Abstract
In the early events of human immunodeficiency virus type 1 (HIV-1) infection, immature dendritic cells (DCs) expressing the DC-specific intercellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN) receptor capture small amounts of HIV-1 on mucosal surfaces and spread viral infection to CD4+ T cells in lymph nodes (22, 34, 45). RNA interference has emerged as a powerful tool to gain insight into gene function. For this purpose, lentiviral vectors that express short hairpin RNA (shRNA) for the delivery of small interfering RNA (siRNA) into mammalian cells represent a powerful tool to achieve stable gene silencing. In order to interfere with DC-SIGN function, we developed shRNA-expressing lentiviral vectors capable of conditionally suppressing DC-SIGN expression. Selectivity of inhibition of human DC-SIGN and L-SIGN and chimpanzee and rhesus macaque DC-SIGN was obtained by using distinct siRNAs. Suppression of DC-SIGN expression inhibited the attachment of the gp120 envelope glycoprotein of HIV-1 to DC-SIGN transfectants, as well as transfer of HIV-1 to target cells in trans. Furthermore, shRNA-expressing lentiviral vectors were capable of efficiently suppressing DC-SIGN expression in primary human DCs. DC-SIGN-negative DCs were unable to enhance transfer of HIV-1 infectivity to T cells in trans, demonstrating an essential role for the DC-SIGN receptor in transferring infectious viral particles from DCs to T cells. The present system should have broad applications for studying the function of DC-SIGN in the pathogenesis of HIV as well as other pathogens also recognized by this receptor.
In addition to classical human immunodeficiency virus type 1 (HIV-1) receptors and coreceptors (CD4, CCR5, and CXCR4), dendritic cells (DCs), such as those derived from blood monocytes as well as those located beneath genital surfaces and within lymphoid tissues, express the DC-specific intercellular adhesion molecule 3 (ICAM-3)-grabbing nonintegrin (DC-SIGN, also known as CD209) receptor (28, 29). DC-SIGN is a type II transmembrane protein with an external C-type lectin domain that specifically recognizes glycan ligands (18). Importantly, DC-SIGN binds the gp120 envelope glycoprotein of HIV-1 (13, 22, 38) and facilitates the transmission of HIV-1 from DC-SIGN transfectants to permissive CD4+ T cells (22, 24, 32, 38, 49). The natural ligands for DC-SIGN are ICAM-3 and ICAM-2, molecules that contribute to transient DC-T-cell attachment (23) and to transmigration of DCs across vascular endothelia (21), respectively.
RNA interference (RNAi) is a sequence-specific posttranscriptional gene silencing mechanism initiated by the introduction of double-stranded RNA (dsRNA) into target cells (42). RNAi is a natural regulatory mechanism conserved among many organisms, including plants, Caenorhabditis elegans, Drosophila, and mammals (42). RNAi is mediated by small interfering RNAs (siRNAs) produced from long dsRNAs of exogenous or endogenous origin by an endonuclease called Dicer. The resulting siRNAs are about 21 to 23 nucleotides long and are incorporated into a nuclease complex, the RNA-induced silencing complex, which then targets and cleaves mRNA that is complementary to the siRNAs. Delivery of siRNAs into mammalian cells by transfection with synthetic siRNAs or DNA plasmids expressing short hairpin RNAs (shRNAs) has been shown to mediate RNAi successfully (9, 16, 36, 46). Importantly, recent reports describe the use of viral vectors to deliver siRNAs successfully into mammalian cells in order to get stable gene silencing (1, 3, 8, 15, 40, 48, 54, 56).
In order to provide a basis for studying in further detail the role of DC-SIGN in HIV pathogenesis, we developed lentiviral vectors conditionally expressing shRNAs targeting DC-SIGN. The objective of the present study was to generate stable cell lines as well as primary DCs deficient in DC-SIGN. Using this approach, we examined the effect of DC-SIGN knockdown on the attachment of the gp120 envelope glycoprotein of HIV-1 as well as the transmission of virus to T cells in trans.
MATERIALS AND METHODS
Vector construction and siRNAs.
The pSUPER, pSUPER-si-GFP (si-GFP), pLV-Th, and pLV-tTR-KRAB-dsRed (LV-KRAB) constructs were described previously (9, 54). Oligonucleotides encoding shRNAs directed against human DC-SIGN mRNA were designed according to the method of Elbashir et al. (17) and purchased from MWG Biotech (Ebersberg, Germany) (see Table 1 for siRNA sequences). These oligonucleotides were annealed and ligated into pSUPER downstream of the H1 promoter, giving rise to pSUPER-siDC-SIGN constructs (referred to hereafter as si-SIGN constructs). Both strands of si-SIGN constructs were sequenced using T3 and T7 primers. The H1 promoter cassette in pLV-TH was replaced by the H1-siRNA cassette excised from si-SIGN constructs, giving rise to LV-si-SIGN lentiviral vectors. Human DC-SIGN (23) and L-SIGN (6) genes were cloned into lentiviral vectors pLOX (LV-DC-SIGN) and pWPXL (LV-L-SIGN), respectively, replacing the green fluorescent protein (GFP) marker gene. Chimpanzee and rhesus macaque DC-SIGN expression plasmids were described previously (20).
TABLE 1.
Human DC-SIGN cDNA target sequences for siRNAsa
| siRNA | Exon | Sequence (5′ to 3′) |
|---|---|---|
| siRNA2 | I | AACAGCTGAGAGGCCTTGGAT |
| siRNA3 | II | AACTCCTCTCCTTCACGCTCT |
| siRNA8 | III | AAGACGCGATCTACCAGAACC |
| siRNA11 | III | AAGGCTGCAGTGGGTGAGCTT |
| siRNA26 | IV | AACTGGCACGACTCCATCACC |
| siRNA35 | VI | AAGACTGCGCGGAATTTAGTG |
Six different siRNAs directed against human DC-SIGN were designed to correspond to distinct parts of the DC-SIGN mRNA sequence. Target sequences of the DC-SIGN cDNA and corresponding exons are shown.
siRNA screening test.
Subconfluent 293T cells were cotransfected with 5 μg of pSUPER control plasmid or si-SIGN constructs and 0.5 μg of human DC-SIGN and pEGFP-N1 (Clontech, Palo Alto, Calif.) expression plasmids by calcium phosphate precipitation. Medium was changed after 16 h, and 24 h later, flow cytometric analysis of GFP and DC-SIGN expression using phycoerythrin (PE)-labeled anti-human DC-SIGN monoclonal antibody (MAb) was performed. To test for the specificity of si-SIGN constructs, the same assay was performed with the human DC-SIGN expression plasmid replaced by a plasmid expressing L-SIGN (PE-labeled anti-human L-SIGN MAb) or chimpanzee or rhesus macaque DC-SIGN (PE-labeled anti-human DC-SIGN MAb).
Production of lentiviral vectors.
All recombinant lentiviruses were produced by transient transfection of 293T cells according to standard protocols (58). Briefly, subconfluent 293T cells were cotransfected with 20 μg of the plasmid vector, 15 μg of pCMV-deltaR8.91, and 5 μg of pMD2G-VSVG by calcium phosphate precipitation. Vector titers were determined by transduction and flow cytometric analysis of GFP expression in HeLa cells. Titers ranged between 2 × 107 and 5 × 108 HeLa-transducing U per ml.
Viral stocks.
Viral stocks were generated by transfection of 293T cells with calcium phosphate-coprecipitated proviral plasmid pR9 which carries a full-length HIV-1 X4 strain (19). Infectious titers of viral stocks were evaluated by limiting dilution on HeLa P4 cells (11) and expressed as infectious units per milliliter. The titer values were also determined by measuring HIV-1 p24gag by using an enzyme-linked immunosorbent assay (ELISA) kit (Beckman Coulter, Paris, France).
Generation of cell lines stably expressing siRNAs.
Raji B cells were cultured as described elsewhere (55). Various Raji cell lines stably expressing siRNAs were generated by transduction with LV-DC-SIGN, LV-si-SIGN8, LV-si-SIGN11, LV-si-SIGN26, LV-L-SIGN, or empty vector. The HeLa P4-R5 cell lines stably expressing siRNAs were generated by transduction with LV-DC-SIGN, LV-si-SIGN8, LV-si-SIGN11, or LV-si-SIGN26. Stable knockdown or expression of the genes of interest by the different cell lines was determined by flow cytometric analysis.
Preparation and transduction of DC progenitors.
Cord blood samples were obtained according to institutional guidelines of the ethical committee. CD34+ cells were purified and cultured as previously described (4). DC progenitors were transduced and analyzed on a FACSCalibur cell analyzer (Becton Dickinson, Mountain View, Calif.) as reported in reference 41 with the following modifications. After 3 to 6 weeks of primary culture, DC progenitors were transduced with LV-si-SIGN lentiviral vectors at a multiplicity of infection (MOI) of 20 for 16 h in the presence of hematopoietic growth factors. Cells were then extensively washed, and 2 × 105 transduced DC progenitors per ml of Iscove's modified Dulbecco medium Glutamax I supplemented with 10% fetal calf serum and antibiotics (Gibco BRL, Paisley, United Kingdom) were induced to differentiate into immature DCs for 6 days with a solution containing 50 ng/ml each granulocyte-macrophage colony-stimulating factor (GM-CSF) and interleukin-4 (IL-4; Strathmann Biotec, Hamburg, Germany) in the presence of 50 μM 2-mercaptoethanol (Sigma, Buchs, Switzerland) or into mature DCs by further addition of 20 ng of lipopolysaccharide (LPS; Escherichia coli strain 055:B5; Difco, Detroit, Mich.)/ml for the last 2 days. DCs were harvested at day 6, analyzed by flow cytometry, and used in subsequent assays.
Cellular sorting.
Immature DCs were sorted on a FACSVantage SE cell sorter (Becton Dickinson) as GFP positive-DC-SIGN negative (LV-si-SIGN11) or GFP positive-DC-SIGN negative (empty vector). Cells were systematically reanalyzed after sorting. Viability and correctness of surface phenotype were excellent (>95%) up to 24 h postsorting. Sorted cells were counted and used for viral assays as described below.
Analysis of DC-SIGN-mediated transfer of HIV-1 infection to target cells.
In order to measure DC-SIGN-mediated trans-enhancement of HIV-1 infection of T cells, aliquots of 5 × 103 Raji transfectants or sorted DCs were incubated for 2 h with HIV-R9 virus at 37°C at a MOI of 0.001, extensively washed, and directly cocultured with activated peripheral blood leukocytes (PBLs). Viral production in the coculture supernatants was monitored by reverse transcriptase assay.
Flow cytometric analysis and MAbs.
Flow cytometric analysis was performed as described previously (5). Cells were analyzed on a FACSCalibur cell analyzer (Becton Dickinson). Data were analyzed using WINMDI software by J. Trotter at Scripps Institute (La Jolla, Calif.).
PE-labeled MAbs were obtained as follows: anti-CD209 (DC-SIGN; mouse immunoglobulin G2b [mIgG2b], clone 120507) and anti-CD209L (L-SIGN; mIgG2b, clone 120604) from R&D Systems (Minneapolis, Minn.); anti-CD4 (mIgG1, clone MT310) from Dako (Glostrup, Denmark); and anti-CD34 (mIgG1, clone 8G12), anti-CD1a (mIgG1, clone HI146), anti-CD83 (mIgG1, clone HB15e), anti-CD206 (macrophage mannose receptor; mIgG1, clone 19.2), and anti-HLA-ABC (mIgG1, clone G46-2.7) from PharMingen (San Diego, Calif.). Allophycocyanin (APC)-labeled MAbs were obtained as follows: anti-CXCR4 (mIgG2a, clone 12G5) from PharMingen and anti-CD209 (DC-SIGN; rat IgG2a, clone eB-h209) from eBiosciences (San Diego, Calif.). Monoclonal isotype controls were obtained as follows: PE-labeled mIgG1 and mIgG2b and APC-labeled mIgG2a from PharMingen and APC-labeled rat IgG2a from eBiosciences.
Fluorescent-bead adhesion assay.
The fluorescent-bead adhesion assay was performed as previously described (25). Carboxylate-modified TransFluorSpheres (488/645 nm; 1.0 μm; Molecular Probes, Eugene, Oreg.) were coated with ICAM-3, monomeric HIV gp120, or Mycobacterium tuberculosis-derived mannose-capped lipoarabinomannan (ManLAM) as described previously (25, 26). Fifty thousand cells were preincubated or not with anti-human DC-SIGN neutralizing MAb AZN-D2 (20 μg/ml) or mannan (1 mg/ml; Sigma) for 45 min at 37°C. Ligand-coated fluorescent beads (20 beads/cell) were added, and the suspension was incubated for 30 min at 37°C. Adhesion was determined by flow cytometric analysis using the FACSVantage SE (Becton Dickinson) by measuring the percentage of cells within the GFP-positive cell population that had bound fluorescent beads.
RT-PCR.
Total RNA was extracted from Raji cell lines by using TRIzol reagent (Life Technologies, Basel, Switzerland). One hundred nanograms of total RNA was subjected to SuperScript one-step reverse transcription (RT)-PCR with Platinum Taq (Invitrogen, Basel, Switzerland) to assess levels of mRNA corresponding to DC-SIGN and the cyclophilin gene as a housekeeping gene. Oligonucleotides with the following sequences (5′ to 3′) were purchased from MWG Biotech: DC-SIGN forward, GAGTGACTCCAAGGAACCAAGACTGCA, and DC-SIGN reverse, GGTAGATCGCGTCTTGCCTGGATTGTT, amplifying a 233-bp PCR product, and cyclophilin forward, GGTCAACCCCACCGTGTTCTTCGACAT, and cyclophilin reverse, GACTTGCCACCAGTGCCATTATGGCGT, amplifying a 228-bp PCR product.
IFN-α/β response.
The amounts of alpha interferon (IFN-α) in the supernatants of Raji cell lines or day-6 transduced immature DCs were measured with an IFN-α ELISA kit (5 to 500 pg/ml; PBL Biomedical Laboratories, New Brunswick, N.J.) according to the manufacturer's instructions. Levels of mRNA corresponding to the classic IFN target gene, the 2′,5′-oligoadenylate synthetase gene, were measured using SuperScript one-step RT-PCR as previously described (7).
RESULTS
siRNA-expressing vectors suppress DC-SIGN expression.
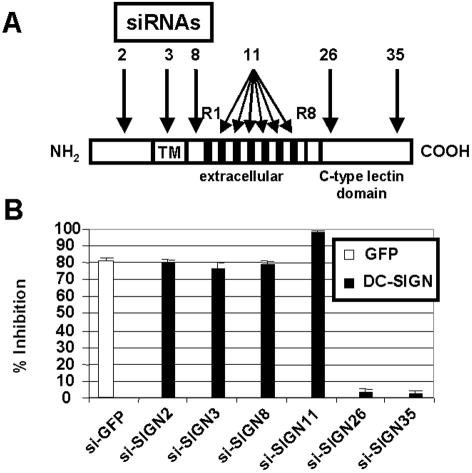
In order to silence human DC-SIGN expression, we followed the design rules of Elbashir et al. (17) and found 41 different siRNAs potentially targeting the DC-SIGN gene. Among them, six siRNAs corresponding to distinct parts of the DC-SIGN cDNA sequence were chosen (Fig. 1A; siRNA sequences are indicated in Table 1). siRNAs were cloned into pSUPER downstream of the H1 promoter, giving rise to si-SIGN constructs.
FIG. 1.
Design and screening of siRNAs targeting human DC-SIGN expression. (A) Design of siRNAs. Six different siRNAs against DC-SIGN were designed to target distinct parts of the DC-SIGN mRNA sequence. siRNA11 targeted tandem repeats (R1 to R8) present in exon III. TM, transmembrane domain. (B) Screening of siRNA-expressing vectors. Subconfluent 293T cells were cotransfected with pSUPER control plasmid, si-GFP, or si-SIGN constructs along with human DC-SIGN and pEGFP-N1 expression plasmids. The mean fluorescence intensity of DC-SIGN and GFP expression was measured in the GFP-positive and the DC-SIGN-positive cell populations, respectively. Histograms show the percentages of inhibition relative to that under the pSUPER control experimental condition. Means ± standard errors of results from four independent experiments are shown.
The ability of each siRNA to inhibit DC-SIGN expression was then tested by cotransfection of 293T cells with pSUPER control plasmid, pSUPER-si-GFP, or si-SIGN constructs along with human DC-SIGN and pEGFP-N1 expression plasmids. This knockdown screening test allowed the identification of four siRNAs effective at inhibiting DC-SIGN expression as assessed by flow cytometric analysis (Fig. 1B). si-SIGN11 was the most effective (97% inhibition) while not affecting GFP expression. si-SIGN11 titration showed dose-dependent inhibition of DC-SIGN expression (data not shown). The same assay was used to test the siRNAs against human L-SIGN, rhesus macaque DC-SIGN, and chimpanzee DC-SIGN. Interestingly, some siRNAs could inhibit human L-SIGN as well as primate DC-SIGN expression despite the presence of mismatches between siRNA and specific mRNA sequences (Table 2). Again, si-SIGN11 was the most effective at inhibiting human L-SIGN and primate DC-SIGN. Interestingly, only si-SIGN8 specifically inhibited human DC-SIGN.
TABLE 2.
siRNA specificitya
| Construct | Inhibition of:
|
|||
|---|---|---|---|---|
| Human DC-SIGN | Human L-SIGN | Chimpanzee DC-SIGN | Rhesus macaque DC-SIGN | |
| si-SIGN2 | + (0) | − (Ø) | + (0) | − (Ø) |
| si-SIGN3 | + (0) | + (1) | + (0) | + (0) |
| si-SIGN8 | + (0) | − (1) | − (1) | − (2) |
| si-SIGN11b | ++ (0/0/0/0/0/0) | ++ (2/2/2/2/2/2) | ++ (0/0/0/0/0/0) | ++ (1/0/0/2/2/5) |
| si-SIGN26 | − (0) | − (1) | − (1) | − (0) |
| si-SIGN35 | − (0) | − (1) | − (0) | − (1) |
Subconfluent 293T cells were cotransfected with pSUPER control plasmid, si-GFP, or si-SIGN constructs along with human DC-SIGN and pEGFP-N1 expression plasmids. Flow cytometric analysis of DC-SIGN expression was performed. The mean fluorescence intensity of DC-SIGN expression was measured in the GFP-positive cell population. To test for the specificity of si-SIGN constructs, the same assay was performed with the human DC-SIGN expression plasmid replaced by plasmids expressing L-SIGN or chimpanzee or rhesus macaque DC-SIGN. Percentages of inhibition relative to that under the pSUPER control experimental condition were then calculated. ++, strong inhibition; +, inhibition; −, no inhibition. The numbers within brackets correspond to mismatches between siRNAs and specific mRNA sequences. Ø, no significative homology.
si-SIGN11 targets six repeated sequences with 100% homology on the human DC-SIGN mRNA.
Lentivirus-mediated delivery of siRNA suppresses DC-SIGN expression in cell lines.
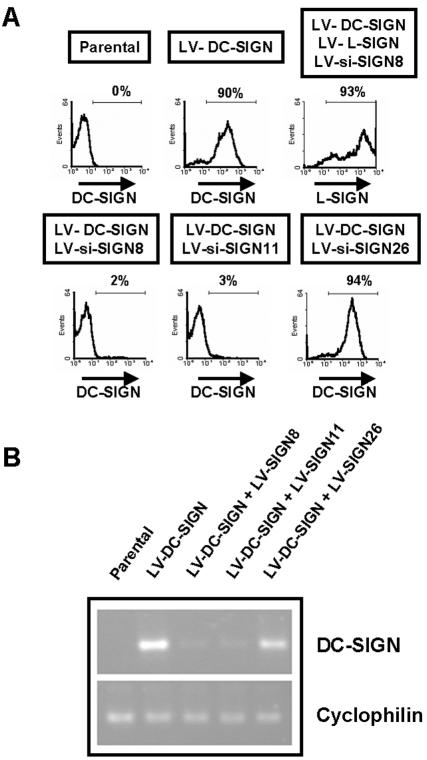
siRNAs were then cloned into lentiviral vectors for stable expression in cell lines (54). In order to evaluate the effectiveness of siRNA-expressing lentiviral vectors (referred to hereafter as LV-si-SIGN) at inhibiting DC-SIGN expression, various Raji cell lines stably expressing siRNAs were generated (Fig. 2A). Transduction with LV-si-SIGN8 or LV-si-SIGN11 along with LV-DC-SIGN induced suppression of DC-SIGN expression. Results of flow cytometric analysis of DC-SIGN expression on Raji stable transfectants correlated with the results obtained using the knockdown screening test described above (Fig. 1B and 2A). LV-si-SIGN8 did not inhibit L-SIGN expression, confirming the specificity of the vector for DC-SIGN downregulation (Fig. 2A and Table 2). It is of note that stable DC-SIGN knockdown could be observed over 6 months (data not shown).
FIG. 2.
Suppression of DC-SIGN expression in cell lines through lentivirus-mediated delivery of siRNAs. (A) Generation of cell lines stably expressing siRNAs. Stable Raji transfectants were generated by transduction with the indicated lentiviral vectors. While both LV-si-SIGN8 and LV-si-SIGN11 suppressed DC-SIGN expression, only LV-si-SIGN11 suppressed L-SIGN expression. One month posttransduction, flow cytometric analysis of GFP, DC-SIGN, and L-SIGN expression was performed. The percentages of cells positive for the indicated cell surface markers are shown. (B) Effect of siRNAs on DC-SIGN mRNA. Total RNA extracted from Raji transfectants was subjected to RT-PCR to assess levels of mRNA corresponding to DC-SIGN and cyclophilin. Both LV-si-SIGN8 and LV-si-SIGN11 downregulated DC-SIGN mRNA expression but not cyclophilin mRNA expression.
To ensure that DC-SIGN downregulation occurred through RNAi and not translational repression through a micro-RNA mechanism, levels of mRNA encoding DC-SIGN in different Raji cell lines stably expressing siRNAs were measured by RT-PCR (Fig. 2B). Very low levels of DC-SIGN mRNA were found in Raji cell lines transduced with LV-si-SIGN8 or LV-si-SIGN11 compared to those in control cell lines, whereas levels comparable to those in the DC-SIGN-expressing cell line were found in cell lines transduced with LV-si-SIGN26.
In addition, we studied the effect of DC-SIGN-specific siRNAs on the expression of the CD4 receptor and the CXCR4 coreceptor involved in HIV binding. For this purpose, various HeLa P4-R5 cell lines stably expressing siRNAs were generated. Whereas transduction with LV-si-SIGN11 induced significant suppression of DC-SIGN expression, CD4 and CXCR4 expression levels were not affected (data not shown). Identical results were obtained using LV-si-SIGN8 (data not shown).
Suppression of DC-SIGN expression by drug-inducible siRNAs.
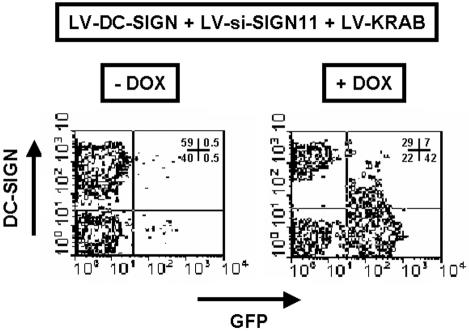
Various HeLa cell lines were generated by transduction with LV-DC-SIGN, LV-si-SIGN11, and LV-KRAB to allow for conditional suppression of DC-SIGN (Fig. 3). Cells cotransduced with these three lentiviral vectors normally expressed DC-SIGN when cultured in the absence of doxycycline, owing to LV-KRAB-mediated suppression of siRNA synthesis (54). In contrast, addition of the drug relieved this inhibition and allowed for DC-SIGN downregulation. Expression of GFP was also subjected to conditional LV-KRAB repression, providing an internal monitoring device. Conditional suppression of DC-SIGN expression could also be obtained using LV-si-SIGN8 (data not shown).
FIG. 3.
Conditional suppression of DC-SIGN expression was evaluated in HeLa cell lines transduced with LV-DC-SIGN, LV-si-SIGN11, and LV-KRAB in the presence (+) or absence (−) of doxycycline (DOX). One week posttransduction, flow cytometric analysis of GFP and DC-SIGN expression was performed. Cell percentages corresponding to each quadrant of two-dimensional plots are shown.
siRNA-expressing lentiviral vectors do not activate IFN pathways.
One concern with siRNA technology is the capacity of dsRNA to trigger IFN-dependent pathways in some cellular systems (7, 14, 43). In order to test whether LV-si-SIGN activated IFN pathways, we measured IFN-α in culture supernatants of Raji cell lines transduced with LV-si-SIGN. Using a highly sensitive ELISA, we could not detect measurable levels of IFN-α (data not shown). Furthermore, we did not observe any significant differences in the levels of mRNA corresponding to IFN target gene the 2′,5′-oligoadenylate synthetase gene by using a semiquantitative RT-PCR (data not shown). While we cannot entirely rule out that highly sensitive tests such as DNA microarrays could detect modifications in cells after transduction with LV-si-SIGN, these results indicate that no significant activation of IFN-dependent pathways occurred in our system.
Transduction of DC progenitors with siRNA-expressing lentiviral vectors allows the generation of DC-SIGN-negative DCs.
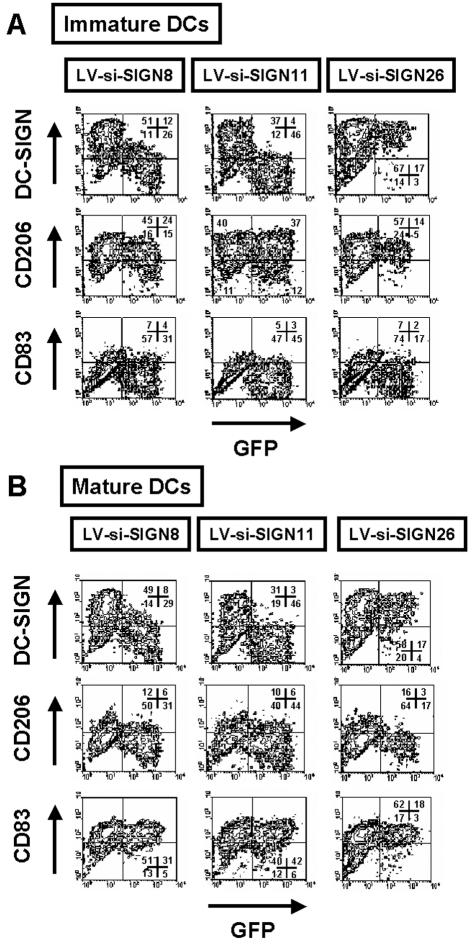
We next evaluated the potential of siRNA-expressing lentiviral vectors to suppress DC-SIGN expression at the surface of DCs. For this purpose, we transduced DC progenitors amplified in vitro from cord blood CD34+ cells with Flt3 ligand, stem cell factor, and thrombopoietin (4, 41). Transduced DC progenitors were then induced to differentiate into immature DCs for 6 days with GM-CSF and IL-4. Flow cytometric analysis of DCs revealed that LV-si-SIGN11 completely suppressed DC-SIGN expression compared to that in control cells or cells transduced with the inefficient LV-si-SIGN26 (Fig. 4A). LV-si-SIGN8 was less potent in inhibiting DC-SIGN expression, although the majority of the cells showed receptor knockdown. CD1a expression levels were comparable between GFP-positive and GFP-negative DCs (data not shown). A similar phenotype for DC-SIGN was obtained using LPS as a maturation factor. Upregulation of HLA-DR and CD83 as well as downregulation of macrophage mannose receptor (CD206) was always observed upon maturation with LPS (Fig. 4B). In addition, transduction with lentiviral vectors did not modulate the expression of HLA-ABC, CD4, or CXCR4 (data not shown). Finally, no IFN-α could be detected in the supernatants of day-6 transduced immature DCs (data not shown). In conclusion, siRNA-expressing lentiviral vectors targeting DC-SIGN allowed for efficient and specific knockdown of this receptor on immature and mature DCs.
FIG. 4.
Generation of DCs with DC-SIGN expression knocked down. DC progenitors were transduced with the indicated lentiviral vectors and differentiated either into immature DCs for 6 days with GM-CSF and IL-4 or into mature DCs by addition of LPS for the last 2 days of culture. Immature or mature DCs harvested at day 6 were analyzed by flow cytometry for the indicated cell surface antigens. LV-si-SIGN8 and LV-si-SIGN11 specifically inhibited DC-SIGN expression in DCs, and control vectors did not. Cell percentages corresponding to each quadrant of two-dimensional plots are shown. Representative results from one experiment out of four are presented.
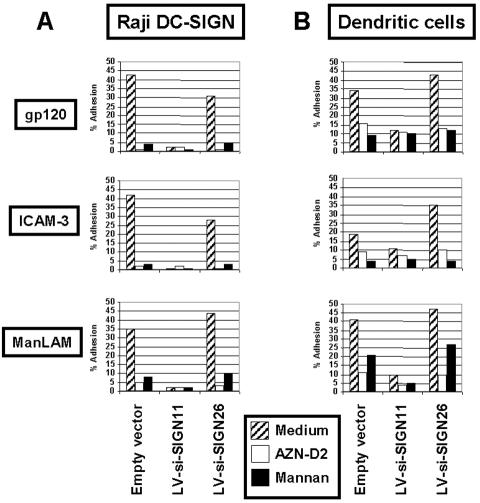
DC-SIGN knockdown impedes HIV-1 gp120 adhesion to target cells.
To test the ability of LV-si-SIGN to inhibit HIV-1 gp120 attachment to Raji transfectants (Fig. 5A) or DC-SIGN-negative DCs (Fig. 5B), we used a well-established fluorescent-bead adhesion assay (23, 25). Beads coated with HIV-1 gp120, M. tuberculosis cell wall component ManLAM, or ICAM-3 were incubated with cells stably expressing siRNAs. HIV-1 gp120, ManLAM, and ICAM-3 binding to cells transduced with LV-si-SIGN11 was severely reduced (Fig. 5). The levels of inhibition obtained using this assay were consistently reproducible with LV-si-SIGN11, but mannan or blocking antibodies showed more intra-assay variation (data not shown).
FIG. 5.
Effect of DC-SIGN knockdown on HIV-1 gp120 adhesion. Raji transfectants (A) or transduced immature DCs (B) were resuspended in adhesion buffer and preincubated with anti-DC-SIGN neutralizing MAb (AZN-D2), mannan, or medium alone for 45 min at 37°C. Carboxylate-modified TransFluorSpheres coated with ICAM-3, gp120, or ManLAM were added, and the suspension was incubated for 30 min at 37°C. Adhesion was determined by measuring the percentages of cells with bound fluorescent beads within the GFP-positive cell population by flow cytometric analysis. The results shown are representative of those from three independent experiments giving similar results.
DC-SIGN knockdown severely reduces trans-enhancement of HIV-1 infection of T cells.
Finally, we investigated whether DC-SIGN-negative DCs were capable of trans-enhancing HIV-1 infectivity in activated PBLs. Similar results were obtained using primary human DCs and Raji DC-SIGN transfectants (Fig. 6). While sorted DCs transduced with a control lentiviral vector could enhance the transfer of small amounts of virus HIV-R9 (MOI of 0.001) to activated PBLs, the same amount of virus in medium alone or added to DC-SIGN-negative DCs could not be transferred to activated PBLs (Fig. 6B). These data confirm the role of DC-SIGN as a crucial HIV receptor that mediates HIV-1 transmission from DCs to T cells.
FIG. 6.
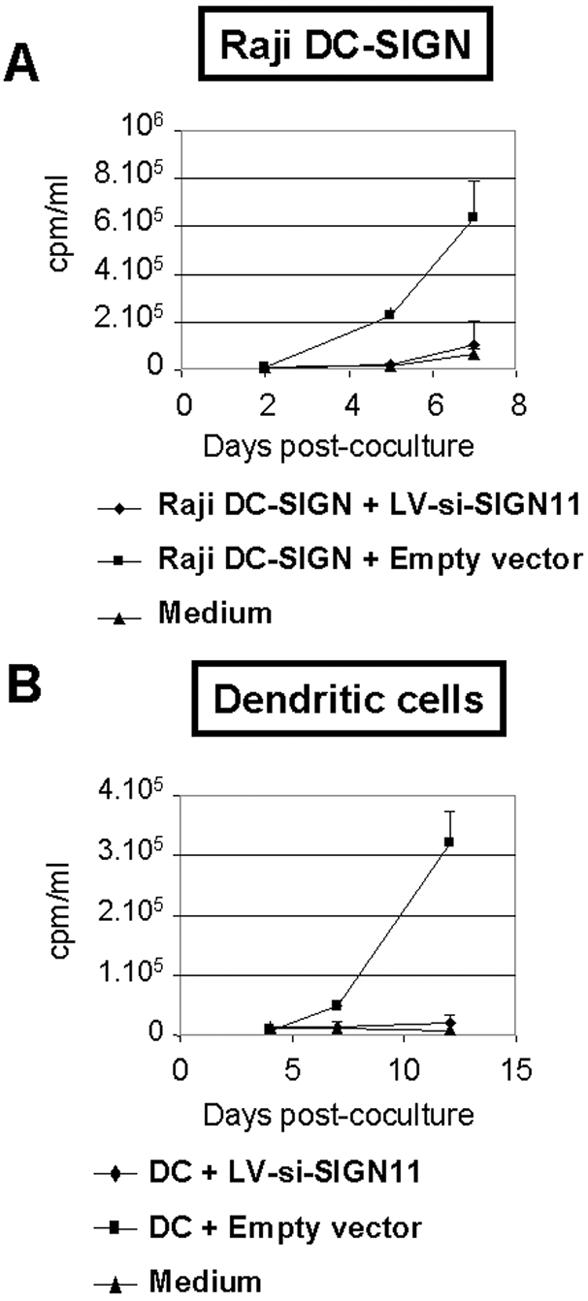
DCs with knocked-down DC-SIGN expression are unable to confer trans-enhancement of HIV-1 infection on T cells. Raji DC-SIGN transfectants (A) or sorted transduced immature DCs (B) were incubated with HIV-1 at a MOI of 0.001 and directly cocultured with activated PBLs. As a control, virus alone was added to cell culture medium (in the absence of Raji DC-SIGN transfectants or sorted DCs) but with addition of activated PBLs (medium). Viral production, expressed in counts per minute per milliliter, was monitored by reverse transcriptase assay of coculture supernatants. Data are means ± standard errors of results from one of three (A) or two (B) sets of duplicate experiments giving similar results.
DISCUSSION
Here we describe a novel system that enables potent suppression of DC-SIGN expression in a conditional manner. In order to disturb DC-SIGN expression, we developed lentiviral vectors conditionally expressing shRNAs targeting DC-SIGN. These tools allowed for the efficient suppression of DC-SIGN expression in various cell lines and in primary human DCs. By silencing DC-SIGN expression, we could inhibit the attachment of the gp120 envelope glycoprotein of HIV-1 to DCs negative for DC-SIGN as well as the transfer of HIV infection from DCs to T cells in trans.
Knockdown of DC-SIGN in DC precursors derived from CD34+ hematopoietic stem cells also revealed that this C-type lectin is not required for DC ontogeny, and this finding is compatible with the main role of DC-SIGN as a pathogen recognition receptor. Indeed, DC-SIGN plays a major role in the capture of HIV and M. tuberculosis on some DC subtypes (34, 35). However, other DC subtypes, such as Langerhans cells, do not express DC-SIGN and may mediate transfer of HIV through additional molecules or may require productive infection to transfer HIV to T cells (30, 37, 50, 51). In addition, the versatility of lentiviral vectors should enable the knockdown of DC-SIGN in a large number of targets, including transgenic animals. Indeed, advances in RNAi technology have provided a rapid loss-of-function method for assessing gene function in mammalian cells (9, 16). Moreover, highly stable and functional systems for silencing of gene expression in animal models are available (31, 40, 44, 53). In particular, a recent report describes a lentivirus-based system to functionally silence genes in transgenic mice by RNAi (40). Therefore, the generation of transgenic animals which would help to elucidate DC-SIGN function in HIV or simian immunodeficiency virus (SIV) pathogenesis is currently possible.
Furthermore, DC-SIGN can bind nonviral pathogens such as Leishmania mexicana (12), Schistosoma mansoni (52), M. tuberculosis (26, 47), Helicobacter pylori (52), and Candida albicans (10), as well as viral pathogens including Ebola virus (2), SIV-1, SIV-2 (38), HIV-2 (38), cytomegalovirus (27), hepatitis C virus (33, 39), and severe acute respiratory syndrome coronavirus (57). Therefore, our system based on lentivirus-mediated delivery of drug-inducible siRNAs targeting DC-SIGN will allow for new insights on this pathogen recognition receptor.
Another important application of our system is to reassess DC-SIGN functions. Most results described so far on the role of DC-SIGN relied on the use of blocking antibodies and mannan (22, 49). While these blocking agents are valuable, significant variations may be observed between results of studies using these tools because the block depends on the affinity of DC-SIGN for its ligand compared to that for the inhibitor used. The system developed here is nonequivocal because it is not affected by the binding properties of inhibitors that are more or less specific on diverse cell types. Lentiviral vectors conditionally expressing shRNAs targeting DC-SIGN could also be used in monocyte-derived DCs. However, we expect a less-efficient downregulation of DC-SIGN, because monocytes are more refractory to transduction than CD34+ cell-derived DC precursors (35; J.-F. Arrighi and V. Piguet, unpublished results). Furthermore, DC-SIGN is expressed at very high levels when monocytes are incubated in GM-CSF and IL-4 for 24 to 48 h (23), potentially limiting the effect of lentivirus-mediated RNAi of this receptor in this system.
Using lentiviral vector-mediated RNAi, we could demonstrate the unique role of DC-SIGN in enhancing transfer of HIV infectivity from DCs to T lymphocytes in trans. Finally, knockdown of DC-SIGN on primary DCs enables us to study unequivocally the function of this receptor in early events of HIV infection as well as its role in the pathogenesis of other viruses recognized by DC-SIGN.
Acknowledgments
We thank D. Wohlwend for his excellent technical assistance concerning fluorescence-activated cell sorter analysis.
This work was supported by the Geneva Cancer League and Swiss National Science Foundation grant no. 3345-67200.01 to V.P. and by a grant from Institut Clayton de la Recherche to D.T. V.P. is the recipient of a “Professor SNF” position (PP00A-68785).
REFERENCES
- 1.Abbas-Terki, T., W. Blanco-Bose, N. Deglon, W. Pralong, and P. Aebischer. 2002. Lentiviral-mediated RNA interference. Hum. Gene Ther. 13:2197-2201. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez, C. P., F. Lasala, J. Carrillo, O. Muniz, A. L. Corbi, and R. Delgado. 2002. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J. Virol. 76:6841-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.An, D. S., Y. Xie, S. H. Mao, K. Morizono, S. K. Kung, and I. S. Chen. 2003. Efficient lentiviral vectors for short hairpin RNA delivery into human cells. Hum. Gene Ther. 14:1207-1212. [DOI] [PubMed] [Google Scholar]
- 4.Arrighi, J. F., C. Hauser, B. Chapuis, R. H. Zubler, and V. Kindler. 1999. Long-term culture of human CD34(+) progenitors with FLT3-ligand, thrombopoietin, and stem cell factor induces extensive amplification of a CD34(−)CD14(−) and a CD34(−)CD14(+) dendritic cell precursor. Blood 93:2244-2252. [PubMed] [Google Scholar]
- 5.Arrighi, J. F., C. Soulas, C. Hauser, S. Saeland, B. Chapuis, R. H. Zubler, and V. Kindler. 2003. TNF-alpha induces the generation of Langerin/(CD207)+ immature Langerhans-type dendritic cells from both CD14-CD1a and CD14+CD1a− precursors derived from CD34+ cord blood cells. Eur. J. Immunol. 33:2053-2063. [DOI] [PubMed] [Google Scholar]
- 6.Bashirova, A. A., T. B. Geijtenbeek, G. C. van Duijnhoven, S. J. van Vliet, J. B. Eilering, M. P. Martin, L. Wu, T. D. Martin, N. Viebig, P. A. Knolle, V. N. KewalRamani, Y. van Kooyk, and M. Carrington. 2001. A dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN)-related protein is highly expressed on human liver sinusoidal endothelial cells and promotes HIV-1 infection. J. Exp. Med. 193:671-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bridge, A. J., S. Pebernard, A. Ducraux, A. L. Nicoulaz, and R. Iggo. 2003. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 34:263-264. [DOI] [PubMed] [Google Scholar]
- 8.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2:243-247. [DOI] [PubMed] [Google Scholar]
- 9.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 10.Cambi, A., K. Gijzen, J. M. de Vries, R. Torensma, B. Joosten, G. J. Adema, M. G. Netea, B. J. Kullberg, L. Romani, and C. G. Figdor. 2003. The C-type lectin DC-SIGN (CD209) is an antigen-uptake receptor for Candida albicans on dendritic cells. Eur. J. Immunol. 33:532-538. [DOI] [PubMed] [Google Scholar]
- 11.Charneau, P., G. Mirambeau, P. Roux, S. Paulous, H. Buc, and F. Clavel. 1994. HIV-1 reverse transcription. A termination step at the center of the genome. J. Mol. Biol. 241:651-662. [DOI] [PubMed] [Google Scholar]
- 12.Colmenares, M., A. Puig-Kroger, O. M. Pello, A. L. Corbi, and L. Rivas. 2002. Dendritic cell (DC)-specific intercellular adhesion molecule 3 (ICAM-3)-grabbing nonintegrin (DC-SIGN, CD209), a C-type surface lectin in human DCs, is a receptor for Leishmania amastigotes. J. Biol. Chem. 277:36766-36769. [DOI] [PubMed] [Google Scholar]
- 13.Curtis, B. M., S. Scharnowske, and A. J. Watson. 1992. Sequence and expression of a membrane-associated C-type lectin that exhibits CD4-independent binding of human immunodeficiency virus envelope glycoprotein gp120. Proc. Natl. Acad. Sci. USA 89:8356-8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diebold, S. S., M. Montoya, H. Unger, L. Alexopoulou, P. Roy, L. E. Haswell, A. Al-Shamkhani, R. Flavell, P. Borrow, and C. Reis e Sousa. 2003. Viral infection switches non-plasmacytoid dendritic cells into high interferon producers. Nature 424:324-328. [DOI] [PubMed] [Google Scholar]
- 15.Dirac, A. M., and R. Bernards. 2003. Reversal of senescence in mouse fibroblasts through lentiviral suppression of p53. J. Biol. Chem. 278:11731-11734. [DOI] [PubMed] [Google Scholar]
- 16.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 17.Elbashir, S. M., J. Harborth, K. Weber, and T. Tuschl. 2002. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 26:199-213. [DOI] [PubMed] [Google Scholar]
- 18.Figdor, C. G., Y. van Kooyk, and G. J. Adema. 2002. C-type lectin receptors on dendritic cells and Langerhans cells. Nat. Rev. Immunol. 2:77-84. [DOI] [PubMed] [Google Scholar]
- 19.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA 94:9825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geijtenbeek, T. B., G. Koopman, G. C. van Duijnhoven, S. J. van Vliet, A. C. van Schijndel, A. Engering, J. L. Heeney, and Y. van Kooyk. 2001. Rhesus macaque and chimpanzee DC-SIGN act as HIV/SIV gp120 trans-receptors, similar to human DC-SIGN. Immunol. Lett. 79:101-107. [DOI] [PubMed] [Google Scholar]
- 21.Geijtenbeek, T. B., D. J. Krooshoop, D. A. Bleijs, S. J. van Vliet, G. C. van Duijnhoven, V. Grabovsky, R. Alon, C. G. Figdor, and Y. van Kooyk. 2000. DC-SIGN-ICAM-2 interaction mediates dendritic cell trafficking. Nat. Immunol. 1:353-357. [DOI] [PubMed] [Google Scholar]
- 22.Geijtenbeek, T. B., D. S. Kwon, R. Torensma, S. J. van Vliet, G. C. van Duijnhoven, J. Middel, I. L. Cornelissen, H. S. Nottet, V. N. KewalRamani, D. R. Littman, C. G. Figdor, and Y. van Kooyk. 2000. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 100:587-597. [DOI] [PubMed] [Google Scholar]
- 23.Geijtenbeek, T. B., R. Torensma, S. J. van Vliet, G. C. van Duijnhoven, G. J. Adema, Y. van Kooyk, and C. G. Figdor. 2000. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell 100:575-585. [DOI] [PubMed] [Google Scholar]
- 24.Geijtenbeek, T. B., and Y. van Kooyk. 2003. DC-SIGN: a novel HIV receptor on DCs that mediates HIV-1 transmission. Curr. Top. Microbiol. Immunol. 276:31-54. [DOI] [PubMed] [Google Scholar]
- 25.Geijtenbeek, T. B., Y. van Kooyk, S. J. van Vliet, M. H. Renes, R. A. Raymakers, and C. G. Figdor. 1999. High frequency of adhesion defects in B-lineage acute lymphoblastic leukemia. Blood 94:754-764. [PubMed] [Google Scholar]
- 26.Geijtenbeek, T. B., S. J. van Vliet, E. A. Koppel, M. Sanchez-Hernandez, C. M. Vandenbroucke-Grauls, B. Appelmelk, and Y. van Kooyk. 2003. Mycobacteria target DC-SIGN to suppress dendritic cell function. J. Exp. Med. 197:7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halary, F., A. Amara, H. Lortat-Jacob, M. Messerle, T. Delaunay, C. Houles, F. Fieschi, F. Arenzana-Seisdedos, J. F. Moreau, and J. Dechanet-Merville. 2002. Human cytomegalovirus binding to DC-SIGN is required for dendritic cell infection and target cell trans-infection. Immunity 17:653-664. [DOI] [PubMed] [Google Scholar]
- 28.Hu, Q., I. Frank, V. Williams, J. J. Santos, P. Watts, G. E. Griffin, J. P. Moore, M. Pope, and R. J. Shattock. 2004. Blockade of attachment and fusion receptors inhibits HIV-1 infection of human cervical tissue. J. Exp. Med. 199:1065-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jameson, B., F. Baribaud, S. Pohlmann, D. Ghavimi, F. Mortari, R. W. Doms, and A. Iwasaki. 2002. Expression of DC-SIGN by dendritic cells of intestinal and genital mucosae in humans and rhesus macaques. J. Virol. 76:1866-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawamura, T., F. O. Gulden, M. Sugaya, D. T. McNamara, D. L. Borris, M. M. Lederman, J. M. Orenstein, P. A. Zimmerman, and A. Blauvelt. 2003. R5 HIV productively infects Langerhans cells, and infection levels are regulated by compound CCR5 polymorphisms. Proc. Natl. Acad. Sci. USA 100:8401-8406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kunath, T., G. Gish, H. Lickert, N. Jones, T. Pawson, and J. Rossant. 2003. Transgenic RNA interference in ES cell-derived embryos recapitulates a genetic null phenotype. Nat. Biotechnol. 21:559-561. [DOI] [PubMed] [Google Scholar]
- 32.Kwon, D. S., G. Gregorio, N. Bitton, W. A. Hendrickson, and D. R. Littman. 2002. DC-SIGN-mediated internalization of HIV is required for trans-enhancement of T cell infection. Immunity 16:135-144. [DOI] [PubMed] [Google Scholar]
- 33.Lozach, P. Y., H. Lortat-Jacob, A. de Lacroix de Lavalette, I. Staropoli, S. Foung, A. Amara, C. Houles, F. Fieschi, O. Schwartz, J. L. Virelizier, F. Arenzana-Seisdedos, and R. Altmeyer. 2003. DC-SIGN and L-SIGN are high affinity binding receptors for hepatitis C virus glycoprotein E2. J. Biol. Chem. 278:20358-20366. [DOI] [PubMed] [Google Scholar]
- 34.McDonald, D., L. Wu, S. M. Bohks, V. N. KewalRamani, D. Unutmaz, and T. J. Hope. 2003. Recruitment of HIV and its receptors to dendritic cell-T cell junctions. Science 300:1295-1297. [DOI] [PubMed] [Google Scholar]
- 35.Neil, S., F. Martin, Y. Ikeda, and M. Collins. 2001. Postentry restriction to human immunodeficiency virus-based vector transduction in human monocytes. J. Virol. 75:5448-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paddison, P. J., A. A. Caudy, E. Bernstein, G. J. Hannon, and D. S. Conklin. 2002. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 16:948-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piguet, V., and A. Blauvelt. 2002. Essential roles for dendritic cells in the pathogenesis and potential treatment of HIV disease. J. Investig. Dermatol. 119:365-369. [DOI] [PubMed] [Google Scholar]
- 38.Pohlmann, S., G. J. Leslie, T. G. Edwards, T. Macfarlan, J. D. Reeves, K. Hiebenthal-Millow, F. Kirchhoff, F. Baribaud, and R. W. Doms. 2001. DC-SIGN interactions with human immunodeficiency virus: virus binding and transfer are dissociable functions. J. Virol. 75:10523-10526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pohlmann, S., J. Zhang, F. Baribaud, Z. Chen, G. J. Leslie, G. Lin, A. Granelli-Piperno, R. W. Doms, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins interact with DC-SIGN and DC-SIGNR. J. Virol. 77:4070-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rubinson, D. A., C. P. Dillon, A. V. Kwiatkowski, C. Sievers, L. Yang, J. Kopinja, D. L. Rooney, M. M. Ihrig, M. T. McManus, F. B. Gertler, M. L. Scott, and L. Van Parijs. 2003. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 33:401-406. [DOI] [PubMed] [Google Scholar]
- 41.Salmon, P., J. F. Arrighi, V. Piguet, B. Chapuis, R. H. Zubler, D. Trono, and V. Kindler. 2001. Transduction of CD34+ cells with lentiviral vectors enables the production of large quantities of transgene-expressing immature and mature dendritic cells. J. Gene Med. 3:311-320. [DOI] [PubMed] [Google Scholar]
- 42.Shi, Y. 2003. Mammalian RNAi for the masses. Trends Genet. 19:9-12. [DOI] [PubMed] [Google Scholar]
- 43.Sledz, C. A., M. Holko, M. J. de Veer, R. H. Silverman, and B. R. Williams. 2003. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 5:834-839. [DOI] [PubMed] [Google Scholar]
- 44.Stein, P., P. Svoboda, and R. M. Schultz. 2003. Transgenic RNAi in mouse oocytes: a simple and fast approach to study gene function. Dev. Biol. 256:187-193. [DOI] [PubMed] [Google Scholar]
- 45.Steinman, R. M., A. Granelli-Piperno, M. Pope, C. Trumpfheller, R. Ignatius, G. Arrode, P. Racz, and K. Tenner-Racz. 2003. The interaction of immunodeficiency viruses with dendritic cells. Curr. Top. Microbiol. Immunol. 276:1-30. [DOI] [PubMed] [Google Scholar]
- 46.Sui, G., C. Soohoo, E. B. Affar, F. Gay, Y. Shi, and W. C. Forrester. 2002. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA 99:5515-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tailleux, L., O. Schwartz, J. L. Herrmann, E. Pivert, M. Jackson, A. Amara, L. Legres, D. Dreher, L. P. Nicod, J. C. Gluckman, P. H. Lagrange, B. Gicquel, and O. Neyrolles. 2003. DC-SIGN is the major Mycobacterium tuberculosis receptor on human dendritic cells. J. Exp. Med. 197:121-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tiscornia, G., O. Singer, M. Ikawa, and I. M. Verma. 2003. A general method for gene knockdown in mice by using lentiviral vectors expressing small interfering RNA. Proc. Natl. Acad. Sci. USA 100:1844-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trumpfheller, C., C. G. Park, J. Finke, R. M. Steinman, and A. Granelli-Piperno. 2003. Cell type-dependent retention and transmission of HIV-1 by DC-SIGN. Int. Immunol. 15:289-298. [DOI] [PubMed] [Google Scholar]
- 50.Turville, S., J. Wilkinson, P. Cameron, J. Dable, and A. L. Cunningham. 2003. The role of dendritic cell C-type lectin receptors in HIV pathogenesis. J. Leukoc. Biol. 74:710-718. [DOI] [PubMed] [Google Scholar]
- 51.Turville, S. G., P. U. Cameron, A. Handley, G. Lin, S. Pohlmann, R. W. Doms, and A. L. Cunningham. 2002. Diversity of receptors binding HIV on dendritic cell subsets. Nat. Immunol. 3:975-983. [DOI] [PubMed] [Google Scholar]
- 52.van Die, I., S. J. van Vliet, A. K. Nyame, R. D. Cummings, C. M. Bank, B. Appelmelk, T. B. Geijtenbeek, and Y. van Kooyk. 2003. The dendritic cell-specific C-type lectin DC-SIGN is a receptor for Schistosoma mansoni egg antigens and recognizes the glycan antigen Lewis x. Glycobiology 13:471-478. [DOI] [PubMed] [Google Scholar]
- 53.Wilson, J. A., and C. D. Richardson. 2003. Induction of RNA interference using short interfering RNA expression vectors in cell culture and animal systems. Curr. Opin. Mol. Ther. 5:389-396. [PubMed] [Google Scholar]
- 54.Wiznerowicz, M., and D. Trono. 2003. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J. Virol. 77:8957-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu, L., T. D. Martin, M. Carrington, and V. N. KewalRamani. 2004. Raji B cells, misidentified as THP-1 cells, stimulate DC-SIGN-mediated HIV transmission. Virology 318:17-23. [DOI] [PubMed] [Google Scholar]
- 56.Xia, H., Q. Mao, H. L. Paulson, and B. L. Davidson. 2002. siRNA-mediated gene silencing in vitro and in vivo. Nat. Biotechnol. 20:1006-1010. [DOI] [PubMed] [Google Scholar]
- 57.Yang, Z. Y., Y. Huang, L. Ganesh, K. Leung, W. P. Kong, O. Schwartz, K. Subbarao, and G. J. Nabel. 2004. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J. Virol. 78:5642-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zufferey, R., D. Nagy, R. J. Mandel, L. Naldini, and D. Trono. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 15:871-875. [DOI] [PubMed] [Google Scholar]