Abstract
Like other positive-strand RNA viruses, hepatitis C virus (HCV) is believed to replicate its RNA in association with host cell cytoplasmic membranes. Because of its association with such membranes, NS4B, one of the virus's nonstructural proteins, may play an important role in this process, although the mechanistic details are not well understood. We identified a putative N-terminal amphipathic helix (AH) in NS4B that mediates membrane association. Introduction of site-directed mutations designed to disrupt the hydrophobic face of the AH abolishes the AH's ability to mediate membrane association. An AH in NS4B is conserved across HCV isolates. Completely disrupting the amphipathic nature of NS4B's N-terminal helix abolished HCV RNA replication, whereas partial disruption resulted in an intermediate level of replication. Finally, immunofluorescence studies revealed that HCV replication complex components were mislocalized in the AH-disrupted mutant. These results identify a key membrane-targeting domain which can form the basis for developing novel antiviral strategies.
Hepatitis C virus (HCV) is a positive-strand RNA virus that belongs to the family Flaviviridae and was classified into a separate genus, Hepacivirus (39). HCV infects over 100 million people worldwide and causes chronic hepatitis that can progress to liver cirrhosis and hepatocellular carcinoma (1, 11). Despite recent progress, current therapies remain inadequate for the majority of patients (30, 33, 50).
HCV's genome is composed of a 9.6-kb positive, single-stranded RNA molecule that encodes an ∼3,000-amino-acid polyprotein, which is proteolytically processed by cellular and viral proteinases into structural proteins (components of the mature virus) and nonstructural proteins (proteins proposed to be involved in the replication of the virus) (2, 8, 38). Like that of other positive-strand RNA viruses (3, 10, 18, 28, 39), HCV's RNA replication is believed to take place on cytoplasmic membranes (15, 21), although the details of the replication complex assembly and maintenance are largely unknown. A better understanding of these mechanistic details should help elucidate a fundamental stage in the viral life cycle and may reveal potential new targets for antiviral therapy.
The introduction of the high-efficiency HCV subgenomic replicon (4, 31) and, recently, of the full-length replicon (5, 6) enables the design and implementation of detailed molecular genetic studies of the HCV replication process. Such replicons contain all the cis and trans elements required for HCV RNA replication and allow studies of engineered HCV mutants (4, 16).
The function of NS4B, one of HCV's nonstructural proteins, is incompletely understood. NS4B is a membrane-associated protein that colocalizes predominantly with endoplasmic reticulum (ER) markers, suggesting an ER or ER-derived membrane localization (24, 26, 32, 42). This membrane association was shown to occur cotranslationally, and NS4B behaves biochemically as an integral membrane protein (24). The precise topology of NS4B with respect to the membrane in which it resides is not clear. It is predicted, however, to harbor at least four transmembrane domains (TMDs) (24, 32) which are believed to be responsible for conferring the protein's membrane association. Lundin et al. recently provided evidence supporting the idea that at least two of the four TMDs traverse the membrane (32), suggesting that these TMDs are involved in enabling at least some of the protein's observed interactions with membranes. Apparent activities in translation inhibition (17, 25), modulation of NS5B enzymatic function (37), and transformation (36) have been reported for NS4B. Their relevance to the HCV life cycle or natural infections awaits further study.
NS4B has also recently been implicated in the perturbation of intracellular membranes and in the formation of the membranous web structures (15) postulated to harbor the HCV replication complex (21). Because membrane-associated RNA replication appears to be central to the HCV life cycle, we sought to provide a more complete description of the membrane-associating elements within NS4B and to examine their role in RNA replication.
Here we report the identification of a new membrane association domain in NS4B which is distinct from the previously described TMDs. This domain is predicted to form an amphipathic alpha helix. We also show that this domain is responsible for the correct localization of other viral NS proteins. Finally we show that the maintenance of an intact amphipathic helix (AH) is vital for HCV RNA replication.
MATERIALS AND METHODS
Cells.
Huh-7 cells were grown in medium containing Dulbecco's minimal essential medium and RPMI medium in a 1:1 ratio in the presence of 10% fetal bovine serum, 2 mM l-glutamine, 100 IU of penicillin/ml, and 100 μg of streptomycin/ml. The cells were grown at 37°C with 5% CO2.
Antibodies.
Monoclonal antibodies against NS5A and NS3 were obtained from Virostat (Portland, Maine). The monoclonal antibody against NS4B was made by X. He, H. Greenberg, and A. Wieczorek (unpublished data). The anti-caveolin antibody was purchased from BD Transduction Laboratories (San Diego, Calif.). A polyclonal antibody against green fluorescent protein (GFP) and all secondary antibodies were purchased from Molecular Probes (Eugene, Ore.). Protein A conjugated to 125I was purchased from Perkin-Elmer (Boston, Mass.)
Plasmid constructions.
Standard recombinant DNA technology was used to construct and purify all plasmids. All mutations were confirmed by automated DNA sequencing. Plasmid DNAs were prepared from large-scale bacterial cultures and purified by use of a QIAGEN (Valencia, Calif.) kit. The positions of all amino acids mutated and deleted in this study refer to the first amino acid of NS4B in the vector pBRTM/HCV 827-3011 (22). Primer sequences used in this study are shown in Table 1.
TABLE 1.
Primers used in for the generation of NS4B mutantsa
| Primer | Sequence |
|---|---|
| ENGFP | 5′-TCCGGCTCCTGGCTAAGGGACAGCAAGGGCGAGGACTGTTCACCGGGGTGG-3′ |
| GFPBG2 | 5′-TTATTTAGATCTTACTTGTACAGCTCGTCCATGCCGAGAGTGATCCCGG-3′ |
| PstXX | 5′-CGCGGTCTGCAGGTCGCCGAGGTCCTTCTGCTTGTCCTGCTCATCGAGCATCATCTCTTGCTCG-3′ |
| 4XF | 5′-GAAATGCAGCTCGACGAACAAGAAATCGGGGAGCTGCAAAC-3′ |
| 4XR | 5′-CTCCCCGATTTCCTTCTGTTTGTCTTGTTCGTCGAGCTGCATTTCCTGTTCGA-3′ |
| 4XF/G10E | 5′-CACCTCCCTTACATCGAACAGGAAATGCAGCTCGCCGAACAATTC-3′ |
| 4XR/G10E | 5′-GAATTGTTCGGCGAGCTGCATTTCCTGTTCGATGTAAGGGAGGTG-3′ |
| 4XF/A14D | 5′-CGAACAGGGAATGCAGCTCGACGAACAATTCAAACAGAAGGC-3′ |
| 4XR/A14D | 5′-GCCTTCTGTTTGAATTGTTCGTCGAGCTGCATTCCCTGTTCG-3′ |
| 4XF/F17D | 5′-GGGAATGCAGCTCGCCGAACAAGACAAACAGAAGGCAATCGGG-3′ |
| 4XR/F17D | 5′-CCCGATTGCCTTCTGTTTGTCTTGTTCGGCGAGCTGCATTCCC-3′ |
| 4XF/A21E | 5′-CGCCGAACAATTCAAACAGAAGGAAATCGGGTTGCTGCAAACAGCC-3′ |
| 4XR/A21E | 5′-GGCTGTTTGCAGCAACCCGATTTCCTTCTGTTTGAATTGTTCGGCG-3′ |
| 4XF/L24E | 5′-CGAACAATTCAAACAGAAGGCAATCGGGGAGCTGCAAACAGCCACCAAGC-3′ |
| 4XR/L24E | 5′-GCTTGGTGGCTGTTTGCAGCTCCCCGATTGCCTTCTGTTTGAATTGTTCG-3′ |
| 4BabF | CCTTACATCGAACAGGGAATGATGCTCGCCGAACAATTCAAACAGAAGGC |
| 4BabR | CCTGTTCGATGTAAGGGAGGTGTTGGGAGCACTCTTCCATCTCATCG |
| 4BX17abF | CTCGCCGAACAAGACAAACAGAAGGCAATCG |
| 4BX17abR | CGATTGCCTTCTGTTTGTCTTGTTCGGCGAG |
Bold characters represent mutations in the NS4B coding sequence.
The plasmid pc4BMA-GFP, coding for NS4B-GFP, was constructed by cloning the NS4B coding region from the vector pBRTM/HCV827-3011 using PCR primers that introduced a methionine and an alanine in the beginning of the NS4B sequence into pcDNA3 (Invitrogen). The GFP coding sequence was cloned by PCR in frame downstream of the C-terminal coding region of NS4B by using the pHygEGFP vector (Clontech) as template and the primers ENGFP and GFPBG2. Subsequent deletion and point mutations were inserted into pc4BMA-GFP.
The plasmid pc4BMAΔ(27-245)-GFP, expressing the deletion mutant NS4BΔ(27-245)-GFP, contains an in-frame deletion of amino acids 27 to 245 of NS4B.
The plasmid pc4BMAxxΔ(27-245)-GFP, expressing the mutated AH along with the deletion of amino acids 27 to 245, was constructed by PCR mutagenesis (19) using primer PstXX. This resulted in the introduction of five point mutations (replacing glycine-10 with glutamate and alanine-14, phenylalanine-17, alanine-21, and leucine-24 with aspartate) in the AH segment of NS4BΔ(27-245)-GFP.
Bart79I, a wild-type replicon that contains a highly adaptive mutation in the NS5A coding region, was described elsewhere (4).
The replicon harboring the five point mutations in the NS4B AH, designated Bart4x79I, was made by PCR mutagenesis (19) of Bart79I using the primer set 4XF-4right and 4XR-1972, in which the codons for glycine-10, alanine-14, phenylalanine-17, alanine-21, and leucine-24 of NS4B were changed to ones that encode glutamate, aspartate, aspartate, glutamate, and glutamate, respectively. The resulting PCR fragment was cut with the restriction enzymes BsrGI and MluI and ligated with the Bart79I plasmid that was cut with the same enzymes. The same cloning strategy was used to construct the partial AH mutant replicons designated Bart4x10-79I, Bart4x14-79I, Bart4x17-79I, Bart4x21-79I, and Bart4x24-79I using the primer sets 4XF/G10E-4XR/G10E, 4XF/A14D-4XR/A14D, 4XF/F17D-4XR/F17D, 4XF/A21E-4XR/A21E, and 4XF/L24E-4XR/L24E, respectively.
The plasmids Bart79Iab and Bart4x17ab were constructed to facilitate the detection of NS4B with a monoclonal antibody raised against the 1a genotype of NS4B. In these plasmids alanine 1, serine 2, and glutamine 11 of NS4B were replaced with serine, glutamine, and methionine, respectively (these amino acid changes reproduce the NS4B peptide immunogen used to generate the anti-NS4B monoclonal antibody). The plasmid Bart79Iab was generated by PCR mutagenesis of Bart79I using the primer sets 1972-4BabR and 4right-4BabF by the same strategy as that described for the other mutant replicons (see above). The plasmid Bart4x17ab was generated by PCR mutagenesis of Bart79Iab using the primer sets 4BX17abF-4right and 1972-4BX17abR by the same strategy as that described for Bart4x17-79I (see above).
Transfection.
DNA constructs were transfected into Huh-7 cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol.
Infection and transfection.
A vaccinia virus that expresses the T7 RNA polymerase was used to infect Huh-7 cells at a multiplicity of infection of 10. Following a 45-min incubation at 37°C, the cells were washed twice with Optimem (Invitrogen) and subjected to transfection with the appropriate construct. The cells were supplemented with growth media and incubated for 5 h at 37°C.
Membrane flotation.
Five hours after the infection-transfection procedure, we collected the cells by scraping in phosphate-buffered saline (PBS) containing protease inhibitors and disrupted the cells mechanically by passing the extract 20 times through a ball bearing homogenizer. We generated the postnuclear supernatants by using 5 min of centrifugation at 1,000 rpm to remove the nuclei. The resulting postnuclear supernatant was overlaid with layers of 30, 25, and 5% OptiPrep (Sigma), and the gradient was centrifuged at 40,000 rpm for 4 h at 4°C in an SW60 rotor. Fractions were collected from the top to the bottom of the density gradient, and the proteins in the collected fractions were precipitated with methanol-chloroform and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (16) followed by immunoblotting using anti-GFP antibody and 125I-protein A as described previously (40). In this gradient, membranes float to the region between the 5% and 25% layers (fraction 2 from the top). Caveolin served as a membrane protein marker (35). The bands were quantified using a Phosphorimager (Molecular Dynamics), and the amount in fraction 2 from the top was divided by that contained in the entire gradient to obtain the percentage of protein floating with the membrane fraction. To express changes in membrane association as a function of the mutation of the wild-type protein, the percentage of protein floating for each mutant relative to the wild-type control (with the latter set at 100) was then calculated.
Fluorescence microscopy.
Cells expressing GFP fusion proteins were fixed with 4% formaldehyde 8 h posttransfection and mounted using a Mowiol mounting medium. Immunofluorescence staining of replicon-encoded proteins was performed as described previously (20). Briefly, cells were plated on coverslips and, following infection and transfection, were fixed in 4% formaldehyde and stained with a primary antibody. Following incubation overnight in the cold, the cells were incubated with a secondary antibody conjugated to Alexa 488 or Alexa 594 and mounted with Mowiol mounting medium. Fluorescence images were captured using a Nikon E600 fluorescence microscope equipped with a SPOT digital camera or a Bio-Rad confocal microscope using a 100× objective (see Fig. 6) and the OpenLab (Improvision, Lexington, Mass.) image acquisition software.
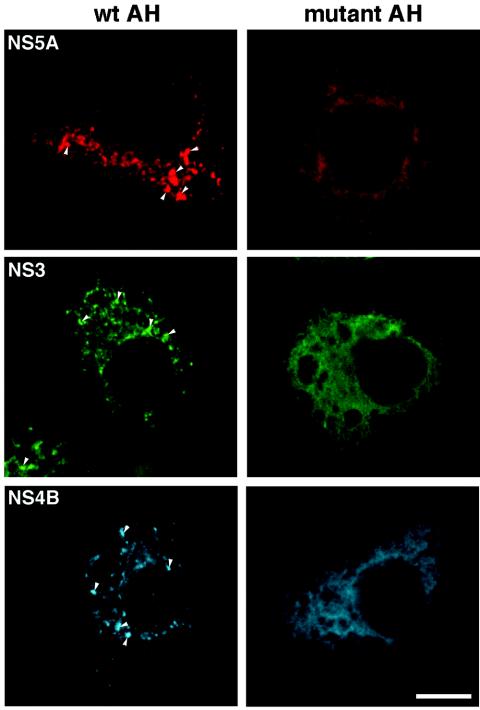
FIG. 6.
Disruption of the NS4B amphipathic helix alters the localization of HCV replication complex components. Huh-7 cells plated on coverslips were infected with a recombinant vaccinia virus expressing T7 RNA polymerase and then transfected with DNA encoding a replicon with an intact NS4B AH (Bart79Iab, left panel) or a replicon harboring a replication-deficient mutation in the NS4B AH (Bart4x17ab, right panel). Following incubation for 5 h at 37°C, the cells were fixed and stained with mouse monoclonal antibodies against either NS4B, NS5A, or NS3 followed by a secondary anti-mouse Alexa494 (against NS4B and NS5A) or anti-mouse Alexa598 (against NS3). The blue color of NS4B staining was introduced artificially using an image analysis software program (Improvision) for clarity of presentation. Note the speckle-like staining pattern (indicated by arrowheads) readily apparent for all three proteins expressed off of the replicon with the intact AH and its absence when the proteins are expressed in the context of the replication-deficient amphipathic helix mutant replicon. Bar = 10 μm.
RNA transcription.
Plasmid DNA of an HCV replicon (Bart79I) or NS4B AH mutant replicons was linearized using ScaI and treated with proteinase K, and then phenol-chloroform extraction and precipitation with ethanol were carried out. The DNA was resuspended in RNase-free water to a final concentration of 1 μg/μl. Four micrograms of DNA was used as template for transcription using the Ribomax RNA production kit (Promega) according to the manufacturer's protocol. The template DNA was digested by addition of 5 U of RQ1 DNase (Promega) followed by incubation for 15 min at 37°C. The unincorporated ribonucleotides were removed by size exclusion using a Micro Bio-Spin P-30 column (Bio-Rad), and the transcribed RNA was extracted with phenol-chloroform and then precipitated in ethanol. The RNA pellet was washed with 70% ethanol and resuspended in H2O. Determination of the RNA concentration was performed by measurement of the optical density at 260 nm. One-percent agarose gel electrophoresis and ethidium bromide staining were used to confirm the integrity and concentration of the RNA.
Electroporation of cultured cells.
In vitro-transcribed RNA was electroporated into Huh-7 cells as described previously (5). Briefly, 5 μg of in vitro-transcribed RNA was mixed with 4 × 106 cells in RNase-free PBS (Biowhittaker) and transferred into a 2-mm-diameter gap cuvette (BTX, San Diego, Calif.). Electroporation was performed using a BTX model 830 electroporator (electroporation conditions were 0.68 kV and five periods of 99 μs at 500-ms intervals). Pulsed cells were left to recover for 10 min at room temperature and then diluted in 10 ml of prewarmed growth medium. Cells were plated in 10-cm3 tissue culture dishes at different dilutions. At 24 h postelectroporation, the cells were supplemented with plain Huh-7 cells to a final density of 106 cells/plate, and 24 h later the medium was supplemented with G-418 to a final concentration of 1 mg/ml. The medium was replaced every 4 days for 3 weeks. Following selection the plates were washed with PBS, incubated in 1% crystal violet made in 20% ethanol for 5 min, and washed five times with H2O to facilitate colony counting. Replication efficiencies of individual replicons were determined by dividing the number of colonies counted on each plate by the number obtained with the polymerase-defective replicon (4) and expressed per microgram of transfected RNA.
RESULTS AND DISCUSSION
An N-terminal AH in NS4B mediates membrane association.
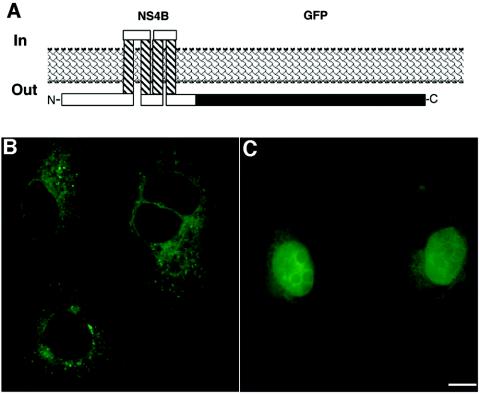
To provide a convenient detection marker and an epitope outside of any future field of mutagenesis, an NS4B protein with a C-terminal in-frame fusion to GFP (47) was constructed (Fig. 1A). Expression of this construct, termed pc4BMA-GFP, in Huh-7 cells yielded a reticular localization pattern accompanied in most cases by distinct foci (Fig. 1B). The GFP signal colocalized with calnexin (data not shown), an ER marker protein, confirming an ER localization previously shown for the native NS4B protein (23, 24, 26, 32, 34, 42). The observed localization of NS4B-GFP, characteristic of membrane association, was different from the diffuse cytoplasmic distribution pattern seen upon expression of GFP, a non-membrane-associated protein, alone (Fig. 1C).
FIG. 1.
NS4B-GFP is localized to intracellular membranes. (A) Schematic of predicted topology of NS4B and its C-terminal GFP fusion with respect to the ER membrane according to the TMpred prediction program and Lundin et al. (32). Crosshatched segments indicate putative TMDs (amino acids 75 to 93, 117 to 138, 140 to 158, and 171 to 191), and the black segment indicates GFP. Huh-7 cells plated on coverslips were transfected with pc4BMA-GFP (B) or pCMV-GFP (C). Eight hours posttransfection, the cells were fixed and imaged with a fluorescence microscope. Note the reticular membrane localization pattern with distinct foci located in the cytoplasm for NS4B-GFP as opposed to the diffuse localization all over the cytoplasm, including the nucleus, for GFP.
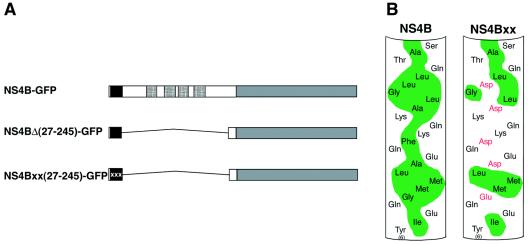
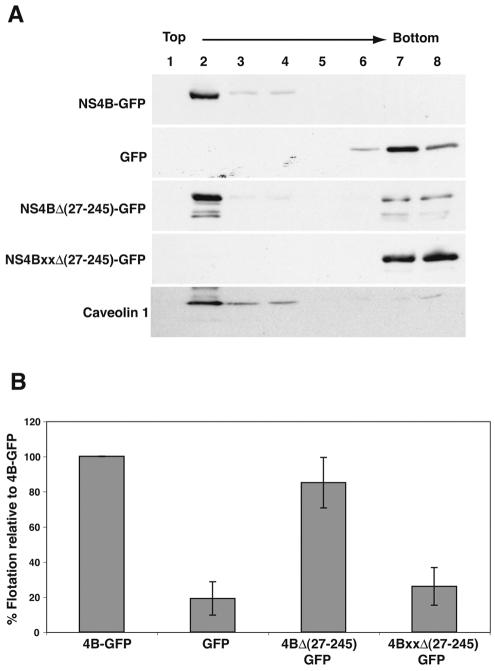
To test the hypothesis that other elements within NS4B besides the TMDs may mediate membrane association, we next constructed a mutant of NS4B—termed pc4BMAΔ(27-245)-GFP—in which all four predicted TMDs were deleted (Fig. 2A). We then examined the biochemical membrane association properties of NS4B-GFP and the deletion mutant using a membrane flotation assay. In this assay, cell extracts are overlaid with a density gradient and subjected to centrifugation. Low-density membrane fractions—and any associated proteins—rise towards the top of the gradient, whereas soluble proteins remain with the denser fractions at the bottom (16, 29, 35). A representative Western blot from a membrane flotation experiment is shown in Fig. 3A. NS4B-GFP (Fig. 3A, top panel), which has a cytoplasmic membrane localization pattern as shown by fluorescence microscopy (Fig. 1B), was found in the low-density fraction (fraction 2 from the top of the gradient) of the flotation gradient. This behavior is consistent with that of a membrane-associated protein, such as caveolin (Fig. 3A, bottom panel) (35). In contrast, GFP is retained at the bottom of the gradient (fractions 7 and 8) in a location characteristic of cytoplasmic soluble proteins (Fig. 3A). Surprisingly, however, deletion of the four predicted TMDs of NS4B did not abolish membrane association of the remaining protein (Fig. 3). Indeed, NS4BΔ(27-245)-GFP floats to the same extent as the full-length NS4B-GFP protein. This result suggested that the four TMDs of NS4B are not required to confer its membrane association and that the remaining part of NS4B in the deletion mutant harbors a membrane-targeting domain. Closer inspection of the amino acid sequence of the extreme N-terminal segment of NS4B revealed the presence of a putative AH domain (Fig. 2B, left panel). A similar domain was shown to confer the membrane association of HCV's NS5A (9, 16) and other viral and cellular proteins (14, 27, 41). To test the hypothesis that the predicted AH in NS4B indeed mediates membrane association, we introduced five site-directed point mutations designed to disrupt the hydrophobic face of the AH in pc4BMAΔ(27-245)-GFP to generate pc4BMAxxΔ(27-245)-GFP (Fig. 2B, right panel). When these mutations were introduced into the full-length NS4B-GFP (or when a truncated version starting at amino acid 24 was used), the resulting protein retained its membrane association (data not shown), presumably due to the protein's TMDs. However, when the four TMDs are deleted along with the introduction of the site-directed mutations in the AH (Fig. 2A, bottom construct), the protein loses its membrane association and behaves like GFP in the membrane flotation assay (Fig. 3). These results show that NS4B can interact with membranes via two types of domains that use different mechanisms of membrane association. It also shows that the TMDs are not necessary to confer membrane association. Recently, it was shown that the N-terminal 93 amino acids of NS4B fractionate with the detergent phase in a Triton X-114 extraction (32), which is a characteristic of proteins tightly associated with membranes (7). In this respect, our results are in agreement with the report by Lundin et al. Moreover, our data pinpoint the AH as a region responsible for this membrane association and show that an intact hydrophobic face of the AH is necessary to mediate interaction of this region with the membrane. It was also reported that the N-terminal region of NS4B adopts a luminal localization after the integration of the protein's TMDs into the membrane (32). The mechanism for such a translocation, however, is not clear. It is possible that the AH domain promotes the insertion of NS4B's N terminus into the ER lumen.
FIG. 2.
NS4B-GFP deletion and AH mutant constructs. (A) Schematic representation of NS4B-GFP and mutants thereof used in this study. The crosshatched segments indicate putative transmembrane domains, the black-filled segments indicate amphipathic helix regions, the segment filled with x's indicates the mutant amphipathic helix, and the gray segments indicate GFP. (B) Helix net diagram representations of the N-terminal amphipathic helices of NS4B (left) and the mutant generated by site-directed mutagenesis (right). In these representations, the cylindrical helix region is cut longitudinally and flattened into the plane of the page. The amino acid sequence of NS4B from amino acids 6 to 29 in the N-terminal to C-terminal direction is shown. The hydrophobic face of the helix is shaded in green; amino acids mutated to disrupt the hydrophobic face are indicated in red.
FIG. 3.
An N-terminal amphipathic helix in NS4B mediates membrane association. (A) Membrane flotation analysis of Huh-7 cells transfected with pc4BMA-GFP (expressing NS4B-GFP), pCMV-GFP (expressing GFP), pc4BMAΔ(27-245)-GFP (NS4B-GFP mutant deleted of all predicted transmembrane domains), and pc4BMAxxΔ(27-245)-GFP (same as the previous mutant but with genetically disrupted amphipathic helix). Cells were mechanically disrupted, and equal amounts of postnuclear supernatants were used for membrane flotation and Western blot analysis with an antibody to GFP (or caveolin, bottom panel) as described in Materials and Methods. Note that NS4B-GFP appears in the same low-density membrane fraction (fraction 2) as caveolin, indicating its membrane association. Representative blots are shown. (B) The distribution of proteins among the gradient fractions from five independent experiments such as those depicted above was quantified by phosphorimager analysis, and the results were expressed as percentages of proteins floating with the membrane fraction relative to the wild-type control (NS4B-GFP), as described in Materials and Methods. Error bars indicate standard errors.
The NS4B N-terminal AH is highly conserved among HCV genotypes.
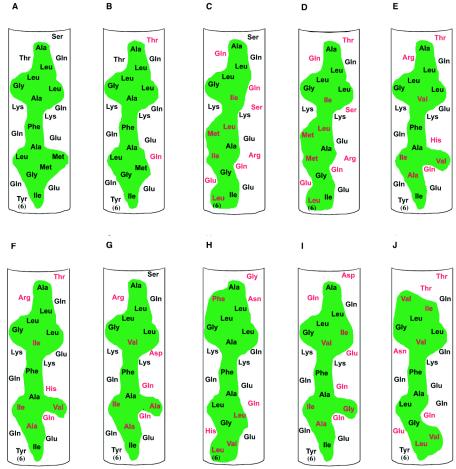
The importance of the AH domain in natural HCV infections may be reflected in the conservation of this domain in different HCV isolates. We examined the predicted amino acid sequences of the N-terminal segment of NS4B in isolates obtained from a variety of patients representing multiple genotypes. As shown in Fig. 4, although there is considerable variability in the specific amino acid sequence in this region, all changes observed conform to a striking constraint—maintenance of a predicted amphipathic helix. In other words, at most amino acid positions a variety of substitutions can be found among the different isolates, yet the hydrophobic (or hydrophilic) nature of the amino acid is preserved. Moreover, when a similar analysis was extended to all sequenced isolates presently available in public databases, the same result was observed (Y. Yang and J. Glass, personal communication). We think it is reasonable to assume that most (if not all) of these sequences maintain the capacity to support a productive viral life cycle, as they were isolated from actively infected patients. We therefore infer that the types of amino acid differences found in these sequenced isolates (compared to those found in a reference isolate) are compatible with genome replication. We would also hypothesize that some of the types of changes not found may be incompatible with efficient replication. Finally, this striking conservation of the AH domain suggests that such a structural motif is indeed important for some essential aspect of the HCV life cycle.
FIG. 4.
Helix net diagrams of the N-terminal amino acid sequence of NS4B obtained from sequenced isolates of HCV from around the world and representing a variety of genotypes. The amphipathic helix of NS4B is conserved across HCV isolates. Note that although there are differences in particular amino acids (shown in red) compared to a reference sequence (A), the amphipathic nature of the helix (indicated by the continuous band of hydrophobic amino acids shaded in green) is conserved. (A) genotype 1a (GenBank accession no. AF009606), (B) genotype 1b (accession no. M58335), (C) genotype 2a (accession no. D00944), (D) genotype 2b (accession no. AB030907), (E) genotype 3a-K (accession no. D28917), (F) genotype 3a-NZL (accession no. D17763), (G) genotype 3b (accession no. D49374), (H) genotype 4a (accession no. Y11604), (I) genotype 10a (accession no. D63821), (J) genotype 11a (accession no. D28917).
Genetic disruption of NS4B's AH inhibits HCV RNA replication.
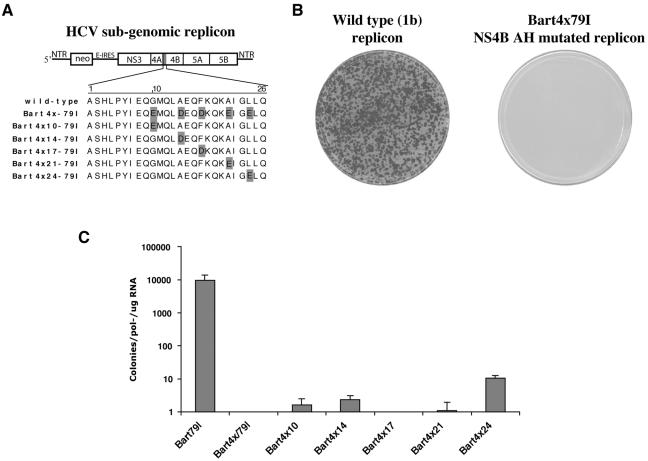
The preservation of an AH in the NS4B proteins of all sequenced HCV clones suggests that the interaction of NS4B's AH with the membrane may have an important role in the HCV life cycle. To test the hypothesis that an intact NS4B AH has a role in HCV RNA replication, five site-directed point mutations within the hydrophobic face of the AH in the same positions that inhibited the membrane association of NS4BxxΔ(75-245)-GFP were introduced into a high-efficiency subgenomic HCV replicon (Fig. 5A) (4). Transfection of the wild-type replicon into Huh-7 cells resulted in numerous G418-resistant colonies (Fig. 5B, left panel). In contrast, the NS4B AH mutant replicon, termed Bart4x79I, was unable to sustain replication, as no colonies were formed (Fig. 5B, right panel). It is unlikely that this dramatic result is simply due to disruption of a critical RNA structural motif, as multiple mutations appear to be tolerated within the mutated segment of NS4B (Fig. 4), including wobble mutations at the amino acid codons mutated in Bart4X79I (M. Elazar, unpublished observation). When each of these five mutations was introduced separately into the wild-type replicon, intermediate levels of replication were observed, except for the mutant F17D, which was replication deficient at the sensitivity of this assay (Fig. 5C). Interestingly, of the five individual mutations, the one at position 17 is predicted to have the greatest effect on disrupting the continuity of the hydrophobic face of the wild-type AH. Taken together, these results suggest that an intact AH is required for efficient HCV RNA replication. Because NS4B membrane association can be provided by the protein's TMDs, these results also suggest that the membrane association conferred by the AH has a role in HCV RNA replication that is beyond simple anchorage of the protein to the ER membrane.
FIG. 5.
Disrupting the amphipathic nature of the NS4B helix impairs HCV RNA replication. (A) (Top) Schematic representation of HCV high-efficiency subgenomic replicons harboring the neomycin resistance gene (neo) and the HCV NS proteins. 5′NTR and 3′NTR represent the nontranslated regions at the ends of the HCV genomic RNA, which contain presumed recognition sequences for the viral replication machinery. The gray region of NS4B represents the AH domain. E-IRES, encephalomyocarditis internal ribosome entry site. (Bottom) The first 26 amino acids of NS4B containing the AH region are shown for the wild-type and mutant replicons. The individual amino acid changes in each mutant are highlighted. (B) Colony formation assays of Huh-7 cells electroporated with RNA from a wild-type replicon (left panel) or from Bart4x79I, an amphipathic-helix-disrupted replicon (right panel), after selection with G418 and staining with crystal violet. Each dot represents a colony of Huh-7 cells that was able to grow in the presence of G418 due to the presence of efficiently replicating intracellular replicons. (C) The replication potential of mutant replicons carrying single amino acid substitutions designed to partially disrupt the NS4B AH (see panel A) was determined by a colony formation assay and presented as the number of colonies obtained for each mutant. As a negative control, a polymerase-defective replicon carrying a lethal mutation (GDD→ AAG) in the active site of the viral RNA-dependent RNA polymerase (4) was used. The bars in this graph represent results ± standard errors from a total of four electroporations performed in two independent experiments.
Disruption of NS4B's AH prevents the correct localization of other HCV NS proteins.
In view of the role of NS4B's involvement in the formation of the HCV membrane structures that presumably harbor the replication complex (15, 21), it is tempting to speculate that the AH domain may be involved in the actual formation of these distinct structures. We hypothesize that NS4B's AH is involved in the functional formation of the replication complex by enabling, directly or indirectly, the correct localization of the replication complex proteins to the replication compartment. To test this hypothesis, we expressed wild-type and AH mutant (F17D) (Fig. 5A) replicon polyproteins in Huh-7 cells and studied the localization of the viral nonstructural proteins. The plasmids Bart79Iab and Bart4x17ab, which express replicon polyproteins with an intact and a disrupted NS4B AH, respectively, and the vaccinia virus expression system were used for these experiments followed by immunofluorescence analysis. When expressed off of the replicon with an intact NS4B AH, NS5A, NS4B, and NS3 display a readily visible speckle-like pattern (Fig. 6, left panels). NS3 shows, in addition to the speckle pattern, more reticular and diffuse staining. These patterns are similar to the staining pattern shown by replicon-expressing cell lines (21, 43). In contrast, when expressed from the replicon harboring a disrupted NS4B AH, the NS proteins' localization is markedly altered. The speckle pattern is lost, and only a reticular staining pattern suggestive of an ER localization is retained (Fig. 6, right panels). This latter localization may be obtained for NS4B through its intact TMDs, whereas NS5A and NS3 presumably retain membrane localization through their natural membrane-associating mechanisms—for NS5A, its membrane-targeting AH (9, 16), and for NS3, its association with NS4A (48). These results clearly show, however, that NS proteins that are considered part of the replication complex (21, 43), and that normally associate with the membranous web structures (15, 21), are unable to be properly sublocalized in the speckled pattern associated with replication-competent replicons in the absence of an intact NS4B AH.
Direct interactions between NS4B and essentially all other HCV NS proteins have been reported (12). Therefore, it is possible that the observed mislocalization of the nonstructural proteins results from interference in the interaction between these proteins and NS4B via disruption of the AH. Because of NS4B's involvement in the formation of the membranous webs, however, we favor an alternate hypothesis, which states that NS4B's AH helps mediate the formation of the membrane structures required for RNA replication and that the altered localization of other NS proteins is a consequence of disrupting this function of the NS4B AH.
This possible role of the AH may be an example of a more general theme. For example, the NS4B protein of bovine viral diarrhea virus (Elazar, unpublished) and the NS protein 2C of both poliovirus and hepatitis A virus all contain an N-terminal AH, which in the case of the latter two has been shown to associate with membranes (13, 14, 27, 46) The poliovirus and hepatitis A virus 2C proteins were also shown to induce the membrane rearrangements associated with RNA replication (44-46).
Our results also show that NS4B appears to share some features recently described for NS5A. Both proteins contain an N-terminal AH, which mediates membrane association (9, 16). In addition, as for NS4B, genetic disruption of the hydrophobic face of the NS5A AH inhibits RNA replication (16). Unlike NS5A, however, NS4B appears to have two distinct mechanisms of membrane association (i.e., TMDs and the AH). Also, while mislocalization as a result of AH disruption appears to be restricted to NS5A itself (unpublished observations), in the case of NS4B AH disruption the localization of other NS proteins is disturbed.
Finally, because of the apparently critical role played by the NS4B AH in the HCV life cycle, strategies designed to disrupt the function of the NS4B AH can be contemplated. These could include agents designed to prevent NS4B's interaction with its membrane target, such as that described for NS5A (16), or compounds which bind the NS4B AH itself. Because increased antiviral responses can be achieved by using multiple agents against independent virus-specific targets (49), anti-NS4B therapies may ultimately help increase the efficacy of anti-HCV regimens.
Acknowledgments
We thank Karla Kirkegaard, Allen Cooper, and Harry B. Greenberg for helpful discussions and critical reading of the manuscript.
M.E. is the recipient of the ALF Postdoctoral Research Fellowship Award. This work was also supported in part by the Eli Lilly-Stanford University HCV initiative, grant no. RO1DK066793, and a Burroughs Welcome Career Award (to J.S.G.).
REFERENCES
- 1.Alter, M. J., D. Kruszon-Moran, O. V. Nainan, G. M. McQuillan, F. Gao, L. A. Moyer, R. A. Kaslow, and H. S. Margolis. 1999. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N. Engl. J. Med. 341:556-562. [DOI] [PubMed] [Google Scholar]
- 2.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 3.Bienz, K., D. Egger, Y. Rasser, and W. Bossart. 1980. Kinetics and location of poliovirus macromolecular synthesis in correlation to virus-induced cytopathology. Virology 100:390-399. [DOI] [PubMed] [Google Scholar]
- 4.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 5.Blight, K. J., J. A. McKeating, J. Marcotrigiano, and C. M. Rice. 2003. Efficient replication of hepatitis C virus genotype 1a RNAs in cell culture. J. Virol. 77:3181-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bordier, C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256:1604-1607. [PubMed] [Google Scholar]
- 8.Branch, A. D. 2000. Hepatitis C virus RNA codes for proteins and replicates: does it also trigger the interferon response? Semin. Liver Dis. 20:57-68. [DOI] [PubMed] [Google Scholar]
- 9.Brass, V., E. Bieck, R. Montserret, B. Wolk, J. A. Hellings, H. E. Blum, F. Penin, and D. Moradpour. 2002. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 277:8130-8139. [DOI] [PubMed] [Google Scholar]
- 10.Chu, P. W., and E. G. Westaway. 1992. Molecular and ultrastructural analysis of heavy membrane fractions associated with the replication of Kunjin virus RNA. Arch. Virol. 125:177-191. [DOI] [PubMed] [Google Scholar]
- 11.Di Bisceglie, A. M. 2000. Natural history of hepatitis C: its impact on clinical management. Hepatology 31:1014-1018. [DOI] [PubMed] [Google Scholar]
- 12.Dimitrova, M., I. Imbert, M. P. Kieny, and C. Schuster. 2003. Protein-protein interactions between hepatitis C virus nonstructural proteins. J. Virol. 77:5401-5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Echeverri, A., R. Banerjee, and A. Dasgupta. 1998. Amino-terminal region of poliovirus 2C protein is sufficient for membrane binding. Virus Res. 54:217-223. [DOI] [PubMed] [Google Scholar]
- 14.Echeverri, A. C., and A. Dasgupta. 1995. Amino terminal regions of poliovirus 2C protein mediate membrane binding. Virology 208:540-553. [DOI] [PubMed] [Google Scholar]
- 15.Egger, D., B. Wolk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elazar, M., K. H. Cheong, P. Liu, H. B. Greenberg, C. M. Rice, and J. S. Glenn. 2003. Amphipathic helix-dependent localization of NS5A mediates hepatitis C virus RNA replication. J. Virol. 77:6055-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Florese, R. H., M. Nagano-Fujii, Y. Iwanaga, R. Hidajat, and H. Hotta. 2002. Inhibition of protein synthesis by the nonstructural proteins NS4A and NS4B of hepatitis C virus. Virus Res. 90:119-131. [DOI] [PubMed] [Google Scholar]
- 18.Froshauer, S., J. Kartenbeck, and A. Helenius. 1988. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 107:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glenn, J. S., J. C. Marsters, Jr., and H. B. Greenberg. 1998. Use of a prenylation inhibitor as a novel antiviral agent. J. Virol. 72:9303-9306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glenn, J. S., J. M. Taylor, and J. M. White. 1990. In vitro-synthesized hepatitis delta virus RNA initiates genome replication in cultured cells. J. Virol. 64:3104-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grakoui, A., C. Wychowski, C. Lin, S. M. Feinstone, and C. M. Rice. 1993. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 67:1385-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, Y., Y. Uchiyama, T. Fujimura, H. Kanamori, T. Doi, A. Takamizawa, T. Hamakubo, and T. Kodama. 2001. A human hepatoma cell line expressing hepatitis c virus nonstructural proteins tightly regulated by tetracycline. Biochem. Biophys. Res. Commun. 281:732-740. [DOI] [PubMed] [Google Scholar]
- 24.Hugle, T., F. Fehrmann, E. Bieck, M. Kohara, H. G. Krausslich, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology 284:70-81. [DOI] [PubMed] [Google Scholar]
- 25.Kato, J., N. Kato, H. Yoshida, S. K. Ono-Nita, Y. Shiratori, and M. Omata. 2002. Hepatitis C virus NS4A and NS4B proteins suppress translation in vivo. J. Med. Virol. 66:187-199. [DOI] [PubMed] [Google Scholar]
- 26.Kim, J. E., W. K. Song, K. M. Chung, S. H. Back, and S. K. Jang. 1999. Subcellular localization of hepatitis C viral proteins in mammalian cells. Arch. Virol. 144:329-343. [DOI] [PubMed] [Google Scholar]
- 27.Kusov, Y. Y., C. Probst, M. Jecht, P. D. Jost, and V. Gauss-Muller. 1998. Membrane association and RNA binding of recombinant hepatitis A virus protein 2C. Arch. Virol. 143:931-944. [DOI] [PubMed] [Google Scholar]
- 28.Lazarus, L. H., and R. Barzilai. 1974. Association of foot-and-mouth disease virus replicase with RNA template and cytoplasmic membranes. J. Gen. Virol. 23:213-218. [DOI] [PubMed] [Google Scholar]
- 29.Lecat, S., P. Verkade, C. Thiele, K. Fiedler, K. Simons, and F. Lafont. 2000. Different properties of two isoforms of annexin XIII in MDCK cells. J. Cell Sci. 113:2607-2618. [DOI] [PubMed] [Google Scholar]
- 30.Liang, T. J., B. Rehermann, L. B. Seeff, and J. H. Hoofnagle. 2000. Pathogenesis, natural history, treatment, and prevention of hepatitis C. Ann. Intern. Med. 132:296-305. [DOI] [PubMed] [Google Scholar]
- 31.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 32.Lundin, M., M. Monne, A. Widell, G. Von Heijne, and M. A. Persson. 2003. Topology of the membrane-associated hepatitis C virus protein NS4B. J. Virol. 77:5428-5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M. H. Ling, S. Cort, J. K. Albrecht, et al. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 34.Moradpour, D., P. Kary, C. M. Rice, and H. E. Blum. 1998. Continuous human cell lines inducibly expressing hepatitis C virus structural and nonstructural proteins. Hepatology 28:192-201. [DOI] [PubMed] [Google Scholar]
- 35.Oliferenko, S., K. Paiha, T. Harder, V. Gerke, C. Schwarzler, H. Schwarz, H. Beug, U. Gunthert, and L. A. Huber. 1999. Analysis of CD44-containing lipid rafts: recruitment of annexin II and stabilization by the actin cytoskeleton. J. Cell Biol. 146:843-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park, J. S., J. M. Yang, and M. K. Min. 2000. Hepatitis C virus nonstructural protein NS4B transforms NIH3T3 cells in cooperation with the Ha-ras oncogene. Biochem. Biophys. Res. Commun. 267:581-587. [DOI] [PubMed] [Google Scholar]
- 37.Piccininni, S., A. Varaklioti, M. Nardelli, B. Dave, K. D. Raney, and J. E. McCarthy. 2002. Modulation of the hepatitis C virus RNA-dependent RNA polymerase activity by the non-structural (NS) 3 helicase and the NS4B membrane protein. J. Biol. Chem. 277:45670-45679. [DOI] [PubMed] [Google Scholar]
- 38.Reed, K. E., and C. M. Rice. 2000. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 242:55-84. [DOI] [PubMed] [Google Scholar]
- 39.Rice, C. M. 1996. Flaviviridae: the viruses and their replication, p. 931-959. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology. Lippincott-Raven Publications, Philadelphia, Penn.
- 40.Sato, S., S. K. Wong, and D. W. Lazinski. 2001. Hepatitis delta virus minimal substrates competent for editing by ADAR1 and ADAR2. J. Virol. 75:8547-8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Segrest, J. P., H. De Loof, J. G. Dohlman, C. G. Brouillette, and G. M. Anantharamaiah. 1990. Amphipathic helix motif: classes and properties. Proteins 8:103-117. [DOI] [PubMed] [Google Scholar]
- 42.Selby, M. J., Q. L. Choo, K. Berger, G. Kuo, E. Glazer, M. Eckart, C. Lee, D. Chien, C. Kuo, and M. Houghton. 1993. Expression, identification and subcellular localization of the proteins encoded by the hepatitis C viral genome. J. Gen. Virol. 74:1103-1113. [DOI] [PubMed] [Google Scholar]
- 43.Shi, S. T., K. J. Lee, H. Aizaki, S. B. Hwang, and M. M. Lai. 2003. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J. Virol. 77:4160-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suhy, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teterina, N. L., K. Bienz, D. Egger, A. E. Gorbalenya, and E. Ehrenfeld. 1997. Induction of intracellular membrane rearrangements by HAV proteins 2C and 2BC. Virology 237:66-77. [DOI] [PubMed] [Google Scholar]
- 46.Teterina, N. L., A. E. Gorbalenya, D. Egger, K. Bienz, and E. Ehrenfeld. 1997. Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J. Virol. 71:8962-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsien, R. Y. 1998. The green fluorescent protein. Annu. Rev. Biochem. 67:509-544. [DOI] [PubMed] [Google Scholar]
- 48.Wölk, B., D. Sansonno, H.-G. Krausslich, F. Dammacco, C. M. Rice, H. E. Blum, and D. Moradpour. 2000. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline-regulated cell lines. J. Virol. 74:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yeni, P. G., S. M. Hammer, C. C. Carpenter, D. A. Cooper, M. A. Fischl, J. M. Gatell, B. G. Gazzard, M. S. Hirsch, D. M. Jacobsen, D. A. Katzenstein, J. S. Montaner, D. D. Richman, M. S. Saag, M. Schechter, R. T. Schooley, M. A. Thompson, S. Vella, and P. A. Volberding. 2002. Antiretroviral treatment for adult HIV infection in 2002: updated recommendations of the International AIDS Society-USA Panel. JAMA 288:222-235. [DOI] [PubMed] [Google Scholar]
- 50.Zeuzem, S., S. V. Feinman, J. Rasenack, E. J. Heathcote, M. Y. Lai, E. Gane, J. O'Grady, J. Reichen, M. Diago, A. Lin, J. Hoffman, and M. J. Brunda. 2000. Peginterferon alfa-2a in patients with chronic hepatitis C. N. Engl. J. Med. 343:1666-1672. [DOI] [PubMed] [Google Scholar]