Abstract
Rotavirus entry is a complex multistep process that depends on the trypsin cleavage of the virus spike protein VP4 into polypeptides VP5 and VP8 and on the interaction of these polypeptides and of VP7, the second viral surface protein, with several cell surface molecules, including integrin αvβ3. We characterized the effect of the trypsin cleavage of VP4 on the binding to MA104 cells of the sialic acid-dependent virus strain RRV and its sialic acid-independent variant, nar3. We found that, although the trypsin treatment did not affect the attachment of these viruses to the cell surface, their binding was qualitatively different. In contrast to the trypsin-treated viruses, which initially bound to the cell surface through VP4, the non-trypsin-treated variant nar3 bound to the cell through VP7. Amino acid sequence comparison of the surface proteins of rotavirus and hantavirus, both of which interact with integrin αvβ3 in an RGD-independent manner, identified a region shared by rotavirus VP7 and hantavirus G1G2 protein in which six of nine amino acids are identical. This region, which is highly conserved among the VP7 proteins of different rotavirus strains, mediates the binding of rotaviruses to integrin αvβ3 and probably represents a novel binding motif for this integrin.
Rotavirus is the leading etiologic agent of severe diarrheal disease in infants worldwide (25). The capsid of this nonenveloped virus is formed by three concentric layers of protein (9), and the initial interactions of the virus with the cell surface are accomplished by the two proteins of the outermost layer: VP7, a glycoprotein that forms the smooth surface of the virion, and VP4, which forms spikes that extend from the surface of the viral particle (34).
Rotavirus infectivity is increased by, and most probably depends on, trypsin treatment of the virus. This proteolytic treatment, which results in the specific cleavage of VP4 (88 kDa) to polypeptides VP8 (28 kDa) and VP5 (60 kDa) (3, 8, 10), is not needed for the virus to attach to the cell surface (5) but for the virion to penetrate into the cells' interior (24). The mechanism through which trypsin enhances virus penetration is not known; however, unlike uncleaved virions, the trypsin-cleaved virus can induce fusion from without in MA104 cells (11, 14).
Rotavirus cell entry is a complex multistep process in which several cellular molecules have been implicated. It has been proposed that rotavirus strains that are sensitive to neuraminidase treatment of cells bind in the first place to a sialic acid-containing receptor. After this initial contact, which is mediated by VP8 (12, 23, 39), a second interaction with integrin α2β1, which is apparently shared by neuraminidase-sensitive and -resistant strains, takes place (4, 38). This interaction is mediated by the integrin-binding motif DGE present at residues 308 to 310 of VP5 (15, 38), and it was recently shown that the I domain of the α2 integrin subunit is both necessary and sufficient for the binding of VP5 (27). In addition to these two interactions, integrins αvβ3 and αxβ2, and the heat shock protein hsc70, have also been shown to be involved at a later step of rotavirus cell entry (16, 17, 19, 28).
Integrins are a family of cell surface receptors that mediate the interaction between the cell surface and the extracellular matrix and also mediate important cell-cell adhesion events; these interactions play a crucial role in the regulation of cell proliferation, migration, differentiation, and survival. Integrins are transmembrane heterodimers composed of noncovalently associated α and β subunits. Human integrins contain at least 18 different α subunits and 8 β subunits, which form 24 different heterodimers. Each integrin heterodimer has distinct ligand-binding specificity and signaling properties. The integrin recognition motifs on several integrin ligands have been described, and it has been established that the integrin recognition sites can often been reduced to small peptide sequences (21, 33). Several viruses and bacteria, which contain canonical integrin-binding motifs in their surface, take advantage of this family of proteins to gain access into the cell (36). In addition, some viruses have been found to interact with integrins through nontypical sequence motifs (32). Thus, although integrin αvβ3 has been shown to be involved in rotavirus cell infection at a postattachment step (17), neither of the virus surface proteins contains the canonical RGD tripeptide-binding motif for this integrin (20, 22); accordingly, it has been previously shown that the interaction between rotaviruses and αvβ3 does not take place through the RGD-binding site of the integrin (17).
We characterized the effect of trypsin treatment on the cell-binding characteristics of the sialic acid-dependent strain RRV and of its sialic acid-independent variant nar3. We found that, although both strains bound to the cell surface efficiently regardless of the cleavage status of VP4, the protein they used to attach to the cell was different depending on the virus strain and on whether VP4 was cleaved or not. The nar3 virus containing an uncleaved VP4 bound to the cell surface by interacting with integrin αvβ3 through a CNP sequence region on VP7. This interaction was also established by trypsin-treated nar3 and by both trypsin-treated and untreated RRV after their initial attachment to the cell surface through VP4.
MATERIALS AND METHODS
Cells and viruses.
MA104 cells were cultured in Eagle minimal essential medium (MEM) supplemented with 10% fetal bovine serum. Rhesus rotavirus (RRV) was obtained from H. B. Greenberg, Stanford University, Stanford, Calif., and rotavirus variant nar3 has been described previously (30). To prepare viruses with cleaved or uncleaved VP4, RRV and nar3 were propagated in MA104 cells as follows. Confluent cell monolayers were washed twice with MEM without serum and virus-containing cell lysates (activated with 10 μg of trypsin/ml for 30 min at 37°C) were adsorbed for 1 h at 37°C. The cells were washed thrice with MEM without serum to remove the excess of trypsin and were left in MEM at 37°C until a cytopathic effect was attained. The virus-infected cells were frozen and thawed twice, and the cell lysate was divided into two parts. One part was treated with 100 μg of trypsin/ml for 30 min at 37°C (Tr; virus treated with trypsin), and the other part was kept on ice (NTr; virus not treated with trypsin). To prepare purified triple-layered particles (TLPs), the virus in the cell lysates were pelleted by centrifugation for 1 h at 25,000 rpm at 4°C in an SW28 rotor (Beckman). The virus pellet was resuspended in TNC buffer (10 mM Tris-HCl [pH 7.5], 140 mM NaCl, 10 mM CaCl2), sonicated once for 20 s, and extracted with Genetron (trichloro-monofluoro-ethane). Then, CsCl was added to obtain a density of 1.36 g/cm3, the mixture was centrifuged for 18 h at 36,000 rpm in an SW50.1 rotor, and the virus bands were collected.
Antibodies.
Monoclonal antibodies (MAbs) 2G4 (specific for VP5), 7A12, 1A9 (specific for VP8), and 159 (specific for VP7), kindly provided by H. B. Greenberg (Stanford University), were used at 50 μg/ml. A polyclonal goat antibody directed to an epitope located at the amino terminus of integrin subunit β3 was obtained from Santa Cruz Biotechnology and used at 20 μg/ml. MAbs to integrins α2 (P1E6), β1 (P4G11), and αvβ3 (LM609) were purchased from Chemicon and used at 10 μg/ml.
Peptides.
Synthetic peptides CNP (NEWLCNPDM), sCNP (WPENNCDLM), and HANTA (NSWACNPPD) were obtained from Research Genetics and used as indicated. Peptide RGD (GRGDSP) was obtained from Gibco. To label the peptides with biotin, three amino acids (KYG) were added to the amino or carboxyl terminus of CNP or sCNP, respectively (peptides KYG-CNP and sCNP-GYK; Research Genetics). Biotinylation was carried out by incubation of 100 μl of a 10-mg/ml solution of the peptide in phosphate-buffered saline (PBS), with a final concentration of 10 μg of Sulfo-NH-LC-Biotin (Pierce)/ml for 2 h at room temperature. The residual free biotin was neutralized by the addition of 50 μl of 100 mM glycine for 2 h at room temperature. The biotinylated peptides were separated from free biotin by spin centrifugation on G10 Sepharose. To verify that biotinylation was attained, a solid-phase assay was performed by coating a 96-well polystyrene enzyme-linked immunosorbent assay (ELISA) plate with the peptides in bicarbonate buffer (50 mM NaHCO3 [pH 9]) and detecting them with streptavidin-peroxidase (see below).
Infectivity assay.
MA104 cells in 96-well plates were washed twice with MEM, and then 2,000 focus-forming units (FFU; multiplicity of infection of ca. 0.04) of RRV or nar3 (preincubated with 10 μg of trypsin/ml for 30 min at 37°C) were adsorbed to the cells for 1 h at 37°C. After the adsorption period, the virus inoculum was removed, the cells were washed twice with MEM, and the cultures were maintained for 14 h at 37°C. The infected cells were fixed and immunostained with an immunoperoxidase focus detection assay, as described previously (2). The FFU were counted by using a Visiolab semiautomatic system (18).
Binding assay.
MA104 cells grown in 48-well plates were washed twice and incubated with MEM without serum for 30 min at 37°C. After this time MEM was removed, and 500 μl of a solution of 1% of bovine serum albumin (BSA) in PBS was added to the cells, followed by incubation for 1 h at room temperature. The cells were then washed with an ice-cold solution of 0.5% BSA-PBS and incubated with the indicated amount of virus, recombinant protein, or peptides, diluted in ice-cold MEM containing 0.5% BSA, for 1 h at 4°C. The cells were then washed three times with ice-cold PBS with 0.5% BSA, and finally 120 μl of lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% Triton X-100) were added. The cells were frozen and thawed twice, and the amount of virus present in the cell lysate was determined by an ELISA. In these experiments, to assure that the saturation observed was due to the saturation of the binding sites in the cell surface and not to the saturation of the detection system, a direct ELISA with increasing amounts of purified viruses was performed in parallel. In this case the optical density (OD) readings of the direct ELISA increased linearly up to 1.2 (data not shown).
Capture ELISA.
To detect the amount of virus bound to cells, goat and rabbit polyclonal sera were used as capture (diluted 1:10,000) and detection (diluted 1:1,500) antibodies, respectively. The ELISA was performed essentially as described by Zárate et al. (39). When the recombinant GST-VP8 and GST-VP5 proteins were used to compete virus binding, the antibody used for the detection step was MAb 159 directed to VP7.
Binding to integrins.
Ninety-six-well polystyrene ELISA plates were coated with 100 ng per well of either integrin αvβ3 or α5β1 (Chemicon) in PBS and incubated overnight at 4°C. To avoid nonspecific binding, 200 μl of 1% BSA in PBS was added to the plate, followed by incubation for 2 h at 37°C. Different amounts of rotavirus nar3 or biotinylated peptides diluted in PBS containing 0.5% BSA were then added, followed by incubation for 1 h at 37°C. After this incubation, the plates were washed twice with PBS with 0.5% BSA, and the binding of biotinylated peptide was detected with streptavidin-peroxidase (1:1,500) in PBS containing 0.5% of BSA; the amount of virus bound to the plate was determined with a rabbit anti-rotavirus sera (diluted 1:1,500) and an anti-rabbit immunoglobulin peroxidase conjugate (diluted 1:1,500). After incubation with the respective peroxidase conjugate for 1 h at 37°C, the substrate was added (0.1 M citrate buffer [pH 4.5], 1 mg o-phenylenediamine [Sigma]/ml, 4 μl of H2O2/10 ml), and the reaction was stopped by the addition of 100 μl of 0.1 M H2SO4 when color was attained. The absorbance at 490 nm was recorded in a Microplate Autoreader EL311 (Bio-Tek Instruments).
RESULTS
Trypsin treatment of RRV and nar3 virions does not affect their binding to the surface of MA104 cells.
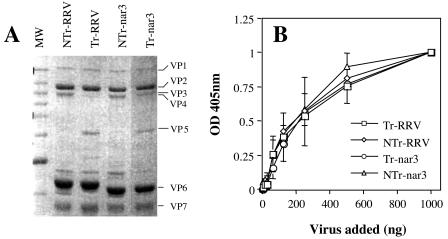
To evaluate the role of trypsin treatment on the binding of RRV and its sialic acid-independent variant nar3 to the cell surface, RRV and nar3 viruses were propagated in MA104 cells in the absence of trypsin, and the cell lysates were then divided in two parts. One was treated with trypsin (Tr virus), whereas the second was mock treated (NTr virus). After the protease treatment, the viruses were purified by CsCl gradients, and the TLPs obtained from each condition were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining (Fig. 1A). NTr viruses had no detectable levels of VP5, whereas in the Tr virions the VP5 protein was clearly visible, and there was scarce, or no VP4, confirming the effectiveness of the protease treatment. The binding of these viral particles to cells was determined by a direct, nonradioactive assay, in which increasing amounts of purified viruses were incubated on a confluent monolayer of MA104 cells. The unbound virus was removed by three washes with ice-cold PBS-BSA, the cell monolayer was frozen and thawed twice, and the virus present in the cell lysate was determined by an ELISA as described previously (39). As shown in Fig. 1B, the binding of Tr- and NTr-RRV and -nar3 viruses to MA104 cells was dose dependent and saturable. Both Tr and NTr viruses bound similarly to the cell surface, suggesting that the trypsin treatment did not affect the attachment of the virions to the cell surface.
FIG. 1.
Binding of trypsin-treated or untreated viral particles to MA104 cells. (A) SDS-PAGE analysis of Tr- or NTr-RRV and -nar3 viruses. Rotaviruses were propagated in MA104 cells in the absence of trypsin; the resultant viral cell lysates were then treated with 100 μg of trypsin/ml for 30 min at 37°C or mock treated, and the virus particles were purified by isopycnic centrifugation. The viral proteins were separated by SDS-10% PAGE and were Coomassie blue stained. The location of the individual proteins is indicated. (B) Binding of Tr- or NTr-RRV and -nar3 viruses to the surface of MA104 cells. The indicated amounts of purified TLPs were added to cells grown in 48-well plates. The cells were incubated with the viruses for 1 h at 4°C, and the amount of cell-bound virus was determined by an ELISA. The total amount of viral particles added in each assay (in nanograms) is plotted against the OD at 405 nm (OD405) reading obtained in the ELISA. A representative result of at least two independent experiments performed in duplicate is shown.
NTr-nar3 virus binds to the cell surface through VP7.
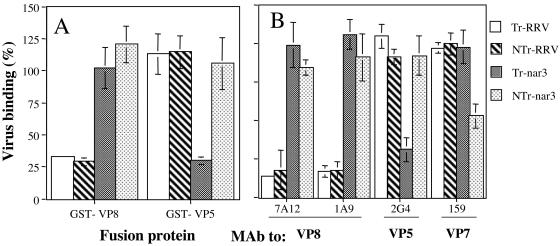
We have previously shown that Tr rotavirus RRV binds initially to the cell surface through the VP8 domain of VP4, and this binding was shown to be blocked by a recombinant glutathione S-transferase (GST)-VP8 protein (39). In contrast, the binding of Tr-nar3 virus was not inhibited by GST-VP8 but was blocked by preincubation of the cells with a GST-VP5 protein (39). To determine whether Tr and NTr viruses use the same protein to attach to the cell surface, we characterized the effect of the GST fusion recombinant polypeptides on the binding of RRV and nar3 virions treated or not treated with trypsin. For this, MA104 cells were preincubated with affinity-purified GST-VP8 or GST-VP5 proteins, and then a fixed amount of Tr or NTr viruses was added and allowed to bind for 1 h at 4°C. The amount of virus bound to the cell surface was determined by an ELISA. As expected, GST-VP8 competed for the binding of Tr-RRV as well as of NTr-RRV, without affecting the binding of Tr- and NTr-nar3 (Fig. 2A). Interestingly, GST-VP5 competed the binding of Tr-nar3, as reported earlier (39), but did not block the attachment of NTr-nar3, suggesting that NTr-nar3 might not be using either VP8 or VP5 to initially bind to the cell surface.
FIG. 2.
Cell binding of Tr- or NTr-RRV and -nar3 viruses in the presence of the recombinant proteins GST-VP8 and GST-VP5 or in the presence of MAbs to the viral proteins. (A) MA104 cells grown in 48-well plates were preincubated with 1.5 μg of affinity-purified GST-VP8 or GST-VP5 proteins. After removal of the excess of protein, 500 ng of the indicated purified virus was added. The cells were further incubated for 1 h at 4°C, and the amount of cell-bound virus was determined as described in Materials and Methods. (B) Purified viral particles (500 ng) were preincubated for 1 h at room temperature with 50 μg of the indicated MAbs to rotavirus proteins/ml. After this incubation, the virus-antibody mixture was added to a monolayer of MA104 cells, followed by incubation for 1 h at 4°C, and the amount of cell-bound virus determined. The data are expressed as the percentage of the virus bound when the cells were preincubated with PBS as a control. The arithmetic means and standard deviations for two independent experiments performed in duplicate are shown.
To further explore this observation, we analyzed the cell binding of NTr viruses in the presence of MAbs directed to VP7, VP8, or VP5 virus surface proteins. In these assays a fixed amount of virus was preincubated with the different MAbs for 1 h at room temperature, this mixture was then added to a cell monolayer, and the cell-bound virus was determined by an ELISA. All MAbs used reacted equally well with cleaved or uncleaved RRV and nar3 viruses as tested by an ELISA (data not shown). Figure 2B shows that MAbs 7A12 and 1A9 (directed to VP8) blocked the binding of both Tr and NTr-RRVs without affecting the binding of Tr- and NTr-nar3. MAbs 2G4 (directed to VP5) and 159 (directed to VP7) did not affect the binding of RRV. The binding of Tr-nar3 was blocked by MAb 2G4, whereas this antibody did not affect the binding of NTr-nar3. Interestingly, MAb 159 to VP7 decreased the binding of NTr-nar3 by ca. 50%, whereas it did not affect the binding of any other virus. Altogether, these results suggest that unlike Tr-nar3, NTr-nar3 does not bind to the cell surface through VP5 but might use VP7 to initially attach to the cell.
An antibody to integrin subunit β3 blocks the binding of NTr-nar3 virus.
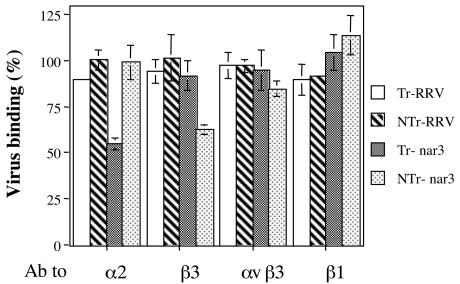
We and others have recently shown that integrin αvβ3 acts as postattachment receptor during rotavirus cell entry (15, 17). To test whether the cellular molecule recognized by NTr-nar3 virions during cell attachment was αvβ3, we studied the effect of antibodies directed to integrin subunits β3, α2, or β1, or to integrin αvβ3, on the binding of Tr- or NTr-RRV and -nar3 viruses. Cells were preincubated with the different antibodies for 1 h at 4°C, and then a fixed amount of purified virus was added and allowed to bind to the cells. As expected, the binding of RRV virions (either Tr or NTr) was not significantly affected by any of the integrin antibodies tested (Fig. 3). On the other hand, the binding of Tr-nar3, but not of NTr-nar3, was reduced by ca. 50% by the MAb to integrin subunit α2 (Fig. 3), a finding consistent with the previous report that Tr-nar3 attaches to the cell surface by using integrin α2β1 (38). Two antibodies to β3 were tested; one of them recognizes an epitope conformed by the two subunits of the αvβ3 heterodimer, while the other only recognizes the β3 subunit. The antibody to αvβ3 did not block the binding of any of the viruses tested, whereas the antibody to the β3 subunit blocked the binding of NTr-nar3 by ca. 40%, though not affecting the cell attachment of the other three viruses. The MAb to integrin subunit β1, which was used as a negative control, did not modify the binding of any virus. These results indicate that, in contrast to Tr-nar3, the binding of NTr-nar3 is not only independent of VP5 but is also independent of integrin α2β1; also, these data suggest that the initial interaction of NTr-nar3 with the cell surface is mediated by the integrin subunit β3.
FIG. 3.
Cell binding of Tr- or NTr-RRV and -nar3 viruses in the presence of antibodies to different integrin subunits. MA104 cells grown in 48-well plates were preincubated with antibodies to the indicated integrin subunits for 1 h at 4°C, the excess of antibody was washed, and a fixed amount of purified virus particles (500 ng) was added to the cells, followed by further incubation for 1 h at 4°C. The cell-bound virus was determined as indicated in Materials and Methods. The antibodies used included polyclonal goat antibody to β3 (20 μg/ml), MAb P4G11 to β1 (10 μg/ml), MAb LM609 to αvβ3 (10 μg/ml), and MAb P1E6 to α2 (10 μg/ml). The data are expressed as the percentage of the virus bound when the cells were preincubated with PBS as a control. The arithmetic means and standard deviations for two independent experiments performed in duplicate are shown.
A peptide that mimics a conserved region of VP7 blocks the binding of NTr-nar3 virus.
As mentioned, the interaction of rotaviruses with integrin αvβ3 was shown to be independent of the canonical motif RGD present in the natural ligands of this integrin (17). Hantavirus cell infection has also been shown to be mediated by β3 integrins and, as in the case of rotaviruses, this interaction is RGD independent (13). In an attempt to identify the protein domain through which rotaviruses and hantaviruses interact with αvβ3, the amino acid sequence of the surface proteins of these two types of viruses was compared. Pairwise alignments of the VP4 and VP7 proteins of rotavirus RRV and the G1G2 surface protein of hantavirus L99 identified a region in VP7 (amino acids [aa] 161 to 169), which has 6 of 9 aa identical to a region in the G1G2 protein of hantavirus comprised between aa 759 and 767 (Fig. 4). To study the relevance of this region, two peptides, one corresponding to the rotavirus VP7 region (named CNP) and a second corresponding to the G1G2 protein of the hantavirus L99 (named HANTA), were synthesized. As a control, a third peptide with the same amino acid composition as the CNP peptide but with different sequence (scrambled-CNP [sCNP]) was also prepared.
FIG. 4.
Alignment of rotavirus RRV protein VP7 (aa 161 to 169) with the G1G2 protein of hantavirus L99 (aa 759 to 767).
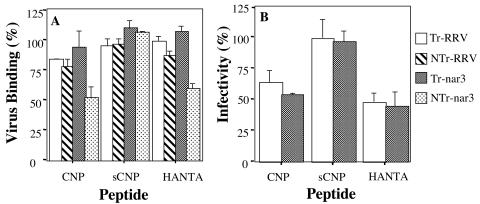
Since our previous experiments suggested that NTr-nar3 virions bound to the cell surface through VP7, we sought to determine whether this attachment was mediated by the protein region containing the CNP peptide. For this, cells were preincubated with the synthetic CNP peptide, its control sCNP, or the HANTA peptide for 1 h at 4°C. After removing the excess of peptide, the Tr- or NTr-RRV and -nar3 viruses were added, followed by incubation with the cells for 1 h at 4°C. The cell-bound virus was determined by an ELISA. As shown in Fig. 5A, both the CNP and the HANTA peptides decreased by ca. 50% the binding of NTr-nar3 virus but only slightly affected the binding of the other viruses. The control sCNP peptide did not affect the binding of any of the viruses tested.
FIG. 5.
Effect of the synthetic peptides on the binding and infectivity of RRV and nar3 rotaviruses. (A) MA104 cells in 48-well plates were preincubated with 4 mg of each peptide/ml for 1 h at 4°C, the excess of peptide was removed, and 500 ng of Tr- or NTr-RRV and -nar3 viruses was then added, followed by incubation for 1 h at 4°C. The amount of bound virus was determined by an ELISA. The data are expressed as the percentage of the virus bound when the cells were preincubated with PBS as a control. The arithmetic means and standard deviations for two independent experiments performed in duplicate are shown. (B) MA104 cells grown in 96-well plates were preincubated for 1 h at 37°C with 4 mg of peptides CNP, sCNP, or HANTA/ml; the excess peptide was removed, and 2,000 FFU per well of either RRV or nar3 virus were then added and adsorbed for 1 h at 37°C. The excess inoculum was removed, and the infection was allowed to proceed for 14 h at 37°C. Finally, the cells were fixed and immunostained as described in Materials and Methods. The data are expressed as the percentage of virus infectivity obtained when the cells were preincubated with MEM as a control. The average numbers of foci representing 100% infectivity were 157 and 207 for RRV and nar3, respectively. The arithmetic means and standard deviations of three independent experiments performed in duplicate are shown.
Peptide CNP blocks rotavirus infectivity.
To determine whether the region represented by peptide CNP was relevant for the infectivity of rotaviruses, the effect of this peptide and of peptide HANTA on the infectivity of RRV and nar3 was tested. For these assays only Tr viruses were used, since the virions must be activated by trypsin to be infectious. Preincubation of the cells with the CNP peptide inhibited the infectivity of RRV and nar3 by ca. 35 and 45%, respectively (Fig. 5B). Of interest, an inhibition of ca. 50% in the infectivity of both viruses was attained when the cells were preincubated with the HANTA peptide; the scrambled peptide used as a control for these experiments did not have any effect. The results of these experiments suggest that the region of VP7 represented by the CNP peptide might mediate the interaction between rotaviruses and integrin αvβ3. Since the hantavirus-derived peptide was also able to interfere with rotavirus binding and infectivity, it is possible that both CNP and HANTA peptides bind to the same region of integrin αvβ3, thus preventing the rotavirus-αvβ3 interaction needed for the cell entry of the virus.
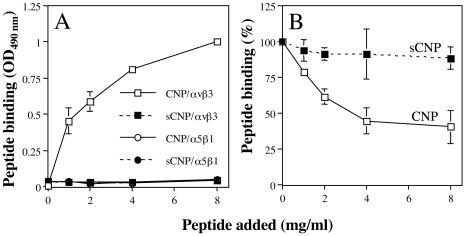
The CNP peptide binds to purified αvβ3 integrin.
To determine whether the CNP peptide was able to interact directly with integrin αvβ3, a solid-phase assay was performed. In this assay, the binding of biotinylated CNP or sCNP peptides to purified, immobilized integrins αvβ3 and α5β1 (both of which recognize the RGD sequence in their natural ligands) was detected with a peroxidase-coupled streptavidin conjugate. Peptide CNP, but not sCNP, bound to integrin αvβ3 in a concentration-dependent manner (Fig. 6A), although neither peptide bound to integrin α5β1. In addition, nonbiotinylated CNP was shown to compete the binding of the biotinylated CNP peptide to αvβ3, whereas sCNP did not displace its binding (Fig. 6B), indicating that peptide CNP binds to integrin αvβ3 in a specific manner.
FIG. 6.
Direct binding of peptide CNP to purified αvβ3 integrin. (A) ELISA plates coated with αvβ3 or α5β1 integrins (100 ng per well) were incubated for 1 h at 37°C with increasing amounts of biotinylated peptides. The amount of peptide bound to the plate was detected by a streptavidin-peroxidase assay. The amount of peptide added is plotted against the OD490 reading obtained in the ELISA. (B) The indicated amounts of nonbiotinylated CNP or sCNP peptide were added to a 96-well plate coated with integrin αvβ3 for 1 h at 37°C. After this incubation, 2 mg of biotinylated-CNP peptide/ml was added, and the plates were further incubated for 1 h at 37°C. The biotinylated peptide bound to the well was determined with streptavidin-peroxidase as described above. The data are expressed as the percentage of peptide bound when the wells were preincubated with PBS as a control. The arithmetic means and standard deviations for two independent experiments performed in duplicate are shown.
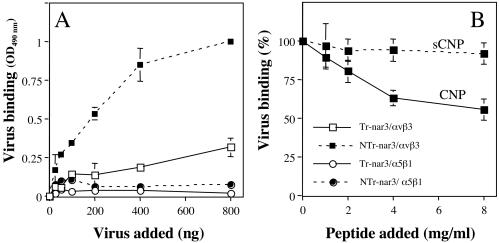
The binding of NTr-nar3 to integrin αvβ3 is blocked by the CNP peptide.
To determine whether rotavirus nar3 either treated or not treated with trypsin binds directly to αvβ3 integrin, an ELISA plate coated with either αvβ3 or α5β1 integrin was incubated with increasing amounts of Tr- or NTr-nar3 virus, and the amount of virus bound to the plate was detected by using a rabbit polyclonal anti-rotavirus sera. NTr-nar3 virus was found to bind to purified integrin αvβ3 in a concentration-dependent manner but not to integrin α5β1, whereas Tr-nar3 did not bind to any of the integrins tested (Fig. 7A). To establish whether the binding of NTr-nar3 to αvβ3 was mediated by the region represented by peptide CNP, the ability of this peptide to compete for the virus-αvβ3 interaction was evaluated. In this case, a plate coated with integrin αvβ3 was preincubated with different amounts of CNP or sCNP peptides, and then a fixed amount of NTr-nar3 virus was added. Increasing concentrations of peptide CNP, but not sCNP, were found to decrease the amount of NTr-nar3 bound (Fig. 7B). Altogether, these results suggest that NTr-nar3 rotavirus interacts directly with integrin αvβ3, most probably through the CNP region of VP7.
FIG. 7.
Binding of NTr-nar3 rotavirus to purified integrin αvβ3. (A) ELISA plates coated with αvβ3 or α5β1 integrins (100 ng per well) were incubated for 1 h at 37°C with increasing amounts of Tr- or NTr-nar3 viral particles. The virus bound to the plates was detected with a rabbit serum against rotavirus. The amount of virus added to the plate is plotted against the OD490 reading obtained for the virus bound. (B) Different amounts of nonbiotinylated CNP or sCNP peptide were added to a 96-well plate coated with αvβ3 integrin for 1 h at 37°C. After this incubation, NTr-nar3 virus (300 ng/well) was added, followed by further incubation for 1 h at 37°C. The amount of virus bound to the plate was detected with specific antibodies to rotavirus. The data are expressed as the percentage of virus bound when the αvβ3 integrin was preincubated with PBS as a control. The arithmetic means and standard deviations of two independent experiments performed in duplicate are shown.
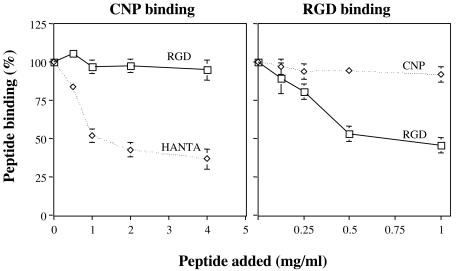
The CNP-binding site on integrin αvβ3 is different from the RGD-binding site.
To test whether the site of interaction of peptide CNP with integrin αvβ3 is different from that used by the RGD motif, we assayed whether these two peptides competed for binding to αvβ3 in a solid-phase assay. For this, an ELISA plate coated with integrin αvβ3 was preincubated with increasing amounts of either peptide HANTA or a synthetic peptide that contains the RGD sequence; a fixed amount of biotinylated CNP peptide was then added, and its binding was detected by using streptavidin-peroxidase. We found that whereas increasing amounts of HANTA peptide competed for the binding of peptide CNP to the integrin, the RGD peptide did not affect the binding of biotinylated CNP at any of the concentrations tested (Fig. 8, left panel). These results were further confirmed in a reciprocal assay, in which the binding of a biotinylated RGD peptide in the presence of nonbiotinylated CNP or RGD peptides was evaluated. As expected, peptide CNP did not block the binding of the RGD peptide to αvβ3, whereas the nonbiotinylated RGD did displace its binding (Fig. 8, right panel). Taken together, these results indicate that the binding sites on αvβ3 integrin for the RGD and CNP peptides are different and suggest that the CNP peptide might represent a novel sequence motif for interaction with this integrin.
FIG. 8.
The binding of CNP to αvβ3 integrin is independent of the RGD-binding site. Left panel, Different amounts of RGD or HANTA peptides were added to a 96-well plate coated with αvβ3 integrin for 1 h at 37°C. After incubation, a fixed amount (1 mg/ml) of biotinylated CNP peptide was added, followed by incubation for 1 h at 37°C. The biotinylated peptide bound to the well was determined by a streptavidin-peroxidase assay. Right panel, An αvβ3 integrin coated plate was preincubated with the indicated amounts of CNP or RGD peptides, and a fixed amount (1 mg/ml) of biotinylated RGD peptide was then added, followed by incubation for 1 h at 37°C. The amount of biotinylated RGD peptide was determined by a streptavidin-peroxidase assay. The data are expressed as the percentage of the peptide bound when the wells were preincubated with PBS as a control. The arithmetic means and standard deviations of two independent experiments performed in duplicate are shown.
DISCUSSION
Although it is well established that trypsin treatment enhances, and probably is required for, the infectivity of rotaviruses, previous studies have shown that the attachment of the virus to the cell surface is independent of this proteolytic treatment (5, 24). Recently, it was shown that the VP4 spikes of non-trypsin-treated particles of the simian rotavirus strain SA11 4F could not be visualized by cryoelectron microscopy, whereas these spikes became visible upon treatment of the virions with trypsin, suggesting that the cleavage of VP4 yields icosahedrally ordered spikes (6) and demonstrating that the spikes are structurally different in trypsin-cleaved and uncleaved virions.
Previous studies with trypsin-activated viruses have shown that the sialic acid-dependent virus RRV attaches to the cell surface by interacting with a sialic acid-containing cell receptor (26, 30). On the other hand, the sialic acid-independent RRV variant nar3 was found to attach to the cell surface by interacting with integrin α2β1 through a DGE motif located in the VP5 cleavage domain of VP4 (15, 38). Sequence analysis of the RRV and nar3 genes encoding the virus surface proteins VP4 and VP7 showed that, although the amino acid sequence of VP7 was identical between the two viruses, the VP4 protein of nar3 differed by 3 aa from the parental RRV VP4 sequence. These amino acid changes were suggested to induce a slight conformational change in VP4, which allows the nar3 virus to bypass the interaction with sialic acid and bind directly to α2β1 (31, 39). In the present study we characterized the cell binding of RRV and nar3 viruses containing cleaved or uncleaved VP4 proteins. The cleavage status of VP4 did not modify the initial interaction of RRV with the cell surface, since both trypsin-treated and untreated virus bound to the cell through VP8, presumably by interacting with a sialic acid-containing cell molecule. On the other hand, the initial interaction of nar3 with the cell surface was dramatically different depending on whether the virus had been treated or not with trypsin. NTr-nar3 did not bind through the DGE domain of VP5, as Tr-nar3 did (Fig. 2) (39), and in fact did not use the spike protein VP4 to attach to the cell but instead used VP7 to achieve this task. Altogether, these results suggest that the cleavage of VP4 is necessary to expose or stabilize the DGE domain present in VP5 and that in the NTr-nar3 particles the conformation of the VP4 spikes (which could be disordered and more flexible) might allow the virions to initially interact with the cell through VP7. These results also point to the remarkable flexibility of rotavirus particles to attach to the cell surface since, depending on the virus strain and the cleavage status of VP4, the virus can interact with a very similar efficiency (Fig. 1B) through VP8, VP5, or VP7. The relevance of the initial virus binding to the cell through VP7 during a natural infection is unclear, since the virions usually contain cleaved VP4 (29); however, in the present study it helped in the dissection of the virus-cell interactions during the process of virus entry.
Rotavirus cell entry is a complex process that involves several interactions between the surface viral proteins and the cell (1, 28). Among the several cell molecules that have been involved as receptors or coreceptors during rotavirus infectivity is the αvβ3 integrin. Rotaviruses have been shown to interact with this integrin at a postattachment step, through a site in the integrin that is different from the site that binds the RGD motif present in the physiological ligands of the integrin (17). In the present study we found that the attachment of NTr-nar3 virus to the cell surface is mediated by the interaction of a CNP region (see below) in VP7 with integrin αvβ3, and evidence was provided that this interaction occurs at a postattachment step, during the cell infection of trypsin-treated RRV and nar3 viruses. These findings confirm and extend those of Graham et al., who recently showed, by using reassortant viruses with one of the surface proteins derived from an integrin-dependent strain and the second surface protein from an integrin-independent virus, that the interaction of the virus with αvβ3 correlates with the presence of a VP7 derived from the integrin-using virus (15).
Similar to rotaviruses, hantaviruses have been reported to interact with integrin αvβ3 during cell infection in an RGD-independent manner; thus, we explored the possibility that these viruses could interact with αvβ3 through a motif in common, one that is different from RGD. Comparison of the amino acid sequence of rotavirus and hantavirus surface proteins identified a 9-aa region in VP7, which shared a 67% identity with the G1G2 protein of hantavirus L99. The sequence of this region in VP7, which we named CNP, is highly conserved among different rotavirus strains, since 586 of 621 reported VP7 sequences in the GenBank are identical, whereas the remaining 35 have one (30) or two (5) amino acid differences in this region. A synthetic peptide containing this conserved region (the CNP peptide) was shown to block the infectivity of RRV and nar3 strains, and the binding of NTr-nar3 to MA104 cells, as well as to block the direct interaction of NTr-nar3 with αvβ3 in a solid-phase assay. Altogether, these results indicate that rotaviruses interact with αvβ3 through the CNP region located at amino acids 161 to 169 of VP7. It remains to be established whether hantaviruses also use the CNP region to bind to integrin subunit β3 and whether this region is important for hantavirus infectivity. Of interest, peptide CNP was found to bind to αvβ3 through a site different from the RGD-binding site; thus, the CNP peptide sequence could represent a new integrin αvβ3 binding motif.
Based on the cryoelectron microscopy of virions complexed with Fab fragments of MAbs directed to VP8 (MAb 7A12) and to VP5 (MAb2G4), Tihova et al. (35) proposed a model in which the carboxy-terminal region of VP5 is located either close to or below the VP7 layer. The crystal structure of the sialic acid-binding domain of VP8 supports this model, since the structure of this domain fits the size and shape of the globular heads of the rotavirus VP4 spikes, as seen in a cryoelectron microscopy-based reconstruction (7). These findings indicate that the region of VP8 outside the characterized domain, as well as VP5, should form the “body” of VP4. Also in agreement with the proposed model, we have recently described that RRV and nar3 bind, at a postattachment step, to the heat shock protein hsc70, through a VP5 domain that is located toward the carboxy-terminal region of the protein (between aa 642 and 658) (37). This information, together with the fact that rotaviruses contact integrin αvβ3 through VP7, suggest that after the initial contact of rotavirus with the cell surface through either VP8 or the DGE region of VP5 (which has been mapped close to the tip of the spike) or both, VP4 might experience a conformational change; this change could, in turn, allow regions of VP4 situated more distal to the globular heads of the spike (next to the smooth layer formed by VP7), and VP7 itself, to become closer to and have contact with cell surface molecules. Direct evidence for this conformational change, however, remains to be obtained. Also, it will be of interest to determine whether the interaction of VP7 with αvβ3 integrin favors directly the internalization of the viral particle or triggers a signaling pathway that in turn facilitates the entry of the viral particle into the cell's cytoplasm.
Acknowledgments
We thank Pavel Isa for critically reading the manuscript. We also appreciate the expert assistance of Shirley Ainsworth in the virtual library.
This study was partially supported by grants 55003662 and 55000613 from the Howard Hughes Medical Institute and grant G37621N from the National Council for Science and Technology of Mexico.
REFERENCES
- 1.Arias, C. F., C. A. Guerrero, E. Mendez, S. Zárate, P. Isa, R. Espinosa, P. Romero, and S. Lopez. 2001. Early events of rotavirus infection: the search for the receptor(s). John Wiley & Sons, Inc., New York, N.Y. [DOI] [PubMed]
- 2.Arias, C. F., M. Lizano, and S. López. 1987. Synthesis in Escherichia coli and immunological characterization of a polypeptide containing the cleavage site associated with trypsin enhancement of rotavirus SA11 infectivity. J. Gen. Virol. 68:633-642. [DOI] [PubMed] [Google Scholar]
- 3.Arias, C. F., P. Romero, V. Alvarez, and S. López. 1996. Trypsin activation pathway of rotavirus infectivity. J. Virol. 70:5832-5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciarlet, M., and M. K. Estes. 1999. Human and most animal rotavirus strains do not require the presence of sialic acid on the cell surface for efficient infectivity. J. Gen. Virol. 80:943-948. [DOI] [PubMed] [Google Scholar]
- 5.Clark, S. M., J. R. Roth, M. L. Clark, B. B. Barnett, and R. S. Spendlove. 1981. Trypsin enhancement of rotavirus infectivity: mechanism of enhancement. J. Virol. 39:816-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crawford, S. E., S. K. Mukherjee, M. K. Estes, J. A. Lawton, A. L. Shaw, R. F. Ramig, and B. V. V. Prasad. 2001. Trypsin cleavage stabilizes the rotavirus VP4 spike. J. Virol. 75:6052-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dormitzer, P. R., Z. Y. J. Sun, G. Wagner, and S. C. Harrison. 2002. The rhesus rotavirus VP4 sialic acid-binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J. 21:885-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Espejo, R. T., S. Lopez, and C. Arias. 1981. Structural polypeptides of simian rotavirus SA11 and the effect of trypsin. J. Virol. 37:156-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Estes, M. K. 2001. Rotaviruses and their replication, p. 1747-1785. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa. [Google Scholar]
- 10.Estes, M. K., D. Y. Graham, and B. B. Mason. 1981. Proteolytic enhancement of rotavirus infectivity: molecular mechanisms. J. Virol. 39:879-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falconer, M. M., J. M. Gilbert, A. M. Roper, H. B. Greenberg, and J. S. Gavora. 1995. Rotavirus-induced fusion from without in tissue culture cells. J. Virol. 69:2629-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fiore, L., H. B. Greenberg, and E. R. Mackow. 1991. The VP8 fragment of VP4 is the rhesus rotavirus hemagglutinin. Virology 181:553-563. [DOI] [PubMed] [Google Scholar]
- 13.Gavrilovskaya, I. N., M. Shepley, R. Shaw, M. H. Ginsberg, and E. M. Mackow. 1998. β3 integrins mediate the cellular entry of hantaviruses that cause respiratory failure. Proc. Natl. Acad. Sci. USA 95:7074-7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert, J. M., and H. B. Greenberg. 1998. Cleavage of Rhesus rotavirus VP4 after arginine 247 is essential for rotavirus-like-particle-induced fusion from without. J. Virol. 72:5323-5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graham, K. L., P. Halasz, Y. Tan, M. J. Hewish, Y. Takada, E. R. Mackow, M. K. Robinson, and B. S. Coulson. 2003. Integrin-using rotaviruses bind α2β1 integrin α2 I domain via VP4 DGE sequence and recognize αXβ2 and αVβ3 by using VP7 during cell entry. J. Virol. 77:9969-9978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerrero, C. A., D. Bouyssounade, S. Zarate, P. Isa, T. Lopez, R. Espinosa, P. Romero, E. Mendez, S. Lopez, and C. F. Arias. 2002. Heat shock cognate protein 70 is involved in rotavirus cell entry. J. Virol. 76:4096-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guerrero, C. A., E. Mendez, S. Zarate, P. Isa, S. Lopez, and C. F. Arias. 2000. Integrin αvβ3 mediates rotavirus cell entry. Proc. Natl. Acad. Sci. USA 97:14644-14649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guerrero, C. A., S. Zárate, G. Corkidi, S. López, and C. F. Arias. 2000. Biochemical characterization of rotavirus receptors in MA104 cells. J. Virol. 74:9362-9371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hewish, M. J., Y. Takada, and B. S. Coulson. 2000. Integrins α2β1 and α4β1 can mediate SA11 rotavirus attachment and entry into cells. J. Virol. 74:228-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horton, M. A. 1997. The αvβ3 integrin “vitronectin receptor.” Int. J. Biochem. Cell Biol. 29:721-725. [DOI] [PubMed] [Google Scholar]
- 21.Hynes, R. O. 2002. Integrins: bidirectional, allosteric signaling machines. Cell 110:673-687. [DOI] [PubMed] [Google Scholar]
- 22.Hynes, R. O. 1992. Integrins: versatility, modulation, and signaling in cell adhesion. Cell 69:11-25. [DOI] [PubMed] [Google Scholar]
- 23.Isa, P., S. Lopez, L. Segovia, and C. F. Arias. 1997. Functional and structural analysis of the sialic acid-binding domain of rotaviruses. J. Virol. 71:6749-6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaljot, K. T., R. D. Shaw, D. H. Rubin, and H. B. Greenberg. 1988. Infectious rotavirus enters cells by direct cell membrane penetration, not by endocytosis. J. Virol. 62:1136-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kapikian, A. Z., and R. M. Chanock. 2001. Rotaviruses, p. 1787-1833. In S. E. Straus (ed.), Virology, vol. 2. Raven Press, New York, N.Y. [Google Scholar]
- 26.Keljo, D. J., and A. K. Smith. 1988. Characterization of binding of simian rotavirus SA-11 to cultured epithelial cells. J. Pediatr. Gastroenterol. Nutr. 7:249-256. [DOI] [PubMed] [Google Scholar]
- 27.Londrigan, S. L., K. L. Graham, Y. Takada, P. Halasz, and B. S. Coulson. 2003. Monkey rotavirus binding to α2β1 integrin requires the α2 I domain and is facilitated by the homologous beta1 subunit. J. Virol. 77:9486-9501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez, S., and C. F. Arias. 2003. Attachment and post-attachment receptors for rotavirus, p. 143-163. In J. Gray (ed.), Viral gastroenteritis. Elsevier Science, Amsterdam, Amsterdam.
- 29.Ludert, J. E., A. A. Krishnaney, J. W. Burns, P. T. Vo, and H. B. Greenberg. 1996. Cleavage of rotavirus VP4 in vivo. J. Gen. Virol. 77(Pt. 3):391-395. [DOI] [PubMed] [Google Scholar]
- 30.Mendez, E., C. F. Arias, and S. Lopez. 1993. Binding to sialic acids is not an essential step for the entry of animal rotaviruses to epithelial cells in culture. J. Virol. 67:5253-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mendez, E., C. F. Arias, and S. Lopez. 1996. Interactions between the two surface proteins of rotavirus may alter the receptor-binding specificity of the virus. J. Virol. 70:1218-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nemerow, G. R., and P. L. Stewart. 2001. Antibody neutralization epitopes and integrin binding sites on nonenveloped viruses. Virology 288:189-191. [DOI] [PubMed] [Google Scholar]
- 33.Plow, E. F., T. A. Haas, L. Zhang, J. Loftus, and J. W. Smith. 2000. Ligand binding to integrins. J. Biol. Chem. 275:21785-21788. [DOI] [PubMed] [Google Scholar]
- 34.Prasad, B. V., J. W. Burns, E. Marietta, M. K. Estes, and W. Chiu. 1990. Localization of VP4 neutralization sites in rotavirus by three-dimensional cryo-electron microscopy. Nature 343:476-479. [DOI] [PubMed] [Google Scholar]
- 35.Tihova, M., K. A. Dryden, R. Bellamy, H. B. Greenberg, and M. Yeager. 2001. Localization of membrane permeabilization and receptor binding sites on the VP4 hemagglutinin of rotavirus: implications for cell entry. J. Mol. Biol. 314:985-992. [DOI] [PubMed] [Google Scholar]
- 36.Triantafilou, K., Y. Takada, and M. Triantafilou. 2001. Mechanisms of integrin-mediated virus attachment and internalization process. Crit. Rev. Immunol. 21:311-322. [PubMed] [Google Scholar]
- 37.Zarate, S., M. A. Cuadras, R. Espinosa, P. Romero, K. O. Juarez, M. Camacho-Nuez, C. F. Arias, and S. Lopez. 2003. Interaction of rotaviruses with Hsc70 during cell entry is mediated by VP5. J. Virol. 77:7254-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zarate, S., R. Espinosa, P. Romero, C. A. Guerrero, C. F. Arias, and S. Lopez. 2000. Integrin α2β1 mediates the cell attachment of the rotavirus neuraminidase-resistant variant nar3. Virology 278:50-54. [DOI] [PubMed] [Google Scholar]
- 39.Zarate, S., R. Espinosa, P. Romero, E. Mendez, C. F. Arias, and S. Lopez. 2000. The VP5 domain of VP4 can mediate attachment of rotaviruses to cells. J. Virol. 74:593-599. [DOI] [PMC free article] [PubMed] [Google Scholar]