Abstract
Mutations critical for the central nervous system (CNS) attenuation of the Sabin vaccine strains of poliovirus (PV) are located within the viral internal ribosome entry site (IRES). We examined the interaction of the IRESs of PV type 3 (PV3) and Sabin type 3 (Sabin3) with polypyrimidine tract-binding protein (PTB) and a neural cell-specific homologue, nPTB. PTB and nPTB were found to bind to a site directly adjacent to the attenuating mutation, and binding at this site was less efficient on the Sabin3 IRES than on the PV3 IRES. Translation mediated by the PV3 and Sabin3 IRESs in neurons of the chicken embryo spinal cord demonstrated a translation deficit for the Sabin3 IRES that could be rescued by increasing PTB expression in the CNS. These data suggest that the low levels of PTB available in the CNS, coupled to a reduced binding of PTB on the Sabin3 IRES, leads to its CNS-specific attenuation. This study also demonstrates the use of the chicken embryo to easily investigate translation of RNA within a neuron in the CNS of an intact living organism.
Poliovirus (PV) infection worldwide has been successfully controlled through the use of two highly effective vaccines. The first, developed by Salk and colleagues, was prepared by formalin treatment of strains from the three major serotypes of PV to generate a killed-virus vaccine. The second, a live, attenuated virus vaccine developed by Sabin and colleagues, was prepared by serial passage of the three major PV strains in nonhuman primates and in cultured primate cells until strains with reduced neurovirulence were obtained (46). These Sabin strains are significantly attenuated in the central nervous system (CNS) but replicate at wild-type (WT) levels in the gut; similarly, these strains do not grow as well in cultured neuroblastoma cells as they do in cultured HeLa cells (25, 26). The Sabin strains are therefore considered to have a neural cell-specific attenuation.
PV has a single-stranded, positive-sense RNA genome of greater than 7,500 nucleotides (nt). An internal ribosome entry site (IRES) located in the 5′ untranslated region (UTR) of the picornavirus RNA genome mediates efficient translation initiation of the viral polyprotein (reviewed in references 18 and 41). Studies of recombinant PVs made between attenuated vaccine strains and their neurovirulent progenitors have shown that a major determinant of neuroattenuation maps to a single point mutation located within the viral IRES at nt 480, 481, or 472 in the case of Sabin type 1 (Sabin1), Sabin2, and Sabin3, respectively (21, 35, 49). In addition, the Sabin3 vaccine strain has been found to revert in humans to neurovirulence, and this reversion is associated with a mutation that restores the WT sequence at nt 472 (6). Studies have demonstrated a translational deficiency of the Sabin3 IRES in Krebs-2 cells and neuroblastoma cells that is not present in HeLa cells (26, 47, 48). These findings have led to the proposal that the attenuating point mutation located in the IRES of the Sabin vaccine strains inhibits IRES-mediated translation of the viral polyprotein in neurons, but not in nonneural cells, leading to neuroattenuation.
The picornavirus IRES mediates translation through its interaction with host cell RNA-binding proteins (1). Several of the proteins that interact with the PV IRES have been identified, including eukaryotic initiation factors 2 (8), 4G (38), and 4B (37); poly(C)-binding protein 1 (PCBP1) and PCBP2 (2); La (31); upstream of N-ras (unr) (15); and polypyrimidine tract-binding protein (PTB) (14, 40). PTB, a protein involved in cellular mRNA splicing, has been shown to play an important role in the translational activity of the PV IRES. Depletion of PTB from cellular extracts inhibits translation mediated by the PV IRES in a cell-free system (14-16), and overexpression of PTB in cultured cells enhances translation mediated by the PV IRES (10). Examination of the interaction of PTB with the PV IRES has demonstrated impaired binding by PTB on the Sabin IRES in comparison with the WT IRES (12, 38). Interestingly, CNS cells express PTB at very low levels but contain high levels of a neural or brain-enriched homologue of PTB, nPTB (22, 28, 30, 44). nPTB has >70% amino acid identity with PTB and is physically associated with two other neuron-specific proteins, Nova-1 and -2 (44).
In this study, we mapped the interaction of PTB and nPTB on the IRES of Sabin3 and its neurovirulent parent, the Leon type 3 strain. We found that the region surrounding nt 472 in the Sabin IRES demonstrated a change in local secondary structure (compared to the Leon IRES) that was associated with decreased binding of PTB and nPTB at an adjacent binding site. In order to compare the translation mediated by these IRES elements in a neuronal cell within the CNS, we electroporated the chicken embryo spinal cord with a bicistronic construct that contained either the Sabin3 or the Leon 5′ UTR. Translation in the chicken embryo spinal cord mediated by the Sabin3 IRES was less efficient than translation mediated by the Leon IRES and was rescued by overexpression of PTB. These data suggest that low levels of PTB that are available in the CNS, coupled to a reduced binding of PTB on the Sabin3 IRES, lead to its CNS-specific attenuation. This study also demonstrates the value of the chicken embryo spinal cord model system in evaluating translation and other functions of viral genes in neuronal cells within the CNS of an intact organism.
MATERIALS AND METHODS
Plasmids.
A plasmid containing the full-length genome of the Leon strain of PV was provided by A. J. Macadam (National Institute for Biological Standards and Control, Potters Bar, Hertfordshire, United Kingdom). The eukaryotic expression vector for PTB, pRc/CMV-PTB, and a vector containing a frameshift mutation that leads to translation termination at residue 86 of PTB, pfs-PTB, were previously described (10) and provided by Stanley Lemon (University of Texas Medical Branch at Galveston). The unr expression construct used was previously described (4) and provided by H. Jacquemin-Sablon (Laboratoire de Pharmacologie des Agents Anticancéreux, Institut Bergonié, Bordeaux, France). The bicistronic reporter construct used (see Fig. 5A) was based on a previously described pCREL plasmid provided by Ann-Catherine Pratts (Institut National de la Sant é et de la Recherche Médicale U397, Toulouse, France) (7). An SpeI site was added to pCREL via a QuikChange (Stratagene, La Jolla, Calif.) reaction. The resulting plasmid, pCRELS2, was digested with BamHI, treated with Klenow enzyme to create a blunt end, and then digested with SpeI. The 5′ UTR of the Leon or Sabin3 full-length clone was amplified by PCR, and the amplified products were digested with EcoRV and SpeI and ligated into the BamHI/SpeI-digested pCRELS2 vector to generate the Leon or Sabin3 bicistronic reporter construct.
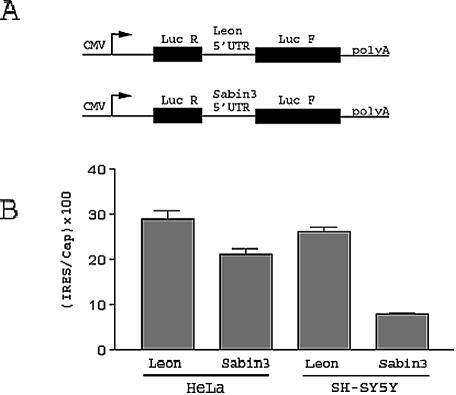
FIG. 5.
The Sabin3 IRES has a translation defect in cultured SH-SY5Y neuroblastoma cells demonstrated by transfection with bicistronic reporter constructs. (A) Schematic representation of the bicistronic constructs containing either the Leon or the Sabin3 5′ UTR. The CMV promoter is represented by an arrow. The Renilla and firefly luciferase genes are represented by black boxes. The Leon or Sabin3 5′ UTR is located in the intercistronic region between the Renilla and firefly luciferase genes. (B) HeLa and SH-SY5Y cells were transiently transfected with bicistronic constructs. Cells were harvested 24 h posttransfection, and luciferase activities in the extracts were measured. IRES activity is shown as the ratio of firefly luciferase (IRES-driven translation) to Renilla luciferase (cap-driven translation) activity times 100.
Bacterial expression constructs for PTB, nPTB, and Nova have been previously described (43). To generate the eukaryotic nPTB expression vector, the nPTB coding sequence was amplified by PCR from its bacterial expression vector, and the resulting PCR product was digested with HindIII and ligated to the large-fragment product of HindIII-digested pRc/CMV-PTB. To generate the eukaryotic expression vector for PCBP2, the PCBP2 coding sequence was excised from its bacterial expression vector by digestion with EcoRI and HindIII. The resulting fragment was ligated into EcoRI/HindIII-digested pcDNA3.1(−) (Invitrogen, Carlsbad, Calif.).
The vector expressing hairpin short interfering RNA (siRNA) directed against PTB mRNA was based on a previously described vector (7), mU6pro, provided by David Turner (University of Michigan Medical Center). The mU6pro vector was digested with Bbs1 and Xba1. The large fragment of this digestion was gel purified and ligated to annealed primers 5′-TTTGCCTGAACGGGCACAAGCTGCACGAATCGTGCAGCTTGTGCCCGTTCAGGTTTTT-3′ and 5′-CTAGAAAAACCTGAACGGGCACAAGCTGCACGATTCGTGCAGCTTGTGCCCGTTCAGG-3′ to create the CU-5 plasmid. The mismatch PTB expression construct, mmPTB, which has silent mutations preventing recognition by the hairpin siRNA, was generated via a QuikChange reaction with pRc/CMV-PTB and primers 5′-CCAGCTGGCCATGAGCCATTTAAATGGCCATAAGCTGCACGGGAAGCC-3′ and 5′-GGCTTCCCGTGCAGCTTATGGCCATTTAAATGGCTCATGGCCAGCTGG-3′.
Purification of recombinant proteins.
Recombinant proteins were expressed in Escherichia coli BL21 (Novagen, San Diego, Calif.). Recombinant PTB was purified as previously described (39). Recombinant nPTB was purified by Ni2+-nitrilotriacetic acid-agarose (QIAGEN, Valencia, Calif.) and poly(U)-Sepharose (Amersham Pharmacia Biotech, Piscataway, N.J.) column chromatography. Recombinant Nova-1 was purified by Ni2+-nitrilotriacetic acid-agarose and monoQ HiTrap (Amersham Pharmacia Biotech) column chromatography.
Immunofluorescent staining of chicken embryo spinal cord.
Chicken embryos were dissected at 24 h postelectroporation, fixed in 4% paraformaldehyde for 2 h, and then incubated overnight in phosphate-buffered saline (PBS) at 4°C. Prior to mounting in O.C.T compound (Fisher, Pittsburgh, Pa.), embryos were again placed in 4% paraformaldehyde for 10 min and then in 30% sucrose for 2 h. Mounted tissues were sectioned on a cryostat (5 to 10 μm) and placed onto glass slides. Sections were fixed with 4% paraformaldehyde for 10 min, washed three times in PBS for 5 min each time, and permeabilized with a PBS solution containing 0.3% Triton X-100. Sections were placed for 1 h at room temperature in blocking solution (4% bovine serum albumin and 0.15% Tween in PBS) prior to incubation for 24 h at 4°C with anti-PTB antibody (Clone #1; Zymed, San Francisco, Calif.) diluted 1:100 in blocking solution. Sections were washed in PBS three times for 5 min each time and then incubated for 1 h at room temperature with Cy5-conjugated anti-mouse antibody (Jackson Immunochemicals, West Grove, Pa.) diluted 1:200 in blocking solution. Sections were washed in PBS three times for 5 min each time and mounted in antifade mounting solution (Fisher) prior to imaging. Images were collected on an IX 70 confocal microscope (Olympus, Melville, N.Y.) with Fluoview 200 software (Olympus).
RNA footprinting.
Plasmids containing the full-length Sabin3 or Leon RNA genome were linearized by digestion with AvrII, and the resulting DNA was used in a T7 Ribomax (Promega, Fitchburg, Wis.) reaction to generate RNA transcripts spanning nt 1 to 1250. RNA transcripts were incubated for 10 min at 37°C with greater-than-equal molar amounts of PTB, nPTB, and Nova-1 to form complexes and then probed with RNase ONE (Promega) or RNase V1 (Amersham Pharmacia Biotech) essentially as previously described (23, 42). After treatment, the RNA was deproteinized, precipitated, dissolved in water, and divided into aliquots for subsequent primer extension with different primers. Cleaved sites were identified by primer extension as previously described (42), with primers complementary to nt 542 to 559 (5′-GACACCCAAAGTAGTCGG-3′), nt 665 to 682 (5′-GTGGATCCAACAAACAAG-3′), and nt 815 to 834 (5′-AATTGTGGTGTAGTTGATCG-3′).
Transfection of cells.
HeLa cells (American Type Culture Collection [ATCC; Manassas, Va.] catalog number CCL-2) and SH-SY5Y neuroblastoma cells (ATCC catalog number CRL-2266) were obtained from ATCC. Cells were grown in six-well tissue culture plates (Becton Dickinson, Bedford, Mass.) and, when 70 to 80% confluent, transfected at 1 μg of DNA per well with Lipofectamine 2000 (Invitrogen) in accordance with the manufacturer's instructions. Cells were harvested at 24 h posttransfection in 100 μl of passive lysis buffer (PLB; Promega) per well. Lysates were collected and used in a dual-luciferase assay (Promega) in accordance with the manufacturer's instructions.
Chicken embryo spinal cord electroporation.
Two days before electroporation, pathogen-free fertilized White Leghorn chicken eggs (Charles River Spafas, Roanoke, Ill.) were transferred to an egg incubator (39°C, 60 to 70% humidity) for development of the embryos to stage 11-12 (Hamburger-Hamilton stage). On the day of electroporation, a window (approximately 10 mm in diameter) was cut into the shell to expose the embryo. Embryos were kept moist by adding 0.5 to 1.0 ml of L-15 medium (Invitrogen) on top of the vitelline membrane. The plasmid DNA (1 mg/ml) was mixed with 1% Fast Green at a 10:1 ratio to visualize the injection. Colored DNA was injected into the central canal of the spinal cord via a glass microinjection pipette (approximately 5-mm tip). Electrodes were placed across the embryo, and a square wave electroporator (BTX, Holliston, Mass.) was used to administer four pulses of current at 25 V for 50 ms each. The open window was then covered with tape, and the eggs were returned to the incubator. Embryos were dissected from the egg at 24 h postelectroporation, placed in 100 μl of PLB, and homogenized with a Kontes pestle (Fischer, Hampton, N.H.). Homogenates were microcentrifuged (Eppendorf centrifuge 5417C) for 1 min at 14,000 rpm. The supernatant was removed and tested in a dual-luciferase assay.
siRNA in cultured BHK-21 cells.
BHK-21 (baby hamster kidney) cells, obtained from ATCC, were grown in six-well plates and, when 60 to 90% confluent, transfected with 450 ng of CU-5 DNA, 100 ng of bicistronic reporter DNA, and either 450 ng of pcDNA3 DNA or 450 ng of mmPTB DNA per well with FuGene6 (Roche, Indianapolis, Ind.) in accordance with the manufacturer's instructions. Cells were harvested at 3 days posttransfection by scraping in 100 μl of PLB. Ten microliters of cell lysate was tested in a dual-luciferase assay.
siRNA in cultured HeLa cells.
HeLa cells were grown in six-well plates and, when 60 to 70% confluent, transfected with 1.0 μg of Leon or Sabin bicistronic reporter DNA per well along with 0.5 μg of CU-5 or mU6pro DNA per well with Effectene transfection reagent (QIAGEN) in accordance with the manufacturer's instructions. Cells were harvested 4 days posttransfection by scraping in 100 μl of 1× PLB. Five microliters of cell lysate was tested in a dual-luciferase assay.
RESULTS
PTB and nPTB bind the Leon and Sabin3 IRESs at a site adjacent to nt 472.
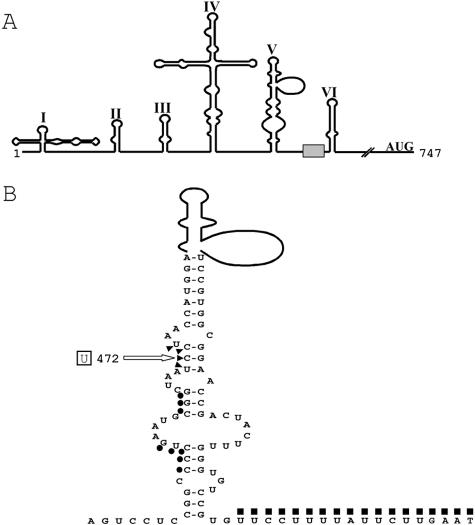
Figure 1A depicts the predicted secondary structure of the PV IRES on the basis of computer modeling and biochemical data (42). Of particular interest is domain V (Fig. 1B), which includes nt 472 of Sabin3. An earlier study showed that PTB binds to three separate regions of the PV IRES; however, the precise locations of these binding sites were not identified (13). In order to map the locations of the PTB-binding sites, to identify and map nPTB-binding sites, and to compare these sites in the Leon IRES with those in the Sabin3 IRES, we examined protection of the Leon and Sabin3 5′ UTRs from cleavage by RNases in the presence of recombinant PTB or nPTB.
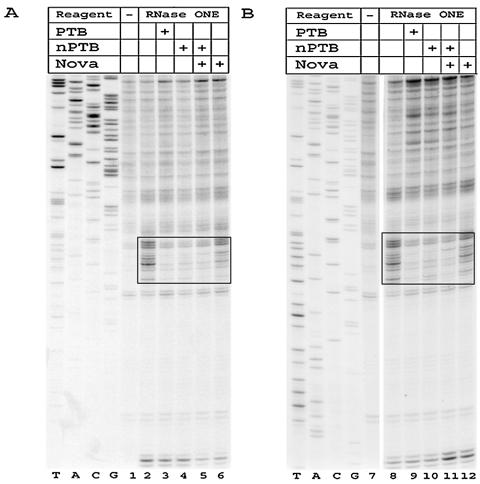
FIG. 1.
The PV 5′ UTR. (A) Schematic representation of the predicted secondary structure of the PV 5′ UTR. The shaded box represents the oligopyrimidine tract. (B) Detailed view of domain V and the oligopyrimidine tract for the Leon strain of PV. The mutation involved in attenuation of Sabin3 is boxed. Nucleotides susceptible to RNase ONE cleavage in the Sabin3, but not the Leon, IRES are indicated by triangles (see Fig. 2A). Nucleotides protected by PTB or nPTB from V1 cleavage are indicated by circles (see Fig. 2B), and nucleotides protected by PTB or nPTB from RNase ONE cleavage are indicated by squares (see Fig. 3).
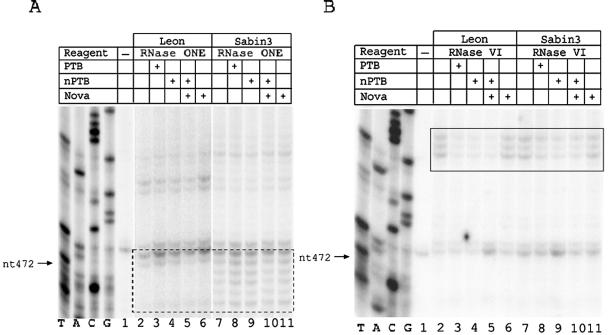
Figure 2 shows results of an RNase protection assay on the Leon and Sabin3 IRESs in the region surrounding nt 472. In order to see full protection, PTB and nPTB were added to protection reaction mixtures in greater-than-equal molar amounts compared to the RNA. A prominent difference in the cleavage pattern between the Leon and Sabin3 IRESs was observed following digestion with RNase ONE, an RNase that cleaves single-stranded RNA. The Leon IRES showed no significant cleavage by RNase ONE in the region surrounding nt 472, whereas the Sabin3 IRES displayed several RNase ONE cleavages in this region (dashed-boxed region in Fig. 2A and triangles in Fig. 1B). These results suggest that the secondary structure in this region changes from double stranded in the Leon IRES to single stranded in the Sabin3 IRES.
FIG. 2.
PTB and nPTB bind the Leon and Sabin3 IRESs at a site adjacent to nt 472. Enzymatic footprint of the Leon and Sabin3 IRESs in the region surrounding nt 472 with RNase ONE (A) and RNase V1 (B). In this figure and the subsequent two figures, the table above the gel indicates whether RNA was incubated alone or with proteins and whether RNA-protein complexes were digested with an enzyme. RNA probes were extended with a primer complementary to nt 542 to 559. The region displaying enhanced cleavage by RNase ONE on the Sabin3 strain is indicated by the dashed box (see also triangles in Fig. 1B). The location of the PTB- or nPTB-binding site is indicated by the solid box (see also circles in Fig. 1B).
Digestion of the Leon and Sabin3 IRESs with RNase V1, an RNase that cleaves double-stranded RNA, identified a region immediately upstream of nt 472 that was protected by both PTB and nPTB (solid-boxed region in Fig. 2B and circles in Fig. 1B); no evidence for protection by Nova-1 in this region was found, and addition of Nova-1 to nPTB did not alter the pattern or degree of nPTB protection. Protection at this site by PTB and nPTB was consistently greater on the Leon IRES than on the Sabin3 IRES (compare lanes 3 to 5 with lanes 8 to 10 in Fig. 2B). These data demonstrate that binding of PTB or nPTB at the binding site adjacent to nt 472 is more efficient on the Leon IRES than on the Sabin3 IRES, presumably because of the change in the local secondary structure in this region of the Sabin3 IRES.
In summary, we found that the secondary structure adjacent to the site of a critical attenuating mutation is altered from double stranded in the Leon IRES to single stranded in the Sabin3 IRES and that both PTB and nPTB bind the Leon and Sabin3 IRESs directly adjacent to this site; however, binding to the Leon IRES is greater than that to the Sabin3 IRES.
A second PTB- or nPTB-binding site in the PV IRES is located downstream of nt 472 in an oligopyrimidine tract.
RNase protection assays performed on the IRES downstream of nt 472 demonstrated that a second site was protected by PTB or nPTB on both the Leon and Sabin3 IRESs (solid boxed region in Fig. 3 and squares in Fig. 1B). This site of protection is located in an oligopyrimidine tract that lies between domains V and VI of the IRES. There were no significant differences in the degree of protection seen in the presence of either PTB or nPTB on both the Leon and Sabin3 IRESs. No evidence for protection in this region was found following addition of Nova-1 alone, and addition of Nova-1 to nPTB did not significantly alter the pattern or degree of nPTB protection. These data demonstrate that PTB or nPTB binds the Leon and Sabin3 IRESs at a site downstream of nt 472 and that binding to this site is not affected by the mutation present in the Sabin3 IRES at nt 472.
FIG. 3.
A second PTB- or nPTB-binding site in the PV IRES is located downstream of nt 472 in an oligopyrimidine tract. Enzymatic footprint of the Leon (A) and Sabin3 (B) IRESs in the region surrounding the oligopyrimidine tract. RNA probes were extended with a primer complementary to nt 665 to 682. The location of the PTB- or nPTB-binding site is indicated by the solid box (see also squares in Fig. 1B).
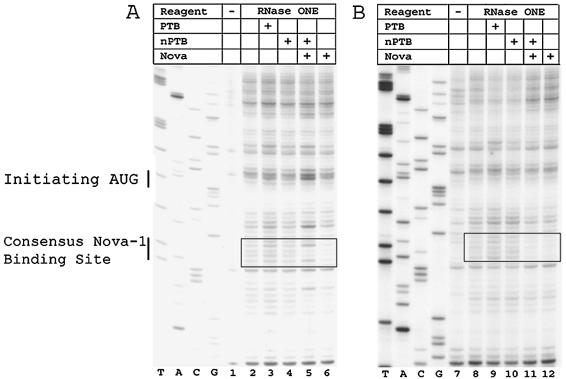
A binding site for Nova-1 is located immediately downstream of the polyprotein's translation start site.
We examined the binding of Nova-1 to the PV IRES because Nova-1 is physically associated with nPTB. Addition of Nova-1 protein alone did not protect the Leon or Sabin3 IRES from cleavage at any site within the 5′ UTR. Surprisingly, there was protection from RNase ONE cleavage by Nova-1 (as well as Nova-1 plus nPTB) on both the Leon and Sabin3 RNAs at a site downstream from the 5′ UTR and 12 nt downstream from the polyprotein's initiation codon (Fig. 4). This site of protection covered nt 755 to 763 and contained a sequence identical to a consensus Nova-1-binding site (UCAU) (5). There was protection at this site by Nova-1 (as well as Nova-1 plus nPTB) on both the Leon and Sabin3 RNAs; some differences in the pattern of protection were noted in the case of Nova-1 versus Nova-1 plus nPTB on the Leon IRES.
FIG. 4.
A binding site for Nova-1 is located immediately downstream of the polyprotein's translation start site. Enzymatic footprint of the Leon (A) and Sabin3 (B) IRESs in the region surrounding the initiation codon for the viral polyprotein. RNA probes were extended with a primer complementary to nt 815 to 834. The location of the Nova-1-binding site downstream of the initiation codon is indicated by the solid boxes. The locations of the initiating AUG and the consensus Nova-1-binding site are shown on the left.
The Sabin3 IRES has a translation defect in cultured neural cells.
The relative decrease in binding of PTB on the Sabin3 IRES compared to the Leon IRES coupled to the low level of PTB expression in neural cells suggested that the Sabin3 IRES was likely to demonstrate a translation defect in these cells. To initially examine this issue, we investigated Leon and Sabin3 IRES activity in HeLa cells and compared it to the activity in neuroblastoma cells. For these studies, we transfected bicistronic constructs that contained a cap-dependent Renilla luciferase gene in the first cistron, followed by the Leon or Sabin3 5′ UTR (containing the IRES) in the intercistronic region and then by firefly luciferase in the second cistron (Fig. 5A). Calculating the ratio of firefly luciferase to Renilla luciferase provided a reproducible and quantitative measurement of IRES function in a cell.
Transfection of the bicistronic construct containing either the Leon or the Sabin3 IRES demonstrated a similar IRES activity in HeLa cells; however, compared to the Leon IRES, the Sabin3 IRES had an approximately threefold decrease in activity in neuroblastoma cells (Fig. 5B). These results are similar to those previously described (12).
The Sabin3 IRES has a translation defect in the CNS.
There are shortcomings to the use of continuously cultured cells because they bear relatively little relationship to an authentic neuron and therefore do not allow one to investigate translation efficiency in a cell that possesses authentic neuron-specific cellular factors and properties. In order to more reliably assess the activity of the Sabin3 IRES in a neuron in situ, we took advantage of a method involving electroporation of DNA into the spinal cords of developing chicken embryos (34, 36). In this method, DNA is microinjected into the neural tube of a chicken embryo, followed by the placement of electrodes on each side of the neural tube and subsequent delivery of current. Electroporation results in delivery of the gene into approximately 50% of the neurons on one side of the caudal half of the spinal cord.
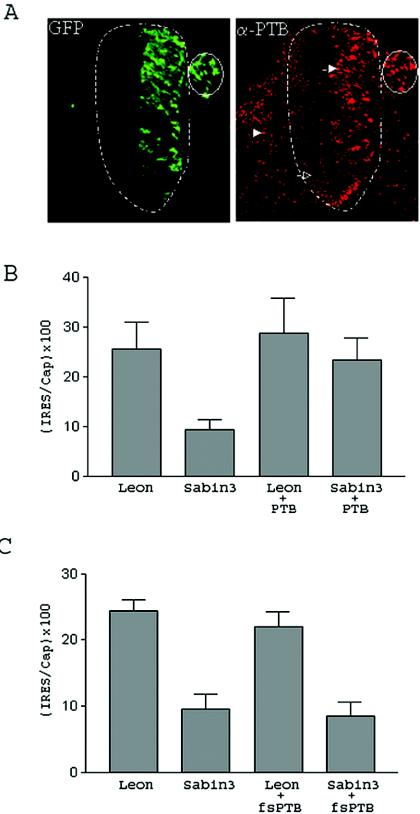
Figure 6A shows a section of a 3-day-old chicken embryo that was electroporated 24 h previously with a PTB expression construct along with a green fluorescent protein (GFP) expression construct (to help identify cells that received the electroporated DNA). The section was stained for PTB expression (shown in red). Neurons on the side of the spinal cord that did not receive the PTB expression construct (open arrow) had lower levels of PTB than did nonneural cells (arrowhead), confirming the low level of PTB in the CNS that has previously been reported (22, 28, 30, 44); although immunofluorescent staining of this endogenous PTB was localized to the nucleus, there is evidence that PTB shuttles to the cytoplasm (32). Electroporation of the PTB expression construct increased expression of PTB in cells on one side of the spinal cord (closed arrow) up to a level comparable to that seen in the surrounding tissues.
FIG. 6.
The Sabin3 IRES has a translation defect in the CNS that can be rescued by coelectroporation of PTB. (A) Two-day-old chicken embryos were electroporated with a vector that expresses PTB and a vector that expresses GFP. Embryos were harvested at 24 h postelectroporation, sectioned, and overlaid with an anti-PTB mouse monoclonal antibody, followed by a Cy5-conjugated anti-mouse antibody. The section of the chicken embryo was then visualized under UV light with a filter for GFP or Cy5. The left panel shows GFP fluorescence in green, while the right panel shows the same field with anti-PTB staining in red. The spinal cord is outlined with a dotted line. As shown, neurons that received the electroporated GFP (and PTB expression construct) have migrated throughout one side of the spinal cord and the dorsal root ganglion (which is surrounded by a solid line). Cells that were electroporated have an increased PTB signal (closed arrow) compared to neurons on the other side of the spinal cord (open arrow) or nonneural cells (arrowhead). (B and C) Two-day-old chicken embryos were electroporated with the Leon or Sabin3 bicistronic construct with or without coelectroporation of the PTB expression construct (B) or with or without coelectroporation of a vector that expresses a truncated, nonfunctional fragment of PTB (pfs-PTB) (C).
A bicistronic construct containing either the Leon or the Sabin3 IRES was electroporated into 2-day-old chicken embryo spinal cords. Translation mediated by the Sabin3 IRES was found to be approximately 2.5- to 4-fold less efficient than translation mediated by the Leon IRES (Fig. 6B). These results suggest that the Sabin3 IRES has a translation defect within cells in the CNS.
Coelectroporation of PTB rescues the translation defect of the Sabin3 IRES in the CNS.
To examine the possibility that the translation defect of the Sabin3 IRES in the CNS is a result of low levels of PTB in this tissue, we coelectroporated a PTB expression construct along with the Leon or Sabin3 IRES bicistronic construct into the chicken embryo spinal cord. Coelectroporation of the PTB construct significantly and consistently enhanced translation mediated by the Sabin3 IRES (P = 0.0001, n = 9) and, importantly, brought it up to the levels seen with the Leon IRES (Fig. 6B). Coelectroporation of a construct encoding a truncated form of PTB that is known to lack activity (10) failed to rescue the translational defect of the Sabin3 IRES (Fig. 6C). Together, these results suggest that overexpression of PTB within the CNS can rescue the observed translation defect of the Sabin3 IRES in this tissue.
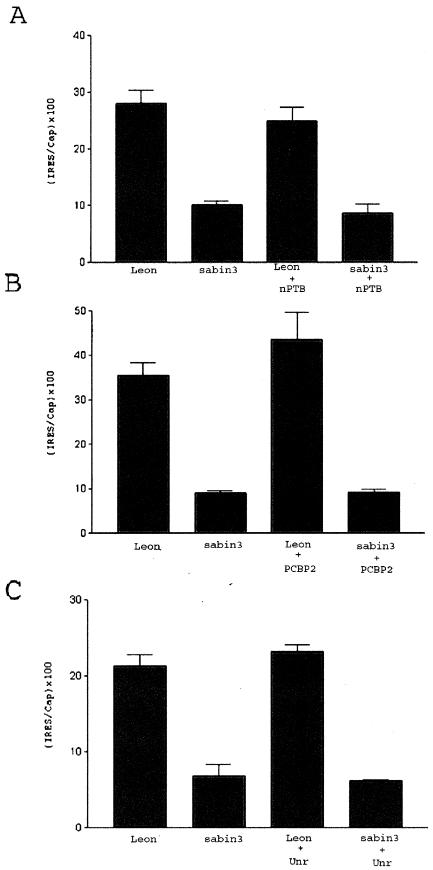
CNS cells are deficient in PTB but contain relatively high levels of nPTB (22, 28, 30, 44). In order to examine the effect of nPTB on translation mediated by the Sabin3 IRES, we coelectroporated an nPTB expression construct along with the Sabin3 bicistronic construct into the chicken embryo spinal cord. Coelectroporation of the nPTB construct did not increase the relatively low level of Sabin3 IRES-mediated translation in the spinal cord (Fig. 7A) and therefore failed to rescue the translational defect of the Sabin3 IRES. These results suggest that nPTB cannot substitute for the function of PTB on the Sabin3 IRES in the CNS.
FIG. 7.
Coelectroporation of nPTB, PCBP2, or unr does not rescue the translation defect of the Sabin3 IRES in the CNS. Two-day-old chicken embryos were electroporated with the Leon or Sabin3 bicistronic construct with or without coelectroporation of an nPTB expression vector (A), a PCBP2 expression vector (B), or a unr expression vector (C).
Several proteins in addition to PTB are known to bind the PV IRES and are thought to play a role in PV IRES-mediated translation. The cellular protein PCBP2 binds to domain IV of the PV IRES; in addition, PCBP2-depleted HeLa cell extracts poorly translate a reporter driven by the PV IRES, while translation activity is partially restored to these extracts by addition of recombinant PCBP2 (3). The cellular protein unr has also recently been shown to play a role in efficient translation by the PV IRES (4). PCBP2 and unr are expressed at similar levels in various tissues, including the CNS (19, 27, 29). In order to examine the effect of PCBP2 and unr expression on translation mediated by the Leon and Sabin3 IRESs in neural cells, we coelectroporated a PCBP2 or unr expression construct along with the bicistronic construct into the chicken embryo spinal cord. Coelectroporation of the PCBP2 or unr construct did not increase the relatively low level of Sabin3 IRES-mediated translation (Fig. 7B and C) and therefore failed to rescue the translation defect of the Sabin3 IRES.
In summary, we found that PTB rescues the translation deficit of the Sabin3 IRES in the CNS; however, overexpression of nPTB, PCBP2, or unr within the CNS had no significant effect on the translation defect of the Sabin3 IRES in this tissue.
PTB knockdown decreases PV IRES translation activity.
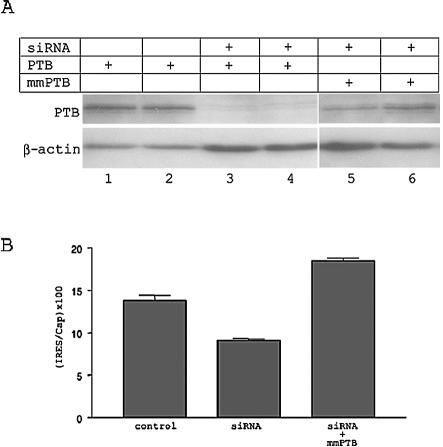
Our present study, along with previous studies, suggested a key role for PTB in translation mediated by the PV IRES. In order to confirm the importance of PTB in PV IRES-mediated translation, we reduced PTB expression in cultured cells by RNA interference. Cells were transfected with a construct that expresses hairpin siRNA complementary to nt 1179 to 1201 of PTB mRNA and has been reported to knock down PTB expression (52). In order to confirm that this construct interferes with PTB expression in transfected cells, we cotransfected a PTB expression construct along with the siRNA construct. Cells transfected with both of these constructs showed significantly reduced PTB protein expression compared to that of cells transfected with the PTB expression construct alone (Fig. 8A). Cotransfection of the siRNA construct along with the Leon bicistronic construct resulted in a reduction in IRES-mediated translation of up to 50% (Fig. 8B). We prepared a mismatch PTB expression construct (mmPTB) that contains silent mutations and therefore prevents recognition of PTB RNA by the hairpin siRNA (Fig. 8A). Coelectroporation of the mmPTB construct along with the siRNA construct, which allowed cells to express PTB even in the presence of the siRNA, prevented any decrease in Leon IRES translation in these cells (Fig. 8B). These results further demonstrate a role for PTB in translation mediated by the PV IRES.
FIG. 8.
Knockdown of PTB expression in cultured cells by RNA interference demonstrates the importance of PTB in PV3 IRES translation. (A) Western immunoblot analysis of lysates from BHK-21 cells transfected with the plasmids indicated in the table above the gel. Lysates were subjected to electrophoresis, followed by immunostaining with anti-PTB antibody. The same membrane was then stripped and reprobed with anti-β-actin antibody. (B) BHK-21 cells were transfected with the Leon bicistronic reporter construct along with one of the following: control (control vector plus pfs-PTB), siRNA (siRNA construct plus pfs-PTB), or siRNA plus mmPTB (siRNA construct plus mmPTB).
We also compared the effect of knockdown of PTB expression on Leon versus Sabin3 IRES activity. Transfection of the PTB siRNA construct into HeLa cells decreased Leon translation by 23% (P < 0.01) and decreased Sabin3 translation by 38% (P < 0.001). These results suggest that lowering the PTB concentration has a greater effect on Sabin3 IRES activity than on Leon IRES activity.
DISCUSSION
The Sabin vaccine strains have led to the development of an effective live, attenuated PV vaccine. These vaccine strains replicate at levels similar to those of their parental counterparts in HeLa cells; however, they are severely attenuated in the CNS and are unable to induce the paralytic disease associated with the parental strains. A major determinant for this neuroattenuation is linked to a single mutation located in the IRES of the viral genome in the case of the three Sabin vaccine strains (21, 35, 49). This mutation is believed to disrupt the interaction of the IRES with a cellular factor, leading to poor translation of the viral polyprotein with subsequent attenuation of the virus. The identity of the IRES-binding factor(s) and the reasons that the effect of the mutation is only apparent in cells of the CNS have remained unclear and have posed long-standing questions in virology. A clarification of the mechanisms of attenuation of the Sabin strains may lead to a better understanding of the pathogenesis of poliomyelitis, as well as provide direction with respect to the development of new vaccines.
Several lines of evidence led us to focus our investigations on the importance of PTB in PV attenuation. PTB binds to the PV IRES and has been reported to cross-link specifically with domain V of the IRES, the site of the attenuating mutation in the Sabin vaccine strains. PTB has also been reported to play an essential role in IRES-mediated translation of the PV polyprotein; our siRNA knockdown data from the present study support a role for PTB in PV IRES-mediated translation. Furthermore, the expression profile for PTB is unique among proteins known to bind the PV IRES; i.e., PTB is expressed abundantly outside the CNS but at low levels in the CNS (22, 28, 30, 44). The fact that CNS tissue expresses relatively low levels of PTB suggests that PTB may have a role in the attenuation of the Sabin strains.
In this study, we examined the binding of PTB, its neural homolog nPTB, and Nova-1 on the IRES of the Sabin3 strain, as well as on the IRES of the parental type 3 strain, Leon. In order to examine the role of PTB, nPTB, and Nova-1 in PV IRES-mediated translation within the CNS, we made use of a technique involving electroporation of the chicken embryo spinal cord.
PTB, nPTB, and Nova-1 binding on the Leon and Sabin PV3 IRESs.
Previous studies of the Leon IRES by UV cross-linking have demonstrated PTB binding to domain V but have not finely localized the binding sites (13). The RNase protection studies of domain V presented here demonstrated two interesting differences between Leon and Sabin3: (i) the Sabin3, but not the Leon, IRES was sensitive in the region of the attenuating mutation to RNase ONE, which recognizes single-stranded RNA, and (ii) PTB binding to the Sabin3 IRES at a site adjacent to nt 472 was significantly decreased compared to that seen with the Leon IRES. These data suggested that an altered RNA secondary structure results from the attenuating mutation in the Sabin3 IRES, which leads to a decrease in PTB binding at the binding site adjacent to the mutation. A second binding site for PTB was found in an oligopyrimidine tract located between domains V and VI of the PV3 IRES. Comparison of the RNase cleavage pattern of this region for the Leon and Sabin3 IRESs showed no differences in RNA secondary structure. In addition, PTB bound similarly to this site in the case of both the Leon and Sabin3 IRESs. Previous studies with a cell-free system identified three separate regions of the PV IRES that cross-link with PTB (13). Both of the binding sites identified in the present study are located within one of these three regions. Our inability to detect binding in the other two regions may result from the absence of other PV IRES-binding proteins in our in vitro assay, which used purified PTB (and nPTB).
Our previous study of the IRES of the GDVII strain of Theiler's murine encephalomyelitis virus showed that nPTB binds to the same binding sites as PTB (43). Mutation of any one of several individual PTB- or nPTB-binding sites on the GDVII IRES led to a decrease in PTB or nPTB binding at that site and to neuroattenuation of the virus. Although both PTB and nPTB bound similarly to the WT GDVII IRES, the mutated GDVII IRES showed a more reduced binding to nPTB than to PTB at a binding site other than the mutated site. These results highlighted the importance of nPTB binding in the neuroattenuation of mutant GDVII strains.
The present study demonstrated that nPTB binds similarly to PTB on the PV IRES and at the same two binding sites; these findings suggest that nPTB (or other tissue-specific variants of PTB) may have binding sites in common with PTB on the IRESs of other picornaviruses besides PV and Theiler's murine encephalomyelitis virus. We found that there was a similar decreased binding of nPTB and PTB at the site adjacent to the attenuating mutation (nt 472) in the case of the Sabin3 IRES, as was the case with the GDVII IRES. We did not observe any differences in PTB versus nPTB binding at the binding site located downstream in the oligopyrimidine tract of the Sabin3 IRES. These results suggest that attenuation of the Sabin vaccine strains in neural cells is not due to a differential in the binding of nPTB compared to PTB on the Sabin3 IRES. We cannot rule out, however, the possibility that there are differences in PTB versus nPTB binding at binding sites that were not detected in our system.
In addition to investigating PTB and nPTB, we examined Nova-1 binding to the PV IRES, since Nova-1 is an RNA-binding protein that interacts with nPTB. Nova-1, like PTB and nPTB, plays a role in regulating tissue-specific splicing of particular mRNAs in the CNS (20, 24). Nova-1 is also the target antigen in a human paraneoplastic neurological motor disorder (51). Interestingly, Nova-1 bound PV RNA within the coding region for the viral polyprotein downstream of the viral 5′ UTR (and downstream of the IRES) and the polyprotein start site. Nucleotides downstream of a polyprotein-initiating codon have been found to have IRES function in other viruses, such as encephalomyocarditis virus (17) and hepatitis C virus (45). The region bound by Nova-1 was found to contain a consensus binding site for Nova-1 (UCAU) (5).
Examination of almost 200 enterovirus sequences present in the database for the presence of a Nova-1-binding site in a location similar to that found in PV3 RNA showed that this feature is conserved among all of the PVs for which the sequence has been determined but in very few enteroviruses of group C besides PV. The presence in PV of the UCA in the Nova-1-binding site, which codes for serine, is of interest since UCA only constitutes 48% of all serine codons, suggesting that there are conservation forces other than the amino acid coding sequence working to maintain this site in PV strains. In order to further investigate the role of this sequence, we mutated the Nova-1 site of the PV3 genome in our bicistronic reporter but found no difference in translational activity in HeLa cells or in chicken embryo spinal cord neurons (data not shown), suggesting that Nova-1 does not have a role in PV translation in either of these cell types under the conditions tested. There may be other situations, however, in which the Nova-1 sequence has a function for PV.
PV IRES function within the CNS.
Previous studies examining translation mediated by the PV IRES have used cell lysates or cultured cells. In order to study PV IRES-mediated translation in vivo, we adopted a method used by developmental neurobiologists involving electroporation of DNA into the chicken embryo spinal cord. We carried out the electroporation at a time when the majority of the cells present within the spinal cord and lining the neural tube are neurons. Electroporation was very efficient, resulting in gene delivery into approximately 50% of the neurons on one side of the caudal half of the spinal cord. Electroporation of bicistronic constructs provided us with a ratio of IRES- to cap-dependent translation that was highly reproducible from one experiment to another despite variation in the number of cells that were electroporated. Within 24 h of electroporation of bicistronic constructs, IRES-mediated translation led to levels of firefly luciferase activity that were 1,000-fold above background, demonstrating that the PV IRES functioned efficiently in chicken embryo spinal cord neurons. The latter finding was not unexpected, since PV has been found to replicate in chicken embryo cells (9).
We used the chicken embryo system to compare the function of the Sabin3 IRES with that of its neurovirulent counterpart and found that the Sabin3 IRES had a translation defect in neurons within the chick spinal cord. Our experiments with chicken embryos are the first to measure PV IRES-mediated translation within the CNS and to demonstrate that the Sabin3 IRES translation defect exists in authentic neurons in situ. Since our binding studies had shown decreased binding of PTB on the Sabin3 IRES compared to that on the Leon IRES, we suspected that the decreased efficiency of Sabin3 RNA translation in chicken embryo neurons was related to the low level of PTB in CNS cells that had previously been reported (22, 28, 30, 44) and was supported by our immunofluorescence studies. We found that coelectroporation of the PTB expression construct along with our bicistronic construct enhanced translation mediated by the Sabin3 IRES, bringing it up to a level seen with the Leon IRES. These results suggest that attenuation of Sabin3 within CNS cells is due to a decrease in the translation efficiency of Sabin3 relative to that of Leon resulting from the decreased amount of PTB within the CNS coupled to a decrease in PTB binding. In a similar way, the increase in the translational efficiency of the Sabin3 IRES in HeLa cells versus SH-SY5Y cells presumably results from the increased amount of PTB expressed in HeLa cells compared to that expressed in neuroblastoma cells (unpublished data; 33).
We questioned whether nPTB could also rescue the translation defect of Sabin3 since nPTB binds to the same sites on the IRES as PTB and because nPTB is highly expressed in neurons. We found that coelectroporation of an nPTB expression construct failed to rescue the translation defect of the Sabin3 IRES. There is more than 70% amino acid homology between PTB and nPTB, with the greatest similarity located in the RNA-binding domains of the two proteins (44). It is likely that regions outside of the RNA-binding domains are important for mediating PV3 translation and that differences in these regions lead to differences in IRES activity following nPTB versus PTB binding. Our recent studies of chimeric PTB-nPTB cDNAs in chicken embryo spinal cord neurons have shown that analogous regions of nPTB and PTB have different activities with respect to their IRES function and therefore cannot substitute for one another (data not shown).
Our study suggests the following explanation for the CNS-specific attenuation of the Sabin3 vaccine strain of PV. The attenuating mutation located in the Sabin3 IRES changes the local secondary structure and results in decreased binding of PTB or nPTB. High levels of PTB in HeLa cells are able to overcome this binding defect, so that Sabin3 IRES-mediated translation is similar in these cells to that seen with the Leon IRES. The low levels of PTB that are present in the CNS, however, are unable to overcome the binding defect, resulting in inefficient IRES-mediated translation and attenuation of Sabin3 within the CNS. Our siRNA studies also suggest that Sabin3 IRES-mediated translation is more sensitive than Leon IRES-mediated translation to low levels of PTB. Our findings also demonstrate that PV RNA and presumably other viral RNAs have various translation efficiencies in different cell types with very different requirements regarding cell-specific proteins and factors. These various requirements can have substantial effects on virus tropism, as demonstrated by experiments in which one virus IRES has been exchanged for that of another (11, 50), and neurovirulence, as in the case of the Sabin strains.
The recognition that PV translated efficiently in chicken embryo spinal cord neurons raised a question as to whether we could electroporate full-length PV RNA with the generation of infectious virus. We found that PV was produced following electroporation of full-length PV RNA into chicken embryo spinal cord neurons (unpublished data). Significant differences in the amount of infectious virus were noted 10 h after electroporation of Leon strain RNA (104.0 50% tissue culture infective doses per ml) versus Sabin strain RNA (102.6 50% tissue culture infective doses per ml). We were unable, however, to determine the effect of addition of the PTB expression construct on the yield of Leon versus Sabin PV since significant expression of PTB protein following electroporation occurred after virus growth peaked.
Our study demonstrates the potential value of electroporation of the chicken embryo spinal cord in investigations of the function of genome segments of PV and other neurotropic viruses. DNA constructs containing a viral IRES or a viral gene can be rapidly and efficiently introduced into a bona fide neuronal cell in situ within the CNS of a living organism. Use of the chicken embryo system avoids difficulties associated with isolation, culture, and transfection of primary neuronal cells and represents a more physiologically relevant context than that afforded by cultured neuroblastoma cells. In addition, this system allows one to easily investigate translation of RNA within a neuron in the CNS of an intact living organism. Our finding that full-length RNA can be efficiently electroporated into the chicken embryo spinal cord with the generation of infectious virus opens the door to studying postentry aspects of the virus life cycle in vivo within cells of the CNS even in the absence of the viral receptor.
Acknowledgments
This work was partially funded by the NIH and the MS Society.
We thank Adam Light and Vytas Bindokas for technical assistance and the following individuals for reagents: Jeffrey Macadam, Stanley Lemon, Hélène Jacquemin-Sablon, Ann-Catherine Pratts, David Turner, and Bert Semler.
REFERENCES
- 1.Belsham, G. J., and N. Sonenberg. 1996. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol. Rev. 60:499-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blyn, L. B., K. M. Swiderek, O. Richards, D. C. Stahl, B. L. Semler, and E. Ehrenfeld. 1996. Poly(rC) binding protein 2 binds to stem-loop IV of the poliovirus RNA 5′ noncoding region: identification by automated liquid chromatography-tandem mass spectrometry. Proc. Natl. Acad. Sci. USA 93:11115-11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blyn, L. B., J. S. Towner, B. L. Semler, and E. Ehrenfeld. 1997. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 71:6243-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boussadia, O., M. Niepmann, L. Creancier, A. C. Prats, F. Dautry, and H. Jacquemin-Sablon. 2003. Unr is required in vivo for efficient initiation of translation from the internal ribosome entry sites of both rhinovirus and poliovirus. J. Virol. 77:3353-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buckanovich, R. J., and R. B. Darnell. 1997. The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo. Mol. Cell. Biol. 17:3194-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cann, A. J., G. Stanway, P. J. Hughes, P. D. Minor, D. M. Evans, G. C. Schild, and J. W. Almond. 1984. Reversion to neurovirulence of the live-attenuated Sabin type 3 oral poliovirus vaccine. Nucleic Acids Res. 12:7787-7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Creancier, L., D. Morello, P. Mercier, and A. C. Prats. 2000. Fibroblast growth factor 2 internal ribosome entry site (IRES) activity ex vivo and in transgenic mice reveals a stringent tissue-specific regulation. J. Cell Biol. 150:275-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.del Angel, R. M., A. G. Papavassiliou, C. Fernandez-Tomas, S. J. Silverstein, and V. R. Racaniello. 1989. Cell proteins bind to multiple sites within the 5′ untranslated region of poliovirus RNA. Proc. Natl. Acad. Sci. USA 86:8299-8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eggers, H. J. 1990. Notes on the pathogenesis of enterovirus infections. Observations, experiments, and speculations. Med. Microbiol. Immunol. 179:297-306. [DOI] [PubMed] [Google Scholar]
- 10.Gosert, R., K. H. Chang, R. Rijnbrand, M. Yi, D. V. Sangar, and S. M. Lemon. 2000. Transient expression of cellular polypyrimidine-tract binding protein stimulates cap-independent translation directed by both picornaviral and flaviviral internal ribosome entry sites in vivo. Mol. Cell. Biol. 20:1583-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gromeier, M., L. Alexander, and E. Wimmer. 1996. Internal ribosomal entry site substitution eliminates neurovirulence in intergeneric poliovirus recombinants. Proc. Natl. Acad. Sci. USA 93:2370-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutierrez, A. L., M. Denova-Ocampo, V. R. Racaniello, and R. M. del Angel. 1997. Attenuating mutations in the poliovirus 5′ untranslated region alter its interaction with polypyrimidine tract-binding protein. J. Virol. 71:3826-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hellen, C. U., T. V. Pestova, M. Litterst, and E. Wimmer. 1994. The cellular polypeptide p57 (pyrimidine tract-binding protein) binds to multiple sites in the poliovirus 5′ nontranslated region. J. Virol. 68:941-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hellen, C. U., G. W. Witherell, M. Schmid, S. H. Shin, T. V. Pestova, A. Gil, and E. Wimmer. 1993. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl. Acad. Sci. USA 90:7642-7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunt, S. L., J. J. Hsuan, N. Totty, and R. J. Jackson. 1999. unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes Dev. 13:437-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunt, S. L., and R. J. Jackson. 1999. Polypyrimidine-tract binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2 RNA. RNA 5:344-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunt, S. L., A. Kaminski, and R. J. Jackson. 1993. The influence of viral coding sequences on the efficiency of internal initiation of translation of cardiovirus RNAs. Virology 197:801-807. [DOI] [PubMed] [Google Scholar]
- 18.Jackson, R. J., and A. Kaminski. 1995. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA 1:985-1000. [PMC free article] [PubMed] [Google Scholar]
- 19.Jeffers, M., R. Paciucci, and A. Pellicer. 1990. Characterization of unr; a gene closely linked to N-ras. Nucleic Acids Res. 18:4891-4899. [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen, K. B., K. Musunuru, H. A. Lewis, S. K. Burley, and R. B. Darnell. 2000. The tetranucleotide UCAY directs the specific recognition of RNA by the Nova K-homology 3 domain. Proc. Natl. Acad. Sci. USA 97:5740-5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawamura, N., M. Kohara, S. Abe, T. Komatsu, K. Tago, M. Arita, and A. Nomoto. 1989. Determinants in the 5′ noncoding region of poliovirus Sabin 1 RNA that influence the attenuation phenotype. J. Virol. 63:1302-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kikuchi, T., M. Ichikawa, J. Arai, H. Tateiwa, L. Fu, K. Higuchi, and N. Yoshimura. 2000. Molecular cloning and characterization of a new neuron-specific homologue of rat polypyrimidine tract binding protein. J. Biochem. (Tokyo) 128:811-821. [DOI] [PubMed] [Google Scholar]
- 23.Kolupaeva, V. G., C. U. Hellen, and I. N. Shatsky. 1996. Structural analysis of the interaction of the pyrimidine tract-binding protein with the internal ribosomal entry site of encephalomyocarditis virus and foot-and-mouth disease virus RNAs. RNA 2:1199-1212. [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar, D. V., A. Nighorn, and P. A. St John. 2002. Role of Nova-1 in regulating α2N, a novel glycine receptor splice variant, in developing spinal cord neurons. J. Neurobiol. 52:156-165. [DOI] [PubMed] [Google Scholar]
- 25.La Monica, N., J. W. Almond, and V. R. Racaniello. 1987. A mouse model for poliovirus neurovirulence identifies mutations that attenuate the virus for humans. J. Virol. 61:2917-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.La Monica, N., and V. R. Racaniello. 1989. Differences in replication of attenuated and neurovirulent polioviruses in human neuroblastoma cell line SH-SY5Y. J. Virol. 63:2357-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leffers, H., K. Dejgaard, and J. E. Celis. 1995. Characterisation of two major cellular poly(rC)-binding human proteins, each containing three K-homologous (KH) domains. Eur. J. Biochem. 230:447-453. [PubMed] [Google Scholar]
- 28.Lillevali, K., A. Kulla, and T. Ord. 2001. Comparative expression analysis of the genes encoding polypyrimidine tract binding protein (PTB) and its neural homologue (brPTB) in prenatal and postnatal mouse brain. Mech. Dev. 101:217-220. [DOI] [PubMed] [Google Scholar]
- 29.Makeyev, A. V., A. N. Chkheidze, and S. A. Liebhaber. 1999. A set of highly conserved RNA-binding proteins, αCP-1 and αCP-2, implicated in mRNA stabilization, are coexpressed from an intronless gene and its intron-containing paralog. J. Biol. Chem. 274:24849-24857. [DOI] [PubMed] [Google Scholar]
- 30.Markovtsov, V., J. M. Nikolic, J. A. Goldman, C. W. Turck, M. Y. Chou, and D. L. Black. 2000. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol. 20:7463-7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meerovitch, K., J. Pelletier, and N. Sonenberg. 1989. A cellular protein that binds to the 5′-noncoding region of poliovirus RNA: implications for internal translation initiation. Genes Dev. 3:1026-1034. [DOI] [PubMed] [Google Scholar]
- 32.Michael, W. M., H. Siomi, M. Choi, S. Pinol-Roma, S. Nakielny, Q. Liu, and G. Dreyfuss. 1995. Signal sequences that target nuclear import and nuclear export of pre-mRNA-binding proteins. Cold Spring Harbor Symp. Quant. Biol. 60:663-668. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell, S. A., K. A. Spriggs, M. J. Coldwell, R. J. Jackson, and A. E. Willis. 2003. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol. Cell 11:757-771. [DOI] [PubMed] [Google Scholar]
- 34.Momose, T., A. Tonegawa, J. Takeuchi, H. Ogawa, K. Umesono, and K. Yasuda. 1999. Efficient targeting of gene expression in chicken embryos by microelectroporation. Dev. Growth Differ. 41:335-344. [DOI] [PubMed] [Google Scholar]
- 35.Moss, E. G., R. E. O'Neill, and V. R. Racaniello. 1989. Mapping of attenuating sequences of an avirulent poliovirus type 2 strain. J. Virol. 63:1884-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muramatsu, T., Y. Mizutani, Y. Ohmori, and J. Okumura. 1997. Comparison of three nonviral transfection methods for foreign gene expression in early chicken embryos in ovo. Biochem. Biophys. Res. Commun. 230:376-380. [DOI] [PubMed] [Google Scholar]
- 37.Ochs, K., L. Saleh, G. Bassili, V. H. Sonntag, A. Zeller, and M. Niepmann. 2002. Interaction of translation initiation factor eIF4B with the poliovirus internal ribosome entry site. J. Virol. 76:2113-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ochs, K., A. Zeller, L. Saleh, G. Bassili, Y. Song, A. Sonntag, and M. Niepmann. 2003. Impaired binding of standard initiation factors mediates poliovirus translation attenuation. J. Virol. 77:115-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pestova, T. V., C. U. Hellen, and I. N. Shatsky. 1996. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16:6859-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pestova, T. V., C. U. Hellen, and E. Wimmer. 1991. Translation of poliovirus RNA: role of an essential cis-acting oligopyrimidine element within the 5′ nontranslated region and involvement of a cellular 57-kilodalton protein. J. Virol. 65:6194-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pestova, T. V., V. G. Kolupaeva, I. B. Lomakin, E. V. Pilipenko, I. N. Shatsky, V. I. Agol, and C. U. Hellen. 2001. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. USA 98:7029-7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pilipenko, E. V., V. M. Blinov, L. I. Romanova, A. N. Sinyakov, S. V. Maslova, and V. I. Agol. 1989. Conserved structural domains in the 5′-untranslated region of picornaviral genomes: an analysis of the segment controlling translation and neurovirulence. Virology 168:201-209. [DOI] [PubMed] [Google Scholar]
- 43.Pilipenko, E. V., E. G. Viktorova, S. T. Guest, V. I. Agol, and R. P. Roos. 2001. Cell-specific proteins regulate viral RNA translation and virus-induced disease. EMBO J. 20:6899-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Polydorides, A. D., H. J. Okano, Y. Y. Yang, G. Stefani, and R. B. Darnell. 2000. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc. Natl. Acad. Sci. USA 97:6350-6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reynolds, J. E., A. Kaminski, H. J. Kettinen, K. Grace, B. E. Clarke, A. R. Carroll, D. J. Rowlands, and R. J. Jackson. 1995. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 14:6010-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sabin, A. B. 1985. Oral poliovirus vaccine: history of its development and use and current challenge to eliminate poliomyelitis from the world. J. Infect. Dis. 151:420-436. [DOI] [PubMed] [Google Scholar]
- 47.Svitkin, Y. V., N. Cammack, P. D. Minor, and J. W. Almond. 1990. Translation deficiency of the Sabin type 3 poliovirus genome: association with an attenuating mutation C472-U. Virology 175:103-109. [DOI] [PubMed] [Google Scholar]
- 48.Svitkin, Y. V., S. V. Maslova, and V. I. Agol. 1985. The genomes of attenuated and virulent poliovirus strains differ in their in vitro translation efficiencies. Virology 147:243-252. [DOI] [PubMed] [Google Scholar]
- 49.Westrop, G. D., K. A. Wareham, D. M. A. Evans, G. Dunn, P. D. Minor, D. I. Magrath, F. Taffs, S. Marsden, M. A. Skinner, G. C. Schild, and J. W. Almond. 1989. Genetic basis of attenuation of the Sabin type 3 oral poliovirus vaccine. J. Virol. 63:1338-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yanagiya, A., S. Ohka, N. Hashida, M. Okamura, C. Taya, N. Kamoshita, K. Iwasaki, Y. Sasaki, H. Yonekawa, and A. Nomoto. 2003. Tissue-specific replicating capacity of a chimeric poliovirus that carries the internal ribosome entry site of hepatitis C virus in a new mouse model transgenic for the human poliovirus receptor. J. Virol. 77:10479-10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, Y. Y., G. L. Yin, and R. B. Darnell. 1998. The neuronal RNA-binding protein Nova-2 is implicated as the autoantigen targeted in POMA patients with dementia. Proc. Natl. Acad. Sci. USA 95:13254-13259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zeng, Y., E. J. Wagner, and B. R. Cullen. 2002. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell 9:1327-1333. [DOI] [PubMed] [Google Scholar]