Abstract
Using a yeast two-hybrid screen of a T-cell cDNA library to identify cellular proteins that bind to the human immunodeficiency virus type 2 (HIV-2) Gag polyprotein, we identified PRP4, a serine-threonine protein kinase. Specific interaction of PRP4 and HIV-2 Gag was confirmed in in vitro and in vivo assays. The interacting region of HIV-2 Gag is located in the conserved matrix and capsid domains, while both the RS (arginine-serine-rich) domain and the KS (kinase) domain of PRP4 are able to bind to HIV-2 Gag. PRP4 is not incorporated into virus particles. HIV-2 Gag is able to inhibit PRP4-mediated phosphorylation of the splicing factor SF2. This is also observed with Gag from simian immunodeficiency virus, a closely related virus, but not with Gag from human T-cell lymphotropic virus type 1. Our results provide evidence for a novel interaction between Gag and a cellular protein kinase involved in the control of constitutive splicing in two closely related retroviruses. We hypothesize that as Gag accumulates in the cell, down regulation of splicing occurs through reduced phosphorylation of SF2. At late stages of infection, this interaction may replace the function of the early viral regulatory protein Rev.
Retroviruses utilize host cell mechanisms throughout their life cycle, from entry and integration through to packaging and membrane budding. Both viral genomic RNA and proviral DNA have been shown to interact with cellular proteins, and viral protein-cell protein interaction is now well documented. Examples include the effector domain of the Rev nuclear export signal, which has been identified as interacting with eukaryotic initiation factor 5A (eIF-5A), an essential cofactor involved in mRNA export (47). Mutations in eIF-5A are able to block Rev activity, halting human immunodeficiency virus type 1 (HIV-1) replication in trans in T cells (3, 25), as well as the functionally equivalent Rex protein of human T-cell lympotrophic virus type 1 (HTLV-1) (12). Similarly, the HIV-1 viral accessory protein Vif binds to cellular enzyme SSAT in vitro and incorporates it into virions. This interaction may alter levels of cellular polyamines modulating RNA, DNA, deoxynucleoside triphosphates (dNTPs), and protein and has an effect on viral infectivity (31). The HIV-1 Gag polyprotein is known to colocalize with Vif (49, 50), binding directly with the NC subunit of Gag (4). Both HIV-1 viral proteins Vif and Gag bind to cellular ATP binding protein HP68 (64). This interaction is thought to assist capsid assembly and protect viral RNA from degradation (31). Vif appears to bind APOBEC3G, thus inhibiting cytidine deamination of the genome (48). Gag binding to F-actin has been suggested to act as a platform for retroviral assembly (44).
Retroviral Gag proteins play a crucial role in the virus life cycle, from mediating virion assembly to association with viral genomic RNA and virus release (53). The Gag protein of HIV-1 is synthesized as a polyprotein precursor, which, upon activation of the viral protease during the budding process, is cleaved to yield the mature virion components: matrix (MA), capsid (CA), nucleocapsid (NC), p6 and two spacer peptides, p2 and p1. Gag proteins contain conserved domains required for membrane targeting (M), interaction between Gag polyproteins (I), and late stages of virus particle assembly and release from the cell (L) (53).
The HIV-1 preintegration complex is known to contain viral and host proteins, as well as viral DNA (5, 41). The N-terminal matrix protein of Gag is a component of the preintegration complex (39) along with integrase (16) and binds with cellular protein virion-associated nuclear shuttling protein (VAN), enhancing nuclear-cytoplasmic shuttling capacity (23). Cellular protein EF1α associates with the HIV-1 Gag cleavage product matrix (MA), inhibiting translation in vitro (9). This allows for the release of RNA from polysomes, allowing for its assembly into virions (9). Cyclophilin A is incorporated into virions of HIV-1 through binding to a proline loop in the capsid domain (34), where it may promote viral core disassembly (18, 34) and may also modulate the response to cellular restriction factors (56).
In the case of HIV-2, we and others have shown previously that TSG-101 binds specifically with the PTAPP region of the Gag polyprotein L domain of both HIV-1 and HIV-2 and is responsible for recruitment of host cell ubiquitination machinery (43, 60).
Although HIV-1 and HIV-2 are both members of the lentivirus subfamily of retroviruses and share similar genetic organization, there is limited homology at the nucleotide and amino acid level (24). HIV-2 differs from HIV-1 in the mechanism it uses to select its genomic RNA for encapsidation, suggesting that HIV-2 Gag utilizes a distinct pathway for selection of unspliced RNA into progeny virions (26). In this report we describe the identification of the serine-threonine kinase PRP4 as an HIV-2 Gag-interacting protein. PRP4 interacts with HIV-2 Gag in vitro and in vivo, but is not incorporated into HIV-2 virus particles. Through the use of in vitro pull-down assays, we were able to map the interacting domains of both HIV-2 Gag and the full-length PRP4 protein. Finally, we have also determined a possible functional role for this interaction as a global inhibitor of splicing in the infected cell.
MATERIALS AND METHODS
Yeast two-hybrid plasmid construction.
Yeast expression plasmids pLexA (bait) and pB42AD (prey) containing HIS3 and TRP1 selection markers, respectively, were used (BD Clontech). The HIV-2 ROD Gag coding sequence (24) from pGexGag2 (described below) was removed and cloned within the EcoRI and SalI restriction sites to generate pLexAGag2 and pB42ADGag2 (43). pBSK-PRP4, which contains human PRP4 cDNA, was kindly donated by Martin Leutzelberger (Braunschweig, Germany). Sequence corresponding to that identified in the screening (1,535 to 3,020 nucleotides) was Pfu PCR amplified from this plasmid with EcoRI and XhoI ends, using the primers PRP4F (5′-GCGCGAATTCATGAAAGTTGAGCAGGAATCTTCG-3′) and PRP4R (5′GGCCCTCGAGTTAAATTTTTTCCTGGATGAAGGCG-3′), and cloned into both bait and prey plasmids, using these restriction sites to generate pB42ADPRP4 and pLexAPRP4.
In vitro binding assay plasmid construction.
Gag sequences of HIV-1 HXB2 and HIV-2 ROD were PCR amplified with Pfu DNA polymerase (Stratagene) with 1GAGF and 1GAGR primers for HIV-1 Gag or 2GAGF and 2GAGR primers for HIV-2 Gag as described previously (43). The resulting PCR products were digested with EcoRI and SalI and cloned into these sites in pGex4T-1 (Amersham Pharmacia Biotech) to construct pGexGag1 and pGexGag2, respectively. PRP4 sequence corresponding to the region identified in the yeast two-hybrid assay (nucleotides 1535 to 3020) was PCR amplified with the primers described above. The resulting product was digested with EcoRI and XhoI and cloned into the EcoRI-XholI site of pGex4T-1. The full-length PRP4 transcript in plasmid pMEHAPRP4 incorporating an N-terminal hemagglutinin (HA) epitope tag was kindly provided by Masatoshi Hagiwara. Bacterial expression plasmids expressing subdomains of PRP4 were created with the following primers: NPRP4F(5′-ATATGGATCCATGGCCGCCGCGGAG-3′) and CPRP4R (5′-GCCCGCGAATTCTTAACGTTTAT-3′) for pGexPRP4ΔCterm (nucleotides 1 to 2060), PPRP4F (5′-GCGGGATCCATGAAAGTTGAGC-3′) and CPRP4Rev (5′-GCGCGGAATTCTTAAATTTTTTCCTGGATGAAGGCG-3′) for pGexΔNterm (nucleotides 1538 to 3020), and PPRPF and CPRP4R to generate pGexPRP4ΔN-Cterm (nucleotides 1538 to 2060). PCR products generated were digested with BamHI and EcoRI and cloned into these sites of pGex4T-1. The in vitro translation plasmid pRSETGag2 was generated by cloning a partial digest from pMALCGag2 (described below) using EcoRI and HindIII restriction sites, into EcoRI-HindIII of pRSETc (Invitrogen). Bacterial expression vectors containing subdomains of HIV-2 Gag were generated via Pfu PCR amplification of pSVR (described below) using the following primers: 2GAGF and 2MAR for pRSETMA2 (nucleotides 546 to 950), 2CAF and 2CAR for pRSETCA2 (nucleotides 951 to 1640), and 2NCF and 2NCR for pSETNC2 (nucleotides 1692 to 1838) as previously described (43). The pRSETGag2Δp2-p6 plasmid was created by cloning the EcoRI-HindIII fragment (nucleotides 541 to 1458) from pGexGag2 into these sites of pRSETCA2, the resulting plasmid containing HIV-2 nucleotides 546 to 1640 and lacking the p2, NC, p1 and p6 domains. pRSETGag2Δp6 was created by Pfu PCR amplification of HIV-2 sequence with primers 2CAF and 2NCR. The resulting product was digested with HindIII (nucleotide 1458) and SalI (nucleotide1839) and cloned into these sites in pRSETGag2. This resulting plasmid contains HIV-2 sequence 546 to 1839 and lacks the p1 and p6 domains. Maltose-binding protein (MBP) fusions were produced for use in the kinase assays. pMALCGag2 was generated by digesting the full-length Gag sequence flanked by EcoRI and SalI restriction sites in pGexGag2 and cloning into these sites located in pMAL-c2 (New England Biolabs). For pMALCSIVGag, the Gag sequence from nucleotides 1054 to 2586 was digested from pGEXSIVGag with EcoRI and SalI restriction enzymes and cloned into pMAL-c2 digested with EcoRI-SalI. pMALCHTLV1Gag was generated by Pfu polymerase PCR (Promega) of proviral clone XMTDM2 using the following primers: HTLV-1F (5′-GCGCCCGAATTCATGGGCCAAATCTTTTCC-3′) and HTLV-1R (5′-GGCCGCGTCGACTTAAACCTCCCCCC-3′) to amplify the full-length Gag sequence (nucleotides 803 to 2092). The product was digested with EcoRI and SalI as previously described and cloned into pMALC2. pET9cSF2 was kindly provided by Adrian Krainer (Cold Spring Harbor Laboratories, Cold Spring Harbor, N.Y.). Full-length human SF2 sequence was subcloned into pSL1190 (Amersham Pharmacia Biotech) by digestion with NdeI and BamHI restriction enzymes, yielding a 750-bp fragment. The resulting plasmid was then digested with PvuII and SalI sites in the polylinker, and the fragment was cloned into pGex4T-3 digested with SmaI and SalI to generate pGexSF2.
Mammalian expression plasmid construction.
pSVR is an infectious proviral clone of HIV-2 ROD containing a simian virus 40 origin of replication (36). Restriction sites are described with respect to the first nucleotide of the viral RNA. pSVR?NB has been previously described (21) and contains a deletion of 550 bp in the env gene between NsiI and BstXI (nt 6369 to 6919). To construct an additional PRP4 expression plasmid, full-length PRP4 cDNA was amplified from pMEHAPRP4 by Pfu DNA polymerase (Strategene) with the following primers: KPRP4F (5′GAAGGTACCATGGCCGCCGCGG-3′) and XPRP4R (5′GCGCTCTAGAAATTTTTTCCTGGATGAAGGC-3′). The resulting product was digested with KpnI and XbaI restriction enzymes and cloned into pCDNA3.1V5HisA (Invitrogen) digested with KpnI-XbaI. The resulting plasmid, pCDNAPRP4HisFL, contains the entire PRP4 coding sequence (3 kb) with an in-frame polyhistidine tag at the C terminus. The integrity of the PCR-amplified sequence was confirmed by DNA sequencing. All HIV-2-based plasmids were propagated in TOPF10′ (Invitrogen) at 30°C to minimize recombination. All other plasmids were propagated in DH5α.
Yeast two-hybrid library screening.
The Matchmaker LexA two-hybrid system was used in conjunction with a Jurkat cDNA library. These were obtained from Clontech. This system identifies bait-interacting proteins through selection for leucine prototrophy and a screen for β-galactosidase activity in Saccharomyces cerevisiae EGY48 (ura3 his3 trp1 LexAop-leu2). A total of 2 × 106 colonies were screened as per the recommended manufacturer's instructions. The putative bait-interacting clones were amplified with primers B42ADF and B42ADR as described previously (43) PCR products were purified with QIAQuick PCR purification columns (QIAGEN), and each library insert was sequenced and compared with entries in the GenBank database.
In vitro binding assays.
Glutathione S-transferase (GST) fusion proteins were prepared as described previously by Smith and Johnson (43), with the exception that the bacterial cultures were incubated for 3 h and then induced with 50 mM isopropyl-β-d-thiogalactopyranoside for a further 3 h. In vitro-translated and radiolabeled protein was generated with the Quick TNT kit (Promega). In vitro transcription-translation reactions were performed to generate [35S]methionine-labeled protein as described by the manufacturer (Promega). GST fusion protein beads (500 ng of protein in a total of 30 μl) were preincubated with bovine serum albumin at room temperature at a final concentration of 0.2 mg/ml and mixed for 5 min. The samples were then mixed with 2 to 5 μl of in vitro-translated test protein in 190 μl of EBC buffer (140 mM NaCl, 0.5% Nonidet P-40, 100 mM NaF, 200 μM orthovanadate, 50 mM Tris-HCl [pH 8.0], 0.5 mM phenylmethylsulfonyl fluoride) and incubated with mixing for 1 h. The beads were centrifuged at 2,000 rpm for 2 min in an MSE Falcon 6/300 centrifuge and washed four times with 1 ml of NETN buffer (100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, 20 mM Tris-HCl [pH 8.0], 0.5 mM phenylmethylsulfonyl floride) and boiled for 5 min in 4× sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis sample buffer. Bound proteins were resolved on SDS-PAGE (12.5% polyacrylamide) gels and detected by autoradiography.
Cell cultures and transfections.
Cell culture conditions were maintained as described previously for 293Tcells (43). Transient transfection of 293T cells was performed by the calcium phosphate method as previously described (43). The cell supernatant was harvested 48 h later and treated as previously described for purification of viral particles (43). Cells were washed with ice-cold phosphate-buffered saline (PBS), resuspended in 150 μl of 4× SDS-PAGE buffer, and sonicated. Cells and virions were analyzed by Western blotting.
Western blotting.
Western blotting analysis was performed with Hybond-C Ultra transfer membrane (Amersham Pharmacia Biotech) and SDS-PAGE (12.5% polyacrylamide) gels as per the manufacturer's instructions. Monoclonal antibody to HIV-2 p26 (Chemicon) was used at 1:1,000 in PBS (5% Marvel, 0.05%Tween 20), and monoclonal antibody to V5 epitope (Clontech) was used at a 1:1,250 dilution. Horseradish peroxidase-conjugated donkey anti-mouse immunoglobulin G secondary antibody (Chemicon) was used at a 1:5,000 dilution. These antibodies were detected by enhanced chemiluminescence Western blotting detection reagent (Amersham Pharmacia Biotech), as specified by the manufacturer.
In vitro kinase assay.
Forty-eight hours following transfection, cells were harvested, washed in cold PBS, and incubated for 1 h at 4°C in 1 ml of EBC buffer. Cell supernatant was centrifuged at 13,000 rpm at 4°C for 20 min. Supernatant was incubated overnight in 200 μl of a 1:1 suspension of protein A-protein G beads (Pharmacia Biotech), and 10 μg of anti-HA epitope monoclonal antibody (New England Biolabs). Beads were centrifuged 7,000 rpm for 2 min and washed three times with 1 ml of EBC buffer. The wash steps were repeated three times with 1 ml of kinase buffer I (20 mM HEPES [pH 7.9], 50 mM KCl, 3 mM MgCl2, 5% glycerol). To each reaction mixture, kinase buffer I, amylose beads, and increasing molar amounts of MBP fusion protein competitor beads were added to make a total volume of 20 μl, using the amylose beads to equalize the amount of beads in all tubes. A 30-μl total volume of kinase buffer II (20 mM HEPES [pH 9], 50 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol), 5 μCi of [γ-32P]ATP, 10 mM ATP, and 100 ng of GST-SF2 fusion beads were added and mixed well. Assay tubes were incubated for 30 min, mixing every 5 min. Beads were washed three times in 1 ml of kinase buffer II. Beads were boiled in 4× SDS-PAGE buffer, run on SDS-PAGE (12.5% polyacrylamide) gels, and detected by autoradiography.
Immunoprecipitation and Western blotting.
Cos-1 cells were maintained in modified Eagle's medium (Gibco-BRL) supplemented with 10% fetal calf serum, penicillin, and streptomycin. Transient transfection of Cos-1 cells was performed by the calcium phosphate method as previously described for 293T cells (43). After a 48-h incubation, medium was discarded and cells were washed twice and harvested in ice-cold PBS. Cell pellets for the Western blot only were suspended in 300 μl of SDS-PAGE buffer and sonicated. Cell pellets for immunoprecipitation were lysed in 1 ml of EBC buffer for 1 h at 4°C. Cleared lysate was incubated for 16 h at 4°C with 6 μg of anti-p26 monoclonal antibody (Chemicon; MAB8838). Fifty microliters of a 1:1 suspension of protein A-protein G beads (Pharmacia Biotech) was added to each lysate sample and incubated 1 h at 4°C. Protein beads were washed four times with 1 ml of NETN buffer and suspended in 10 μl of 4× SDS-PAGE buffer. Both cell pellets for Western blot and immunoprecipitation beads for Western blot were boiled for 5 min, and 10 μl of each was loaded onto an SDS-PAGE (12.5% polyacrylamide) gel and blotted as previously described. Monoclonal antibody to HIV-2 Gag p26 (Chemicon; no. 8838) or monoclonal antibody to the PRP4 HA tag (New England Biolabs; no. 2362) was diluted to 1:1,000 and used in the detection, along with the ECL enhanced chemiluminescence reagent as described previously.
RESULTS
Identification of an interaction between HIV-2 Gag and PRP4.
Uncleaved HIV-2 Gag was used as bait in a yeast two-hybrid screen to identify interacting proteins encoded by a Jurkat cDNA library. Approximately 2 × 106 transformants were screened, and several positive clones were identified. Comparison of the cDNA sequences with the GenBank database revealed that one of the clones was human elongation factor 1 alpha, which has been reported to interact with HIV-1 Gag (9). Another was human Tsg101, an inactive homologue of ubiquitin ligase E2, and another was the human homologue of yeast serine-threonine kinase PRP4.
PRP4 interacts specifically with HIV-2 Gag.
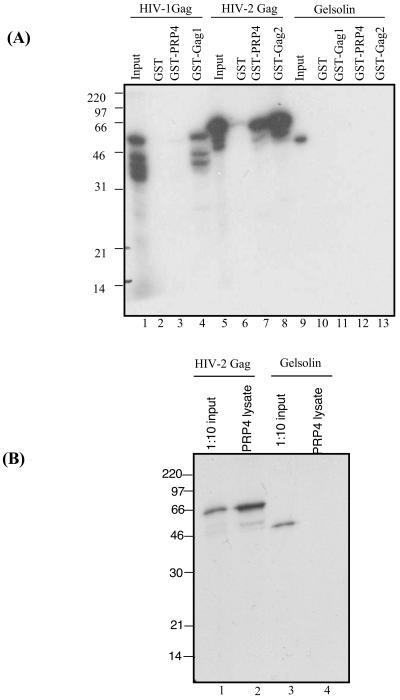
To confirm the specificity of the interaction between HIV-2 Gag and PRP4, we carried out in vitro binding assays using GST-PRP4 fusion proteins and in vitro-translated HIV Gag. Figure 1A shows that PRP4 interacts specifically with HIV-2 Gag, but not HIV-1 Gag. There was no specific binding of HIV-2 Gag or HIV-1 Gag to GST alone, confirming the specificity of the interaction between HIV-2 Gag and PRP4. The cellular protein gelsolin, used as a nonspecific control, did not bind to GST, GST-PRP4, GST Gag1, or GST Gag2. GST Gag1 and GST Gag2 function as positive controls for binding in this assay, as they are known to self-associate in vitro and in vivo. This interaction was further confirmed in an immunoprecipitation in vitro assay. 293T cells were transfected with full-length PRP4 fused to a V5 epitope tag. Cells were harvested and lysed, and supernatant was incubated with V-5 monoclonal antibody (Invitrogen) and an equal volume of protein A-protein G beads (Amersham Pharmacia Biotech). These beads were then used in an in vitro binding assay with radiolabeled HIV-2 Gag and gelsolin cellular protein; the results are shown in Fig. 1B. HIV-2 Gag specifically binds PRP4 protein beads and does not bind gelsolin.
FIG. 1.
PRP4 binds specifically with HIV-2 Gag. (A) In vitro binding of PRP4 to HIV-1 and HIV-2 Gag proteins. In vitro-transcribed and -translated HIV-1 Gag (lanes 2 to 4), HIV-2 Gag (lanes 6 to 8), or gelsolin (lanes 10 to 13) was analyzed by GST fusion pull-down assays for the ability to bind GST, GST-PRP4, GST-Gag1, or GST-Gag2 immobilized on glutathione-Sepharose beads. Inputs were 10% of the amount of HIV-1 Gag (lane 1), HIV-2 Gag (lane 5), or gelsolin (lane 9) used in each assay. (B) PRP4 cellular lysate binding to HIV-2 Gag protein. In vitro-transcribed and -translated HIV-2 Gag (lane 2) or gelsolin (lane 4) was analyzed by pull-down assay for the ability to bind PRP4 protein extract from transfected 293T cells immobilized on protein A-protein G beads. Inputs were 10% of the amount of HIV-2 Gag (lane 1) or gelsolin (lane 3) used in each assay. The positions of marker proteins are indicated in kilodaltons.
PRP4 binds to the N terminus of HIV-2 Gag.
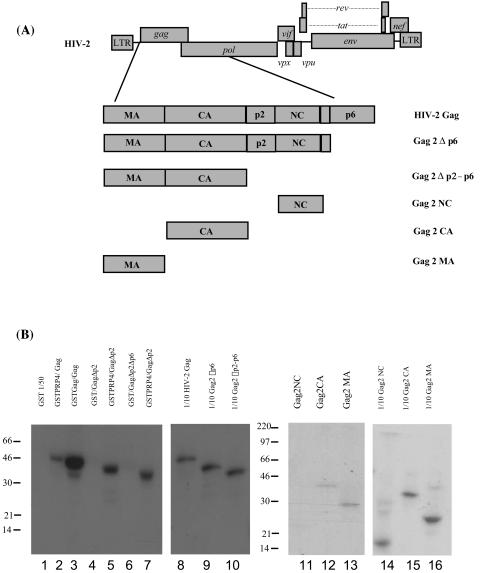
To map the region of the HIV-2 Gag polyprotein required for interaction with PRP4, HIV-2 Gag proteins with N- or C-terminal truncations were in vitro translated and radiolabeled and used in binding assays with the region of PRP4 identified in the yeast two-hybrid library screening fused to GST. The truncations generated are shown in Fig. 2A. The in vitro binding assay result is shown in Fig. 2B and indicates that removing the N-terminal portion of Gag reduces the ability of PRP4 to bind the HIV-2 Gag polyprotein. GST fusion protein alone did not bind any of the truncated HIV-2 Gag proteins. Expression of individual subdomains of Gag showed evidence of binding to MA and CA but no evidence of interaction with the NC region. This together with the C-terminal truncations suggests binding sites for PRP4 in the N terminus of Gag.
FIG. 2.
Determination of the region of HIV-2 Gag involved in binding PRP4. (A) Schematic diagram of HIV-2 Gag domain proteins prepared for in vitro binding assays. 35S-labeled HIV-2 Gag truncations were prepared by TNT in vitro translation. (B) In vitro binding of HIV-2 Gag truncations to PRP4 protein. C-terminal protein truncations were analyzed by GST fusion pull-down assay for the ability to bind GST or GST-PRP4 (lanes 1 to 7) immobilized on glutathione-Sepharose beads. Individual Gag subdomains (MA, CA, and NC) were also assayed (lanes 11 to 13) Inputs (lanes 8 to 10 and 14 to 16) were 10% of the amount of the relevant 35S-labeled HIV-2 Gag protein used in each assay. The positions of marker proteins are in kilodaltons.
HIV-2 Gag binds to the C terminus of PRP4.
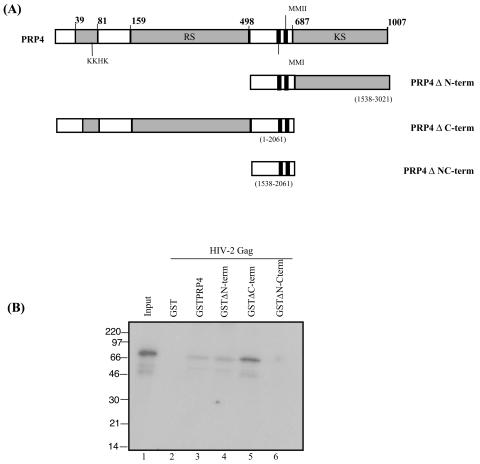
The human PRP4 protein contains a serine-threonine protein kinase located at the C terminus. It was of interest to map the interacting region of PRP4 with HIV-2 Gag, as binding of the C terminus has previously been implicated in cellular protein binding and phosphorylation (29). GST fusion proteins of various PRP4 protein truncations were generated. These are shown in Fig. 3A. Full-length HIV-2 Gag protein was in vitro translated and radiolabeled as previously described (43). The results are shown in Fig. 3B. Deletion of the C terminus of PRP4 has little effect on the binding, while removal of the N terminus reduces binding, but does not eliminate it. Only when both N- and C-terminal portions of PRP4 are removed is the binding to HIV-2 Gag completely abolished. The apparent increase in binding to Gag with the ΔC terminus deleted may relate to the truncated protein revealing its binding site more, although the assay, while it will show the presence or absence of binding, is not accurately quantitative between different positive binding affinities.
FIG. 3.
Determination of the region of PRP4 involved in binding to HIV-2 Gag. (A) Schematic diagram of PRP4 domains expressed as GST fusion proteins. Numbers in parentheses indicate nucleotide positions. Amino acid residues are shown in bold. Conserved domains are shaded. KKHK, KKHK box; RS, arginine-serine-rich domain; KS, kinase domain; MMI and MMII, conserved domains. (B) In vitro binding of PRP4 truncations to full-length HIV-2 Gag protein. PRP4 protein truncations (lanes 3 to 6) or GST (lane 2) were immobilized on glutathione-Sepharose beads and analyzed by GST fusion pull-down assay for the ability to bind in vitro-translated and radiolabeled HIV-2 Gag. Input (lane 1) is 10% of the amount of the 35S-labeled HIV-2 Gag protein used in the assay. Marker lane proteins are indicated in kilodaltons.
PRP4 is not incorporated into virions.
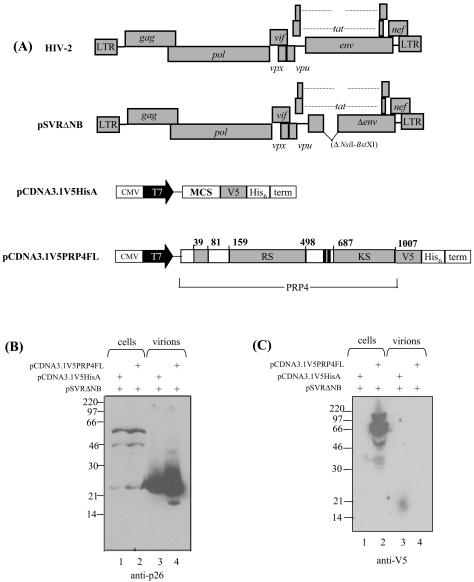
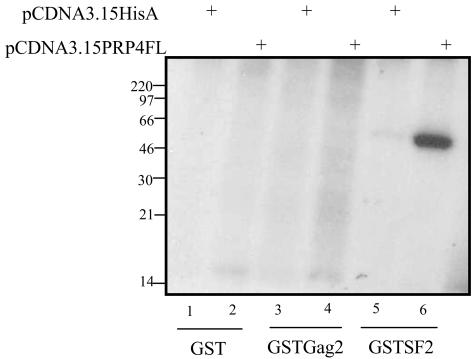
Having shown a specific interaction between PRP4 and HIV-2 Gag, we then asked whether PRP4 was incorporated into HIV-2 virus particles. 293T cells were cotransfected with an HIV-2 proviral expression construct and either PRP4 expression constructs or empty vector constructs. The expression constructs used in this experiment are shown in Fig. 4A. Cell and virion samples were analyzed by Western blotting (Fig. 4B and C). Figure 4B shows Gag polyprotein in the cells and mature p26 capsid in the virions. In Fig. 4C, a 175-kDa PRP4 protein was detected in the transfected cells. PRP4 was not detected in virions.
FIG. 4.
PRP4 is not incorporated into HIV-2 virus particles. (A) Schematic diagram of HIV-2 Gag and PRP4 expression constructs. CMV, cytomegalovirus immediate-early inhancer; T7, promoter/priming site; MCS, multiple cloning site; V5, V5 epitope; His6, polyhistidine tag; RS, arginine-serine-rich domain; KS, kinase domain. (B and C) 293T cells were transfected with PRP4 plasmid (pCDNA3.1V5PRP4FL) or control plasmid (pCDNA3.1V5HisA) and an env-deleted HIV-2 provirus as indicated. Cell and virus particle samples were prepared as described in Materials and Methods and subjected to analysis by Western blotting using monoclonal antibodies to either p26 (HIV-2 capsid) (B) or V5-tagged PRP4 (C). The positions of marker proteins are indicated in kilodaltons.
HIV-2 Gag is not phosphorylated by PRP4 in vitro.
We were interested in determining whether HIV-2 Gag was phosphorylated by PRP4. 293T cells were transfected with a PRP4 expression plasmid or an empty expression plasmid, and the proteins were immunoprecipitated (IP) from the cell supernatant. The PRP4-IP beads were used as the active protein in the kinase assay. Fusion proteins of full-length HIV-2 Gag and SF2 protein fused to GST were generated, as well as GST protein alone, and these were used as targets for PRP4 phosphorylation. The results in Fig. 5 show a clear phosphorylation of SF2, as observed in vivo. PRP4 is not capable of phosphorylating HIV-2 Gag or the negative control GST.
FIG. 5.
PRP4 phosphorylation observed in kinase assays is unique to SF2 protein. 293T cells were transfected with either empty vector (pCDNA3.1V5HisA) or vector containing full-length PRP4 fused to a V5 epitope tag (pCDNA3.1V5PRP4FL). Cells were lysed and protein was immunoprecipitated as described in Materials and Methods. Empty vector protein (lanes 1, 3, and 5) and PRP4 protein (lanes 2, 4, and 6) were used in a kinase assay as phosphorylating agents. GST fusion proteins were used as targets of phosphorylation: GST alone (lanes 1 and 2), GST fused to HIV-2 Gag (lanes 3 and 4), and GST fused to SF2 protein (lanes 5 and 6).
Interaction between HIV-2 Gag and PRP4 inhibits phosphorylation of SF2.
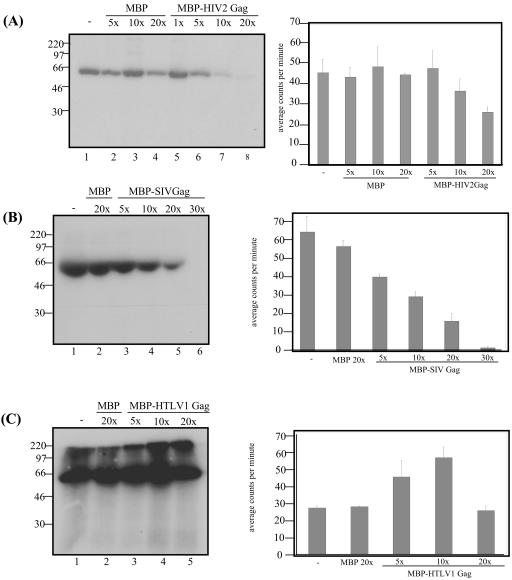
Since PRP4 was not observed to phosphorylate HIV-2 Gag, we wanted to determine whether Gag was instead acting as a competitor to SF2 phosphorylation, sequestering PRP4 and inhibiting the phosphorylation in vitro. We repeated the transfection of 293T cells to produce PRP4-IP bead-bound fusion protein and prepared SF2 protein fused to GST as a target for phosphorylation as previously described. Increasing molar amounts of HIV-2 Gag polyprotein fused to MBP were added as a competitor in each reaction. MBP was used as a negative control competitor. Figure 6A shows a marked decrease in the phosphorylation of SF2 with increasing molar amounts of MBP-HIV-2 Gag. There is no effect on SF2 phosphorylation with increasing MBP.
FIG. 6.
Competitive effect of lentiviral Gag protein on the kinase activity of PRP4. (A) PRP4 protein incorporating a V5 epitope tag immunoprecipitated from transfected 293T cells phosphorylates SF2 protein fused to glutathione-Sepharose beads (lane 1). Increasing molar excess of MBP protein (lanes 2, 3, and 4) or MBP-HIV-2 Gag protein (lanes 5, 6, and 7) are titrated against a constant molar amount of PRP4. (B) PRP4 protein phosphorylates SF2 alone (lane 1) and when a 20× molar excess of MBP is added (lane 2). MBP-SIV Gag protein is titrated against PRP4 protein at 5, 10, 20, and 30× the amount of PRP4 (lanes 3, 4, 5, and 6). (C) PRP4 phosphorylates SF2 (lane 1) and when 20× molar excess of MBP protein is added (lane 2). MBP-HTLV-1 Gag is titrated against the phosphorylation at 5, 10, and 20× the molar excess of PRP4 (lanes 3, 4, and 5). The data in each graph to the right represent the mean phosphorylation activity of three independent kinase assays for each lentivirus. Data were analyzed with NIH Image (A) and the Packard Instant Imager (B and C).
Retroviral Gag proteins differ in their ability to affect phosphorylation of SF2 by PRP4.
To determine whether the inhibition of SF2 phosphorylation was unique to HIV-2 Gag, similar kinase assays were carried out with full-length Gag polyprotein from simian immunodeficiency virus of macaques (SIVmac; a lentivirus closely related to HIV-2) and HTLV-1 (a distantly related retrovirus). Kinase assay SF2 and PRP4-IP beads were prepared as previously described. Gag proteins were prepared as GST fusions and added in increasing molar ratios as with HIV-2 Gag. The SIVmac Gag competitor displayed a notable inhibition of phosphorylation, as observed with HIV-2 Gag, while HTLV-1 Gag had no effect on the phosphorylation of SF2. These results are shown in Fig. 6B and C.
HIV-2 Gag and PRP4 interact in vivo.
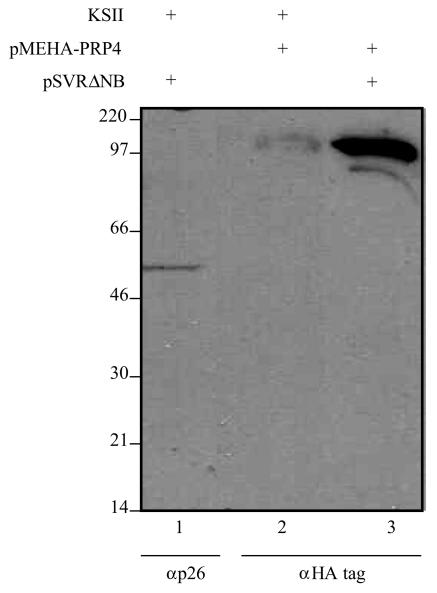
To confirm the interaction between the HIV-2 Gag polyprotein and cellular protein PRP4 occurs in vivo, we carried out an immunoprecipitation assay. Cos-1 cells were cotransfected with the HIV-2 proviral construct pSVRΔNB and a full-length PRP4 expression plasmid incorporating an HA tag on the N terminus. These plasmids were also transfected singly to check their expression levels, using pBluescript DNA (Stratagene) to equalize the total amount of DNA in all transfections. Protein-IP bead fusions were generated with an anti-p26 monoclonal antibody, and the resulting beads were run in a Western blot along with cell pellet extracts from the single transfections. Samples were blotted with either anti-p26 or anti-HA monoclonal antibody. Following a 1-min exposure, PRP4 protein can clearly be detected in both the immunoprecipitate and the cellular lysate. Full-length HIV-2 Gag is also detected in cellular lysate. These results are summarized in Fig. 7.
FIG. 7.
PRP4 and HIV-2 Gag protein interact in vivo. Cos-1 cells were transfected with an HIV-2 proviral construct (lane 1), a full-length PRP4 expression vector incorporating an HA tag (lane 2), or both (lane 3). Cell pellet extracts were blotted either with anti-p26 (lane 1) or anti-HA (lane 2) antibody to determine the amount of protein present. Cells were cotransfected with these plasmids, and HIV-2 Gag protein was immunoprecipitated with anti-p26 and then blotted with anti-HA monoclonal antibody to detect PRP4 (lane 3). Marker lane proteins are indicated in kilodaltons.
DISCUSSION
This is the first report of an interaction between a serine/threonine protein kinase and the HIV-2 Gag polyprotein. Phosphorylation of the P6 cleavage product of Gag has previously been documented (42).
The interaction between HIV-2 Gag and human PRP4 was detected in a yeast two-hybrid screen of a Jurkat cDNA library, and the specificity of the interaction was confirmed in vitro and in vivo. In HIV-1 a kinase interaction has been recognized; phosphorylation of the CA (p24) protein on three serine residues is necessary for complete reverse transcription and subsequent infectivity (8). Mitogen-activated protein kinase ERK-2, a serine/threonine kinase, and an unknown 53-kDa protein kinase are incorporated into virus particles, suggesting that one of these may phosphorylate these serine residues (7).
PRP4 interacts with the N-terminal portion of HIV-2 Gag. Despite considerable amino acid homology between the N-terminal Gag domains of these retroviruses, we did not observe this interaction between PRP4 and HIV-1 Gag in the same in vitro binding assays. Using in vitro kinase assays, we have demonstrated that the HIV-2 Gag polyprotein itself is not phosphorylated by PRP4, but rather acts as an inhibitor of phosphorylation of SF2, the usual target of PRP4. We also observed this with SIVmac Gag polyprotein, but not with HTLV-1 Gag.
Both PRP4 and ASF/SF2 are members of the cellular SR protein family of splicing factors. PRP4 was originally identified in yeast, through its role in pre-mRNA splicing, when pre-mRNA from intron-containing genes was found to accumulate when a temperature-sensitive mutant was maintained at the restrictive temperature (1, 46, 58). PRP4 is ubiquitously expressed in multiple tissues (29) and belongs to the serine-arginine-rich protein-specific kinases, recognizing serine-arginine-rich substrates (22). Previous attempts at isolating a mammalian form of PRP4 only uncovered partial clones, with Kaufer et al. reporting the first human PRP4 cDNA cloned from a HeLa cDNA library to be 55 kDa (22). Later, Kojima et al. identified the full-length, 170-kDa form, with 1,007 amino acids (29). The N-terminal portion of PRP4 contains an arginine-serine-rich domain and putative nuclear localization signals, and the protein colocalizes in a nuclear speckled pattern with splicing factor ASF/SF2 (29). ASF/SF2 was originally identified as an alternative splicing factor of simian virus 40 (SV40) pre-mRNA in nuclear extracts (19). It is now known that ASF/SF2 is an integral part of the spliceosome machinery and is highly phosphorylated in vivo (38, 63). SR proteins are active in virtually all stages of spliceosome assembly, especially in early splice site recognition (14, 28, 30, 51, 55) and alternative splicing (15, 20, 35, 59). The RS domains of SR proteins undergo cycles of phosphorylation/dephosphorylation throughout spliceosome assembly and pre-mRNA splicing, having the ability to enhance or repress splicing in vitro (6, 38). The gene coding for ASF/SF2 is responsible for the regulation of alternative splicing in eukaryotic (10, 17, 32) and viral (54, 61, 62) pre-mRNAs in vitro and in vivo and is an essential gene (61).
Alternative splicing greatly increases coding potential of genes in mammalian cells and viruses and is an important regulatory mechanism in gene expression (19). Viral late gene expression is often dependent on alternative splicing and polyadenylation, both cis- and trans-acting factors being necessary. It is suggested that alternatively spliced mRNAs are controlled temporally by SR proteins, with distinct species accumulating at different time points during infection. This has been observed in both adenovirus (40) and papillomavirus (33) infection, where ASF/SF2 regulates the early-to-late shift in viral mRNA splicing and expression.
In HIV, gene transcription is a temporal cascade mediated by the use of alternate splice sites. The expression of unspliced and multiply spliced HIV viral mRNA is posttranscriptionally controlled during splicing and nuclear export (13, 27), with transport of singly and unspliced mRNAs mediated by the Rev protein. During early HIV infection, small, multiply spliced RNAs encoding Tat and Rev are generated (11, 45). When a threshold level of Rev protein is reached, singly and unspliced mRNA is transported to the cytoplasm via interaction of Rev with the Rev-responsive element (RRE). As Rev increases the steady-state levels of these mRNAs in the cytoplasm, Gag, Pol, and Env proteins are efficiently synthesized, along with the accumulation of full-length genomic RNA for packaging. Rev protein not only transports full-length mRNA containing an RRE to the cytoplasm, but also causes it to bypass splicing machinery. In this situation, Rev is acting in a negative feedback loop, limiting its own production through inhibiting export of spliced messages. We hypothesize that the increase in the Gag polyprotein further enhances this down-regulation in splicing, by binding to PRP4 and sequestering it away from splicing factor ASF/SF2 in the nucleus. This disrupts the splicing machinery and effects a general down-regulation of cellular splicing during late infection. This would be useful in a cell where Rev is limited and must shuttle back into the nucleus to transport additional unspliced and singly spliced mRNA, as it replaces one of the functions that Rev provides in early infection. It is interesting to note that, unlike HIV-2 and SIV, HTLV-1 Gag does not inhibit PRP4-dependent phosphorylation. HTLV-1 may have alternative mechanisms for augmenting the Rex/RexRE-mediated splicing suppression (57).
We have shown that HIV Gag protein can inhibit the activity of a component of the splicing machinery, while others have shown that Gag RNA can affect the spliceosome in simple retroviruses. Arrigo and Beemon (2) and Stoltzfus and Fogarty (52) showed that a segment of avian sarcoma leukosis virus Gag RNA was able to act as a negative regulator of splicing. This was further supported by later work identifying the same Gag RNA binding to ASF/SF2 and functioning as a “mini-exon.” This results in a complex that inhibits normal splicing at the 5′ and 3′ splice sites (37). Taken together, our results suggest that retroviruses have several mechanisms with which they influence splicing and both effect and maintain the early to late switch in gene expression.
Acknowledgments
We thank Jane Greatorex and Nick Bennett for critical reading and helpful suggestions.
This work was funded by the Wellcome Trust and the Medical Research Council.
REFERENCES
- 1.Alahari, S. K., H. Schmidt, and N. F. Kaufer. 1993. The fission yeast prp4+ gene involved in pre-mRNA splicing codes for a predicted serine/threonine kinase and is essential for growth. Nucleic Acids Res. 21:4079-4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arrigo, S., and K. Beemon. 1988. Regulation of Rous sarcoma virus RNA splicing and stability. Mol. Cell. Biol. 8:4858-4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bevec, D., H. Jaksche, M. Oft, T. Wohl, M. Himmelspach, A. Pacher, M. Schebesta, K. Koettnitz, M. Dobrovnik, R. Csonga, F. Lottspeich, and J. Hauber. 1996. Inhibition of HIV-1 replication in lymphocytes by mutants of the Rev cofactor eIF-5A. Science 271:1858-1860. [DOI] [PubMed] [Google Scholar]
- 4.Bouyac, M., M. Courcoul, G. Bertoia, Y. Baudat, D. Gabuzda, D. Blanc, N. Chazal, P. Boulanger, J. Sire, R. Vigne, and B. Spire. 1997. Human immunodeficiency virus type 1 Vif protein binds to the Pr55Gag precursor. J. Virol. 71:9358-9365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukrinsky, M. I., N. Sharova, T. L. McDonald, T. Pushkarskaya, W. G. Tarpley, and M. Stevenson. 1993. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl. Acad. Sci. USA 90:6125-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao, W., S. F. Jamison, and M. A. Garcia-Blanco. 1997. Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA 3:1456-1467. [PMC free article] [PubMed] [Google Scholar]
- 7.Cartier, C., M. Deckert, C. Grangeasse, R. Trauger, F. Jensen, A. Bernard, A. Cozzone, C. Desgranges, and V. Boyer. 1997. Association of ERK2 mitogen-activated protein kinase with human immunodeficiency virus particles. J. Virol. 71:4832-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cartier, C., P. Sivard, C. Tranchat, D. Decimo, C. Desgranges, and V. Boyer. 1999. Identification of three major phosphorylation sites within HIV-1 capsid. Role of phosphorylation during the early steps of infection. J. Biol. Chem. 274:19434-19440. [DOI] [PubMed] [Google Scholar]
- 9.Cimarelli, A., and J. Luban. 1999. Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 73:5388-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cramer, P., J. F. Caceres, D. Cazalla, S. Kadener, A. F. Muro, F. E. Baralle, and A. R. Kornblihtt. 1999. Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol. Cell 4:251-258. [DOI] [PubMed] [Google Scholar]
- 11.Cullen, B. R. 1992. Mechanism of action of regulatory proteins encoded by complex retroviruses. Microbiol. Rev. 56:375-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elfgang, C., O. Rosorius, L. Hofer, H. Jaksche, J. Hauber, and D. Bevec. 1999. Evidence for specific nucleocytoplasmic transport pathways used by leucine-rich nuclear export signals. Proc. Natl. Acad. Sci. USA 96:6229-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frankel, A. D., and J. A. Young. 1998. HIV-1: fifteen proteins and an RNA. Annu. Rev. Biochem. 67:1-25. [DOI] [PubMed] [Google Scholar]
- 14.Fu, X. D. 1993. Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature 365:82-85. [DOI] [PubMed] [Google Scholar]
- 15.Fu, X. D. 1995. The superfamily of arginine/serine-rich splicing factors. RNA 1:663-680. [PMC free article] [PubMed] [Google Scholar]
- 16.Gallay, P., S. Swingler, J. Song, F. Bushman, and D. Trono. 1995. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell 83:569-576. [DOI] [PubMed] [Google Scholar]
- 17.Gallego, M. E., R. Gattoni, J. Stevenin, J. Marie, and A. Expert-Bezancon. 1997. The SR splicing factors ASF/SF2 and SC35 have antagonistic effects on intronic enhancer-dependent splicing of the beta-tropomyosin alternative exon 6A. EMBO J. 16:1772-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gamble, T. R., F. F. Vajdos, S. H. Yoo, D. K. Worthylake, M. Houseweart, W. I. Sundquist, and C. P. Hill. 1996. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87:1285-1294. [DOI] [PubMed] [Google Scholar]
- 19.Ge, H., and J. L. Manley. 1990. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell 62:25-34. [DOI] [PubMed] [Google Scholar]
- 20.Graveley, B. R. 2000. Sorting out the complexity of SR protein functions. RNA 6:1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffin, S. D. C., J. F. Allen, and A. M. L. Lever. 2001. The major human immunodeficiency virus type 2 (HIV-2) packaging signal is present on all HIV-2 RNA species: co-translational RNA encapsidation and limitation of Gag protein confer specificity. J. Virol. 75:12058-12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gross, T., M. Lutzelberger, H. Weigmann, A. Klingenhoff, S. Shenoy, and N. F. Kaufer. 1997. Functional analysis of the fission yeast Prp4 protein kinase involved in pre-mRNA splicing and isolation of a putative mammalian homologue. Nucleic Acids Res. 25:1028-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta, K., D. Ott, T. J. Hope, R. F. Siliciano, and J. D. Boeke. 2000. A human nuclear shuttling protein that interacts with human immunodeficiency virus type 1 matrix is packaged into virions. J. Virol. 74:11811-11824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guyader, M., M. Emerman, P. Sonigo, F. Clavel, L. Montagnier, and M. Alizon. 1987. Genome organization and transactivation of the human immunodeficiency virus type 2. Nature 326:662-669. [DOI] [PubMed] [Google Scholar]
- 25.Junker, U., D. Bevec, C. Barske, C. Kalfoglou, S. Escaich, M. Dobrovnik, J. Hauber, and E. Bohnlein. 1996. Intracellular expression of cellular eif-5a mutants inhibits HIV-1 replication in human T cells: a feasibility study. Hum. Gene Ther. 7:1861-1869. [DOI] [PubMed] [Google Scholar]
- 26.Kaye, J. F., and A. M. L. Lever. 1999. Human immunodeficiency virus types 1 and 2 differ in the predominant mechanism used for selection of genomic RNA for encapsidation. J. Virol. 73:3023-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kjems, J., and P. Askjaer. 2000. Rev protein and its cellular partners. Adv. Pharmacol. 48:251-298. [DOI] [PubMed] [Google Scholar]
- 28.Kohtz, J. D., S. F. Jamison, C. L. Will, P. Zuo, R. Luhrmann, M. A. Garcia-Blanco, and J. L. Manley. 1994. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature 368:119-124. [DOI] [PubMed] [Google Scholar]
- 29.Kojima, T., T. Zama, K. Wada, H. Onogi, and M. Hagiwara. 2001. Cloning of human PRP4 reveals interaction with Clk1. J. Biol. Chem. 276:32247-32256. [DOI] [PubMed] [Google Scholar]
- 30.Krainer, A. R., G. C. Conway, and D. Kozak. 1990. Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes Dev. 4:1158-1171. [DOI] [PubMed] [Google Scholar]
- 31.Lake, J. A., J. Carr, F. Feng, L. Mundy, C. Burrell, and P. Li. 2003. The role of Vif during HIV-1 infection: interaction with novel host cellular factors. J. Clin. Virol. 26:143-152. [DOI] [PubMed] [Google Scholar]
- 32.Lemaire, R., A. Winne, M. Sarkissian, and R. Lafyatis. 1999. SF2 and SRp55 regulation of CD45 exon 4 skipping during T cell activation. Eur. J. Immunol. 29:823-837. [DOI] [PubMed] [Google Scholar]
- 33.Liu, X., A. Mayeda, M. Tao, and Z.-M. Zheng. 2003. Exonic splicing enhancer-dependent selection of the bovine papillomavirus type 1 nucleotide 3225 3′ splice site can be rescued in a cell lacking splicing factor ASF/SF2 through activation of the phosphatidylinositol 3-kinase/Akt pathway. J. Virol. 77:2105-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luban, J. 1996. Absconding with the chaperone: essential cyclophilin-Gag interaction in HIV-1 virions. Cell 87:1157-1159. [DOI] [PubMed] [Google Scholar]
- 35.Manley, J. L., and R. Tacke. 1996. SR proteins and splicing control. Genes Dev. 10:1569-1579. [DOI] [PubMed] [Google Scholar]
- 36.McCann, E. M., and A. M. L. Lever. 1997. Location of cis-acting signals important for RNA encapsidation in the leader sequence of human immunodeficiency virus type 2. J. Virol. 71:4133-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNally, L. M., and M. T. McNally. 1996. SR protein splicing factors interact with the Rous sarcoma virus negative regulator of splicing element. J. Virol. 70:1163-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mermoud, J. E., P. T. Cohen, and A. I. Lamond. 1994. Regulation of mammalian spliceosome assembly by a protein phosphorylation mechanism. EMBO J. 13:5679-5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller, M. D., C. M. Farnet, and F. D. Bushman. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molin, M., and G. Akusjärvi. 2000. Overexpression of essential splicing factor ASF/SF2 blocks the temporal shift in adenovirus pre-mRNA splicing and reduces virus progeny formation. J. Virol. 74:9002-9009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moroianu, J. 1999. Nuclear import and export: transport factors, mechanisms and regulation. Crit. Rev. Eukaryot. Gene Expr. 9:89-106. [DOI] [PubMed] [Google Scholar]
- 42.Müller, B., T. Patschinsky, and H. G. Kräusslich. 2002. The late-domain-containing protein p6 is the predominant phosphoprotein of human immunodeficiency virus type 1 particles. J. Virol. 76:1015-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myers, E. L., and J. F. Allen. 2002. Tsg101, an inactive homologue of ubiquitin ligase E2, interacts specifically with human immunodeficiency virus type 2 Gag polyprotein and results in increased levels of ubiquitinated Gag. J. Virol. 76:11226-11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rey, O., J. Canon, and P. Krogstad. 1996. HIV-1 Gag protein associates with F-actin present in microfilaments. Virology 220:530-534. [DOI] [PubMed] [Google Scholar]
- 45.Rosen, C. A. 1992. HIV regulatory proteins: potential targets for therapeutic intervention. AIDS Res. Hum. Retrovir. 8:175-181. [DOI] [PubMed] [Google Scholar]
- 46.Rosenberg, G. H., S. K. Alahari, and N. F. Kaufer. 1991. prp4 from Schizosaccharomyces pombe, a mutant deficient in pre-mRNA splicing isolated using genes containing artificial introns. Mol. Gen. Genet. 226:305-309. [DOI] [PubMed] [Google Scholar]
- 47.Ruhl, M., M. Himmelspach, G. M. Bahr, F. Hammerschmid, H. Jaksche, B. Wolff, H. Aschauer, G. K. Farrington, H. Probst, D. Bevec, et al. 1993. Eukaryotic initiation factor 5A is a cellular target of the human immunodeficiency virus type 1 Rev activation domain mediating trans-activation. J. Cell Biol. 123:1309-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 49.Simon, J. H. M., E. A. Carpenter, R. A. M. Fouchier, and M. H. Malim. 1999. Vif and the p55Gag polyprotein of human immunodeficiency virus type 1 are present in colocalizing membrane-free cytoplasmic complexes. J. Virol. 73:2667-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simon, J. H. M., R. A. M. Fouchier, T. E. Southerling, C. B. Guerra, C. K. Grant, and M. H. Malim. 1997. The Vif and Gag proteins of human immunodeficiency virus type 1 colocalize in infected human T cells. J. Virol. 71:5259-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Staknis, D., and R. Reed. 1994. SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol. Cell. Biol. 14:7670-7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stoltzfus, C. M., and S. J. Fogarty. 1989. Multiple regions in the Rous sarcoma virus src gene intron act in cis to affect the accumulation of unspliced RNA. J. Virol. 63:1669-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly and processing of viral protein, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 54.Tange, T. O., and J. Kjems. 2001. SF2/ASF binds to a splicing enhancer in the third HIV-1 tat exon and stimulates U2AF binding independently of the RS domain. J. Mol. Biol. 312:649-662. [DOI] [PubMed] [Google Scholar]
- 55.Tarn, W. Y., and J. A. Steitz. 1995. Modulation of 5′ splice site choice in pre-messenger RNA by two distinct steps. Proc. Natl. Acad. Sci. USA 92:2504-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Towers, G. J., T. Hatziioannou, S. Cowan, S. P. Goff, J. Luban, and P. D. Bieniasz. 2003. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 9:1138-1143. [DOI] [PubMed] [Google Scholar]
- 57.Toyoshima, H., M. Itoh, J.-I. Inoue, M. Seiki, F. Takaku, and M. Yoshida. 1990. Secondary structure of the human T-cell leukemia virus type 1 rex-responsive element is essential for rex regulation of RNA processing and transport of unspliced RNAs. J. Virol. 64:2825-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Urushiyama, S., T. Tani, and Y. Ohshima. 1996. Isolation of novel pre-mRNA splicing mutants of Schizosaccharomyces pombe. Mol. Gen. Genet. 253:118-127. [DOI] [PubMed] [Google Scholar]
- 59.Valcarcel, J., and M. R. Green. 1996. The SR protein family: pleiotropic functions in pre-mRNA splicing. Trends Biochem. Sci. 21:296-301. [PubMed] [Google Scholar]
- 60.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, J., and J. L. Manley. 1995. Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA 1:335-346. [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, J., S. H. Xiao, and J. L. Manley. 1998. Genetic analysis of the SR protein ASF/SF2: interchangeability of RS domains and negative control of splicing. Genes Dev. 12:2222-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiao, S. H., and J. L. Manley. 1997. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 11:334-344. [DOI] [PubMed] [Google Scholar]
- 64.Zimmerman, C., K. C. Klein, P. K. Kiser, A. R. Singh, B. L. Firestein, S. C. Riba, and J. R. Lingappa. 2002. Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature 415:88-92. [DOI] [PubMed] [Google Scholar]