Abstract
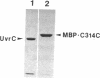
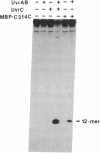
The UvrC protein is one of three subunits of the Escherichia coli repair enzyme (A)BC excinuclease. This subunit is thought to have at least one of the active sites for nucleophilic attack on the phosphodiester bonds of damaged DNA. To localize the active site, mutant UvrC proteins were constructed by linker-scanning and deletion mutagenesis. In vivo studies revealed that the C-terminal 314 amino acids of the 610-amino acid UvrC protein were sufficient to confer UV resistance to cells lacking the uvrC gene. The portion of the uvrC gene encoding the C-terminal half of the protein was fused to the 3' end of the E. coli malE gene (which encodes maltose binding protein), and the fusion protein MBP-C314C was purified and characterized. The fusion protein, in combination with UvrA and UvrB subunits, reconstituted the excinuclease activity that incised the eighth phosphodiester bond 5' and the fourth phosphodiester bond 3' to a psoralen-thymine adduct. These results suggest that the C-terminal 314 amino acids of UvrC constitute a functional domain capable of interacting with the UvrB-damaged DNA complex and of inducing the two phosphodiester bond incisions characteristic of (A)BC excinuclease.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Caron P. R., Grossman L. Incision of damaged versus nondamaged DNA by the Escherichia coli UvrABC proteins. Nucleic Acids Res. 1988 Aug 25;16(16):7855–7865. doi: 10.1093/nar/16.16.7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N. Y., Zhang J. J., Paulus H. Chromosomal location of the Bacillus subtilis aspartokinase II gene and nucleotide sequence of the adjacent genes homologous to uvrC and trx of Escherichia coli. J Gen Microbiol. 1989 Nov;135(11):2931–2940. doi: 10.1099/00221287-135-11-2931. [DOI] [PubMed] [Google Scholar]
- Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu Rev Biochem. 1988;57:199–233. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- Lambert B., Jones B. K., Roques B. P., Le Pecq J. B., Yeung A. T. The noncovalent complex between DNA and the bifunctional intercalator ditercalinium is a substrate for the UvrABC endonuclease of Escherichia coli. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6557–6561. doi: 10.1073/pnas.86.17.6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin J. D., Johnson A. W., Demple B. Homogeneous Escherichia coli endonuclease IV. Characterization of an enzyme that recognizes oxidative damage in DNA. J Biol Chem. 1988 Jun 15;263(17):8066–8071. [PubMed] [Google Scholar]
- Lin J. J., Sancar A. A new mechanism for repairing oxidative damage to DNA: (A)BC excinuclease removes AP sites and thymine glycols from DNA. Biochemistry. 1989 Oct 3;28(20):7979–7984. doi: 10.1021/bi00446a002. [DOI] [PubMed] [Google Scholar]
- Lin J. J., Sancar A. Reconstitution of nucleotide excision nuclease with UvrA and UvrB proteins from Escherichia coli and UvrC protein from Bacillus subtilis. J Biol Chem. 1990 Dec 5;265(34):21337–21341. [PubMed] [Google Scholar]
- Luckow B., Renkawitz R., Schütz G. A new method for constructing linker scanning mutants. Nucleic Acids Res. 1987 Jan 26;15(2):417–429. doi: 10.1093/nar/15.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy S. R., Nunn W. D. Selection for loss of tetracycline resistance by Escherichia coli. J Bacteriol. 1981 Feb;145(2):1110–1111. doi: 10.1128/jb.145.2.1110-1111.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orren D. K., Sancar A. Formation and enzymatic properties of the UvrB.DNA complex. J Biol Chem. 1990 Sep 15;265(26):15796–15803. [PubMed] [Google Scholar]
- Orren D. K., Sancar A. The (A)BC excinuclease of Escherichia coli has only the UvrB and UvrC subunits in the incision complex. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5237–5241. doi: 10.1073/pnas.86.14.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Rupp W. D. A novel repair enzyme: UVRABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. Cell. 1983 May;33(1):249–260. doi: 10.1016/0092-8674(83)90354-9. [DOI] [PubMed] [Google Scholar]
- Sancar A., Sancar G. B. DNA repair enzymes. Annu Rev Biochem. 1988;57:29–67. doi: 10.1146/annurev.bi.57.070188.000333. [DOI] [PubMed] [Google Scholar]
- Sancar G. B., Sancar A., Rupp W. D. Sequences of the E. coli uvrC gene and protein. Nucleic Acids Res. 1984 Jun 11;12(11):4593–4608. doi: 10.1093/nar/12.11.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P. Hazards and their exploitation in the applications of molecular biology to structure-function relationships. Biochemistry. 1990 Oct 16;29(41):9495–9502. doi: 10.1021/bi00493a001. [DOI] [PubMed] [Google Scholar]
- Selby C. P., Sancar A. Noncovalent drug-DNA binding interactions that inhibit and stimulate (A)BC excinuclease. Biochemistry. 1991 Apr 23;30(16):3841–3849. doi: 10.1021/bi00230a006. [DOI] [PubMed] [Google Scholar]
- Selby C. P., Sancar A. Structure and function of the (A)BC excinuclease of Escherichia coli. Mutat Res. 1990 Sep-Nov;236(2-3):203–211. doi: 10.1016/0921-8777(90)90005-p. [DOI] [PubMed] [Google Scholar]
- Snowden A., Kow Y. W., Van Houten B. Damage repertoire of the Escherichia coli UvrABC nuclease complex includes abasic sites, base-damage analogues, and lesions containing adjacent 5' or 3' nicks. Biochemistry. 1990 Aug 7;29(31):7251–7259. doi: 10.1021/bi00483a013. [DOI] [PubMed] [Google Scholar]
- Thomas D. C., Levy M., Sancar A. Amplification and purification of UvrA, UvrB, and UvrC proteins of Escherichia coli. J Biol Chem. 1985 Aug 15;260(17):9875–9883. [PubMed] [Google Scholar]
- Toth M. J., Schimmel P. Deletions in the large (beta) subunit of a hetero-oligomeric aminoacyl-tRNA synthetase. J Biol Chem. 1990 Jan 15;265(2):1000–1004. [PubMed] [Google Scholar]
- Van Houten B., Gamper H., Sancar A., Hearst J. E. DNase I footprint of ABC excinuclease. J Biol Chem. 1987 Sep 25;262(27):13180–13187. [PubMed] [Google Scholar]
- di Guan C., Li P., Riggs P. D., Inouye H. Vectors that facilitate the expression and purification of foreign peptides in Escherichia coli by fusion to maltose-binding protein. Gene. 1988 Jul 15;67(1):21–30. doi: 10.1016/0378-1119(88)90004-2. [DOI] [PubMed] [Google Scholar]