Abstract
The X gene of hepatitis B virus (HBV) is one of the major factors in HBV-induced hepatocarcinogenesis and is essential for the establishment of productive HBV replication in vivo. Recent studies have shown that the X gene product targets mitochondria and induces calcium flux, thereby activating Ca+-dependent signal transduction pathways. However, regulatory mechanisms of X gene expression have remained unclear. Previous studies had localized a minimal promoter activity to a 21-bp GC-rich sequence located 130 bp upstream of the X protein coding region and showed that there was a cellular protein bound to this DNA. Interestingly, the 21-bp sequence identified as an X gene minimal promoter does not contain any previously identified core promoter elements, such as a TATA box. To better understand the mechanisms of transcriptional initiation of the X gene, we set out to biochemically purify the binding protein(s) for the 21-bp DNA. We report here the identification of the X gene minimal promoter-binding activity as nuclear respiratory factor 1 (NRF1), a previously known transcription factor that activates the majority of nucleus-encoded mitochondrial genes and various housekeeping genes. Primer extension analyses of the X mRNAs show that mutations at the binding site specifically inactivate transcription from this promoter and that a dominant-negative NRF1 mutant and short interfering RNAs inhibit transcription from this promoter. Therefore, NRF1 specifically binds the 21-bp minimal promoter and positively contributes to transcription of the X gene. Simultaneous activation of the X gene and mitochondrial genes by NRF1 may allow the X protein to target mitochondria most efficiently.
Chronic hepatitis B virus (HBV) infection is a major, global cause of hepatocellular carcinoma (HCC). The X gene of this virus has been indicated to be a viral oncogenic factor (reviewed in references 5 and 27). The X gene product exerts a pleiotropic effect on diverse cellular functions including transcriptional regulation (27), apoptosis (13, 35, 37, 40), DNA repair (18), and the proteasome function (34, 44). These effects of the X gene product are thought to be mediated by interaction with cellular proteins and activation of signal transduction pathways (27). Recent findings also established a connection between the X gene and mitochondrial function. The X protein was shown to cause calcium ion (Ca+) flux by targeting mitochondria, consequently activating proline-rich tyrosine kinase 2 (Pyk2) and then the Ca+-dependent Src kinase signaling pathway (2). Consistent with this finding, the X protein has been shown to be colocalized with mitochondria (26) and to change mitochondrial membrane potential, resulting in release of cytochrome c from mitochondria. These observations may explain why the X gene deregulates cellular control of apoptosis (35, 36). In the natural viral life cycle, the X gene is essential for the establishment of productive HBV replication and infection in vivo (6, 45).
To understand mechanisms of HBV-induced hepatocarcinogenesis, it is important to also understand regulation of X gene expression. However, analysis of X gene transcription is not straightforward since the expression levels of X mRNA are generally low and the X mRNA-transcribed region is overlapped by all of the other HBV RNA-transcribed regions. Consequently, transcription start sites of X mRNA have remained obscure. Hence, although there is evidence from reverse transcription-PCR analyses of X mRNA expression in HBV-infected liver tissues (both tumor and nontumor tissues from either hepatitis B surface antigen-positive or -negative HBV patients) (24), mechanisms of transcriptional regulation have previously not been extensively studied. The study of X protein expression by immunostaining has indicated that X protein is expressed preferentially in HCC and the surrounding parenchyma. However, X protein expression could only be detected in a small number of parenchymal and malignant cells (32). It is possible that the X gene is expressed only under some specific physiological conditions or cell cycle phase. These observations from clinical samples highlight the importance of clarifying the mechanisms of X gene transcriptional control to better understand the extraordinary link between HBV infection and HCC.
To gain a complete picture of X gene transcription mechanisms, it is necessary to identify both the core (or basal) promoter and regulatory elements during transcription of the gene. In the remaining part of this report, we will use the term “core promoter” defined as “the minimal stretch of contiguous DNA sequence that is sufficient to direct accurate initiation of transcription by the RNA polymerase II machinery” (4, 31). One of the promoters of HBV that control transcription of the 3.5-kb RNA that encodes the HBV nucleocapsid and/or functions as the pregenome RNA is also called the core promoter, but this promoter will not be discussed in this report. Previous studies of the X promoter region by EMSA (electrophoretic mobility shift assay) and S1 mapping analyses indicated that there was an unidentified cellular protein whose binding site was at a position consistent with a potential function as a core (or basal) promoter-binding protein (22, 43). The binding site of this protein was also shown to overlap the minimal region required for the promoter activity (minimal promoter) analyzed by chloramphenicol acetyltransferase (CAT) reporter assays (33). In this study, we biochemically purified this binding protein, termed, X promoter-binding protein (X-PBP) and found that it is identical to NRF1 (nuclear respiratory factor 1), a protein that has been shown to activate mitochondrion-related and many other housekeeping genes. Transient transfection of X gene templates with a dominant-negative NRF1 mutant gene or NRF1 short interfering RNA (siRNA) expression DNAs confirmed that NRF1 plays an essential role in transcription from this promoter. Mutations at the NRF1-binding site specifically inactivated transcription from this promoter. Our primer extension analyses also showed that there was another transcriptional start site in the X promoter region, but this second start site did not appear to be dependent on NRF1 as strongly as the initially identified start site. Therefore, our data suggest that NRF1 is an essential factor for transcription from one of the major transcriptional start sites of the X mRNA.
MATERIALS AND METHODS
EMSA and DNA footprinting.
Mobility shift assays and DNA footprinting were performed essentially as previously described (33, 43). EMSA probe MP4 for X-PBP binding was made by annealing synthetic 20-mers (5′-AGCTTCGGCGCATGCGTGGA-3′ and 5′-AGCTTCCACGCATGCGCCGA-3′) that contain nucleotides (nt) 1101 to 1116 of HBV DNA and HindIII sites at both ends. As a control, mutant probe MP4m was made by annealing 5′-AGCTTCGGCGCTTTTGTGGA-3′ and 5′-AGCTTCCACAAAAGCGCCGA-3′. For the competition assays shown in Fig. 5, double-stranded oligonucleotides of three additional mutants, 1 to 3 (MP2m, MP4m2, and Sph16), were prepared by annealing synthetic 34-mers (5′-ATAGGCCATCGGCGCTTTTGTGGAACCTTTGTGG-3′ and 5′-CCACAAAGGTTCCACAAAAGCGCCGATGGCCTAT-3′ for MP2m, 5′-GCCATCGGCGCATGCATGCAACCTTTGTGGCTCC-3′ and 5′-GGAGCCACAAAGGTTGCATGCATGCGCCGATGGC-3′ for MP4m2, and 5′-TAGGCCATCGGCGCAGATCTGCGTGGAACCTTTG-3′ and 5′-CAAAGGTTCCACGCAGATCTGCGCCGATGGCCTA for Sph16) that cover nt 1093 to 1126 of HBV DNA.
FIG. 5.
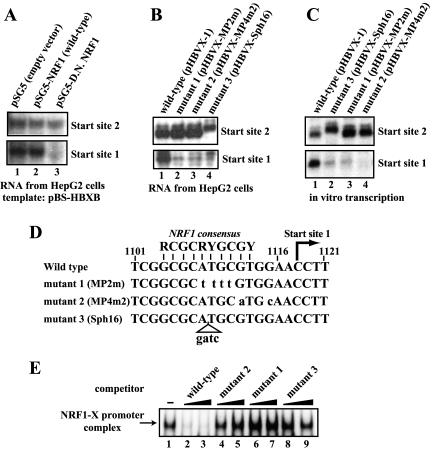
NRF1 is essential for transcription from one of the two major start sites in the X promoter region. (A) Dominant-negative NRF1 abolishes transcription from start site 1. An enhancer-containing X gene template plasmid (pBS-HBXB) was transfected into HepG2 cells with an empty plasmid (lane 1), the wild-type NRF1 expression plasmid (lane 2), or a dominant-negative NRF1 mutant expression plasmid (lane 3). The X transcripts were examined by primer extension analyses. (B) Mutations at the NRF1-binding site disrupt transcription from start site 1 in HepG2 cells. An enhancer-containing X gene template plasmid (pHBVX-1) (lane 1) and its derivatives (lane 2 to 4) that contain mutations at the NRF1-binding site were transfected into HepG2 cells. The X transcripts were analyzed by primer extension. Since mutant 3 had a 4-bp insertion mutation at the NRF1-recognition sequence (see panel D), the primer extension product of the transcript from start site 2 of mutant 3 migrates slower (lane 4) but the start site of this transcript has been confirmed to be the same start site, 2. (C) In vitro transcription assays also show that mutations at the NRF1-binding site disrupt transcription from start site 1. In vitro transcription was performed with HepG2 NE and the same set of plasmids as for panel B. The in vitro transcripts were analyzed by primer extension. (D) Nucleotide sequences of the wild-type and mutated 21-bp minimal promoter region. Mutated residues are in lowercase. The previously reported NRF1-binding consensus sequence, the position of start site 1, and the nucleotide numbers of the HBV DNA are also shown. (E) The mutant promoters are unable to bind NRF1. The 32P-labeled 16-bp wild-type DNA probe (MP4) was mixed with HepG2 NE and poly(dA-dT)/poly(dA-dT) in the presence of a 3-fold (lanes 2, 4, 6, and 8) or a 10-fold (lanes 3, 5, 7, and 9) molar excess of the indicated DNA as a competitor. The NRF1-X promoter DNA (MP4) complex was analyzed by EMSA.
Purification of X-PBP.
Nuclear extract (NE) was prepared from a 70-liter culture of HeLa cells essentially as previously described (7). The NE was equilibrated to 0.35 M NaCl-HGED (20 mM HEPES [pH 7.6], 10% glycerol, 0.25 mM EDTA, 1 mM dithiothreitol) and applied to a Poros HS column. The X-PBP activity, as monitored by mobility shift assay, was eluted from the Poros HS column with a 0.35 to 0.8 M NaCl gradient with peak activity occurring between 0.6 and 0.7 M NaCl. The peak fractions were pooled and subjected to a 20 to 55% ammonium sulfate cut. The 20 to 55% ammonium sulfate precipitates were resuspended in 0.2 M NaCl-HGED and applied to a Superose 6 gel filtration column equilibrated with 0.2% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS)-0.4 M NaCl-HGED. The X-PBP activity was eluted with 400- to 500-kDa proteins. The X-PBP fractions were pooled and further applied to a Poros HQ column and eluted with a 0.2 to 0.8 M NaCl gradient. The peak fractions of X-PBP activity were pooled and further purified by DNA affinity chromatography.
DNA affinity purification.
Biotinylated 48-mer DNAs that contained three tandem repeats of the X-PBP binding sequence (wild type or mutant) were synthesized, high-performance liquid chromatography purified, annealed, and used for affinity purification of X-PBP. The partially purified X-PBP fraction (∼150 μg of protein) from the Poros HQ column was equilibrated with 0.3% NP-40-0.2 M NaCl-HGED and mixed with the biotinylated 48-bp wild-type X-PBP binding DNA (∼3 μg) at 30°C for 20 min in the presence of the carrier DNA [poly(dA-dT)/poly(dA-dT)]. The biotinylated X promoter DNA-X-PBP complexes were then collected by binding to streptavidin beads (Immunopure Ultralink beads; Pierce) at room temperature for 1 h. The beads were washed with 1 mg of lysozyme per ml-0.25 M NaCl-0.5% NP-40-HGED once and then with 0.5% NP-40-0.2 M NaCl-HGED five times to remove nonspecifically bound proteins. After washing, sequence-specifically bound proteins were eluted from the beads by incubation with an excess amount of the wild-type or mutant nonbiotinylated X promoter DNA at 30°C for 1 h. The eluates from the wild-type DNA column were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with a 7 to 15% gradient polyacrylamide gel, blotted onto a nitrocellulose membrane, and stained with Ponceau S, and the protein bands were excised. The nitrocellulose strips were digested with Lys-C, and the peptides produced from the digestion were separated by reversed-phase high-performance liquid chromatography and subjected to Edman microsequencing. For reference, a small part of the pooled X-PBP fraction from the Poros HQ column was separately mixed with the biotinylated mutant DNA and bound to the streptavidin beads. After extensive washing, the bound proteins were analyzed by SDS-PAGE followed by silver staining to identify any proteins that nonspecifically bound to the beads.
Plasmids.
An HBV DNA fragment that contains the core domain of the enhancer 1 and the X promoter region through the translational initiation codon of the X protein (nt 989 to 1253) were cloned into pSV00CAT (1). This reporter plasmid is equivalent to pXStNcCAT (11). The minimal promoter construct pXMP2CAT has been described previously (33). The NRF1 expression plasmid pSG5-NRF1 (39) was kindly provided by R. Scarpulla. An expression plasmid of a dominant-negative mutant form of NRF1 (deletion mutant form of the transcriptional activation domain located between amino acids 305 and 503) was constructed by PCR and standard cloning procedures. An X gene template plasmid (pHBVX-1) that contains the core domain of enhancer 1, the X promoter, and the X open reading frame through the poly(A) addition signal (nt 989 to 1858) in the pBR322 vector has been described previously (43). Derivatives of pHBVX-1 (pHBVX-MP2m, pHBVX-MP4m2, and pHBVX-Sph16) were constructed with a QuikChange site-directed mutagenesis kit (Stratagene) and the 34-mer oligonucleotides described under the EMSA procedures in Materials and Methods. Another X gene template plasmid (pBS-HBXB) that contains the whole enhancer 1 region, the X promoter, and the X open reading frame through the poly(A) addition signal (nt 123 to 1858) in the pBluescript vector was constructed for part of the X mRNA analyses.
DNA transfection, CAT assay, and RNA analysis.
HepG2 human hepatoblastoma cells were maintained with Dulbecco's modified minimal essential medium supplemented with 10% fetal bovine serum. The X gene template or the reporter plasmid was transfected into HepG2 cells with NRF1 expression plasmid pSG5NRF1 by the calcium phosphate-DNA coprecipitation method as described previously (33). Plasmid pCMV-β was cotransfected to monitor transfection efficiency. Forty-eight hours after transfection, the cells were harvested and cell extracts were assayed for CAT and β-galactosidase activities as previously described (33). To analyze X mRNAs expressed from the X gene templates, total RNA was extracted with TRIZOL reagent (Invitrogen), poly(A)+ RNA was selected with Oligotex beads (QIAGEN) as needed, and the X mRNA was detected by primer extension with a 32P-labeled oligonucleotide corresponding to nt 1252 to 1229 (nucleotide numbering based on reference 19). The resultant 32P-labeled cDNAs were analyzed on 6% acrylamide gels containing 7 M urea. Sequence ladders were made by the dideoxy termination method with the same set of primers and template plasmids as used for transfection and the primer extension reactions.
For analysis of the transcript from the minimal promoter CAT plasmid, primer extension was carried out with the CAT primer (28).
RNA interference.
NRF1-siRNA expression units (NRF1-siRNA cassettes) were constructed by connecting human U6.1 promoter and NRF1 siRNA coding regions with the RNA polymerase III transcriptional termination signal. The siRNA coding regions (78 bp in length) contained DNA sequences that can form hairpin structures with a 21-bp double-stranded region homologous to part of the NRF1 protein coding region (nt 140 to 160, 214 to 234, or 396 to 416 of the sequence with GenBank accession no. L22454). These NRF1-siRNA cassettes were transiently transfected into HepG2 cells with the X minimal promoter-CAT plasmid pXMP2CAT. The siRNA expression cassette DNA without the NRF1-siRNA coding sequence was used as the negative control. Two days after transfection, total cell extracts were prepared and analyzed for CAT activity.
In vitro transcription.
The standard transcription reaction mixtures contained 15 mM HEPES (pH 7.6), 5% glycerol, 6 mM MgCl2, 60 mM KCl, 1 mM dithiothreitol, ∼2.5 mg of HepG2 NE per ml, and 6.7 μg of template DNA per ml. The transcription reaction mixtures were incubated for 30 min at 30°C before ribonucleotides were added to a final concentration of 0.5 mM. Following the transcription reaction with nucleotides, proteins were digested with proteinase K and the mixture was extracted with phenol-chloroform and ethanol precipitated. The transcripts were detected by primer extension as described above for detection of the X mRNA from transfected HepG2 cells.
RESULTS
Purification and identification of X-PBP from HeLa cells.
Previous studies with the CAT reporter system indicated that a GC-rich 21-bp sequence located about 130 bp upstream of the X protein coding region contained a minimal promoter activity and that a ubiquitously expressed but unidentified cellular protein, X-PBP, could bind to this DNA (22, 43). Mutations at the X-PBP binding site diminished the interaction between the 21-bp DNA and this protein and also diminished the promoter activity, suggesting that this cellular protein plays a significant role in transcription of the X gene. As part of our ongoing effort to characterize the regulatory mechanisms of expression control of the HBV X gene, we set out to purify and identify this protein.
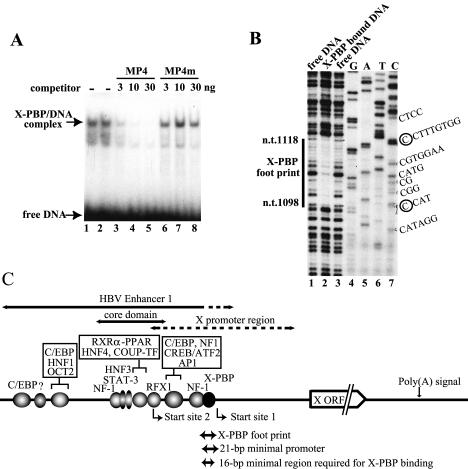
Since X-PBP has been reported to be expressed ubiquitously and because HeLa cells have often been used successfully by many investigators as the starting material to identify new transcription factors, we first checked for the presence and DNA-binding specificity of X-PBP in a HeLa cell NE by EMSAs and DNA footprinting analyses. As shown in Fig. 1A and B, our HeLa cell NEs showed the same EMSA and DNA footprinting patterns (nt 1097 or 1098 to 1118 or 1119) as those previously reported for hepatoma cell lines (22, 43). The X-PBP binding was detected only when poly(dA-dT)/poly(dA-dT) or sonicated salmon sperm DNA but not poly(dI-dC)/poly(dI-dC) was used as the carrier (data not shown) as reported. Since we also found that a 16-bp DNA subfragment (nt 1101 to 1116) from the 21-bp (nt 1101 to 1121) region was sufficient for X-PBP binding, we decided to use the 16-bp DNA fragment for EMSAs during purification of the X-PBP binding activity from HeLa cells. The locations of the X-PBP footprinting region, the 21-bp minimal promoter, and the 16-bp sequence required for X-PBP binding within the enhancer 1-X promoter region are schematically shown in Fig. 1C.
FIG. 1.
EMSA and DNase I footprinting assays showing that HeLa cell NE contains X-PBP. (A) An EMSA was performed with HeLa NE (3 μg), a 32P-labeled X promoter probe (MP4) (∼0.3 ng), and poly(dA-dT)/poly(dA-dT) (2.5 μg) in the presence or absence of the indicated amount of nonlabeled MP4 itself (lanes 3 to 5) or a mutated probe (MP4m) (lanes 6 to 8) as a competitor. (B) DNase I footprinting of X-PBP with HeLa NE. The binding reaction mixture containing the 32P-labeled X promoter DNA and HeLa NE was treated with a limited amount of DNase I and then applied to an EMSA gel. After exposure for autoradiography, the X-PBP-DNA complex in the gel was excised and DNA was purified and analyzed on a sequencing gel with sequencing ladders. (C) Diagram of the X gene expression unit in the HBV genome. HBV enhancer 1, the enhancer core region, the X promoter region, and the two major transcriptional start sites (nt 1029 ± 1 nt and nt 1118 ± 1 nt) are shown along with the X-PBP footprint region (nt 1098 ± 1 nt to nt 1118 ± 1 nt), the previously reported 21-bp minimal promoter region (nt 1101 to 1121), and the 16-bp region (nt 1101 to 1116) required for X-PBP binding. ORF, open reading frame.
To purify X-PBP, NE was prepared from HeLa cells and fractionated as shown in the purification scheme (Fig. 2A) by following the X-PBP activity by EMSA. As an example, the X-PBP activity assays (EMSAs) of the Poros HQ column fractions are shown in Fig. 2B. After the fractionation by conventional column chromatography, the peak fractions of the X-PBP activity (fractions 20 to 22 of the Poros HQ column) were pooled and used for DNA affinity purification with biotinylated 48-bp synthetic DNAs that contain three copies of the 16-bp minimal X-PBP binding region. In parallel, a small part of the pooled X-PBP peak fraction was mixed with 48-bp synthetic DNAs bearing three copies of the mutated 16-bp X-PBP binding region to examine sequence-nonspecific DNA binding. The DNA-X-PBP complexes were removed from the solution by binding to streptavidin beads, and the remaining part of the solution (unbound fraction) was assayed for X-PBP activity by EMSA. As shown in Fig. 2C, X-PBP was depleted by the wild-type DNA but not by the mutant DNA. The patterns of proteins present in the input, bound, and unbound fractions from the wild-type or mutant DNA were also assayed by SDS-PAGE and visualized by silver staining (Fig. 2D, lanes 1, 4, 5, 6, and 7). After washing the X-PBP-DNA-streptavidin bead complex, proteins specifically bound to the wild-type DNA were eluted with an excess amount of nonbiotinylated synthetic DNAs containing either the wild-type or mutant minimal promoter DNA sequences. As shown in Fig. 2D (lanes 2 to 5), a 68-kDa protein was found to be specifically bound to the wild-type DNA (lane 5) and to be specifically eluted from the wild-type DNA resin by the wild-type X promoter DNA (lane 3) but not by the mutant DNA (lane 2). Therefore, the 68-kDa protein band was isolated and subjected to microsequencing analyses. As shown in Fig. 3, microsequencing results indicated that the purified protein was NRF1, a 68-kDa protein that had previously been demonstrated to regulate a number of nucleus-encoded mitochondrion-related genes, translation- or protein turnover-related genes, DNA synthesis or repair genes, and cell proliferation- or apoptosis-related genes (8, 12, 39). The reported consensus sequence of NRF1-binding sites, RCGCRYGCGY (R denotes A or G; Y denotes T or C) (8, 12, 39), was found to be located within the X-PBP-footprinted region. The size of the region footprinted by the X-PBP factor was comparable to those reported by previous investigators for NRF1 (9, 17).
FIG. 2.
Purification of X-PBP. (A) Scheme of X-PBP purification. AmSO4, ammonium sulfate. (B) Mobility shift assay of Poros HQ column fractions. FT, flowthrough. (C) Mobility shift assay of input and unbound fractions from the DNA affinity purification. The unbound fraction from the mutant DNA resin retained as high a level of X-PBP activity as the input. In contrast, the unbound fraction from the wild-type DNA resin was depleted of X-PBP activity. (D) Silver staining analysis of input, unbound, and eluted fractions of DNA affinity purification. A 68-kDa protein was specifically bound to the wild-type DNA resin and eluted specifically with the wild-type X minimal promoter DNA. M, mutant; W, wild type.
FIG. 3.
Identification of X-PBP as NRF1. The amino acid sequence of human NRF1 is shown. The positions of the polypeptide sequences obtained by microsequencing are in bold and underlined. The numbers on the left are amino acid positions.
Functional characterization of NRF1 in transcription of the X gene.
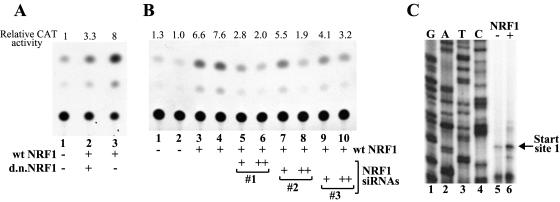
To determine whether the NRF1 binding to the X gene promoter was functionally relevant, we transfected HepG2 cells with an NRF1 expression plasmid and then examined the effects of NRF1 overexpression on transcription of the X gene. When the X gene minimal promoter-CAT construct pXMP2CAT was cotransfected with the NRF1 expression plasmid, 5- to ∼10-fold augmentation of the CAT activity was observed (Fig. 4A, lane 1 versus lane 3, and B, lanes 1 and 2 versus lanes 3 and 4). To confirm that this activation occurred at the level of transcription and that the activated transcription was initiated from the accurate start site, we examined the levels and start site(s) of the transcript expressed in transfected HepG2 cells by primer extension. As shown in Fig. 4C, the minimal promoter construct pXMP2CAT displayed transcription from nt 1118 ± 1 nt (numbering based on reference 19), which is consistent with one of two major start sites reported by Yaginuma et al. (43) and very close to one of the start sites reported by Siddiqui et al. (30). We found that transcription from this start site was indeed activated by NRF1 (Fig. 4C, lane 5 versus lane 6). We further found that this activation could be inhibited by overexpression of the dominant-negative NRF1 mutant (14, 42) that retained the DNA-binding domain but lacked the major transcriptional activation domain (Fig. 4A, lane 2 versus lane 3) or by expression of siRNAs directed against the NRF1 message (Fig. 4B, lanes 3 and 4 versus lanes 5 to 10).
FIG. 4.
NRF1 activates transcription from the 21-bp X gene minimal promoter, and NRF1 activation is inhibited by dominant-negative mutant NRF1 or NRF1 siRNAs. (A) HepG2 was transfected with the X minimal promoter-CAT construct pXMP2CAT plus an empty plasmid (lane 1) or a wild-type (wt) NRF1 expression plasmid (lanes 2 and 3) with (lane 2) or without (lane 3) a dominant-negative (d.n.) NRF1 mutant expression plasmid. Two days after transfection, cells were harvested and the CAT activity of the cell lysate was assayed. Relative CAT activity was calculated on the basis of the conversion rate of 14C-chloramphenicol into the acetylated forms. (B) HepG2 was transfected with the X minimal promoter-CAT construct pXMP2CAT plus an empty plasmid (lanes 1 and 2) or a wild-type NRF1 expression plasmid (lanes 3 to 10) with (lanes 5 to 10) or without (lanes 3 and 4) three different NRF1 siRNA expression DNAs (numbered 1 to 3) that target different parts of the X mRNA. NRF1 siRNA expression DNAs (0.5 μg/6-cm-diameter dish [lanes 5, 7, and 9] or 3.5 μg/6-cm dish [lanes 6, 8, and 10]) were transfected. CAT assays were performed as described above. (C) Analysis of the transcripts expressed from pXMP2CAT. HepG2 cells were transfected with pXMP2CAT plus the NRF1 expression plasmid (lane 6) or pXMP2CAT plus an empty plasmid (lane 5). Two days after transfection, poly(A)+ RNAs were purified and the transcripts from pXMP2CAT were detected by primer extension with a 32P-labeled CAT primer.
However, when the enhancer 1-X promoter-containing reporter plasmid (pXStNcCAT) was cotransfected with the NRF1 expression plasmid, augmentation of CAT activity by NRF1 was not very obvious (data not shown). There are potential explanations for the difference in transcriptional response between the minimal promoter context and the enhancer 1-containing context. First, the endogenous level of NRF1 in HepG2 cells may be sufficient for the enhancer-X promoter context to be efficiently activated by NRF1 and therefore additional overexpression of NRF1 did not result in further augmentation of transcription. In contrast, the X minimal promoter construct that does not contain other activator-binding sites may have weaker NRF1 protein-binding affinity because there would be no cooperative binding between NRF1 and other transcription activators. Hence, a minimal promoter construct may need a higher level of NRF1 than endogenous NRF1. Therefore, additional expression of NRF1 resulted in activation of transcription from the minimal promoter. If this is the case, inhibition of NRF1 may result in reduced transcription from the enhancer 1-containing constructs. Second, it is possible that there is an additional transcriptional start site(s) for the X mRNA (15, 30, 38, 43) and transcription from only one of them may be dependent on NRF1, making the effect(s) of NRF1 unclear in CAT assays.
To determine whether NRF1 is really important for X gene transcription in the enhancer-1 X promoter context, we transiently transfected HepG2 cells with enhancer-containing X gene plasmid pBS-HBXB together with the wild-type or dominant-negative NRF1 expression plasmid and analyzed X mRNA by primer extension. Two major transcriptional start sites (nt 1118 ± 1 nt and nt 1029 ± 1 nt) were reproducibly detected under the conditions used (Y. Tokusumi and S. Takada, unpublished data; for approximate position, see Fig. 1C). We will refer to nt 1118 ± 1 nt as start site 1 and nt 1029 ± 1 nt as start site 2 in this report. Start site 1 (nt 1118 ± 1 nt) is the same start site as that of minimal promoter plasmid pXMP2CAT. Start site 2 (nt 1029 ± 1 nt) has not been previously reported. We found that overexpression of NRF1 did not result in significant changes in transcription from either start site (Fig. 5A, lane 1 versus lane 2). However, when the dominant-negative form of NRF1 was overexpressed, transcription from start site 1 was markedly inhibited (Fig. 5A, lane 1 versus lane 3, lower column). Transcription from start site 2 appears to be slightly inhibited by the dominant-negative NRF1 but not as remarkably as start site 1 (Fig. 5A, lane 1 versus lane 3, upper column). These results suggest that NRF1 indeed contributes to activation of X gene transcription from start site 1 but that the endogenous level of NRF1 in HepG2 cells is not limiting for transcription in the enhancer-X promoter context.
To further assess the function of NRF1 in X gene transcription, we examined the transcripts expressed from the mutant X gene template that contain mutations at the NRF1-binding site shown in Fig. 5D. Figure 5B shows the X transcripts from the wild-type or mutant templates expressed in transfected HepG2 cells. As expected, transcription from start site 1 in the mutant templates was largely reduced compared to that in the wild-type template (Fig. 5B, lanes 2 to 4 versus lane 1), indicating again that the NRF1-binding site is very important for transcription from start site 1. Transcription from start site 2 of mutant 3 may be somewhat decreased but not as much as that from start site 1. Similar results were obtained from in vitro transcription assays with the mutant X gene templates (Fig. 5C). To confirm that these mutant DNAs actually have reduced NRF1-binding activity, we carried out an EMSA with these mutant DNAs as competitors. As expected, the three mutant DNAs did not inhibit binding between the wild-type DNA and NRF1 (Fig. 5E, lanes 4 to 9 versus lane 1) under conditions in which the nonlabeled wild-type DNA (MP4) clearly showed inhibition (Fig. 5E, lanes 2 and 3 versus lane 1). Taken together, our data suggest that NRF1 binds to the X minimal promoter DNA region and positively contributes to transcription of the X mRNA in a start site-specific manner.
DISCUSSION
NRF1 plays a critical role in core (or basal) promoter function for transcription from one of the major start sites of the X mRNA.
To understand the role of the X protein in transformation of hepatocytes, it is essential to understand not only the function(s) of the X gene product but also regulatory mechanisms of X gene expression. Recent studies examining RNA polymerase II core (or basal) promoters have pointed out the importance of promoter-enhancer specificity. These studies indicate that in many cases different core promoters respond differently to enhancer elements (3, 23). Such a mechanism may be at play in the regulation of HBV transcription, where the HBV genome contains two enhancers (enhancer 1 and 2) and at least five promoters (preS1, preS2, preC, C, and X). Transcription from all of the HBV promoters is under the control of these two enhancers, but the transcription patterns of the five HBV mRNAs are different (20). Therefore, it is critical to characterize the X gene promoter. Since the X gene promoter region contains no previously defined RNA polymerase II core (or basal) promoter elements, it was especially of interest to find out which protein(s) interacts with the X promoter region. As a first step toward understanding the regulatory mechanisms of X gene transcription, here we identified NRF1 as an important binding protein required for transcription of one of the major forms of X mRNA.
NRF1 has been most often described as a transcriptional activator whose binding site is located relatively close (less than 1 kb upstream) to a transcriptional start site(s). About 20% of NRF1-binding sites are within a −50- to ∼+40-bp region relative to transcription start sites (39), similar to the NRF1-binding site we defined in the X gene promoter. Interestingly, a search for genes containing NRF1-binding sites that are no more than 300 bp upstream of transcription initiation sites has shown that the overwhelming majority of the identified genes contain TATA-less promoters and that they code for housekeeping enzymatic activities expressed in most tissues (12). The fact that NRF1-binding sites are often found in core (or basal) promoter regions (i.e., generally within the range of −50 to ∼+50 bp relative to transcription start sites) is suggestive of a role for NRF1 in the regulation of core promoter function in TATA-less promoters. Our observation that NRF1 activated transcription from start site 1 rather than start site 2 of the X mRNA with some start site selectivity is also consistent with the idea that NRF1 is involved in core promoter function. Even though transcription from start site 2 may be also moderately modulated by NRF1 under some unidentified conditions, it still appears that there is an additional specific connection between NRF1 and transcriptional initiation from start site 1. The difference between start sites 1 and 2 in response to NRF1 also suggests functional diversity between different core promoters. Further studies are necessary to determine whether NRF1 activation of the X promoter utilizes additional cofactors or whether the mechanism NRF1 contributes to X gene transcription is different from those of many other enhancer-bound transcription regulators that activate transcription from outside the core promoter region.
Mechanisms of NRF1 activation of transcriptional initiation from start site 1 of the X mRNA.
In our transfection experiments with HepG2 cells, NRF1 binding to the 21-bp minimal promoter appears to be essential for transcription from start site 1 of the X mRNA because mutations at the NRF1-binding site disrupted transcription from start site 1 and because the dominant-negative NRF1 protein and NRF1 siRNAs inhibited transcription from start site 1. However, considering the fact that the NRF1-binding sites in the previously reported promoters have been so far described as regulatory elements rather than core (or basal) promoter elements, there is still a possibility that the NRF1-binding site by itself is not sufficient for the complete core promoter function and that there is an additional DNA element(s) in the 21-bp core (or basal) promoter region.
The GC box and the CAAT box are also often found in the proximal promoter regions of many genes, including TATA-less genes (31), but similar to the NRF1-binding site at the X gene, do not have core promoter activity by themselves. The binding factors for these elements, Sp1 family and CBP family proteins, are not general transcription factors (GTFs) but have been shown to directly interact with GTFs, such as TAF3, TBP, and TFIIB (10, 16). Because the location of NRF1-binding sites is somewhat similar to that of these elements, it will be interesting to see if there is any direct interaction between NRF1 and GTFs. Recent in vitro studies have shown that the transcription machinery (i.e., GTFs and RNA polymerase II) could be at least recruited to the core (or basal) promoter in the presence of an activator and the mediator even if the TATA box sequence was mutated, suggesting that recruitment of RNA polymerase II can occur through a mechanism that is activator dependent but TFIID (and/or TATA box) independent (41). To determine if transcription from start site 1 of the X gene utilizes such activator-dependent RNA polymerase II (and GTF) recruitment mechanisms, further DNA-protein and protein-protein interaction studies, as well as in vitro reconstituted transcription studies, are necessary.
NRF1 has been also previously shown to interact with PGC-1 (peroxisome proliferator-activated receptor γ coactivator 1) and/or PRC (PGC1-related coactivator) (29). As PGC-1 has been shown to directly interact with RNA polymerase II and RNA splicing factors (21, 25), it is a possibility that NRF1 indirectly interacts with RNA polymerase II through this bridging interaction(s) with PGC-1 and could potentially contribute to recruitment of RNA polymerase II to transcription start site 1 of the X gene. To date, our attempts to observe coactivation of X gene transcription by overexpression of PGC-1 or PRC together with NRF1 in HepG2 cells have been unsuccessful (S. Takada, unpublished data). However, the effects of PGC-1-PRC at the promoter may be masked since there is endogenous PRC expression in HepG2 cells. Further studies will examine siRNA knockdown of endogenous PGC-1-PRC levels.
Curiously, Yaginuma et al. (43) previously reported a function of the X-PBP binding site in regulation of X mRNA transcriptional start site selection. They observed that disruption of the X-PBP binding site gave rise to induction of transcription from random sites in the upstream region instead of simple loss of transcription from the normal site. However, both their observation and our observation are likely to reflect the role of NRF1 in core (or basal) promoter function (or regulation) because expression patterns of NRF1 and its cofactors may vary among different cell types or different physiological states and because different cofactor availability could result in a slightly different transcription response. Identification of additional players in transcription of the X gene will benefit further understanding of regulatory mechanisms of X gene expression.
Recent revelations provided by analyses of the human genome sequence showed that there are a substantial number of genes that are regulated by as yet unidentified core (or basal) promoter elements. This category of genes includes (but is not restricted to) many cell growth- or death regulation-related genes and housekeeping genes, as well as mitochondrial function-related genes. Defective expression of these genes could result in disruption of cellular functions, consequently leading to a variety of human diseases, including cancer. The knowledge we obtain from study of the X gene promoter is not only beneficial to understand the basis of HBV-induced liver diseases but also will likely be applicable to understanding transcription of important classes of TATA-less genes in eukaryotic cells.
Finally, since expression of NRF1 is responsive to environmental and/or physiological stresses, the finding that NRF1 acts to regulate expression of the HBV X gene opens an additional pathway by which X gene regulation may be responsive to stress and/or changes in the physiological conditions of infected cells. Importantly, our findings suggest that the X gene is likely to be activated in concert with mitochondrial genes since NRF1 is known to be a key activator driving the expression of this class of genes. This linkage strengthens the recently found connection between the function of the X protein and mitochondria.
Acknowledgments
We are grateful to Katsuro Koike at the Japanese Foundation for Cancer Research for HBV plasmids and to Richard Scarpulla for NRF1 expression plasmids. We also thank Robert Tjian at the University of California, Berkeley, for supporting the NRF1 purification described in this report. Special thanks go to Raymond Jacobson and Sharon Dent at the University of Texas M. D. Anderson Cancer Center (UT MDACC) for reading and editing the manuscript.
This research was supported in part by an institutional research grant at the UT MDACC and by an American Gastroenterological Association/Elsevier Research Initiative Award. DNA sequencing analyses were performed by the UT MDACC DNA sequencing core facility, which was supported by the core grant CA16672.
REFERENCES
- 1.Araki, E., F. Shimada, M. Shichiri, M. Mori, and Y. Ebina. 1988. pSV00CAT: low background CAT plasmid. Nucleic Acids Res. 16:1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouchard, M. J., L. H. Wang, and R. J. Schneider. 2001. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science 294:2376-2378. [DOI] [PubMed] [Google Scholar]
- 3.Butler, J. E., and J. T. Kadonaga. 2001. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 15:2515-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butler, J. E., and J. T. Kadonaga. 2002. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 16:2583-2592. [DOI] [PubMed] [Google Scholar]
- 5.Caselman, W. 1998. Transactivators of HBV, signal transduction and tumorigenesis. Imperial College Press, London, United Kingdom.
- 6.Chen, H.-S., S. Kaneko, R. Girones, R. W. Anderson, W. E. Hornbuckle, B. C. Tennant, P. J. Cote, J. L. Gerin, R. H. Purcell, and R. H. Miller. 1993. The woodchuck hepatitis virus X gene is important for establishment of virus infection in woodchucks. J. Virol. 67:1218-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Efiok, B. J., J. A. Chiorini, and B. Safer. 1994. A key transcription factor for eukaryotic initiation factor-2 alpha is strongly homologous to developmental transcription factors and may link metabolic genes to cellular growth and development. J. Biol. Chem. 269:18921-18930. [PubMed] [Google Scholar]
- 9.Evans, M. J., and R. C. Scarpulla. 1990. NRF-1: a trans-activator of nuclear-encoded respiratory genes in animal cells. Genes Dev. 4:1023-1034. [DOI] [PubMed] [Google Scholar]
- 10.Frontini, M., C. Imbriano, A. diSilvio, B. Bell, A. Bogni, C. Romier, D. Moras, L. Tora, I. Davidson, and R. Mantovani. 2002. NF-Y recruitment of TFIID, multiple interactions with histone fold TAF(II)s. J. Biol. Chem. 277:5841-5848. [DOI] [PubMed] [Google Scholar]
- 11.Fukai, K., S. Takada, O. Yokosuka, H. Saisho, M. Omata, and K. Koike. 1997. Characterization of a specific region in the hepatitis B virus enhancer I for the efficient expression of X gene in the hepatic cell. Virology 236:279-287. [DOI] [PubMed] [Google Scholar]
- 12.Gomez-Cuadrado, A., M. Martin, M. Noel, and A. Ruiz-Carrillo. 1995. Initiation binding repressor, a factor that binds to the transcription initiation site of the histone h5 gene, is a glycosylated member of a family of cell growth regulators. Mol. Cell. Biol. 15:6670-6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottlob, K., M. Fulco, M. Levrero, and A. Graessmann. 1998. The hepatitis B virus HBx protein inhibits caspase 3 activity. J. Biol. Chem. 273:33347-33353. [DOI] [PubMed] [Google Scholar]
- 14.Gugneja, S., C. M. Virbasius, and R. C. Scarpulla. 1996. Nuclear respiratory factors 1 and 2 utilize similar glutamine-containing clusters of hydrophobic residues to activate transcription. Mol. Cell. Biol. 16:5708-5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gustin, K., M. Shapiro, W. Lee, and R. D. Burk. 1993. Characterization of the role of individual protein binding motifs within the hepatitis B virus enhancer I on X promoter activity using linker scanning mutagenesis. Virology 193:653-660. [DOI] [PubMed] [Google Scholar]
- 16.Hoey, T., R. O. Weinzierl, G. Gill, J. L. Chen, B. D. Dynlacht, and R. Tjian. 1993. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell 72:247-260. [DOI] [PubMed] [Google Scholar]
- 17.Jacob, W. F., T. A. Silverman, R. B. Cohen, and B. Safer. 1989. Identification and characterization of a novel transcription factor participating in the expression of eukaryotic initiation factor 2 alpha. J. Biol. Chem. 264:20372-20384. [PubMed] [Google Scholar]
- 18.Jia, L., X. W. Wang, and C. C. Harris. 1999. Hepatitis B virus X protein inhibits nucleotide excision repair. Int. J. Cancer. 80:875-879. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi, M., and K. Koike. 1984. Complete nucleotide sequence of hepatitis B virus DNA of subtype adr and its conserved gene organization. Gene 30:227-232. [DOI] [PubMed] [Google Scholar]
- 20.Kosovsky, M. J., I. Qadri, and A. Siddiqui. 1998. The regulation of hepatitis B virus gene expression: an overview of the cis- and trans-acting components. Imperial College Press, London, United Kingdom.
- 21.Monsalve, M., Z. Wu, G. Adelmant, P. Puigserver, M. Fan, and B. M. Spiegelman. 2000. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol. Cell 6:307-316. [DOI] [PubMed] [Google Scholar]
- 22.Nakamura, I., and K. Koike. 1992. Identification of a binding protein to the X gene promoter region of hepatitis B virus. Virology 191:533-540. [DOI] [PubMed] [Google Scholar]
- 23.Ohtsuki, S., M. Levine, and H. N. Cai. 1998. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev. 12:547-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paterlini, P., K. Poussin, M. Kew, D. Franco, and C. Brechot. 1995. Selective accumulation of the X transcript of hepatitis B virus in patients negative for hepatitis B surface antigen with hepatocellular carcinoma. Hepatology 21:313-321. [PubMed] [Google Scholar]
- 25.Puigserver, P., G. Adelmant, Z. Wu, M. Fan, J. Xu, B. O'Malley, and B. M. Spiegelman. 1999. Activation of PPARγ coactivator-1 through transcription factor docking. Science 286:1368-1371. [DOI] [PubMed] [Google Scholar]
- 26.Rahmani, Z., K. W. Huh, R. Lasher, and A. Siddiqui. 2000. Hepatitis B virus X protein colocalizes to mitochondria with a human voltage-dependent anion channel, HVDAC3, and alters its transmembrane potential. J. Virol. 74:2840-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossner, M. T. 1992. Review: hepatitis B virus X gene product: a promiscuous transcriptional activator. J. Med. Virol. 36:101-117. [DOI] [PubMed] [Google Scholar]
- 28.Ryu, S., S. Zhou, A. G. Ladurner, and R. Tjian. 1999. The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature 397:446-450. [DOI] [PubMed] [Google Scholar]
- 29.Scarpulla, R. C. 2002. Transcriptional activators and coactivators in the nuclear control of mitochondrial function in mammalian cells. Gene 286:81-89. [DOI] [PubMed] [Google Scholar]
- 30.Siddiqui, A., S. Jameel, and J. Mapoles. 1987. Expression of the hepatitis B virus X gene in mammalian cells. Proc. Natl. Acad. Sci. USA 84:2513-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smale, S. T., and J. T. Kadonaga. 2003. The RNA polymerase II core promoter. Annu. Rev. Biochem. 72:449-479. [DOI] [PubMed] [Google Scholar]
- 32.Su, Q., C. H. Schroder, W. J. Hofmann, G. Otto, R. Pichlmayr, and P. Bannasch. 1998. Expression of hepatitis B virus X protein in HBV-infected human livers and hepatocellular carcinomas. Hepatology 27:1109-1120. [DOI] [PubMed] [Google Scholar]
- 33.Takada, S., N. Kaneniwa, N. Tsuchida, and K. Koike. 1996. Hepatitis B virus X gene expression is activated by X protein but repressed by p53 tumor suppressor gene product in the transient expression system. Virology 216:80-89. [DOI] [PubMed] [Google Scholar]
- 34.Takada, S., H. Kido, A. Fukutomi, T. Mori, and K. Koike. 1994. Interaction of hepatitis B virus X protein with a serine protease, tryptase TL2 as an inhibitor. Oncogene 9:341-348. [PubMed] [Google Scholar]
- 35.Takada, S., Y. Shirakata, N. Kaneniwa, and K. Koike. 1999. Association of hepatitis B virus X protein with mitochondria causes mitochondrial aggregation at the nuclear periphery, leading to cell death. Oncogene 18:6965-6973. [DOI] [PubMed] [Google Scholar]
- 36.Terradillos, O., A. de La Coste, T. Pollicino, C. Neuveut, D. Sitterlin, H. Lecoeur, M. L. Gougeon, A. Kahn, and M. A. Buendia. 2002. The hepatitis B virus X protein abrogates Bcl-2-mediated protection against Fas apoptosis in the liver. Oncogene 21:377-386. [DOI] [PubMed] [Google Scholar]
- 37.Terradillos, O., T. Pollicino, H. Lecoeur, M. Tripodi, M. L. Gougeon, P. Tiollais, and M. A. Buendia. 1998. p53-independent apoptotic effects of the hepatitis B virus HBx protein in vivo and in vitro. Oncogene 17:2115-2123. [DOI] [PubMed] [Google Scholar]
- 38.Treinin, M., and O. Laub. 1987. Identification of a promoter element located upstream from the hepatitis B virus X gene. Mol. Cell. Biol. 7:545-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Virbasius, C. A., J. V. Virbasius, and R. C. Scarpulla. 1993. NRF-1, an activator involved in nuclear-mitochondrial interactions, utilizes a new DNA-binding domain conserved in a family of developmental regulators. Genes Dev. 7:2431-2445. [DOI] [PubMed] [Google Scholar]
- 40.Wang, X. W., M. K. Gibson, W. Vermeulen, H. Yeh, K. Forrester, H. W. Sturzbecher, J. H. Hoeijmakers, and C. C. Harris. 1995. Abrogation of p53-induced apoptosis by the hepatitis B virus X gene. Cancer Res. 55:6012-6016. [PubMed] [Google Scholar]
- 41.Wu, S. Y., T. Zhou, and C. M. Chiang. 2003. Human mediator enhances activator-facilitated recruitment of RNA polymerase II and promoter recognition by TATA-binding protein (TBP) independently of TBP-associated factors. Mol. Cell. Biol. 23:6229-6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu, Z., P. Puigserver, U. Andersson, C. Zhang, G. Adelmant, V. Mootha, A. Troy, S. Cinti, B. Lowell, R. C. Scarpulla, and B. M. Spiegelman. 1999. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98:115-124. [DOI] [PubMed] [Google Scholar]
- 43.Yaginuma, K., I. Nakamura, S. Takada, and K. Koike. 1993. A transcription initiation site for the hepatitis B virus X gene is directed by the promoter-binding protein. J. Virol. 67:2559-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, Z., N. Torii, A. Furusaka, N. Malayaman, Z. Hu, and T. J. Liang. 2000. Structural and functional characterization of interaction between hepatitis B virus X protein and the proteasome complex. J. Biol. Chem. 275:15157-15165. [DOI] [PubMed] [Google Scholar]
- 45.Zoulim, F., J. Saputelli, and C. Seeger. 1994. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J. Virol. 68:2026-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]