Abstract
The genome of some icosahedral RNA viruses plays an essential role in capsid assembly and structure. In T=3 particles of the nodavirus Pariacoto virus (PaV), a remarkable 35% of the single-stranded RNA genome is icosahedrally ordered. This ordered RNA can be visualized at high resolution by X-ray crystallography as a dodecahedral cage consisting of 30 24-nucleotide A-form RNA duplex segments that each underlie a twofold icosahedral axis of the virus particle and interact extensively with the basic N-terminal region of 60 subunits of the capsid protein. To examine whether the PaV genome is a specific determinant of the RNA structure, we produced virus-like particles (VLPs) by expressing the wild-type capsid protein open reading frame from a recombinant baculovirus. VLPs produced by this system encapsidated similar total amounts of RNA as authentic virus particles, but only about 6% of this RNA was PaV specific, the rest being of cellular or baculovirus origin. Examination of the VLPs by electron cryomicroscopy and image reconstruction at 15.4-Å resolution showed that the encapsidated RNA formed a dodecahedral cage similar to that of wild-type particles. These results demonstrate that the specific nucleotide sequence of the PaV genome is not required to form the dodecahedral cage of ordered RNA.
Analysis of the three-dimensional structure of icosahedral viruses has provided insights into the function and biology of virus particles. The techniques used rely on the symmetry of the particles, and therefore less is known about the structure of the encapsidated genome. The nucleic acid is seen only where it is arranged with symmetry that correlates with that of the icosahedrally ordered protein shell. Nevertheless, a portion of the genome has been visualized in several icosahedral viruses with positive-sense RNA genomes (reviewed in reference 22). Although these viruses contain single-stranded genomes, the RNA visualized by X-ray crystallography in both T=1 and T=3 virus particles predominantly resembles A-form duplex RNA, suggesting that the conformation of the RNA encapsidated in these particles is influenced by factors other than the genome sequence. In the 3-Å X-ray crystallographic structure of Pariacoto virus (PaV), a T=3 nodavirus, an unprecedented 1,500 nucleotides (nt) of the 4,322-nt genome were visible (27), providing an opportunity to examine the factors involved in ordering of the RNA.
The structure and assembly of viruses from the Alphanodavirus genus of the Nodaviridae family have been studied extensively (reviewed in references 10 and 25). Nodaviruses are positive-sense RNA viruses that have 30-nm nonenveloped particles with T=3 icosahedral symmetry. Each particle is assembled from 180 copies of a capsid protein precursor (alpha), which is autocatalytically cleaved following assembly into the two mature capsid proteins beta and gamma (7). A copy of each of the two genome segments, RNA1 (3 kb) and RNA2 (1.3 kb), are coencapsidated in each virus particle (16, 21). Authentic nodavirus particles do not encapsidate nonviral RNAs or the subgenomic RNA3 (0.4 kb), which is synthesized during replication, indicating that there is specific selectivity for genomic RNA during particle assembly. Early data suggested that a stem-loop in RNA2 was the signal for encapsidation into the particles of the nodavirus Flock House virus (FHV) (31). However, expression of the capsid protein of FHV from RNA2 in the absence of RNA1 results in the assembly of virus-like particles (VLPs) that are morphologically indistinguishable from wild-type (wt) FHV particles (24); these particles encapsidate only 19% of the wt levels of RNA2, the rest of the RNA being heterologous in origin (15). In addition, replicating FHV RNAs are encapsidated more specifically than nonreplicating RNAs, and it has been suggested that this may be related to temporal regulation of capsid protein synthesis, RNA conformation, or factors related to the local environment within the cell (15). These data indicate that selective coencapsidation of nodavirus RNAs is a complex process.
Structural studies of two alphanodaviruses, FHV and black beetle virus, indicated that the genomic RNA forms an integral part of the capsid structure (5, 29), with a 10-nt duplex of RNA visible in the crystal structures forming part of a molecular switch that produces flat contacts between the asymmetric protein units along each twofold axis. The 3-Å crystal structure of wt PaV particles shared many features with earlier structures of nodavirus particles (27). In addition, 35% of the genomic RNA (a total of 1,500 nt) was visible and was shown to interact extensively with the N-terminal region of the 60 subunits of the capsid protein that surrounded the 12 fivefold symmetry axes. The RNA duplexes at the twofold axis of PaV particles were 24 nt long with an additional unpaired nucleotide at each end of the duplex, and together they formed a dodecahedral cage. Electron cryomicroscopy (cryoEM) and image reconstruction also clearly showed the dodecahedral RNA cage (27). The RNA duplex fitted well with a model based on A-form double-stranded RNA (dsRNA). However, the genome of PaV consists of two single-stranded segments, a 3,011-nt RNA1 that encodes the replicase and a 1,311-nt RNA2 that encodes the capsid proteins (13, 14, 30), and the primary sequence of the genome is incompatible with the degree of Watson-Crick base pairing implied by the observed dsRNA. Therefore, it seemed likely that interaction(s) with the capsid proteins was involved in the formation of the RNA structure.
To investigate the sequence-specific contribution of the genomic RNA to its three-dimensional arrangement in particles, we analyzed PaV VLPs that had a wt protein content but encapsidated predominantly nonviral RNAs. To minimize the amount of viral sequences present in our system we expressed only the 401-amino-acid capsid protein open reading frame (ORF) (without the RNA2 5′ and 3′ untranslated regions) from a recombinant baculovirus. We purified VLPs and determined that they encapsidated total amounts of RNA similar to those of wt PaV particles, but only about 6% of this RNA was derived from viral sequences. Image reconstruction from cryoEM showed that the degree of icosahedral order imposed on the encapsidated heterologous RNA was similar to that of viral genomic RNA in wt virus particles at comparable resolution.
MATERIALS AND METHODS
Cells and virus.
Spodoptera exigua (Se-1) cells (8) were maintained in static culture at 28°C in Excell-401 medium (JRH Biologicals) supplemented with 10% heat-inactivated fetal bovine serum and antibiotics. The baby hamster kidney-derived cell line BSR/T7-5 (2) was grown at 37°C as described previously (11). Clonal wt PaV was purified from inoculated Galleria mellonella larvae (11).
Baculovirus construction and growth.
To produce VLPs the PaV capsid protein was expressed by using the Bac-to-Bac expression system (Invitrogen). To generate the recombinant baculovirus bCAP, the PaV capsid protein ORF was amplified by PCR with Pfu-turbo (Stratagene) from the plasmid PaV2(0,0) (14) using two oligonucleotides. The first (5′ AAACTGCAGTTACATGTACATTCCTGTTAGTTGATG 3′) annealed to nucleotides 1202 to 1228 of RNA2 (underlined), which encode the C terminus of the capsid protein and the stop codon next to a PstI site. The second oligonucleotide (5′ GAATTCAGCAAGATGGTATCAAGAACTAAGAATCG 3′) included an EcoRI site followed by the nucleotides AGC and annealed to the nucleotides complementary to nucleotides 20 to 45 of RNA2 (underlined), which encode the start codon and the N terminus of the capsid protein. The EcoRI/PstI-digested PCR-amplified fragment was ligated into pFASTBAC-DUAL so that the capsid ORF was under the control of the polyhedrin promoter with the sequence context surrounding the translation initiation codon identical to the efficiently translated GP64 (3). The sequence of PaV capsid ORF was verified to be identical to that of wt PaV (14).
Following transposition of the expression cassette into the bacmid carried by E. coli DH10Bac (Invitrogen), bacmid DNA was prepared and an 80% confluent monolayer of Se-1 cells was transfected with bacmid DNA, using Cellfectin (Invitrogen). After 5 h of incubation at 28°C the transfection mix was removed and replaced by ExCell-401 containing serum. Three days posttransfection the baculovirus containing cell supernatant was recovered and clarified by low-speed centrifugation. The resulting recombinant baculovirus bCAP was passaged once in Se-1 cells and was titrated by plaque assay on Se-1 cells.
Purification of VLPs.
Se-1 cells were seeded in 75-cm2 flasks and, upon reaching 80% confluency (approximately 3 × 107 cells/flask), were infected with bCAP at a multiplicity of infection of 20 PFU/cell. Following 4 days of incubation at 28°C, cells were lysed by addition of NP-40 to a final concentration of 0.5%. After 30 min at room temperature the cell lysates were frozen and thawed, clarified by centrifugation at 9,000 × g for 15 min, and treated with RNase A (10 ng/ml) for 2 h at room temperature. VLPs were then pelleted through a 30% sucrose cushion and purified on sucrose gradients as previously described (12). The yield of purified VLPs was about 200 μg per flask. Proteins were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and on Western blots as previously described (11).
Preparation of radiolabeled PaV and VLPs.
To radiolabel PaV, confluent BSR-T7/5 cells in 35-mm-diameter dishes were transfected with 50 ng of PaV virion RNA as previously described (11). After 3 days of incubation at 28°C the medium was replaced with medium containing 10 μg of actinomycin-D/ml for 20 min. The products of RNA replication were then metabolically labeled by incorporation of [3H]uridine (20 μCi/ml) for 5 h in medium containing 10 μg of actinomycin-D/ml. After a 24-h chase in complete medium, the cells were lysed by addition of NP-40 and were stored at −20°C.
To radiolabel VLPs, Se-1 cells were infected with bCAP and 3 days later the cell medium was removed and the products of protein synthesis were labeled by incorporation of [35S]Met-Cys (5 μCi/ml) for 6 h at 28°C in Sf900II medium lacking Met and Cys. After an 18-h chase by addition of complete ExCell-401 medium the cells were lysed by addition of NP-40 and stored at −20°C.
Cell lysates containing labeled PaV and VLPs were treated with RNase A, pelleted through 30% sucrose cushions, resuspended in 50 mM sodium phosphate buffer (pH 7.2), and stored at −20°C.
Analysis of labeled particles on gradients.
35S-labeled VLPs and 3H-labeled PaV were cosedimented on sucrose gradients as described previously (12) and were cobanded on CsCl gradients following the method described by Scotti (26). Briefly, radiolabeled particles were fixed in 3.7% formaldehyde at 4°C overnight, and then 200 μl of particles in 50 mM sodium phosphate buffer (pH 7.2) was mixed with 5.9 ml of 27% (wt/wt) CsCl in 50 mM sodium phosphate buffer (pH 7.2), layered on 5.9 ml of 42% (wt/wt) CsCl in 50 mM sodium phosphate buffer (pH 7.2), and centrifuged at 130,000 × g for 24 h at 20°C.
Following centrifugation, gradients were harvested on a piston gradient fractionator (Biocomp). The top 8 mm of the gradient was removed, and then 30 fractions each of 2 mm (about 280 μl) were collected with an air rinse for 2 s between each fraction. Aliquots of 100 μl from each fraction were diluted with 300 μl of water, mixed with 3.5 ml of Budget-solve scintillation cocktail (RPI), and counted by double-label liquid scintillation spectrometry in two windows: 5 to 200 for 3H and 240 to 655 for 35S.
RNA extraction and analysis.
RNA was extracted using an RNAgents kit (Promega), and samples were resolved on denaturing 1% agarose-formaldehyde gels and visualized by staining with Sybr Gold nucleic acid gel stain (Molecular Probes). For Northern blot analysis RNAs were transferred to Nytran nylon membranes and were hybridized with 32P-labeled RNA probes using conditions described previously (23). Probes designed to hybridize to PaV genomic RNA were generated by in vitro transcription using a MAXIscript kit (Ambion) with SP6 polymerase from PCR templates that included the SP6 promoter adjacent to RNA1 (nt 2598 to 3011) or RNA2 (nt 908 to 1311) sequences. The RNA2 probe included 47 nt of heterologous sequence at the 3′ end. Hybridized membranes were exposed to a phosphor screen and were visualized by a digital radioactivity imaging system (Molecular Dynamics Storm).
RNase protection assay.
RNase protection assays were performed using the RPA III system (Ambion) following the manufacturer's instructions. Briefly, 32P-labeled probes were made for RNA2 as described above and were gel purified on 5% acrylamide-urea gels. Encapsidated RNAs were extracted from VLPs and PaV particles, and the concentration of RNA samples was calculated from absorbance at an optical density of 260 nm (OD260). The RNAs were diluted and the RNA concentration was confirmed by triplicate measurements of the absorbance at OD260 prior to hybridization of the RNA with a molar excess of probe overnight at 42°C. The samples were digested with RNase A/T1 (1/150 dilution of supplied enzyme mix) for 30 min at 37°C. Following precipitation, the protected probe was resolved by electrophoresis on denaturing acrylamide gels. Fixed and dried gels were exposed to a phosphor screen, and the protected RNA products were quantitated using ImageQuant software (Amersham Biosciences Products).
cryoEM and image reconstruction.
The VLPs were freeze-hydrated on holey electron microscopy grids and were examined by cryoEM as described previously (27). Briefly, a 5-μl droplet of VLP sample was applied onto a previously glow-discharged copper grid coated with a holey carbon film. After removal of the excess solution by blotting, the grid was plunged into ethane slush and transferred into liquid nitrogen. The grid was inserted into a Phillips CM120 electron microscope that used a Gatan 626 cryostage. Focal pairs of electron micrographs were recorded under the low-dose condition at a magnification of 45,000×. Electron micrographs were digitized with a Zeiss scanner. The original step size for scanning was 7 μm, and the images were averaged to yield a pixel size of 4.67 Å. A total of 3,346 particles were boxed from the closer-to-focus micrographs with the program EMAN (19) and were used in image reconstruction with the program SPIDER (6). As the particles were oriented preferentially with the icosahedral threefold axes perpendicular to the grids (Fig. 1A), we tilted the grids by 20° to compensate for the uneven angular distribution. Among the particles used in reconstruction, 2,052 were tilted images. The contrast transfer function was not corrected, as the positions of the first zero of the contrast transfer functions computed from both tilted and nontilted particles were beyond the resolution of the reconstruction. The resolution was 15.4 Å as estimated with the Fourier shell correlation method using a criterion of 0.5. To facilitate comparison with the cryoEM structure of wt PaV (27), the 15.4-Å VLP structure was computationally filtered to yield a 22-Å-resolution structure. The images shown in Fig. 5A to C were contoured at 1 σ above background. The difference maps shown in panels D, E, and F were contoured at 1.7, 1.0, and 0.8 σ above background, respectively.
FIG. 1.
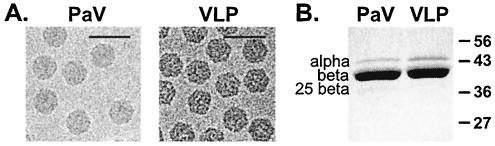
Analysis of VLPs produced from bCAP in Se-1 cells. (A) Electron cryomicrographs of gradient purified PaV and VLPs recorded at underfocus of 2.9 and 2.6 μm, respectively. The scale bar represents 500 Å. (B) Protein composition of wt PaV and VLPs. The proteins in purified particles were resolved by electrophoresis on an SDS-10% polyacrylamide gel and visualized by Coomassie blue staining. The wt PaV capsid proteins are indicated on the left (the cleaved form of capsid protein initiated at Met25 is indicated as 25 beta), and molecular mass markers are shown to the right in kilodaltons.
FIG. 5.
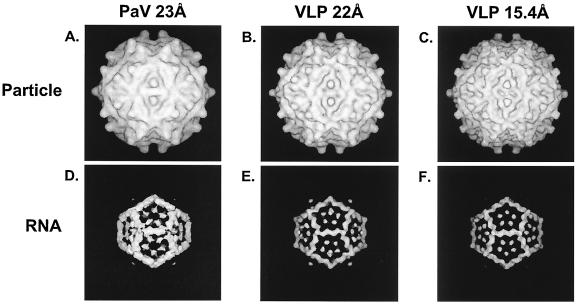
Three-dimensional reconstructed images of wt PaV and VLPs. CryoEM maps of PaV and VLP are shown for the total densities (A to C) and difference maps (D to F) generated by subtraction of the capsid protein density as determined by the crystal structure of PaV. The PaV reconstructions are shown at 23-Å resolution (A and D) (27), and the VLP map was determined at 15.4-Å resolution (C and F) and is also shown computationally filtered at 22 Å (B and E). The RNA cage identified in wt PaV is also visible in the VLPs.
RESULTS
VLPs produced from a recombinant baculovirus.
To examine the RNA encapsidated in PaV VLPs, we expressed the PaV capsid protein precursor from a recombinant baculovirus (bCAP) in which the entire ORF of the PaV capsid protein was cloned under control of the polyhedrin promoter. The recombinant baculovirus bCAP was used to infect Se-1 cells, and abundant capsid protein was expressed in the absence of PaV RNA1 and RNA replication. Electron microscopy of VLPs purified from bCAP-infected cells showed abundant uniform particles with morphology similar to that of wt PaV virions (Fig. 1A). Their protein components were examined by SDS-polyacrylamide gel electrophoresis and visualized by staining with Coomassie blue (Fig. 1B). The protein profiles of wt PaV and VLPs were similar, with the major protein in the VLPs comigrating with the mature cleaved form of the major capsid protein in wt PaV, protein beta. Both wt PaV and VLPs contained a small amount of the uncleaved capsid protein precursor (alpha), although this protein was slightly more abundant in the VLPs. The VLPs also contained a small amount of a 36-kDa protein that comigrated with a minor capsid protein which is derived from translational initiation at the second AUG of the capsid protein ORF (12). These results validated the synthesis, assembly, and maturational cleavage of the PaV capsid protein precursor expressed from bCAP in Se-1 cells.
Physical properties of VLPs.
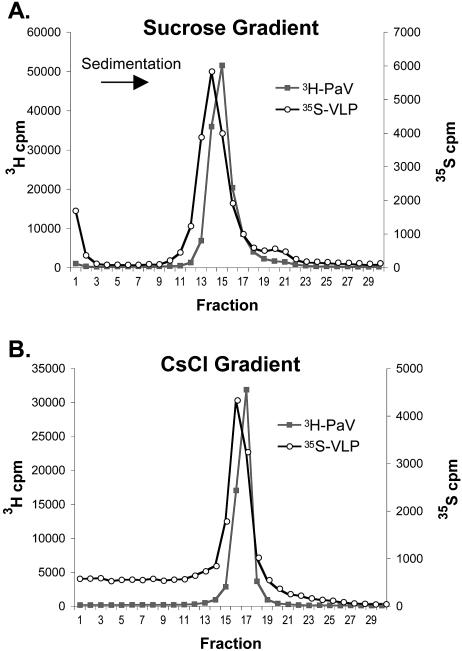
To compare their physical properties, wt virions and VLPs were differentially radiolabeled and analyzed by sedimentation on sucrose velocity gradients and by buoyant density on isopycnic CsCl gradients. VLPs were labeled by incorporation of [35S]Met-Cys into their proteins, whereas wt PaV particles were labeled by incorporation of [3H]uridine into their RNA. Samples of semipurified wt PaV and VLPs were mixed and cosedimented through a 15 to 45% sucrose velocity gradient. Gradient fractions were assayed by double-label scintillation spectrometry. To minimize the 35S spillover into the 3H window, about 10-fold more 3H counts were loaded on each gradient. wt PaV particles sedimented as a single sharp peak near the middle of the gradient centered on fraction 15 (Fig. 2A). The major peak of VLPs was in a similar position but reproducibly sedimented one fraction slower than wt PaV. The major VLP peak was slightly broader than that of wt virions, raising the possibility that the VLPs were more heterogeneous in size and/or mass. VLP preparations also reproducibly contained a second very minor peak that sedimented about six fractions faster than the main peak (fraction 21). Western blot analysis on fractions from VLP gradients (data not shown) showed that the minor peak contained capsid proteins, and a similar minor protein peak was detected by Western blot of PaV gradient fractions (data not shown). The ratio of uncleaved to cleaved protein did not appear to vary across the fractions. We attribute the 35S near the top of the gradient to label not incorporated in VLPs.
FIG. 2.
Ultracentrifugation analysis of radiolabeled PaV and VLPs. Particles were labeled with either [35S]Met-Cys (VLPs) or [3H]uridine (PaV) and purified by pelleting through 30% sucrose cushions. The particles were mixed and sedimented through a 15 to 45% sucrose gradient (A) or centrifuged to equilibrium on a CsCl gradient (B). Thirty fractions were collected, and the radioactive counts in each fraction were determined. The top of each gradient is on the left.
To compare the buoyant density of VLPs with that of wt PaV, differentially radiolabeled samples were mixed and subjected to isopycnic CsCl density gradient ultracentrifugation. Initial experiments indicated that, like some other nodaviruses (18, 21), PaV particles were unstable when analyzed on CsCl gradients (data not shown). Therefore, we fixed the particles with formaldehyde before CsCl gradient analysis. Formaldehyde fixation had no effect on the sedimentation of particles in sucrose gradients (data not shown). Fixed, differentially labeled wt PaV and VLPs were mixed with 27% CsCl, layered onto an equal volume of 42% CsCl, and centrifuged to equilibrium at 130,000 × g. The gradients were fractionated, and the fractions were assayed by double-label scintillation spectrometry (Fig. 2B). A single sharp 3H peak was detected centering on fraction 17 with a density of 1.31 g/ml. The majority of the 35S VLP signal was found in a single peak that equilibrated within one fraction of wt PaV (fraction 16; density, 1.30 g/ml). We interpreted the similarity in buoyant density of the two types of particles as indicating that they contained a similar amount of RNA. The VLP peak was slightly broader than that of wt PaV, suggesting that the VLPs may be more heterogeneous in density (20). When the wt PaV and VLP samples were premixed with the 42% CsCl and subjected to centrifugal flotation, the relative peak positions were similar to those shown in Fig. 2B (data not shown). The baseline of the 35S signal was reproducibly elevated in the half of the gradient that the particles were loaded; we attribute this signal to unincorporated label. As an independent measure, the RNA:protein ratios were calculated from ODs of the particles measured at 260 and 280 nm using the equations described by Layne (17). In agreement with the isopycnic gradient analysis, these calculations suggested that the RNA:protein ratio was slightly lower in VLPs than in PaV virions. We interpreted these results as indicating that the VLPs contained amounts of RNA similar to those of wt PaV virions.
RNA encapsidated in VLPs.
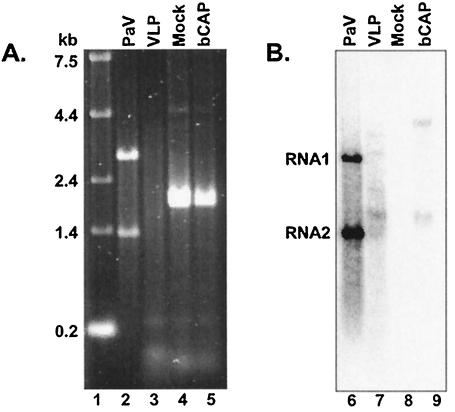
To characterize the RNAs encapsidated by VLPs we extracted RNAs from sucrose gradient-purified unlabeled wt PaV and VLPs and examined them by electrophoresis on denaturing agarose gels alongside total cellular RNA from mock- or bCAP-infected Se-1 cells (Fig. 3A). The mock- and bCAP-infected cell samples appeared very similar (Fig. 3A, lanes 4 and 5), showing a major doublet of rRNA of about 2 kb with a background smear of RNAs ranging between about 0.1 to 5.0 kb. As expected, wt PaV (Fig. 3A, lane 2) contained two RNAs of about 3 and 1.3 kb that correspond to genomic RNA1 and -2. In contrast, the VLPs contained a smear of RNAs ranging in size from less than 0.2 kb to about 4 kb with no predominant species (Fig. 3A, lane 3).
FIG. 3.
Analysis of encapsidated RNAs. Duplicate samples of RNA extracted from purified PaV (lanes 2 and 6), VLPs (lanes 3 and 7), or Se-1 cells that were mock infected (lanes 4 and 8) or infected with bCAP (lanes 5 and 9) were resolved by electrophoresis on a denaturing 1% agarose-formaldehyde gel. The amount of RNA loaded was 1 μg in lanes 2 and 3; 2 μg in lanes 4 and 5; 50 ng in lanes 6 and 7; and 500 ng in lanes 8 and 9. The gel was divided in two, and RNA was visualized by staining with Sybr-gold (A) or was transferred to Nytran nylon membranes and hybridized with probes for PaV RNA1 and -2 (B). The sizes of RNA markers (lane 1) and the positions of PaV RNA1 and -2 are shown to the left of panels A and B, respectively.
The size distribution of RNAs that contained PaV RNA2-related sequences was examined by Northern blot hybridization of duplicate RNA samples loaded on the same gel as that used for Fig. 3A. The RNA probe was complementary to the last 322 nt of the capsid protein ORF of PaV RNA2. A probe complementary to the 3′ 413 nt of RNA1 was included to visualize RNA1 in the wt PaV RNA sample as a size marker (Fig. 3B, lane 6). The PaV RNA2 probe detected two RNAs of about 1.8 and 4.4 kb in bCAP-infected Se-1 cells (Fig. 3B, lane 9). The 1.8-kb RNA is predicted to be the polyadenylated bCAP transcript that includes the PaV capsid protein ORF. The origin of the 4.4-kb RNA is not clear, but it may represent a readthrough transcript that terminated downstream. No PaV-related sequences were detected in the mock-infected Se-1 cellular RNA sample (lane 8). The 1.8-kb RNA was the major species among several RNA2-related molecules encapsidated in VLPs, but the 4.4-kb RNA was not detected (Fig. 3B, lane 7). A probe complementary to the 5′ region of RNA2 yielded an identical pattern of bands (data not shown), indicating that the RNAs detected in Fig. 3B, lane 7, contained both ends of the capsid protein ORF. Because the banding pattern of VLP encapsidated RNAs observed by hybridization (Fig. 3B, lane 7) was not evident in the stained gel (Fig. 3A, lane 3), we concluded that not all the RNA encapsidated in the VLPs contained PaV RNA2 sequences.
Quantitation of PaV RNA encapsidated in VLPs.
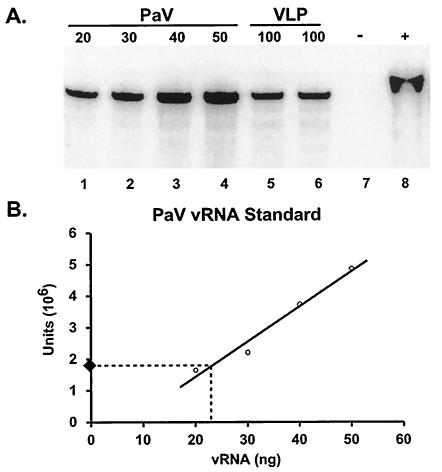
To measure the relative amount of RNA2-related sequences encapsidated in VLPs compared to that in wt PaV, RNAs extracted from the two types of particles were analyzed with a quantitative RNase protection assay. An RNA probe complementary to the 3′ 322 nt of the capsid protein ORF was hybridized in excess with the extracted RNA, unprotected probe was digested with a mixture of RNases, the protected RNA fragments were resolved on acrylamide gels, and the products were visualized by phosphorimaging. In the absence of RNase the probe remained intact (Fig. 4A, lane 8), whereas in the absence of template and the presence of RNase the probe was degraded (Fig. 4A, lane 7). A dilution series of PaV virion RNA (20 ng in lane 1, 30 ng in lane 2, 40 ng in lane 3, and 50 ng in lane 4) provided a standard curve for comparison of duplicate 100-ng samples of VLP RNA (lanes 5 and 6). For each sample a single major protected fragment was evident that was slightly smaller than the full-length probe due to removal of the unprotected noncomplementary 47 nt of the probe. An example of quantitation of the protected RNA in the wt PaV RNA samples (Fig. 4A, lanes 1 to 4) is graphed as a standard curve in Fig. 4B. The standard curve was repeated for each independent repetition of the experiment. Data pooled from several experiments (including two pools of RNA extracted independently from particles) showed that VLPs contained 22% (±1.4%) of the RNA2 content of wt PaV particles. Therefore, a maximum of one in four VLPs contained PaV-derived RNA.
FIG. 4.
Quantitation of RNA2 sequences encapsidated in VLPs. RNA was extracted from purified PaV and VLPs and was analyzed by RNase protection. (A) RNAs were quantitated by spectrometry and annealed with an excess of a 32P-labeled RNA2 probe. In the absence of template (lane 7) the probe was completely digested by RNase A/T1, and in the absence of RNase the probe remained intact (lane 8). The protected RNA in 20-, 30-, 40-, and 50-ng samples of wt virion RNA (lanes 1 to 4) was compared to that protected in 100-ng samples of VLP RNA (lanes 5 and 6). A phosphorimage of the dried gel is shown. (B) Quantitation of the protected RNAs from the gel shown in panel A is shown graphically in panel B. Each circle represents a data point from which the standard curve line was calculated. The black diamond represents the value obtained for protected RNA in 100 ng of VLP RNA (the average of the samples in lanes 5 and 6). The dotted line shows the intercept with the standard curve and an estimate for the equivalent amount of RNA2 sequences protected in wt virus RNA. Data pooled from this and several other experiments indicated that VLPs contained 22% (±1.4%) of the RNA2 content of wt PaV particles.
The similar buoyant densities of the VLPs and wt PaV particles (Fig. 2) suggested that they encapsidated a similar total amount of RNA. In wt PaV virions, the coencapsidated RNA1 (3,011 nt) and RNA2 (1,311 nt) segments constitute 70 and 30% of the total 4,322-nt RNA content, respectively. Because there was no RNA1 present in the VLP system, any RNA not related to the PaV capsid protein ORF (1,206 nt, 28% of genome) must have been of cellular or baculovirus origin. These considerations lead to the conclusion that only about 6% of the RNA encapsidated in VLPs consisted of PaV RNA sequence (i.e., 22 of 28%), with the remaining RNA being of heterologous origin.
Structure of RNA in VLPs.
To examine the three-dimensional arrangement of the RNA in the VLPs, sucrose gradient-purified particles were analyzed by image reconstruction from cryoEM. Three-dimensional reconstruction of the VLPs was computed, and the resolution was estimated to be 15.4 Å (Fig. 5C). The surface view of these particles resembled the 23-Å cryoEM reconstruction of wt PaV virions (27), with the major features, such as the protrusions at the quasi-threefold axis, being easily discernible in both reconstructions. The higher resolution VLP image showed greater surface detail than the PaV reconstruction (Fig. 5A), and the differences may be a consequence of the different sizes of the data sets and the resolution of the reconstruction. The VLP reconstruction was computationally filtered at 22-Å resolution (Fig. 5B), and the surface view more closely resembled that of the 23-Å PaV map (Fig. 5A). For example, the surface pits on the twofold axis in the 15.4-Å VLP structure were almost invisible at 22-Å resolution.
Because the structure of wt PaV was determined by X-ray crystallography at 3-Å resolution (27), the electron density attributable to the capsid proteins could be subtracted from the PaV reconstruction to reveal the icosahedrally ordered components of the encapsidated RNA. When this approach was applied to authentic PaV, the results showed an internal dodecahedral cage that fitted well with the atomic model of the duplex RNA underlying the icosahedral twofold axes derived from the crystal structure (27). Using a similar approach, the arrangement of the mostly heterologous RNAs encapsidated by the VLPs was examined by using the wt capsid protein density to calculate a difference map for the VLP reconstruction. The VLP difference map revealed a cage of density (Fig. 5E and F) that was similar in shape to the dodecahedral cage of RNA duplexes seen in authentic virions (Fig. 5D). As noted for the surface views, the internal VLP image was more refined than that of wt PaV, but we attribute this to the difference in resolution as before. The ratio of average RNA to average protein density was computed by masking the RNA and protein regions of the VLP and wt PaV reconstructions (data not shown). These calculations suggested that the relative RNA:protein density ratio was equal or greater for the VLP reconstruction compared to that of the wt PaV reconstruction.
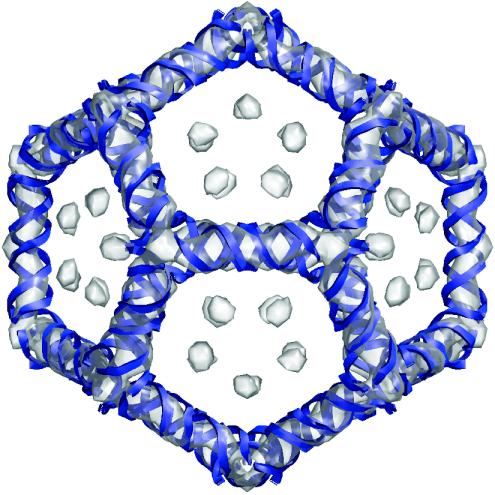
The evident removal of the capsid in the difference map is indicative of the high quality of the VLP reconstruction. At the higher resolution of the VLP difference image, the helical conformation of the RNA duplexes was more clearly visible. Superimposition of the 15.4-Å resolution VLP difference map with the RNA model from the PaV crystal structure (27) showed that the helical structure of the RNA seen in VLPs is consistent with the structure of RNA seen in wt PaV particles (Fig. 6). From these results, we conclude that a portion of the RNA encapsidated in VLPs formed a dodecahedral cage composed of duplexes situated under the twofold axes, and this cage was equivalent at this resolution to that seen in authentic PaV virions.
FIG. 6.
Superimposition of VLP and wt PaV RNA. The 15.4-Å VLP difference map (semitransparent) is shown superimposed on the model of the RNA cage (blue ribbons) from the PaV crystal structure (27).
DISCUSSION
Thirty-five percent of the viral RNA encapsidated in wt PaV particles is icosahedrally ordered (27). This ordered RNA was visualized by X-ray crystallography as a discontinuous cage formed from 30 24-nt A-form RNA duplex segments, with each duplex underlying a twofold icosahedral axis of the viral particle. At 3-Å resolution the electron density attributable to the ordered RNA becomes indistinct near and at the threefold axes where the duplexes meet, suggesting that this is the location of most or all of the connections between the icosahedrally ordered RNA and the remaining 65% of the genome. The ordered RNA interacts extensively with the basic N-terminal region of the quasi-equivalent A subunit of the capsid protein and neutralizes its charge. Together the RNA and the N terminus of the A subunit form a molecular switch which is integral to the formation of the T=3 icosahedral virions of PaV and other nodaviruses (5, 9, 27). The importance of RNA to particle formation is also demonstrated by the lack of formation of empty nodavirus capsids (5).
In this study we examined the specific contribution of the genomic RNA to formation of the icosahedrally ordered RNA conformation. To this end, VLPs were assembled from PaV capsid proteins expressed from a recombinant baculovirus. The PaV RNA sequence in the system was limited to the minimum required for capsid protein expression, i.e., the capsid protein ORF. The RNA-to-protein ratio of VLPs was compared to that of wt PaV virions using two independent approaches. The comparative buoyant density and RNA:protein ratios calculated from OD260 and OD280 indicated that the VLPs had a slightly lower ratio of RNA to protein than wt PaV virions. Because both types of particles were assembled from wt capsid proteins, we interpreted this to indicate that there was slightly less RNA encapsidated in VLPs than PaV virions. However, because neither of these methods directly measures the amount of RNA encapsidated, we concluded that the VLPs contain amounts of RNA similar to that of wt PaV. Quantitation of the encapsidated RNAs showed that the VLPs encapsidated only about 6% of the wt amount of PaV-specific RNAs. Thus, if VLPs and wt PaV virions contained similar total amounts of RNA, then only about 6% of the encapsidated RNA was PaV specific, the remaining RNA being of either cellular or baculovirus origin.
The three-dimensional arrangement of the VLP-encapsidated RNA was examined by cryoEM image reconstruction. To more clearly visualize the RNA, a difference map was produced by subtracting the icosahedrally ordered protein components seen by X-ray crystallography of native PaV particles (27). Implicit in this approach was the assumption that the structure of the capsid proteins in VLPs was indistinguishable from that in native particles. This assumption seemed reasonable, given that the vector-expressed protein was wt in sequence and the VLP protein profile was very similar to that of wt PaV (Fig. 1A). Strikingly, the difference map showed a dodecahedral cage of helical RNA density similar to that seen in wt PaV and was shown to fit well with the X-ray crystallographic model of A-form duplex RNA (Fig. 6). The level of RNA density in the VLP images was at least as great as that in wt virions. Because 35% of the encapsidated RNA was visible in wt virions and only about 6% of the RNA encapsidated in the VLPs was RNA2-related, we conclude that the observed three-dimensional arrangement reflects that of the predominant encapsidated RNA, which was heterologous in origin. These observations indicate that non-PaV RNA encapsidated in VLPs adopted a similar three-dimensional structure to that formed by the genomic RNAs in wt PaV particles.
There are several ways that RNA could contribute to its three-dimensional conformation in virions through either RNA-RNA or RNA-protein interactions. Both the generic ribose phosphate backbone and sequence-specific bases can be involved in these interactions. The data presented here imply that generic RNA interactions suffice to stabilize the dodecahedral cage of RNA duplexes within particles of PaV. Evidently the inner surface of the PaV capsid can confer the dodecahedral arrangement of duplex segments on RNA of generic sequence, presumably by relying primarily on interactions that involve the RNA backbone. Because the X-ray structure of PaV represents an averaging of all 30 RNA duplexes, it does not resolve individual specific bases and we therefore cannot rule out the possibility that the ordered RNA has a conserved sequence motif. However, it seems most likely that the dodecahedral cage is formed with little or no contribution from nucleotide sequence-specific interactions. Other structural elements that are not icosahedrally ordered and therefore not seen in this type of analysis may play important roles in particle assembly and stability.
The PaV RNA visualized by X-ray crystallography conforms to the geometry of A-form dsRNA (27). However, the nucleotide sequences of PaV RNA1 and -2 preclude the possibility of perfect Watson-Crick base pairing for more than a small fraction of the observed duplex (14). For example, the longest perfect inverted repeat anywhere in the PaV genome contains only 14 consecutive nucleotides. This strongly suggests that the RNA duplexes are stabilized less by Watson-Crick base pairing than by other interactions that are largely independent of the nucleotide sequence, such as base stacking and interactions between the ribose phosphate backbone and the capsid proteins. The formation of a similar helical structure from heterologous RNA in the PaV VLPs analyzed here supports this interpretation.
For the related nodavirus FHV, a shorter 10-nt duplex RNA was visualized in the X-ray crystal structure (5) and by cryoEM image reconstruction (4). The capsid protein sequences of FHV and PaV share only 41% identical amino acids (14). Before the present study, it was not clear whether the greater icosahedral order of the RNA in PaV capsids was determined by the capsid protein, programmed by the genomic RNA, or due to a combination of these factors. The clearly visible ordered RNA in PaV VLPs indicates that the direct contribution of the genomic sequence to the icosahedral RNA structure is minimal and suggests that the capsid protein is the main determinant in forming the 24-nt RNA duplexes. It appears that the capsid protein can organize the three-dimensional structure of heterologous RNAs with few, if any, nucleotide sequence constraints. This conclusion is consistent with the finding that synthetic FHV VLPs which were examined by X-ray scattering at low resolution (28) appeared to have RNA structure similar to that of wt FHV particles analyzed by cryoEM. Like the PaV VLPs, those from FHV contained predominantly heterologous RNAs (15, 24).
Although the icosahedral order of the RNA appears to be independent of the nucleotide sequence, the RNA encapsidated in nodavirus particles nevertheless plays an essential role in particle integrity. FHV VLPs that contained mainly nonviral RNA (15, 24) were more sensitive to protease digestion than wt particles, indicating that the authentic RNA plays a stabilizing role that is not fulfilled by heterologous RNAs (1). It is not yet known whether this difference is specifically related to the sequences, secondary structures, sizes, or other properties of the genomic RNAs.
During natural infection nodaviruses show high fidelity for specific encapsidation of their genomic RNAs. The determinants of specificity for PaV RNA encapsidation have not yet been investigated, and the experiments described here were not designed to address this question. Indeed, the RNA-protein interactions in fully assembled particles may not accurately reflect those responsible for RNA selection during the dynamic process of assembly. However, our results show that whatever the determinants of specificity are, PaV RNA itself is not necessary for formation of the structurally important RNA cage. Instead, it appears more likely that RNA duplex formation is a consequence of induced fit with the capsid proteins, whose juxtaposition creates specific binding sites for A-form duplex RNA. However, our results do not exclude the possibility that part of the selective mechanism for RNA encapsidation in PaV virions may be a requirement for the RNA to have the potential to form duplex elements and that not all RNAs have this potential. However, if such a requirement exists, it can evidently be satisfied by a wide variety of heterologous RNAs.
In conclusion, heterologous RNA encapsidated in PaV VLPs has icosahedrally ordered three-dimensional arrangement similar to that of genomic RNA encapsidated in wt virions. This demonstrates that factors other than the specific nucleotide sequence and other attributes of the genomic RNA are the main determinants of the formation of the structure. The significance of this RNA structure predicts that it may not be possible to form T=3 PaV capsids in the absence of the dodecahedral RNA cage. That is, mutations in the capsid protein that disrupt the formation of the RNA duplexes may also disrupt assembly so that particle formation is not achieved.
ADDENDUM IN PROOF
Recently Tihova et al. (M. Tihova, K. A. Dryden, T. L. Le, S. C. Harvey, J. E. Johnson, M. Yeager, and A. Schneemann, Virol. 78:2897-2905, 2004) showed that the RNA structure seen in VLPs of the nodavirus Flock house virus was also independent of the nucleotide sequence.
Acknowledgments
We thank our colleagues for discussions and critical reading of the manuscript.
This work was supported by grants to L.A.B. (NIH R01AI18270) and J.E.J. (NIH R01GM034220).
REFERENCES
- 1.Bothner, B., A. Schneemann, D. Marshall, V. Reddy, J. E. Johnson, and G. Siuzdak. 1999. Crystallographically identical virus capsids display different properties in solution. Nat. Struct. Biol. 6:114-116. [DOI] [PubMed] [Google Scholar]
- 2.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, M. J., J. Kuzio, and G. W. Blissard. 1999. Modulation of translational efficiency by contextual nucleotides flanking a baculovirus initiator AUG codon. Virology 259:369-383. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, R. H., V. S. Reddy, N. H. Olson, A. J. Fisher, T. S. Baker, and J. E. Johnson. 1994. Functional implications of quasi-equivalence in a T=3 icosahedral animal virus established by cryo-electron microscopy and X-ray crystallography. Structure 2:271-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fisher, A. J., and J. E. Johnson. 1993. Ordered duplex RNA controls capsid architecture in an icosahedral animal virus. Nature (London) 361:176-182. [DOI] [PubMed] [Google Scholar]
- 6.Frank, J., M. Radermacher, P. Penczek, J. Zhu, Y. Li, M. Ladjadj, and A. Leith. 1996. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116:190-199. [DOI] [PubMed] [Google Scholar]
- 7.Gallagher, T. M., and R. R. Rueckert. 1988. Assembly-dependent maturation cleavage in provirions of a small icosahedral insect ribovirus. J. Virol. 62:3399-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gelernter, W. D., and B. A. Federici. 1986. Continuous cell line from Spodoptera exigua (Lepidoptera: Noctuidae) that supports replication of nuclear polyhedrosis viruses from Spodoptera exigua and Autographa californica. J. Invertebr. Pathol. 48:199-207. [Google Scholar]
- 9.Hosur, M. V., T. Schmidt, R. C. Tucker, J. E. Johnson, T. M. Gallagher, B. H. Selling, and R. R. Rueckert. 1987. Structure of an insect virus at 3.0 Angstrom resolution. Proteins 2:167-176. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, J. E., and V. Reddy. 1998. Structural studies of nodaviruses and tetraviruses, p. 171-223. In L. K. Miller and L. A. Ball (ed.), The insect viruses. Plenum Publishing Corporation, New York, N.Y.
- 11.Johnson, K. N., and L. A. Ball. 2001. Recovery of infectious Pariacoto virus from cDNA clones and identification of susceptible cell lines. J. Virol. 75:12220-12227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson, K. N., and L. A. Ball. 2003. Virions of Pariacoto virus contain a minor protein translated from the second AUG codon of the capsid protein open reading frame. J. Gen. Virol. 84:2847-2852. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, K. N., K. L. Johnson, R. Dasgupta, T. Gratsch, and L. A. Ball. 2001. Comparisons among the larger genome segments of six nodaviruses and their encoded RNA replicases. J. Gen. Virol. 82:1855-1866. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, K. N., J. Zeddam, and L. A. Ball. 2000. Characterization and construction of functional cDNA clones of Pariacoto virus, the first Alphanodavirus isolated outside Australasia. J. Virol. 74:5123-5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krishna, N. K., D. Marshall, and A. Schneemann. 2003. Analysis of RNA packaging in wild-type and mosaic protein capsids of Flock House virus using recombinant baculovirus vectors. Virology 305:10-24. [DOI] [PubMed] [Google Scholar]
- 16.Krishna, N. K., and A. Schneemann. 1999. Formation of an RNA heterodimer upon heating of nodavirus particles. J. Virol. 73:1699-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Layne, E. 1957. Spectrophotometric and turbidimetric methods for measuring proteins. Methods Enzymol. 3:447-454. [Google Scholar]
- 18.Longworth, J. F., and G. P. Carey. 1976. A small RNA virus with a divided genome from Heteronychus arator (F.) [Coleoptera: Scarabaeidae]. J. Gen. Virol. 33:31-40. [DOI] [PubMed] [Google Scholar]
- 19.Ludtke, S. J., P. R. Baldwin, and W. Chiu. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82-97. [DOI] [PubMed] [Google Scholar]
- 20.Meselson, M., F. W. Stahl, and J. Vinograd. 1957. Equilibrium sedimentation of macromolecules in density gradients. Proc. Natl. Acad. Sci. USA 43:581-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newman, J. F. E., and F. Brown. 1977. Further physicochemical characterization of Nodamura virus. Evidence that the divided genome occurs in a single component. J. Gen. Virol. 38:83-95. [DOI] [PubMed] [Google Scholar]
- 22.Prasad, B. V., and P. E. Prevelige, Jr. 2003. Viral genome organization. Adv. Protein Chem. 64:219-258. [DOI] [PubMed] [Google Scholar]
- 23.Price, B. D., M. Roeder, and P. Ahlquist. 2000. DNA-directed expression of functional Flock House virus RNA1 derivatives in Saccharomyces cerevisiae, heterologous gene expression, and selective effects on subgenomic mRNA synthesis. J. Virol. 74:11724-11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneemann, A., R. Dasgupta, J. E. Johnson, and R. R. Rueckert. 1993. Use of recombinant baculoviruses in synthesis of morphologically distinct viruslike particles of flock house virus, a nodavirus. J. Virol. 67:2756-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneemann, A., V. Reddy, and J. E. Johnson. 1998. The structure and function of nodavirus particles: a paradigm for understanding chemical biology. Adv. Virus Res. 50:381-446. [DOI] [PubMed] [Google Scholar]
- 26.Scotti, P. D. 1985. The estimation of virus density in isopycnic cesium chloride gradients. J. Virol. Methods 12:149-160. [DOI] [PubMed] [Google Scholar]
- 27.Tang, L., K. N. Johnson, L. A. Ball, T. Lin, M. Yeager, and J. E. Johnson. 2001. The structure of Pariacoto virus reveals a dodecahedral cage of duplex RNA. Nat. Struct. Biol. 8:77-83. [DOI] [PubMed] [Google Scholar]
- 28.Tsuruta, H., V. S. Reddy, W. R. Wikoff, and J. E. Johnson. 1998. Imaging RNA and dynamic protein segments with low-resolution virus crystallography: experimental design, data processing and implications of electron density maps. J. Mol. Biol. 284:1439-1452. [DOI] [PubMed] [Google Scholar]
- 29.Wery, J.-P., V. S. Reddy, M. V. Hosur, and J. E. Johnson. 1994. The refined three-dimensional structure of an insect virus at 2.8 Å resolution. J. Mol. Biol. 235:565-586. [DOI] [PubMed] [Google Scholar]
- 30.Zeddam, J. L., J. L. Rodriguez, M. Ravallec, and A. Lagnaoui. 1999. A noda-like virus isolated from the sweetpotato pest Spodoptera eridania (Cramer) (Lep.; Noctuidae). J. Invertebr. Pathol. 74:267-274. [DOI] [PubMed] [Google Scholar]
- 31.Zhong, W., R. Dasgupta, and R. Rueckert. 1992. Evidence that the packaging signal for nodaviral RNA2 is a bulged stem-loop. Proc. Natl. Acad. Sci. USA 89:11146-11150. [DOI] [PMC free article] [PubMed] [Google Scholar]