Abstract
The cis-replicating RNA elements in the 5′ and 3′ nontranslated regions (NTRs) of the hepatitis C virus (HCV) genome have been thoroughly studied before. However, no cis-replicating elements have been identified in the coding sequences of the HCV polyprotein until very recently. The existence of highly conserved and stable stem-loop structures in the RNA polymerase NS5B coding sequence, however, has been previously predicted (A. Tuplin, J. Wood, D. J. Evans, A. H. Patel, and P. Simmonds, RNA 8:824-841, 2002). We have selected for our studies a 249-nt-long RNA segment in the C-terminal NS5B coding region (NS5BCR), which is predicted to form four stable stem-loop structures (SL-IV to SL-VII). By deletion and mutational analyses of the RNA structures, we have determined that two of the stem-loops (SL-V and SL-VI) are essential for replication of the HCV subgenomic replicon in Huh-7 cells. Mutations in the loop and the top of the stem of these RNA elements abolished replicon RNA synthesis but had no effect on translation. In vitro gel shift and filter-binding assays revealed that purified NS5B specifically binds to SL-V. The NS5B-RNA complexes were specifically competed away by unlabeled homologous RNA, to a small extent by 3′ NTR RNA, and only poorly by 5′ NTR RNA. The other two stem-loops (SL-IV and SL-VII) of the NS5BCR domain were found to be important but not essential for colony formation by the subgenomic replicon. The precise function(s) of these cis-acting RNA elements is not known.
Plus-strand RNA viruses belong to many different families, but they all have similar strategies for replicating their genomes. The nonstructural viral proteins in concert with cellular proteins form a complex with the viral RNA on membranous vesicles, where RNA replication takes place. The input RNA is first transcribed into a minus strand, which in turn serves as the template for production of progeny plus strands. For many years it was generally believed that the specificity of genome translation and replication resides in cis-acting RNA elements located in the 5′ and 3′ nontranslated regions (NTRs) of viral genomes. However, recently the importance of cis-replicating elements in the coding sequences of the viral polyprotein of a number of plus-strand RNA viruses has been demonstrated.
Hepatitis C virus (HCV) is the major causative agent of transfusion-related and sporadic non-A, non-B hepatitis (9). HCV, a member of the Flaviviridae family, is an enveloped plus-stranded RNA virus with a genome length of about 9,600 nucleotides (nt) (reviewed in reference 52). The viral RNA contains a long 5′ NTR, a single open reading frame, and a 3′ NTR (Fig. 1A). The 5′ NTR contains the IRES (internal ribosomal entry site), which promotes the translation of a large polyprotein. The virus-encoded polypeptide is processed by host and viral proteinases into at least 10 distinct proteins: NH2-C-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B-COOH.
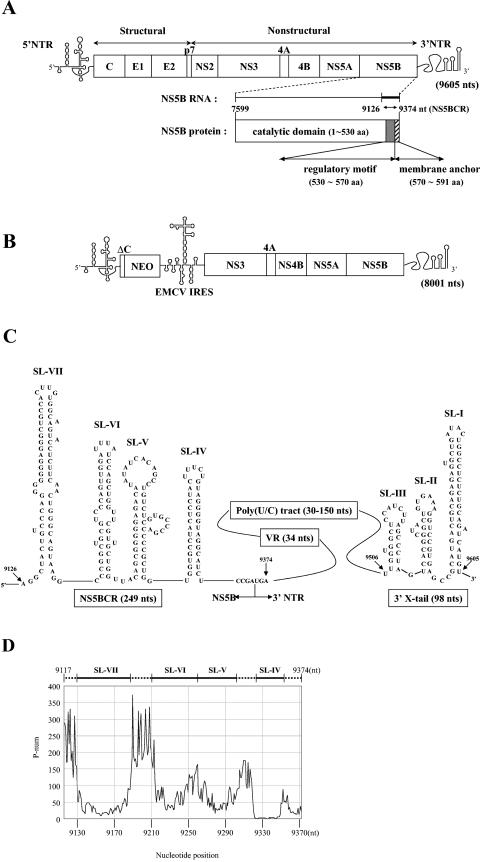
FIG.1.
(A) Genomic structure of full-length HCV RNA. The single-stranded RNA genome of HCV is shown with the 5′ NTR (single line), the structural and nonstructural proteins of the polyprotein (boxed), and the 3′ NTR (single line) indicated. The NS5B domain is shown enlarged both as a coding sequence for the NS5B protein and as an RNA sequence for potential cis-replicating RNA elements. (B) Genomic structure of the subgenomic HCV replicon RNA. The single-stranded RNA genome of the subgenomic replicon contains the HCV 5′ NTR (single line), the neo gene (boxed), the EMCV IRES (single line), the nonstructural proteins of HCV from NS3 to NS5B (boxed), and the 3′ NTR (single line). (C) Predicted RNA stem-loop structures in the HCV 3′ NTR and the C-terminal coding domain of NS5B. Illustrated are the predicted structures of stem-loops IV to VII contained in a 249-nt-long segment of the NS5B coding region (nt 9126 to 9374 of HCV RNA), designated NS5BCR. The single-stranded stretch of nucleotides between stem-loops VI and VII is designated L. Downstream from NS5BCR is the 3′ NTR, which contains a variable region, a poly(U/C) tract, and the 3′ X tail. The highly conserved 3′ X domain is predicted to consist of three stem-loops, I to III (15, 68, 69). (D) P-num values of a 257-nt-long segment (nt 9117 to 9374) of the NS5B C-terminal coding sequence, including all of the NS5BCR sequence, were obtained for HCV-J1 from the website www.virology.wisc.edu/acp/RNAFolds. Stem-loops were identified by consistently low P-num values. aa, amino acids.
Sequences of the HCV genome, the identities of viral proteins, and the basic steps in replication have been known for several years. Studies of HCV replication have been hindered, however, by the low level of virus in biological samples, the lack of an efficient tissue culture system, and the lack of an adequate small-animal model. Recently a selectable subgenomic replicon of subtype 1b has been developed that replicates in the human hepatoma cell line Huh-7 (37). This dicistronic replicon, which in its basic architecture resembles a dicistronic poliovirus previously described (42), consists of the HCV 5′ NTR, directing translation of the neomycin phosphotransferase gene (neo), the IRES of encephalomyocarditis virus (EMCV), HCV nonstructural proteins NS3 to NS5B, and the HCV 3′ NTR (Fig. 1B). Following transfections of such RNAs into Huh-7 cells and selection with neomycin (G418), cell clones have been identified that carry persistently replicating HCV replicons. In these cell clones, a number of adaptive mutations were identified that, when transferred into the original HCV replicon, significantly increased replication efficiency (5, 38, 39).
The replication of HCV follows the general pathway used by other plus-strand RNA viruses. Although these basic steps in replication have been well established, very little is known about the details of these processes except that both cis-replicating RNA elements and viral proteins, possibly also host proteins, are involved. The RNA structures that make up the 5′ NTR and 3′ NTR as cis-replicating elements for replication and/or translation have been the subject of numerous studies. The 5′ NTR contains 341 nt of which the first 40 are essential for RNA replication with no role in translation (14). Although the first 125 nt of the 5′ NTR are sufficient for RNA replication, full efficiency of replication also requires sequences located within the IRES (14). The 3′ NTR contains three distinct domains, a highly conserved 3′-terminal 98-nt segment (3′ X), an upstream poly(U/C) tract, and a variable region. Of the 225-nt-long 3′ NTR, the 3′-most 125-nt segment [3′ X and 52 nt of the poly(U/C) tract] was found to be essential for RNA replication while the remainder had only enhancing activity (15, 25, 68, 69).
The viral proteins required for replication include NS3, NS4A, NS4B, NS5A, and NS5B. NS3 is composed of two domains with distinct enzymatic activities (reviewed in references 11 and 52). The N-terminal domain contains a proteinase that is required for the proper processing of the NS3-to-NS5B region. The C-terminal part of the NS3 region contains NTPase and helicase activities that are essential for replication. NS4A is an important cofactor for NS3 proteinase, while the function of NS4B is not known. Protein NS5A is a phosphoprotein that is believed to play a role in replication since most of the cell culture-adapted mutations identified in the subgenomic replicon map to the central domain of NS5A. The protein primarily responsible for HCV replication is the RNA-dependent RNA polymerase NS5B (3).
HCV NS5B belongs to the large family of nucleic acid polymerases that possess the structure of a right hand consisting of finger, thumb, and palm domains (6, 33). Recombinant HCV NS5B has been expressed and purified from bacterial (48, 65) and insect (3) cells, and the availability of the purified enzyme has facilitated its biochemical characterization. Oligomerization of the protein was demonstrated both by in vitro methods and by yeast two-hybrid analysis, and two extensive interfaces have been identified in the crystal lattice (48, 65). As an oligomeric structure, the polymerase catalyzes RNA synthesis in a cooperative manner (48). The protein also interacts with other viral (NS2, NS3, NS4A, and NS5A) (10, 24, 57) and cellular proteins (nucleolin) (21) and eukaryotic initiation factor 4AII (32). Interestingly, the core protein also binds to NS5B and a 97-amino-acid C-terminal domain of NS5B is required for this interaction (62). In vitro, the enzyme possesses two types of synthetic activities: de novo initiation and elongation of an oligonucleotide primer on a suitable template (3, 23, 49, 71). Full-length NS5B contains 591 amino acids, but the C-terminal 60 amino acids can be deleted without loss of enzyme function (6). Recently it was shown that HCV NS5B belongs to a class of membrane proteins called “tail-anchored” proteins and that the last 21 amino acids, comprising a highly hydrophobic domain, are responsible for membrane attachment of the protein (26, 54). Two recent reports indicate that the C-terminal region, upstream from the membrane anchor domain, contains a regulatory motif that inhibits RNA binding and polymerase activity (1, 34). Deletion of this fragment of NS5B actually enhanced RNA synthesis in vitro up to 50-fold (34).
Despite numerous studies dealing with the function of the 5′ and 3′ NTRs of HCV RNA in replication, there have been no reports of cis-replicating elements in the coding sequences of the genome until very recently (70). Interestingly, several independent groups have predicted the existence of highly conserved stem-loop structures in the HCV coding sequences, suggesting the possibility that these have a function in vivo. The function(s) of these conserved RNA elements in RNA replication, if any, has not been determined. Further evidence suggesting the possibility that some of these RNA structures are biologically relevant to HCV replication is the observation by Cheng et al. (8) that partially purified NS5B specifically binds to the C-terminal coding sequence of NS5B RNA. Interestingly, two independent RNA-binding domains of NS5B were identified, one from amino acids 83 to 194 and the second from residues 196 to 298.
To determine whether the highly conserved and stable stem-loops located near the C-terminal domain of the NS5B coding sequence have a function as cis-replicating elements in HCV RNA replication, we have undertaken a deletion and mutational analysis of a 249-nt-long RNA segment located just upstream of and including the hydrophobic membrane-binding domain of NS5B. With an RNA folding program, we have predicted the presence of four stem-loops (SL-IV to SL-VII) within this segment and a single-stranded region (L) between stem-loops VI and VII. An examination of the effects of deletions and silent mutations on HCV RNA replication, with the HCV subgenomic replicon system and real-time RT-PCR analyses, revealed that stem-loops V and VI are essential for function while the integrity of SL-IV and SL-VII is less important. Interestingly, we observed specific binding of purified NS5B to stem-loop V with in vitro gel shift and filter-binding assays.
MATERIALS AND METHODS
Cell cultures.
Monolayers of the human hepatoma cell line Huh-7 were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 2 mM l-glutamine, nonessential amino acids, 100 U of penicillin, 100 μg of streptomycin, and 10% fetal bovine serum. In cell lines carrying HCV replicons, 500 μg of G418 (Geneticin; Life Technologies) per ml was added to the growth medium.
Plasmids.
Plasmid pFastBAC NS5B, which contains the full-length NS5B sequence, was a gift of L. Tomei. This construct contains a cDNA fragment encoding amino acids 2420 to 3010 of the HCV-BK polyprotein cloned between the BamHI and PstI sites of the pFastBac1 baculovirus expression vector. The plasmid of the subgenomic replicon (pFK-I389neo/NS3-3′/5.1, abbreviated as NK5.1) was generously provided by R. Bartenschlager. It is a dicistronic construct containing the HCV 5′ NTR, 16 amino acids of the core coding region, the neo gene, and the EMCV IRES for the translation of HCV sequences NS3 to NS5B, followed by the 3′ NTR. To introduce mutations into the replicon, site-directed mutagenesis was used with the oligonucleotides listed in Table 1. The nucleotide positions refer to the HCV subtype 1b nucleotide sequence (National Center for Biotechnology Information accession no. AJ238799). A subclone, pHCV(Eco-Spe) (EcoRI [nt 6699] to SpeI [nt 9609]), of the HCV replicon in plasmid pFastBac1 was constructed and used as the template for all mutagenesis. The mutated fragments, EcoRI/SpeI cleaved, were transferred back into the original replicon, NK5.1. All PCR fragments and final constructs were sequenced with the ABI Prism DNA sequencing kit.
TABLE 1.
Oligonucleotides used for PCR-based mutagenesis
| No. | Positiona (sense) | Sequence (5′ to 3′) | Construct |
|---|---|---|---|
| 1 | 8717-8739 (+)b | G CAC GAT GCA TCT GGC AAA AGG G | |
| 2 | 9588-9617 (−)b,d | TGC ACT AGT AGT ACT TGA TCT GCA GAG AGG | |
| 3 | 9309-9330 (+)b | GCC AAG CTT TGG TTC ATG TGG TGC CTA CTC C | MT (ΔSL-V) |
| 4 | 9261-9282 (+)b | GCC AAG CTT TAC AGC GGG GGA GAC ATA TAT C | MT (ΔSL-VI + L) |
| 5 | 9213-9233 (+)b | GCC AAG CTT ATC CCG GCT GCG TCC CAG TTG | MT (ΔL) |
| 6 | 9241-9259 (−)b | GCC AAG CTT ACC AGC AAC GAA CCA GCT GG | MT (ΔSL-V) |
| 7 | 9170-9188 (−)b | GCC AAG CTT TAC TGC CCA GTT GAA GAG G | MT (ΔSL-VI + L), MT (ΔL) |
| 8 | 9326-9363 (+)c | T TTG TTG TTA TCA GTC GGT GTA GGC ATC TAT CTA CTC C | MT (IV) |
| 9 | 9310-9344 (−)c | ACC GAC TGA TAA CAACAAAAG GCA CCA CAT GAA CC | MT (IV) |
| 10 | 9237-9266 (+)c | CTG AGT TCG TGG TTC GTT GCT GGT TAC AGC | MT (VI) |
| 11 | 9217-9247 (−)c | CA CGA ACT CAG ATC CAA CTG GGA CGC AGC CG | MT (VI) |
| 12 | 9285-9314 (+)c | C TCT TTA AGC AGG GCC CGA CCC CGC TGG TTC | MT (V) |
| 13 | 9267-9296 (−)c | CCT GCT TAA AGA GTG ATA TAT GTC TCC CCC | MT (V) |
| 14 | 9158-9191 (+)c | A TGC GGG AAA TAT CTG TTC AAC TGG GCA GTA AGG | MT (VII) |
| 15 | 9140-9173 (−)c | CAG ATA TTT CCC GCA TGT GGC AGC CCT CCC CCC C | MT (VII) |
| 16 | 9209-9242 (+)c | CCA ATC CCG GCT GCG AGC CAG TTG GGA TTT ATC C | SL-VI (UC/AG) |
| 17 | 9209-9242 (−)c | GGA TAA ATC CAA CTG GCT CGC AGC CGG GAT TGG | SL-VI (UC/AG) |
Nucleotide positions in the complete HCV type 1b genome (National Center for Biotechnology Information accession no. AJ238799) are shown.
Oligonucleotide contains an enzyme restriction site, which is underlined.
Oligonucleotide contains nucleotide changes, which are in bold.
Nucleotide positions over 9605 represent the nucleotide sequence of the vector (pBR322) (38).
Constructs containing deletions in the C-terminal NS5B coding region (NS5BCR).
PCR mutagenesis was used to make deletions in the cDNA of subclone pHCV(Eco-Spe) with primers 1 to 6 (Table 1). Two PCR fragments were made, one containing NsiI/HindIII restriction sites, the other containing HindIII/SpeI sites. The fragments were cut with NsiI/HindIII and HindIII/SpeI, respectively. The two fragments were cloned into the NsiI/SpeI-restricted pHCV(Eco-Spe) cDNA. Finally, an EcoRI/SpeI fragment of the subclone was transferred into the similarly restricted cDNA of subgenomic replicon NK5.1.
Constructs containing silent mutations in NS5BCR.
The silent mutations in NS5BCR were introduced by overlapping PCR mutagenesis. Primer pairs 8 to 17 (Table 1) were used to generate two PCR products with pHCV(Eco-Spe) plasmid DNA as the template. These PCR products were used as templates in the second PCR with primers 1 and 2 (Table 1). The resulting PCR fragment was cleaved with NsiI/SpeI and ligated into the similarly restricted pHCV(Eco-Spe) subclone cDNA. The EcoRI/SpeI fragment of the subclone, containing the mutations, was transferred back into the subgenomic replicon cDNA (NK5.1).
In vitro translations.
In vitro translations were carried out with rabbit reticulocyte lysates (Promega) as described previously by Friebe et al. (14). The reaction mixtures contained 12.5 μl of lysate, 0.75 μl of an amino acid mixture without methionine, 0.75 μl of Translabel (ICN Biochemicals) containing [35S]methionine, and 800 ng of in vitro-transcribed RNA in a final volume of 20 μl. Following incubation for 90 min at 30°C, the samples were analyzed by sodium dodecyl sulfate (SDS)-12.5% polyacrylamide gel electrophoresis (PAGE).
Replicon RNA quantitation by real-time RT-PCR.
After RNA transfection, 4 × 105 cells were plated in 35-mm-diameter plates. The cells were maintained without G418 selection and harvested at 24-h intervals for 5 days. TRIzol reagent (Gibco-BRL) was used for purification of total RNA from cells in accordance with the manufacturer's protocol. At each time point, a 500-ng aliquot of total RNA was used for quantitation of the HCV replicon copy number with a LightCycler system (Roche). Real-time reverse transcription (RT)-PCR amplifications were done with the LightCycler RNA amplification kit SYBR Green I (Roche) and a primer pair specific for HCV NS5B (5′ CCATAGTTACTCTCCAGGTGAGATC 3′ [plus-strand sequence] and 5′ GTGTTTAGCTCCCCGTTCA 3′ [minus-strand sequence]). Glyceraldehyde 3-phosphate dehydrogenase was used as an internal control. The primers used to amplify the mRNA were 5′ GGAAGGTGAAGGTCGGAGTCAACGG 3′ (plus-strand sequence) and 5′ TCCTGGAAGATGGTGATGGGATTTC 3′ (minus-strand sequence) (12). RT was carried out at 50°C for 30 min. The PCR protocol consisted of 40 cycles of 95°C for 10 s, 50°C for 10 s, and 72°C for 15 s. Transcript HCV RNA standards of known concentrations were used with each set of reaction mixtures, and these were used to determine a standard curve. The real-time PCR signals were analyzed with the LightCycler software, version 3.5 (Roche).
In vitro gel shift and filter-binding assays.
Labeled RNA probes were generated by in vitro transcription, in the presence of [α-32P]UTP, of templates obtained with primers encoding the T7 promoter (5′ GATTGTAATACGACTCACTATAGGGGCTACTGTCCCAGGGGGGG 3′, T7 promoter plus nt 9127 to 9146) and a minus-strand primer containing the HCV sequence from nt 9348 to 9374. To prevent nonspecific binding of the proteins to the RNA probes, a 1,000-fold excess of nonspecific RNA (5 μg of tRNA per 30-μl reaction mixture) was added to all binding reaction mixtures. The 32P-labeled RNAs (2 × 104 cpm) were denatured by incubation at 45°C for 30 min in 20 mM HEPES (pH 7.9)-2 mM MgCl2-60 mM KCl. Purified NS5B protein was added to the RNA, and the mixture was incubated at 30°C for 20 min in binding buffer (5 mM HEPES [pH 7.9], 2 mM MgCl2, 25 mM KCl, 5% glycerol). One-third of the mixture (10 μl) was loaded onto a nondenaturing 0.5× Tris-borate-EDTA-6% polyacrylamide gel containing 5% glycerol. The gels were run at 30 mA for 2 h 30 min in 0.5× Tris-borate-EDTA buffer. The gel was dried and subjected to autoradiography. Filter-binding assays were performed in duplicate. One-third of the binding mixture (10 μl) was spotted onto a prewetted nitrocellulose membrane (0.45-μm pore size; Schleicher & Schuell). Beneath the nitrocellulose membrane, a nylon membrane (Boehringer Mannheim) and two pieces of 3MM cellulose membrane were placed. Filters were washed once with 20 mM HEPES (pH 6.8)-1 mM Mg acetate-10 mM β-mercaptoethanol and dried. Radioactivity was measured by liquid scintillation counting in the presence of EcoLite scintillation cocktail (ICN).
In vitro transcription, electroporation, and selection of G418-resistant cells.
Wild-type (WT) and mutant NK5.1 plasmid DNAs were linearized with ScaI and transcribed into RNA with T7 RNA polymerase. The template DNA was removed by digestion with RNase-free DNase I for 1 h at 37°C. The RNA was purified with an RNeasy mini kit (QIAGEN). Subconfluent monolayers of Huh-7 cells were detached from the culture dish by trypsin treatment, washed three times with phosphate-buffered saline, and resuspended in Cytomix (120 mM KCl, 0.15 mM CaCl2, 10 mM potassium phosphate [pH 7.6], 25 mM HEPES [pH 7.6], 2 mM EGTA [pH 7.6], 5 mM MgCl2) (39). Two to 5 μg of replicon RNA was mixed with the cell suspensions, and the mixtures were transferred into an electroporation cuvette. Electroporation of the RNA was carried out with a Bio-Rad Gene Pulser II at 270 V and 960 μF in a cuvette with a 0.4-cm gap (Bio-Rad). The cells were immediately transferred into 12 ml of complete DMEM (1.25% dimethyl sulfoxide) and seeded into a 10-cm-diameter culture dish. At 24 h, the medium was replaced with complete DMEM supplemented with 500 μg of G418 (Geneticin; Gibco Life Sciences) per ml. The growth medium was changed two to three times a week, and 2 to 3 weeks after transfection, colonies were stained with Coomassie brilliant blue. For each replicon, three to five independent transfections were performed.
Expression and purification of the NS5B protein.
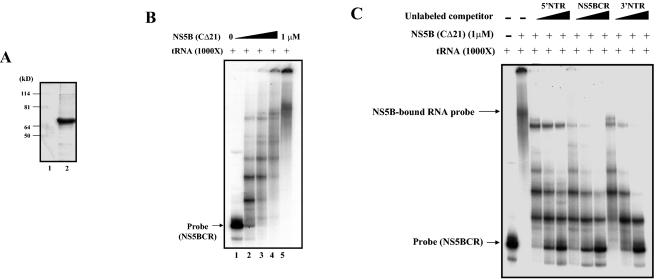
Construction of a recombinant baculovirus carrying the NS5B sequence with the C-terminal 21 amino acids deleted and infection of SF9 cells were carried out as described by the manufacturer (Invitrogen). The protein was purified by the method of Tomei et al. (60), with some modifications. After infection with recombinant baculovirus, the cells were harvested and washed once with cold phosphate-buffered saline. The pellet was resuspended in cold lysis buffer (50% glycerol, 20 mM Tris [pH 7.5], 10 mM dithiothreitol, 0.5 M NaCl, 1 mM EDTA, 1 tablet of complete protease inhibitor [Roche]). The cells were broken up in a Dounce homogenizer, and Triton X-100 was added to a final concentration of 2%. After the addition of MgCl2 (10 mM) and DNase I (20 U/ml), the mixture was rotated for 15 min at 4°C. The extract was centrifuged at 11,000 × g for 30 min, and the supernatant was diluted with elution buffer (20% glycerol, 20 mM Tris [pH 7.5], 10 mM dithiothreitol, 1 mM EDTA, 0.5% Triton X-100, 1 mM MgCl2) to adjust the final NaCl concentration to 300 mM. The diluted extract was centrifuged at 20,000 × g for 15 min. The extract was loaded onto a DEAE-Sepharose fast-flow column equilibrated with elution buffer containing 300 mM NaCl. The flowthrough of the DEAE-Sepharose fast-flow column was loaded on a heparin-Sepharose column, and the protein was eluted with a salt gradient (0.3 to 1.0 M NaCl) in 20 mM Tris buffer (pH 7.5). The peak fractions (0.7 to 0.8 M NaCl) were diluted to 0.2 M NaCl, and the sample was applied to a poly(U)-Sepharose column. The NS5B protein was eluted with a 0.2 to 2.0 M NaCl gradient in 20 mM Tris buffer (pH 7.5). The peak fractions (0.8 to 1.0 M NaCl) were pooled and dialyzed against 0.2 M NaCl-Tris (pH 7.5). This preparation was >90% pure and was used in all of the RNA-binding experiments (see Fig. 5A).
FIG. 5.
Effects of silent mutations in SL-V and VI on in vitro translation of replicon RNA. Transcripts of WT and mutant RNAs were translated in rabbit reticulocyte lysate as described in Materials and Methods. The samples were analyzed by SDS-12.5% PAGE. The control reaction mixture (C) contained no RNA. The positions of the major translation products (NS3 and HCV core-Neo fusion protein) are indicated. The values on the left indicate the positions of molecular size standards.
Computer-based prediction of RNA structures.
RNA secondary structures were predicted by using the MFOLD program designed by M. Zucker (www.bioinfo.rpi.edu/application/mfold).
RESULTS
Although various computational analyses and phylogenetic comparisons have predicted the existence of highly conserved and stable stem-loop structures in the HCV NS5B coding region, until very recently (70) no effort has been made to characterize these structures as potential cis-replicating elements. Several different groups of investigators (20, 22, 30, 58, 61) have observed such structures in the NS5B coding sequence, particularly near the C terminus, suggesting the possibility that one or more of these elements possess biological functions. The first group of reports predicted, either by simple inspection of the sequences or by RNA folding programs, the presence of stable stem-loops near the 3′ end of the NS5B coding sequence (20, 30, 58). Subsequently, Hofacker et al. (22) identified 13 highly conserved structures in the HCV genome by a combination of thermodynamic structure prediction and phylogenetic comparison. Of these, two were located in the NS5B coding sequence around nt 8000 and 9100. In the most detailed study by Tuplin et al. (61), various methods of computational analysis were used to predict the existence of eight highly conserved stem-loop structures within the coding region of the HCV, six in the NS5B domain and two in the core coding region. The function(s) of these conserved RNA elements in RNA replication, if any, has not been determined.
Prediction of RNA secondary structures in the HCV NS5B C-terminal coding region.
With the knowledge of previous reports of highly conserved and stable RNA structures in the NS5B C-terminal coding sequence, we have undertaken a search for these RNA elements in a 249-nt-long segment (nt 9126 to 9374) of the full-length HCV sequence. This domain consists of the C-terminal NS5B coding sequences including the membrane anchor domain. With the MFOLD program of M. Zuker, we have predicted the presence of four stable stem-loop structures in this domain, which we have designated SL-IV (nt 9318 to 9355), SL-V (nt 9262 to 9311), SL-VI (nt 9215 to 9260), and SL-VII (nt 9129 to 9189) (Fig. 1C). This nomenclature is used to distinguish the RNA elements from those in the 3′ X domain of the 3′ NTR (SL-I to SL-III) (Fig. 1C). The RNA stem-loops were also identified as regions displaying unusually low P-num values (Fig. 1D). We will refer to the region encompassing all four stem-loops as NS5BCR. Three of the four stem-loops, with the exception of SL-V, which is predicted by us, correspond to those previously proposed by Tuplin et al. (61), Han and Houghton (20), and Smith and Simmonds (58).
Deletion analyses of the HCV NS5B C-terminal coding region.
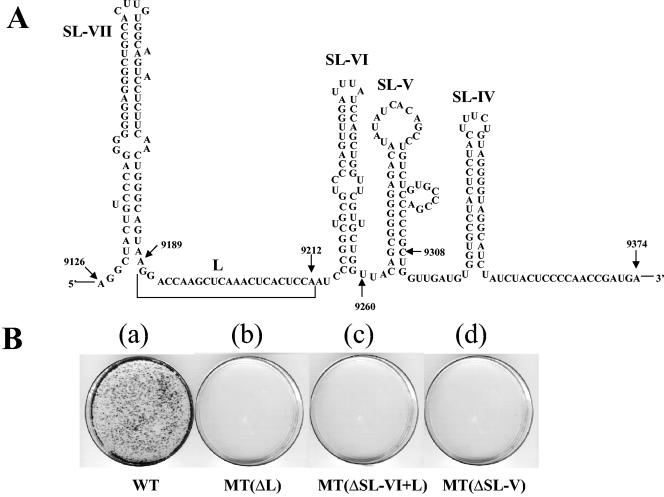
As the first step in testing the potential function of the NS5BCR RNA sequence in HCV replication, we have made deletions of those sequences and structures that do not fall into the essential catalytic or membrane-binding domains of the NS5B polymerase (Fig. 1A). These sequences include SL-V, SL-VI, and a single-stranded stretch of nucleotides between SL-VI and SL-VII designated L (linker) (Fig. 2A). To investigate the potential role of stem-loops V and VI in replication, we determined the effect of their deletion on the colony-forming ability of the subgenomic replicon. It should be noted that deletions of stem-loops V and VI are expected to have no effect on the enzymatic activity of purified NS5B because they are located within the 60-amino-acid C-terminal domain of NS5B, which is known to be dispensable for enzymatic activity in vitro (6). We designed three in-frame deletions in this domain of the subgenomic replicon: (i) deletion of SL-V, (ii) deletion of SL-VI and L, and (iii) deletion of L (Fig. 2A). With the RNA folding program, we confirmed that none of the deletions had an apparent effect on the secondary structure of the remaining portion of NS5BCR (data not shown). Deletion of SL-IV could not be tested in these experiments because its sequence overlaps the coding region of the essential membrane-binding domain of NS5B. Similarly, the effect of deleting SL-VII could not be examined because these sequences are contained within the catalytic domain of the RNA polymerase. Following transfection of RNA transcripts containing these deletions into Huh-7 cells and selection with G418, we observed that the colony-forming ability of the subgenomic replicons was totally abolished (Fig. 2B). No drug-resistant colonies could be observed within 2 weeks after transfection of the RNAs into Huh-7 cells. These results suggest that these RNA sequences contain cis-replicating elements, that the deleted amino acid sequence of NS5B is important for replication, or both.
FIG. 2.
Effects of deletions in NS5BCR on the colony-forming ability of the subgenomic HCV replicon. (A) Illustrated are the predicted stem-loop structures in the NS5BCR domain of NS5BCR and the deletions that were made: SL-V (nt 9261 to 9308), SL-VI plus L (nt 9189 to 9260), and L (nt 9189 to 9212). SL-V, SL-VI, and L are contained within the noncatalytic portion of the NS5B protein. (B) RNA transcripts of the constructs containing the deletions were transfected into Huh-7 cells, and the efficiency of colony formation was compared with that of the WT replicon (see Materials and Methods).
Mutational analyses of the HCV NS5B C-terminal coding region.
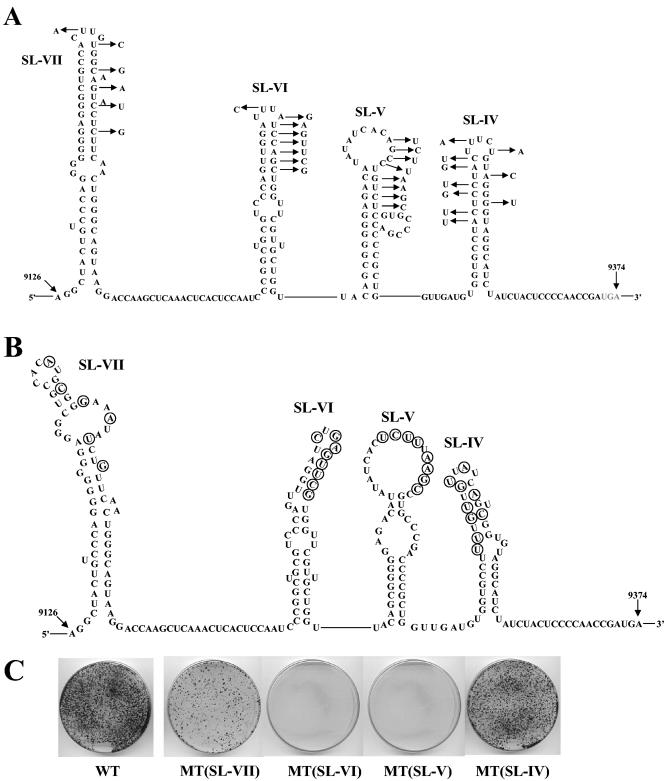
To ascertain that the effect of the deletions is due to RNA sequence or structure rather than NS5B function, we introduced several silent mutations into the upper half of SL-V and SL-VI (Fig. 3A). SL-IV and VII, which could not be tested by deletion analyses, were also included in the mutational studies. The substitutions in each individual stem-loop were designed so as to induce changes in the secondary structure of the mutated element (Fig. 3B) without an effect on the structure of the neighboring stem-loops of NS5BCR (data not shown). The mutant cDNAs were transcribed, and the RNAs were transfected into Huh-7 cells. The lack of colony formation by the mutated SL-V and SL-VI replicons (Fig. 3C) indicated that these are essential RNA sequences or structures required for HCV replication. Mutations of SL-IV had only a mildly detrimental effect on replication, and substitutions in SL-VII resulted in significantly reduced colony formation compared to that of the WT (Fig. 3B). These results suggest that SL-IV and SL-VII are important but not essential for RNA replication. Segment L (Fig. 2A) was not included in this analysis.
FIG. 3.
Effects of silent mutations in NS5BCR RNA on the colony-forming ability of the subgenomic replicon. (A) Illustrated are the silent mutations that were introduced into each of the four predicted stem-loops of NS5BCR. (B) Predicted secondary structures of each the mutated RNA elements. (C) RNA transcripts of the constructs containing mutations in one of the stem-loops were transfected into Huh-7 cells, and the colony-forming efficiency of the replicons was measured as described in Materials and Methods. The colony-forming efficiency of the mutant replicons is compared to that of the WT replicon (see Materials and Methods).
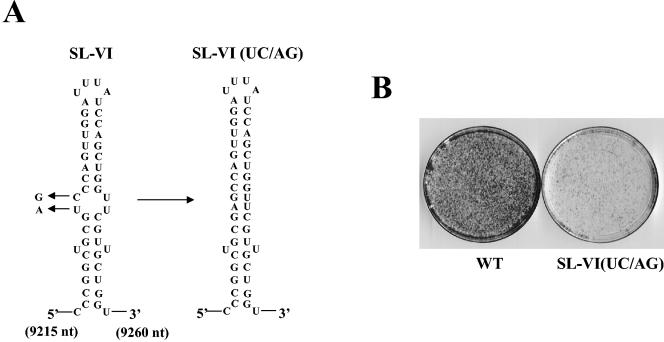
RNA structures that serve as cis-acting elements in translation or replication usually consist of a hairpin that contains a loop and a stem, frequently with an internal bulge that is important for function. To assess the importance of the central bulge in the stem of SL-VI, an additional mutant was designed in which two silent mutations (UC→AG) were predicted to close the bulge (Fig. 4A). Although the nucleotide substitutions did not result in a lethal phenotype, the colony-forming ability of the mutant replicon was significantly reduced (Fig. 4B). These results further confirmed the importance of SL-VI in RNA replication and identified the bulge as a sequence and/or structure required for optimal function.
FIG. 4.
Effects of silent mutations in the central bulge of stem-loop VI of NS5BCR RNA. (A) Illustrated are the two mutations that were introduced into the predicted central bulge of SL-VI. (B) RNA transcripts containing the mutations were transfected into Huh-7 cells, and the colony-forming efficiency of the mutants was compared to that of the WT replicon (see Materials and Methods).
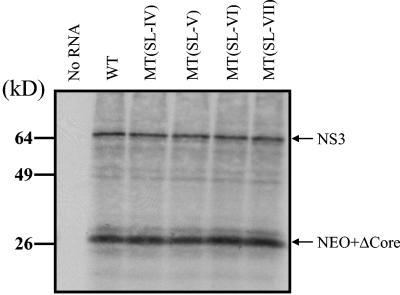
Stem-loops V and VI of NS5BCR RNA are not required for translation.
To rule out the possibility that stem-loops V and VI of NS5BCR function at the level of translation rather than replication, we prepared in vitro translation reaction mixtures with rabbit reticulocyte lysates of the WT and mutant replicon RNAs. Since the replicon is a dicistronic construct, the translation efficiency of the neo gene under the control of the HCV IRES could be determined independently of the translation efficiency of the downstream cistron (NS3 to NS5B), which is under the control of the EMCV IRES. As shown on Fig. 5, there was no difference in the amounts of core-Neo fusion protein and NS3 translated from the WT and mutant RNAs. These results indicate that stem-loops V and VI of NS5BCR have no effect on translation efficiency. It should be noted that under these translation conditions only some unprocessed precursors and NS3 are clearly visible on the gel (14). The lack of efficient polyprotein processing has been attributed to a lack of microsomal membranes or to poor stability of the cleavage products in the translation reaction mixtures (14).
Stem-loops V and VI of NS5BCR are required for RNA replication.
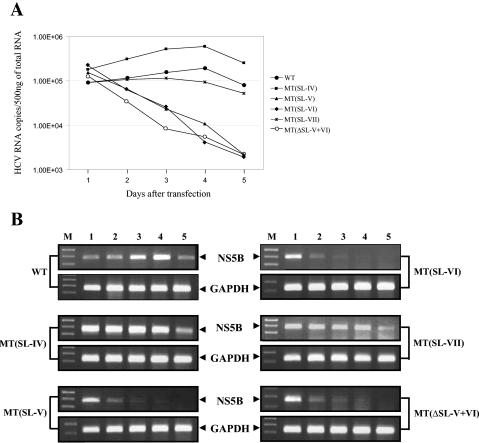
To assess the impact of NS5BCR deletions and mutations directly on RNA replication, we compared the levels of replicon RNA in cells transfected with the WT and mutant replicon RNAs. Recently it has been shown that real-time RT-PCR is a powerful tool in the determination of replicon RNA copy number in transfected cells (31). Therefore we carried out a real-time RT-PCR analysis to quantitate the levels of plus-strand RNA following RT with a primer complementary to the C-terminal NS5B coding sequence. Samples were taken for 5 days and analyzed. As shown in Fig. 6A and B, on the first day following transfection similar levels of replicon RNA were present in all of the samples examined. This suggested that the deletions and mutations in the replicon RNA did not have a significant effect on the stability of the input RNA. On days 1 to 4, the level of WT replicon RNA increased but the amount of mutant SL-V and SL-VI RNA progressively decreased. During this time period, mutants SL-IV and SL-VII exhibited only slightly reduced levels of replicon RNA synthesis compared to that of the WT. By day 5, the amount of WT replicon RNA decreased and this was most likely due to the fact that by this time the cells had reached more than 100% confluency, which inhibits normal cell growth. It has been recently demonstrated that prevention of cell growth by various means results in substantial inhibition of HCV replicon RNA synthesis (55). The results of the real-time RT-PCR analyses and measurement of replicon RNA levels indicate that SL-V and SL-VI are essential cis-replicating elements and fully agree with those measuring the colony-forming efficiency of the mutated replicons.
FIG. 6.
Effects of deletions and mutations in NS5BCR on HCV replicon RNA levels. Transfected cells were harvested daily for 5 days, and the total RNA was isolated. Five hundred nanograms of total RNA was analyzed by real-time RT-RCR as described in Materials and Methods. (A) Quantitation of the plus-strand replicon RNA present in cells on days 1 to 5 by real-time PCR. Similar results were obtained in three separate experiments. (B) Agarose gel electrophoresis of the final PCR products obtained in the real-time RT-PCR experiments. The PCR products collected after 40 cycles of PCR on days 1 to 5 (lanes 1 to 5) were analyzed by 2% native agarose gel electrophoresis. The upper bands are specific for HCV NS5B, and the lower bands are specific for glyceraldehyde 3-phosphate dehydrogenase, as indicated by the arrows. Lane M, DNA molecular weight marker.
Gel shift analyses of NS5B binding to RNA stem-loop structures of NS5BCR.
It was previously reported that NS5B is a nonspecific RNA-binding protein, which was demonstrated both with RNA homopolymers and with transcripts of HCV 3′ NTR RNA (36). Similar studies by Kim et al. (29) have indicated that although NS5B binds to its own C-terminal coding RNA sequence, it does so in a sequence-nonspecific manner. In contrast, Cheng et al. (8) observed that partially purified NS5B specifically interacts with an RNA fragment that contains 295 nt of the C-terminal NS5B coding sequence and 45 nt downstream from the stop codon. To determine whether the two stem-loops (SL-V and SL-VI) are important for replication function as cis-replicating elements by virtue of their ability to bind the RNA polymerase specifically, we have carried out gel shift analyses with purified NS5B protein and 32P-labeled transcript RNAs of the stem-loops as probes. NS5B, lacking the C-terminal 21 amino acids, was expressed in insect cells and purified to near homogeneity (Fig. 7A). This enzyme preparation, which is highly active in oligonucleotide elongation reactions on suitable templates (data not shown), was used for studies of binding in the presence of a 1,000-fold excess of nonspecific tRNA. Our results indicated the formation of increasingly larger complexes of NS5BCR RNA-NS5B as the protein concentration was increased from 0 to 1 μM (Fig. 7B, lanes 2 to 5). The presence of large complexes in the binding mixture was not surprising in view of previous reports demonstrating that NS5B oligomerizes and acts as a functional oligomer (48, 65). The formation of the large complexes could be significantly reduced by the addition of unlabeled NS5BCR RNA (Fig. 7C, lanes 6 to 8), indicating the specificity of the RNA-protein interaction. Cheng et al. (8) have previously reported that NS5B has the ability to weakly interact with the 3′ NTR but not with the 5′ NTR. In agreement with these findings, we observed that transcripts of the HCV 3′ NTR exhibited weak competition of the NS5B-NS5BCR interaction (Fig. 7C, lanes 9 to 11) and the addition of 5′ NTR RNA had almost no effect (Fig. 7B, lanes 3 to 5).
FIG. 7.
Gel shift analysis of purified NS5B binding to RNA transcripts of NS5BCR. (A) Purification of HCV NS5B. The enzyme was expressed in baculovirus and purified to near homogeneity (see Materials and Methods). The purity of the enzyme was tested by SDS-PAGE, followed by Coomassie staining. Protein sizes are indicated on the left. (B) Gel shift analysis of purified NS5B (0 to 1 μM) binding to NS5BCR RNA was carried out in the presence of a 1,000-fold excess of tRNA as described in Materials and Methods. (C) Competition of NS5B (1 μM) binding to NS5BCR RNA (7 nM, 2 × 104 cpm) by increasing amounts (0.5 to 2.0 μg) of unlabeled RNA transcripts of the 5′ NTR (5 to 20 μM), NS5BCR (6 to 24 μM), and the 3′ NTR (7 to 28 μM). Gel shift analysis was carried out in the presence of tRNA as described in Materials and Methods.
SL-V is the primary determinant of NS5B binding to NS5BCR.
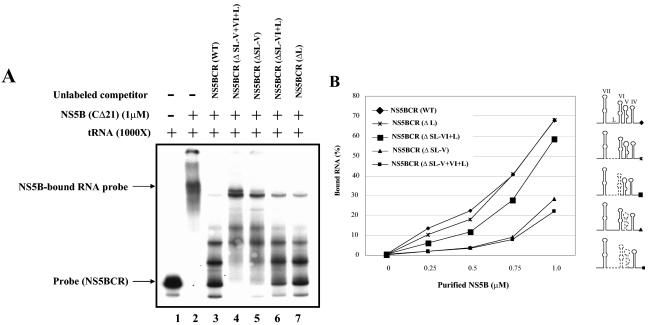
Our gel shift assays showed specific binding of NS5B to NS5BCR but did not identify the exact determinant of the interactions. Therefore we carried out competition experiments with unlabeled fragments of NS5BCR containing deletions of various lengths. As shown in Fig. 8A, addition of unlabeled full-length NS5BCR RNA to the binding mixture strongly reduced the formation of complexes between NS5B and the homologous RNA (Fig. 8A, compare lanes 2 and 3). On the other hand, RNA transcripts lacking SL-V plus SL-VI plus L (Fig. 8A, compare lanes 2 and 4) or just SL-V (Fig. 8A, compare lanes 2 and 5) could not efficiently compete the interaction between NS5B and the labeled full-length NS5BCR probe. Deletion of SL-VI plus L (Fig. 8A, compare lanes 2 and 6) or of just L (Fig. 8A, compare lanes 2 and 7) from competitor RNAs had a strong inhibitory effect on the formation of the NS5B-NS5BCR complexes. These results suggest that SL-V is the primary determinant of the interaction between the RNA polymerase and NS5BCR.
FIG. 8.
Specificity of NS5B binding to NS5BCR RNA fragments. (A) Gel shift analysis of NS5B (1 μM) binding to full-length NSBCR RNA (7 nM, 2 × 104 cpm) in the presence of tRNA (see Materials and Methods). Unlabeled NS5BCR RNA fragments containing deletions (Δ) were used as competitors as indicated (NS5BCR WT, 13 μM; NS5BCR [ΔV+ΔVI+ΔL], 25 μM; NS5BCR [ΔV], 15 μM; NS5BCR [ΔVI+L], 17 μM; NS5BCR [ΔL], 14 μM). (B) Filter-binding assays of NS5B binding to full-length NS5BCR RNA (7 nM, 2 × 104 cpm) or to NS5BCR RNAs containing deletions. The assays were done in the presence of tRNA (see Materials and Methods). The data are expressed as a percentage of the total NS5BCR RNA that is protein bound. The deleted regions of the RNA probes are indicated by dotted lines in the drawings on the right.
To confirm the importance of SL-V in the RNA-protein interaction, we repeated the analyses with filter-binding assays that measure the total amount of protein bound to the 32P-labeled RNA probe. As shown in Fig. 8B, the amount of protein bound by full-length NS5BCR RNA increased to about 70% of the input probe as the concentration of NS5B was increased from 0 to 1 μM. Probes that lacked SL-V or SL-V plus SL-VI plus L exhibited poor binding of NS5B. Deletion from NS5BCR of L had no effect, and that of SL-VI plus L had only a slight effect, on the protein-RNA interaction. These observations confirm our previous conclusion that the ability of NS5B to bind NS5BCR RNA is primarily due to the presence of SL-V.
DISCUSSION
During the last few years, a great deal of information has accumulated on the requirement of cis-acting RNA elements for the replication of plus-strand RNA viruses. Although most of these cis-replicating elements were first identified in the 5′ and 3′ NTRs of viral RNAs, recently the presence of such elements in the coding sequences of a variety of viral RNA genomes has also been demonstrated. Many of these structures were initially only identified as regions of highly conserved RNA sequences presenting low P-num values. Some of these RNA elements are found in picornaviruses such as poliovirus (18, 45, 53), human rhinoviruses 2 (17) and 14 (40), Theiler's virus (35), and mengovirus (35). In addition to picornaviruses, important cis-replicating RNA elements have also been demonstrated in the coding sequences of another human virus (hepatitis D virus [64]), an animal virus (murine coronavirus [28]), an insect virus (flock house virus [2]), plant viruses (tobacco etch virus [19], cucumber necrosis virus [7], tomato bushy stunt tombusvirus [7], brome mosaic virus RNA3 [13], alfalfa mosaic virus [63], and turnip crinkle virus [44]), and coliphage Qβ (41, 56). In picornaviruses (entero- and rhinoviruses), these internal RNA structures were found to be the template for the initiation of protein-primed RNA synthesis by the viral polymerases (17, 45, 53, 67). In some of the plant and animal viruses and bacteriophages, these structures have been proposed to be a part of the site on the genome that is recognized by the viral RNA polymerase prior to replication (41, 56, 59) or they were found to be either enhancers (44, 50, 51, 63) or silencers (46) of plus- or minus-strand RNA synthesis.
For HCV, several different groups of investigators have previously predicted the presence of stable and highly conserved RNA structures in the NS5B coding sequence. These computational studies, however, have not been followed up by biochemical and genetic characterization of the RNA elements until very recently (70). The recently developed HCV subgenomic replicon system has made it possible to investigate the involvement of such RNA elements in HCV translation and/or replication. The aim of our studies was to determine whether the predicted structures in the NS5B coding sequences are important for the replication of the HCV subgenomic replicon in Huh-7 cells. We have selected a 249-nt-long RNA segment near the C terminus of the NS5B coding sequence for analysis and predicted the formation of four stable structures by this RNA sequence (SL-IV to SL-VII). By deletion analysis of two of the structures (SL-V and SL-VI plus L), which are located in a nonessential catalytic domain of the NS5B protein, we now show that stem-loop V and stem-loop VI plus segment L are important for the colony-forming efficiency of the subgenomic replicon. To ascertain that SL-V and SL-VI function in RNA replication as RNA sequences or structures rather than as part of the NS5B coding sequence, we carried out a mutational analysis of these elements and measured the effects of the mutations on the colony-forming efficiency of the subgenomic replicon. These studies indicated that SL-V and VI are cis-acting elements essential for replication of the subgenomic replicon. This conclusion was confirmed by real-time RT-PCR analyses of total WT and mutant replicon RNA present in transfected cells. The introduction of 10 silent mutations into the upper half of SL-IV or 6 silent mutations into SL-VII, on the other hand, only slightly reduced colony formation, suggesting that these structures are important but not essential for replication.
cis-acting RNA elements consisting of stem-loop structures frequently contain internal bulges within the stem that are important for function. To test the importance of the internal bulge in SL-VI, we introduced two silent mutations that resulted in closing of the stem and elimination of the bulge. The 2-nt substitutions were found to be highly detrimental for the colony formation of the subgenomic replicon, indicating the importance of the structure and/or sequence of the bulge in SL-VI.
The function of cis-acting RNA elements in both translation and replication is frequently dependent on the binding of viral or cellular proteins. To test the possibility that the function of the NS5BCR stem-loops in the replication of the subgenomic replicon is dependent on NS5B binding, we carried out gel shift, filter-binding, and competition experiments. These experiments provided a clear correlation between NS5B binding to stem-loop V and the importance of this RNA structure for colony formation by the subgenomic replicon. Our results extend those of Cheng et al. (8), who demonstrated NS5B binding to a fragment of the NS5B C-terminal coding sequences. Another viral system in which replicase binding to an internal RNA stem-loop was found to be essential is that of bacteriophage Qβ. That interaction was shown to be important both for template recognition and for minus-strand synthesis in vitro (56).
The precise biochemical function(s) of the stem-loops contained in the NS5BCR domain of the NS5B coding sequence, particularly that of SL-V and SL-VI, in colony-formation by the subgenomic replicon is not known. This function could be either in translation, in replication, or in both and might involve RNA-RNA interactions and/or association of the RNA elements with viral and cellular proteins. Our studies indicate that SL-V and SL-VI of the NS5BCR RNA segments are not involved in translation but rather in replication. These results, however, do not rule out the possibility that these RNA elements are involved in regulating the switch from translation to replication. The passage of ribosomes through the C-terminal NS5B coding sequence is expected not only to disrupt the secondary structure of the stem-loops but also to prevent minus-strand RNA synthesis. After translation of the polyprotein is complete, reformation of the secondary structure of the stem-loops and subsequent binding of NS5B could initiate the switch between translation and replication. The importance of transient disruption of important RNA structures by translation for genome amplification has been previously proposed for plus-strand RNA viruses, such as poliovirus (16), Qβ (27), MS2 (4, 47), and tobacco etch virus (19).
Another possible function of the NS5BCR stem-loop structures is to serve as the site, or part of the site, of recognition by the viral RNA polymerase prior to minus-strand RNA synthesis. Since in vivo RNA transcription by the RNA polymerase of plus-strand RNA viruses is confined to viral sequences and not to amplification of cellular mRNAs, the specificity of this reaction is usually attributed to the sequences near or in the 3′ NTR. It is generally believed that binding by the RNA polymerase to the 3′-terminal sequences of the genomic RNA is an important step in the recognition of the viral template and initiation of minus-strand RNA synthesis. Therefore, the binding of NS5B to stem-loop V of NS5BCR might contribute to the recognition of the HCV genome as the template for transcription in vivo. In this respect, it is interesting that Yamashita et al. (66) have demonstrated that the ability of NS5B to interact with the RNA template is correlated with its in vitro activity in RNA synthesis. They have shown that the 3′ X RNA or the poly(U/C) stretch of the HCV 3′-terminal domain possessed very poor, if any, template activity in vitro for RNA synthesis by NS5B. In contrast, an RNA segment containing the C-terminal NS5B coding sequence (430 nt) plus 44 nt of the 3′ NTR was highly efficient as a template and yielded a product with the size of the input RNA.
An alternative function of the NS5BCR stem-loops could be to act as regulatory elements in RNA synthesis. For example, NS5B binding to stem-loop V might down regulate the amount of minus-strand RNA synthesized. It has been well established that during the replication of plus-strand RNA viruses, a significantly smaller amount of minus-strand than plus-strand RNA is synthesized. Recently it has been shown with tomato bushy stunt virus that one way of achieving asymmetric RNA replication is with a silencer element upstream of the promoter for minus-strand RNA synthesis (46). A regulatory role for the stem-loops of NS5BCR in plus-strand RNA synthesis is less likely. The template for plus-strand RNA synthesis is believed to be the minus-strand RNA contained within a double-stranded replicative form in which the secondary structure of the stem-loops is disrupted. Therefore, the stem-loops could function in plus-strand RNA synthesis only when the replicative form is nearly unwound and the progeny RNA strand is almost complete. Alternatively, the stem-loops of NS5BCR would have to function at some early step of plus-strand RNA synthesis that occurs prior to minus-strand RNA synthesis. Such a two-step mechanism has been proposed, for example, for protein-primed plus-strand RNA synthesis by the RNA polymerase of poliovirus (43).
Another unanswered question concerning the function of the essential stem-loops of the NS5B C-terminal coding sequence is whether they function as an independent unit or simply as a 5′ extension of the RNA structures contained within the 3′ NTR. If the latter possibility were correct, one would expect either direct RNA-RNA or RNA-protein-RNA interactions between the two structural elements. By computer analysis, we have predicted no RNA-RNA interaction between the stem-loops of the 3′ NTR and the NS5BCR RNA segment (Fig. 1C) and do not know if NS5B or any other protein serves as a protein bridge between the two RNA elements.
In a very recent publication, You et al. (70) reported the identification of a stem-loop in the NS5B coding sequence that is required for RNA replication. This RNA structure is identical to SL-V, which is described in this report, and our results are in full agreement. However, our findings differ from theirs in that we have identified an additional essential hairpin structure just upstream of SL-V in the NS5B coding sequence, SL-VI. We have provided evidence that SL-VI is required for RNA synthesis and for replication of the replicon in Huh-7 cells.
Since the C-terminal NS5B coding sequence is highly conserved among different HCV isolates (22, 58), detection of this sequence may offer a useful alternative method to diagnose HCV infections in clinical samples. In addition, the identification of essential cis-replicating elements in HCV RNA that function in replication provides an additional target for the development of new drugs to control or cure chronic HCV infections.
Acknowledgments
The subgenomic replicon of HCV was a generous gift of R. Bartenschlager. Plasmid pFastBAC NS5B was kindly provided by L. Tomei. We thank E. Mejia for expert technical help.
This work was supported by a grant from the NIH (NIAID no. 5R37AI15122).
REFERENCES
- 1.Adachi, T., H. Ago, N. Habuka, K. Okuda, M. Komatsu, S. Ikeda, and K. Yatsunami. 2002. The essential role of C-terminal residues in regulating the activity of hepatitis C virus RNA-dependent RNA polymerase. Biochim. Biopsy. Acta 1601:38-48. [DOI] [PubMed] [Google Scholar]
- 2.Ball, L. A., and Y. Li. 1993. cis-acting requirements for the replication of flock house virus. J. Virol. 67:3544-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behrens, S. E., L. Tomei, and R. de Francesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 4.Berkhout, B., B. F. Schmiht, A. van Strien, J. H. van Boom, J. van Westrenen, and J. van Duin. 1987. Lysis gene of bacteriophage MS2 is activated by translation termination at the overlapping coat gene. J. Mol. Biol. 195:517-524. [DOI] [PubMed] [Google Scholar]
- 5.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 6.Bressanelli, S., L. Tomei, A. Roussel, I. Incitti, R. L. Vitale, M. Mathieu, R. de Francesco, and F. A. Rey. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. USA 96:13034-13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, Y. C., M. Borja, H. B. Scholthof, A. O. Jackson, and T. J. Morris. 1995. Host effects and sequences essential for accumulation of defective interfering RNAs of cucumber necrosis and tomato bushy stunt tombusviruses. Virology 210:41-53. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, J.-C., M.-F. Chang, and S. C. Chang. 1999. Specific interaction between the hepatitis C virus NS5B RNA polymerase and the 3′ end of the viral RNA. J. Virol. 73:7044-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choo, Q.-L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 10.Dimitrova, M., I. Imbert, M. P. Kieny, and C. Schuster. 2003. Protein-protein interactions between hepatitis C virus nonstructural proteins. J. Virol. 77:5401-5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubuisson, J., F. Penin, and D. Moradpour. 2002. Interaction of hepatitis C virus proteins with host cell membranes and lipids. Trends Cell Biol. 12:517-523. [DOI] [PubMed] [Google Scholar]
- 12.Franscini, N., N. Blau, R. B. Walter, A. Schaffner, and G. Schoedon. 2003. Critical role of interleukin-1β for transcriptional regulation of endothelial 6-pyruvoyltetrahydropterin synthase. Arterioscler. Thromb. Vasc. Biol. 23:50-53. [DOI] [PubMed] [Google Scholar]
- 13.French, R., and P. Ahlquist. 1987. Intercistronic as well as terminal sequences are required for efficient amplification of brome mosaic virus RNA3. J. Virol. 61:1457-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friebe, P., V. Lohmann, N. Krieger, and R. Bartenschlager. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friebe, P., and R. Bartenschlager. 2002. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 76:5326-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gamarnick, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerber, K., E. Wimmer, and A. V. Paul. 2001. Biochemical and genetic studies of the initiation of human rhinovirus 2 RNA replication: identification of a cis-replicating element in the coding sequence of protein 2Apro. J. Virol. 75:10979-10990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodfellow, I., Y. Chaudry, A. Richardson, J. Meredith, J. W. Almond, W. Barcley, and D. J. Evans. 2000. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 74:4590-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haldeman-Cahill, R., J.-A., Daros, and J. C. Carrington. 1998. Secondary structures in the capsid protein coding sequence and 3′ nontranslated region involved in amplification of tobacco etch virus genome. J. Virol. 72:4072-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han, J. H., and M. Houghton. 1992. Group specific sequences and conserved secondary structures at the 3′ end of HCV genome and its implication for viral RNA replication. Nucleic Acids Res. 20:3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirano, M., S. Kaneko, T. Yamashita, H. Luo, W. Qin, Y. Shirota, T. Nomura, K. Kobayashi, and S. Murakami. 2003. Direct interaction between nucleolin and hepatitis C virus NS5B. J. Biol. Chem. 278:5109-5115. [DOI] [PubMed] [Google Scholar]
- 22.Hofacker, I. L., M. Fekete, C. Flamm, M. A. Huynen, S. Rauscher, P. E. Stolorz, and P. F. Stadler. 1998. Automatic detection of conserved RNA structure elements in complete RNA virus genomes. Nucleic Acids Res. 26:3825-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong, Z., C. E. Cameron, M. P. Walker, C. Castreo, N. Yao, J. Y. N. Lau, and W. Zhong. 2001. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology 285:6-11. [DOI] [PubMed] [Google Scholar]
- 24.Ishido, S., T. Fujita, and H. Hotta. 1998. Complex formation of NS5B with NS3 and NS4A proteins of hepatitis C virus. Biochem. Biophys. Res. Commun. 244:35-40. [DOI] [PubMed] [Google Scholar]
- 25.Ito, T., and M. M. Lai. 1997. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. J. Virol. 71:8698-8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ivashkina, N., B. Wolk, V. Lohmann, R. Bartenschlager, H. E. Blum, F. Penin, and D. Moradpour. The hepatitis C virus RNA-dependent RNA polymerase membrane insertion sequence is a transmembrane segment. J. Virol. 76:13088-13093. [DOI] [PMC free article] [PubMed]
- 27.Jacobson, A. B., R. Arora, M. Zuker, C. Priano, C. H. Lin, and D. R. Mills. 1998. Structural plasticity in RNA and its role in the regulation of protein translation in coliphage Qβ. J. Mol. Biol. 275:589-600. [DOI] [PubMed] [Google Scholar]
- 28.Kim, Y.-N., and S. Makino. 1995. Characterization of a murine coronavirus defective interfering RNA internal cis-acting replication signal. J. Virol. 69:4963-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, M., H. Kim, S.-P. Cho, and M.-K. Min. 2002. Template requirements for de novo RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase on the viral X RNA. J. Virol. 76:6944-6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolykhalov, A. A., S. M. Feinstone, and C. Rice. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 70:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komurian-Pradel, F., M. Perret, B. Deiman, M. Sodoyer, V. Lotteau, G. Paranhos-Baccala, and P. Andre. 2004. Strand specific quantitative real-time PCR to study replication of hepatitis C virus genome. J. Virol. Methods 116:103-106. [DOI] [PubMed] [Google Scholar]
- 32.Kyono, K., M. Miyashiro, and I. Taguchi. 2002. Human eukaryotic initiation factor 4AII associates with hepatitis C virus NS5B protein in vitro. Biochem. Biophys. Res. Commun. 292:659-666. [DOI] [PubMed] [Google Scholar]
- 33.Lesberg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 34.Leveque, V. J.-P., R. B. Johnson, S. Parsons, J. Ren, C. Xie, F. Zhang, and Q. M. Wang. 2003. Identification of a C-terminal regulatory motif in hepatitis C virus RNA-dependent RNA polymerase: structural and biochemical analysis. J. Virol. 77:9020-9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lobert, P.-E., N. Escriou, J. Ruelle, and T. Michiels. 1999. A coding RNA sequence acts as a replication signal in cardioviruses. Proc. Natl. Acad. Sci. USA 96:11560-11565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lohmann, V., F. Korner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lohmann, V., F. Korner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 38.Lohmann, V., F. Korner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lohmann, V., S. Hoffman, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKnight, K. L., and S. M. Lemon. 1998. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA 4:1569-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miranda, G., D. Schuppli, I. Barrera, C. Hausherr, J. M. Sogo, and H. Weber. 1997. Recognition of bacteriophage Qβ plus strand RNA as a template by Qβ replicase: role of RNA interactions mediated by ribosomal proteins S1 and host factor. J. Mol. Biol. 267:1089-1103. [DOI] [PubMed] [Google Scholar]
- 42.Molla, A., S. K. Jang, A. V. Paul, Q. Reuer, and E. Wimmer. 1992. Cardioviral internal ribosomal entry site is functional in a genetically engineered dicistronic poliovirus. Nature 356:255-257. [DOI] [PubMed] [Google Scholar]
- 43.Murray, K. E., and D. J. Barton. 2003. Poliovirus CRE-dependent VPg-uridylylation is required for plus-strand RNA synthesis but not for negative-strand RNA synthesis. J. Virol. 77:4739-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagy, P. D., J. Pogany, and A. E. Simon. 2001. In vivo and in vitro characterization of an RNA replication enhancer in a satellite RNA associated with turnip crinkle virus. Virology 288:315-324. [DOI] [PubMed] [Google Scholar]
- 45.Paul, A. V., E. Rieder, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 74:10359-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pogany, J., M. R. Fabian, K. A. White, and P. D. Nagy. 2003. A replication silencer element in a plus strand RNA virus. EMBO J. 22:5602-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poot, R. A., N. V. Tsareva, I. V. Boni, and J. van Duin. 1997. RNA folding kinetics regulates translation of phage MS2 maturation gene. Proc. Natl. Acad. Sci. USA 94:10110-10115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qin, W., H. Luo, T. Nomura, N. Hayashi, T. Yamashita, and S. Murakami. 2002. Oligomeric interaction of hepatitis C virus NS5B is critical for catalytic activity of RNA-dependent RNA polymerase. J. Biol. Chem. 277:2132-2137. [DOI] [PubMed] [Google Scholar]
- 49.Ranjith-Kumar, C. T., Y.-C., Kim, L. Gutshall, C. Silverman, S. Khandekar, R. T. Sarisky, and C. C. Kao. 2003. Mechanism of de novo initiation by the hepatitis C virus RNA-dependent RNA polymerase: role of divalent metals. J. Virol. 76:12513-12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ranjith-Kumar, C. T., X. Zhang, and C. C. Kao. 2003. Enhancer-like activity of a brome mosaic virus RNA promoter. J. Virol. 77:1830-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ray, D., and K. A. White. 1999. Enhancer-like properties of an RNA element that modulates Tombus virus RNA accumulation. Virology 256:162-171. [DOI] [PubMed] [Google Scholar]
- 52.Reed, K. E., and C. M. Rice. 2000. Overview of hepatitis C virus genome structure, polyprotein processing and protein properties. Curr. Top. Microbiol. Immunol. 242:55-84. [DOI] [PubMed] [Google Scholar]
- 53.Rieder, E., A. V. Paul, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J. Virol. 74:10371-10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmidt-Mende, J., E. Bieck, T. Hugle, F. Penin, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. Determinants for membrane association of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 276:44052-44063. [DOI] [PubMed] [Google Scholar]
- 55.Scholle, F., K. Li, F. Bodola, M. Ikeda, B. A. Luxon, and S. M. Lemon. 2004. Virus-host interactions during hepatitis C virus RNA replication: impact of polyprotein expression on the cellular transcriptome and cell cycle association with viral RNA synthesis. J. Virol. 78:1513-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schuppli, D., G. Miranda, S. Qiu, and H. Weber. 1998. A branched stem-loop structure in the M-site of bacteriophage Qβ RNA is important for template recognition by Qβ replicase holoenzyme. J. Mol. Biol. 283:585-593. [DOI] [PubMed] [Google Scholar]
- 57.Shirota, Y., H. Luo, W. Qin, S. Kaneko, T. Yamashita, K. Kobayashi, and S. Murakami. 2002. Hepatitis C virus (HCV) NS5A binds RNA-dependent RNA polymerase (RDRP) NS5B and modulates RNA-dependent RNA polymerase activity. J. Biol. Chem. 277:11149-11155. [DOI] [PubMed] [Google Scholar]
- 58.Smith, D. B., and P. Simmonds. 1997. Characteristics of nucleotide substitution in the hepatitis C virus genome: constraints on sequence change in coding regions at both ends of the genome. J. Mol. Evol. 45:238-246. [DOI] [PubMed] [Google Scholar]
- 59.Sullivan, M. L., and P. Ahlquist. 1999. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J. Virol. 73:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tomei, L., R. L. Vitale, I. Incitti, S. Serafini, S. Altamura, A. Vitelli, and R. De Francesco. 2000. Biochemical characterization of a hepatitis C virus RNA-dependent RNA polymerase mutant lacking the C-terminal hydrophobic sequence. J. Gen. Virol. 81:759-767. [DOI] [PubMed] [Google Scholar]
- 61.Tuplin, A., J. Wood, D. J. Evans, A. H. Patel, and P. Simmonds. 2002. Thermodynamic and phylogenetic prediction of RNA secondary structures in the coding region of hepatitis C virus. RNA 8:824-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Uchida, M., N. Hino, T. Yamanaka, H. Fukushima, T. Imanishi, Y. Uchiyama, T. Kodama, and T. Doi. 2002. Hepatitis C virus core protein binds to a C-terminal region of NS5B RNA polymerase. Hepatol. Res. 22:297-306. [DOI] [PubMed] [Google Scholar]
- 63.van Rossum, C. M. A., C. B. E. M. Reusken, F. T. Brederode, and J. F. Bol. 1997. The 3′ untranslated region of alfalfa mosaic virus RNA3 contains a core promoter for minus strand synthesis and an enhancer element. J. Gen. Virol. 78:3045-3049. [DOI] [PubMed] [Google Scholar]
- 64.Wang, H.-W., H.-L. Lu, D.-S. Chen, and P.-J. Chen. 1997. Identification of the functional regions required for hepatitis D virus replication and transcription by linker-scanning mutagenesis of viral genome. Virology 239:119-131. [DOI] [PubMed] [Google Scholar]
- 65.Wang, Q. M., M. A. Hockman, K. Staschke, R. B. Johnson, K. A. Case, J. Lu, S. Parsons, F. Zhang, R. Rathnachalam, K. Kirkegaard, and J. M. Colacino. 2002. Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 76:3865-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamashita, T., S. Kaneko, Y. Shirota, W. Qin, T. Nomura, K. Kobayashi, and S. Murakami. 1998. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J. Biol. Chem. 273:15479-15486. [DOI] [PubMed] [Google Scholar]
- 67.Yang, Y., R. Rijnbrand, K. L. McKnight, E. Wimmer, A. Paul, A. Martin, and S. L. Lemon. 2002. Sequence requirements for viral RNA replication and VPg uridylylation directed by the internal cis-acting replication element (cre) of human rhinovirus type 14. J. Virol. 76:7485-7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yi, M., and S. M. Lemon. 2003. 3′ nontranslated RNA signals required for replication of hepatitis C virus RNA. J. Virol. 77:3557-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yi, M., and S. M. Lemon. 2003. Structure-function analysis of the 3′ stem-loop of hepatitis C virus genomic RNA and its role in viral RNA replication. RNA 9:331-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.You, S., D. D. Stump, A. D. Branch, and C. M. Rice. 2004. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus replication. J. Virol. 78:1352-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhong, W., A. S. Uss, E. Ferrari, J. Y. N. Lau, and Z. Hong. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]