Abstract
The cytosine deaminase APOBEC3G, in the absence of the human immunodeficiency virus type 1 (HIV-1) accessory gene HIV-1 viral infectivity factor (vif), inhibits viral replication by introducing G→A hypermutation in the newly synthesized HIV-1 DNA negative strand. We tested the hypothesis that genetic variants of APOBEC3G may modify HIV-1 transmission and disease progression. Single nucleotide polymorphisms were identified in the promoter region (three), introns (two), and exons (two). Genotypes were determined for 3,073 study participants enrolled in six HIV-AIDS prospective cohorts. One codon-changing variant, H186R in exon 4, was polymorphic in African Americans (AA) (f = 37%) and rare in European Americans (f < 3%) or Europeans (f = 5%). For AA, the variant allele 186R was strongly associated with decline in CD4 T cells (CD4 slope on square root scale: −1.86, P = 0.009), The 186R allele was also associated with accelerated progression to AIDS-defining conditions in AA. The in vitro antiviral activity of the 186R enzyme was not inferior to that of the common H186 variant. These studies suggest that there may be a modifying role of variants of APOBEC3G on HIV-1 disease progression that warrants further investigation.
In the adaptive struggle between host and pathogen, mammalian species have evolved an arsenal of strategies for acquired and innate immunity to viral pathogens, which in turn have evolved mechanisms to evade this surveillance. It has long been observed that most T-cell lines support high replication rates of both wild-type (wt) human immunodeficiency virus type 1 (HIV-1) and HIV-1 with deletion of the accessory gene encoding the viral infectivity factor (Δvif) but that primary T cells, macrophages, and certain nonpermissive T-cell lines do not support replication of Δvif HIV-1 (9, 30). Sheehy et al. identified a cellular factor, CEM15, later shown to be the cytidine deaminase APOBEC3G (27), a member of a family of RNA editing enzymes that mediates HIV-1 Δvif suppression. APOBEC3G has been shown to deaminate cytidine residues in the newly synthesized DNA negative strand, causing G→A hypermutation on the RNA genome of HIV-1 (13, 17, 21, 33). APOBEC3G deamination of the minus strand of cDNA also makes the newly synthesized DNA vulnerable to degradation by uracil N-glycosylation (23). It has been demonstrated that Vif potently reduces the amount of APOBEC3G encapsulated in HIV-1 virions by associating with the cellular enzyme and triggering its proteasomal degradation (23, 28, 29). HIV-1 Vif does not prevent encapsidation of APOBEC3G proteins from mouse, African green monkey, or rhesus monkey cells, suggesting that species specificity of the Vif-APOBEC3G interaction plays a role in restricting HIV-1 infection to humans (22). Furthermore, APOBEC3G was found to act on other retroviruses in addition to HIV-1, suggesting that targeted DNA deamination is a general innate defense mechanism against retroviruses (13, 21, 22).
Since APOBEC3G is an important host factor that may confer an intrinsic block to HIV-1 and possibly other human pathogens that have an obligatory reverse transcription step, we screened the APOBEC3G gene for both regulatory and coding region variants that could modify APOBEC3G transcription or amino acid sequence. We analyzed the effects of six single nucleotide polymorphisms (SNPs) and their haplotypes in five United States-based natural history cohorts and the Swiss HIV cohort study for their influence on progression.
MATERIALS AND METHODS
Study participants.
The United States-based cohort study groups consisted of 965 HIV-1 seroconverters, 763 seroprevalents, and 702 seronegatives, for a total of 2,430 participants (1,481 European Americans [EA] and 949 African Americans [AA]) enrolled in five natural history HIV-AIDS cohorts: AIDS Link to the Intravenous Experience (ALIVE) (31), Multicenter AIDS Cohort Study (MACS) (26), the San Francisco City Clinic Study (SFCC) (4), Hemophilia Growth and Development Study (HGDS) (14), and the Multicenter Hemophilia Cohort Study (MHCS) (11). The date of seroconversion after study enrollment was estimated as the midpoint between the last seronegative and first seropositive HIV-1 antibody test; only individuals with less than 2 years' elapsed time between the two tests were included in the seroconverter progression analysis. The censoring date was the earlier of the date of the last recorded visit or 31 December 1995 for the MACS, MHCS, HGDS, or SFCC cohorts or 31 July 1997 for the ALIVE cohort to avoid potential confounding by highly effective antiretroviral therapy (HAART); because administration of HAART was delayed in the ALIVE cohort (5, 31), the later censoring date was used. The MACS, MHCS, SFCC, and ALIVE cohorts consisted of both seroconverter (infected after study enrollment) and seroprevalent (infected before study enrollment) individuals: because of the potential for frailty bias among seroprevalents, only seroconverters enrolled in the MACS, MHCS, and SFCC cohorts were used in the survival analysis. Longitudinal CD4 T-cell counts used for the random effects model were available for each semiannual visit for 225 AA seroconverters (from enrollment in 1988-1989 to 2003 or the censoring date) in the ALIVE cohorts and 643 seroprevalents (median follow-up time, 3.1 years) enrolled in the Swiss HIV cohort (www.shcs.ch). High-risk exposed uninfected (HREU) individuals were those with documented high-risk behaviors or documented exposure to contaminated blood products as previously described (2). DNA samples from 129 healthy blood donor Han Chinese were included in the study for allele frequencies. The Institutional Review Board of participating institutes approved study protocols and consent procedures.
Identification of DNA polymorphisms.
Nucleotide polymorphisms in APOBEC3G were discovered using a DNA panel consisting of 92 EA and 92 AA. A nonisotopic RNA cleavage assay (NIRCA) was performed to detect polymorphisms (12). Overlapping PCR primers were designed to cover the putative 5′ regulatory region, eight exons, exon-intron junctions, intron 1, and the 3′ untranslated region of the APOBEC3G gene according to GenBank sequences AL022318 and AL078641. Gene-specific primers for APOBEC3G were designed to avoid amplification of pseudogenes, other phorbolin-like genes, or APOBEC genes (15). PCR primers and conditions are available upon request. PCR products that revealed aberrant bands by NIRCA analysis were purified and sequenced.
Genotyping of genetic variants.
Genotypes were determined by PCR-restriction fragment length polymorphism or TaqMan assays. PCR primer sequences, TaqMan probes and primers, PCR conditions, and restriction enzymes used in genotyping assays for each SNP are listed in Table S1 in the supplemental material. PCR was carried out with 35 cycles of denaturing at 94°C for 30 s, annealing at 54 to 60°C for 30 s, and extension at 72°C for 45 s. TaqMan assays were performed according to the manufacturer's manual (Perkin-Elmer). Primers are available on Table S1 in the supplemental material.
In vitro analysis.
CD4 T cells from 128 healthy Caucasian blood donors were isolated by anti-CD4 magnetic beads (Miltenyi Biotech, Bergisch Glabach, Germany), cultured, and infected (106 cells) with 1,000 pg of p24 antigen of R5 clone HIV NL4-3BaLenv. In addition, peripheral blood lymphocytes from 22 AA donors were similarly infected. Samples from four donors were also infected with the Vif-defective HIV-1 strain R9Δvif (32). After 7 days, p24 antigen production was monitored by enzyme-linked immunosorbent assay (Abbott Laboratories). Viral RNA was extracted from cells and supernatant and subjected to sequence analysis to identify potential changes in the frequency of deamination.
Analysis of antiviral activity of APOBEC3G variants.
The Vif-defective HIV-1 proviral clone R9Δvif was previously described (32). The plasmid expressing a hemagglutinin-tagged form of APOBEC3G (27) was a kind gift from M. Malim (University College, London, United Kingdom). The APOBEC3G 186R mutant was constructed with the QuickChange Mutagenesis kit (Stratagene) and sequenced. HIV-1 was produced by transient transfection of 293T cells with Fugene (Roche) in six-well plates. For this, 0.7 μg of R9Δvif was used with either an empty vector or different doses of the APOBEC3G-expressing vector. Thirty-six hours after transfection, supernatant was collected and filtered. Virus titer was scored by infecting HeLa-CD4-LacZ reporter cells. Virion infectivity was normalized for particle amount, as measured by reverse transcriptase (RT) activity assay.
Statistical analysis.
The genetic effects of SNPs on HIV-1 infection susceptibility were assessed by comparing genotypic frequencies between HIV-1 HREU individuals and HIV-1 seroconverters with use of Fisher's exact test. Kaplan-Meier survival statistics and the Cox proportional hazards model were used to assess the effects of SNPs and haplotypes on the rate of progression to AIDS. Four end points were evaluated: time to less than 200 CD4+ cells/mm3 (CD4 < 200); AIDS-1987 and the expanded AIDS-1993 Centers for Disease Control and Prevention case definitions (6, 7), and AIDS-related death. The significance of genotypic associations and relative hazards (RH) for dominant and recessive genetic models were determined with unadjusted and adjusted Cox model analysis. All P values are two-tailed. Genetic factors previously shown to affect progression to AIDS in EA and AA groups were included as confounding covariates in the adjusted Cox model analysis: CCR5 Δ32, CCR2-64I, CCR5-P1, HLA-B*27, HLA-B*57, HLA-B*35Px, and HLA class I homozygosity for EA (8, 10, 16, 24); CCR5-P1, RANTES-In1.1C, HLA-B*57, and HLA class I homozygosity for AA (1, 8, 16, 24). CCR2-64I, HLA-B*27, and HLA-B*35Px were not considered as covariates in AA due to absent or weak effects in our AA participants, and CCR5 Δ32 was not considered due to its rarity in AA. Participants were stratified by sex and by age at seroconversion: 0 to 20, >20 to 40, and over 40 years (25).
A random effects linear model of CD4 T-cell trajectories from HIV-1 seroconverters in the ALIVE study and HIV-1 seroprevalents in the Swiss cohort study was used to provide estimates of mean CD4 T-cell measurements over time while accounting for the correlation of repeated measurements within each individual. The CD4 slope of each patient was modeled using a repeat measures random effects model in STATA (Stata v8.0; College Station, Tex.) for the ALIVE cohort or the Mlwin software for the Swiss cohort. The square root CD4 gradient was modeled as a linear function of estimated time since midpoint (ALIVE) or imputed (Swiss) seroconversion dates. Additional covariates included sex, age, and time since seroconversion for the ALIVE cohort and sex, age (dichotomized at 40 years), intravenous drug use, race, and CCR5 Δ32 (3) for the Swiss cohort. Differences in CD4 gradient were compared to the group with the most common genetic variant by use of a Wald test of the null hypothesis that the difference is zero and the likelihood ratio test for models with and without genetic variables. The change in mean square root CD4 T cells over time was compared for APOBEC3G 186R/R (recessive genetic model) and R/H plus R/R (dominant model). Additionally, to account for the potential confounding effects of HAART in the ALIVE cohort, we fit the random effects model (i) by censoring at 31 July 1997, before HAART was available to this population, and (ii) by restricting the analysis to follow-up after 31 July 1997 and stratifying the model to individuals who reported use or no use of HAART (31). These analyses were done only for the ALIVE cohort because no significant effects of the H186R polymorphism were observed in the predominantly European Swiss cohort. Only these two cohorts had the complete CD4 T-cell follow-up data available to our group for this analysis.
We quantified linkage disequilibrium (LD) between all pairs of biallelic loci by use of the absolute value of Lewontin's D′ (18). Absolute values of D′ range from 0 for independence to 1 for complete LD between the pairs. P values represent the null hypothesis of independence. Haplotypes were inferred by the expectation maximization algorithm (19).
RESULTS
Identification of APOBEC3G variants.
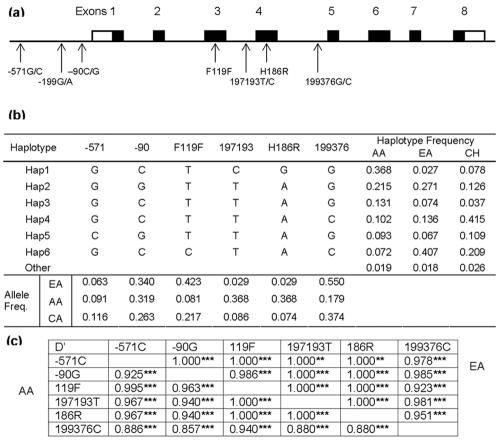
The genomic structure of APOBEC3G was predicted from a comparison of human DNA clone CTA-150C2 (GenBank sequence AL022318) and clone RP3-494G10 (AL078641) with its mRNA sequence (NM_021822). The human APOBEC3G gene spans ∼9 kb on 22q13.1-13.2 and has eight exons and seven introns. NIRCA and DNA resequencing were employed to detect SNPs (15). Seven SNPs were identified (Fig. 1a): three in the putative 5′ regulatory region, F119F in exon 3, H186R in exon 4, and two within introns. The genotype of each SNP was determined for 2,430 study participants enrolled in the United States-based HIV-AIDS prospective cohorts, 643 participants in the Swiss HIV cohort, and 129 healthy blood donor Han Chinese. The allele frequencies are listed in Fig. 1b. The single codon-changing variant, H186R, was rare in EA and European Caucasians (f = 0.03 and 0.05) but frequent in AA (f = 0.37).
FIG. 1.
Gene map, LD, and haplotypes of APOBEC3G. (a) Exon distribution (not to scale) and genomic locations of the SNPs. Coding exons are marked by shaded blocks, and the 5′ and 3′ untranslated regions are marked by white blocks. The SNPs were numbered according to the first base of the translation start site for those located in the putative promoter region, of reference sequence AL022318 for intronic SNPs, and of the amino acid sequence (NP_068594). (b) The six most frequent haplotypes and their frequencies in AA, EA, and Chinese (CH). (c) Detailed LD parameter D′ for the six SNPs in AA (below the diagonal) and EA (above the diagonal); **, P < 0.001; ***, P < 0.0001.
LD and haplotypes of APOBEC3G variants.
Pairwise tests of LD were performed among all APOBEC3G SNPs except for the rare −199G/A SNP. Strong LD was observed between all APOBEC3G SNPs (1.0 ≤ D′ > 0.923, P < 1.1 × 10−3 in EA and 1.0 ≤ D′ > 0.880, P < 1.1 × 10−5 in AA, respectively) (Fig. 1c). There were 13 and 14 haplotypes in AA and EA, respectively. The six most frequent haplotypes were shared in both races and accounted for more than 98% of AA or EA chromosomes (Fig. 1b).
Effect of APOBEC3G SNPs and haplotypes on susceptibility to HIV-1 infection.
We compared APOBEC3G allele, genotype, and haplotype frequencies for HREU individuals or HIV-1 seronegatives to HIV-1-infected seroconverters for dominant and recessive genetic models. No associations were observed for any SNP or haplotype (see Table S2 in the supplemental material), suggesting that APOBEC3G variants have no obvious effect on HIV-1 transmission.
Effect of APOBEC3G SNPs and haplotypes on AIDS progression.
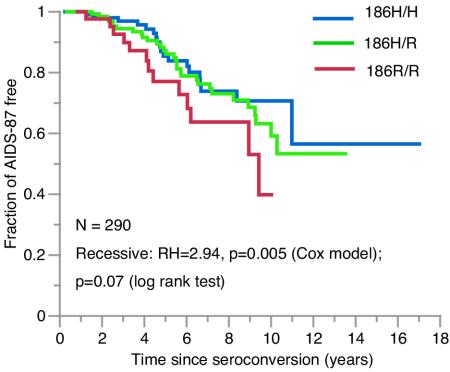
The influences of APOBEC3G SNP alleles and haplotypes on AIDS progression were evaluated for dominant and recessive genetic models among EA (n = 673) and AA (n = 292) HIV-1 seroconverters by use of the Cox proportional hazards model. Survival analyses with and without adjustments for the influence of AIDS-restriction gene covariates are listed in Table 1 and shown in Fig. 2. In the unadjusted analysis, the 186R/R genotype tended to be associated with disease progression to AIDS-1993, AIDS-1987, and death (RH = 1.43 to 1.82, P = 0.059 to 0.16). After adjusting for the potentially confounding effects of other genetic factors, the 186R/R genotype was associated with accelerated rate of progression to AIDS-1993, AIDS-1987, and death (RH = 1.94, 2.94, and 3.38, respectively; P = 0.024, 0.005, and 0.008, respectively). To understand why the 186R/R association was masked in the unadjusted analysis, each of the genetic covariates was sequentially used in the adjusted Cox model. The covariate RANTES-In1.1C alone revealed nearly the full strength of the association (data not shown); however, no interaction was observed between APOBEC3G 186R and RANTES-In1.1C (P = 0.35), consistent with the current understanding of the functional pathways of the two genes.
TABLE 1.
Cox model analysis of the effects of APOBEC3G SNPs on AIDS progressiona
| SNP | Race | Model | CD4 200
|
AIDS-1993
|
AIDS-1987
|
Death
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RH | 95% CI | P | RH | 95% CI | P | RH | 95% CI | P | RH | 95% CI | P | |||
| Unadjustedb | ||||||||||||||
| 197193Cc | EA | Dom | 1.00 | 0.62-1.61 | 0.990 | 0.91 | 0.59-1.41 | 0.680 | 1.03 | 0.64-1.67 | 0.890 | 0.84 | 0.48-1.47 | 0.550 |
| AA | Dom | 1.29 | 0.84-1.97 | 0.240 | 1.23 | 0.84-1.81 | 0.290 | 1.28 | 0.74-2.20 | 0.370 | 1.16 | 0.57-2.38 | 0.680 | |
| Rec | 1.34 | 0.82-2.19 | 0.240 | 1.47 | 0.94-2.30 | 0.089 | 1.88 | 1.03-3.44 | 0.040 | 1.96 | 0.91-4.21 | 0.087 | ||
| 186R | EA | Dom | 0.98 | 0.62-1.56 | 0.930 | 0.90 | 0.59-1.37 | 0.610 | 1.04 | 0.65-1.66 | 0.880 | 0.87 | 0.51-1.50 | 0.620 |
| AA | Dom | 1.34 | 0.87-2.04 | 0.180 | 1.32 | 0.90-1.94 | 0.160 | 1.37 | 0.79-2.38 | 0.270 | 1.12 | 0.55-2.31 | 0.750 | |
| Rec | 1.28 | 0.77-2.10 | 0.340 | 1.43 | 0.91-2.26 | 0.120 | 1.82 | 0.98-3.39 | 0.059 | 1.76 | 0.79-3.92 | 0.170 | ||
| 199376C | EA | Dom | 0.99 | 0.76-1.28 | 0.930 | 1.02 | 0.80-1.30 | 0.870 | 1.10 | 0.83-1.47 | 0.520 | 1.12 | 0.82-1.53 | 0.460 |
| Rec | 1.04 | 0.82-1.32 | 0.730 | 1.28 | 1.04-1.58 | 0.021 | 1.36 | 1.07-1.73 | 0.013 | 1.37 | 1.06-1.77 | 0.016 | ||
| AA | Dom | 1.16 | 0.76-1.77 | 0.500 | 1.19 | 0.81-1.75 | 0.390 | 1.01 | 0.58-1.77 | 0.970 | 0.90 | 0.42-1.92 | 0.780 | |
| Adjustedd | ||||||||||||||
| 197193Cc | EA | Dom | 0.95 | 0.58-1.56 | 0.830 | 0.95 | 0.61-1.49 | 0.820 | 1.10 | 0.67-1.82 | 0.700 | 0.87 | 0.48-1.56 | 0.630 |
| AA | Dom | 1.15 | 0.70-1.90 | 0.570 | 1.09 | 0.71-1.69 | 0.690 | 1.37 | 0.75-2.50 | 0.310 | 1.50 | 0.65-3.45 | 0.340 | |
| Rec | 1.59 | 0.83-3.03 | 0.160 | 2.03 | 1.16-3.57 | 0.013 | 3.06 | 1.49-6.29 | 0.002 | 3.76 | 1.60-3.84 | 0.002 | ||
| 186R | EA | Dom | 0.91 | 0.56-1.48 | 0.720 | 0.91 | 0.59-1.41 | 0.670 | 1.07 | 0.66-1.74 | 0.770 | 0.88 | 0.50-1.55 | 0.670 |
| AA | Dom | 1.14 | 0.70-1.88 | 0.600 | 1.14 | 0.74-1.78 | 0.550 | 1.47 | 0.79-2.73 | 0.220 | 1.45 | 0.63-3.37 | 0.380 | |
| Rec | 1.46 | 0.75-2.84 | 0.270 | 1.94 | 1.09-3.44 | 0.024 | 2.94 | 1.39-6.21 | 0.005 | 3.38 | 1.38-8.27 | 0.008 | ||
| 199376C | EA | Dom | 1.08 | 0.82-1.42 | 0.580 | 1.11 | 0.87-1.43 | 0.390 | 1.21 | 0.90-1.63 | 0.200 | 1.16 | 0.84-1.59 | 0.370 |
| Rec | 1.11 | 0.87-1.42 | 0.400 | 1.28 | 1.03-1.60 | 0.024 | 1.34 | 1.05-1.73 | 0.020 | 1.38 | 1.05-1.80 | 0.020 | ||
| AA | Dom | 1.06 | 0.64-1.76 | 0.810 | 1.02 | 0.65-1.60 | 0.940 | 0.87 | 0.46-1.64 | 0.670 | 0.74 | 0.31-1.80 | 0.510 | |
Analysis was done in seroconverters (n = 656 and 292 in EA and AA, respectively) for dominant (Dom) or recessive (Rec) model; Recessive model was not assessed in groups with five or fewer individuals carrying the genotype; results for the other three SNPs are listed in Tables 3 in the supplemental material.
Results were unadjusted for covariates.
Missing 197193T/C genotype for one patient compared to H186R.
Results were adjusted with covariates CCR5 Δ32 and four HLA alleles (Zyg_N, B27, B35Px, and B57) for EA and covariates Zyg_N. B57, CCR5-P1, and RANTES-In1.1C for AA.
FIG. 2.
Kaplan-Meier survival curves of HIV-1 progression to AIDS-1987 after seroconversion for the three genotypes of APOBEC3G-H186R in AA. RH and Wald P values are from an adjusted Cox model analysis, and log rank P value is for the Kaplan-Meier statistic.
A weak but statistically significant protective tendency was observed for the intron 4 APOBEC3G-199376C allele for AIDS-1993, AIDS-1987, and death in the recessive model for EA, both without and with adjustment (RH = 1.36 to 1.58, P = 0.01 to 0.003) for potential confounding genetic factors (Table 1). Results for SNPs with no significant associations are listed in Table S3 in the supplemental material. APOBEC3G-197193C, located in intron 2, is in complete and nearly absolute LD with 186R, thus showing very similar associations (Table S3 in the supplemental material).
Since the alleles within the APOBEC3G gene are in complete or near-complete LD, we considered the combination of alleles that are inherited together on a single chromosomal segment (haplotype) since these more closely represent the natural condition. Since the 186R allele is found only on haplotype 1 (Hap 1), the associations observed for Hap 1 were quite similar to that of the 186R allele (Tables 1 and 2; Fig. 2). The slight differences observed in RH and P values are due to individuals missing one or more genotypes. Modest accelerating effects to AIDS-1987 and death were observed for Hap 4 in EA. Conversely, Hap 3, which contains neither 186R nor 199376C, was slightly protective in both AA and EA for the early outcomes, CD4 < 200 or AIDS-1993 (P = 0.01 to 0.10) (Table 2).
TABLE 2.
Cox model analysis of APOBFC3G haplotypes on AIDS progression, adjusting for covariatesa
| Group and haplotype | Model | CD4 < 200
|
AIDS-1993
|
AIDS-1987
|
Death
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RH | 95% CI | P | RH | 95% CI | P | RH | 95% CI | P | RH | 95% CI | P | ||
| AA (n = 268)b | |||||||||||||
| Hap 1 | Rec | 1.80 | 0.92-3.53 | 0.088 | 2.01 | 1.10-3.66 | 0.022 | 2.94 | 1.33-6.49 | 0.008 | 3.60 | 1.46-8.87 | 0.005 |
| Dom | 1.41 | 0.83-2.40 | 0.200 | 1.25 | 0.78-1.99 | 0.351 | 1.72 | 0.88-3.36 | 0.111 | 1.89 | 0.79-4.52 | 0.153 | |
| Hap 2 | Dom | 1.00 | 0.62-1.61 | 0.990 | 1.01 | 0.66-1.57 | 0.953 | 0.54 | 0.29-1.02 | 0.057 | 0.39 | 0.16-0.96 | 0.040 |
| Hap 3 | Dom | 0.46 | 0.25-0.85 | 0.013 | 0.52 | 0.31-0.88 | 0.015 | 0.90 | 0.47-1.72 | 0.747 | 0.89 | 0.39-2.06 | 0.791 |
| Hap 4 | Dom | 1.19 | 0.66-2.17 | 0.564 | 0.96 | 0.56-1.67 | 0.900 | 0.88 | 0.40-1.93 | 0.759 | 0.93 | 0.31-2.77 | 0.890 |
| Hap 5 | Dom | 0.64 | 0.30-1.37 | 0.252 | 0.95 | 0.51-1.77 | 0.878 | 2.11 | 0.95-4.66 | 0.065 | 1.14 | 0.37-3.54 | 0.816 |
| Hap 6 | Dom | 1.49 | 0.83-2.66 | 0.183 | 1.65 | 0.97-2.80 | 0.063 | 1.04 | 0.45-2.40 | 0.930 | 0.98 | 0.31-3.10 | 0.970 |
| EA (n = 605)c | |||||||||||||
| Hap 1 | Dom | 0.94 | 0.57-1.56 | 0.818 | 0.94 | 0.60-1.48 | 0.795 | 1.04 | 0.63-1.73 | 0.866 | 0.83 | 0.46-1.49 | 0.534 |
| Hap 2 | Dom | 1.00 | 0.79-1.27 | 1.000 | 0.94 | 0.76-1.16 | 0.566 | 0.87 | 0.68-1.12 | 0.274 | 0.86 | 0.66-1.13 | 0.277 |
| Hap 3 | Dom | 0.73 | 0.50-1.06 | 0.099 | 0.76 | 0.54-1.06 | 0.102 | 0.85 | 0.58-1.25 | 0.421 | 0.73 | 0.47-1.13 | 0.153 |
| Hap 4 | Dom | 1.25 | 0.96-1.63 | 0.103 | 1.36 | 1.07-1.73 | 0.011 | 1.35 | 1.02-1.78 | 0.034 | 1.58 | 1.18-2.13 | 0.003 |
| Hap 5 | Dom | 1.11 | 0.79-1.57 | 0.536 | 0.95 | 0.69-1.30 | 0.752 | 0.99 | 0.68-1.43 | 0.959 | 1.04 | 0.69-1.55 | 0.864 |
| Hap 6 | Dom | 1.05 | 0.83-1.34 | 0.707 | 1.08 | 0.86-1.35 | 0.503 | 1.15 | 0.88-1.49 | 0.304 | 1.06 | 0.80-1.40 | 0.694 |
The rate of progression to four AIDS end points in patients carrying one or two copies of the haplotype was compared with that in patients without that haplotype. CI, RH, and significance were calculated with the use of the Cox model. Both Dom (dominant) and Rec (recessive) genetic models were tested, but results for the recessive model were not shown if they were not significant. The analyses were adjusted unless indicated as unadjusted.
Adjusted for HLA Zyg_N, B57, CCR5-P1, and RANTES-In1.1.
Adjusted for CCR5-P1, CCR5 Δ32, and four HLA alleles (Zyg_N, B27, B35Px, and B57).
Effect of H186R on overall CD4 T-cell slope gradients.
We evaluated the effect of APOBEC3G-H186R on CD4 slope using the random effects model for the ALIVE and Swiss cohorts (Table 3). Among 225 AA seroconverters enrolled in the ALIVE study, individuals carrying at least one copy of APOBEC3G-186R had a −1.86 (95% confidence interval [CI], −3.25, −0.45; P = 0.009) lower mean CD4 T-cell trajectory than did the reference 186H/H group. Analysis restricted to 217 individuals who were censored at 31 July 1997 had a similar decrease in mean CD4 T-cell trajectory (−1.51; 95% CI, −2.96, −0.65; P = 0.041) which persisted after increased availability of HAART (−2.25 lower; 95% CI, −4.13, −0.37; P = 0.019). For the dominant genetic model in the Swiss seroprevalent cohort, 186R (H/R, n = 57; R/R, n = 5) was found to be marginally associated with protection with a gradient difference of +0.29 (95% CI, −0.04, 0.62; P = 0.08) relative to the reference 186H/H genotype (n = 581). The low number of homozygotes precluded an analysis of the recessive model. Estimates were not modified significantly after adjusting for the potentially confounding effects of other genetic factors.
TABLE 3.
Effects of APOBEC3G H186R on overall CD4 T-cell slope gradients in the ALIVE seroconverter and Swiss seroprevalent cohortsa
| Genetic model and cohort | Total no. of:
|
CD4 T-cell slope | P (95% CI) | |
|---|---|---|---|---|
| Individuals | Observations | |||
| APOBEC3G 186 H/R and R/R | ||||
| ALIVE seroconverters (all)b | 225 | 2,852 | −1.86 | 0.009 (−3.26, −0.45) |
| ALIVE SC censored 7/97c | 217 | 1,741 | −1.51 | 0.041 (−2.96, −0.65) |
| ALIVE SC 7/97-1/03d | 151 | 1,120 | −2.25 | 0.019 (−4.13, −0.37) |
| Swiss seroprevalents | 643 | 4,501 | +0.29 | 0.080 (−0.04, 0.62) |
| APOBEC3G 186 R/R only | ||||
| ALIVE seroconverters (all)b | 225 | 2,852 | −1.62 | 0.09 (−3.48, 0.24) |
| ALIVE SC censored 7/97c | 217 | 1,741 | −2.21 | 0.11 (−4.93, 0.49) |
| ALIVE SC 7/97-1/03d | 151 | 1,120 | −2.02 | 0.14 (−4.72, 0.68) |
For the ALIVE seroconverters (SC) all analyses were adjusted for age and sex. For the Swiss seroprevalent cohort, analyses were adjusted for age, sex, race, CCR5 Δ32, and intravenous drug use. Only five homozygotes were observed in the Swiss cohort; therefore, a recessive genetic model was not tested.
Observation period for all seroconverters not censored for HAART.
Observation period before onset of HAART (July 1997).
Observation period after onset of HAART therapy (July 1997 to January 2003).
In vitro antiviral activity of 186R enzyme.
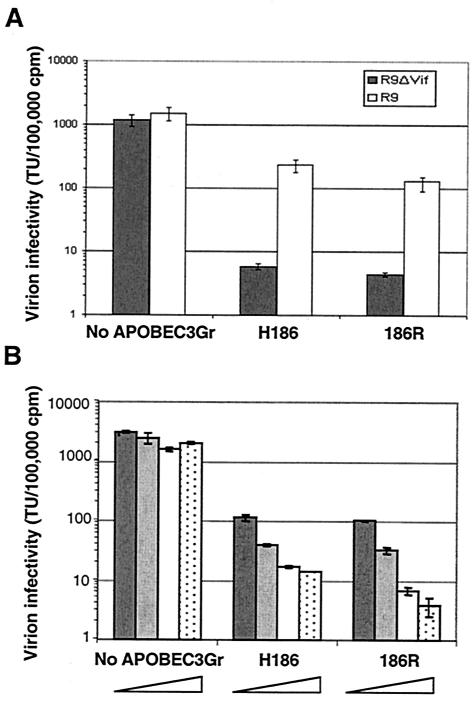
We tested whether mutating histidine to arginine at position 186 could influence the inhibitory potential of APOBEC3G. A single-round assay was used to measure the infectivity of wt and Δvif particles produced by transient transfection of 293T cells, in the presence of different doses of the wt or mutated cellular factor (Fig. 3). We found that the 186R mutant was fully able to inhibit viral infectivity.
FIG. 3.
Antiviral activity of APOBEC3G 186H and 186R variants. (A) 293T cells were transfected with R9 HIV-1 or with the Vif-defective R9Δvif proviral clone in the presence of empty vector, wt hAPOBEC3G (H186R), or 186R APOBEC3G-expressing vector. Supernatant was filtered, and viral titer was scored by infecting HeLa-CD4-LacZ reporter cells not expressing APOBEC3G. Virion infectivity is normalized for particle amount, as measured by RT activity assay. (B) In a second experiment, 293T cells were transfected with R9Δvif in the presence of four different doses of either an empty vector, wt hAPOBEC3G, or 186R APOBEC3G-expressing vector. The four ratios of APOBEC3G to R9Δvif are 0.5 (dark grey), 1 (light grey), 2 (white), and 3 (stippled). Analyses were done in duplicate, with bars representing the standard deviations.
The in vitro assessment of the role of H186R variants in influencing viral infectivity or deamination was completed by using peripheral mononuclear cells collected from healthy blood donors representing the various genotypes. Cells were infected with the R5-tropic clone HIV NL4-3BaLenv and by the Vif-deficient R9Δvif. There were no measurable differences in cell permissiveness to HIV infection in the in vitro system for the various H186R genotypes: mean and standard deviation of log p24 picograms per milliliter after 7 days of culture were 4.96 ± 4.79 for cells from 112 Caucasian donors representing the common H186 genotype versus 4.98 ± 4.78 for cells from 13 Caucasian donors carrying the 186R variant, P = 0.7. Similarly, there were no measurable differences in cell permissiveness to HIV infection in cells from AA donors: mean and standard deviation of log p24 picograms per milliliter after 7 days of culture were 4.64 ± 0.83 for cells from 11 AA donors representing the common H186 genotype versus 4.93 ± 0.27 for cells from 11 AA donors homozygous for the 186R variant, P = 0.49. This higher viral level with 186R, though not significant, warrants further elucidation in a large number of samples. The Vif-deficient R9Δvif strain was confirmed noninfectious for the various donor cells. No specific pattern of deamination of cytidine residues reflecting changes associated with the enzyme alleles was observed in the viral progeny emerging after 7 days of replication in the various cell populations (data not shown).
DISCUSSION
We have investigated the role of APOBEC3G SNPs and haplotypes on HIV-1 infection and progression in six longitudinal HIV-1 cohorts. With use of survival analysis, the 186R/R genotype was shown to be significantly associated with rapid progression to AIDS and death in AA. With use of the random effects model, 186R carriers had a more rapid decline of CD4 T-cell slope than did individuals homozygous for H186.
The discrepancies between results from the two analyses may reflect the fact that the random effects model has increased resolution with many more observation outcomes. Although the effects of 186R do not reach significance in the Cox model analysis with use of a CD4 cell count of <200 as an outcome in AA, the tendency towards more rapid CD4 loss in 186R carriers is consistent with the CD4 trajectory in this group. In contrast, the effects of 186R on CD4 slope in the Swiss Caucasian seroprevalent cohort tended to be slightly protective, possibly because of the modifying influence of as-yet-undetected polymorphisms, differences in cohort design, or the low allele frequency of 186R in this group. The intronic 199376C allele carried on Hap 4 and Hap 6 was associated with a modest accelerating effect to AIDS and death. Hap 4 was associated with rapid progression in only EA, and Hap 6 showed a similar tendency only in AA. These results suggest that 199376G/C is not itself causal but may be tracking by LD an allele as yet undetected.
It has been demonstrated that a moderate amount of human APOBEC3G causes a threefold decrease in infectivity of the wt HIV-1, suggesting that alterations that affect APOBEC3G activity or expression level may influence HIV-1 replication (22). H186R is located in the leucine-rich region possibly involved in protein-protein interaction, either with complementation factors and/or in homodimer formation (15). Disruption of this region could modify editing functions as a result of altered assembly of the enzyme (20). However, there is no functional evidence for a defect in the antiviral activity of APOBEC3G-186R in our preliminary in vitro experiments. It would be interesting to determine if 186R increases G-to-A mutation and breadth of the quasispecies variation, as this would promote the emergence of immune escape mutants.
APOBEC3G haplotypes are distributed differently among individuals of Asian, European, and African ancestry. This observation indicates that APOBEC3G haplotypes were possibly shaped through different evolutionary or historical events in geographic populations. The different genetic backgrounds in allele frequencies, LD, and haplotype architecture observed among world populations may account for, at least in part, population-specific genetic effects on HIV-1 disease.
Supplementary Material
Acknowledgments
We gratefully acknowledge the participants, their families, and their clinicians who participated in the ALIVE, MACS, HGDS, MHCS, SFCC, and SHCS studies. We thank Beth Binns-Roemer, Yuchun Zhou, Zhengmei Liu, and Kui Gong for excellent technical assistance and Daniele Fallin for helpful comments.
This study was funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. NO1-CO-124000; the National Institute on Drug Abuse, National Institutes of Health, under grant DA04334; and the Swiss National Science Foundation and Leenaards Foundation.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.An, P., G. W. Nelson, L. Wang, S. Donfield, J. J. Goedert, J. Phair, D. Vlahov, S. Buchbinder, W. L. Farrar, W. Modi, S. J. O'Brien, and C. A. Winkler. 2002. Modulating influence on HIV/AIDS by interacting RANTES gene variants. Proc. Natl. Acad. Sci. USA 99:10002-10007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An, P., D. Vlahov, J. B. Margolick, J. Phair, T. R. O'Brien, J. Lautenberger, S. J. O'Brien, and C. A. Winkler. 2003. A tumor necrosis factor-alpha-inducible promoter variant of interferon-gamma accelerates CD4+ T cell depletion in human immunodeficiency virus-1-infected individuals. J. Infect. Dis. 188:228-231. [DOI] [PubMed] [Google Scholar]
- 3.Bleiber, G., M. May, C. Suarez, R. Martinez, C. Marzolini, M. Egger, and A. Telenti. 2004. MDR1 genetic polymorphism does not modify either cell permissiveness to HIV-1 or disease progression before treatment. J. Infect. Dis. 189:583-586. [DOI] [PubMed] [Google Scholar]
- 4.Buchbinder, S. P., M. H. Katz, N. A. Hessol, P. M. O'Malley, and S. D. Holmberg. 1994. Long-term HIV-1 infection without immunologic progression. AIDS 8:1123-1128. [DOI] [PubMed] [Google Scholar]
- 5.Celentano, D. D., N. Galai, A. K. Sethi, N. G. Shah, S. A. Strathdee, D. Vlahov, and J. E. Gallant. 2001. Time to initiating highly active antiretroviral therapy among HIV-infected injection drug users. AIDS 15:1707-1715. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control. 1987. Revision of the CDC surveillance case definition for acquired immunodeficiency syndrome. Council of State and Territorial Epidemiologists; AIDS Program, Center for Infectious Diseases. Morb. Mortal. Wkly. Rep. 36(Suppl. 1):1S-15S. [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 1992. 1993 revised classification system for HIV infection and expanded surveillance case definition for AIDS among adolescents and adults. Morb. Mortal. Wkly. Rep. 41:1-19. [PubMed] [Google Scholar]
- 8.Dean, M., M. Carrington, C. Winkler, G. A. Huttley, M. W. Smith, R. Allikmets, J. J. Goedert, S. P. Buchbinder, E. Vittinghoff, E. Gomperts, S. Donfield, D. Vlahov, R. Kaslow, A. Saah, C. Rinaldo, R. Detels, and S. J. O'Brien. 1996. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science 273:1856-1862. [DOI] [PubMed] [Google Scholar]
- 9.Gabuzda, D. H., K. Lawrence, E. Langhoff, E. Terwilliger, T. Dorfman, W. A. Haseltine, and J. Sodroski. 1992. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J. Virol. 66:6489-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao, X., G. W. Nelson, P. Karacki, M. P. Martin, J. Phair, R. Kaslow, J. J. Goedert, S. Buchbinder, K. Hoots, D. Vlahov, S. J. O'Brien, and M. Carrington. 2001. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N. Engl. J. Med. 344:1668-1675. [DOI] [PubMed] [Google Scholar]
- 11.Goedert, J. J., C. M. Kessler, L. M. Aledort, R. J. Biggar, W. A. Andes, G. C. White II, J. E. Drummond, K. Vaidya, D. L. Mann, M. E. Eyster, et al. 1989. A prospective study of human immunodeficiency virus type 1 infection and the development of AIDS in subjects with hemophilia. N. Engl. J. Med. 321:1141-1148. [DOI] [PubMed] [Google Scholar]
- 12.Goldrick, M. M., G. R. Kimball, Q. Liu, L. A. Martin, S. S. Sommer, and J. Y. Tseng. 1996. NIRCA: a rapid robust method for screening for unknown point mutations. BioTechniques 21:106-112. [PubMed] [Google Scholar]
- 13.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 14.Hilgartner, M. W., S. M. Donfield, A. Willoughby, C. F. Contant, Jr., B. L. Evatt, E. D. Gomperts, W. K. Hoots, J. Jason, K. A. Loveland, S. M. McKinlay, et al. 1993. Hemophilia growth and development study. Design, methods, and entry data. Am. J. Pediatr. Hematol. Oncol. 15:208-218. [DOI] [PubMed] [Google Scholar]
- 15.Jarmuz, A., A. Chester, J. Bayliss, J. Gisbourne, I. Dunham, J. Scott, and N. Navaratnam. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79:285-296. [DOI] [PubMed] [Google Scholar]
- 16.Kaslow, R. A., M. Carrington, R. Apple, L. Park, A. Munoz, A. J. Saah, J. J. Goedert, C. Winkler, S. J. O'Brien, C. Rinaldo, R. Detels, W. Blattner, J. Phair, H. Erlich, and D. L. Mann. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2:405-411. [DOI] [PubMed] [Google Scholar]
- 17.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 18.Lewontin, R. C. 1964. The interaction of selection and linkage. II. Optimum models. Genetics 50:757-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long, J. C., R. C. Williams, and M. Urbanek. 1995. An E-M algorithm and testing strategy for multiple-locus haplotypes. Am. J. Hum. Genet. 56:799-810. [PMC free article] [PubMed] [Google Scholar]
- 20.MacGinnitie, A. J., S. Anant, and N. O. Davidson. 1995. Mutagenesis of apobec-1, the catalytic subunit of the mammalian apolipoprotein B mRNA editing enzyme, reveals distinct domains that mediate cytosine nucleoside deaminase, RNA binding, and RNA editing activity. J. Biol. Chem. 270:14768-14775. [PubMed] [Google Scholar]
- 21.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 22.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 23.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398-1403. [DOI] [PubMed] [Google Scholar]
- 24.Martin, M. P., M. Dean, M. W. Smith, C. Winkler, B. Gerrard, N. L. Michael, B. Lee, R. W. Doms, J. Margolick, S. Buchbinder, J. J. Goedert, T. R. O'Brien, M. W. Hilgartner, D. Vlahov, S. J. O'Brien, and M. Carrington. 1998. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science 282:1907-1911. [DOI] [PubMed] [Google Scholar]
- 25.O'Brien, S. J., G. W. Nelson, C. A. Winkler, and M. W. Smith. 2000. Polygenic and multifactorial disease gene association in man: lessons from AIDS. Annu. Rev. Genet. 34:563-591. [DOI] [PubMed] [Google Scholar]
- 26.Phair, J., L. Jacobson, R. Detels, C. Rinaldo, A. Saah, L. Schrager, and A. Munoz. 1992. Acquired immune deficiency syndrome occurring within 5 years of infection with human immunodeficiency virus type-1: the Multicenter AIDS Cohort Study. J. Acquir. Immune Defic. Syndr. 5:490-496. [PubMed] [Google Scholar]
- 27.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 28.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 29.Stopak, K., C. de Noronha, W. Yonemoto, and W. C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12:591-601. [DOI] [PubMed] [Google Scholar]
- 30.Strebel, K., D. Daugherty, K. Clouse, D. Cohen, T. Folks, and M. A. Martin. 1987. The HIV ′A' (sor) gene product is essential for virus infectivity. Nature 328:728-730. [DOI] [PubMed] [Google Scholar]
- 31.Vlahov, D., N. Graham, D. Hoover, C. Flynn, J. G. Bartlett, J. B. Margolick, C. M. Lyles, K. E. Nelson, D. Smith, S. Holmberg, and H. Farzadegan. 1998. Prognostic indicators for AIDS and infectious disease death in HIV-infected injection drug users: plasma viral load and CD4+ cell count. JAMA 279:35-40. [DOI] [PubMed] [Google Scholar]
- 32.von Schwedler, U., J. Song, C. Aiken, and D. Trono. 1993. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J. Virol. 67:4945-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.