et al. 2015), 90MX contained higher amount of organic acids while CE had higher quantities of highly bioactive compounds such as phenolic acids, flavonols, and flavanols (see Table 1). More specific, components with a high antioxidant capacity such as chlorogenic acid, epicatechin, and quercetin were found in high concentrations in CE. The total PACs concentration was 18 times higher in CE than 90MX (51,000 vs. 2800 mg/100 g; [Martín et al. 2015]). In HepG2 cells, both CEs decreased ROSs and protected the cells from t‐BOOH‐induced cytotoxicity, albeit with CE being much more potent. Interestingly, stress‐related signaling through the JNK pathway by t‐BOOH was not averted by CE, indicating another mechanism such as redox regulation, might be primary in the protection from oxidative stress by CE in HepG2 cells.
The CEs significantly decreased expression of TNFα, CAT, and SOD1 mRNA (Fig. 1) relative to the vehicle control in LPS‐stimulated THP‐1 cells. In addition, the microarray studies showed mRNA for acute phase response genes (IL33, FOS, IL36G, IL1A, FN1, SERPINA1, SERPINE1) and interferon‐signaling genes (IFIT3, IFIT1, OAS1, MX1, STAT1; Table 4) were decreased in these cells. These responses are consistent with several other studies showing an anti‐oxidant and anti‐inflammatory effect of cranberries and their bioactive molecules, in particular the PACs. In rats fed an atherogenic diet, C‐reactive protein (CRP) and IL‐1β were significantly lower in the cranberry powder groups compared to those in control rats (Kim et al. 2011). A CE (OptiBerry) significantly inhibited basal MCP‐1 and inducible NF‐κB transcription as well as the inflammatory biomarker IL‐8 in a similar in vivo model (Zafra‐Stone et al. 2007). Treatment of Caco‐2 cells with these CEs prevented iron/ascorbate‐mediated lipid peroxidation and counteracted LPS‐mediated inflammation as evidenced by the decrease in proinflammatory cytokines (TNF‐α and IL‐6), cyclo‐oxygenase‐2 (COX‐2) and prostaglandin E2 (PGE2, [Denis et al. 2015]). Networks of genes involved in the antimicrobial and inflammatory responses and infectious disease (Table 3) were identified as being coordinately affected by CE, adding support for the associations seen in observational studies. The moderate increase in IL‐6 mRNA observed in the present studies, in the absence of increases in other proinflammatory markers such as COX‐2 and IL‐1α, may be the result of several factors. First, this suggests that CE is not causing generalized oxidative stress since other mRNAs would be coordinately affected. Second, this increase in IL6 mRNA may not be indicative of altered activity due to the small increase observed (2.5‐fold), the fact that protein level of this factor was not examined and IL6‐responsive genes (such as IL1) were not altered. Finally, it is obvious that CE, due to the complex mixture of bioactive molecules, has multiple mechanisms of action.
The mechanism of this anti‐inflammatory effect has not been extensively studied, although PACs from cranberries and other sources have been proposed to act through oxygen free radical scavenging (Li et al. 2001) as well as metal chelation (Arola‐Arnal and Bladé 2011), a contributor to ROS production. PACs affect the activity of signaling molecules associated with the inflammatory response, including the redox‐sensitive transcription factor NF‐κB (Liu et al. 2014), 5′‐AMP activated protein kinase (AMPK)(Cui et al. 2012), p38 mitogen‐activated protein kinase (MAPK, [Dinh et al. 2014]) and JNK (Guha et al. 2013; Liu et al. 2014). Data presented herein support the premise that CE decreases activity of NF‐κB, p38 MAPK and NFAT in LPS‐challenged macrophages. Quercetin, a polyphenol found in high concentration in CE (400 mg/100 g; [Martín et al. 2015]), induces metallothionein (MT) expression by activating the phosphorylation of JNK, p38 and PI3K/Akt as well as by enhancing Nrf2 DNA‐binding activity in HepG2 cells (Weng et al. 2011). In CE‐treated THP‐1 cells, MT1E and 1F mRNA were among the most robustly induced transcripts, indicating that similar signaling pathways are affected in this cell line and that CEs may protect against metal‐induced oxidative stress and toxicity.
Several in vitro studies suggest that cranberry bioactives are potential therapeutic agents for the treatment of cancer (Neto 2007; Zafra‐Stone et al. 2007). For example, CEs and PACs decrease proliferation of the human breast (Ferguson et al. 2004; Sun and Liu 2006), glioblastoma multiforme (U87), colon (HT‐29), prostate (DU145), and oral cancer cell lines (Seeram et al. 2004; Ferguson et al. 2006). Cell cycle arrest and apoptosis was induced by a cranberry PAC‐rich extract in human esophageal adenocarcinoma (EAC) cells (Kresty et al. 2008) and prostate cells (MacLean et al. 2011). Although cell cycle and proliferation was not studied herein, several genes involved in checkpoint control such as PCNA, RFC4, ATR, CDK1, CDC25A, RFC3, and NBN (Tables 3, 5) were induced at the mRNA level by CE in THP‐1 cells. Interestingly, cranberry ethanolic extract (CEE) prevents the DNA damage produced by benzo[a]pyrene (B[a]P) in an in vivo mouse peripheral blood micronucleus assay (Madrigal‐Santillán et al. 2012). Our current study showed that several CE inducible genes are involved in G2/M DNA Damage checkpoint regulation, including GADD45A and CCNB1 (Tables 4, 5).
Taken together, these studies support the role of cranberry and its complex and rich phytochemical composition as an adjuvant for the treatment or prevention of chronic diseases with an inflammatory component. CEs decreased expression of several markers of inflammation and ROS signaling in the human monocyte cell line THP‐1 that had been challenged with LPS. In addition, through the use of comprehensive gene expression analysis, the potential for cranberry or extracts for the prevention of metal‐induced toxicity as well as cancer is supported. A limitation of the studies presented is that they are exclusively in vitro, using surrogate model systems for inflammation and oxidative stress. Although predictive of the potential of CE to affect human health, there must be consideration of availability of the bioactive constituents and the concentration achieved in vivo. In support of an in vivo response and hence bioavailability of active constituents, rats and mice‐fed CEs exhibited reduced CRP, proinflammatory interleukins, and increase NO synthesis (Wang et al. 2012; Blumberg et al. 2013; Foxman et al. 2015). The range of doses of the cranberry phenolic powders was selected according to previous studies showing that concentrations in the μg/mL range with 30–40 μmol/L of cranberry phytochemicals have been found in plasma after cranberry juice intake (Martín et al. 2015). Although the identity of the specific cranberry‐derived bioactive molecule or the molecular pathway involved has not been definitively identified, this study supports a wide range of in vitro, animal as well as nutritional invention studies showing this berry have unique and powerful health benefits.
Conflict of Interest
JVH is an employee of Penn State University and has a financial stake with Indigo Biosciences Inc., which may constitute a conflict of interest.
Supporting information
Table S1. Genes significantly regulated by CE (P < 0.05, twofold).
Table S2. PANTHER over‐representation test.
Figure S1. Cell proliferation network affected by CE. Genes significantly affected by CE were examined by Ingenuity Pathway Analysis (Qiagen). Red filled symbols denotes increased, while green filled symbols denote decreased, expression relative to control (DMSO‐treated cells).
Figure S2. Inflammation network affected by CE. See legend to Figure S1.
Acknowledgments
This work was conducted under a contract between Ocean Spray and Indigo Biosciences Inc.
References
- Arola‐Arnal, A. , and Bladé C.. 2011. Proanthocyanidins modulate microRNA expression in human HepG2 cells. PLoS ONE 6:e25982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagchi, D. , Sen C. K., Bagchi M., and Atalay M.. 2004. Anti‐angiogenic, antioxidant, and anti‐carcinogenic properties of a novel anthocyanin‐rich berry extract formula. Biochemistry (Mosc) 69: 75–80, 1 p preceding 75. [DOI] [PubMed] [Google Scholar]
- Basu, A. , and Lyons T. J.. 2012. Strawberries, blueberries, and cranberries in the metabolic syndrome: clinical perspectives. J. Agric. Food Chem. 60:5687–5692. [DOI] [PubMed] [Google Scholar]
- Basu, A. , Betts N. M., Ortiz J., Simmons B., Wu M., and Lyons T. J.. 2011. Low‐energy cranberry juice decreases lipid oxidation and increases plasma antioxidant capacity in women with metabolic syndrome. Nutr. Res. 31:190–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg, J. B. , Camesano T. A., Cassidy A., Kris‐Etherton P., Howell A., Manach C., et al. 2013. Cranberries and their bioactive constituents in human health. Adv. Nutr. 4:618–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy, A. , O'Reilly É. J., Kay C., Sampson L., Franz M., Forman J. P., et al. 2011. Habitual intake of flavonoid subclasses and incident hypertension in adults. Am. J. Clin. Nutr. 93:338–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton, P.W. , Pothecary M. R., Lees D. M., Khan N. Q., Wood E. G., Shoji T., et al. 2010. Regulation of vascular endothelial function by procyanidin‐rich foods and beverages. J. Agric. Food Chem. 58:4008–4013. [DOI] [PubMed] [Google Scholar]
- Côté, J. , Caillet S., Doyon G., Sylvain J. F., and Lacroix M.. 2010. Bioactive compounds in cranberries and their biological properties. Crit. Rev. Food Sci. Nutr. 50:666–679. [DOI] [PubMed] [Google Scholar]
- Crozier, A. , Del Rio D., and Clifford M. N.. 2010. Bioavailability of dietary flavonoids and phenolic compounds. Mol. Aspects Med. 31:446–467. [DOI] [PubMed] [Google Scholar]
- Cui, X. , Liu X., Feng H., et al. 2012. Grape seed proanthocyanidin extracts enhance endothelial nitric oxide synthase expression through 5′‐AMP activated protein kinase/Surtuin 1‐Krüpple like factor 2 pathway and modulate blood pressure in ouabain induced hypertensive rats. Biol. Pharm. Bull. 35:2192–2197. [DOI] [PubMed] [Google Scholar]
- Del Rio, D. , Rodriguez‐Mateos A., Spencer J. P. E., M. Tognolini , Borges G., and Crozier A.. 2013. Dietary (poly)phenolics in human health: structures, bioavailability, and evidence of protective effects against chronic diseases. Antioxid. Redox Signal. 18:1818–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis, M.‐C. , Desjardins Y., Furtos A., Marcil V., S. Dudonné , Montoudis A., et al. 2015. Prevention of oxidative stress, inflammation and mitochondrial dysfunction in the intestine by different cranberry phenolic fractions. Clin. Sci. (Lond.) 128:197–212. [DOI] [PubMed] [Google Scholar]
- Dinh, J. , Angeloni J. T., Pederson D. B., Wang X., Cao M., and Dong Y.. 2014. Cranberry extract standardized for proanthocyanidins promotes the immune response of Caenorhabditis elegans to Vibrio cholerae through the p38 MAPK pathway and HSF‐1. PLoS ONE 9:e103290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohadwala, M. M. , Holbrook M., Hamburg N. M., Shenouda S. M., Chung W. B., Titas M., et al. 2011. Effects of cranberry juice consumption on vascular function in patients with coronary artery disease. Am. J. Clin. Nutr. 93:934–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, P. J. , Kurowska E., Freeman D. J., Chambers A. F., and Koropatnick D. J.. 2004. A flavonoid fraction from cranberry extract inhibits proliferation of human tumor cell lines. J. Nutr. 134:1529–1535. [DOI] [PubMed] [Google Scholar]
- Ferguson, P. J. , Kurowska E. M., Freeman D. J., Chambers A. F., and Koropatnick J.. 2006. In vivo inhibition of growth of human tumor lines by flavonoid fractions from cranberry extract. Nutr. Cancer 56:86–94. [DOI] [PubMed] [Google Scholar]
- Foxman, B. , Cronenwett A. E. W., Spino C., Berger M. B., and Morgan D. M.. 2015. Cranberry juice capsules and urinary tract infection after surgery: results of a randomized trial. Am. J. Obstet. Gynecol. 213:194.e1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, L. , Kelm M. A., Hammerstone J. F., Beecher G., J. Holden , Haytowitz D., et al. 2004. Concentrations of proanthocyanidins in common foods and estimations of normal consumption. J. Nutr. 134:613–617. [DOI] [PubMed] [Google Scholar]
- Guha, S. , Cao M., Kane R. M., Savino A. M., Zou S., and Dong Y.. 2013. The longevity effect of cranberry extract in Caenorhabditis elegans is modulated by daf‐16 and osr‐1. Age (Dordr) 35:1559–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M. J. , Ohn J., Kim J. H., and Kwak H.‐K.. 2011. Effects of freeze‐dried cranberry powder on serum lipids and inflammatory markers in lipopolysaccharide treated rats fed an atherogenic diet. Nutr. Res. Pract. 5:404–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kresty, L. A. , Howell A. B., and Baird M.. 2008. Cranberry proanthocyanidins induce apoptosis and inhibit acid‐induced proliferation of human esophageal adenocarcinoma cells. J. Agric. Food Chem. 56:676–680. [DOI] [PubMed] [Google Scholar]
- Lee, Y. , Thompson J. T., and Vanden Heuvel J. P.. 2009. 9E,11E‐conjugated linoleic acid increases expression of the endogenous antiinflammatory factor, interleukin‐1 receptor antagonist, in RAW 264.7 cells. J. Nutr. 139: 1861–1866. [DOI] [PubMed] [Google Scholar]
- Li, W. G. , Zhang X. Y., Wu Y. J., and Tian X.. 2001. Anti‐inflammatory effect and mechanism of proanthocyanidins from grape seeds. Acta. Pharmacol. Sin. 22:1117–1120. [PubMed] [Google Scholar]
- Liu, H. , Liu H., Wang W., Khoo C., Taylor J., and Gu L., 2011. Cranberry phytochemicals inhibit glycation of human hemoglobin and serum albumin by scavenging reactive carbonyls. Food Funct. 2:475–482. [DOI] [PubMed] [Google Scholar]
- Liu, C.‐M. , Ma J.‐Q., Liu S.‐S., Zheng G.‐H., Feng Z.‐J., and Sun J.‐M.. 2014. Proanthocyanidins improves lead‐induced cognitive impairments by blocking endoplasmic reticulum stress and nuclear factor‐κB‐mediated inflammatory pathways in rats. Food Chem. Toxicol. 72:295–302. [DOI] [PubMed] [Google Scholar]
- MacLean, M. A. , Scott B. E., Deziel B. A., Nunnelley M. C., Liberty A. M., Gottschall‐Pass K. T., et al. 2011. North American cranberry (Vaccinium macrocarpon) stimulates apoptotic pathways in DU145 human prostate cancer cells in vitro. Nutr. Cancer 63:109–120. [DOI] [PubMed] [Google Scholar]
- Madrigal‐Santillán, E. , Fragoso‐Antonio S., Valadez‐Vega C., Solano‐Solano G., Pérez C. Z., Sánchez‐Gutiérrez M., et al. 2012. Investigation on the protective effects of cranberry against the DNA damage induced by benzo[a]pyrene. Molecules 17:4435–4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín, M. A. , Ramos S., Mateos R., Marais J. P., Bravo‐Clemente L., Khoo C., and Goya L.. 2015. Chemical characterization and chemo‐protective activity of cranberry phenolic powders in a model cell culture. Response of the antioxidant defenses and regulation of signaling pathways. Food Res. Int. 71:68–82. [Google Scholar]
- McKay, D. L. , Chen C.‐Y. O., Zampariello C. A., and J. B. Blumberg . 2015. Flavonoids and phenolic acids from cranberry juice are bioavailable and bioactive in healthy older adults. Food Chem. 168:233–240. [DOI] [PubMed] [Google Scholar]
- Neto, C. C. 2007. Cranberry and blueberry: evidence for protective effects against cancer and vascular diseases. Mol. Nutr. Food Res. 51:652–664. [DOI] [PubMed] [Google Scholar]
- Novotny, J. A. , Baer D. J., Khoo C., Gebauer S. K., and Charron C. S.. 2015. Cranberry juice consumption lowers markers of cardiometabolic risk, including blood pressure and circulating C‐reactive protein, triglyceride, and glucose concentrations in adults. J. Nutr. 145:1185–1193. [DOI] [PubMed] [Google Scholar]
- Seeram, N. P. , Adams L. S., Hardy M. L., and Heber D.. 2004. Total cranberry extract versus its phytochemical constituents: antiproliferative and synergistic effects against human tumor cell lines. J. Agric. Food Chem. 52:2512–2517. [DOI] [PubMed] [Google Scholar]
- Shmuely, H. , Ofek I., Weiss E. I., Rones Z., and Houri‐Haddad Y.. 2012. Cranberry components for the therapy of infectious disease. Curr. Opin. Biotechnol. 23:148–152. [DOI] [PubMed] [Google Scholar]
- Simão, T. N. C. , Lozovoy M. A. B., Simão A. N. C., Oliveira S. R., Venturini D., Morimoto H. K., et al. 2013. Reduced‐energy cranberry juice increases folic acid and adiponectin and reduces homocysteine and oxidative stress in patients with the metabolic syndrome. Br. J. Nutr. 110:1885–1894. [DOI] [PubMed] [Google Scholar]
- Stevenson, D. E. , and Hurst R. D.. 2007. Polyphenolic phytochemicals–just antioxidants or much more? Cell. Mol. Life Sci. 64:2900–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, J. , and Liu Hai. 2006. R. Cranberry phytochemical extracts induce cell cycle arrest and apoptosis in human MCF‐7 breast cancer cells. Cancer Lett. 241:124–134. [DOI] [PubMed] [Google Scholar]
- Vinson, J. A. , Su X., Zubik L., and Bose P.. 2001. Phenol antioxidant quantity and quality in foods: fruits. J. Agric. Food Chem. 49:5315–5321. [DOI] [PubMed] [Google Scholar]
- Wang, C.‐H. , Fang C.‐C., Chen N.‐C., Liu S. S.‐H., Yu P.‐H., Wu T.‐Y., et al. 2012. Cranberry‐containing products for prevention of urinary tract infections in susceptible populations: a systematic review and meta‐analysis of randomized controlled trials. Arch. Intern. Med. 172:988–996. [DOI] [PubMed] [Google Scholar]
- Watson, R. R. , Preedy V. R., and Zibadi S.. 2014. Polyphenols in human health and disease
- Wedick, N. M. , Pan A., Cassidy A., Rimm E. B., Sampson L., Rosner B., et al. 2012. Dietary flavonoid intakes and risk of type 2 diabetes in US men and women. Am. J. Clin. Nutr. 95:925–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng, C.‐J. , Chen M.‐J., Yeh C.‐T., and Yen G.‐C.. 2011. Hepatoprotection of quercetin against oxidative stress by induction of metallothionein expression through activating MAPK and PI3K pathways and enhancing Nrf2 DNA‐binding activity. N. Biotechnol. 28:767–777. [DOI] [PubMed] [Google Scholar]
- Wilson, T. , Luebke J. L., Morcomb E. F., Carrell E. J., Leveranz M. C., Kobs L., et al. 2010. Glycemic responses to sweetened dried and raw cranberries in humans with type 2 diabetes. J. Food Sci. 75:H218–H223. [DOI] [PubMed] [Google Scholar]
- Wu, X. , Beecher G. R., Holden J. M., Haytowitz D. B., Gebhardt S. E., and Prior R. L.. 2004. Lipophilic and hydrophilic antioxidant capacities of common foods in the United States. J. Agric. Food Chem. 52:4026–4037. [DOI] [PubMed] [Google Scholar]
- Yan, X. , Murphy B. T., Hammond G. B., Vinson J. A., and Neto C. C.. 2002. Antioxidant activities and antitumor screening of extracts from cranberry fruit (Vaccinium macrocarpon). J. Agric. Food Chem. 50:5844–5849. [DOI] [PubMed] [Google Scholar]
- Zafra‐Stone, S. , Yasmin T., Bagchi M., Chatterjee A., Vinson J. A., and Bagchi D.. 2007. Berry anthocyanins as novel antioxidants in human health and disease prevention. Mol. Nutr. Food Res. 51:675–683. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Kris‐Etherton P. M., Thompson J. T., and J. P. Vanden Heuvel . 2010. Effect of pistachio oil on gene expression of IFN‐induced protein with tetratricopeptide repeats 2: a biomarker of inflammatory response. Mol. Nutr. Food Res. 54 (Suppl. 1): S83–S92. [DOI] [PubMed] [Google Scholar]
- Zhu, M. , Hu J., Perez E., Phillips D., Kim W., Ghaedian R., et al. 2011. Effects of long‐term cranberry supplementation on endocrine pancreas in aging rats. J. Gerontol. A Biol. Sci. Med. Sci. 66:1139–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Genes significantly regulated by CE (P < 0.05, twofold).
Table S2. PANTHER over‐representation test.
Figure S1. Cell proliferation network affected by CE. Genes significantly affected by CE were examined by Ingenuity Pathway Analysis (Qiagen). Red filled symbols denotes increased, while green filled symbols denote decreased, expression relative to control (DMSO‐treated cells).
Figure S2. Inflammation network affected by CE. See legend to Figure S1.


